In this study, Hurtz et al. used genetic mouse models and patient-derived leukemia cells to show that the transcriptional repressor BCL6 functions as a central driver of drug resistance in MLL-rearranged B-cell leukemia. Their genetic and ChIP-seq analyses demonstrated that MLL-AF4 and MLL-ENL fusions directly bound to the BCL6 promoter and up-regulated BCL6 expression, thus providing new insights into a previously unrecognized requirement of malignant transformation by oncogenic MLL fusions.
Keywords: B cells, BCL6, BIM, MLL
Abstract
Chromosomal rearrangements of the mixed lineage leukemia (MLL) gene occur in ∼10% of B-cell acute lymphoblastic leukemia (B-ALL) and define a group of patients with dismal outcomes. Immunohistochemical staining of bone marrow biopsies from most of these patients revealed aberrant expression of BCL6, a transcription factor that promotes oncogenic B-cell transformation and drug resistance in B-ALL. Our genetic and ChIP-seq (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing) analyses showed that MLL-AF4 and MLL-ENL fusions directly bound to the BCL6 promoter and up-regulated BCL6 expression. While oncogenic MLL fusions strongly induced aberrant BCL6 expression in B-ALL cells, germline MLL was required to up-regulate Bcl6 in response to physiological stimuli during normal B-cell development. Inducible expression of Bcl6 increased MLL mRNA levels, which was reversed by genetic deletion and pharmacological inhibition of Bcl6, suggesting a positive feedback loop between MLL and BCL6. Highlighting the central role of BCL6 in MLL-rearranged B-ALL, conditional deletion and pharmacological inhibition of BCL6 compromised leukemogenesis in transplant recipient mice and restored sensitivity to vincristine chemotherapy in MLL-rearranged B-ALL patient samples. Oncogenic MLL fusions strongly induced transcriptional activation of the proapoptotic BH3-only molecule BIM, while BCL6 was required to curb MLL-induced expression of BIM. Notably, peptide (RI-BPI) and small molecule (FX1) BCL6 inhibitors derepressed BIM and synergized with the BH3-mimetic ABT-199 in eradicating MLL-rearranged B-ALL cells. These findings uncover MLL-dependent transcriptional activation of BCL6 as a previously unrecognized requirement of malignant transformation by oncogenic MLL fusions and identified BCL6 as a novel target for the treatment of MLL-rearranged B-ALL.
Chromosomal translocations involving the mixed lineage leukemia (MLL) gene account for ∼70% of B-cell lineage acute lymphoblastic leukemia (B-ALL) in infants and ∼10% in older children and adults (Tkachuk et al. 1992; Ayton and Cleary 2001; Winters and Bernt 2017). With overall survival rates of <50% (Issa et al. 2017; Sun et al. 2018), MLL rearrangements define a group of patients with particularly poor clinical outcome. MLL-rearranged B-ALL clones carry very few additional genetic lesions (Andersson et al. 2015), and it is currently unclear how oncogenic MLL fusions promote drug resistance and, ultimately, poor outcomes in patients.
MLL belongs to the family of histone methyltransferases and plays a critical role in maintaining hematopoietic stem cells (Jude et al. 2007). More than 80 fusion partners have been identified in MLL rearrangements, including AF4 and ENL as the most frequently rearranged MLL fusion partners (Meyer et al. 2018). MLL fusion proteins retain the MLL N-terminal (MLLN) DNA-binding domains (including AT hook and CXXC) and the capacity to interact with Menin and to translocate to the nucleus. Most MLL fusion partners are nuclear proteins involved in the transcriptional elongation regulation (Shilatifard et al. 1996). Together with the positive transcription elongation factor b (P-TEFb) and the H3K79 methyltransferase DOT1L, the fusion partners form a large elongation machinery called the superelongation complex (SEC) (Luo et al. 2012). Aberrant transactivation of MLL target genes (for example, HOXA9 and HOXA10) induced by recruitment of SEC to MLL-binding sites is linked to development of MLL-rearranged leukemias (Krivtsov et al. 2008; Lin et al. 2010; Bernt et al. 2011; Smith et al. 2011).
The BCL6 proto-oncogene was initially discovered in diffuse large B-cell lymphoma (DLBCL) (Baron et al. 1993; Kerckaert et al. 1993; Ye et al. 1993) as part of the BCL6-IGH rearrangement owing to the t(3;14)(q27;q32) translocation, which is frequently found in DLBCL and other germinal center B-cell-derived lymphomas. BCL6 is also essential for proliferation and survival of normal germinal center B cells during antibody affinity maturation (Bunting et al. 2016). During early stages of B-cell development, BCL6 promotes self-renewal of B-cell precursors and enables the formation of a diverse polyclonal B-cell repertoire (Duy et al. 2010). We previously identified BCL6 as a novel mediator of drug resistance to tyrosine kinase inhibitors in Philadelphia chromosome-positive (Ph+) ALL (Duy et al. 2011) and found that MLL fusion transcription factors can bind to BCL6 (Geng et al. 2012). Analyzing gene expression data from 207 children with high-risk ALL, we found that BCL6 is a predictor of poor clinical outcome in B-ALL, particularly in cases with MLL rearrangements. Thus, while the mechanisms of drug resistance in MLL-rearranged B-ALL are largely unknown, we studied here a potential role of BCL6 in promoting the aggressive and refractory phenotype in this subtype of B-ALL.
Results
BCL6 as a predictor of poor clinical outcome in MLL-rearranged B-ALL
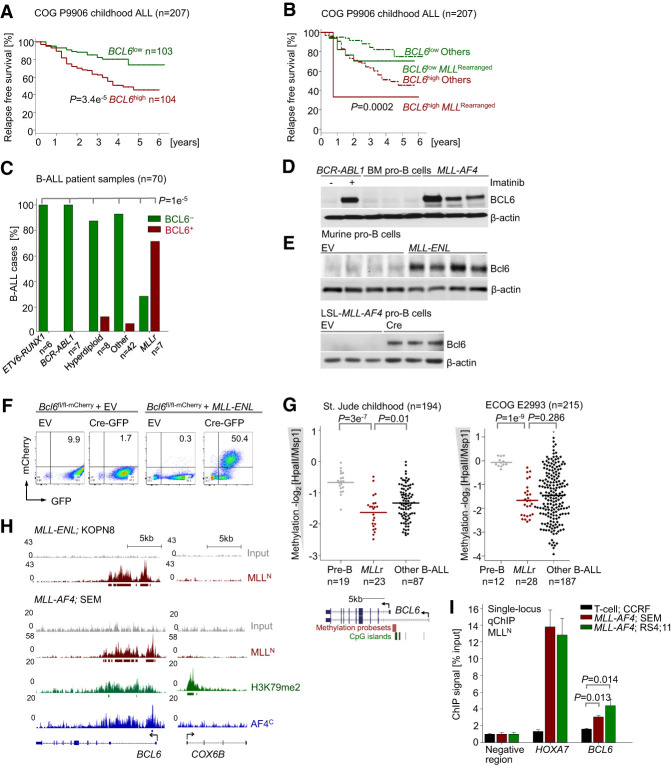
Studying gene expression data from a pediatric clinical trial for high-risk B-ALL (Children's Oncology Group [COG] P9906; n = 207), including MLL-rearranged B-ALL (Harvey et al. 2010), higher than median expression levels of BCL6 at the time of diagnosis were associated with shorter relapse-free survival (RFS) (Fig. 1A) and overall survival (Supplemental Fig. S1A). In addition, comparing matched sample pairs from 49 patients at the time of diagnosis and subsequent relapse, BCL6 mRNA levels were significantly higher in the relapse samples (P = 5.5 × 10−05) (Supplemental Fig. S1B). While these results suggest that high BCL6 mRNA levels predict poor outcome across multiple cytogenetic subtypes of high-risk B-ALL, multivariate analyses showed that this was the case in particular for patients with MLL-rearranged B-ALL (Fig. 1B). For instance, RFS at 4 yr for patients with low BCL6 and lacking MLL rearrangements was 85% (95% combination index [CI], 79%–91%) compared with 37% (95% CI, 27%–46%) for patients with high BCL6 and rearranged MLL. These findings prompted us to study potential interactions between BCL6 and MLL function in MLL-rearranged B-ALL.
Figure 1.
BCL6 is up-regulated in MLL-rearranged ALL and correlates with poor clinical outcome. (A) Patients in a pediatric high-risk ALL trial (COG P9906; n = 207) were segregated into two groups based on whether BCL6 mRNA levels were higher (BCL6high) or lower (BCL6low) than the median expression value. RFS was assessed in the two groups by Kaplan-Meier analysis. Log-rank test, P = 3.39 × 10−05. (B) Multivariate analysis of RFS in pediatric B-ALL patients from the COG P9906 clinical trial. n = 207. Patients were segregated into four groups based on higher or lower than median expression levels of BCL6 and MLL status (rearranged or other). Log-rank test, P = 0.000208. (C) Immunohistochemical staining (Supplemental Fig. 2A) in bone marrow biopsies from B-ALL patients of different subtypes (n = 70) (Supplemental Table 2), including ETV6-RUNX1 (n = 6), BCR-ABL1 (n = 7), hyperdiploid (n = 8), MLL-rearranged (n = 7), and others (n = 42). Shown are percentages of different subtypes of B-ALLs that express (red) or do not express (green) BCL6. (D) BCL6 protein levels in CD19+ B cells from healthy donors (n = 3) and patient-derived MLL-rearranged B-ALL cells (n = 3). As a positive control for BCL6 expression, human Ph+ ALL (BV173) cells were treated with 10 µmol/L imatinib for 24 h. (E) Western blot analyses were performed to measure protein levels of Bcl6 upon overexpression of MLL-ENL (top panel) and Cre-mediated inducible activation of LSL-MLL-AF4 upon excision of a loxP-flanked Stop cassette (bottom panel) in murine pre-B cells. (F) A conditional Bcl6 knockout/mCherry reporter (Bcl6fl/fl-mCherry) mouse model in which exons 5–10 of Bcl6 are flanked by LoxP sites was developed (Geng et al. 2015). Cre-mediated deletion results in expression of a truncated Bcl6 protein fused to mCherry, allowing for simultaneous inducible ablation of Bcl6 and measurement of transcriptional activity of the Bcl6 promoter. Murine pre-B cells from Bcl6fl/fl-mCherry mice were transduced with MLL-ENL or an empty vector (EV) control, followed by transduction with a Cre-GFP expression vector or EV. Using the reporter capability, significantly higher transcriptional activation of Bcl6 in MLL-ENL transduced cells was observed, as reflected by increases in proportions of mCherry-positive cells. Transcriptional activation of Bcl6 was increased in concert with Cre-mediated deletion of Bcl6. (G) DNA methylation values of the BCL6 promoter region obtained from the HELP assays in pre-B cells from healthy donors, MLL-rearranged ALL patient samples, and other subtypes of B-ALL (St. Jude Childhood and Eastern Cooperative Oncology Group [ECOG] E2993). For DNA methylation at the BCL6 locus, methylation probe sets and CpG islands are shown. (H) ChIP-seq (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing) tracks on the BCL6 promoter region using an antibody specific for MLLN (red) in human B-ALL cell lines with MLL rearrangement: KOPN8 (MLL-ENL) and SEM (MLL-AF4). Gene models are shown in University of California at Santa Cruz Genome Browser view (hg18). ChIP-seq tracks for H3K79me2 (green) and AF4 C-terminal antibody (AF4C; blue) in the promoter region of BCL6 in SEM cells are also shown. The Y-axis represents the number of reads for peak summit normalized by the total number of reads per track (set to 1 Gb for each track). COX6B served as a negative control. (I) Quantitative single-locus ChIP validation of MLL binding to the promoter of BCL6 in SEM and RS4;11 cells was performed using HOXA7 (a known target of MLL fusion) as a positive control. ALL cells (CCRF-CEM) with no MLL rearrangement and an intergenic region with no binding enrichment were used as negative controls. Shown are mean values ± SD. n = 3.
