Abstract
Inflammatory bowel disease (IBD) is a chronic disorder of the gastrointestinal tract. The molecular mechanisms underlying IBD are poorly characterized and treatment options are limited. To gain insight into the pathogenesis of chronic colonic inflammation (colitis), we performed a multi-omic analysis that integrates RNA microarray, total protein mass spectrometry (MS), and phosphoprotein MS measurements from a mouse model of the disease. Because we collected all three types of data from individual samples, we could track information flow from RNA to protein to phosphoprotein to identify signaling molecules that were coordinately or discordantly regulated. With this information, we identified pathways that had complex in vivo regulation. For example, the genes encoding acute phase proteins were expressed in the liver, but the proteins were detected by MS in colons during inflammation. We also used the multi-dimensional dataset to ascertain which types of data best described particular facets of chronic inflammation. Using gene set enrichment analysis and trans-omic co-expression network analysis, we found that each data set provides a unique viewpoint on the molecular pathogenesis of colitis. Combining human transcriptomic data with the mouse multi-omic data implicated increased p21-activated kinase (Pak) signaling as a driver of colitis and chemical inhibition of Pak1 and Pak2 with FRAX597 suppressed active colitis in mice. These studies provide translational insights into the mechanisms contributing to colitis and identify Pak as a therapeutic target in IBD.
One Sentence Summary:
Transcriptomics, proteomics, and phospho-proteomics reveals therapeutic targets in colitis.
Introduction
Inflammatory bowel disease (IBD), composed of Crohn’s disease (CD) and ulcerative colitis (UC), affects more than 5 million people worldwide. Sufferers experience a variety of debilitating gastrointestinal symptoms that require medical and, eventually, surgical intervention. The ultimate target of medical treatment is mucosal healing, which improves outcomes. Nevertheless, this goal remains elusive in many patients. Although there have been advances in therapeutics, the current treatment options for IBD are limited and include general immunomodulators and targeted biologics, such as antibodies targeting tumor necrosis factor alpha (TNF-α), α4β7 integrin, and IL-12/23 (1). All of these therapies suffer from variable efficacy and non-durable response, as well as a spectrum of negative side effects. A better understanding of the molecular pathogenesis of IBD would lead to new therapeutic strategies that could lead to more effective treatments.
Genetic and epidemiological studies in human patients (2, 3), as well as experimental studies in animal models (4), have identified numerous genetic and environmental risk factors for CD and UC, but have not identified clear driver mutations that could lead to therapeutic opportunities. To account for the complexity in IBD etiology, researchers have taken transcriptomic (5, 6), proteomic (7, 8), metabolomic (9, 10) and metagenomic (11, 12) approaches in an attempt to understand global disease networks and to identify genes, proteins, and metabolites that may be involved in disease pathogenesis. Although these approaches have provided valuable insight, they have fallen short of identifying potential high value therapeutic targets in IBD.
In this study, we generated a multi-omic dataset in which transcriptomic, proteomic, and phosphoproteomic measurements were made from individual colons of mice with and without colitis. We used this dataset to understand the relationships between RNA abundance, protein abundance, and protein phosphorylation and to determine what each type of data reveals about gut inflammation. Because all three types of data were collected from each individual sample, we could identify discrepancies between transcriptomic and proteomic measurements, thus we could predict post-translational protein regulation and identify changes in gene expression that originated at distant organ sites. Finally, we performed co-expression network analysis to identify signaling pathways that were coordinately or uniquely dysregulated in the different data sets and we computationally inferred kinase activation from global phosphoproteomic mass spectrometry (pMS) data by collating kinase substrate lists and using them with the gene set enrichment (GSEA) algorithm. These complementary computational approaches implicated Pak signaling as a potential driver of colitis. We validated the role of Pak signaling in a preclinical therapeutic study in mice, and analysis of gene expression data from humans indicated that this finding was applicable to IBD patients. Together, these studies provide an unprecedented view of dysregulated signaling in colitis and identify a previously unrecognized pathogenic signaling pathway that represents a viable therapeutic opportunity.
Results
Collection of multi-omic data from mouse colon
The initial goal of this study was to quantify global transcriptomic and proteomic changes that occur during chronic colitis. Because this requires a large amount of starting material, we chose to use a mouse model of IBD, namely the adoptive transfer mouse model of CD, known for high penetrance and relatively short latency (13). Rag1 null animals on a C57BL/6J genetic background were injected with 400,000 CD45RBhi naïve T cells or, as a negative control, 200,000 regulatory T cells (Tregs) from isogenic wild-type (WT) animals and then weighed bi-weekly and assessed for symptoms related to the onset of colitis, such as diarrhea and rectal prolapse. Animals were sacrificed following sustained weight loss of greater than 1.5 grams for one week, which was indicative of severe colitis. Control animals were sacrificed concomitantly. Upon sacrifice, 3mm of tissue from the medial colon was removed and fixed for histological assessment (fig. S1). The remaining colon was opened longitudinally and approximately 1/8th was snap frozen for microarray analysis, while the remaining matched tissue was snap frozen for mass spectrometry (fig. S2). With this tissue processing strategy, we obtained RNA, total protein, and phospho-protein from an individual colon, enabling relative quantification among control and experimental animals.
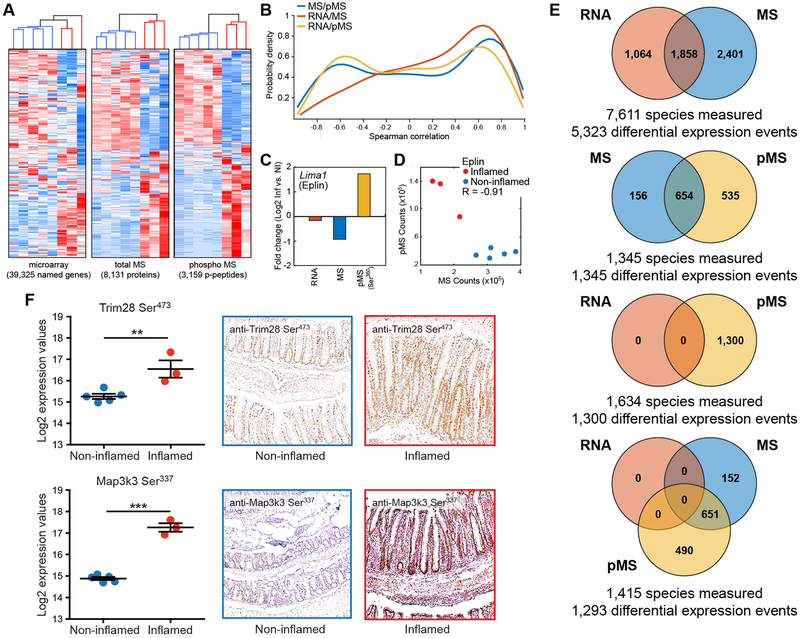
Following sample processing and data collection, we quantified 39,325 named (38,666 unique) RNA transcripts, 7,951 proteins, and 3,159 phosphopeptides representing 3,325 unique phosphorylation sites on 1,711 proteins (tables S1–S3). Unsupervised hierarchical clustering indicated that each of the three measurements segregated the inflamed mice from the non-inflamed (Fig. 1A). Of the 7,951 proteins measured by mass spectrometry (hereafter, we refer to total protein mass spectrometry as MS), 7,611 (96%) were represented in the RNA data set. RNA transcripts were measured for 1,634 (95%) of the 1,711 proteins in the pMS data set. Total protein MS data were obtained for 1,474 (86%) of the 1,711 proteins measured by pMS and 1,415 species were measured in all three data sets.
Fig. 1.
Multi-omic analyses of murine colitis. (A) Unsupervised clustering of microarray, mass spectrometry (MS), and phospho mass spectrometry (pMS) data. Non-inflamed samples are marked in blue in the dendrograms above the heat maps. Inflamed samples are marked in red. (B) Probability distributions of Spearman correlations for each pairwise comparison between–omic data sets. (C) Lima1 (Eplin) average fold changes between inflamed and non-inflamed colons. (D) Scatter plot of phospho-Ser360 and total Eplin protein counts in individual samples. (E) Venn diagrams summarizing the unique and overlapping differential expression events between the RNA, MS, and pMS data sets. Species refers to RNAs, proteins, or phosphopeptides that were detected in each dataset from the comparison. Expression events refers to the direct comparison between the datasets for a given species. (F) Cellular localization of representative up-regulated phospho-signals in colitis. Left panels show abundance in individual samples, as detected by MS, for Trim28 Ser473 (top) and Map3k3 Ser337 (bottom). ** p < 0.01, *** p < 0.001 in an unpaired T test. Other panels show immunohistochemistry for phospho-Trim28 and phospho-Map3k3 in non-inflamed and inflamed colons. In all panels, N = 5 for control (non-inflamed) samples and N = 3 for inflamed samples.
