In this study, Comoglio et al. devised a novel quantitative framework discriminating different types of nucleosome remodeling events in micrococcal nuclease ChIP-seq data sets and used it to analyze nucleosome dynamics at stimulus-regulated cis-regulatory elements. Their results provide a systematic view of the impact of stimulation on nucleosome organization and genome accessibility in mammalian cells.
Keywords: chromatin, macrophages, nucleosome
Abstract
Accessibility of the genomic regulatory information is largely controlled by the nucleosome-organizing activity of transcription factors (TFs). While stimulus-induced TFs bind to genomic regions that are maintained accessible by lineage-determining TFs, they also increase accessibility of thousands of cis-regulatory elements. Nucleosome remodeling events underlying such changes and their interplay with basal positioning are unknown. Here, we devised a novel quantitative framework discriminating different types of nucleosome remodeling events in micrococcal nuclease ChIP-seq (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing) data sets and used it to analyze nucleosome dynamics at stimulus-regulated cis-regulatory elements. At enhancers, remodeling preferentially affected poorly positioned nucleosomes while sparing well-positioned nucleosomes flanking the enhancer core, indicating that inducible TFs do not suffice to overrule basal nucleosomal organization maintained by lineage-determining TFs. Remodeling events appeared to be combinatorially driven by multiple TFs, with distinct TFs showing, however, different remodeling efficiencies. Overall, these data provide a systematic view of the impact of stimulation on nucleosome organization and genome accessibility in mammalian cells.
Controlled access to the cis-regulatory information contained in mammalian genomes is critical for the accurate execution of transcriptional programs. During development, a discrete and largely cell type-specific fraction of a total repertoire of about half a million cis-regulatory elements (Dunham et al. 2012) is exposed and thus made available to transcription factors (TFs) and machineries such as RNA polymerases. Such selective exposure of distinct components of the cis-regulatory repertoire determines the unique gene regulatory networks in place in different cell types.
Access to the regulatory information in mammalian genomes is controlled by two counteracting forces. On the one hand, many regulatory elements have an intrinsic and DNA sequence-driven propensity to assemble nucleosomes (Tillo et al. 2010; Barozzi et al. 2014), which implies that most functional TF motifs are occluded by default. On the other hand, nucleosome displacement actively enforced by chromatin remodelers enables the selective exposure of the fraction of the regulatory information available for transcriptional control in a given cell type (Barozzi et al. 2014). Such a process is instructed by sequence-specific DNA-binding proteins able to recognize motifs in nucleosomal DNA (Zhu et al. 2018) and instigate nucleosome displacement or remodeling (Zaret and Carroll 2011). While their mode of binding to nucleosomal DNA can be extremely diversified (Zhu et al. 2018), such TFs have been collectively termed “pioneers” and coincide at least in part with lineage-determining TFs, whose expression is triggered by microenvironmental cues in the developmental niche of individual tissues. This is typified by M-CSF (macrophage colony-stimulating factor) induction of the myeloid lineage-determining TF PU.1 in the bone marrow (Mossadegh-Keller et al. 2013). Importantly, the maintenance of nucleosome depletion at most enhancers requires the constitutive binding of lineage-determining TFs, whose depletion results in rapid nucleosome reassembly (Barozzi et al. 2014). A notable exception to this scheme is represented by CpG islands, whose low nucleosome occupancy is imposed by their peculiar nucleotide composition (Ramirez-Carrozzi et al. 2009). Indeed, the relationship between G+C content and nucleosome assembly is typically bimodal, with both very low and very high G+C content being anticorrelated with nucleosome occupancy (Valouev et al. 2011; Fenouil et al. 2012; Barozzi et al. 2014). Overall, the combination of DNA sequence composition and trans-acting factors determines both the degree of nucleosome occupancy of each genomic region and the accuracy of nucleosome positioning and thus, eventually, the accessibility of the underlying regulatory DNA (Lai and Pugh 2017).
Importantly, while nucleosomes are reproducibly maintained at a fixed position at some genomic sites, at others they tend to occupy a range of overlapping yet distinct positions in different cells of the same population (Lai et al. 2018), thus resulting in fuzzy signals in micrococcal nuclease (MNase)-based population analyses. In general, compared with nucleosomes at inert genomic regions, nucleosomes at active cis-regulatory elements tend to be more reproducibly positioned across different cells in the same population (Lai et al. 2018). Maintenance of well-positioned nucleosomes involves a combination of DNA sequence determinants favoring or disfavoring nucleosome assembly and trans-acting factors; namely, TFs working in concert with chromatin remodelers to restrain lateral nucleosome movements (Tillo et al. 2010; Valouev et al. 2011; Barozzi et al. 2014). The eviction or shifting of well-positioned nucleosomes therefore requires that such cis-acting and trans-acting determinants be overruled, while less constrained nucleosomes might provide opportunities for more rapid and less energy-demanding remodeling. Finally, from a biological point of view, inaccurate positioning may generate opportunities for stochastic exposure of the underlying DNA sequence and thus for TF binding and gene activation. Techniques such as ATAC-seq (assay for transposase-accessible chromatin [ATAC] using high-throughput sequencing) (Buenrostro et al. 2013) and DNaseI-seq (Boyle et al. 2008) enable the exploration of the accessible nucleosome-depleted genome and thus are informative of how the regulatory information is exposed in different cells or conditions. However, these techniques do not provide direct information on the nucleosomal changes associated with (and, in fact, determining) variations in the accessibility of the genomic DNA in different conditions and time windows—from developmental transitions to acute responses to microenvironmental stimuli.
When considering acute responses to stimulation, a general paradigm inferred from the analysis of a wealth of genomic data is that stimulus-regulated TFs (such as NF-kB and estrogen or androgen receptors) bind to genomic regions that are constitutively occupied and kept accessible by lineage-determining TFs such as PU.1 in myeloid cells and FOXA1/2 in cells of endodermal origin (Ghisletti et al. 2010; Heinz et al. 2010; Wang et al. 2011; Barozzi et al. 2014; Glass and Natoli 2016; Monticelli and Natoli 2017). However, this simple scheme does not account for the common observation that acute stimulation increases accessibility of hundreds to thousands of cis-regulatory regions (Scott-Browne et al. 2016; Mueller et al. 2017; Park et al. 2017). Moreover, in the case of T lymphocyte differentiation in response to polarizing cytokines such as IFNγ and IL-4, stimulus-activated TFs are required to establish the enhancer landscape characteristic of individual T lymphocyte subsets (Vahedi et al. 2012). These observations raise several important questions: (1) How are nucleosomes associated with cis-regulatory elements affected by an acute stimulus? (2) How is the pre-existing organization restored after stimulus termination? (3) Is the pre-existing organization of nucleosomes at rapidly inducible enhancers designed to enable acute changes in accessibility? (4) Are individual TFs endowed with a different ability to disrupt nucleosome organization both qualitatively (e.g., at distinct classes of cis-regulatory elements) and quantitatively (with different efficiency)?
To address these questions, in this study, we investigated the interplay between DNA accessibility and nucleosome organization on a genome-wide scale in both basal and stimulated conditions by combining ATAC-seq and ChIP-seq (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing) on nucleosomal MNase-digested chromatin from basal and lipopolysaccharide (LPS)-treated macrophages. We devised and implemented a quantitative framework to detect and classify remodeling events and applied it to determine which changes in nucleosome organization occur at regions showing alterations in either accessibility or TF binding upon stimulation. By integrating these data with a comprehensive panel of time-resolved ChIP-seq profiles for several TFs, we inferred the nucleosome remodeling potential of various TFs regulated by stimulation.
Results
The nucleosomal landscape of unstimulated macrophages
Nucleosome preparations were obtained from formaldehyde-fixed untreated or LPS-treated (30, 60, 120, and 240 min) mouse bone marrow-derived macrophages. MNase digestion was calibrated to generate a prevalence of mononucleosomes over dinucleosomes (∼80% and 20%, respectively) (Barozzi et al. 2014), with negligible amounts of polynucleosomes. Two biological replicates were used as a source of chromatin for ChIP assays with antibodies recognizing histone modifications preferentially associated with poised and active promoters (H3K4me3), poised putative enhancers (H3K4me1), and active cis-regulatory elements (H3K27Ac) (Supplemental Table 1). During the window of time of these experiments, the global levels of the histone modifications analyzed, as judged by Western blot analysis, were stable (data not shown). MNase-ChIP-seq samples were sequenced to an average depth of 42 million reads. A high-depth (800 million reads) MNase-seq data set generated previously in untreated macrophages (Barozzi et al. 2014) was used as a reference for comparisons. To estimate the nucleosome midpoints and for data visualization, the minus strand coverage was subtracted from the plus strand coverage as described (Haberle et al. 2014). For each nucleosome, the midpoint coincides with the transition between the plus and the minus strand signals.