Oncogenic MLL fusion proteins drive aberrant expression of BCL6
Immunohistochemical staining of bone marrow biopsies from B-ALL patients (n = 70) revealed that the majority of MLL-rearranged B-ALL (five out of seven; 71%) showed aberrant BCL6 expression. In contrast, BCL6 expression was rarely found in other B-ALL samples (four out of 63; 6%; P = 1 × 10−06), including hyperdiploid (one out of eight; 13%), ETV6-RUNX1 (zero out of six; 0%), BCR-ABL1 (zero out of seven; 0%), and B-ALL with hypodiploid or normal karyotype (three out of 42; 7%) (Fig. 1C; Supplemental Fig. S2A; Supplemental Table S2). BCL6 protein levels were substantially elevated in patient-derived MLL-rearranged B-ALL cells when compared with normal CD19+ bone marrow pro-B cells from healthy donors (Fig. 1D). To determine whether oncogenic MLL fusion proteins mediate aberrant BCL6 expression, IL7-dependent murine pro-B cells were retrovirally transduced with MLL-ENL, which induced 10-fold to 25-fold up-regulation of Bcl6 protein levels (Fig . 1E). Likewise, Cre-mediated excision of a Stop cassette and transcriptional activation of an MLL-AF4 knock-in allele (Krivtsov et al. 2008) substantially increased Bcl6 levels in murine pro-B cells (Fig. 1E). Studying the effects of MLL-ENL in pro-B cells from a conditional Bcl6 mCherry reporter (Bcl6fl/fl-mCherry) mouse model developed recently by our group (Geng et al. 2015) further confirmed strong transcriptional activation of the Bcl6 locus by MLL-ENL (Fig. 1F). Collectively, these findings show that oncogenic MLL-ENL and MLL-AF4 fusions induce aberrant transcriptional activation of BCL6.
MLL fusions interact with H3K79 methyltransferase DOT1L to activate transcription of target genes (Shilatifard 2006; Bernt et al. 2011; Biswas et al. 2011). Target recognition by MLL fusions is achieved through binding of the N-terminal MLL CXXC domain to unmethylated CpG sites (Cierpicki et al. 2010). Studying the methylation status of the BCL6 promoter in patient-derived MLL-rearranged B-ALL samples, BCL6 promoter regions were substantially hypomethylated in MLL-rearranged B-ALL samples (n = 23) compared with normal bone marrow pro-B cells from healthy donors (n = 13) (Fig. 1G). ChIP-seq (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing) analyses in human B-ALL cell lines carrying MLL-ENL (KOPN8) and MLL-AF4 (SEM) fusions revealed binding of MLLN to hypomethylated BCL6 promoter sequences (Fig. 1H). Direct binding of the MLL-AF4 fusion protein was further confirmed by ChIP-seq using an AF4 C-terminal antibody (AF4C) (Fig. 1H). Finally, single-locus quantitative ChIP (ChIP-qPCR) using the MLLN antibody confirmed binding of MLL-AF4 to the BCL6 promoter in two B-ALL cell lines (SEM and RS4;11) carrying MLL-AF4 (Fig. 1I). MLL fusions can induce the H3K79me2 chromatin mark through recruitment of DOT1L (Bernt et al. 2011), which correlates with active MLL-induced transcription (Schubeler et al. 2004). Consistent with the scenario that MLL fusions drive aberrant expression of BCL6 through binding to hypomethylated BCL6 promoter sequences, H3K79me2 was strongly coenriched with MLLN and AF4C at the BCL6 promoter (Fig. 1H).
Germline MLL is required for up-regulation of BCL6 expression during normal B-cell development
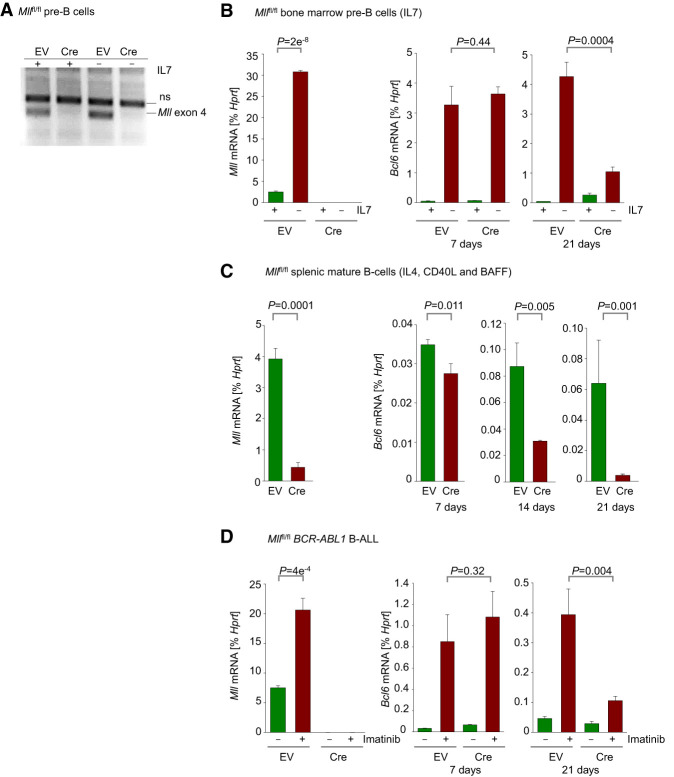
In addition to oncogenic MLL fusions, germline-encoded MLL may also interact with BCL6. Germline MLL is essential for the development of hematopoietic stem cells (Jude et al. 2007) and is crucial for the proliferation and survival of MLL-AF9-driven acute myeloid leukemia (Thiel et al. 2010). Like BCL6, high mRNA levels of germline-encoded MLL were associated with poor clinical outcome in children with B-ALL (COG P9906; n = 207) (Supplemental Figs. 1C, 3A). In multivariate analyses, high expression levels of MLL and BCL6 independently correlated with poor clinical outcome. The interaction between MLL and BCL6 mRNA levels was comparable with interaction between MLL mRNA levels and white blood cell counts (WBCs) as an established independent predictive factor. Both high BCL6 mRNA levels and WBCs compounded the association between MLL and unfavorable outcomes (COG P9906) (Supplemental Figs. 1D, 3B,C). Interestingly, BCL6 and germline MLL mRNA levels were positively correlated in both pediatric (COG P9906) and adult (Eastern Cooperative Oncology Group [ECOG] E2993) B-ALL patient samples (P = 0.002 and P = 0.00001, respectively) (Supplemental Figs. 3D,E), raising the possibility that germline-encoded MLL could transcriptionally activate BCL6 in ways similar to MLL fusions. Hence, we studied the effects of genetic ablation of Mll on expression of Bcl6 in normal pre-B cells and transformed B-ALL cells as well as mature splenic B cells from Mllfl/fl mice. Strong up-regulation of BCL6 in pre-B cells can be induced by withdrawal of IL7 from cell culture (Duy et al. 2010), treatment of BCR-ABL1 B-ALL cells with imatinib (Duy et al. 2011), and activation of splenic B cells with IL4 and CD40L. Cre-mediated ablation of Mll (Fig. 2A–D) did not have an immediate effect on Bcl6 expression levels in any of these three situations. At early time points following Cre-mediated deletion of Mll, IL7 withdrawal, BCR-ABL1 kinase inhibition and activation by IL4 and CD40L induced Bcl6 up-regulation at levels similar to those in B cells retaining Mll-floxed alleles (Fig. 2B–D). After 14 and 21 d following induction of Cre, however, IL7 withdrawal in pre-B cells, BCR-ABL1 inhibition in B-ALL, and activation by IL4 and CD40L in mature B cells failed to elicit significant up-regulation of Bcl6 (Fig. 2B–D).
Figure 2.
Germline MLL is required for up-regulation of BCL6 expression during normal B-cell development. (A) Verification of deletion of Mll by genotyping. (B) Mllfl/fl pre-B cells expressing EV or Cre were cultured in the presence of IL-7. qRT-PCR was performed to measure levels of Mll (day 7) and Bcl6 (days 7 and 21) following Cre-mediated deletion of Mll with or without IL-7 withdrawal. Withdrawal of IL-7 was carried out 24 h prior to RNA extraction. (C) Mll (day 7) and Bcl6 (days 7, 14, and 21) mRNA levels were measured in Mllflfl spleen cells cultured in the presence of IL4, CD40L, and BAFF following Cre-mediated deletion of Mll. (D) qRT-PCR measurements of Mll (day 7) and Bcl6 (days 7 and 21) levels in Mllfl/fl BCR-ABL1 B-ALL cells following Cre-mediated deletion of Mll. Cells were treated with vehicle control or 1 µmol/L imatinib 24 h prior to RNA extraction. All qRT-PCR measurements were performed using Hprt as a reference. Shown are mean values ± SD. n = 3.