Because we measured RNA, protein, and phosphoprotein from individual samples, we could perform one-to-one matched correlation of individual genes across measurements (Fig. 1B). The probability density functions for Spearman correlations showed a correlation landscape for RNA/MS comparison that was distinct from RNA/pMS and MS/pMS. Most RNA/MS gene pairs were positively correlated with each other, with only a few species showing an inverse correlation (Fig. 1B). The RNA/pMS probability density function showed a bimodal distribution, indicating that there were similarly sized sub-groups for which there was negative correlation, no correlation, or positive correlation between RNA abundance and phospho-peptide abundance. The MS/pMS probability distribution was similar to the RNA/pMS distribution, but there were more positively correlated MS to pMS species than RNA/pMS species (Fig. 1B). We hypothesized that the inverse correlation between some MS/pMS species (higher MS to pMS) represented proteins that are marked for degradation by phosphorylation. Eplin (encoded by the Lima1 gene) has a ubiquitin-priming phosphorylation site and exemplified this type of regulation in our data. Lima1 transcripts were essentially unchanged (1.1-fold reduced) in inflamed versus non-inflamed tissue, yet the protein abundance was ~two-fold decreased (Fig. 1C). We detected a 3.3-fold increase in phosphorylation at Ser360 (Fig. 1C and D), which targets the protein for ubiquitination and degradation (14).
Differential RNA expression and protein and phosphoprotein abundance analysis
We determined the RNAs, proteins, and phosphopeptides that had significantly different abundance in inflamed colons relative to non-inflamed colons. Overall, 7,752 of 38,666 RNA transcripts, 4,443 of 7,951 proteins, and 2,346 of 3,325 phosphopeptides had differential abundance (table S4). Of the 7,611 species for which we had both RNA and MS data, 1,858 showed changes in abundance that were similar (either both increased or both decreased), 1,064 RNAs and 2,401 proteins had differential abundance in only their respective data set, and the remaining 2,288 species were not differentially changed (Fig. 1E). All 1,345 proteins detected in the pMS and MS data had differential abundance in either one data set or both (Fig. 1E), while none of the 1,634 RNAs with co-measured phospho-peptides in the pMS data were differentially expressed, resulting in limited differential abundance similarity between all three data sets (Fig. 1E). This comparison demonstrated that the pMS data are critical for exploring the molecular pathogenesis of colitis.
Because our goal was to characterize changes to the tissue-level signaling network during colitis, we isolated RNA and protein from the whole colon, which includes the epithelium, lamina propria (including immune, stromal, and vascular cells), and muscularis as the starting material for our RNA and protein datasets. The cellular representation of an inflamed colon is different from that of a normal colon, especially with respect to the influx of inflammatory cells (fig. S1). To explore whether the changes that we detected through transcriptomic and proteomic analyses reflected the influx of inflammatory cells, we used immunohistochemistry to analyze the cellular localization of signals that were up-regulated in animals with colitis. We selected phosphorylation of Trim28 (Ser473) and Map3k3 (Ser337) because they were among the few sites that met the criteria of being upregulated during colitis and having phospho-specific antibodies that worked for immunohistochemistry. Both exhibited substantially greater phosphorylation during colitis (Fig. 1F); however, their distribution in the colon differed. Trim28 phosphorylation was increased in the colonic epithelium, whereas Map3k3 phosphorylation was increased in all components of the colonic environment (Fig. 1F). These observations indicated that the changes in RNA, protein, and phosphoprotein abundance that occur during colitis reflect both a major change in the cellular composition of the tissue and changes in the tissue-level signaling network.
Pathway enrichment analysis
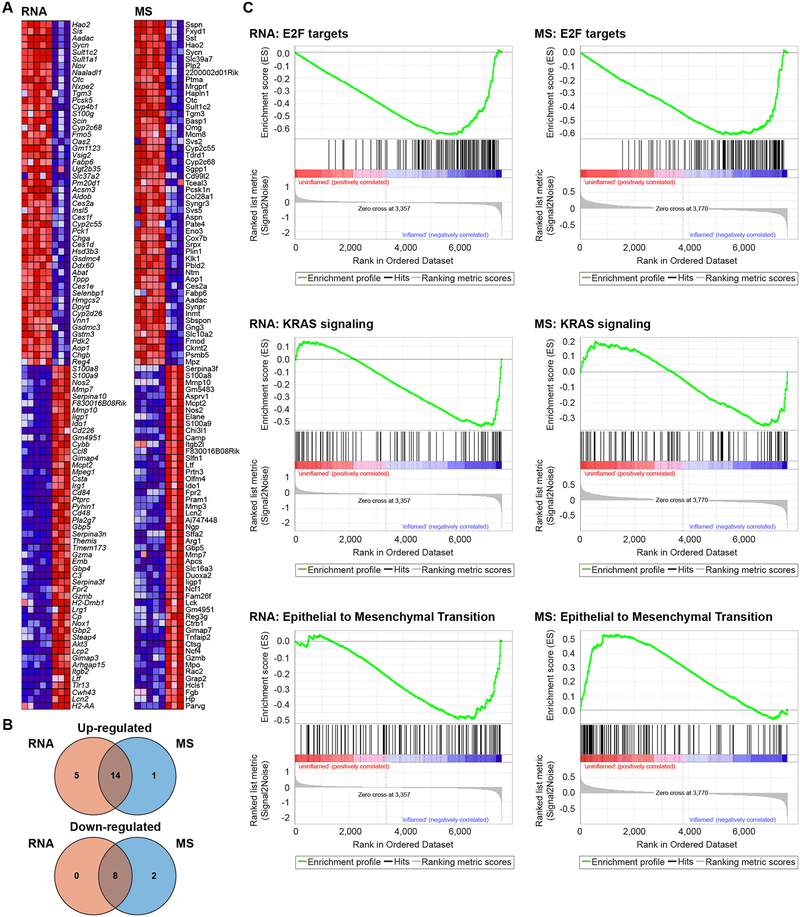
Although these analyses described how the -omics data sets related to one another, we wanted to determine whether the differences between inflamed and non-inflamed samples in each data set represented similar pathways and high-level functional categories. We performed GSEA (15) on the 7,611 genes represented in both the RNA and MS data sets to identify differentially regulated pathways between inflamed and non-inflamed mice. At the RNA level, 19 pathways were positively enriched and 8 pathways were negatively enriched in inflamed mice (Fig. 2A and table S5). Most were the same as the 15 positively enriched and 10 negatively enriched pathways in inflamed mice at the total protein level, suggesting strong functional concordance between the transcriptomic and proteomic data (Fig. 2B and table S5). The finding that 14/20 positively enriched and 8/10 negatively enriched pathways were similarly regulated in inflamed mice was consistent with the Spearman correlation histogram of the RNA/MS data (Fig. 1B). Coordinately positively enriched pathways included those involved in the inflammatory response and tumor necrosis factor alpha signaling, as well as pathways controlling epithelial proliferation, such as E2F targets, KRAS signaling, and MYC targets (Fig. 2C). The “epithelial to mesenchymal transition” gene set was positively enriched at the RNA level and negatively enriched at the protein level, suggesting complex regulation of this particular pathway (Fig. 2C). This observation highlights the potential for RNA analysis to provide misleading information about the role of a particular pathway in driving a disease.
Fig. 2.
Differential RNA expression, differential protein abundance, and pathway analysis. (A) Heatmaps of RNA and total protein measurements most strongly contributing to pathway enrichment scores of GSEA. (B) Venn diagrams summarizing the unique and overlapping up and down-regulated pathway enrichment in the RNA and MS data sets. (C) GSEA plots from RNA and total protein MS data set.
Analysis of non-correlated RNA and protein signals
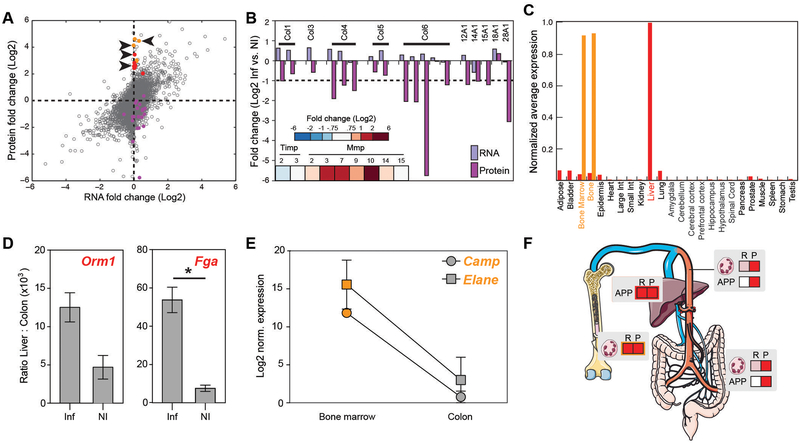
Comparative analysis of the changes in RNA and protein abundance presents an opportunity to explore if and how each data set provides unique information on the regulatory events associated with colitis, especially when those data sets do not change in the same direction. For example, our GSEA analysis identified the “interferon alpha response” as positively enriched in the MS data set, but not in the RNA data set (table S5). To expand upon this observation, we examined the correlation between RNA and protein abundance for the 7,611 species for which we collected both types of data. Although we observed general concordance in fold-change differences (control vs. colitis) between RNA and protein measurements, a small number of species were altered at the protein level, but exhibited no change at the RNA level (Fig. 3A). To determine if these discordant species were present in particular functional categories, we performed Gene Ontology (GO) enrichment analysis for all species that showed greater than 22-fold change in protein abundance and less that 20.75-fold-change in RNA abundance. We found enrichment for genes involved in defense response (positively enriched in the MS dataset) and extracellular matrix (negatively enriched in the MS dataset). The extracellular matrix category of proteins was composed primarily of collagens (Fig. 3A, purple dots) and, when we surveyed all of the collagens shared between the microarray and total MS data sets, we found that 12/17 collagens were reduced at least 2-fold in protein abundance, but unchanged or weakly increased in abundance at the RNA level (Fig. 3B). Moreover, we observed an increase in several extracellular matrix metalloproteinases (MMPs), notably MMPs 3, 7, 9 and 10, in the MS data set. All of these enzymes function to degrade extracellular matrix components such as collagen and fibronectin (16). This illustrates how comparative RNA/protein abundance analysis can infer genes for which the corresponding protein is regulated by degradation.