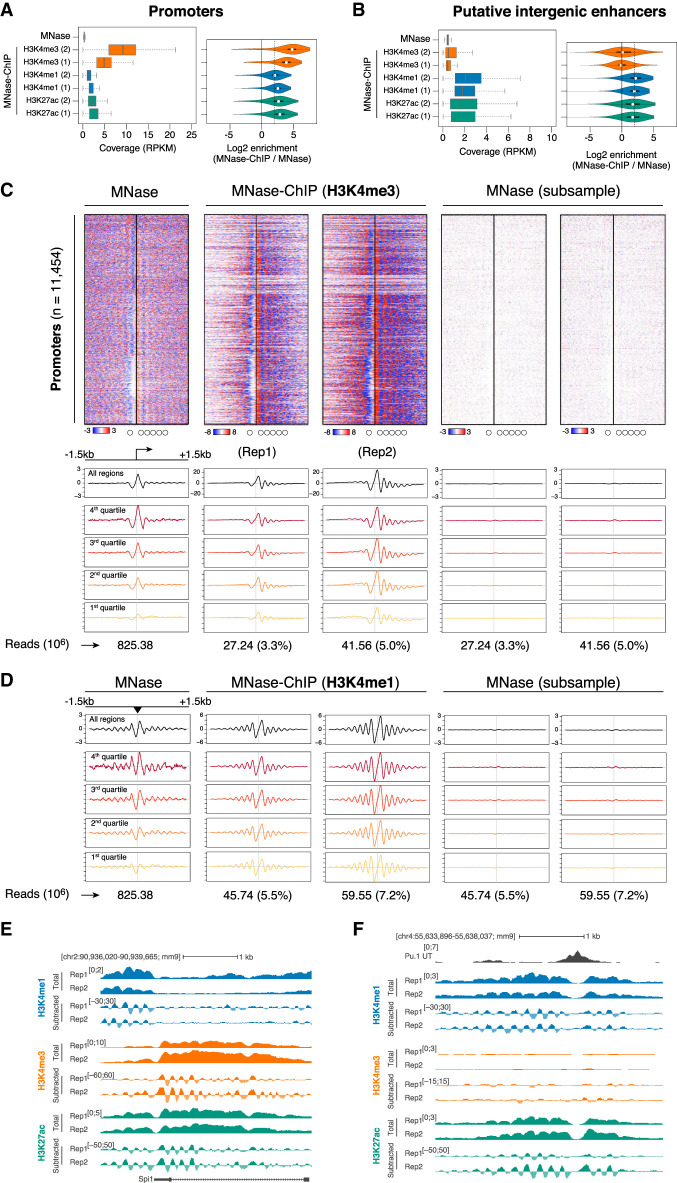
Compared with MNase-seq, isolation of covalently modified nucleosomes by MNase-ChIP-seq resulted in a consistent enrichment of read coverage at both promoters and intergenic enhancers (log2 enrichment range: 1.8–4.9) (Fig. 1A,B). Patterns of nucleosome positioning in the two MNase-ChIP-seq replicates were highly reproducible (Supplemental Figure 2A,B).
Figure 1.
Analysis of nucleosome positioning and occupancy by MNase-ChIP-seq. (A,B) Normalized coverage of H3K4me3, H3K4me1, and H3K27ac MNase-ChIP-seq data at promoters (A) and putative intergenic enhancers (B). (Right) Violin plots show the MNase-ChIP-seq signal enrichment relative to MNase-seq (log2). (Dashed line) Fourfold enrichment; (RPKM) reads per kilobase per million mapped reads. (C) Nucleosome organization at active promoters. Genomic regions were centered on the main mapped transcription start site (TSS), extended by 1.5 kb on each side, and clustered based on H3K4me3 MNase-ChIP-seq signal in the central region (±150 bp). For comparison, heat maps based on total and downsampled MNase-seq data are shown. The plots below the heat maps show the average signal for the entire set (black) or for the regions split in quartiles of decreasing signal based on intensity of the central 1 kb in the MNase-seq experiment. (D) Same as C (average plots) but for putative extragenic enhancers, using H3K4me1 MNase-ChIP-seq data and centering on the summit of the PU.1 peak, extended by 1.5 kb on each side. (E) Total and subtracted signals from a representative TSS-proximal region. (F) Total and subtracted signals from a representative TSS-distal putative enhancer region.
Nucleosome maps at 11,454 H3K4me3-positive promoters and 19,417 H3K4me1- and PU.1-positive putative intergenic enhancers are shown in Figure 1C and D, and Supplemental Figure 1. Representative snapshots are illustrated in Figure 1E and F, and an extended genomic region is shown in Supplemental Figure 2F. Since PU.1 plays a central role in the organization of macrophage-specific enhancers, where it is also required to maintain nucleosome depletion (Barozzi et al. 2014), the summit of PU.1 peaks was used as a proxy for the identification of enhancer core regions. Therefore, enhancer maps were centered on the summit of PU.1 peaks (±1.5 kb), while promoter maps were centered on the main annotated transcription start site (TSS; ±1.5 kb).
Together, these results indicate that the relatively shallow sequencing of MNase-ChIP-seq libraries yielded high-resolution and reproducible nucleosome positioning maps at cis-regulatory elements in a mammalian genome.
Two classes of enhancers with distinct nucleosomal symmetry
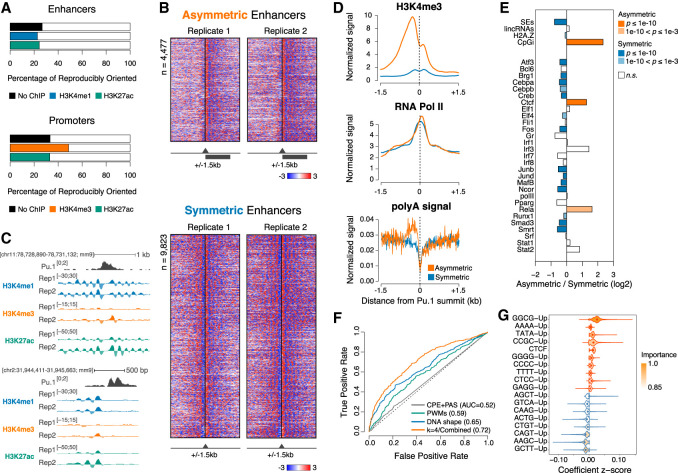
Visual exploration of the data hinted at the existence of two distinct types of enhancers distinguished by their nucleosomal symmetry; namely, by the relative enrichment of H3K4me1-positive nucleosomes upstream of and downstream from the enhancer core. Since enhancers are thought to act independently of their orientation, the existence of asymmetric enhancers was intriguing and warranted additional investigations. To systematically classify these two types of enhancers, we computed the ratio between H3K4me1 signals upstream of and downstream from the enhancer core regions using a window size of 500 bp (based on initial tests to identify the optimal window) (data not shown) and classified as asymmetric those enhancers with an absolute ratio ≥1.5 in both replicates (Fig. 2A). The correlation between upstream and downstream signals among replicates was consistently high (SCC = 0.68–0.74) and was further confirmed in MNase-seq replicates (SCC = 0.77) (Supplemental Fig. 2C–E). Although the exact number of asymmetric enhancers depended on the window size used for signal summarization, 19%–35% of enhancers were consistently classified as asymmetric by our analysis using either H3K4me1 or H3K27ac (Fig. 2A,B). By comparison, 49%–62% of promoters were found to be asymmetric based on H3K4me3 signals (Fig. 2A). These results were not affected by differences in repeat content or sequence mappability (Supplemental Fig. 3A–C).
Figure 2.
Identification of two classes of enhancers with distinct nucleosomal symmetry. (A) The percentage of reproducibly oriented elements, defined as showing concordant normalized signal bias with respect to the core element in both replicates (±500 bp), for both MNase-seq and MNase-ChIP-seq targeting the indicated modified nucleosomes. (B) Asymmetric (top) and symmetric (bottom) H3K4me1 MNase-ChIP-seq signals at enhancers centered on the summit of the PU.1 peak. Genomic regions were clustered based on signals in the central region (±500 bp). (C) Representative genomic regions showing a symmetric (top) and an asymmetric (bottom) enhancer. (D) Cumulative distributions of H3K4me3, RNA polymerase II (Pol II), and polyA signals at symmetric and asymmetric enhancers. (E) Enrichment of selected genomic features at symmetric and asymmetric enhancers. P-values are from a two-sided Fisher's exact test. (F) Receiver operating characteristic (ROC) curve and mean area under the ROC curve (AUC) values for least absolute shrinkage and selection operator (lasso) logistic regression models trained on the indicated sets of features. The ROC curve closest to the mean AUC from 100 models is shown. (CPE) Core promoter elements; (PAS) polyA signal. (G) Top-ranked features selected by bootstrap lasso for the combined model in F. Violin plots are color-coded according to coefficient signs.
Overall, when using a window size of ±500 bp relative to the enhancer core, 4477 enhancers (32.7%) exhibited an asymmetric distribution of H3K4me1 nucleosomes, while 9823 (67.3%) were symmetric (Fig. 2A,B; Supplemental Table 2). Representative genomic regions are shown in Figure 2C. Compared with symmetric enhancers, asymmetric enhancers shared higher similarity with promoters, including higher levels of H3K4me3, moderate enrichment for CpG islands and CTCF sites (P < 1 × 10−10; Fisher's exact test), and an enrichment for polyadenylation sites (PASs) opposite to the side exhibiting well-organized nucleosomes and RNA polymerase II (Pol II) occupancy (Fig. 2D,E; Supplemental Fig. 3D). The two groups of enhancers showed no or minimal differences in terms of evolutionary conservation, relation to nearest active genes, associated functional categories, association with long noncoding RNAs (lncRNAs), and distance from the boundaries of TADs (topologically associating domains) (Fig. 2E; Supplemental Material).
These observations prompted us to test whether DNA sequence features could discriminate symmetric from asymmetric nucleosomal patterns at enhancers. To this end, we considered three sets of features: (1) k-mer frequencies (with 2 ≤ k ≤ 4), (2) DNA shape features (Chiu et al. 2016), and (3) motif scores from a curated collection of >1700 TF motifs (Diaferia et al. 2016). These features were used alone or in combination to train classifiers (Comoglio et al. 2015, 2018) using cross-validation and to evaluate the prediction accuracies on a test set. This analysis revealed that models combining all of the considered feature sets could achieve a fair classification accuracy (mean area under the receiver operating characteristic curve [AUC] = 0.72) but did not outperform models based solely on 4-mers (Fig. 2F), indicating a high feature redundancy. To identify the most predictive sequence features, we then performed feature selection using a procedure that assigns high importance to indispensable features (Comoglio and Paro 2014). We found that GC-rich, polyA, and TATA sequences upstream of the enhancer core, along with CTCF motifs at the site, were most predictive for asymmetric enhancers, whereas GCTT, AAGC, and CAGT sequences were predictive for symmetric nucleosomal patterns (Fig. 2G).
Together, these results indicate the existence of two distinct classes of enhancers distinguished by the symmetry of their nucleosomal patterns. Moreover, they suggest that such distinctive patterns are determined primarily by DNA sequence features.