These results, based on three different experimental situations, showed that Mll is essential for transcriptional activation of Bcl6 in pre-B, B-ALL, and mature B cells. However, significant effects on Bcl6 expression were not observed until >1 wk following Mll ablation. Protection from methylation by MLL maintains accessibility of target genes to transcriptional activation, and the known time frame for acquisition of promoter methylation following Mll deletion is 1–4 wk (Erfurth et al. 2008). To examine whether the delay in Bcl6 down-regulation could be due in part to protection of CpG islands within the Bcl6 promoter region from methylation by germline MLL, we performed bisulfite conversion followed by methylation-specific PCR to determine whether increases in methylation in the Bcl6 promoter region were observed following Mll deletion. After 7 and 21 d following induction of Cre in BCR-ABL1 B-ALL cells, no changes in the methylation status of the Bcl6 promoter region were observed (Supplemental Figs. 2B,C), suggesting that other effects on transcriptional programming might be involved.
BCL6 is essential for positive regulation of MLL in pre-B cells
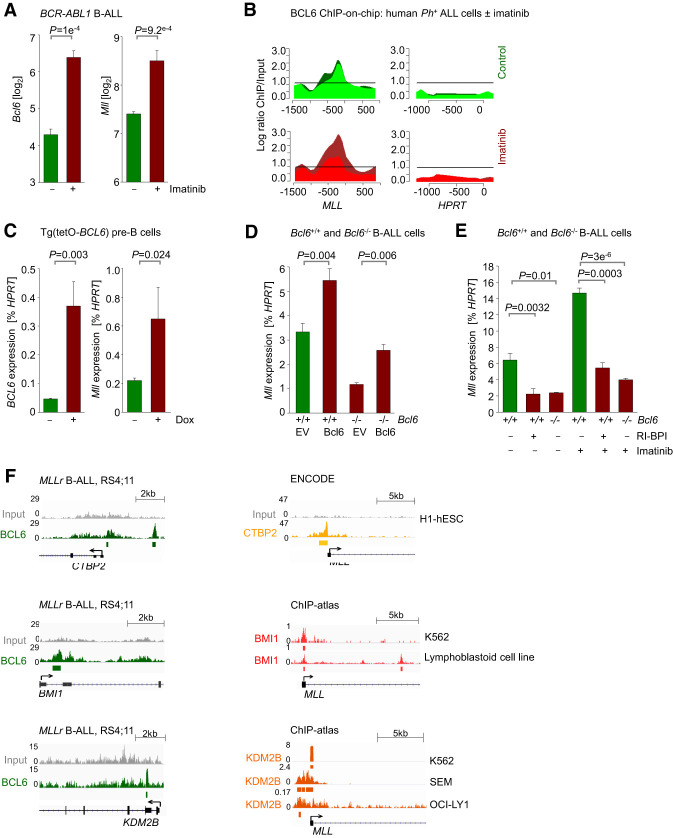
We demonstrated that MLL fusions drive aberrant expression of BCL6 and that germline MLL is essential for maintaining transcriptional activation of BCL6 in B-ALL and normal B-cell subsets. Since BCL6 and MLL mRNA levels were positively correlated in B-ALL samples from two clinical cohorts (Supplemental Figs. 3D,E), we tested potential reciprocal feedback regulation between BCL6 and MLL. Inhibition of BCR-ABL1 tyrosine kinase signaling in Ph+ B-ALL cells by imatinib up-regulated not only BCL6 but also MLL mRNA levels (GSE21664) (Fig. 3A). Interestingly, ChIP and DNA microarray (ChIP-on-chip) analysis (GSE24426) indicated that BCL6 was enriched in the MLL promoter in human B-ALL cells (Fig. 3B). To examine potential transcriptional regulation of MLL by BCL6, we studied gain and loss of function of Bcl6 in mouse pre-B and B-ALL cells. For gain-of-function studies, we used a transgenic mouse model in which expression of the human BCL6 transgene is induced by doxycycline [Dox; Tg(tetO-BCL6)] Baron et al. 2004, 2012) and studied the impact of acute activation of BCL6 on Mll expression in mouse pre-B cells. Inducible expression of BCL6 increased Mll mRNA levels (Fig. 3C), corroborating a scenario in which BCL6 plays a role in positively regulating MLL expression and vice versa. For loss-of-function studies, we measured Mll levels upon genetic ablation of Bcl6 or pharmacological inhibition of Bcl6 activity using a specific retro–inverso peptide inhibitor (RI-BPI) (Cerchietti et al. 2009). Genetic ablation of Bcl6 led to reduced Mll levels, which were restored by reconstitution of Bcl6 (Fig. 3D). Similar to effects observed upon deletion of Bcl6, treatment with RI-BPI decreased Mll expression (Fig. 3E). In addition, imatinib-mediated up-regulation of Mll mRNA levels was abrogated following treatment with RI-BPI or Bcl6 deletion (Fig. 3E). These results collectively show that BCL6 is involved in positive regulation of MLL expression and identified a novel positive feedback loop between MLL and BCL6. Other examples of—likely indirect—transcriptional activation by BCL6 include positive regulation of pre-B-cell receptor components by BCL6 in pre-B cells (Geng et al. 2015) and positive regulation of MED24 and ZEB1 in breast cancer cell lines (Walker et al. 2015; Yu et al. 2015). Nevertheless, since BCL6 mostly functions as a repressor, it is possible that BCL6 may repress expression of negative transcriptional regulators of MLL and thus indirectly increase MLL mRNA levels. To identify BCL6 targets that may allow for an indirect effect on up-regulating MLL expression, we performed ChIP-seq analysis in human MLL-rearranged B-ALL cells. We observed binding of BCL6 to promoter regions of genes that encode negative regulators of transcription, including CTBP2, BMI1, and KDM2B (Fig. 3F). CTBP2 is a transcriptional corepressor (Furusawa et al. 1999) and is a component of the CtBP/LSD1/CoREST repressor complex (Li et al. 2017). Ring finger protein BMI1 is a core component of the polycomb-repressive complex 1 (PRC1) (Gray et al. 2016), which plays a role in epigenetic regulation of gene silencing. Histone demethylase KDM2B links PRC1 to CpG islands to repress lineage-specific genes during development (He et al. 2013). Notably, analysis of ChIP-seq data showed that CTBP2, BMI1, and KDM2B bind to the MLL locus (Fig. 3F), suggesting that BCL6 may indirectly promote MLL expression through repression of CTBP2, BMI1, and KDM2B. Interestingly, pharmacological inhibition of BCL6 function using the BCL6 peptide inhibitor RI-BPI or FX1, a recently developed BCL6 small molecule inhibitor (Cardenas et al. 2016), reduced expression of MLL-AF4 in MLL-rearranged B-ALL cell lines (Supplemental Fig. 3F). Therefore, in addition to playing a positive role in promoting expression of the germline MLL, BCL6 may be involved in modulating expression of the MLL fusion allele.
Figure 3.
BCL6 positively regulates expression of MLL in pre-B and BCR-ABL1 pre-B-ALL cells. (A) Microarray analysis of Bcl6 and Mll expression in murine BCR-ABL1-driven B-ALL cells treated with either vehicle control or 10 µmol/L imatinib for 16 h. n = 3; GSE20987. (B) Human Ph+ ALL cells (Tom1) were treated with vehicle control or 10 µmol/L imatinib for 16 h and then subjected to ChIP-on-chip analysis using a BCL6 antibody (GSE24426). The Y-axis indicates enrichment versus input, while the X-axis indicates the location of probes within the respective locus relative to the transcriptional start site. The dark-green and light-green (vehicle control-treated) or red (imatinib-treated) tracings depict two replicates. Recruitment to MLL and HPRT (negative control) is shown. (C) Pre-B cells from a Tg(tetO-BCL6) mouse were cultured in the presence of IL-7 and treated with either vehicle control or 1 µg/mL Dox for 24 h to induce BCL6 expression. qRT-PCR was performed to measure mRNA levels of BCL6 and Mll relative to Hprt. (D) Bcl6+/+ and Bcl6−/− BCR-ABL1 B-ALL cells transduced with either a control or a BCL6-ER-overexpressing vector. qRT-PCR was performed to measure mRNA levels of Bcl6 and Mll relative to Hprt. (E) Bcl6+/+ and Bcl6−/− BCR-ABL1 B-ALL cells were treated with 10 µmol/L RI-BPI for 4 h, 1 µmol/L imatinib for 4 h, or a combination of both. Cells were then subjected to qRT-PCR to measure mRNA levels of Mll relative to Hprt. (F) ChIP-seq analyses of human MLL-rearranged B-ALL (RS4;11) cells revealed binding of BCL6 to the loci of CTBP2, BMI1, and KDM2B (GSE38403). ChIP-seq tracks showing binding of CTBP2 to the MLL promoter in human H1-hESC cells (ENCODE), binding of BMI1 to the MLL promoter in K562 (GSM937872) and a lymphoblastoid cell line (GSM3384454), and binding of KDM2B to the MLL promoter in K562 (GSM1812033), SEM (GSM2212235), and OCI-LY1 (GSM2171650) cell lines.