Fig. 3.
Differential regulation of RNA and protein. (A) Scatter plot of fold change (inflamed vs. non-inflamed) in RNA expression plotted against fold change in protein abundance for species that were present in both data sets. Colored dots represent ECM proteins (purple), acute-phase proteins (red), and neutrophil proteins (orange), with the arrowheads indicating genes that were further investigated in panels D and E. (B) Collagen expression in the RNA data and abundance in the protein data. Differential abundance of MMPs and TIMPs in the MS data are indicated in the heatmap inset with a key. (C) Tissue expression patterns of acute phase (red) and neutrophil (orange) transcripts. Each gene was normalized to a maximum of one, and all of the genes from each category averaged to generate bars. (D) Induction of acute phase RNA in the liver during inflammation. Bars represent the ratio of liver expression to colon expression for Orm1 and Fga from N = 2 each of inflamed and non-inflamed animals. Assays were performed in duplicate. * p < 0.05 in an unpaired T test. p = 0.085 for Orm1. (E) Loss of neutrophil gene expression by colonic neutrophils. Abundance of the RNA for Camp and Elane from bone marrow neutrophils relative to that in neutrophils isolated from colon is shown. The plot represents log-transformed data from N = 2 inflamed animals, normalized to the smallest expression value for each gene. For both genes, p < 0.05 in a paired T test. (F) Model depicting RNA expression (R) and protein abundance (P) for acute phase proteins (APP) and neutrophil proteins (represented by a polymorphonuclear cell symbol) in the colon and distant organ sites.
Within the defense response GO category, we focused on the acute phase proteins (Fig. 3A, red dots) and a set that are specific to neutrophils (Fig. 3A, orange dots). The liver coordinates the acute phase response and therefore we hypothesized that these genes may be transcribed and translated in the liver and then travel to the colon through the blood stream or possibly through bile. According to the publicly available mouse gene atlas (17), all are highly expressed in the liver, with minimal expression in other tissues (Fig. 3C). To confirm this expression pattern in our experimental model, we assayed the RNA abundance of two of the genes, Fga and Orm1, in the colons and livers of animals with and without colitis. For both genes, there was at least 5,000-fold higher expression in the liver than in the colon (Fig. 3D). In inflamed animals, this ratio increased to ~12–50,000-fold (Fig. 3D), indicating that the transcriptional events that ultimately resulted in increased colonic abundance of acute phase proteins most likely originated in the liver.
We selected the five neutrophil-specific genes with discordant RNA and protein changes (Fig. 3A, orange dots) because this group of genes was unexpected: The adoptive transfer model of IBD exhibits strong neutrophil recruitment to the colon and there were many other neutrophil-specific genes that were increased in abundance at the RNA and protein levels in animals with colitis. According to the mouse gene atlas (17), these five genes exhibit bone marrow-specific expression in normal mice (Fig. 3C). However, these genes belong to other groups of genes that show decreasing RNA expression as neutrophils travel from the bone marrow to the circulating blood (18). To assess whether this expression pattern underlies the discordant colonic RNA and protein abundance in our model, we isolated neutrophils by flow cytometry from the bone marrow and colons of animals with colitis. We assessed the abundance of the transcripts for Camp and Elane in neutrophils from these two tissues, which revealed an average of 2,257- and 6,142-fold higher expression in the bone marrow derived neutrophils compared to colonic neutrophils from inflamed animals (Fig. 3E). These data suggested that neutrophils transcribe and translate these proteins in the bone marrow, and that the RNA message is degraded by the time the neutrophils reach the site of inflammation, leaving only the protein. Together, these examples demonstrate how transcriptomic and proteomic data covering the same genes can be used to infer regulation of expression in distant organ sites. In both cases, the message and protein were initially expressed in a distant organ but carried to the colon through the blood (acute phase proteins) or within cells recruited to the colon (neutrophil proteins) (Fig. 3F).
Trans-omic co-expression network analysis
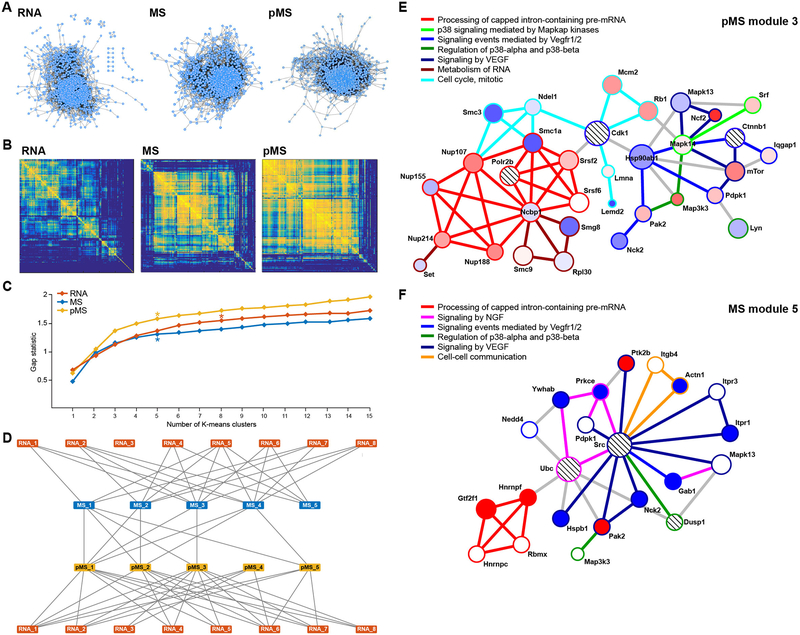
To gain further insight about the relative information content of the three data sets, we assessed the co-abundance changes of the 1,415 species (1,429 RNA transcripts, 1,452 proteins, 3,080 phosphopeptides) that were present in all data sets. Spearman correlation coefficients were calculated for all pairs of variables within each data set and dichotomized. Correlations greater than 0.9 or less than −0.9 were set to 1 and all others were set to 0. We visualized both the correlation networks and the two-step generalized topological overlap matrix (GTOM2) to explore the correlation landscape, modularity, and existence of highly correlated subsets of variables of each data set (Fig. 4A and B). GTOM2 relates the interconnectedness of two genes by computing the number of shared-two step network neighbors. Clustering the GTOM2 matrix facilitates identification of groups of highly correlated genes called modules (19, 20). At the RNA level, we noted several small clusters of highly correlated transcripts, whereas the protein abundance and phospho-peptide abundance changes were less modular (Fig. 4B). To estimate the true number of clusters in the RNA, MS, and pMS data sets, we computed the gap statistic for up to 15 clusters, by k-means clustering, of the GTOM2 matrix (21). The gap statistic measures the within-cluster variation of each cluster in a dataset compared to a null distribution of expected within-cluster variation. The cluster at which the gap curve begins to level off or decrease as a function of the number of clusters indicates the number of clusters in each data set. Based upon the gap curves and clustered GTOM2 matrices, we estimated that there were 8 RNA clusters, 5 MS clusters, and 5 pMS clusters in our data (Fig. 4C).
Fig. 4.
Co-expression and co-abundance network landscapes of RNA, MS, and pMS measurements. (A) Correlation networks for RNA, MS, and pMS data sets. Nodes indicate genes or proteins and edges connect two genes or proteins if the Spearman correlation between the expression of two genes or abundance of the two proteins is greater than 0.9 or less than −0.9. (B) Two-step generalized topological overlap matrices (GTOM2) of the RNA, MS, and pMS data sets clustered by unsupervised hierarchical clustering. Correlations greater than 0.9 or less than −0.9 were set to 1 (yellow) and all others were set to 0 (blue). Square regions indicate highly connected clusters of genes or proteins. (C) Plot of the gap statistic versus the number of clusters in each data set. Clustering cutoff points are marked with a star for each data set based upon the gap statistic and GTOM2 topology. (D) Network visualization of module overlap. Nodes indicate particular modules; edges are present if there was significant overlap in genes or proteins between the two modules (Fisher Exact p<0.05, FDR q<0.25). (E) YourCrosstalker network module for pMS cluster 3. Nodes are colored by differential phosphorylation status in inflamed relative to uninflamed colons (red = hyper-phosphorylated, blue = de-phosphorylated, WMW p<0.05, FDR q<0.25) and edges are colored by pathway membership of the interaction. Gray edges indicate PPI’s that exist but are not part of an enriched pathway in the subnetwork. Pathways with higher statistical significance determine the interaction pathway association for interactions in multiple pathways. Striped nodes were recruited by the algorithm during the random walk procedure as significantly traversed “crosstalker” nodes. (F) YourCrosstalker network for MS cluster 5. Nodes are colored by total protein differential abundance status in inflamed relative to uninflamed mice (red = increased abundance, blue = decreased abundance, WMW p<0.05, FDR q<0.25). Edges and striped nodes are defined as in panel E.