A quantitative framework to measure dynamic nucleosomal changes
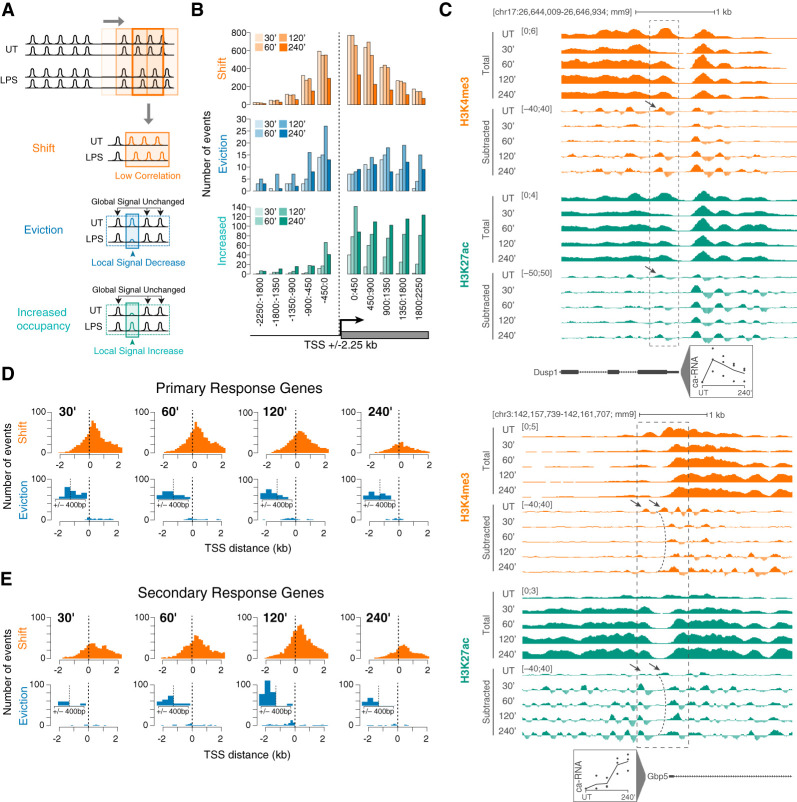
To analyze LPS-induced changes in nucleosomal organization at promoters and enhancers, we first devised a quantitative approach aimed at detecting different types of remodeling events. Since MNase-ChIP was carried out using antibodies for different histone modifications, a critical issue was to discriminate bona fide remodeling events (loss, gain, or shift of nucleosomes) induced by LPS stimulation from losses or gains of a given histone modification. We reasoned that since histone modifications tend to occur at many consecutive nucleosomes, a local stimulus-induced signal change occurring within a broader unaffected region would be indicative of the remodeling of individual nucleosomes. Therefore, we set out to identify local signal changes in windows of 450 bp lying within broader regions of 4.5 kb in which the histone modification analyzed was instead stable (see the Materials and Methods for a complete description). This procedure allowed us to score quantitative changes in MNase-ChIP-seq signals associated with one to three nucleosomes within regions in which the overall signal of the modification did not change. Such quantitative changes were interpreted as evidence of nucleosome evictions in the case of a signal loss or increased nucleosome occupancy in the case of a signal gain (Fig. 3A). This strategy was complemented with a different one aimed at identifying local changes in correlation between coverage profiles across conditions (untreated and multiple time points of LPS stimulation). In this case, a loss in correlation in local nucleosomal signals was interpreted as a shift in the nucleosome position (Fig. 3A). Importantly, since these approaches rely on the presence of an overall stable signal before and after stimulation, they were not suitable to identify nucleosomal changes at regions showing massive gains (or losses) of histone modifications—notably a subset of LPS-inducible gene promoters undergoing H3K4me3 gain upon stimulation.
Figure 3.
A quantitative approach for detecting inducible nucleosome remodeling events. (A) Schematic representation of the approach used to detect different types of nucleosomal changes induced by stimulation. A sliding window of 450 nucleotides (step size 150 bp) was used to identify local nucleosomal changes within a larger windows (4.5 kb) whose overall signal was not significantly affected by stimulation. Nucleosome shifts were identified as significant drops in correlation between profiles, while evictions and locally increased occupancy were identified as significant decreases/increases that were instead not identified when considering a 4.5-kb window. (B) H3K4me3 was used to determine the distribution of the three distinct types of nucleosome remodeling events at the TSS of LPS-inducible genes at different time points after LPS stimulation. Events are shown in 10 consecutive windows of 450 bp each. (C) Representative genomic regions showing remodeling of TSS-proximal nucleosomes. (D,E) Distributions of nucleosome remodeling events centered on the TSS of primary (D) and secondary (E) response genes at different time points after LPS stimulation. H3K4me3 was used to score nucleosome remodeling. Insets show an enlargement of the distributions of eviction events throughout the central 900 bp around the TSS.
We used H3K4me3 to score nucleosomal changes at promoters and H3K4me1 for enhancers. While H3K4me3 pre-exists at a large fraction of primary LPS-inducible genes, it is induced at the promoters of many secondary response genes (Foster et al. 2007; Ramirez-Carrozzi et al. 2009). H3K4me1 is instead a very stable modification that is not significantly affected by stimulation at most LPS-activated enhancers (Kaikkonen et al. 2013; Ostuni et al. 2013). Because of the high magnitude of the acetylation changes induced by LPS at inducible promoters and enhancers and its overall lower levels before stimulation, H3K27ac data were not suitable to identify remodeling events using these approaches.
In the interpretation of these data, it should also be noted that many inducible genes (such as IL12b and interferon-stimulated genes [ISGs]) are activated in only a fraction of the LPS-stimulated cells (Weinmann et al. 2001; Shalek et al. 2013). Because of this heterogeneity, remodeling events occurring in a small fraction of cells will be diluted by unaffected nucleosomes, thus reducing sensitivity. Therefore, the events identified by our analysis likely underestimate the set of remodeling events occurring at LPS-induced genes. Moreover, as we restricted our analysis to those genomic regions that showed consistent H3K4me3 signal before and after LPS, we were able to analyze nucleosome remodeling events at only a subset of LPS-activated gene promoters (1544 out of 1879 LPS-inducible genes).
Nucleosome remodeling at stimulus-regulated promoters
We first applied our analytical framework to the promoters of LPS-inducible genes to identify nucleosome remodeling events otherwise undetectable by a conventional ChIP-seq analysis. We used a sliding window of 450 bp to identify at each time point of LPS stimulation local nucleosomal changes within 4.5-kb regions centered on the TSS (Fig. 3B). Remodeling events identified downstream from the TSS often extended further downstream into the body of LPS-inducible genes, likely reflecting the remodeling activity of the elongating RNA Pol II. Conversely, remodeling events upstream of the TSS were generally limited to a proximal region spanning a few nucleosomes. Nucleosome shifts were by far more common than evictions. As an example, the Dusp1 gene was constitutively marked by H3K4me3 and was rapidly induced by LPS, with a peak of transcription at 30 min after stimulation, as indicated by nascent transcript analysis (Fig. 3C). The overall H3K4me3 signal intensity was only moderately affected by stimulation. However, the +1 nucleosome underwent a near-complete eviction within 30 min of LPS stimulation, followed by a progressive gain in signal that correlated with the gradual reduction in transcriptional activity (Fig. 3C). At the Gbp5 gene promoter, one H3K4me3-positive nucleosome upstream of the TSS (Fig. 3C, bottom panel, left arrow) was evicted, while the downstream nucleosome underwent a sustained shift (Fig. 3C, bottom panel).
The frequency of remodeling events peaked between 30 and 60 min at primary response genes (Fig. 3D), while it peaked at 120 min after stimulation at secondary response genes (Fig. 3E). This result is consistent with the average timing of induction of the genes in the two groups (Bhatt et al. 2012). Similar profiles were obtained by dividing inducible genes into two kinetic classes (early vs. late induction) (Supplemental Fig. 4A,B). While drops in correlation were detected on both sides of the TSS (with a higher frequency downstream from the TSS), evictions were by far more common upstream of the TSS of secondary response genes (Fig. 3E).
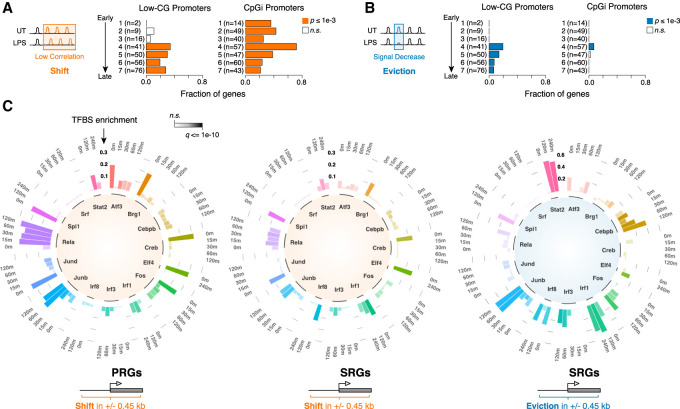
Overall, when considering a conservative window of 900 bp centered on the TSS of the 1544 LPS-inducible genes in which the H3K4me3 signal was present already in unstimulated cells, we detected H3K4me3-positive nucleosome remodeling events at the promoters of 437 genes (28.3%) (Supplemental Table 3). Considering the full set of up-regulated genes, a significant enrichment of nucleosomal shifts was observed at their TSSs (P-value = 6.3 × 10−12; Fisher's exact test; compared with expressed genes unaffected by LPS) but not at those of down-regulated genes (P-value = 0.33). Similar results were obtained for evictions (P-value = 2.5 × 10−12 for up-regulated genes; P-value = 0.16 for down-regulated genes). Previous studies demonstrated that LPS-inducible genes with a CpG island in their promoter, which tend to be activated earlier in the LPS response, do not require chromatin remodeling for activation due to the poor propensity of very high G+C content regions to assemble stable nucleosomes (Ramirez-Carrozzi et al. 2009). Conversely, the induction of genes with lower G+C content, which tend to show slower activation kinetics, is dependent on Swi/Snf-mediated nucleosome remodeling (Ramirez-Carrozzi et al. 2009). Therefore, we divided LPS-induced genes from seven increasingly slower kinetic classes (Bhatt et al. 2012) based on the presence or absence of a CpG island and analyzed chromatin remodeling events within each class (Fig. 4A,B). Importantly, although CpG islands are overall depleted of nucleosomes, the regions surrounding them show high occupancy, which in principle makes the detection of changes in nucleosome organization—and specifically evictions—possible (data not shown). Nucleosome evictions, which preferentially occurred upstream of the TSS (Fig. 3D,E), were nearly exclusively observed at the promoters of slowly inducible non-CpG island genes (classes 5–7), with the exception of class 4 (Fig. 4A,B; Supplemental Fig. 4C for validations). Conversely, nucleosome shifts were detected across all classes and likely reflected remodeling events associated with RNA Pol II recruitment and elongation (note that low G+C promoters in classes 1–3, the groups showing limited enrichment, include only 2, 9, and 16 genes, respectively) (Fig. 4A). Of note, eviction events were mainly found upstream of a small subset of TSSs and appeared transient. This is particularly pronounced in a small group of both primary and secondary response genes (Ccl3, Ccl4, Rsad2, Tnf, Gbp5, Gbp3, Gbp2, Cxcl10, Oasl1, and Tnip3) that are highly depleted for CpG islands (Fig. 4B; Supplemental Fig. 4A).