BCL6 is required for survival of MLL-rearranged B-ALL cells
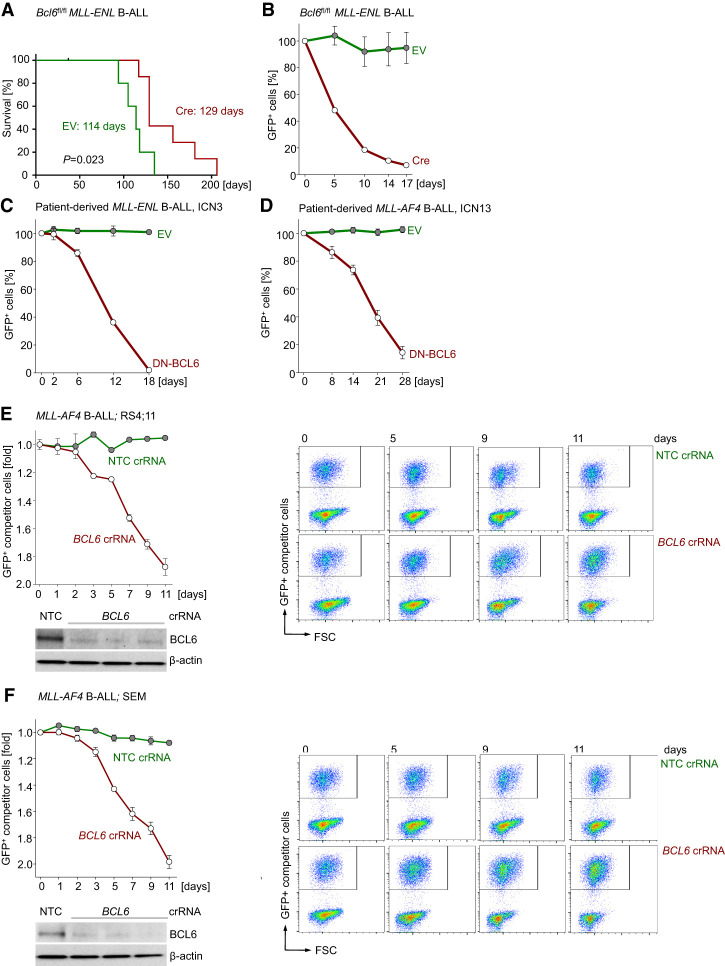
Given that MLL-rearranged B-ALL expressed high levels of BCL6 and that higher than median expression levels of BCL6 correlated with poor clinical outcome (Fig. 1; Supplemental Fig. 1), we hypothesized that positive feedback regulation between BCL6 and MLL contributes to leukemogenesis in MLL-rearranged-B-ALL. To elucidate the role of BCL6 in MLL-rearranged B-ALL, we transduced pro-B cells from Bcl6fl/fl mice with GFP-tagged retroviral MLL-ENL and tamoxifen-inducible Cre (Cre-ERT2) or an ERT2 empty vector (EV) control. When injected into sublethally irradiated NSG mice, MLL-ENL transduced Bcl6fl/fl pro-B cells carrying ERT2 gave rise to fatal leukemia within 4 mo of transplantation (Fig. 4A). Tamoxifen-induced activation of Cre-ERT2 in MLL-ENL transduced Bcl6fl/fl pro-B cells did not prevent leukemia but significantly prolonged overall survival of recipient mice (log-rank test; P = 0.023) (Fig. 4A; Supplemental Fig. 4A). The efficiency of Cre-mediated deletion of Bcl6floxed alleles was >95%. However, genotyping of samples from lethal MLL-ENL B-ALL in the Cre-ERT2 group revealed that fatal leukemia in these animals developed from a small number of B-ALL cells that escaped deletion of Bcl6 and retained Bcl6floxed alleles (Supplemental Fig. 4A). The strong selection for the few clones that evaded Cre-mediated Bcl6 deletion argues for a central role of BCL6 in MLL-rearranged leukemogenesis. However, it is also possible that clones with complete deletion of Bcl6 would emerge after extended disease latency and eventually develop fatal disease. To conclusively address the role of Bcl6 in MLL-rearranged B-ALL, we studied the effects of acute Bcl6 deletion under in vitro conditions.
Figure 4.
BCL6 is required for initiation and progression of MLL-rearranged B-ALL. (A) One million Bcl6fl/fl MLL-ENL B-ALL cells expressing EV or Cre were injected (intravenously) into sublethally irradiated NSG recipient mice. Cells were induced with 4-OHT for 24 h prior to injection. Overall survival of mice was compared by Kaplan-Meier analysis. Log-rank test, P = 0.023. Median survival for each group is indicated. (B) Bcl6fl/fl pre-B cells expressing MLL-ENL were transduced with a GFP-tagged 4-OHT-inducible Cre or EV control. Following 4-OHT induction, enrichment or depletion of GFP+ cells was monitored by flow cytometry. n = 3. (C,D) Patient-derived MLL-rearranged B-ALL cells (ICN3 [C] and ICN13 [D]) were transduced with a GFP-tagged 4-OHT-inducible dominant-negative form of BCL6 (DNBCL6-ERT2-GFP) or EV control. (B–D) Relative changes of GFP+ cells were monitored over time following induction. n = 3. GFP+ cells (percentage) were normalized to EV of each day. (E,F) Electroporation of Cas9 ribonucleoproteins (RNPs), complexes of recombinant Cas9 with nontargeting (NT) crRNAs or crRNAs targeting BCL6, and tracrRNA was performed to transfect MLL-rearranged B-ALL cells (RS4;11 and SEM). The efficiency of CRISPR/Cas9-mediated deletion of BCL6 was assessed by Western blot analyses. RS4;11 (E) and SEM (F) cells transfected with Cas9/RNPs carrying NT or BCL6 crRNAs were mixed with GFP+ RS4;11 and GFP+ SEM competitor cells, respectively. Enrichment or depletion of GFP+ competitor cells was monitored by flow cytometry. n = 3.
Acute Cre-mediated genetic ablation of Bcl6 resulted in rapid depletion of murine Bcl6fl/fl MLL-ENL B-ALL cells from culture in competitive growth assays (Fig. 4B; Supplemental Fig. 4B). To examine the effects of genetic BCL6 inhibition in patient-derived leukemia, patient-derived xenografts (PDXs) from one MLL-ENL (ICN3) and one MLL-AF4 (ICN13) B-ALL sample were transduced with a 4-OHT-inducible dominant-negative BCL6 mutant lacking the BTB domain (DNBCL6-ΔBTB) (Shaffer et al. 2000). Upon 4-OHT-mediated induction, DNBCL6-ΔBTB-expressing MLL-rearranged B-ALL cells from both PDX samples were rapidly depleted from cell culture in competitive growth assays (Fig. 4C,D; Supplemental Fig. 4C). To further validate our observations, we used Cas9 ribonucleoproteins (RNPs) and guide RNAs targeting BCL6 for genetic deletion in MLL-rearranged B-ALL cell lines (RS4;11 and SEM). Consistent with genetic experiments in murine (Bcl6fl/fl) and patient-derived (DNBCL6-ΔBTB) MLL-rearranged B-ALL cells, CRISPR-mediated deletion of BCL6 resulted in depletion of cells from cell culture in competitive growth assays (Fig. 4E,F). Taken together, these findings suggest that BCL6 function represents a previously unrecognized vulnerability in MLL-rearranged-driven B-ALL.
Validation of BCL6 as a therapeutic vulnerability in MLL-rearranged B-ALL cells
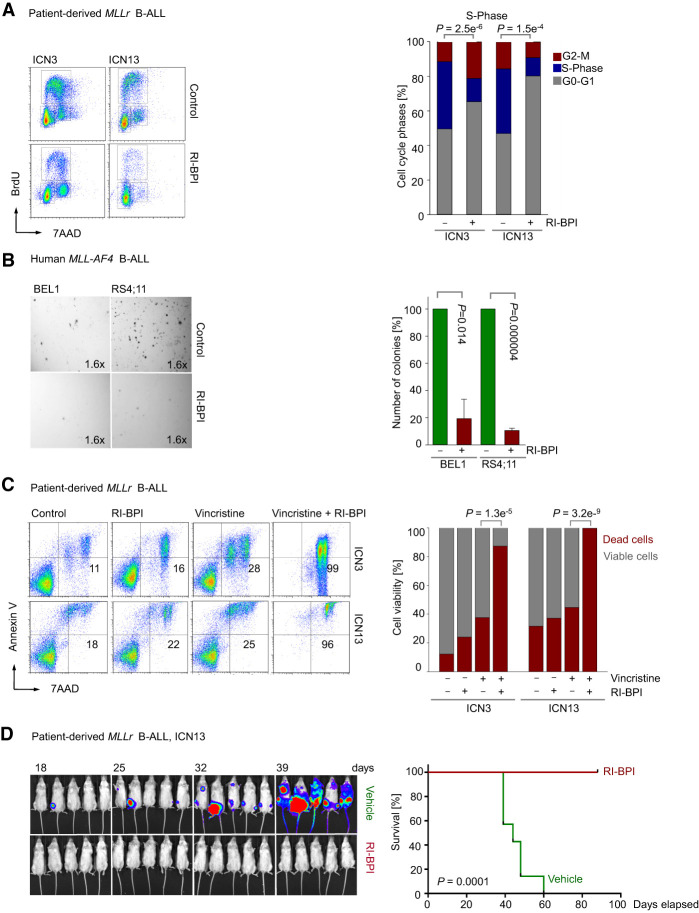
To study whether BCL6 may represent a potential therapeutic target in MLL-rearranged B-ALL, we treated patient-derived MLL-rearranged B-ALL (ICN3 and ICN13) cells with the retro–inverso BCL6 peptide inhibitor RI-BPI. Treatment of patient-derived MLL-rearranged B-ALL cells with RI-BPI inhibited proliferation and caused cell cycle arrest in the G0/G1 phase as measured by BrdU staining (P = 1.5 × 10−04) (Fig. 5A). We therefore studied colony-forming capacity in four human B-ALL cell lines that carry MLL rearrangements. Pharmacological inhibition of BCL6 activity reduced the colony-forming ability by approximately fourfold to 10-fold (Fig. 5B; Supplemental Fig. 5A). However, RI-BPI treatment did not induce substantial cell death in patient-derived B-ALL cells carrying MLL-ENL (ICN3) and MLL-AF4 (ICN13) gene rearrangements, even at high concentrations (5 µmol/L) (Fig. 5C). Since RI-BPI induced cell cycle arrest and suppressed colony formation but failed to induce cell death, we studied whether RI-BPI could cooperate with antimitotic drugs. Vincristine is a central component of the vast majority of current chemotherapy regimens for B-ALL patients and functions as an antimitotic drug that prevents tubulin dimers from polymerizing to form microtubules and the mitotic spindle. While ICN3 and ICN13 MLL-rearranged B-ALL cells were largely resistant to vincristine, concurrent treatment with RI-BPI overcame vincristine resistance, a frequent complication in MLL-rearranged B-ALL, and dramatically sensitized MLL-rearranged B-ALL cells (Fig. 5C). To test the efficacy of RI-BPI treatment on MLL-rearranged B-ALL cells in vivo, luciferase-labeled patient-derived MLL-rearranged B-ALL (ICN13) cells were injected intrafemorally into sublethally irradiated (2.5 Gy) NOD/SCID mouse recipients. Transplant recipient mice were treated with intraperitoneal injections of vehicle or 25 mg/kg RI-BPI five times, and in vivo expansion of leukemic cells was monitored by luciferase bioimaging (Fig. 5D). Recipient mice in the vehicle group developed fatal leukemia within 8 wk of transplantation. In contrast, RI-BPI-treated mice developed no signs of disease and were sacrificed after 90 d for minimal residual disease studies (Fig. 5D). Unlike mice in the vehicle group, no minimal residual disease was detected (Supplemental Fig. 5B).
