Abstract
Objective:
Intrauterine growth restriction (IUGR) is a complication of pregnancy that has both short- and long-term sequelae for affected mothers and offspring. The pathophysiology of disease stems from poor nutrient and oxygen provision to the fetus, resulting in increased oxidative stress within the placenta. As the milieu within the local microenvironment alters macrophage differentiation, we hypothesized that macrophage plasticity may be altered in placentas associated with IUGR, and that they would show hallmarks of lipid peroxidation including altered aldehyde metabolism.
Methods;
In human placentas taken from normal pregnancies resulting in appropriate-for-gestational-age (AGA) newborns and placentas associated with IUGR, placental macrophages were evaluated by immunohistochemistry and shown in IUGR to resemble pro-inflammatory activated M1-type macrophages. To link oxidative stress to macrophages, the expression of aldehyde dehydrogenase (ALDHs) isozymes ALDH1, ALDH2, and ALDH3 was assessed.
Results:
All three isozymes displayed preferential staining for distinct cellular populations within the term human placenta. ALDH1 and ALDH2 were strongly expressed in macrophages and decidual stromal cells. ALDH3, in contrast, was exclusively present in extravillous trophoblasts. Comparing AGA and IUGR-associated placentas, ALDH1 and ALDH2 trended to have greater expression in macrophage populations but lower expression in decidual cell populations in IUGR-associated placentas. ALDH3 had higher expression in IUGR-associated placentas but localized specifically to extravillous trophoblast populations.
Conclusion:
Therefore, we speculate that specific ALDH isozymes have cell-specific functions related to differentiation, inflammation, or oxidative stress responses that are altered in IUGR-associated term human placentas. This family of isozymes may be a novel method to identify human placentas affected by placental insufficiency/IUGR.
Keywords: aldehyde dehydrogenase (ALDH), placenta, oxidative stress, intrauterine growth restriction, placental insufficiency, macrophage polarization
1. Introduction:
Human pregnancy is a condition where high levels of reactive oxygen species (ROS) occur routinely in uncomplicated, healthy pregnancies, and the placenta, at the maternal-fetal interface, is particularly well-equipped to function in this high oxidative stress environment (1, 2). The presence of ROS reflects the changing intrauterine oxidative environment, and leads to localized inflammation that allows tissue remodeling (3), but is balanced by anti-inflammatory mediators and endogenous antioxidants (4). However, excessive generation of ROS, unchecked by antioxidant mechanisms, can damage various molecules, including DNA, lipids, and proteins important for both metabolism and cellular signaling (5), and it is predicted to be a cause for placental insufficiency (6, 7). Placental insufficiency encompasses a spectrum of pregnancy-related disorders such as preterm birth, intrauterine growth restriction (IUGR) and pre-eclampsia, all thought to result from disordered placental development or long-standing placental insufficiency leading to poor nutrient and oxygen provision to the growing fetus (1). Intrauterine growth restriction can occur early or late in the pregnancy, and the timing of onset may be related to early structural lesions secondary to inappropriate spiral artery remodeling, or placental villous insufficiency, respectively. Regardless of timing, placentas associated with these disorders likely represent an extreme, where there is an imbalance between increased generation of damaging levels of ROS and decreased availability or activity of antioxidants that prevent these harmful effects (6–8). As the burden of maternal and neonatal morbidity and mortality associated with these disorders is significant (1, 6), evaluation of oxidative stress markers may be clinically meaningful in differentiating degrees of disease severity and in understanding potential mechanisms and pathways affecting vulnerability to oxidative stress in these conditions.
Immune dysregulation potentially contributes to placental insufficiency. Throughout pregnancy, macrophages of both maternal and fetal origin contribute to the successful course of pregnancy. These macrophages exhibit considerable plasticity and heterogeneity throughout implantation and pregnancy and are commonly divided into the M1 and M2 subtypes. M1 skewing refers to the classically activated macrophage, which presents antigens to the adaptive immune system (9, 10). In contrast, markers such as CD163, CD206, CD209, and IL-10 characterize M2 populations, and M2-like macrophages have immunosuppressive capacities and contribute to tissue remodeling. In many diseases such as cancer and atherosclerosis, macrophage populations are altered in response to oxidative stress, altering the balance of M1/M2 skewing (5). Throughout pregnancy, macrophages demonstrate significant plasticity (11), highlighting that macrophages both affect the progression of pregnancy and respond to alterations/insults at the maternal fetal interface. Especially in the early establishment of pregnancy, maternal decidual macrophages in extravillous sites have been shown to mediate maternal–fetal tolerance, promote spiral artery remodeling, and regulate trophoblast invasion (9). During implantation, maternal macrophages show enhanced M1 polarization but they subsequently shift to an M2 profile until the end of pregnancy in order to allow fetal development to occur (9). Hofbauer cells, or placental macrophages of fetal origin, also play an important role in maintaining pregnancy, affecting immune regulation, placental morphogenesis, control of stromal water content, and transport of ions and serum proteins across the maternal-fetal interface (12). Abnormalities in the homeostatis of this heterogeneous population of macrophages has been associated with placental disease including infection, inflammation, and abnormal placental development. Some reports suggest that in normal pregnancy, these Hofbauer cells demonstrate an M2-like phenotype (13), whereas in conditions of placental disorders, this M1:M2 balance may be disturbed. We thus predict that this M1:M2 ratio and expression of associated pro- and anti-inflammatory cytokines are altered in IUGR placentas.
Oxidative stress is coupled to lipid peroxidation, a process that yields many toxic aldehydes (10). To help maintain cellular homeostasis, a family of proteins known as the aldehyde dehydrogenases (ALDHs) are often upregulated to metabolize both endogenous and exogenous aldehydes. However, little is known about the role and expression of ALDH isozymes within the placenta generally and in macrophages specifically. To date, there are 19 identified subtypes in humans, with most isoforms widely distributed throughout the body, but specific to different cellular compartments (14, 15). Thus far, only two of the ALDH isozymes have been described during pregnancy. ALDH1 expression has been shown in stem/stromal cells derived from the maternal decidua as well as the chorionic villi (16, 17), while ALDH3 activity has been proposed to regulate trophoblast differentiation during murine placental development(18). With regards to macrophages, both ALDH1 and ALDH2 have been implicated in their biology (19, 20), but this effect has not been described during placentation. We thus hypothesized that ALDH activity may be high in the macrophages of IUGR placentas, important for handling ROS, and that they may serve as a marker of oxidative stress in disorders of placental insufficiency.
2. Materials and Methods
2.1. Human placental collection and processing
Collection and processing of the placentas used for this study have been described previously (21). Briefly, informed consent was obtained by study investigators from women with normal term or near-term (delivered after 36 weeks’ gestation) singleton pregnancies (with appropriate-for-gestational-age newborns, defined as birthweight between the 10th and 90th percentiles for gestational age, based on WHO guidelines) (n=21) or with late-onset singleton IUGR, as defined by newborn weight ≤ 10th percentile for gestational age (based upon WHO guidelines) and a trajectory of fetal growth deceleration in utero, diagnosed by prenatal ultrasound (n=11). All IUGR cases were deemed idiopathic (constitutional) IUGR, unrelated to fetal infection and without chromosomal abnormalities reported in the neonate. Of the cases that had available histology reports, samples with evidence of chronic villitis or villitis of unknown etiology were excluded. Samples with histologically confirmed chorioamnionitis were excluded.