Figure 4.
Nucleosome remodeling at different classes of LPS-inducible genes. (A,B) Fraction of genes showing nucleosome shift (A) or eviction (B) at their promoter region (±450 bp), grouped by kinetics of induction upon LPS stimulation (Bhatt et al. 2012) and G+C content. H3K4me3 MNase-ChIP-seq data were used to determine nucleosome changes. (CpGi) CpG island. P-values are from a two-sided hypergeometric test. (C) Circular stacked bar plots showing TF binding enrichment at the promoters (TSS ± 450 bp) of LPS-inducible genes exhibiting remodeling events. The analysis is based on ChIP-seq data at the indicated time points of LPS stimulation. Three different groups are shown: shift at primary response genes (PRGs), shift at secondary response genes (SRGs), and evictions at SRGs. The enrichment is calculated as the difference between the fraction of remodeled sites showing binding for the indicated TF and the fraction of total sites associated with TF binding. The q-value of the enrichment is color-coded. P-values are from a two-sided χ2 test, corrected for multiple hypothesis testing using Benjamini-Hochberg.
To determine how TF binding at promoters correlates with nucleosome organization, we collected 67 TF ChIP-seq profiles, including 40 newly generated data sets, encompassing 27 TFs in untreated and LPS-treated macrophages. The quality of the TF ChIP-seq data sets used was assessed by determining the fraction of reads in peaks (FRIP) (Supplemental Table 1) and characterizing the overrepresented motifs in each data set (Supplemental Fig. 6). In addition to typical stimulus-activated TFs (NF-kB, AP-1, CREB, IRF [interferon regulatory factor], STAT, and SRF) we included two myeloid lineage TFs (PU.1 and C/EBPβ) (Feng et al. 2008; Goode et al. 2016) and the ETS family member ELF4, which is selectively associated with active cis-regulatory elements, contributing to their basal activity (Curina et al. 2017). We also included total RNA Pol II and BRG1, one of the two active subunits of the Swi/Snf chromatin remodeling complexes.
Basal or LPS-induced TF binding enrichment was measured at the promoters (TSS ± 450 nucleotide [nt]) of both primary and secondary response genes showing remodeling. Next, in order to identify those TFs whose recruitment correlated more strongly with remodeling, we measured the enrichment of individual TFs at remodeled promoters relative to all other promoters. Figure 4C shows the difference in the recruitment of individual TFs at different classes of promoters showing remodeling versus all other promoters (see also Supplemental Fig. 5A; Supplemental Table 4). This analysis revealed that (1) constitutive binding of PU.1 (SPI1) was associated with all types of remodeling events; (2) constitutive binding of ELF4 was associated with nucleosome shifts, likely reflecting the high transcriptional activity of ELF4-associated promoters (Curina et al. 2017); (3) basal and inducible IRF1 binding along with inducible IRF3 and STAT2 binding were selectively associated with remodeling events (especially evictions) at the TSS of secondary response genes, which include genes activated in a paracrine/autocrine manner by early released IFNβ; and (4) inducible NF-kB binding was associated with shifts but not evictions, and this association was stronger for primary than for secondary response gene promoters. The correlation between basal, inducible, and reduced TF-binding events and nucleosome remodeling at different classes of genes is highlighted in Supplemental Figure 5B.
Overall, these results indicate that recruitment of different TFs to the promoters of LPS-inducible genes correlated with distinct types of nucleosome remodeling events, which in turn correlated with the DNA sequence features of the promoters.
Nucleosome remodeling at stimulus-regulated enhancers
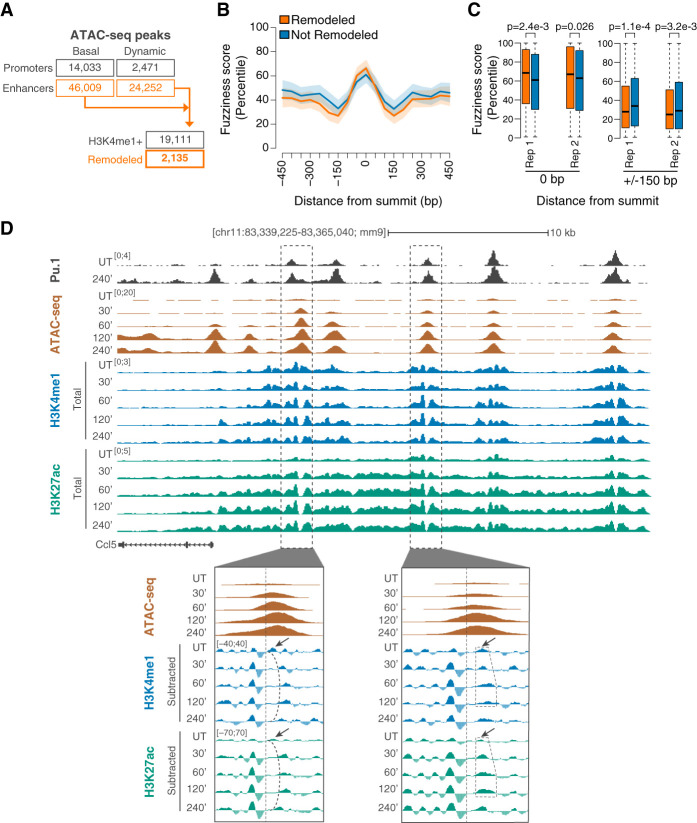
We next investigated nucleosome remodeling at genomic regions showing either constitutive or LPS-induced accessibility. To this end, we generated ATAC-seq profiles at multiple time points of LPS stimulation (30 min, 1 h, 2 h, and 4 h) (Supplemental Fig. 7). While ∼60,000 ATAC-seq peaks were accessible prior to stimulation, the accessibility of 24,252 TSS-distal and 2471 TSS-proximal peaks was affected by LPS treatment (Fig. 5A; Supplemental Fig. 7A,D–G). ATAC-seq signal gains and losses exhibited broad kinetic complexity, including peaks that were selectively gained or lost at either early or late time points (Supplemental Fig. 7B,C). We thus focused on 19,111 TSS-distal extragenic regions marked by H3K4me1 that were either basally accessible or differentially accessible at one or more time points after LPS stimulation (Fig. 5A). We then applied our analytical framework (Fig. 3A) to identify nucleosome remodeling events at these 19,111 putative enhancers. While no significant nucleosome eviction or locally increased occupancy was detected within these regions, 2135 enhancers (11.2%) exhibited one or more nucleosomal shifts (Fig. 5A; Supplemental Table 5). These events showed a significant enrichment for symmetric sites as compared with the asymmetric ones (P-value < 2.2 × 10−16, χ2 test). Nucleosomes occupying these remodeled regions were significantly fuzzier (as determined by the DANPOS2 pipeline) (Chen et al. 2013) than nucleosomes at regions where remodeling was not detected (Fig. 5B,C, left panel), although the magnitude of the difference in fuzziness was limited. These remodeled accessible regions were instead flanked by more positioned nucleosomes (Fig. 5B,C, right panel). Representative examples of asymmetrically remodeled nucleosomes in the Ccl5 locus are shown in Figure 5D.
Figure 5.
Nucleosome remodeling events at putative enhancers. (A) Summary of nucleosome remodeling events at accessible regulatory elements identified by ATAC-seq. Nucleosome remodeling was determined using H3K4me1 MNase-ChIP-seq data. Accessible regions either in basal conditions or after LPS stimulation and either proximal or distal to the annotated gene TSS are indicated. (B) Nucleosome fuzziness at TSS-distal remodeled (orange) and not remodeled (blue) regions. Data are shown in a window of ±450 nt centered on the summit of ATAC-seq peaks. Confidence intervals were estimated from a spline fit of the median fuzziness of two biological replicates. (C) Distribution of fuzziness scores (percentiles) at the ATAC-seq peak summit (0 bp) and 150 bp upstream or downstream (±150 bp) for remodeled and not remodeled regions and each replicate. P-values are from a two-sided Wilcoxon rank-sum test. (D) Snapshots highlighting representative nucleosome remodeling events at two LPS-inducible putative enhancers upstream of Ccl5.
These observations suggest that nucleosomes with intrinsically fuzzier positioning located at enhancer cores may be more susceptible to remodeling, while those with a stronger positioning are often unperturbed by the landing of TFs in their close proximity.
Dynamic TF occupancy and nucleosome remodeling events
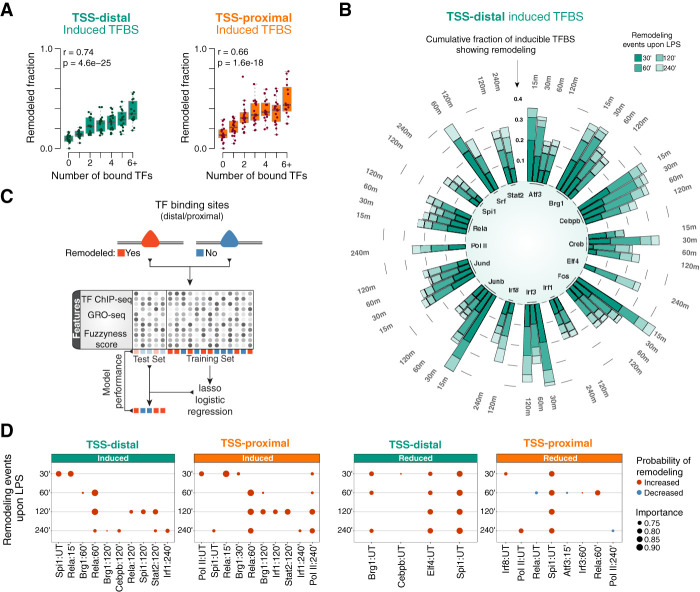
To investigate the correlation between TF binding and changes in nucleosome organization at enhancers, we analyzed the panel of TF ChIP-seq profiles described above (Supplemental Table 1). We first defined master sets of regulatory regions showing at least one LPS-induced (or LPS-reduced) TF-binding event at both TSS-distal and TSS-proximal sites (Supplemental Table 6). We then correlated the number of distinct TFs that bind each regulatory region and the occurrence of remodeling (Fig. 6A; Supplemental Fig. 8A). We found a significant positive correlation between the number of recruited TFs and changes in nucleosome organization at both putative enhancers (Spearman's rank correlation coefficient of 0.74, P-value = 4.6 × 10−25) and promoters (r = 0.66, P-value = 1.6 × 10−18). This result suggests that the higher the number of TFs recruited to enhancers, the higher the probability of remodeling, which in turn indicates that remodeling is a combinatorial process driven by multiple collaborating TFs.