Figure 5.
Inhibition of BCL6 compromises proliferation, colony formation, and leukemia initiation of human MLL-rearranged B-ALL cells. (A) Patient-derived MLL-rearranged B-ALL cells (ICN3 and ICN13) were treated with 5 µmol/L RI-BPI or vehicle control for 2 h and then subjected to cell cycle analysis by measuring BrdU incorporation in combination with 7AAD staining. Representative FACS plots from three independent experiments are shown. Percentages of cells in the G0/G1, S, and G2/M phases are indicated. (B) Ten-thousand human MLL-rearranged B-ALL cells (BEL1 and RS4;11) were treated with 5 µmol/L RI-BPI or vehicle control and plated on semisolid methylcellulose. Colonies were counted after 14 d. The bar graphs show the mean values of the number of colonies ± SD. n = 3. (C) ICN3 and ICN13 cells were treated with vehicle control, 1 nmol/L vincristine, 5 µmol/L RI-BPI, or a combination of both for 5 d. Viability was measured by Annexin V/7AAD staining. Shown are the mean values from three independent experiments. (D) ICN13 cells (5 × 105) labeled with firefly luciferase were injected (intrafemorally) into sublethally irradiated (2.5 Gy) NOD/SCID mice that were treated with intraperitoneal injections of vehicle or 25 mg/kg RI-BPI five times. Leukemia engraftment and progression were monitored by luciferase bioimaging at the times indicated. Kaplan-Meier analysis showing overall survival for each group (n = 7) of the recipient mice. Log-rank test, P = 0.0001.
BCL6 suppresses expression of proapoptotic BIM (BCL2L11) in MLL-rearranged B-ALL
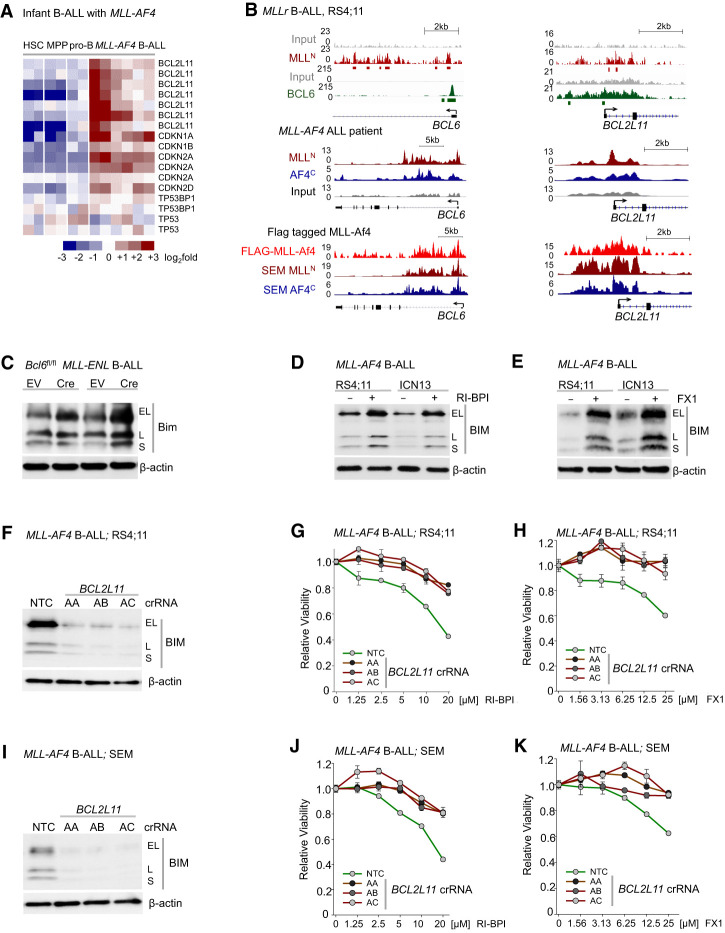
During early B-cell development, BCL6 mediates pro-B-cell survival through transcriptional repression of cell cycle checkpoint regulators (CDKN1A/p21, CDKN1B/p27, and CDKN2A/Arf) (Duy et al. 2010) and protects BCR-ABL1 B-ALL cells from Arf/p53-mediated apoptosis (Duy et al. 2011). For this reason, we examined whether BCL6 promotes MLL-rearranged-driven leukemogenesis through transcriptional repression of proapoptotic molecules. Recently, reverse-phase protein array (RPPA) analyses revealed that MLL-rearranged B-ALL is associated with increased expression of the proapoptotic BH3-only protein BIM (BCL2L11) (Benito et al. 2015). Our analysis of gene expression profiles of patients with MLL-rearranged B-ALL confirmed up-regulation of BIM (BCL2L11) in MLL-rearranged B-ALL compared with normal pro-B cells (Fig. 6A) and B-ALL lacking MLL rearrangement (Supplemental Fig. 6). Interestingly, ChIP-seq analyses of human MLL-rearranged B-ALL cells showed that both MLL and BCL6 bind to the BCL2L11 locus (Fig. 6B), suggesting that BCL6 promotes survival of MLL-rearranged B-ALL cells through regulation of BIM (BCL2L11) expression. For this reason, we tested the effects of inducible deletion of Bcl6 and pharmacological inhibition of BCL6 function on Bim expression in MLL-rearranged B-ALL cells. Bim protein levels and activity were increased upon Cre-mediated deletion of Bcl6 in MLL-ENL B-ALL cells (Fig. 6C). Similarly, treatment with RI-BPI (Fig. 6D) or the BCL6 small molecule inhibitor FX1 (Fig. 6E) derepressed BIM expression in human MLL-rearranged B-ALL cells. In addition, CRISPR/Cas9-mediated genetic deletion of BIM (BCL2L11) in MLL-rearranged B-ALL cells (RS4;11 and SEM) substantially desensitized cells to treatment with RI-BPI and FX1 (Fig. 6F–K). These results suggest that genetic ablation of BIM circumvents the essential function of BCL6, highlighting the mechanistic contribution of BCL6-mediated transcriptional repression of BIM in MLL-rearranged B-ALL cells.
Figure 6.
BCL6-mediated transcriptional repression of BIM represents a vulnerability in MLL-rearranged B-ALL. (A) Gene expression analysis of proapoptotic and cell cycle checkpoint molecules in hematopoietic stem cells, myeloid hematopoietic progenitor cells, pre-B cells, and MLL-AF4+ blasts from infant ALL (GSE79450). (B, top) ChIP-seq analyses of human MLL-rearranged ALL (RS4;11) cells showed that MLL and BCL6 bind to the loci of BCL6 and BCL2L11 (BIM) (GSE38403). (Middle) ChIP-seq tracks showing binding of MLLN and AF4C to the loci of BCL6 and BCL2L11 in a MLL-AF4 ALL patient (GSE83671). (Bottom) MLL-Af4-Flag ChIP-seq peaks in MLL-Af4 leukemia cells and MLLN and AF4C ChIP-seq peaks in MLL-rearranged ALL (SEM) cells (GSE84116). (C) Protein levels of Bim upon 4-OHT-inducible Cre-mediated deletion of Bcl6 in pre-B cells expressing MLL-ENL. (D) Effects of vehicle control or 10 µmol/L RI-BPI for 48 h on BIM protein levels in human MLL-rearranged ALL cells. (E) Protein levels of BIM upon treatment with 50 µmol/L FX1 in human MLL-rearranged ALL cells. (F–K) Electroporation of Cas9 RNPs, complexes of recombinant Cas9 with nontargeting (NT) crRNAs or crRNAs targeting BIM (BCL2L11), and tracrRNA was performed to transfect MLL-rearranged B-ALL cells (RS4;11 [F–H] and SEM [I–K]). (F,I) The efficiency of CRISPR/Cas9-mediated deletion of BIM was assessed by Western blot analysis. RS4;11 (G,H) and SEM (J,K) cells transfected with Cas9/RNPs carrying NT or BIM crRNAs were treated with increasing concentrations of RI-BPI (G,J) or FX1 (H,K). Shown is average relative viability. n = 3.