For placental sample processing, the extreme ends of the basal plate and chorionic surface and membranes were removed by sharp dissection, and placental fragments were obtained at the middle of the initial placental depth. Sections were divided into two horizontal segments, with the basal plate on the bottom (maternal side) and the chorionic surface on the top (fetal side). Samples taken from the fetal and maternal side were snap frozen and stored at −80C, or processed for immunohistochemistry. Additional samples were obtained from archived specimens stored within the Department of Pathology at UCLA (n=10/group), and these sections were processed for immunohistochemistry as detailed below. For all samples utilized for this study, available pertinent clinical data is presented in Table 1 (22). Patient data for samples used in individual experiments is presented as Supplemental File 1, as not all samples were used for all studies.
2.2. Histology and immunohistochemistry
All tissues were rinsed in PBS, fixed in cold 10% neutral buffered formalin for 24 hours and then transferred to cold 70% ethanol. Samples were processed routinely and embedded in paraffin, sectioned at 5 μm and stained with hematoxylin and eosin by the Tissue Procurement Laboratory at UCLA. Sections were processed for ALDH1, ALDH2, and ALDH3 immunohistochemistry as previously described (23). Briefly, samples were deparaffinized and dehydrated in alcohol. Antigen retrieval was performed using 0.1mol/L citrate, pH 6.0, at 95°C for 20 minutes. The slides were then incubated with antibody or control IgG at the same dilution overnight. The antibody signal was visualized using the appropriate species Vector ABC kit (Vector Labs, PK6101; Burlingame, CA) followed by the DAB substrate according to the manufacturer’s instructions. Mouse anti-ALDH1 (cat #611194, BD Biosciences, San Jose, CA, USA) was used at a dilution of 1:200; ALDH2 (cat # ab108306, Abcam, Cambridge, MA, USA) was used at a dilution of 1:100; ALDH3 (cat #ab103895, Abcam) was used at a dilution of 1:100. To confirm staining populations, CD11c (clone D3V1E, Cell Signaling, Danvers, MA) for macrophages, rabbit anti-Vimentin (clone D21H3, Cell Signaling, Danvers, MA) for stromal cells, and rabbit anti-cytokeratin (clone Z0622, DAKO, Carpinteria, CA) for trophoblasts were used in consecutive sections within a 10 micron distance.
Sections were similarly processed for CD68 (general monocyte marker; clone PG_M1), MHCII (M1 type marker; polyclonal) and CD11c (M1 type marker), and CD206 (M2 type marker; polyclonal) expression as previously described (23), in order to better characterize the macrophage population expressing ALDH. CD68 (cat# M087629–2, DAKO, Carpinteria, CA) was used at a dilution of 1:50, MHCII (cat #NBP1–76296SS, Novus, Littleton, CO) at a dilution of 1:200, CD11c (clone D3V1E, Cell Signaling, Danvers, MA) at a dilution of 1:100, and CD206 (cat #ab64693, Abcam, Cambridge, MA, USA) was used at a dilution of 1:250.
2.3. Image Calibration
Staining was captured using a 10 or 20 or 40 × objective (×100 or ×200 or x400 magnification) from at least four non-overlapping microscopic fields per well by an AxioCam CCD digital camera (Carl Zeiss) mounted to an inverted epifluorescence microscope (AxioVert 135; Carl Zeiss). Files were saved in a 12-bit tagged image file format (.tif).
2.4. Statistical analysis of immunohistochemistry data
We conducted semi-quantitative analysis of ALDH1, ALDH2, and ALDH3 staining, given the pattern of distribution of positive staining (n=4–13/group). Semi-quantitative analysis of ALDH staining in the placental sections was conducted by two pathologists (PS and BC) blinded to clinical information. The percentage of positively stained cells was estimated, and staining intensities of 0 to 3 (0=below the level of detection, 1=weak, 2=moderate, 3=strong) were scored and are referred to as the Histological score (H-score). Samples with sub-optimal staining, or that could not be analyzed due lack of the appropriate anatomic area of the placenta represented in the section, were excluded from analysis.
Quantitative analysis of CD68 staining was performed by a blinded observer counting the number of positively stained cells per region within the placental villi, performed in triplicate on randomly selected fields using a 20X objective. Average cell counts were calculated for each placental sample (n=5–8/group). Analysis of CD206 (confirmed by CD11c staining) and MHCII was conducted as described above for the ALDH isoforms using a Histologic score (H-score) for staining intensities and overall amount of positively stained cells per region of interest (n=8–9/group).
2.5. Quantitative Real Time PCR
Total cellular RNA was isolated from frozen placental samples using the RNeasy mini kit (Qiagen, Valencia, CA, USA), as previously described (19). Complementary DNA was generated from 1.5 μg of total RNA from whole human placental tissue samples (n=5–10/group) by reverse transcription (RT) using the Superscript III Reverse Transcriptase kit (Invitrogen, cat#18080–400, San Diego, CA, USA). Amplification was performed in triplicate using Taqman-based detection (Taqman Fast Advanced Master Mix, Applied Biosystems, cat#4444557) on a Step One real-time quantitative PCR thermocycler (Applied Biosystems). For IL6, the gene expression assay (Applied Biosystems, cat#4453320, ID Hs00985639_m1) was used according to the manufacturer’s instructions. For IL10, the IL-10 Taqman Gene expression Assay (Mm01288386_m1) was used. For MPO, the Taqman gene expression assay (cat#4453320, ID Hs00165162_m1) was used. Relative gene expression was calculated using the comparative CT method with 18S (Applied Biosystems, #4319413E) expression used as the internal control for normalization. The amplification cycles consisted of: 50°C for 2 minutes, 95°C for 20 seconds, then 40 cycles of 95°C for 1 second (denaturation), and the 60°C for 20 seconds (annealing), followed by 72°C for 5 minutes (extension).
2.6. Western immunoblotting
Briefly, placentas were weighed, divided into a maternal or fetal aspect, frozen, then homogenized using a mortar and pestle. Tissue was resuspended in RIPA buffer containing protease and phosphatase inhibitors (Millipore Sigma) at 1mg/ml. 5 micrograms of each lysate was loaded per lane. Protein concentration was determined by using the Bio-Rad dye-binding assay. Western blotting was performed as described previously (n=5–7/group) (23). Briefly, the solubilized protein homogenates were subjected to electrophoresis on 10% SDS-polyacrylamide gels and transferred to nitrocellulose membranes (Transblot; Bio-Rad, Hercules, CA). The following primary antibodies were used for signal detection: ALDH1 antibody (cat# 611195, BD Biosciences, City, San Jose, CA, USA; ~55 kDA; 1:1000 dilution) and ALDH2 antibody (cat #ab108306, Abcam, Cambridge, MA; ~56 kDA; 1:1000 dilution). Anti-beta-actin antibody (cat #A0760–40, US Biological, Salem, MA; ~42kDA; 1:20,000 dilution) was used to detect endogenous beta actin expression, which served as an internal control for inter-lane loading variability. The quantification of protein bands was performed by densitometry using ImageJ software, as previously described (23).
2.7. Statistical Analysis
Statistical analyses were performed using GraphPad Prism software (version 5, GraphPad Software Inc, La Jolla, CA, USA). Non-parametric statistical testing was completed using the Mann-Whitney U test for analysis of the following data sets: scoring of CD206, MHCII, ALDH1, ALDH2, and ALDH3 intensity. Parametric statistical testing was completed using the Student’s t-test for analysis of the following data sets: protein expression quantification using Western blot (ALDH1, ALDH2, ALDH3), RNA expression quantification using PCR (IL6, IL10, MPO), CD68 cell counts by immunohistochemistry. Chi-square testing to compare proportions was used for demographic information. P-values <0.05 were considered statistically significant. Data were expressed as means ± SEM, when data is quantitative (e.g. PCR, western data values) and normally distributed (as tested using the D’Agostino and Pearson normality test). Data are expressed as median when data is non-normally distributed or results were semi-quantitative. Sample sizes were calculated based upon the ability to detect a 20% increase in macrophage cell counts between groups, assuming a mean of 100 macrophages per high-powered field with SD of 20 in our control AGA group. Power analysis shows that a sample size of 7 per group provides 80% power to detect a 20% difference in macrophage populations.