We extracted the genes in each of these clusters (table S6) and compared the gene lists between clusters (table S7). We visualized the significant overlap between modules in a network diagram and found that most RNA modules shared a significant overlap with multiple MS and pMS modules (Fig. 4D). Although some MS and pMS modules overlapped, the connections were sparser and the partially isolated modules (RNA_3, MS_5, and pMS_4) might present interesting cases of specific trans-omic regulation in colitis. To interpret the function of the individual modules in each data set and to examine function across data sets, we used the network analysis tool YourCrosstalker (22, 23) to identify the enriched pathways, filter out network edges that were not associated with protein-protein interactions, and identify topologically relevant genes that were not in our original cluster gene lists. We performed the YourCrosstalker random walk on the STRING protein-protein interaction network and performed pathway enrichment with a combination of Reactome and the National Cancer Institute Pathway Interaction Database (NCI-PID) (24–27).
The correlation structure of module RNA_3 was selectively associated with the phosphorylation modules pMS_3 and pMS_4. The 54 genes in RNA_3 showed enrichment only for nerve growth factor (Ngf) signaling. The 121 genes in pMS_4 showed enrichment only for processing of capped intron-containing pre-mRNAs. In contrast, we found a greater diversity of pathway enrichments in the YourCrosstalker modules of the 397 genes in pMS_3 (Fig. 4E). Here we found a central network that connected multiple Vegf signaling, p38 signaling, cell cycle, and pre-mRNA processing pathways (Fig. 4E). We analyzed the 221 proteins in the module MS_5, a protein module that did not significantly associate with any pMS modules but was significantly connected to RNA modules 2, 4, 5, and 6 (Fig. 4F). Similar pathways were implicated by the genes in MS_5 as in pMS_3. In particular, we noted the shared MS_5-pMS_3 pathways of Vegf, pre-RNA processing, cell cycle, and p38 signaling, as well as the presence of Ngf signaling shared with RNA_3 (Fig. 4F). This observation suggests that the signaling network at the intersection of these pathways plays a role in colitis and that the components of this network are dysregulated in distinct ways in the different omics data sets. This discovery was enabled by the integrated picture provided by our multi-omic data set and trans-omic analysis approach.
Inferring kinase activity from phosphoproteomic data
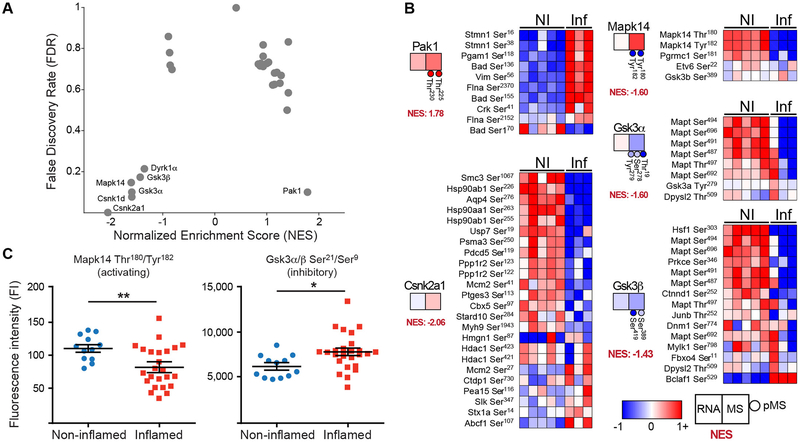
Our overarching goal is to understand how dysregulated signaling contributes to the pathogenesis of colitis and to determine whether there are signaling pathways that could represent novel therapeutic targets for IBD. To this end, we reasoned that our pMS data set would most accurately reflect therapeutically tractable changes in signal transduction that occur during colitis. Our pMS analysis identified 2,346 differentially phosphorylated peptides in animals with colitis and, although 80% of the phosphopeptides had been previously identified, only 1.68% were functionally annotated (fig. S3). In essence, the biological importance of most of the measured phosphorylation events is presently unknown. We sought to overcome this deficiency in prior knowledge by using known kinase-substrate relationships to computationally infer kinase activity from the pMS data. Lists of kinase-substrate relationships for 348 kinases across a range of kinase families were curated from PhosphoSitePlus (table S8) (28). These were entered as ‘gene’ lists into the GSEA algorithm, enabling us to perform a “kinase activation” analysis from pMS data. Although many kinases showed substrate enrichment in inflamed colons (table S9 and Fig. 5A), only Pak1, which had an uncertain role in IBD pathogenesis, reached statistical significance as defined by a false discovery rate (FDR) less than 0.25 (Fig. 5A,B). Both Pak1 and Pak2 were part of several of the YourCrosstalker modules that we identified in our co-expression network analysis (Fig 4E and F and table S6), providing a cross validation of the different computational analyses.
Fig. 5.
Inferring kinase activity from pMS measurements. (A) Volcano plot of Normalized Enrichment Score (NES) versus False Discovery Rate (FDR). Kinases with positive or negative enrichment and an FDR < 0.25 are specified. (B) Heatmaps of phospho-peptides corresponding to known kinase substrates from non-inflamed (NI) and inflamed (Inf) animals. The kinase is indicated to the left of each set of substrates with the Log2 differential abundance (inflamed vs. non-inflamed) for RNA (left box), protein (right box), and phosphorylation (circles). Normalized Enrichment Score (NES) is specified for each kinase. All of the kinases shown had FDR < 0.25 and are predicted to be either up-regulated (positive NES) or down-regulated (negative NES) in colitis. (C) Validation of Mapk14 and Gsk3α/β phosphorylation in colon samples from inflamed and noninflamed animals.. Fluorescence intensity (FI) was measured using Luminex assays specific to each phosphorylation site. N = 12 samples from individual non-inflamed animals and N = 25 samples from individual inflamed animals. * p < 0.05 and ** p < 0.01 in an unpaired T test.
In addition to the predicted activation of Pak1 during colitis, GSEA predicted that six kinases – Casein Kinase 2A1 (Csnk2a1), Gsk3α/β, p38α (Mapk14), Casein Kinase 1D (Csnk1d), and Dyrk1α – were less active in the animals with colitis than in the control animals (Fig. 5A and B). Of these kinases, only Mapk14, one of the stress-induced mitogen-activated protein kinases (MAPKs), has been linked to inflammation (29). To confirm the predicted kinase activation, we measured the phosphorylation of sites on Gsk3α/β and Mapk14 that regulate their activation states. We performed a Luminex-based phosphorylation assay on samples from an independent cohort of animals with adoptive transfer-induced colitis. This analysis revealed that animals with colitis had decreased phosphorylation of Mapk14 at activating sites (Thr180/Tyr182) and increased phosphorylation of Gsk3α/β at inhibitory sites (Ser21/Ser9) (Fig. 5C). As such, this experiment validated the reduction in Mapk14 and Gsk3α/β activity as predicted by GSEA of the pMS dataset.
Evidence of Pak signaling in human IBD patients
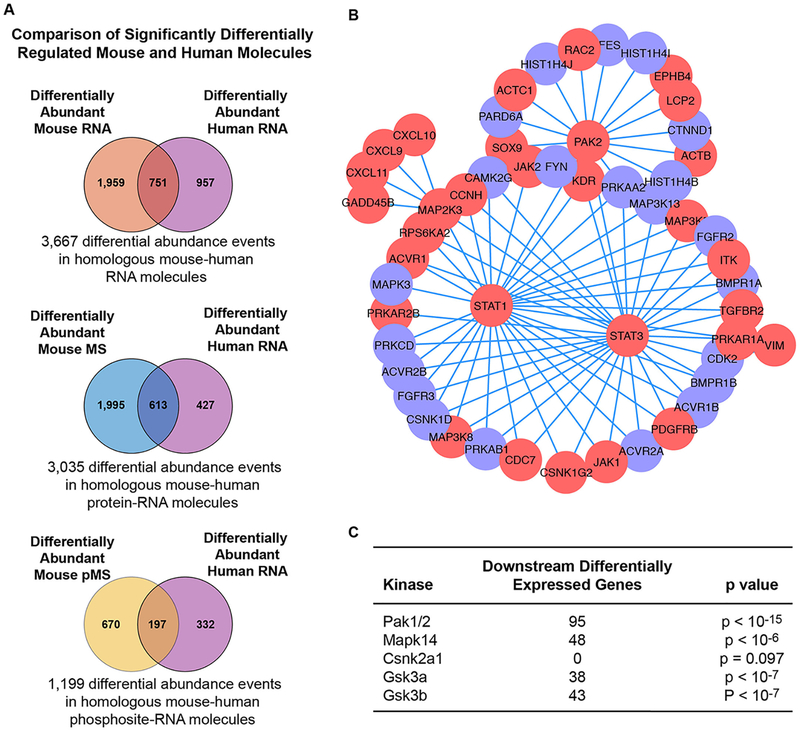
An important consideration with any experimental model system is the generalizability of the findings of that system to the human in vivo context. To compare our mouse data to human, we obtained a publicly available gene expression dataset of inflamed (n=12) and uninflamed (n=16) IBD patient colonic biopsies (6). Our aim was to assess global concordance between human IBD differential gene expression and, in particular, whether the identification of increased Pak1 and Pak2 activity was conserved in patients. We performed differential expression analysis on the entire human dataset and compared differential abundance of human RNA to mouse RNA, MS, and pMS. There were 1,708 genes differentially expressed between inflamed and uninflamed human samples. All 1,708 human RNAs were represented in the mouse RNA data, 1,040 genes were represented in the mouse MS data, and 529 genes were found in the mouse pMS data. Of the 2,710 homologous mouse RNA transcripts that were differentially expressed, 751 were also differentially expressed in humans (Fig. 6A). Of the 2,608 homologous mouse proteins with differential abundance, 613 were also represented by differentially expressed transcripts in the human RNA data (Fig. 6A). Finally, of the 867 homologous differentially phosphorylated proteins measured in the mouse pMS data, 197 were differentially expressed in the human RNA data (Fig. 6A). In general, more molecular species (RNA, MS, and pMS) tended to exhibit differential abundance in the mouse relative to the human RNA data. The larger set of differentially abundant mouse RNA, MS, and pMS species represented 43.9%, 58.9%, and 37.2% of the possible homologous differential expression events. This suggests that the mouse MS dataset offers a reasonable experimental representation of the human disease context. Furthermore, similar to the generally weak correspondence between RNA and phospho-peptide differential activity in the mouse, we observed that the products of genes differentially expressed in the human tended to not be differentially phosphorylated in the mouse.