Rationale for dual targeting of BCL2 and BCL6 in MLL-rearranged B-ALL
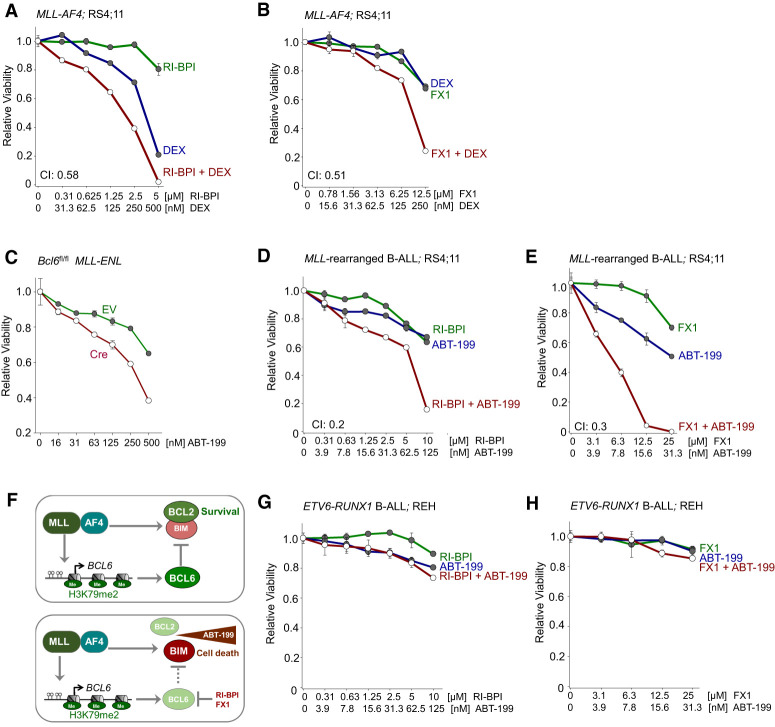
The genetic and pharmacological studies suggest that BCL6 transcriptionally represses the BH3-only apoptosis facilitator BIM, which is otherwise constitutively up-regulated in MLL-rearranged B-ALL cells. Interestingly, apoptosis induced by glucocorticoids, a central component of the chemotherapy regimen for ALL, is dependent on up-regulation of BIM expression (Jing et al. 2015). Given the essential role of BCL6-mediated transcriptional repression of BIM in MLL-rearranged B-ALL cells, we studied the interactions between glucocorticoids and RI-BPI or FX1. Treatment of human MLL-rearranged B-ALL cells with RI-BPI (Fig. 7A) or FX1 (Fig. 7B) synergized with dexamethasone (Dex), suggesting that Dex and pharmacological inhibition of BCL6 may represent a relevant drug combination (Supplemental Table S8). Bim is typically sequestered by members of the antiapoptotic BCL2 family proteins (Cheng et al. 2001). Thus, targeted restriction of antiapoptotic activity of BCL2 proteins will allow Bim to induce mitochondrial apoptosis (Cheng et al. 2001). MLL-rearranged B-ALL cells are uniquely sensitive to treatment with the BCL2-selective inhibitor ABT-199 (venetoclax), which disrupts the sequestration of BIM by BCL2 (Cheng et al. 2001; Benito et al. 2015). Given the constitutively high levels of BIM expression in MLL-rearranged B-ALL, our findings suggest that MLL-rearranged B-ALL cells are selectively dependent on BCL6 activity and its ability to curb expression and activity of BIM. Based on this rationale, we tested whether loss of Bcl6 function can selectively sensitize to inhibition of Bcl2 in MLL-rearranged B-ALL cells. Inducible deletion of Bcl6 rendered MLL-ENL B-ALL cells more sensitive to ABT-199 treatment (Fig. 7C). These findings support the scenario that oncogenic MLL fusions transcriptionally activate expression of Bim, hence creating a disease-specific dependency on both BCL2 and BCL6 to curb proapoptotic activity of Bim (Fig. 7F). While BCL2-mediated sequestration of BIM can be targeted by ABT-199 (Benito et al. 2015), BCL6-mediated transcriptional repression of BIM can be disrupted by RI-BPI (Cerchietti et al. 2009) or FX1 (Cardenas et al. 2016) (Fig. 6D,E). For this reason, we tested drug interactions between ABT-199 and the BCL6 peptide inhibitor RI-BPI and the BCL6 small molecule inhibitor FX1. Treatment of human MLL-rearranged B-ALL cells with either RI-BPI or FX1 strongly synergized with ABT-199 (Fig. 7D,E; Supplemental Table S8). In contrast, ABT-199 did not synergize with RI-BPI or FX1 in ETV6-RUNX1 B-ALL cells (Fig. 7G,H), suggesting a selective vulnerability in MLL-rearranged B-ALL.
Figure 7.
Dual targeting of BCL2 and BCL6 in MLL-rearranged B-ALL. (A) Human MLL-rearranged B-ALL cells were treated with Dex, RI-BPI, or a combination of both. CI values for ED50 are shown. Relative viability was assessed. n = 3. (B) Human MLL-rearranged B-ALL cells were treated with Dex, FX1, or a combination of both. Relative viability and CI were assessed. n = 3. (C) Relative viability (n = 3) was assessed upon treatment with increasing concentrations of ABT-199 in pre-B cells expressing MLL-ENL following 4-OHT Cre-mediated deletion of Bcl6. (D) Human MLL-rearranged B-ALL cells were treated with ABT-199, RI-BPI, or ABT-199 in combination with RI-BPI. Relative viability and CI were assessed. (E) Relative viability and CI were examined following treatment of human MLL-rearranged B-ALL cells with ABT-199, FX1, or a combination of both. (F) MLL-rearranged fusion proteins induce aberrant expression of BCL6, which contributes to MLL-rearranged-driven leukemogenesis by restricting expression of BIM, a proapoptotic molecule. The BCL2 inhibitor ABT-199 disrupts the interaction between BCL2 and BIM. Pharmacological inhibition of BCL6 (RI-BPI or FX1) synergizes with ABT-199 in eradicating MLL-rearranged B-ALL cells. (G,H) Relative viability was determined upon treatment with increasing concentrations of ABT-199, RI-BPI (G), FX1 (H), or a combination of ABT-199 with RI-BPI (G) or FX1 (H) in ETV6-RUNX1 B-ALL cells.
Discussion
In summary, these findings show that germline-encoded MLL is required for transactivation of BCL6 in normal B-cell development, while oncogenic MLL fusions in MLL-rearranged B-ALL drive aberrant expression of BCL6, which in turn plays a—likely indirect—role in transcriptional activation of MLL. Oncogenic MLL fusions and germline-encoded MLL positively regulated BCL6. Several lines of evidence suggest that oncogenic MLL fusions cooperate with germline-encoded MLL in MLL-rearranged leukemia. For instance, in MLL-AF9-driven acute myeloid leukemia, germline MLL promotes proliferation and survival (Thiel et al. 2010) and cooperates with MLL-AF9 for efficient transactivation of the HOXA9 locus (Milne et al. 2010). Other studies suggest that germline MLL2 rather than MLL is essential for MLL-AF9 leukemogenesis; however, MLL contributes to leukemia cell survival through collaboration with MLL2 (Chen et al. 2017). Germline MLL is more active than MLL-AF4 in protecting CpGs from methylation, while MLL-AF4 is a more potent transcriptional activator (Erfurth et al. 2008). These findings suggest that germline MLL and MLL fusion proteins cooperate in regulating gene expression required for MLL-rearranged leukemogenesis. Two mechanisms are likely involved; namely, (1) maintenance of demethylation of regulatory elements, mediated mainly by germline MLL, and (2) target gene transactivation, achieved largely by MLL fusion proteins (Milne et al. 2010). Our genetic studies revealed a delay in Bcl6 down-regulation following genetic ablation of Mll (Fig. 2). However, bisulfite conversion followed by methylation-specific PCR showed that the methylation status of the Bcl6 promoter region was not impacted upon Mll deletion (Supplemental Fig. 2B). These findings suggest that other effects on transcriptional programming, rather than acquisition of methylation, might be involved in down-regulation of Bcl6 expression. Our findings demonstrated that BCL6 plays a key role in MLL-rearranged-driven leukemogenesis through a reciprocal positive feedback loop between BCL6 and MLL. While BCL6 is an established transcriptional repressor, it is possible that BCL6 may indirectly promote MLL expression through repression of negative transcriptional regulators of MLL. Through ChIP-seq analysis, we identified transcriptional corepressor CTBP2, ring finger protein BMI1 (a component of the PRC1), and histone demethylase KDM2B as potential targets of BCL6 in MLL-rearranged B-ALL cells (Fig. 3F). Importantly, CTBP2, BMI1, and KDM2B bind to the MLL locus as revealed by analysis of ChIP-seq data obtained in various cell types (Fig. 3F). Our results suggest that transcriptional activation of MLL may result from BCL6-mediated transcriptional repression of CTBP2, BMI1, and KDM2B.
Various strategies have been devised to suppress the oncogenic activity of MLL fusion proteins, including inhibition of MEN1, LEDGF, BRD4, and DOT1L (Marschalek et al. 2015) as well as approaches targeting germline MLL for degradation (Liang et al. 2017; Zhao et al. 2019). Given its critical role in MLL-rearranged B-ALL, we propose BCL6 as therapeutic target in the MLL-rearranged subtype of B-ALL. Genetic (Bcl6fl/fl, DNBCL6-ΔBTB) as well as pharmacological peptide (RI-BPI) inhibition of BCL6 had profound effects in MLL-rearranged B-ALL cells and potentiated the efficacy of conventional chemotherapy agents (e.g., vincristine). As shown by genetic experiments in murine (Bcl6fl/fl) (Fig. 4A,B) and patient-derived (DNBCL6-ΔBTB) (Fig. 4C,D) MLL-rearranged B-ALL, the positive feedback loop between BCL6 and MLL represents a central vulnerability in this B-ALL subset. BCL6 binds to its own promoter and represses its own expression through a negative autoregulatory circuit (Pasqualucci et al. 2003). The DNBCL6-ΔBTB mutant binds to DNA but lacks the ability to act as a transcriptional repressor (Shaffer et al. 2000). Consequently, while induction of the DNBCL6-ΔBTB mutant, which lacks the BTB domain, results in antileukemia effects, it is unlikely that the mutant suppresses transcription of BCL6.