3. Results
3.1. Study Population
A total of 52 placental samples were used for all studies. Basic demographic information including maternal characteristics and newborn anthropometric data is included in Table 1 for all samples, and information for sample subsets used for individual experiments is available in Supplemental File 1.
3.2. Characterization of placental macrophages
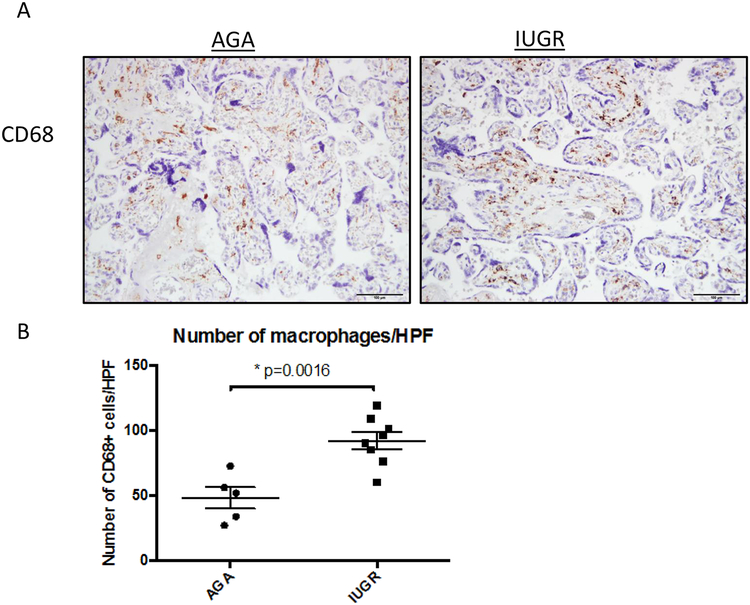
To initially quantitate macrophage populations, placentas were stained with anti-CD68 antibodies. An overall semi-quantitative increase in CD68+ cell numbers in IUGR-associated placentas was observed compared to the AGA placentas (p=0.0016 by Student’s t-test) (Fig 1).
Figure 1. CD68+ expression in human placentas from AGA and IUGR pregnancies.
Term human placentas were stained for CD68 expression (brown staining), a pan-marker of macrophage populations, and representative sections pictured here. Scale bars represent 100 μm. (A) The mean number of CD68+ cells per high-powered field (measured in triplicate) was increased in IUGR placentas (AGA: 48.15±18.23, n=5; IUGR: 92±18.7, n=8; p=0.0016) (B). Data are represented in scatter plots with horizontal lines representing means and standard error mean (SEM) values. Asterisk indicates p<0.05 by Student’s t-test.
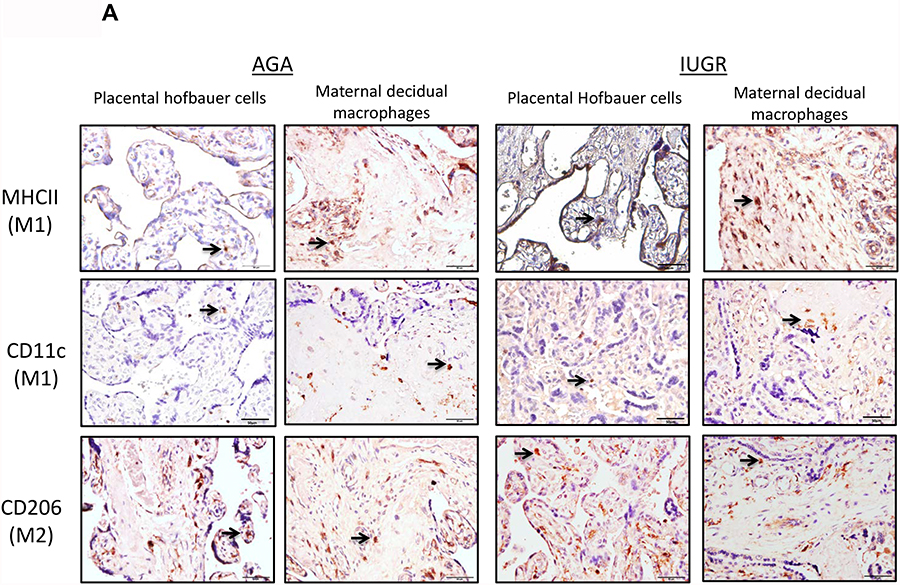
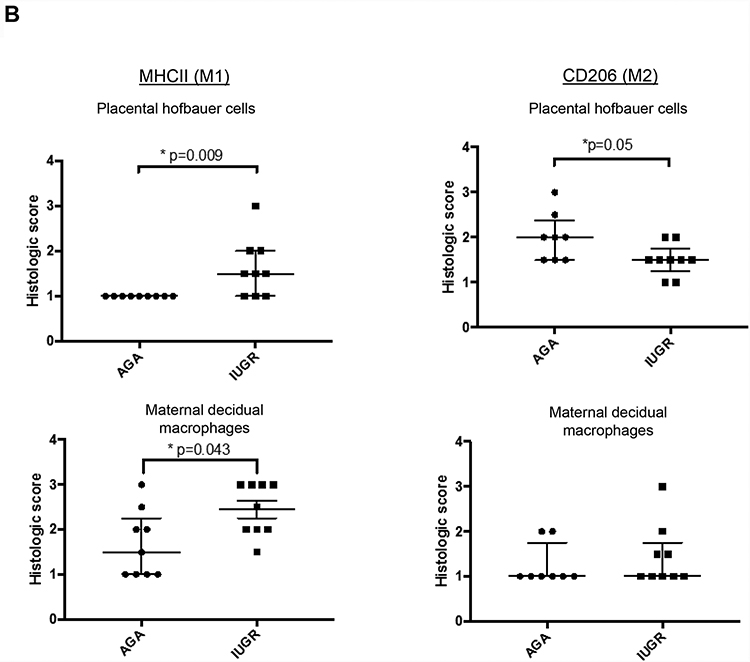
To further characterize the macrophage populations in these samples, MHCII and CD11c (indicating M1 polarization) and CD206 (indicating M2 polarization) expression was determined (Fig 2A) (24, 25). The MHC II positive staining was increased in IUGR associated placentas in the villous (p=0.009 by Mann-Whitney U testing) (presumed Hofbauer cells) and extravillous areas (p=0.043 by Mann-Whitney U testing) (presumed maternal decidual macrophages), and there was a corresponding decrease in the amount of CD206 positive cells in the villous area (p=0.05) (presumed Hofbauer cells). To confirm staining of MHC II in macrophage populations, CD11c was stained in a representative set of samples in consecutive sections, and a similar staining pattern was observed. In contrast to M1 macrophage populations, however, no significant change was observed in CD206 cells in extravillous areas (p=0.56) (Fig 2B) (presumed maternal decidual macrophages).
Figure 2: M1 versus M2 type polarization of macrophages in human AGA and IUGR placentas by MHCII, CD11c and CD206+ staining.