Figure 6.
TF binding and inducible nucleosome remodeling at promoters and enhancers. (A) The fraction of putative enhancers (left) and promoters (right) undergoing detectable nucleosome remodeling correlates with the number of inducible TF-binding events within a ±450-nt window. (B) Circular stacked bar plots showing the cumulative fraction of inducible TF binding at different time points following LPS stimulation that overlap nucleosome remodeling events. (C) Schematic illustration of the modeling framework. TF-binding sites either showing or devoid of remodeling events were randomly assigned to either balanced training or the test sets. Lasso logistic regression models were then trained in cross-validation, and the prediction accuracies were evaluated on a test set. The matrix shows the value (grayscale) of each feature (examples of which are indicated at the left) across the sites. (D) Top-ranked dynamic TF-binding events (e.g., Rela:60′ denotes binding of Rela occurring at 60 min after LPS) associated with remodeling events at a given time point after LPS stimulation. (UT) Untreated.
Next, for each TF, we measured the fraction of LPS-inducible changes in TF binding (induction or reduction) that was associated with detectable nucleosome remodeling at the TF recruitment site at different time points. For the majority of TFs, most remodeling events occurred within 60 min of LPS, with additional events becoming progressively less common over time (Fig. 6B; Supplemental Fig. 8B–D; Supplemental Table 7). This is consistent with the genome-wide pattern of LPS-inducible accessibility at putative regulatory elements (Supplemental Fig. 7D–F). A fine-grained dissection of remodeling events by time points (Supplemental Fig. 9A) revealed that while these occurred throughout the entire stimulation kinetics, they became overall less common at the last time point analyzed (240 min), which we interpret as a reflection of the overall reduction in signaling strength at later time points after LPS stimulation. Importantly, the analysis of ChIP-seq profiles at the early time points (15–30 min after LPS) indicates that, in many cases (e.g., IRF3, RELA, and JUNB), remodeling events associated with early TF recruitment tended to persist over time.
Finally, we set out to test whether and to what extent the recruitment of different TFs could predict the observed remodeling events. To this end, we trained lasso (least absolute shrinkage and selection operator) logistic regression models at distal and proximal sites based on TF ChIP-seq signals and estimated the relative importance of each TF with respect to remodeling events at a given time point (Fig. 6C,D). We found that model performances were generally modest but significantly higher at distal sites (AUC = 0.64–0.70; AUPRC = 0.61–0.67) than at proximal sites (AUC = 0.58–0.64; AUPRC = 0.55–0.62). Inclusion of additional features such as global run-on sequencing (GRO-seq) signals, nucleosome fuzzyness scores, pairwise interactions between TFs, and feature discretization (presence or absence of TF binding) did not further improve model performances in a significant manner.
These results indicate that some relevant features are likely missing from our analysis. However, feature importance analysis identified TF-binding events associated with increased or decreased probability of occurrence of a remodeling event. LPS-inducible binding of NF-kB (RELA) within 60 min of stimulation was the most predictive feature of remodeling (selected >90% of the times) at both TSS-distal and TSS-proximal sites. Moreover, RELA recruitment at 60 min predicted remodeling also at later time points, indicating that nucleosome alterations induced by NF-kB are sustained over time. Late (120–240 min) binding of IRF1 and STAT2 was also moderately associated with remodeling (selected >75% of the times) at these regions. In contrast, PU.1 (SPI1) binding in resting conditions was strongly associated (selected >90% of the times) with remodeled regions showing reduced TF-binding upon LPS stimulation (Fig. 6D). As an example, the snapshot in Supplemental Figure 9B shows the nucleosomal changes and TF recruitment at regulatory regions upstream of the LPS-inducible gene Nos2. The region highlighted in the inset shows a nucleosome remodeling event identified by our pipeline and associated with a gain in the ATAC-seq signal. The H3K4me1-marked nucleosomes on both sides showed increased positioning after remodeling of the central fuzzy nucleosome.
Overall, these data provide a comprehensive overview of the impact of signal-induced recruitment of individual TFs on nucleosome remodeling. They show that only a fraction of TF recruitment events is associated with detectable nucleosome remodeling and indicate that individual TFs, while acting combinatorially to displace nucleosomes, appear to have different remodeling efficiencies.
Discussion
In mammalian cells, nucleosome organization at cis-regulatory elements is determined by a complex interplay between TFs bound to chromatin and DNA sequence features that either facilitate or disfavor nucleosome assembly (Tillo et al. 2010; Valouev et al. 2011; Barozzi et al. 2014; Lai and Pugh 2017). A general paradigm is that the unique nucleosome profile characteristic of a given cell type is largely driven by two opposing forces: On the one hand, enhancers have an intrinsically high propensity to assemble nucleosomes, thus generally making their regulatory information inaccessible unless appropriate combinations of TFs are expressed (Tillo et al. 2010; Barozzi et al. 2014); on the other hand, lineage-determining TFs actively maintain a fraction of the genomic regulatory repertoire in a nucleosome-depleted state, regulating its accessibility in a cell type-specific manner (Barozzi et al. 2014). The barrier effect generated by lineage-determining TFs bound to enhancers represents a major force driving the precise positioning of nucleosomes at the two sides of accessible enhancer cores.
In this study, we set out to investigate a critical and unaddressed issue in this area; namely, the interplay between pre-existing nucleosome organization and the recruitment to cis-regulatory elements of TFs activated by acute stimulation. A widely accepted model is that TFs activated by stimulation land at enhancers that are premarked and thus are kept accessible by the lineage-determining TFs characteristic of a given cell type (Glass and Natoli 2016). In this way, transcriptional responses to otherwise identical stimuli are contextualized and rendered cell type-specific. In this model, binding of stimulus-activated TFs is highly opportunistic and in fact restricted to genomic regions constitutively bound by lineage-determining TFs, which displace nucleosomes and maintain the underlying DNA recognition motifs exposed and accessible. A corollary of this model is that since inducible binding of stimulus-activated TFs occurs within accessible regions, it should not significantly impact nucleosome depletion. However, this inference is not easily reconciled with the observation that acute stimulation also increases accessibility of a subset of enhancers, as determined by techniques such as ATAC-seq or DNaseI-seq (Novakovic et al. 2016; Park et al. 2017). The nature of chromatin alterations leading to such changes in accessibility induced by stimulation is unknown. In particular, it is unclear whether the recruitment of stimulus-activated TFs suffices to displace well-positioned nucleosomes in order to promote a further expansion of the accessible repertoire of regulatory elements. Our data indicate that nucleosomes undergoing remodeling in response to stimulation are significantly less positioned than nonremodeled nucleosomes. These fuzzy nucleosomes are frequently flanked by well-positioned nucleosomes that are refractory to the nucleosome remodeling activity of stimulus-inducible TFs recruited in their immediate vicinity.
These data support a model in which stimulus-responsive cis-regulatory elements are frequently flanked by immobile well-positioned nucleosomes, while their core is wrapped into intrinsically mobile and/or partially accessible nucleosomes amenable to rapid displacement or reorganization upon inducible recruitment of TFs. Therefore, the overall nucleosomal organization of stimulus-responsive enhancers is extremely robust to extrinsic perturbations, a feature that might prevent deviations from the differentiated state when cells are exposed to perturbations. Such resilience of the nucleosomal landscape likely reflects the dominance of the nucleosome-organizing activity of lineage-determining TFs such as PU.1 (Barozzi et al. 2014), whose overall genomic distribution is only marginally affected by stimulation (Ostuni et al. 2013; Mancino et al. 2015).
Our study also revealed that cobinding of multiple TFs to cis-regulatory elements is tightly associated with remodeling. This result suggests that even remodeling of fuzzy nucleosomes might be a highly cooperative process in which multiple collaborating TFs provide an important contribution. Nevertheless, not every TF appears to elicit remodeling with the same efficiency. Indeed, some TFs bound with a similar frequency to remodeled and unaffected nucleosomes, whereas others were strongly associated with remodeling events. The most unexpected finding in this regard relates to NF-kB, whose DNA-binding domain is structurally incapable of accommodating nucleosomal DNA (Natoli et al. 2005; Lone et al. 2013). Nevertheless, when recruited to DNA, NF-kB appeared to strongly associate with the disruption of local nucleosome organization, consistently predicting nucleosome remodeling after LPS stimulation. As NF-kB plays a pivotal role in the induction of the inflammatory gene expression program, its association with enhanced chromatin accessibility suggests that this may also depend on its ability to control a feed-forward mechanism acting at promoters and enhancers, whereby NF-kB binding augments local chromatin accessibility, thus favoring the subsequent recruitment of additional TFs. It is important to remark, however, that our results are, in principle, compatible with the possibility that recruitment of NF-kB occurs at previously remodeled chromatin that was driven by other TFs. Therefore, additional genetic analyses are warranted to address this issue.
Overall, the data reported in this study contribute to clarify how the exposure of mammalian cells to external stimuli impacts the accessible landscape of cis-regulatory elements by remodeling mainly poorly positioned nucleosomes. The remarkable resilience of the overall nucleosomal landscape in the face of even very strong stimuli lends support to the notion that maintenance of the nucleosomal organization in a changing environment represents an essential feature of cell differentiation.
Materials and methods
Cell cultures
Macrophages were derived from bone marrows of C57/BL6 mice (Harlan) as described (Curina et al. 2017).