Fig. 6.
Mouse model omic data overlap with human IBD biopsy transcripts. (A) Venn diagrams representing the differential expression analysis of human IBD colonic biopsies in inflamed and uninflamed phenotypes (Wilcoxon Mann Whitney p<0.05, FDR q<0.25) compared to differentially expressed RNA, protein abundance, and phospho-peptide abundance between inflamed and uninflamed mouse colons. (B) Human IBD differentially expressed genes in the PAK signaling network neighborhood. Genes are colored by differential expression direction (red = up-regulated, blue = down-regulated) in inflamed relative to un-inflamed human colonic biopsies. (C) Assessment of the overlap between the genes regulated by kinases significantly associated with colitis in the mouse and human genes differentially expressed in the kinase regulated network (Hypergeometric test p<0.05).
We next sought to determine the extent to which the Pak1 and Pak2 signaling network neighborhood was differentially active in human IBD. We assembled a human protein-protein interaction (PPI) network by querying PAK1 and PAK2 in the Pathway Commons database (26). The PAK network neighborhood contained 529 unique genes and 3,666 interactions. We filtered the human expression data and PAK network for overlapping genes. After filtering for expression array coverage, the final PAK neighborhood contained 431 genes and 2,534 interactions (fig. S4). When we overlaid the differentially expressed genes from human IBD patients onto the Pak network neighborhood, we found that 95 genes were differentially expressed (Fig. 6B) and the largest connected network of these genes implicated PAK2, STAT1, and STAT3 as key hub nodes. This analysis suggested that the inferred PAK signaling mechanism from the mouse model translates to the human disease and may be a viable therapeutic target in IBD.
To assess the relative importance of PAK signaling compared to that of other kinases implicated by phosphopeptide based GSEA (Fig. 5), we repeated the analysis of constructing a network neighborhood of the kinase and searching for differentially expressed human genes. A hypergeometric test was applied to assess the significance of the overlap between differentially expressed human genes and the network neighborhood of the kinases MAPK14, CSNK2A1, GSK3A, and GSK3B. The PAK1 neighborhood contained 95 differentially expressed genes and was the most significant kinase (p < 10−15) (Fig. 6C). Although MAPK14, GSK3A, and GSK3B also have significantly active network neighborhoods, the PAK1 neighborhood is statistically the most significant of the kinases (Fig. 6C).
Validation of Pak as a therapeutic target in colitis
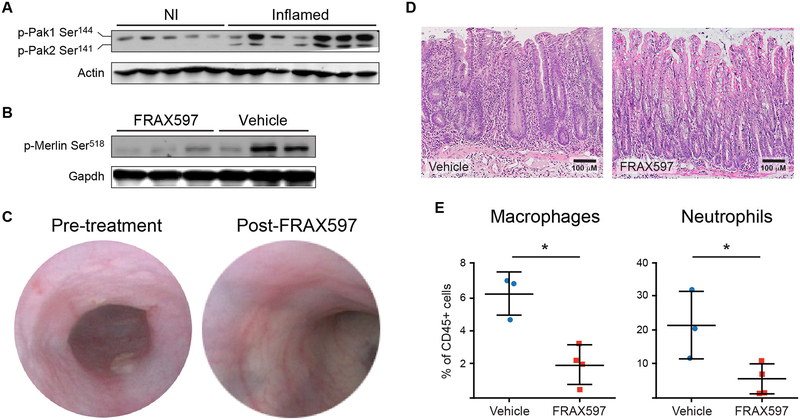
With multiple lines of evidence pointing toward dysregulation of Pak signaling in colitis, we investigated whether pathway activation was a cause or consequence of the disease. First, we sought to validate, in an independent cohort of animals, that Pak was activated during colitis. Phosphorylation of Pak1/2 on Ser144/Ser141 is associated with kinase activation (30) and western blotting of colonic protein lysates from the new cohort confirmed increased phosphorylation in inflamed colons (Fig. 7A). This observation validated the MS analysis and kinase inference that predicted Pak1 activation based on increased phosphorylation of its substrates in inflamed colons (Fig. 5A and B). Note that Pak2 was not predicted by GSEA to be activated because not enough of its known substrates (table S8) were represented in the pMS dataset. We also found that Pak1 auto-phosphorylation was increased in animals with acute colitis induced with dextran sodium sulfate, indicating that Pak activation is not specific to the adoptive transfer mouse model of colitis (fig. S5).
Fig. 7.
Validation of Pak as a therapeutic target in colitis. (A) Validation of Pak activation in the colons of animals with induced colitis (Inflamed). Phosphorylated Pak1 and Pak2 were detected by western blotting. Each lane represents a sample from a different animal. .NI, not inflamed. (B) Inhibition of Pak activity by FRAX597. Merlin phosphorylation on Ser518, a Pak substrate, was assessed by western blotting in the colons of inflamed animals treated for 24 hrs with FRAX597 (100 mg/kg single dose) or polyethylene glycol and polyvinylpyrrolidone (vehicle). (C) Colonoscopic monitoring of colitis. Colonoscopy images of a representative mouse with adoptive transfer-induced colitis. Animal was imaged before and 7 days after FRAX597 treatment (100 mg/kg per day). (D) Histological effects of FRAX597 treatment on the colon of mice with induced colitis. (E) Immunological effects of FRAX597 on the colon of mice with induced colitis. The percentages of macrophages and neutrophils were quantified by flow cytometry in the colons of inflamed animals after 7d of FRAX597 or vehicle treatment. * denotes p < 0.05 in one-tailed Mann-Whitney test. N = 3 for vehicle-treated animals and N = 4 for FRAX597-treated animals.
Next, we investigated whether inhibition of Pak signaling could suppress colitis. We chose to focus on animals that already had severe inflammation because our goal was to determine whether inhibition of the pathway could be effective in patients with active disease. We found that colons from animals with colitis that were treated acutely with FRAX597, a Pak1/2 inhibitor (31), exhibited reduced phosphorylation of Merlin on Ser518, a Pak1/2 substrate (Fig. 7B). Next, we treated sick animals with FRAX597 for 7 days (100 mg/kg/day) and assessed their phenotype by endoscopic monitoring. Following treatment with the Pak inhibitor FRAX597, the animals exhibited decreased mucosal thickness, return of both small and large vessel visible vascular markings, and resolution of contact friability and bleeding (Fig. 7C), all of which are signs that the active colitis had been diminished after Pak inhibition.
At the histologic level, animals treated with FRAX597 exhibited reduced immune cell infiltrate in their colons and a return to more normal epithelial crypt morphology (Fig. 7D). The colitis that arises in the T cell transfer model is characterized by increased numbers of colonic macrophages and neutrophils in the lamina propria (32). Pak has a direct role in neutrophil migration by promoting chemotaxis (33). To determine whether Pak inhibition altered the immune milieu in the colon, we used fluorescence-activated cell sorting (FACS) to quantify immune cells from the colon. Consistent with the reduction in the clinical and histologic presentation of colitis, we found that animals treated with FRAX597 had reduced numbers of colonic macrophages and neutrophils relative to vehicle-treated controls (Fig. 7E). Together, our computational and experimental studies implicate Pak signaling as a driver of chronic inflammation in the colon.
Discussion
Here we present a case in which RNA microarray, total proteomic, and phosphoproteomic measurements have been analyzed and integrated in matched tissue from single animals in a mouse model of colitis. This “all measurements from a single animal” approach enabled us to determine how changes in gene expression are carried through to protein expression and modification. We found that genes that were differentially expressed at the RNA level showed similar patterns of differential regulation in the MS data set, but that differential expression at the RNA level did not predict protein phosphorylation status. Furthermore, though most genes differentially expressed at the total protein level were also differentially phosphorylated at the phosphoprotein level, the pMS data contained many differentially expressed phosphopeptides that were unchanged at the RNA and total protein level (Fig. 1E). Although there are typically a greater number of molecular species measured through transcriptomics, our finding demonstrates that there are additional layers of molecular regulation that are not represented within transcriptomic datasets, underscoring the importance of proteomic measurement for understanding disease pathogenesis at the molecular level.