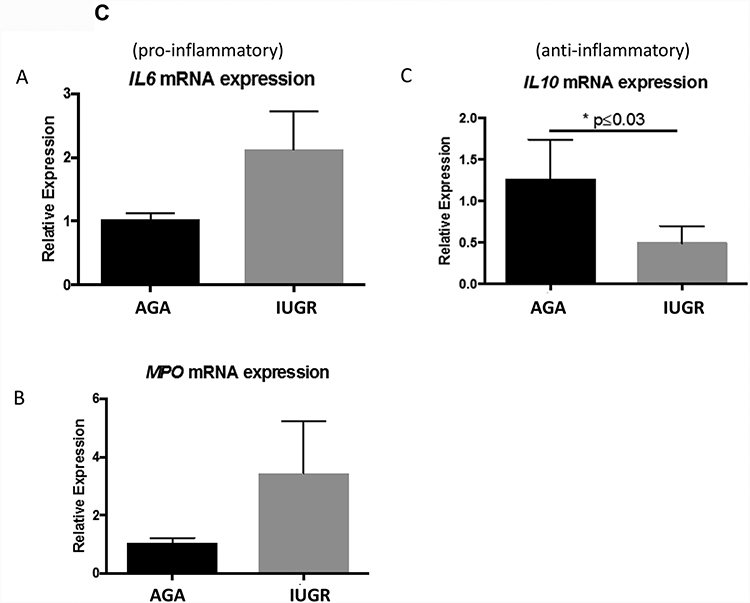
(A) Term human placentas were stained for MHCII (marker for M1-type macrophages), CD11c (M1-type macrophage marker) or CD206 (M2-type macrophage marker), and representative sections are pictured here at 200X magnification. Scale bars represent 50 μm. Black arrows indicate positively stained cells. (B) IUGR-associated placentas show enhanced MHCII staining in Hofbauer cells (AGA: median 1, interquartile range: 1–1, n=9; IUGR: 1.5, interquartile range: 1–2, n=9; p=0.009 by Mann-Whitney U testing) and in maternal decidual macrophages (AGA: median=1.5, interquartile range 1–2.25, n=9; IUGR: median=2.5, interquartile range: 2–3, n=9; p=0.043 by Mann-Whitney U testing) compared to AGA placentas. Concordantly, a reduction in M2-type marker staining (CD206) in the Hofbauer cells (AGA: median=2, interquartile range=1.5–2.375, n=8; IUGR: median=1.5, interquartile range: 1.25–1.75, n=9; p=0.05) was found, though there was no difference in the maternal decidual macrophages (p=0.56). Data are represented in scatter plots with horizontal lines representing median and interquartile range values. Asterisk indicates p<0.05 by Mann-Whitney U testing. (C) Gene expression. qRT-PCR analysis for gene expression of pro- (IL6: AGA: 1.03±0.098, n=7; IUGR: 2.12±0.61, n=5; p=0.06) (MPO: AGA: 1.037±0.18, n=3; IUGR: 3.442±1.773, n=2; p=0.17) and anti-inflammatory cytokines (IL10: AGA: 1.257±0.48, n=4; IUGR: 0.49±0.21, n=4; p=0.027) in whole placental samples. Asterisk indicates statistical significance with indicated p value, by Student’s t-test. Error bars indicate SEM values.
The data above suggested an increase in M1 Hofbauer cells within IUGR placentas. To confirm the functional changes associated with an increase in these cells, qRT-PCR of pro- and anti-inflammatory signaling genes: IL6 (pro-inflammatory cytokine), myeloperoxidase (MPO; produced by activated macrophages) and IL10 (anti-inflammatory cytokine produced during pregnancy) were evaluated. Within whole placental samples, patients with IUGR exhibited an overall pro-inflammatory signaling pattern. Pro-inflammatory IL6 (p=0.06 by Student’s t-test) and MPO (p=0.17 by Student’s t-test) gene expression were both increased in IUGR groups, though this did not reach statistical significance. In contrast, the anti-inflammatory IL10 signaling was significantly decreased in the human IUGR-associated placentas (AGA: p=0.027 by Student’s t-test) (Fig 2C), suggesting that IUGR correlates with the presence of M1-type Hofbauer cells.
3.3. ALDH expression within the placenta
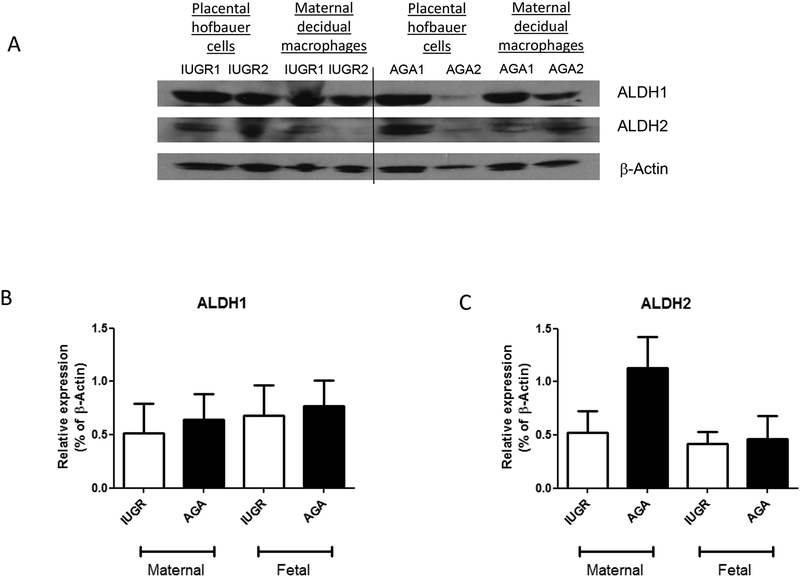
To understand the relationship of ALDH isozymes within the placenta to IUGR generally, whole sample protein expression was evaluated for ALDH1, ALDH2, and ALDH3 by Western blotting. ALDH1 and ALDH2 protein expression (expressed as a % of beta-Actin) was not different between AGA- and IUGR-associated placental samples. In AGA vs. IUGR placental samples, the mean expression of ALDH1 was not different (p=0.63 by Student’s t-test). For ALDH2, though there was a trend towards decreased ALDH2 expression in the IUGR-associated placental samples, no statistically significant differences were detected (p=0.28 by Student’s t-test) (Fig 3). ALDH3 expression could not be detected by Western immunoblotting.
Figure 3. ALDH1 and ALDH2 expression in human placental whole samples from normal AGA and IUGR pregnancies.
Western blotting was performed in whole placental samples to quantitate placental ALDH isoform expression in AGA and IUGR pregnancies. ALDH1 and ALDH2 are expressed within the placenta, but no statistical differences were observed between AGA and IUGR pregnancies (ALDH1: AGA 0.71±0.2 vs. IUGR 0.58±0.2; n=5/group; p=0.63 by Student’s t-test) (ALDH2: AGA 0.80±0.2 vs. IUGR 0.54±0.1; p=0.28 by Student’s t-test). ALDH3 expression was not detected by western blot analysis.