MNase-ChIP-seq and ChIP-seq
Macrophages (2 × 107) were fixed with 1% HCOH, quenched with 0.125 mM Tris, and washed twice with cold PBS. Cell pellets were resuspended in a 15 mM NaCl, 15 mM Tris-HCl (pH 7.6), 60 mM KCl, 2 mM EDTA, 0.5 mM EGTA, and 0.3 M sucrose buffer (0.5 mM PMSF, 1 mM DTT, 0.2 mM spermine, 1 mM spermidine) and lysed upon addition of 0.4% NP-40. Nuclei were washed with a 15 mM NaCl, 15 mM Tris-HCl (pH 7.6), 60 mM KCl, and 0.3 M sucrose buffer (0.5 mM PMSF, 1 mM DTT, 0.2 mM spermine, 1 mM spermidine). Digestion was performed with 6 U of MNase (Roche, 10107921001) in 500 µL of a 20 mM Tris-HCl (pH 7.6) and 5 mM CaCl2 digestion buffer for 2 h at 37°C to have ∼80% mononucleosomes and 20% dinucleosomes. The reaction was stopped by adding EDTA to a 50 mM final concentration. Digestion was checked on an agarose gel after decross-linking of a small fraction of the lysate. The samples were diluted in 3 mL of a 10 mM Tris-HCl (pH 8), 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-deoxycholate, and 0.5% Na-laurylsarcosine buffer; gently sonicated to favor nucleus lysis; and then divided into four aliquots (corresponding to 5 million each) for immunoprecipitation with 5 µg of antibody. Antibodies were prebound to G protein-coupled paramagnetic beads (Dynabeads) in PBS/0.5% BSA and incubated with lysates overnight at 4°C. Beads were washed six times in a modified RIPA buffer (50 mM HEPES at pH 7.6, 500 mM LiCl, 1 mM EDTA, 1% NP-40, 0.7% Na-deoxycholate) and once in TE containing 50 mM NaCl. DNA was eluted in TE-2% SDS, and cross-links were reversed by incubation overnight at 65°C. DNA was then purified by MinElute PCR purification kit (Qiagen) and quantified with PicoGreen (Invitrogen). ChIP DNA was prepared for HiSeq 2000 sequencing following standard protocols. The antibodies used included anti-H3K4me1 (Abcam, ab88959), anti-H3K4me3 (Active Motif, 39159), and anti-H3K27ac (Abcam, ab4729).
TF ChIP-seq experiments were carried out as described (Tong et al. 2016) using the following antibodies: anti-Atf3 (Santa Cruz Biotechnology, sc-188, lot no. L2214), anti-Brg1 (Abcam, ab110641), anti-Cebpb (in-house rabbit polyclonal antisera for mouse CEBPβ amino acids 22–195), anti-Creb (Santa Cruz Biotechnology, sc-186, lot no. C0614), anti-Fos (Santa Cruz Biotechnology, sc-253, lot no. I0808), anti-Junb (Santa Cruz Biotechnology, sc-46, lot no. F1814), anti-Jund (Santa Cruz Biotechnology, sc-74, lot no. A3015), and anti-Rela (Santa Cruz Biotechnology, sc-372, lot no. H1114).
ATAC-seq
The original ATACseq protocol (Buenrostro et al. 2013) was modified according to Lara-Astiaso et al. (2014). Briefly, 5 × 104 cells were lysed in 100 µL of 10 mM Tris-HCl (pH 7.4), 10 mM NaCl, 3 mM MgCl2, and 0.1% Igepal CA-630. Nuclei were pelleted by centrifugation at 500g for 20 min at 4°C. The nuclear pellet was resuspended in 25 µL of a 10 mM Tris-HCl (pH 8.4) and 5 mM MgCl2 buffer containing 1 µL of Tn5 transposase (made in-house). The reaction was incubated for 1 h at 37°C and then stopped by adding 5 µL of cleanup buffer (900 mM NaCl, 300 mM EDTA), 2 uL of 5% SDS, and 2 µL of 20 µg/µL Proteinase K and incubating the reaction for 30 min at 40°C. Tagmented DNA was purified using 2× SPRI beads (Agencourt AMPure XP, Beckman Coulter). Finally, 2 µL of 10 µM indexing primers and KAPA HiFi HotStart ReadyMix were used to PCR-amplify the obtained DNA library of tagmented DNA. Fragments <600 bp were isolated by size selection (using 0.65× SPRI beads) and then purified with 1.8× SPRI beads. ATAC-seq library size was assessed using a Tapestation D5000 High-Sensitivity ScreenTape (Agilent). Libraries were sequenced on an Illumina NextSeq 500.
Chromatin-associated RNA
For each time point, 10 × 106 cells were lysed with an ice-cold 10 mM Tris-HCl (pH 7.5), 0.05% NP-40, and 150 mM NaCl buffer for 5 min. The lysate was then layered on 2.5 vol of a chilled sucrose cushion (24% sucrose in lysis buffer) and centrifuged at 14,000 rpm for 10 min at 4°C. The nucleus pellet was gently rinsed with ice-cold PBS/1 mM EDTA and then resuspended in a 20 mM Tris-HCl (pH 7.9), 75 mM NaCl, 0.5 mM EDTA, 0.85 mM DTT, 0.125 mM PMSF, and 50% glycerol buffer by gentle flicking of the tube. An equal volume of a cold 10 mM HEPES (pH 7.6), 1 mM DTT, 7.5 mM MgCl2, 0.2 mM EDTA, 0.3 M NaCl, 1 M urea, and 1% NP-40 buffer was added. The tube was gently vortexed and then centrifuged at 14,000 rpm for 2 min at 4°C. The chromatin pellet was rinsed with cold PBS/1 mM EDTA and then dissolved in TRIzol (Invitrogen). Chromatin-associated RNA was purified with TRIzol with an additional phenol/chloroform extraction step prior to precipitation. Purified RNA was used for library preparation with the Illumina TruSeq version 2 protocol. Libraries were sequenced on an Illumina HiSeq 2000 (50-bp single reads).
Gene, promoter, and enhancer annotations
Models for University of California at Santa Cruz (UCSC) known genes were downloaded from iGenome (https://support.illumina.com/sequencing/sequencing_software/igenome.html) on July 17, 2015. Unless stated otherwise, a TSS-proximal region was defined as ±2.5 kb from a TSS. Enhancers were defined as TSS-distal, Pu.1-bound, and H3K4me1-positive regions (Barozzi et al. 2014). Regions on chromosome M were excluded.
MNase-ChIP-seq data analysis
Reads were aligned to the reference genome (mm9) using Bowtie (version 0.12.7) (Langmead et al. 2009) with parameters -v 2 -m 1, ensuring only unique-mapping aligned reads with two or fewer mismatches were retained. Enriched regions were identified using MACS version 1.4 (Zhang et al. 2008) with parameters –nomodel –nolambda –P value = 1 × 10−5 –bw = 100 –shiftsize = 75 –gsize = mm. Wiggle tracks for visualization on the UCSC genome browser (Kent et al. 2002) were generated using MACS version 1.4 and rescaled to RPM (reads per million sequenced reads; deduplicated reads). SAMtools (version 0.1.19) (Li et al. 2009) was used to split the reads by strand, and then genomeCoverageBed from BEDTools (version 2.19.1) (Quinlan and Hall 2010) was used to generate the strand-specific genome-wide coverage profiles at single-base-pair resolution. The minus profile was then subtracted from the plus profile using wigmath.subtract from Java Genomics Toolkit (https://github.com/timpalpant/java-genomics-toolkit).
For comparative analyses, the derivative of the cumulative signal was calculated from previously published MNase-seq data in untreated macrophages (pool of four biological replicates) (Barozzi et al. 2014). SAMtools (samtools view -s) was used to subsample the total reads.
ATAC-seq data analysis
Reads were aligned to the reference genome as described for the MNase-ChIP-seq profiles. Accessible regions were then identified using MACS version 1.4 (Zhang et al. 2008) with parameters –gsize = mm –bw = 300 –nomodel –nolambda –shiftsize = 150. Wiggle profiles were also generated as described for the MNase-ChIP-seq profiles. First, accessible regions from each sample were split into TSS-distal and TSS-proximal as described above. Master lists of peaks were generated, merging all of the replicates and all time points (but separating TSS-distal and TSS-proximal) using mergeBed from BEDTools. A region was considered for further analyses if it showed a peak in two or more replicates in at least one time point. Raw read counts over these regions were quantified using coverageBed from BEDTools. edgeR (version 3.20.9) was then used for normalization and for estimating differentially accessible regions between each time point and the untreated. estimateCommonDisp and estimateTagwiseDisp were run separately followed by trimmed mean of M-values (TMM) normalization. Differential accessibility was evaluated by exactTest followed by a filter on an absolute fold change of ≥2 and a q-value of ≤0.05 (Benjamini-Hochberg correction) (Benjamini and Hochberg 1995). A master list of intergenic accessible regions was derived from the TSS-distal intervals. Regions showing accessibility in the untreated (either changed or unchanged by LPS) or gaining accessibility upon LPS treatment were considered. A filter was applied to keep only those regions with consistent H3K4me1 between replicates (untreated). This filter ensures a fair estimation of nucleosome remodeling across all of the regions, whether constitutively active or induced by LPS.
TF ChIP-seq data analysis
FASTQ files were downloaded from the Gene Expression Omnibus (GEO) (Barrett et al. 2013) and processed using Trim Galore (-q 20; https://www.bioinformatics.babraham.ac.uk/projects/trim_galore). Reads were aligned to the reference genome as described for the MNase-ChIP-seq profiles. TF-bound locations were identified using MACS version 1.4 (Zhang et al. 2008) with parameters –gsize = mm –bw = 300 –nomodel –shiftsize = 100 and sample-matched input DNA as control. LPS-induced and LPS-reduced events were identified using the same parameters. Those locations showing differential binding and overlapping a significant peak versus input DNA were kept. Unless specified otherwise, only peaks showing a P-value of ≤1 × 10−10 were retained for further analyses. Master lists of TF-binding events (i.e., regions bound by at least one TF at any time point) were derived separately for LPS-induced and LPS-reduced events, independently for TSS-proximal and TSS-distal elements, as follows. First, summits within 450 bp from each other were clustered together using mergeBed from BEDTools. A summit for each cluster was then calculated as the average of all of the summit positions of the peaks included in the cluster.