We observed stronger concordance between the RNA and MS data at the pathway level than at the single gene level (Fig. 2B). The enriched pathways pointed to several dysregulated signaling pathways, such as oxidative phosphorylation and signaling through E2F, KRAS, and MYC, that might provide some therapeutic targets in IBD. Work by Bar et al. demonstrated that mice with DSS- and TNBS-induced colitis are protected against more severe symptoms when oxidative phosphorylation is more active (34). Their work suggested that increasing the activity of this pathway could reduce inflammation and crypt formation in the intestinal epithelium through enhanced nuclear factor κB (NF-κB) signaling (34). Because our analysis identified a link to oxidative phosphorylation in the T cell transfer model of colitis, it appears that the anti-inflammatory activity of this pathway is conserved among the DSS, TNBS, and adoptive transfer models.
Furthermore, whereas the RNA and MS measurements exhibited conserved patterns of expression and higher-order process enrichment, there were also many proteins that were differentially regulated in inflamed versus non-inflamed tissue, but had unchanged RNA expression. Using GO enrichment analysis, we identified differential regulation in defense response and extracellular matrix. This led us to infer post-translational regulation of collagens by matrix metalloproteinases (Fig. 3B). Additionally, we found a subset of neutrophil and acute-phase proteins that were transcribed and translated in other tissues (bone marrow and liver) and transported to the colon in the blood stream or through infiltrating immune cells (Fig. 3F). These analyses leveraged the transcriptomic and proteomic data to produce hypotheses regarding organismal scale gene and protein regulation that could not have been made with either data set alone, underscoring the value of multi-omics measurement. Only multi-omics analysis of an intact organism could identify these instances of physiologic regulation of protein expression.
The collection of multi-omics data enabled us to perform trans-omic co-expression network analysis to examine the correlation structure between all pairs of RNA transcripts, proteins, and phosphopeptides measured in all three data sets (Fig. 4). By clustering the data sets into modules and performing targeted network analysis on the modules with YourCrosstalker, we were able to identify important functional commonalities between the phospho-peptide module pMS_3 and the total protein module MS_5. We found that pMS_3 associated with all 8 RNA modules and was characterized by an intriguing core network of hyper-phosphorylated proteins in the RNA metabolism and pre-RNA processing pathways coupled to a de-phosphorylated network of Vegf signaling. The MS_5 module did not associate with any pMS modules, and yet it was characterized by similar coupling of pre-RNA processing pathway proteins to Vegf signaling proteins through the cell cycle signaling pathway (Fig. 4E and F). Although the characteristic proteins of MS_5 and pMS_3 did not overlap, the same signaling pathways and core network architecture were identified by YourCrosstalker, suggesting an important role for these pathways in colitis. RNA processing and cell cycle are general terms and it is difficult to ascertain specific mechanisms potentially involved in colitis, but Vegf is plays a role in angiogenesis and lymphangiogenesis during colitis (35). Indeed, Vegf therapy is beneficial in mouse models of the disease (36, 37). By extension, Vegf signaling in experimental colitis is likely to be a result of the inflammation as the tissue attempts to repair itself.
The primary goal of this study was to use the multi-dimensional dataset to identify new drivers of colitis and we noted that the pMS analysis provided additional mechanistic insight that was not revealed by the RNA or protein expression studies. Because most of the individual sites identified by pMS were not functionally annotated, we reasoned that phosphoproteomic data coupled with prior knowledge of kinase substrates would enable computational inference of kinase activity. Public databases and software packages provide information on kinase-substrate interactions, kinase recognition motifs, and kinase substrate predictions and various algorithms have been used to determine a kinase activity metric based on these relationships. For example, similar to our analysis, Drake and colleagues compiled substrate sets and used an algorithm analogous to GSEA to quantify enrichment (38). In our study, GSEA predicted one significant positively enriched and six significant negatively enriched kinases from our pMS dataset (Fig. 5A), most of which had unknown or poorly characterized roles in IBD. The activation or inhibition of several of these predicted kinases was confirmed through the measurement of regulatory phosphorylation sites on those kinases (Figs. 5C and 7A). Pak1 was of particular interest because our enrichment analysis indicated that it was activated during colitis, suggesting that inhibition could be a therapeutic strategy. Although Pak2 was not implicated by GSEA, western blotting and YourCrosstalker network analysis revealed that it is also activated in animals with colonic inflammation and in human patients (Fig. 4E and F and 6B). Inhibition of Pak1/2 with FRAX597 suppressed inflammation in animals with active colitis, indicating that this pathway plays an active role in the pathogenesis of the disease (Fig. 7C to E). Thus, we demonstrated that chemical inhibition of Pak signaling can revert inflammatory disease in the colon.
Pak1 and Pak2 are members of the family of group I p21-activated kinases that regulate inflammatory responses, in part by stimulating assembly of the NADPH oxidase complex in neutrophils (39). Pak1 was previously reported to be increased in abundance in epithelial cells during colitis and is thought to be regulated by mTOR signaling (40). Our YourCrosstalker network analysis identified the mTOR pathway as being activated in the T cell transfer model (Fig. 4E), although our GSEA analysis failed to identify enrichment for mTOR substrates in pMS data from animals with colitis (table S9). As such, the mechanism of activation of Pak1 in the T cell transfer model is not clear. Nevertheless, the effect of Pak inhibition validates the prediction made by our computational modeling approaches: that Pak plays a critical role in the pathogenesis of colitis. In sum, our analyses demonstrate the added value of multi-omic measurements by showing how different molecular species in each data set may be acting on similar pathways in distinct ways. We used multi-omics comparisons to obtain mechanistic insight into the pathogenesis of chronic inflammation in the colon, in particular identifying Pak signaling as a bona fide therapeutic target. This work highlights the power of analyzing the global proteome and phosphoproteome to uncover dysregulated signaling pathways that are not revealed by transcriptomic studies alone.
Materials and Methods
T cell transfer model of colitis
T cell transfer (TCT) was performed according to established methods (41). Briefly, splenocytes were isolated from wild-type C57BL/6J animals (Jackson Laboratory) and depleted for red blood cells by treatment with ACK lysis buffer. CD4+ T cells were enriched using a Dynal CD4 untouched kit (Thermo-Fisher). Naïve T cells (CD4+CD45RBhi) and regulatory T cells (CD4+CD25+) were isolated by FACS. Naïve T cells (400,000) or Tregs (200,000) were injected IP in PBS vehicle into C57BL/6J Rag1 null mice (Jackson Laboratory). Recipients were weighed bi-weekly. Animals treated with naïve T were sacrificed after sustained weight loss of 1.5g for one week. Treg-injected animals were sacrificed at these time points. Upon sacrifice, the medial colon was formalin fixed for histology and then matched tissue was snap-frozen for microarray analysis and MS analysis (fig. S2). All animal work was approved by the Institutional Care and Use Committees of Massachusetts General Hospital and Beth Israel Deaconess Medical Center.
Microarray analysis
RNA was isolated from snap-frozen tissue with a Qiagen RNeasy microarray tissue mini kit (Qiagen: 73304). RNA expression was quantified on Affymetrix Mouse Transcriptome 1.0 Arrays and data were processed using the Affymetrix Expression Console software. Subsequent analysis was performed on named transcripts. Microarray data were submitted to Gene Expression Omnibus (GEO accession GSE95705).
Protein digestion and tandem mass tag (TMT) labeling for MS
Excised colon tissue was re-suspended in mammalian cell lysis buffer (75mM NaCl, 50mM HEPES [pH 8.5], 10mM sodium pyrophosphate, 10mM NaF, 10mM β-glycerophosphate, 10mM sodium orthovanadate, 1mM PMSF, 3% SDS, and complete mammalian protease inhibitor tablet [Roche]). Suspensions were mixed with zirconium oxide beads (1mM diameter) and lysed on a mini bead beater (Biospec) four times for 45 seconds, cooling the sample in between. Beads were removed, the lysate was centrifuged at 15,000×g for 5 minutes at 4°C, and insoluble debris was discarded. Ditheiothreitol (DTT) was used to reduce disulfide bonds and free thiols were alkylated with iodoacetamide (IAA) as described previously (42). Reduced and alkylated proteins were then precipitated following the methanol/chloroform method as described previously (43). Precipitated proteins were reconstituted in 300μL of 1M urea in 50mM HEPES, pH 8.5. Vortexing and sonication were used to aid solubility. Proteins were then digested in a two-step process, first with 3μg endoproteinase Lys-C (Wako) for 17 hours at room temperature (RT) and then with 3μg sequencing-grade trypsin (Promega) for 6 hours at 37°C. The digest was acidified with trifluoroacetic acid (TFA). Peptides were desalted over Sep-Pak C18 solid-phase extraction (SPE) cartridges (Waters). The peptide concentration was determined using a BCA assay (Thermo Scientific) and a maximum of 50μg of peptides were aliquoted, then dried under vacuum and stored at −80°C before labeling with TMT reagents. Peptides were labeled with 10-plex TMT reagents (Thermo Scientific). TMT reagents were suspended in dry acetonitrile (44) at a concentration of 20μg/μL. Dried peptides were re-suspended in 30% dry ACN in 200mM HEPES, pH 8.5, and 5μL of the appropriate TMT reagent was added to the sample, which was incubated at room temperature for one hour. The reaction was then quenched by adding 6μl of 5% (w/v) hydroxylamine in 200mM HEPES (pH 8.5) and incubated for 15 min at room temperature. The solutions were acidified by adding 50μl of 1% TFA, combined into one sample, and desalted.