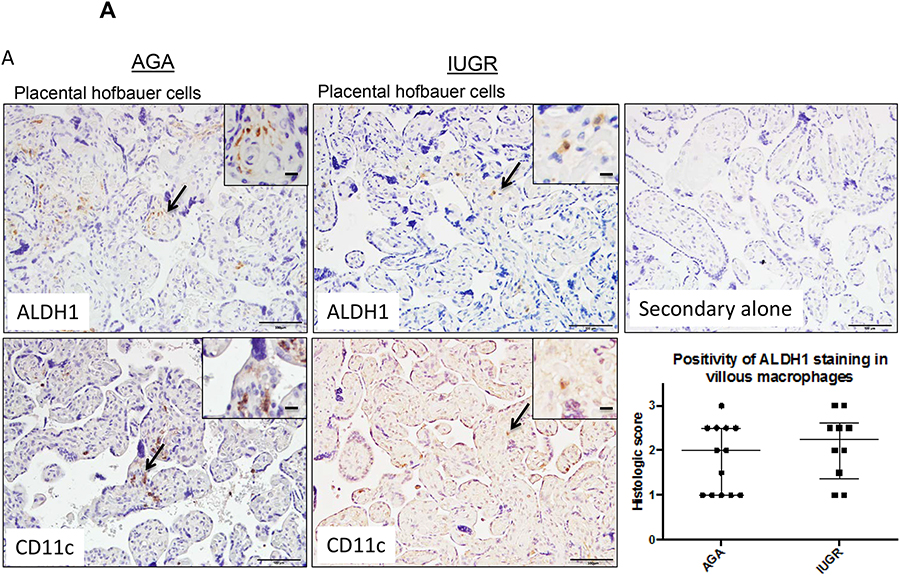
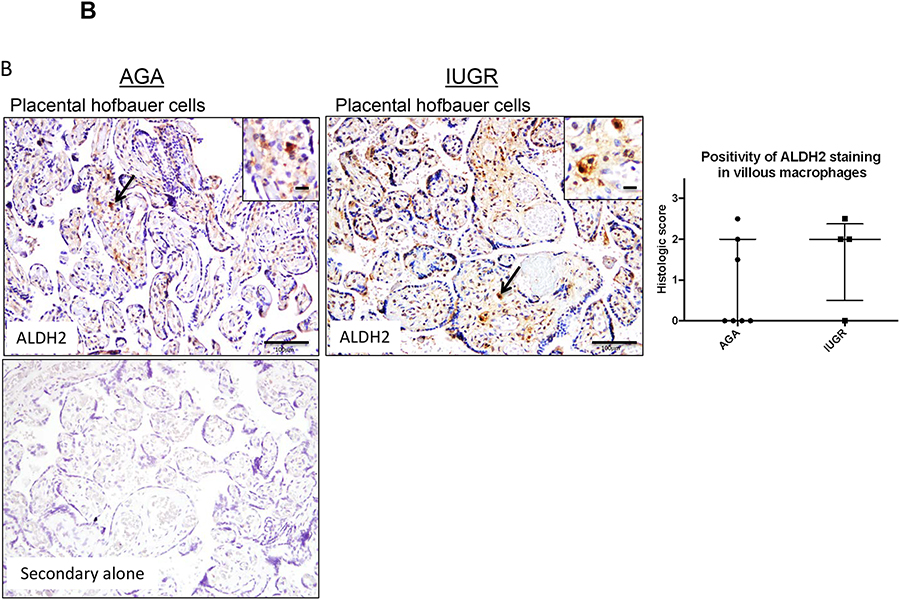
To next understand the relationship of ALDH isozymes in macrophages specifically, immunohistochemistry was performed using antibodies to ALDH1, ALDH2, and ALDH3. ALDH1 expression was localized within the placenta to two main cell populations – placental Hofbauer cells (Fig 4A) and decidual stromal cells. Using CD11c staining to confirm staining of M1 macrophages, there was no difference in ALDH1 expression between AGA-associated and IUGR-associated Hofbauer cells by histologic grading scores (p=0.38; Fig 4A). Sections were stained within 10 microns of each other. Similar to ALDH1, ALDH2 expression was also detected within Hofbauer cells (Fig 4B) and decidual stromal cells. Within Hofbauer cells, the mean ALDH2 expression was increased ~2 fold in IUGR associated compared with AGA placentas (p=0.33 by Mann-Whitney U testing), though this did not reach significance due to sample heterogeneity (Fig 4B).
Figure 4. ALDH1 and ALDH2 are expressed in placental Hofbauer cells.
Term human placentas were stained for ALDH1 and ALDH2 expression, and representative sections of Hofbauer cells pictured here. Scale bars represent 100 μm in the top row and 50 μm in the inset panels. Arrows indicate positively stained cells (brown), with magnified insets included in the upper right hand corners for ALDH1 staining. (A) ALDH1 and (B) ALDH2 expression are both detected in placental Hofbauer cell populations, confirmed by CD11c staining in consecutive sections. (A) Positivity of ALDH1 staining by Histologic scoring in Hofbauer cells did not differ in AGA and IUGR groups (AGA: median=2, interquartile range=1–2.5, n=13; IUGR: median=2.25, interquartile range 1.375–2.625, n=10; p=0.38 by Mann-Whitney U testing. (B) Staining of Hofbauer cells with ALDH2 was twice as high in IUGR placentas compared to the AGA group (AGA: median=0, interquartile range-0–2, n=7; IUGR: median=2, interquartile range 0.5–2.375, n=4, p=0.33 by Mann-Whitney U testing). Data are represented in scatter plots with horizontal lines representing median and interquartile range values.
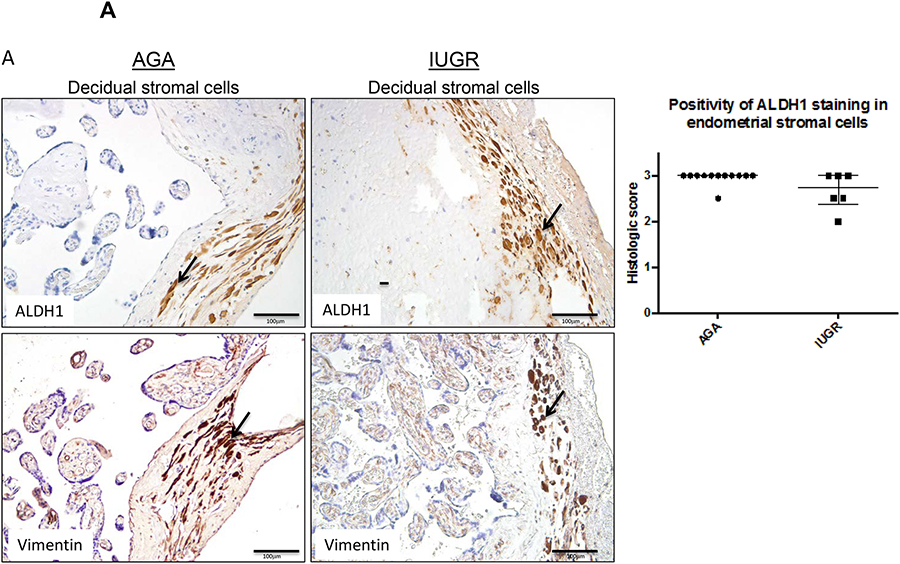
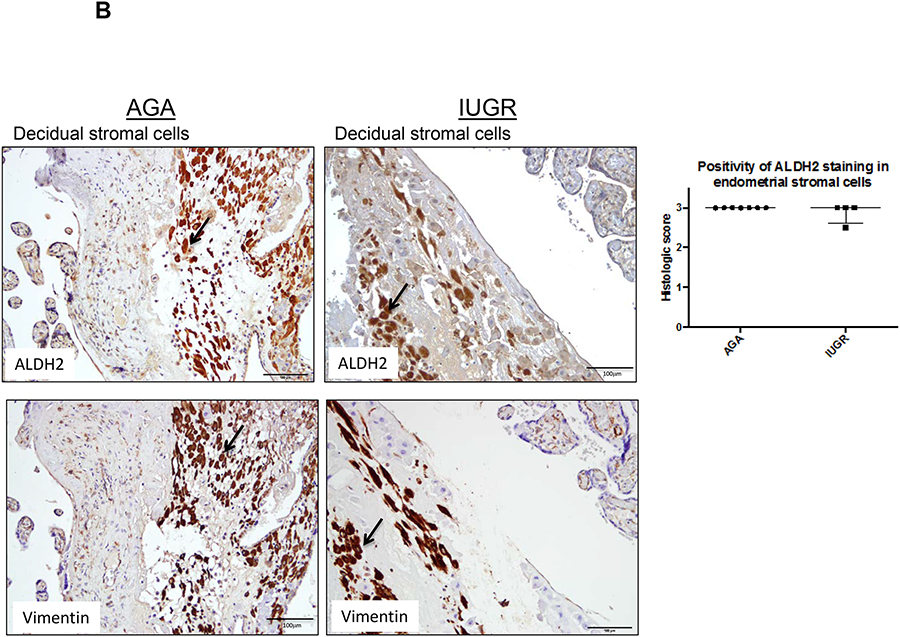
High expression of both ALDH1 and ALDH2 were observed in decidual stromal cells. However, the trends were contrary to what was expected. There was a trend towards decreased positivity of ALDH1 staining intensity in the decidual stromal cell population in IUGR-associated placentas compared to AGA placentas (p=0.06 by Mann-Whitney U testing; Fig. 5A). In decidual stromal cell populations, confirmed by the presence of vimentin, ALDH2 staining intensity did not distinguish between IUGR and AGA-associated placentas though there was a trend towards lower expression in IUGR (p=0.36 by Mann-Whitney U testing; Fig 5B). An independent pathologist confirmed the scoring for ALDH1 and ALDH2 in Hofbauer cell and decidual cell populations, and the same non-significant results were obtained.