Chromatin-associated RNA-seq data analysis
Reads were aligned to the indexed murine cDNA (assembly GRCm38, release 92) using Kallisto quant (version 0.44.0) (Bray et al. 2016) with parameters -b 100 -l 300 -s 100 –single. Sleuth (R package version 0.29.0) (Pimentel et al. 2017) was used to identify differentially expressed genes (DEGs) upon LPS stimulation. Sleuth identifies DEGs by comparing a full to a reduced model. Here, the reduced model considered only replicates, while a full model considered replicates and time of stimulation. Both models were fit using sleuth_fit and compared using sleuth_lrt. DEGs were defined if q-value ≤ 0.05 and an absolute twofold-change. For LPS induction, genes up-regulated as early as 30 or 60 min were considered early response genes as opposed to late response genes; i.e., those showing increased expression only at 120 or 240 min.
Genes were classified as either primary or secondary response genes using published gene expression profiles from stimulated macrophages in the presence of cycloheximide (CHX) and type I IFN receptor (IFNAR)-deficient (Ifnar−/−) stimulated macrophages (Tong et al. 2016). Given a time point, LPS-induced genes down-regulated by ≥70% in either CHX-treated or Ifnar−/− macrophages were considered as secondary response genes. Genes that were not defined as secondary response genes at any time point were classified as primary response genes.
Characterization of symmetric and asymmetric nucleosomal arrays at putative enhancers
The orientation of nucleosomal patterns was inferred by the ratio of normalized signals on the two sides of the Pu.1 summit. These coverages were calculated using coverageBed from BEDTools. Spatially resolved signals were extracted for (1) H3K4me3 and total RNA Pol II ChIP-seq profiles in untreated macrophages (Ostuni et al. 2013), (2) CTCF (ChIP-seq profile in untreated macrophages) (Ghisletti and Natoli, unpubl.), and (3) the PAS using a model as described (Austenaa et al. 2015).
Statistical learning
Lasso logistic regression models were used to discriminate symmetric from asymmetric nucleosomal patterns at intergenic enhancers based on DNA sequence features and predict nucleosome remodeling events associated with TF dynamics during LPS stimulation. Models were trained and evaluated as described (Comoglio et al. 2015, 2018).
For modeling of nucleosomal symmetry patterns at enhancers, DNA sequences were extracted from the reference genome (mm9) and scored as follows. Three sets of features were considered: (1) DNA sequence content encoded as k-mers (2 ≤ k ≤ 4) within 450 bp upstream of and downstream from the PU.1 summit; (2) average DNA shape feature values within the same window, computed using the R package DNAshapeR version 1.5.3 (Chiu et al. 2017)—13 DNA shape features were used in this study (Sagendorf et al. 2017), along with predicted minor groove electrostatic potential (Chiu et al. 2017); (3) transformed FIMO P-values [−10*log10(p)] (Grant et al. 2011) for a curated collection of >1700 position weight matrices (PWMs) representing mammalian TF motifs (Diaferia et al. 2016), core promoter elements, 5′ splice site, and PAS motifs—these were computed within 300 bp of the PU.1 summit as described previously (Barozzi et al. 2014) using FIMO from MEME version 4.11.3.
To train the classifiers, data points were partitioned randomly into 10 balanced training (80%) and test (20%) sets. Lasso logistic regression models were trained in cross-validation (10-fold) using the R package glmnet version 2.0-13 (Friedman et al. 2010). The value of the regularization parameter resulting in a cross-validated misclassification error within one standard error from the minimum was used to predict the class labels of the enhancers in the test set. Model performances were evaluated by computing the AUC using the R package ROCR version 1.0-7 (Sing et al. 2005), and average AUC values were computed. Feature importance analysis was performed via bootstrap-lasso as described previously (Comoglio and Paro 2014). Features with selection probability (importance) of ≥0.85 were considered.
The same approach was used to model nucleosome remodeling events associated with TF dynamics using TF ChIP-seq signals (reads per kilobase per million mapped reads [RPKM]) at dynamic peaks as input features.
Detection and classification of nucleosome remodeling events and downstream analyses
Analyses were run on predefined sets of regions based on the reproducibility of either H3K4me3 (for TSS-proximal regions) or H3K4me1 (for TSS-distal regions) patterns in the untreated condition and in the LPS time point under consideration. These sets considered the peak intersection of the two biological replicates.
Local losses (eviction) or gains in MNase-ChIP-seq signals upon LPS stimulation were identified as local significant changes within globally unaffected broader regions as follows. First, the differences between LPS and untreated were computed using a fixed window of 5.4 kb (450 bp × 12, defining the broad context). Second, each region was scanned using a sliding window of 450 bp (defining the local context), with a step of 150 bp. Sliding windows exhibiting a statistically significant difference occurring within a fixed window displaying no significant difference were classified into losses or gains based on sign. Formally, the difference for each window was assessed using csaw (R package version 1.12.0) (Lun and Smyth 2016). After normalization and estimation of dispersion, a quasilikelihood negative binomial generalized log-linear model was used to fit the count data and estimate the significance of differential signals. A q-value of ≤0.05 (Benjamini-Hochberg correction) (Benjamini and Hochberg 1995) was applied.
To identify nucleosomal shifts upon LPS stimulation, reproducible decreases in signal correlation between two conditions were detected using strand-specific subtracted MNase-ChIP-seq coverage profiles. A sliding window (450 bp in size, with a step size of 150 bp) was used to scan a 2.25-kb region centered on the landmark of interest (i.e., the TSS of a gene or the summit of a peak). For each window, the subtracted MNase-ChIP-seq coverage was extracted and individually summarized at 15-bp resolution (n = 30 bins) for each replicate and condition. Next, the Spearman's rank correlation coefficient of the summarized coverage between the two untreated samples and each of the four possible untreated/LPS-treated sample pairs was computed. We then determined whether the correlation between the summarized coverage of a given untreated replicate and a given LPS-treated replicate was significantly smaller than the correlation between the two untreated replicates for each of the four possible untreated/LPS-treated pairs. The statistical significance of the difference between these dependent correlations was determined using the paired.r function from the R package psych. The maximum P-values from these four tests was used as conservative estimate of significance. P-values from the same 2.25-kb region were combined using the Simes’ method (Sarkar and Chang 1997) and further corrected for multiple hypothesis testing by independent hypothesis weighting (IHW; R package version 1.6.0) (Ignatiadis et al. 2016). The geometric mean of the signal coverage of the four samples (two untreated and two LPS-treated) was used as a covariate.
Nucleosome fuzziness was assessed genome-wide using the function dpos of DANPOS (version 2.2.2) (Chen et al. 2013) with default parameters. Only those estimates overlapping a peak called from the same MNase-ChIP-seq profiles were considered. The fuzziness scores were then converted to percentiles.
Data and software availability
Raw sequencing data were deposited at the Gene Expression Omnibus (GEO) under accession number GSE119693.
Acknowledgments
This study was supported by the European Research Council (ERC AdG no. 692789 to G.N.). F.C. was supported by an EMBO long-term fellowship (1305-2015), Marie Curie Actions (LTFCOFUND2013/GA-2013-609409), and a Swiss National Science Foundation postdoctoral fellowship (P2EZP3_165206). I.B. was funded through an Imperial College Research Fellowship. We thank Boris Lenhard (Imperial College London) for discussions, and Luca Rotta, Thelma Capra, and Salvatore Bianchi (Istituto Europeo di Oncologia and Italian Institute of Technology Center for Genomic Sciences, Milan) for the preparation and processing of part of the next-generation sequencing libraries. F.C. thanks Tony Green and Bas van Steensel for supporting this project.
Author contributions: M.S., S.P., and X.L. generated the data. F.C. and I.B. analyzed and modeled the data. G.N. and S.T.S. acquired funding and supervised the study. All authors contributed to writing, reviewing, and editing the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.326348.119.
Freely available online through the Genes & Development Open Access option.