Proof-of-concept experiments showed that pharmacological inhibition of BCL6 using a RI-BPI compromised MLL-rearranged B-ALL leukemia initiation and subverted vincristine resistance (Fig. 5C,D). A central mechanistic aspect of BCL6 function in MLL-rearranged B-ALL involves transcriptional repression of the proapoptotic BH3-only molecule Bim (BCL2L11). Profiling for BH3-only proteins in various B-ALL subtypes revealed that MLL-rearranged B-ALL is associated with increased expression of the proapoptotic protein BIM (BCL2L11) (Benito et al. 2015). Sequestration of BIM by BCL2 protein prevents oligomerization of BAX/BAK and thereby protects against subsequent mitochondrial apoptosis (Cheng et al. 2001). ABT-199 (venetoclax) is a BH3 mimetic that disrupts the interaction between BCL2 and BIM, leading to BIM release and induction of apoptosis. Various studies have found that MLL-rearranged leukemia cells are sensitive to the BCL2 inhibitor ABT-199 (Benito et al. 2015; Khaw et al. 2016; Frismantas et al. 2017). Besides BCL2, we here identified BCL6 as a central antagonist of proapoptotic BIM function in MLL-rearranged B-ALL cells. In genetic experiments, we showed that oncogenic MLL fusions strongly activated BCL2L11 transcription, reinforcing the notion that constitutively high BIM expression levels represent an important and selective vulnerability in MLL-rearranged B-ALL 2015; (Khaw et al. 2016; Frismantas et al. 2017). We found that BCL6 bound to the BCL2L11 promoter, and BCL6-mediated transcriptional repression was required to curtail BIM activity in MLL-rearranged B-ALL. Hence, we concluded that BCL6 acts as a BCL2L11 repressor and contributes to MLL-rearranged-driven leukemogenesis by limiting MLL-induced BIM activation (Fig. 7F). Both BCL2 and BCL6 represent crucial antagonists of BIM in MLL-rearranged B-ALL: BCL2 mediates BIM sequestration, and BCL6 is required for transcriptional repression of BIM. In support of this scenario, peptide (RI-BPI) and small molecule (FX1) inhibitors of BCL6 strongly synergized with blockade of BCL2-mediated BIM sequestration (ABT-199) in killing MLL-rearranged B-ALL cells. Notably, inhibition of BCL6 did not synergize with ABT-199 in B-ALL cells that were driven by ETV6-RUNX1 instead of an oncogenic MLL fusion, suggesting that BCL6-mediated repression of BIM is a selective vulnerability in MLL-rearranged B-ALL. Oncogenic MLL fusions strongly activated BCL2L11 transcription. Hence, constitutively high BIM expression levels represents an important and selective vulnerability in MLL-rearranged B-ALL. Previous findings demonstrated that BCL6 functions as a transcriptional repressor of BCL2 (Saito et al. 2009), which likely represents another reason for the unique vulnerability of MLL-rearranged B-ALL to BCL2 inhibitors. Here we showed that MLL-rearranged B-ALL cells are selectively dependent on BCL6 in large part because of its ability to curb expression and activity of BIM, supporting a rationale for dual pharmacological targeting of BCL6 and BCL2 in this B-ALL subgroup.
Materials and methods
Murine primary and B-ALL cells
Bone marrow cells were extracted from young age-matched mice (Supplemental Table S3) and processed as described in the Supplemental Material. All mouse experiments were approved by the University of California at San Francisco Institutional Animal Care and Use Committee (IACUC). Bone marrow cells collected were retrovirally transformed with BCR-ABL1 in the presence of 10 ng/mL IL-7 (Peprotech) for Ph+ ALL-like cells or a cocktail of 10 ng/mL IL-3, 25 ng/mL IL-6, and 50 ng/mL SCF (PeproTech) for chronic myeloid leukemia-like cells on RetroNectin-coated (Takara) dishes. See the Supplemental Material for details.
ChIP-qPCR and genomic DNA fragment library for ChIP-seq
ChIP assays were performed as described in the Supplemental Material. Immunoprecipitated DNA sequences were analyzed by qPCR. Antibodies and primer sequences used for qChIP analyses are listed in Supplemental Tables S5 and S6, respectively. ChIP-seq was performed as described in the Supplemental Material.
Western blotting and immunohistochemistry
Primary antibodies used in Western blotting are listed in Supplemental Table S5. Immunohistochemistry was performed at the University of California at San Francisco Immunohistochemistry and Molecular Pathology Core Facility. See the Supplemental Material for details.
A detailed description of experimental methods and patient samples is in the Supplemental Material.
Competing interest statement
S.A.A. has been a consultant and/or shareholder for Epizyme, Inc.; Imago Biosciences; Vitae/Allergan Pharma; Cyteir Therapeutics; C4 Therapeutics; Syros Pharmaceuticals; OxStem Oncology; Accent Therapeutics; and Mana Therapeutics.
Acknowledgments
We thank Lars Klemm, Janet Winchester, Eamon Aghania, and current and former members of the Müschen laboratory for their support. A.M. is supported by the Chemotherapy Foundation and the National Cancer Institute (NCI; R35CA220499 and R01CA198089). S.A.A. is supported by National Institutes of Health grants R01CA176745, R01CA231637, R01CA204639, and R01CA066996. Research in the Müschen laboratory is funded by the NCI through Outstanding Investigator Award R35CA197628 (to M.M.), U10CA180827 (to A.M. and M.M.), and R01CA137060, R01CA157644, R01CA172558, and R01CA213138 (to M.M.); the Howard Hughes Medical Institute HHMI-55108547 (to M.M.); the Norman and Sadie Lee Foundation (for pediatric cancer; to M.M.); the Falk Medical Research Trust Catalyst Award (to M.M.); the Pediatric Cancer Research Foundation (PCRF) and the Cancer Research Institute (CRI) through a Clinic and Laboratory Integration Program (CLIP) grant (to M.M.); and the California Institute for Regenerative Medicine (CIRM) through DISC2-10061. M.M. is a Howard Hughes Medical Institute Faculty Scholar.
Author contributions: M.M. conceived and designed the study. L.N.C., A.M., T.M., and M.M. interpreted data and wrote the paper. C.H., H.K., J.C., and C.-W.C. contributed to the writing and editing of the manuscript. H.G. performed bioinformatics, clinical outcome, and statistical analyses. C.H., L.N.C., E.B., G.X., G.D., and T.M. designed and performed experiments and analyzed data. S.A.A., P.E., and M.M. provided important reagents and genetic mouse models.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.327593.119.
References
- Andersson AK, Ma J, Wang J, Chen X, Gedman AL, Dang J, Nakitandwe J, Holmfeldt L, Parker M, Easton J, et al. 2015. The landscape of somatic mutations in infant MLL-rearranged acute lymphoblastic leukemias. Nat Genet 47: 330–337. 10.1038/ng.3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayton PM, Cleary ML. 2001. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene 20: 5695–5707. 10.1038/sj.onc.1204639 [DOI] [PubMed] [Google Scholar]
- Baron BW, Nucifora G, McCabe N, Espinosa R III, Le Beau MM, McKeithan TW. 1993. Identification of the gene associated with the recurring chromosomal translocations t(3;14)(q27;q32) and t(3;22)(q27;q11) in B-cell lymphomas. Proc Natl Acad Sci 90: 5262–5266. 10.1073/pnas.90.11.5262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron BW, Anatasi J, Montag A, Huo D, Baron RM, Karrison T. 2004. The human BCL6 transgene promotes the development of lymphomas in the mouse. Proc Natl Acad Sci 101: 14198–14203. 10.1073/pnas.0406138101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron BW, Anatasi J, Hyjek EM, Bies J, Reddy PL, Dong J, Joseph L, Thirman MJ, Wroblewski K, Wolff L, et al. 2012. PIM1 gene cooperates with human BCL6 gene to promote the development of lymphomas. Proc Natl Acad Sci 109: 5735–5739. 10.1073/pnas.1201168109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito JM, Godfrey L, Kojima K, Hogdal L, Wunderlich M, Geng H, Marzo I, Harutyunyan KG, Golfman L, North P, et al. 2015. MLL-rearranged acute lymphoblastic leukemias activate bcl-2 through H3K79 methylation and are sensitive to the BCL-2-specific antagonist ABT-199. Cell Rep 13: 2715–2727. 10.1016/j.celrep.2015.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, Feng Z, Punt N, Daigle A, Bullinger L, et al. 2011. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell 20: 66–78. 10.1016/j.ccr.2011.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas D, Milne TA, Basrur V, Kim J, Elenitoba-Johnson KS, Allis CD, Roeder RG. 2011. Function of leukemogenic mixed lineage leukemia 1 (MLL) fusion proteins through distinct partner protein complexes. Proc Natl Acad Sci 108: 15751–15756. 10.1073/pnas.1111498108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting KL, Soong TD, Singh R, Jiang Y, Béguelin W, Poloway DW, Swed BL, Hatzi K, Reisacher W, Teater M, et al. 2016. Multi-tiered reorganization of the genome during B cell affinity maturation anchored by a germinal center-specific locus control region. Immunity 45: 497–512. 10.1016/j.immuni.2016.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas MG, Yu W, Beguelin W, Teater MR, Geng H, Goldstein RL, Oswald E, Hatzi K, Yang SN, Cohen J, et al. 2016. Rationally designed BCL6 inhibitors target activated B cell diffuse large B cell lymphoma. J Clin Invest 126: 3351–3362. 10.1172/JCI85795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerchietti LC, Yang SN, Shaknovich R, Hatzi K, Polo JM, Chadburn A, Dowdy SF, Melnick A. 2009. A peptomimetic inhibitor of BCL6 with potent antilymphoma effects in vitro and in vivo. Blood 113: 3397–3405. 10.1182/blood-2008-07-168773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cierpicki T, Risner LE, Grembecka J, Lukasik SM, Popovic R, Omonkowska M, Shultis DD, Zeleznik-Le NJ, Bushweller JH. 2010. Structure of the MLL CXXC domain–DNA complex and its functional role in MLL-AF9 leukemia. Nat Struct Mol Biol 17: 62–68. 10.1038/nsmb.1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Anastassiadis K, Kranz A, Stewart AF, Arndt K, Waskow C, Yokoyama A, Jones K, Neff T, Lee Y, et al. 2017. MLL2, not MLL1, plays a major role in sustaining MLL-rearranged acute myeloid leukemia. Cancer Cell 31: 755–770.e6. 10.1016/j.ccell.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng EH, Wei MC, Weiler S, Flavell RA, Mak TW, Lindsten T, Korsmeyer SJ. 2001. BCL-2, BCL-XL sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol Cell 8: 705–711. 10.1016/S1097-2765(01)00320-3 [DOI] [PubMed] [Google Scholar]