Figure 5. ALDH1 and ALDH2 expression in decidual stromal cells.
Term human placentas were stained for ALDH1 and ALDH2 expression, and representative sections of the decidual stromal cells pictured here. Vimentin staining in serial sections was used to confirm cell populations of interest. Scale bars =100 μm. Arrows indicate positively stained cells (brown). (A) ALDH1 expression trended lower in decidual stromal cells of IUGR-associated placenta compared to AGA placentas (AGA: median=3, interquartile range=3–3, n=12; IUGR: median=2.75, interquartile range: 2.375–3, n=6; p=0.06 by Mann-Whitney U testing). (B) Positivity of ALDH2 staining in decidual stromal cells was not different between groups (AGA: median=3, interquartile range 3–3, n=7; IUGR: median=3, interquartile range 2.625–3, n=4; p=0.36 by Mann-Whitney U testing). Data are represented in scatter plots with horizontal lines representing median and interquartile range values.
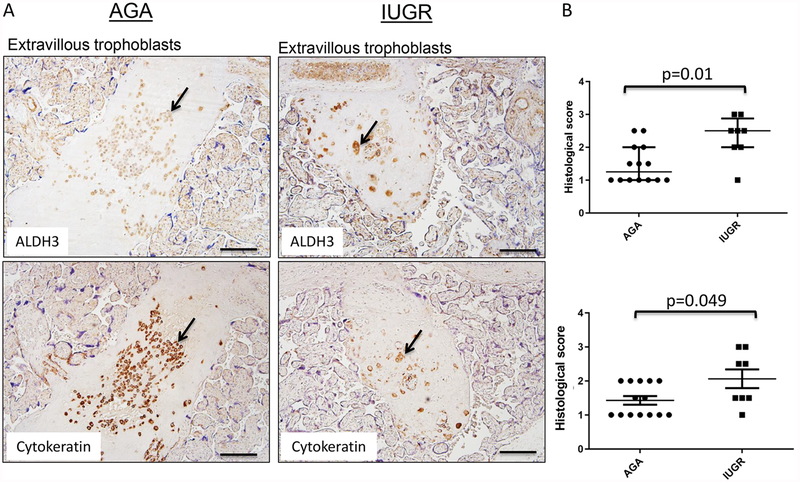
ALDH3 had a distinctly different staining pattern within the human placenta, with consistent positive cytosolic staining localized exclusively to extravillous trophoblast (EVT) (Fig 6). Staining of these cells was confirmed by their pan cytokeratin positivity. While there was ALDH3 staining present in the terminally differentiated EVTs embedded in fibrin matrix, a particularly strong signal was noted in EVT associated with maternal decidual vessels. By Mann-Whitney U analysis, using two independent pathologists to determine the inter-observer variation in scoring, the overall ALDH3 staining intensity was significantly higher in IUGR associated placentas compared to the AGA controls (p=0.009 and p=0.049, for each scorer; Fig. 6). However, no differences were observed in the percentage of ALDH3-positively stained cells between the two groups (data not shown, p=0.42).
Figure 6. ALDH3 expression in extravillous trophoblasts.
Term human placentas were stained for ALDH3 expression, and representative sections of extravillous trophoblasts pictured here. Scale bars represent 100 μm. Arrows indicate positively stained cells (brown). (A) ALDH3 expression is detected exclusively in extravillous trophoblast, especially those found in close proximity to maternal decidual blood vessels. The presence of these cells was confirmed by cytokeratin staining. (B) Positivity of ALDH3 staining in extravillous trophoblast was increased in IUGR placentas scored by two independent pathologists. (PS: AGA: median=1.4, interquartile range 1.0–2.0, n=14; IUGR: median=2.1, interquartile range 1.5–2.9, n=8; p=0.01 by Mann-Whitney U testing); (BC: AGA: median: 1.0; interquartile range 1.0–2, n=14; IUGR: median=2.5, interquartile range 1.0–2.5, n=8; p=0.049 by Mann-Whitney U testing). Data are represented in scatter plots with horizontal lines representing median and interquartile range values.
4. Discussion
It is widely accepted that both maternal and fetally derived macrophages play an important role in all stages of pregnancy. Macrophages are present throughout the maternal-fetal interface, and it has been shown that successful pregnancies require appropriate macrophage activation (26). Macrophage function, which is a manifestation of the cell’s activation state, is ultimately decided by the surrounding milieu. M1 macrophages present antigens to T cells to produce a TH1 or cell mediated immune response (11, 26). In contrast, M2 macrophages promote a more immunosuppressive milieu, driving TH2 or antibody mediated immune responses and tissue remodeling and repair. Within pregnancy, it has been shown that macrophage populations fluctuate during the course of pregnancy. Late in pregnancy, the expression pattern on placental Hofbauer cells, skew towards an M2 phenotype and others have shown that markers such as CX3CR1, IL-7R and CCR7 have low expression (25). A number of M2 markers have been used to identify placental macrophages, but each has limitations of selectivity including CD163, CD206/mannose receptor, CD209, IL10, and folate receptor (FR)-β (25, 27–29). CD68, the “classical” macrophage marker, is generally considered a pan-macrophage marker, and has been described in placental macrophage populations (25). Typically, the expression of these markers is regulated by environmental signals, such as cytokines and hormones, which may be altered in pregnancy complications.
In this study, high numbers of CD68+ tissue macrophages were observed, and the number and intensity of staining was increased in IUGR compared to the age-matched AGA term placentas. These cells skewed towards an M1-macrophage phenotype in IUGR placentas, suggesting the creation of a proinflammatory microenvironment.