References
- Austenaa LM, Barozzi I, Simonatto M, Masella S, Della Chiara G, Ghisletti S, Curina A, de Wit E, Bouwman BA, de Pretis S, et al. 2015. Transcription of mammalian cis-regulatory elements is restrained by actively enforced early termination. Mol Cell 60: 460–474. 10.1016/j.molcel.2015.09.018 [DOI] [PubMed] [Google Scholar]
- Barozzi I, Simonatto M, Bonifacio S, Yang L, Rohs R, Ghisletti S, Natoli G. 2014. Coregulation of transcription factor binding and nucleosome occupancy through DNA features of mammalian enhancers. Mol Cell 54: 844–857. 10.1016/j.molcel.2014.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. 2013. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res 41: D991–D995. 10.1093/nar/gks1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Methodol 57: 289–300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- Bhatt DM, Pandya-Jones A, Tong AJ, Barozzi I, Lissner MM, Natoli G, Black DL, Smale ST. 2012. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell 150: 279–290. 10.1016/j.cell.2012.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. 2008. High-resolution mapping and characterization of open chromatin across the genome. Cell 132: 311–322. 10.1016/j.cell.2007.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L. 2016. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34: 525–527. 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10: 1213–1218. 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Xi Y, Pan X, Li Z, Kaestner K, Tyler J, Dent S, He X, Li W. 2013. DANPOS: dynamic analysis of nucleosome position and occupancy by sequencing. Genome Res 23: 341–351. 10.1101/gr.142067.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu TP, Comoglio F, Zhou T, Yang L, Paro R, Rohs R. 2016. DNAshapeR: an R/Bioconductor package for DNA shape prediction and feature encoding. Bioinformatics 32: 1211–1213. 10.1093/bioinformatics/btv735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu TP, Rao S, Mann RS, Honig B, Rohs R. 2017. Genome-wide prediction of minor-groove electrostatic potential enables biophysical modeling of protein–DNA binding. Nucleic Acids Res 45: 12565–12576. 10.1093/nar/gkx915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comoglio F, Paro R. 2014. Combinatorial modeling of chromatin features quantitatively predicts DNA replication timing in Drosophila. PLoS Comput Biol 10: e1003419. 10.1371/journal.pcbi.1003419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comoglio F, Schlumpf T, Schmid V, Rohs R, Beisel C, Paro R. 2015. High-resolution profiling of Drosophila replication start sites reveals a DNA shape and chromatin signature of metazoan origins. Cell Rep 11: 821–834. 10.1016/j.celrep.2015.03.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comoglio F, Park HJ, Schoenfelder S, Barozzi I, Bode D, Fraser P, Green AR. 2018. Thrombopoietin signaling to chromatin elicits rapid and pervasive epigenome remodeling within poised chromatin architectures. Genome Res 28: 295–309. 10.1101/gr.227272.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curina A, Termanini A, Barozzi I, Prosperini E, Simonatto M, Polletti S, Silvola A, Soldi M, Austenaa L, Bonaldi T, et al. 2017. High constitutive activity of a broad panel of housekeeping and tissue-specific cis-regulatory elements depends on a subset of ETS proteins. Genes Dev 31: 399–412. 10.1101/gad.293134.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaferia GR, Balestrieri C, Prosperini E, Nicoli P, Spaggiari P, Zerbi A, Natoli G. 2016. Dissection of transcriptional and cis-regulatory control of differentiation in human pancreatic cancer. EMBO J 35: 595–617. 10.15252/embj.201592404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, et al. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng R, Desbordes SC, Xie H, Tillo ES, Pixley F, Stanley ER, Graf T. 2008. PU.1 and C/EBPα/β convert fibroblasts into macrophage-like cells. Proc Natl Acad Sci 105: 6057–6062. 10.1073/pnas.0711961105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenouil R, Cauchy P, Koch F, Descostes N, Cabeza J, Innocenti C, Ferrier P, Spicuglia S, Gut M, Gut I, et al. 2012. CpG islands and GC content dictate nucleosome depletion in a transcription-independent manner at mammalian promoters. Genome Res 22: 2399–2408. 10.1101/gr.138776.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster SL, Hargreaves DC, Medzhitov R. 2007. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature 447: 972–978. 10.1038/nature05836 [DOI] [PubMed] [Google Scholar]
- Friedman J, Hastie T, Tibshirani R. 2010. Regularization paths for generalized linear models via coordinate descent. J Stat Softw 33: 1–22. 10.18637/jss.v033.i01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, et al. 2010. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity 32: 317–328. 10.1016/j.immuni.2010.02.008 [DOI] [PubMed] [Google Scholar]
- Glass CK, Natoli G. 2016. Molecular control of activation and priming in macrophages. Nat Immunol 17: 26–33. 10.1038/ni.3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode DK, Obier N, Vijayabaskar MS, Lie ALM, Lilly AJ, Hannah R, Lichtinger M, Batta K, Florkowska M, Patel R, et al. 2016. Dynamic gene regulatory networks drive hematopoietic specification and differentiation. Dev Cell 36: 572–587. 10.1016/j.devcel.2016.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS. 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018. 10.1093/bioinformatics/btr064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle V, Li N, Hadzhiev Y, Plessy C, Previti C, Nepal C, Gehrig J, Dong X, Akalin A, Suzuki AM, et al. 2014. Two independent transcription initiation codes overlap on vertebrate core promoters. Nature 507: 381–385. 10.1038/nature12974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatiadis N, Klaus B, Zaugg JB, Huber W. 2016. Data-driven hypothesis weighting increases detection power in genome-scale multiple testing. Nat Methods 13: 577–580. 10.1038/nmeth.3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, Chun HB, Tough DF, Prinjha RK, Benner C, et al. 2013. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell 51: 310–325. 10.1016/j.molcel.2013.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. 2002. The human genome browser at UCSC. Genome Res 12: 996–1006. 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai WKM, Pugh BF. 2017. Understanding nucleosome dynamics and their links to gene expression and DNA replication. Nat Rev Mol Cell Biol 18: 548–562. 10.1038/nrm.2017.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai B, Gao W, Cui K, Xie W, Tang Q, Jin W, Hu G, Ni B, Zhao K. 2018. Principles of nucleosome organization revealed by single-cell micrococcal nuclease sequencing. Nature 562: 281–285. 10.1038/s41586-018-0567-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Astiaso D, Weiner A, Lorenzo-Vivas E, Zaretsky I, Jaitin DA, David E, Keren-Shaul H, Mildner A, Winter D, Jung S, et al. 2014. Immunogenetics. Chromatin state dynamics during blood formation. Science 345: 943–949. 10.1126/science.1256271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lone IN, Shukla MS, Charles Richard JL, Peshev ZY, Dimitrov S, Angelov D. 2013. Binding of NF-κB to nucleosomes: effect of translational positioning, nucleosome remodeling and linker histone H1. PLoS Genet 9: e1003830. 10.1371/journal.pgen.1003830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun AT, Smyth GK. 2016. csaw: a Bioconductor package for differential binding analysis of ChIP-seq data using sliding windows. Nucleic Acids Res 44: e45. 10.1093/nar/gkv1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancino A, Termanini A, Barozzi I, Ghisletti S, Ostuni R, Prosperini E, Ozato K, Natoli G. 2015. A dual cis-regulatory code links IRF8 to constitutive and inducible gene expression in macrophages. Genes Dev 29: 394–408. 10.1101/gad.257592.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticelli S, Natoli G. 2017. Transcriptional determination and functional specificity of myeloid cells: making sense of diversity. Nat Rev Immunol 17: 595–607. 10.1038/nri.2017.51 [DOI] [PubMed] [Google Scholar]
- Mossadegh-Keller N, Sarrazin S, Kandalla PK, Espinosa L, Stanley ER, Nutt SL, Moore J, Sieweke MH. 2013. M-CSF instructs myeloid lineage fate in single haematopoietic stem cells. Nature 497: 239–243. 10.1038/nature12026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller B, Mieczkowski J, Kundu S, Wang P, Sadreyev R, Tolstorukov MY, Kingston RE. 2017. Widespread changes in nucleosome accessibility without changes in nucleosome occupancy during a rapid transcriptional induction. Genes Dev 31: 451–462. 10.1101/gad.293118.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli G, Saccani S, Bosisio D, Marazzi I. 2005. Interactions of NF-κB with chromatin: the art of being at the right place at the right time. Nat Immunol 6: 439–445. 10.1038/ni1196 [DOI] [PubMed] [Google Scholar]
- Novakovic B, Habibi E, Wang SY, Arts RJW, Davar R, Megchelenbrink W, Kim B, Kuznetsova T, Kox M, Zwaag J, et al. 2016. β-Glucan reverses the epigenetic state of LPS-induced immunological tolerance. Cell 167: 1354–1368.e14. 10.1016/j.cell.2016.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, Natoli G. 2013. Latent enhancers activated by stimulation in differentiated cells. Cell 152: 157–171. 10.1016/j.cell.2012.12.018 [DOI] [PubMed] [Google Scholar]
- Park SH, Kang K, Giannopoulou E, Qiao Y, Kang K, Kim G, Park-Min KH, Ivashkiv LB. 2017. Type I interferons and the cytokine TNF cooperatively reprogram the macrophage epigenome to promote inflammatory activation. Nat Immunol 18: 1104–1116. 10.1038/ni.3818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel H, Bray NL, Puente S, Melsted P, Pachter L. 2017. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods 14: 687–690. 10.1038/nmeth.4324 [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST. 2009. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell 138: 114–128. 10.1016/j.cell.2009.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagendorf JM, Berman HM, Rohs R. 2017. DNAproDB: an interactive tool for structural analysis of DNA–protein complexes. Nucleic Acids Res 45: W89–W97. 10.1093/nar/gkx272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar SK, Chang C-K. 1997. The simes method for multiple hypothesis testing with positively dependent test statistics. J Am Stat Assoc 92: 1601–1608. 10.1080/01621459.1997.10473682 [DOI] [Google Scholar]
- Scott-Browne JP, López-Moyado IF, Trifari S, Wong V, Chavez L, Rao A, Pereira RM. 2016. Dynamic changes in chromatin accessibility occur in CD8+ T cells responding to viral infection. Immunity 45: 1327–1340. 10.1016/j.immuni.2016.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalek AK, Satija R, Adiconis X, Gertner RS, Gaublomme JT, Raychowdhury R, Schwartz S, Yosef N, Malboeuf C, Lu D, et al. 2013. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature 498: 236–240. 10.1038/nature12172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing T, Sander O, Beerenwinkel N, Lengauer T. 2005. ROCR: visualizing classifier performance in R. Bioinformatics 21: 3940–3941. 10.1093/bioinformatics/bti623 [DOI] [PubMed] [Google Scholar]
- Tillo D, Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Field Y, Lieb JD, Widom J, Segal E, Hughes TR. 2010. High nucleosome occupancy is encoded at human regulatory sequences. PLoS One 5: e9129. 10.1371/journal.pone.0009129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AJ, Liu X, Thomas BJ, Lissner MM, Baker MR, Senagolage MD, Allred AL, Barish GD, Smale ST. 2016. A Stringent systems approach uncovers gene-specific mechanisms regulating inflammation. Cell 165: 165–179. 10.1016/j.cell.2016.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahedi G, Takahashi H, Nakayamada S, Sun HW, Sartorelli V, Kanno Y, O'Shea JJ. 2012. STATs shape the active enhancer landscape of T cell populations. Cell 151: 981–993. 10.1016/j.cell.2012.09.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A, Johnson S, Boyd S, Smith C, Fire A, Sidow A. 2011. Determinants of nucleosome organization in primary human cells. Nature 474: 516–520. 10.1038/nature10002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al. 2011. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474: 390–394. 10.1038/nature10006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann AS, Mitchell DM, Sanjabi S, Bradley MN, Hoffmann A, Liou HC, Smale ST. 2001. Nucleosome remodeling at the IL-12 p40 promoter is a TLR-dependent, Rel-independent event. Nat Immunol 2: 51–57. 10.1038/83168 [DOI] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. 2011. Pioneer transcription factors: establishing competence for gene expression. Genes Dev 25: 2227–2241. 10.1101/gad.176826.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol 9: R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F, Farnung L, Kaasinen E, Sahu B, Yin Y, Wei B, Dodonova SO, Nitta KR, Morgunova E, Taipale M, et al. 2018. The interaction landscape between transcription factors and the nucleosome. Nature 562: 76–81. 10.1038/s41586-018-0549-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw sequencing data were deposited at the Gene Expression Omnibus (GEO) under accession number GSE119693.