Basic pH reversed-phase liquid chromatography (bRPLC) sample fractionation
bRPLC was used to perform sample fractionation with concatenated fraction combining. Briefly, samples were re-suspended in 5% formic acid (FA)/5% ACN and separated over a 4.6 mm × 250 mm ZORBAX Extend C18 column (5μm, 80 Å, Agilent Technologies) on an Agilent 1260 HPLC system outfitted with a fraction collector, degasser, and variable wavelength detector. A two buffer system (Buffer A: 5% ACN, 10mM ammonium bicarbonate; Buffer B: 90% ACN, 10mM ammonium bicarbonate) was used for separation, with a 20–35% gradient of Buffer B over 60 minutes at a flow rate of 0.5 mL/minute. A total of 96 fractions were collected, which were combined in a total of 24 fractions. The combined fractions were dried under vacuum, reconstituted with 8μL of 5 % FA/5 % ACN, 3μL of which were analyzed by LC-MS2/MS3.
Phosphopeptide enrichment
For each sample, 450μg of total peptides were subjected to phospho-peptide enrichment using a 4:1 ratio of titanium dioxide beads:peptide (w/w). Peptides were resuspended in 2M lactic acid in 50% ACN and added to 1.8mg of titanium dioxide beads. The mixture was shaken gently for 1 hour. Beads were collected by centrifugation and washed 3 times with 2M lactic acid in 50% ACN and 3 times with 50% ACN/0.1% TFA. Phospho-peptides were eluted with 2 × 200μL of 50mM KH2PO4, pH 10, and acidified with 1% TFA. Eluted phospho-peptides were desalted, lyophilized, and labeled with 2μL of 10-plex TMT reagents 127n-130c as described earlier. The combined sample was enriched for phospho-tyrosine-containing peptides using phospho-tyrosine antibody-conjugated beads (Cell Signaling Technology) according to the manufacturer’s protocol. Unbound peptides (phosphoserine and phosphothreonine peptides) were desalted, lyophilized, and fractionated by bRPLC using a gradient of 5 to 28% Buffer B. A total of 96 fractions were collected, and fractions were combined into 12 fractions. Bound peptides (phospho-tyrosine peptides) were eluted and desalted. All 12 fractions were re-suspended in 5% ACN / 5% formic acid and analyzed on an Orbitrap Fusion mass spectrometer using LC-MS2/MS3 for identification and quantification of the phospho-peptides.
Mass spectrometry acquisition and analysis
TMT-labeled peptides were subjected to multiplexed quantitative proteomics analysis on an Orbitrap Fusion mass spectrometer (Thermo Scientific) coupled to an EASY-nLC 1000 integrated autosampler and HPLC pump system. Peptides were separated over a 100μm inner diameter microcapillary column, packed in-house with 0.5cm of Magic C4 resin (5μm, 100Å, Michrom Bioresources), followed by 0.5cm of Maccel C18 resin (3μm, 200Å, Nest Group), followed by 29cm of GP-C18 resin (1.8μm, 120Å, Sepax Technologies). Samples were eluted over 165 minutes at a flow rate of 300nL/minute over a gradient of 6 to 25% ACN/0.125% formic acid. TMT-labeled peptides were identified using MS2 spectra and quantified using a MultiNotch (Simultaneous Precursor Selection, SPS) MS3 method (42, 45) in a data-dependent mode. Each scan sequence began with acquisition of a full MS spectrum (MS1) acquired in the Orbitrap (m/z range: 500–1,200; resolution: 60,000; AGC target: 5×105; maximum injection time: 100ms). From this spectrum, the ten highest intensity peptide ions were subjected to MS2 analysis, where acquisition time was optimized in an automated fashion (top speed: 5 seconds). Peptides were fragmented by CID (normalized collision energy: 30%), and low-resolution MS2 scans were performed in the linear ion trap (quadrupole isolation width: 0.5 Th; AGC target: 1×104; maximum injection time: 35ms). From each MS2 spectrum, the ten highest intensity fragment ions were selected for SPS MS3 analysis. Fragment ions were restricted to an m/z range of 400 to 2000, an m/z range of −40 to + 15 around the precursor peptide ion m/z was excluded from selecting fragment ions, and “TMT” was selected for Isobaric Tag Loss exclusion settings. This group of MS2 fragment ions was further fragmented by HCD (normalized collision energy: 50%), and high resolution MS3 scans were performed in the Orbitrap (resolution: 60,000; AGC target: 5×104; maximum injection time: 250ms). When analyzing phosphopeptide samples, two MS2 spectra were acquired per peptide, a 15,000 resolution spectrum in the Orbitrap upon HCD fragmentation (normalized collision energy = 40%) and a low resolution CID-MS2 spectrum as described earlier. Precursor ion selection for MS3 spectra was done based on the low resolution MS2 spectral data using the top ten intensity fragment ions. Data analysis was performed on an in-house, SEQUEST-based software platform (46, 47). RAW files were converted into the mzXML format using a modified version of ReAdW.exe. Peptide identification was performed as reported previously (48), searching against a protein sequence database containing all protein sequences in the mouse ORF database (downloaded 01/14/2014), as well as that of known contaminants. For phosphopeptide data, high- and low-resolution spectra were annotated in two separate searches and subsequently combined. Phosphorylation of serine, threonine, and tyrosine residues (79.966331 Da) was set as a variable modification, and up to 3 modifications were allowed. Peptides were quantified based on the TMT reported ion intensities in the collected MS3 spectra, as reported previously (48, 49). Quantified peptides were required to have a summed signal-to-noise value greater than 386 and an isolation specificity greater than 0.75 (42). TMT intensities for all peptides assigned to a protein were summed for protein quantification. Both protein and phosphopeptide quantitative data were normalized in a two-step procedure. First, the average intensity of each species (protein or phospho-peptide) was calculated and normalized to the median of all of these average intensities. Second, to account for any mixing errors, the intensity of each species was normalized to the ratio of the median intensity for a given TMT channel to the median of all species intensities. Total protein and phospho-protein MS datasets were deposited into the Mass Spectrometry Interactive Virtual Environment (MassIVE, accession number MSV000081198).
RT-PCR confirmation of acute phase and neutrophil genes
To confirm distant tissue expression, mice were subjected to TCT-induced colitis as described earlier. Inflamed and negative control animals were sacrificed and whole liver and colon tissue were snap-frozen in liquid nitrogen. In addition, bone marrow was collected from one femur from each animal. In brief, femurs were flushed with Hank’s Balanced Salt Solution (HBSS) and passed through a 45 μm filter. Red blood cells were lysed in ACK buffer for three minutes and remaining cells were stained with APC-Cy7 CD45 (BioLegend) and Alexa-700 LY6G 1A8 (BioLegend) for ten minutes at room temperature. In addition, colons were dissociated in a collagenase solution for 1hr at 37 degrees C with agitation. Tissue was then passed through a 45 μm filter and stained with APC-Cy7 CD45 (BioLegend) and Alexa-700 LY6G 1A8 (BioLegend) as described earlier. Cells were gated on CD45+ and LY6Ghi population (neutrophils) and sorted directly into trizol on the BD Biosciences ARIA flow cytometer and stored at −80°C until processing. RNA was isolated from whole tissue and sorted neutrophils by extraction with Trizol. For whole tissue extraction, liver and colon segments were homogenized in trizol by chopping with a razor blade. After RNA extraction, cDNA was produced using the Taqman High Capacity cDNA Reverse Transcription Kit (Applied Biosystems), and pre-amplification was performed according to manufacturers’ protocols (Applied Biosystems). For liver genes, we assayed Fibronectin alpha chain (Fga, TaqMan Mm00802584_m1) and Alpha-1-acid glycoprotein 1 (Orm1, TaqMan Mm00435456_g1). For neutrophils, we assayed Cathelicidin antimicrobial peptide (Camp, Mm00438285_m1) and Elastase neutrophil expressed (Elane, Mm01168928_g1). 18s rRNA (Mm04277571_s1) was used as a standard in each case and expression levels were determined by the dCT/dCT method. For liver genes, graphs represent the ratio of liver:colon transcripts in inflamed and non-inflamed tissue. For neutrophil genes, all measurements were normalized with the lowest value set to one for each gene. Graph represents Log2 normalized expression.
Western blotting for Pak1 and Merlin
Lysates from additional inflamed and non-inflamed animals (generated using the protocol described earlier) were run on 4–12% Novex tris-gly gels (Invitrogen). After transfer, membranes were probed with primary antibodies as follows: phospho-Merlin (Ser518) from Cell Signaling (13281) used at 1:1,000; phospho-Pak1/2 Ser144/Ser141 from Cell Signaling (2606) used at 1:1,000; Actin from Santa Cruz Biotechnology (sc-1616) used at 1:10,000; Gapdh used at 1:10,000 from Cell Signaling (D16H11). Secondary antibodies were anti-mouse and anti-rabbit IgG HRP-linked as appropriate (Cell Signaling 7076, 7074; used at 1:5,000).