Given that the balance of M1 and M2 macrophages has been linked with an increase in spontaneous preterm labor (11), IL-6/IL-10 levels were quantitated to distinguish between a M1 and M2 associated microenvironment (29, 30). These cytokines are known to distinguish the macrophage microenvironment in chronic disease, and similarly, it has been shown that a pro-inflammatory signature exists in peripheral blood of patients with IUGR with placental insufficiency. IL-6, TNFα, and IL-12 levels were all significantly higher by ELISA while the levels of the anti-inflammatory cytokine IL- 10 were lower in stimulated peripheral blood mononuclear cells in the context of IUGR compared to those without placental insufficiency (31). Similarly, within the placenta itself, our results show a pro-inflammatory bias in IUGR with placental insufficiency compared to AGA controls. Whole placental samples showed increased IL-6 and reduced IL-10 expression, consistent with a pro-inflammatory state that would be associated with M1 polarization.
ALDHs represent a superfamily of NADP(+)-dependent enzymes that protect cells from cellular damage induced by active aldehydes, and they play a vital role in clearing reactive oxygen species (ROS) by inducing oxidation of both exogenous (such as alcohol) and endogenous aldehydes (such as lipid, amino acids) into carboxylic acids (15). If aldehydes accumulate un-metabolized in high levels, they can induce enzyme inactivation, DNA damage and cell death. Given the high ROS observed in IUGR, we hypothesized that ALDH activity in macrophages may be increased and that it may be a marker of oxidative stress in disorders of placental insufficiency.
In our study, ALDH1 and ALDH2 expression was observed in Hofbauer cells. Within this cell population, ALDH1 did not distinguish placental insufficiency in IUGR-associated pregnancies from normal AGA pregnancies, while ALDH2 expression trended to be higher in IUGR-associated pregnancies. While Hofbauer cells were identified more than 100 years ago, little is known about the regulation and function of these cells. Detected by 4 weeks post-conception, Hofbauer cells are present throughout pregnancy, and they are proposed to contribute to a variety of functions including placental morphogenesis, immune regulation, and passage of serum proteins across the maternal-fetal barrier (12, 32). Reports suggest that Hofbauer cells in normal later stages of pregnancy typically display an M2-like phenotype (13), and this is consistent with the results observed here. Moreover, our results suggest that these cells are responsive to putative oxidative stress as in IUGR, and thus may show ALDH2 up-regulation.
We also observed ALDH1 and ALDH2 expression in decidual stromal cells. While the presence of ALDH1 in human-derived decidual (endometrial) stromal cell culture has been previously reported (16, 17, 33), its function in these cells has not been well characterized. Though the functions of this cell population are still debated, decidual stromal and mesenchymal stem cells have been described as important in regulating the immunologic milieu in the placenta (22). Lower ALDH1 expression in this cell type in IUGR may indicate impaired ALDH1 function in this cell population. Interestingly, ALDH1 expression in a number of malignancies is present in both the parenchyma and stroma of the tumor, with higher levels often observed within the stroma allowing for continued tumor cellular growth (34). This data may suggest, similar to the data presented here, that ALDH1 expression in decidual cells may be linked to increased inflammation through separate compensatory mechanisms. Similar to ALDH1, ALDH2 expression was strongly expressed on decidual stromal cells as well as in Hofbauer cells but did not distinguish between AGA and IUGR-associated pregnancies. Aldh2 expression in pregnant mice has been shown to be necessary for fetal genome preservation in Fanca, Fanconi anemia DNA repair pathway gene deficient embryos, and it has been shown to be required for aldehyde catabolism during normal hematopoiesis in utero (35). Therefore, ALDH2 in stromal cell populations may have important function at earlier stages in pregnancy, but not as a stress response late in pregnancy.
Interestingly, our study also detected ALDH3 immunoreactivity specifically in extravillous trophoblast, especially those in close proximity to maternal decidual blood vessels. The expression of ALDH3 in trophoblast has been reported previously in human tissue (36), and it has been implicated as essential for trophoblast differentiation during murine placental development (18). However, in our cohort, comparing IUGR and AGA-associated placentas, it is interesting to note that ALDH3 expression in extravillous trophoblast (EVTs) can distinguish IUGR from AGA by positivity of staining. Given that the placenta is assumed to be a relatively hypoxic environment, the stronger ALDH3 expression in EVTs closely approximated to maternal blood vessels in IUGR are consistent with its putative function as an “oxygen stress” sensor. Further study will be needed to establish the biologic function of ALDH3 in this population, whether it be important for oxidative stress response or differentiation or otherwise in this particular cell population.
Previous studies to try to identify oxidative stress markers in the placentas of women with placental insufficiency have yielded inconsistent results, likely due to differences in sampling and localization (37–40). In addition, many of these studies have largely measured RNA and protein levels in whole tissue samples, limiting cell-specific conclusions as to the role and utility of changes seen in antioxidant levels. Though we had initially hypothesized that ALDH isozyme expression would be important in macrophages as a marker of oxidant stress in inflammatory cell populations in placental insufficiency, what we found was that ALDH expression is isozyme- and cell-population specific, and that various isozymes trend towards differential expression in these distinct populations in IUGR. As we have shown in our study, changes in expression of oxidative stress markers that are only expressed in specific cell populations that represent a small percentage of the total cells in the placenta would be difficult to detect in small patient cohorts by RNA or total protein level measurements of whole placental samples. Our results suggest that ALDH isozymes are expressed and can be used to define discrete cellular populations within the placenta. Moreover, alterations in their expression can be detected in placental pathologies such as in IUGR. Additional work will be needed to determine the specific functions of the ALDH isozymes within these distinct cell populations as related to cell differentiation, inflammatory responses, and oxidant stress mechanisms.
Supplementary Material
Funding:
This work was supported by the NIH UCLA CTSI KL2TR001882 (PI: Wong, to AC), NICHD 1K08HD093874–01 (to AC), NCI R01 CA163971 (to MW) and the American Heart Association Beginning Grant-in-Aid Western Affiliate States 15BGIA25710060 (to AC). These funding sources had no involvement in the study design, collection, analysis, interpretation of data, writing of the report, or decision to submit the article for publication.