- Duy C, Yu JJ, Nahar R, Swaminathan S, Kweon SM, Polo JM, Valls E, Klemm L, Shojaee S, Cerchietti L, et al. 2010. BCL6 is critical for the development of a diverse primary B cell repertoire. J Exp Med 207: 1209–1221. 10.1084/jem.20091299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duy C, Hurtz C, Shojaee S, Cerchietti L, Geng H, Swaminathan S, Klemm L, Kweon SM, Nahar R, Braig M, et al. 2011. BCL6 enables Ph+ acute lymphoblastic leukaemia cells to survive BCR-ABL1 kinase inhibition. Nature 473: 384–388. 10.1038/nature09883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erfurth FE, Popovic R, Grembecka J, Cierpicki T, Theisler C, Xia ZB, Stuart T, Diaz MO, Bushweller JH, Zeleznik-Le NJ. 2008. MLL protects CpG clusters from methylation within the Hoxa9 gene, maintaining transcript expression. Proc Natl Acad Sci 105: 7517–7522. 10.1073/pnas.0800090105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frismantas V, Dobay MP, Rinaldi A, Tchinda J, Dunn SH, Kunz J, Richter-Pechanska P, Marovca B, Pail O, Jenni S, et al. 2017. Ex vivo drug response profiling detects recurrent sensitivity patterns in drug-resistant acute lymphoblastic leukemia. Blood 129: e26–e37. 10.1182/blood-2016-09-738070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa T, Moribe H, Kondoh H, Higashi Y. 1999. Identification of CtBP1 and CtBP2 as corepressors of zinc finger-homeodomain factor δEF1. Mol Cell Biol 19: 8581–8590. 10.1128/MCB.19.12.8581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng H, Brennan S, Milne TA, Chen WY, Li Y, Hurtz C, Kweon SM, Zickl L, Shojaee S, Neuberg D, et al. 2012. Integrative epigenomic analysis identifies biomarkers and therapeutic targets in adult B-acute lymphoblastic leukemia. Cancer Discov 2: 1004–1023. 10.1158/2159-8290.CD-12-0208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng H, Hurtz C, Lenz KB, Chen Z, Baumjohann D, Thompson S, Goloviznina NA, Chen WY, Huan J, LaTocha D, et al. 2015. Self-enforcing feedback activation between BCL6 and pre-B cell receptor signaling defines a distinct subtype of acute lymphoblastic leukemia. Cancer Cell 27: 409–425. 10.1016/j.ccell.2015.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray F, Cho HJ, Shukla S, He S, Harris A, Boytsov B, Jaremko L, Jaremko M, Demeler B, Lawlor ER, et al. 2016. BMI1 regualtes PRC1 architecture and activity through homo- and hetero-oligomerization. Nat Commun 7: 13343. 10.1038/ncomms13343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey RC, Mullighan CG, Wang X, Dobbin KK, Davidson GS, Bedrick EJ, Chen IM, Atlas SR, Kang H, Ar K, et al. 2010. Identification of novel cluster groups in pediatric high-risk B-precursor acute lymphoblastic leukemia with gene expression profiling: correlation with genome-wide DNA copy number alterations, clinical characteristics, and outcome. Blood 116: 4874–4884. 10.1182/blood-2009-08-239681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Shen L, Wan M, Taranova O, Wu H, Zhang Y. 2013. Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nat Cell Biol 15: 373–384. 10.1038/ncb2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa GC, Kantarjian HM, Yin CC, Qiao W, Ravandi F, Thomas D, Short NJ, Sasaki K, Garcia-Manero G, Kadia TM, et al. 2017. Prognostic impact of pretreatment cytogenetics in adult Philadelphia chromosome-negative acute lymphoblastic leukemia in the era of minimal residual disease. Cancer 123: 459–467. 10.1002/cncr.30376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing D, Bhadri VA, Beck D, Thoms JA, Yakob NA, Wong JW, Knezevic K, Pimanda JE, Lock RB. 2015. Opposing regulation of BIM and BCL2 controls glucocorticoid-induced apoptosis of pediatric acute lymphoblastic leukemia cells. Blood 125: 273–283. 10.1182/blood-2014-05-576470 [DOI] [PubMed] [Google Scholar]
- Jude CD, Climer L, Xu D, Artinger E, Fisher JK, Ernst P. 2007. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell 1: 324–337. 10.1016/j.stem.2007.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerckaert JP, Deweindt C, Tilly H, Quief S, Lecocq G, Bastard C. 1993. LAZ3, a novel zinc-finger encoding gene, is disrupted by recurring chromosome 3q27 translocations in human lymphomas. Nat Genet 5: 66–70. 10.1038/ng0993-66 [DOI] [PubMed] [Google Scholar]
- Khaw SL, Suryani S, Evans K, Richmond J, Robbins A, Kurmasheva RT, Billups CA, Erickson SW, Guo Y, Houghton PJ, et al. 2016. Venetoclax responses of pediatric ALL xenografts reveal sensitivity of MLL-rearranged leukemia. Blood 128: 1382–1395. 10.1182/blood-2016-03-707414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov AV, Feng Z, Lemieux ME, Faber J, Vempati S, Sinha AU, Xia X, Jesneck J, Bracken AP, Silverman LB, et al. 2008. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell 14: 355–368. 10.1016/j.ccr.2008.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Liu X, He L, Yang J, Pei F, Li W, Liu S, Chen Z, Xie G, Xu B, et al. 2017. ZNF516 suppresses EGFR by targeting the CtBP/LSD1/CoREST complex to chromatin. Nat Commun 8: 691. 10.1038/s41467-017-00702-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang K, Volk AG, Haug JS, Marshall SA, Woodfin AR, Bartom ET, Gilmore JM, Florens L, Washburn MP, Sullivan KD, et al. 2017. Therapeutic targeting of MLL degradation pathways in MLL-rearranged leukemia. Cell 168: 59–72.e13. 10.1016/j.cell.2016.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A. 2010. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell 37: 429–437. 10.1016/j.molcel.2010.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z, Lin C, Shilatifard A. 2012. The super elongation complex (SEC) family in transcriptional control. Nat Rev Mol Cell Biol 13: 543–547. 10.1038/nrm3417 [DOI] [PubMed] [Google Scholar]
- Marschalek R. 2015. MLL leukemia and future treatment strategies. Arch Pharm (Weinheim) 348: 221–228. 10.1002/ardp.201400449 [DOI] [PubMed] [Google Scholar]
- Meyer C, Burmeister T, Gröger D, Tsaur G, Fechina L, Renneville A, Sutton R, Venn NC, Emerenciano M, Pombo-de-Oliveira MS, et al. 2018. The MLL recombinome of acute leukemias in 2017. Leukemia 32: 273–284. 10.1038/leu.2017.213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne TA, Kim J, Wang GG, Stadler SC, Basrur V, Whitcomb SJ, Wang Z, Ruthenburg AJ, Elenitoba-Johnson KS, Roeder RG, et al. 2010. Multiple interactions recruit MLL1 and MLL1 fusion proteins to the HOXA9 locus in leukemogenesis. Mol Cell 38: 853–863. 10.1016/j.molcel.2010.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Migliazza A, Basso K, Chaganti RS, Dalla-Favera R. 2003. Mutations of the BCL6 proto-oncogene disrupt its negative autoregulation in diffuse large B-cell lymphoma. Blood 101: 2914–2923. 10.1182/blood-2002-11-3387 [DOI] [PubMed] [Google Scholar]
- Saito M, Novak U, Piovan E, Basso K, Sumazin P, Schneider C, Crespo M, Shen Q, Bhagat G, Califano A, et al. 2009. BCL6 suppression of BCL2 via Miz1 and its disruption in diffuse large B cell lymphoma. Proc Natl Acad Sci 106: 11294–11299. 10.1073/pnas.0903854106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O'Neill LP, Turner BM, Delrow J, et al. 2004. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev 18: 1263–1271. 10.1101/gad.1198204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM. 2000. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity 13: 199–212. 10.1016/S1074-7613(00)00020-0 [DOI] [PubMed] [Google Scholar]
- Shilatifard A. 2006. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem 75: 243–269. 10.1146/annurev.biochem.75.103004.142422 [DOI] [PubMed] [Google Scholar]
- Shilatifard A, Lane WS, Jackson KW, Conaway RC, Conaway JW. 1996. An RNA polymerase II elongation factor encoded by the human ELL gene. Science 271: 1873–1876. 10.1126/science.271.5257.1873 [DOI] [PubMed] [Google Scholar]
- Smith E, Lin C, Shilatifard A. 2011. The super elongation complex (SEC) and MLL in development and disease. Genes Dev 25: 661–672. 10.1101/gad.2015411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YN, Hu YX, Gao L, Xiao PF, Lu J, Wu SY, Wang M, Shao XJ, Zhou CY, Ling J, et al. 2018. The therapeutic efficacy of pediatric ALL patients with MLL gene rearrangement treated with CCLG-ALL2008 protocol. Eur Rev Med Pharmacol Sci 22: 6020–6029. [DOI] [PubMed] [Google Scholar]
- Thiel AT, Blessington P, Zou T, Feather D, Wu X, Yan J, Zhang H, Liu Z, Ernst P, Koretzky GA, et al. 2010. MLL-AF9-induced leukemogenesis requires coexpression of the wild-type Mll allele. Cancer Cell 17: 148–159. 10.1016/j.ccr.2009.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkachuk DC, Kohler S, Cleary ML. 1992. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell 71: 691–700. 10.1016/0092-8674(92)90602-9 [DOI] [PubMed] [Google Scholar]
- Walker SR, Liu S, Xiang M, Nicolais M, Hatzi K, Giannopoulou E, Elemento O, Cerchietti L, Melnick A, Frank DA. 2015. The transcriptional modulator BCL6 as a molecular target for breast cancer therapy. Oncogene 34: 1073–1082. 10.1038/onc.2014.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winters AC, Bernt KM. 2017. MLL-rearranged leukemias—an update on science and clinical approaches. Front Pediatr 5: 4. 10.3389/fped.2017.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye BH, Lista F, Lo Coco F, Knowles DM, Offit K, Chaganti RS, Chaganti RS, Dalla-Favera R. 1993. Alterations of a zinc finger-encoding gene, BCL-6, in diffuse large-cell lymphoma. Science 262: 747–750. 10.1126/science.8235596 [DOI] [PubMed] [Google Scholar]
- Yu JM, Sun W, Hua F, Xie J, Lin H, Zhou DD, Hu ZW. 2015. BCL6 induces EMT by promoting the ZEB1-mediated transcription repression of E-cadherin in breast cancer cells. Cancer Lett 365: 190–200. 10.1016/j.canlet.2015.05.029 [DOI] [PubMed] [Google Scholar]
- Zhao Z, Wang L, Volk AG, Birch NW, Stoltz KL, Bartom ET, Marshall SA, Rendleman EJ, Nestler CM, Shilati J, et al. 2019. Regulation of MLL/COMPASS stability through its proteolytic cleavage by taspase1 as a possible approach for clinical therapy of leukemia. Genes Dev 33: 61–74. 10.1101/gad.319830.118 [DOI] [PMC free article] [PubMed] [Google Scholar]