Abbreviations:
- AGA
appropriate for gestational age
- ALDH
aldehyde dehydrogenase
- IUGR
intrauterine growth restriction
- MPO
myeloperoxidase
Footnotes
Declarations of Interest: none
References:
- 1.Jauniaux E, Poston L, & Burton GJ (2006) Placental-related diseases of pregnancy: involvement of oxidative stress and implications in human evolution. Human Reproduction Update 12(6):747–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raijmakers MT, Roes EM, Poston L, Steegers EA, & Peters WH (2008) The transient increase of oxidative stress during normal pregnancy is higher and persists after delivery in women with pre-eclampsia. European journal of obstetrics, gynecology, and reproductive biology 138(1):39–44. [DOI] [PubMed] [Google Scholar]
- 3.Sauer H, Wartenberg M, & Hescheler J (2001) Reactive oxygen species as intracellular messengers during cell growth and differentiation. Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology 11(4):173–186. [DOI] [PubMed] [Google Scholar]
- 4.Rinaldi SF, Hutchinson JL, Rossi AG, & Norman JE (2011) Anti-inflammatory mediators as physiological and pharmacological regulators of parturition. Expert review of clinical immunology 7(5):675–696. [DOI] [PubMed] [Google Scholar]
- 5.Kalyanaraman B (2013) Teaching the basics of redox biology to medical and graduate students: Oxidants, antioxidants and disease mechanisms. Redox biology 1:244–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duley L (2009) The global impact of pre-eclampsia and eclampsia. Seminars in perinatology 33(3):130–137. [DOI] [PubMed] [Google Scholar]
- 7.Menon R (2014) Oxidative stress damage as a detrimental factor in preterm birth pathology. Frontiers in immunology 5:567–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schoots MH, Gordijn SJ, Scherjon SA, van Goor H, & Hillebrands JL (2018) Oxidative stress in placental pathology. Placenta 69:153–161. [DOI] [PubMed] [Google Scholar]
- 9.Zhang YH, He M, Wang Y, & Liao AH (2017) Modulators of the Balance between M1 and M2 Macrophages during Pregnancy. Front Immunol 8:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh S, et al. (2013) Aldehyde dehydrogenases in cellular responses to oxidative/electrophilic stress. Free radical biology & medicine 56:89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown MB, von Chamier M, Allam AB, & Reyes L (2014) M1/M2 Macrophage Polarity in Normal and Complicated Pregnancy. Frontiers in Immunology 5:606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reyes L, Wolfe B, & Golos T (2017) Hofbauer Cells: Placental Macrophages of Fetal Origin. Results and problems in cell differentiation 62:45–60. [DOI] [PubMed] [Google Scholar]
- 13.Mantovani A, Biswas SK, Galdiero MR, Sica A, & Locati M (2013) Macrophage plasticity and polarization in tissue repair and remodelling. The Journal of pathology 229(2):176–185. [DOI] [PubMed] [Google Scholar]
- 14.Xu X, et al. (2015) Aldehyde dehydrogenases and cancer stem cells. Cancer letters 369(1):50–57. [DOI] [PubMed] [Google Scholar]
- 15.Marchitti SA, Brocker C, Stagos D, & Vasiliou V (2008) Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert opinion on drug metabolism & toxicology 4(6):697–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kusuma GD, et al. (2016) Mesenchymal Stem/Stromal Cells Derived From a Reproductive Tissue Niche Under Oxidative Stress Have High Aldehyde Dehydrogenase Activity. Stem cell reviews 12(3):285–297. [DOI] [PubMed] [Google Scholar]
- 17.Kusuma GD, Abumaree MH, Perkins AV, Brennecke SP, & Kalionis B (2017) Reduced aldehyde dehydrogenase expression in preeclamptic decidual mesenchymal stem/stromal cells is restored by aldehyde dehydrogenase agonists. Scientific reports 7:42397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishiyama M, Nita A, Yumimoto K, & Nakayama KI (2015) FBXL12-Mediated Degradation of ALDH3 is Essential for Trophoblast Differentiation During Placental Development. Stem cells (Dayton, Ohio) 33(11):3327–3340. [DOI] [PubMed] [Google Scholar]
- 19.Magnusson MK, et al. (2016) Macrophage and dendritic cell subsets in IBD: ALDH+ cells are reduced in colon tissue of patients with ulcerative colitis regardless of inflammation. Mucosal immunology 9(1):171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kühl AA, Erben U, Kredel LI, & Siegmund B (2015) Diversity of Intestinal Macrophages in Inflammatory Bowel Diseases. Frontiers in immunology 6:613–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janzen C, et al. (2013) Placental glucose transporter 3 (GLUT3) is up-regulated in human pregnancies complicated by late-onset intrauterine growth restriction. Placenta 34(11):1072–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson DM & Burton GJ (2011) A technical note to improve the reporting of studies of the human placenta. Placenta 32(2):195–196. [DOI] [PubMed] [Google Scholar]
- 23.Chu A, et al. (2016) Gestational food restriction decreases placental interleukin-10 expression and markers of autophagy and endoplasmic reticulum stress in murine intrauterine growth restriction. Nutrition research (New York, N.Y.) 36(10):1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abumaree MH, et al. (2013) Human placental mesenchymal stem cells (pMSCs) play a role as immune suppressive cells by shifting macrophage differentiation from inflammatory M1 to anti-inflammatory M2 macrophages. Stem cell reviews 9(5):620–641. [DOI] [PubMed] [Google Scholar]
- 25.Joerink M, Rindsjo E, van Riel B, Alm J, & Papadogiannakis N (2011) Placental macrophage (Hofbauer cell) polarization is independent of maternal allergen-sensitization and presence of chorioamnionitis. Placenta 32(5):380–385. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y-H, He M, Wang Y, & Liao A-H (2017) Modulators of the Balance between M1 and M2 Macrophages during Pregnancy. Frontiers in Immunology 8(120). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang Z, et al. (2013) Decreased levels of folate receptor-beta and reduced numbers of fetal macrophages (Hofbauer cells) in placentas from pregnancies with severe pre-eclampsia. American journal of reproductive immunology (New York, N.Y. : 1989) 70(2):104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gustafsson C, et al. (2008) Gene expression profiling of human decidual macrophages: evidence for immunosuppressive phenotype. PloS one 3(4):e2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Svensson J, et al. (2011) Macrophages at the fetal-maternal interface express markers of alternative activation and are induced by M-CSF and IL-10. Journal of immunology (Baltimore, Md. : 1950) 187(7):3671–3682. [DOI] [PubMed] [Google Scholar]
- 30.Xu Y, et al. (2016) An M1-like Macrophage Polarization in Decidual Tissue during Spontaneous Preterm Labor That Is Attenuated by Rosiglitazone Treatment. Journal of immunology (Baltimore, Md. : 1950) 196(6):2476–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Al-Azemi M, Raghupathy R, & Azizieh F (2017) Pro-inflammatory and anti-inflammatory cytokine profiles in fetal growth restriction. Clin Exp Obstet Gynecol 44(1):98–103. [PubMed] [Google Scholar]
- 32.Tang Z, Abrahams VM, Mor G, & Guller S (2011) Placental Hofbauer cells and complications of pregnancy. Annals of the New York Academy of Sciences 1221:103–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Popovici RM, et al. (2006) Gene expression profiling of human endometrial-trophoblast interaction in a coculture model. Endocrinology 147(12):5662–5675. [DOI] [PubMed] [Google Scholar]
- 34.Kunju LP, et al. (2011) EZH2 and ALDH-1 mark breast epithelium at risk for breast cancer development. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc 24(6):786–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oberbeck N, et al. (2014) Maternal aldehyde elimination during pregnancy preserves the fetal genome. Molecular cell 55(6):807–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marchitti SA, Orlicky DJ, Brocker C, & Vasiliou V (2010) Aldehyde dehydrogenase 3B1 (ALDH3B1): immunohistochemical tissue distribution and cellular-specific localization in normal and cancerous human tissues. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society 58(9):765–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sahay AS, Sundrani DP, Wagh GN, Mehendale SS, & Joshi SR (2015) Regional differences in the placental levels of oxidative stress markers in pre-eclampsia. International journal of gynaecology and obstetrics: the official organ of the International Federation of Gynaecology and Obstetrics 129(3):213–218. [DOI] [PubMed] [Google Scholar]
- 38.Mistry HD, et al. (2010) Differential expression and distribution of placental glutathione peroxidases 1, 3 and 4 in normal and preeclamptic pregnancy. Placenta 31(5):401–408. [DOI] [PubMed] [Google Scholar]
- 39.Hempstock J, et al. (2003) Intralobular differences in antioxidant enzyme expression and activity reflect the pattern of maternal arterial bloodflow within the human placenta. Placenta 24(5):517–523. [DOI] [PubMed] [Google Scholar]
- 40.Gohil JT, Patel PK, & Gupta P (2011) Evaluation of oxidative stress and antioxidant defence in subjects of preeclampsia. Journal of obstetrics and gynaecology of India 61(6):638–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.