Luminex analysis for p38 and Gsk3α/β phosphorylation
BioplexPro (Bio-Rad) phospho-protein measurements were performed according to the manufacturer’s instructions with the following kits: Gsk3α/β Ser21/Ser9 (171-V50007M) and p38 MAPK Thr180/Tyr182 (171-V50014M).
Pak inhibition in vivo
Mice that had received adoptive transfer of naïve T cells were monitored for the development of colitis with regular weight measurements. Once mice developed signs of colitis, they underwent rigid endoscopy to confirm evidence of inflammation, using a validated endoscopic scoring system for colitis (50). Once inflammation was demonstrated, mice were treated with FRAX597 (100 mg/kg QD) or vehicle by oral gavage for 7 days (31). A subset of mice underwent post-treatment endoscopy and then all mice were sacrificed at 7 days. The colons were resected and opened longitudinally; two side portions (1/5th) from the entire length were reserved, one for flow cytometry and one placed in Bioplex lysis buffer. The rest of the tissue was used for flow cytometric analysis.
Flow cytometry
Tissue was homogenized in serum-free DMEM with 2mg/ml collagenase type I C (VWR 234153–100MG) and incubated for 1hr at 37C. After incubation, the sample was strained through a 45μm filter and centrifuged for 5 minutes at 700g. Cells were then stained with the following antibodies from BioLegend (1:300 in FACS buffer) for 10 minutes: FITC CD4 (116004) (BV-421 F4/80 (123131), BV-605 CD4 (100547), BV-510 cd11b (101245), Alexa-700 Ly6G (127622), PE/Cy7 cd11c (117317), APC/Cy7 CD45 (103116), and PE CD45RB (103308). Cells were analyzed on a 5-laser LSR II flow cytometer (Becton Dickinson SORP).
Unsupervised clustering
Unsupervised hierarchical clustering was performed using the clustergram function in Matlab, using default parameters. Input data included log2-transformed RNA, MS, and pMS datasets. For the purposes of visualization, RNA data was masked using the genevarfilter, genelowvalfilter, and geneentropyfilter as described: http://www.mathworks.com/help/bioinfo/examples/gene-expression-profile-analysis.html.
Differential gene expression and pathway analysis
To compare univariate differential expression in each data set, RNA, MS, and pMS data were analyzed using the Wilcoxon-Mann Whitney test with Benjamini-Hochberg False Discovery Rate Correction, p<0.05, q<0.25. Pathway analysis was performed on the RNA and MS data sets using GSEA. GSEA was performed with weighted log2ratio ranking and 1000 gene set permutations using the “Hallmarks Gene Sets” from MSigDb.
Co-expression network analysis
Spearman correlations were calculated for all pairs of species represented in all three data sets. Correlation coefficients less than 0.9 or greater than −0.9 were set to 0 and all others were set to 1 to define the undirected network adjacency matrix. The topological similarity of nodes in each data set was calculated using GTOM2 (19, 20). We then clustered the GTOM2 using k-means clustering and determined the number of GTOM2 clusters in each data set by calculating the gap statistic for 1 to 15 possible clusters. The genes were then extracted and module overlap was computed using Fisher’s exact test with Benjamini-Hochberg False Discovery Rate correction (p<0.05, q<0.25). All analysis was performed in MATLAB R2016a and networks were generated using Cytoscape 3.4.0 (51). GTOM2 visualizations and calculations were performed using the implementation described in: https://www.mathworks.com/matlabcentral/fileexchange/17668-gtom-generalized-topological-overlaping-measure
YourCrosstalker network analysis
Co-expression network modules were analyzed using the YourCrosstalker network analysis tool (www.youromics.com) (22, 23). Given an input set of gene seeds and a protein-protein interaction network database, Crosstalker identifies subnetworks of highly connected genes as scored by a random walk with restarts at the seed genes. As the random walk reaches a steady state, seed genes are removed if they are not topologically related in the protein-protein interaction (PPI) network and additional genes are recruited if significantly traversed (“Crosstalkers”), indicating possible association with the seed genes. Pathway enrichment is then tested using Fisher’s exact test against a database of curated pathways and network edges are colored by pathway. The Reactome and National Cancer Institute Pathway Interaction Database pathway databases and STRING protein-protein interaction network (high confidence interactions, edge weights >0.7) were used in the analysis (24–27).
Differential regulation analysis
We selected 122 genes/proteins as having greater than 2-fold increased or decreased abundance at the protein level and less than 2.75-fold increased or decreased expression at the RNA level. Gene Ontology (GO) enrichment was performed using Gorilla (http://cbl-gorilla.cs.technion.ac.il/) with the background list as all of the shared RNA and protein species. GO terms that were enriched with a p value < 10−3 were deemed significant. Tissue expression of acute phase and neutrophil genes was assessed using the mouse tissue expression atlas. Each gene was normalized to 1 by the highest expression for that gene. After normalization, the average value for all of the genes of a given category was calculated and plotted for each of the indicated tissues.
Gene set enrichment analysis of kinase substrates
Functional annotation was assessed based on previous knowledge obtained from downloads available on PhosphoSitePlus. Identified proteins were obtained from Phosphorylation_site_dataset, which contains all of the published mouse phosphorylation sites found in their database; functional annotation was obatained from Regulatory_sites, and we included all of the phospho-sites that had known effects on function; upstream kinase information was obtained from kinase lists. Kinase substrate lists were obtained from PhosphoSitePlus (http://phosphosite.org). Lists were composed of known mouse, rat, and human kinase substrate sites. Human and rat sites were converted to mouse numbering. This produced a total of 348 kinase substrate sets with a maximum of 558 unique phospho-sites and a median of 6. Enrichment was performed with a rank-based test using GSEA software developed at the Broad Institute (http://www.broadinstitute.org/gsea/index.jsp). Parameters can be found along with the output of the analysis in table S4. Briefly, of 348 kinases sets, 29 met the minimum size threshold of overlap with our dataset (4 phospho-sites). Input data was the ratio of phospho-peptides to total peptides from match samples. The ratio measurement was used because this is more reflective of specific activation of phosphorylation. Phospho-sites were ranked by ‘Log2 ratio of classes’ and permuted 35,000 times to the gene set (because of the limited number of samples, phenotype permutation is not recommended). The kinase substrate lists have been added to the Msigdb so that they can be easily applied to the GSEA software.
Supplementary Material
Fig. S1. Colonic histology from animals analyzed in the study.
Table S5. Pathway analysis for each data set.
Table S6. Modules from trans-omic co-expression network analysis.
Table S7. Statistics for trans-omic co-expression network analysis.
Table S8. Phospho site lists for each kinase used in GSEA.
Table S9. Statistics for GSEA-based kinase enrichment.
Fig. S2. Schematic of experimental design.
Fig. S3. Functional annotation of phosphorylation sites for mammalian proteins.
Fig. S4. Human Pak1 and Pak2 signaling network neighborhood.
Fig. S5. Pak activation during acute colitis.
Table S1. Affymetrix microarray quantification of gene expression in individual samples.
Table S2. MS-based quantification of proteins in individual samples.
Table S3. MS-based quantification of phospho-peptides in individual samples.
Table S4. Differential expression analysis for each data set.
Acknowledgments:
The authors would like to acknowledge Towia Libermann at the BIDMC Genomics Core and Stuart Levine at the MIT BioMicroCenter for microarray processing and instructive conversation. We would also like to thank Marcia Haigis for helpful comments on the manuscript.
Funding: J.L. and L.S-L. were supported by a Research Fellowship Awards from the Crohn’s & Colitis Foundation of America. S.D.S. was supported by a National Science Foundation Graduate Research Fellowship under Grant No. 1122374. K.M.H. was funded by grants from the NIH (R01-GM088827) and the DOD (W81XWH-16-1-0042). J.K. was funded by a grant from the NIH (R01-CA124495). D.A.L. and D.K.B. were funded by grants from Boehringer-Ingelheim as part of the SHINE program and from the Army Research Office Institute for Collaborative Biotechnologies (W911NF-09–0001).
Footnotes
Competing interests: The authors declare no competing financial interests.
Data and materials availability: Microarray data were submitted to Gene Expression Omnibus (GEO accession GSE95705). Total protein and phosphoprotein MS datasets were deposited into the Mass Spectrometry Interactive Virtual Environment (MassIVE, accession number MSV000081198).
References and Notes
- 1.Neurath MF, Current and emerging therapeutic targets for IBD. Nat Rev Gastroenterol Hepatol 14, 688 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Hugot JP JP, Laurent-Puig P, Gower-Rousseau C, Olson JM, Lee JC, Beaugerie L, Naom I, Dupas JL, Van Gossum A, Orholm M, Bonaiti-Pellie C, Weissenbach J, Mathew CG, Lennard-Jones JE, Cortot A, Colombel JF, Thomas G, Mapping of a susceptibility locus for Crohn’s disease on chromosome 16. Nature 379, 821–823 (1996). [DOI] [PubMed] [Google Scholar]
- 3.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, Achkar JP, Brant SR, Bayless TM, Kirschner BS, Hanauer SB, Nuñez G, Cho JH, A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 411, 603–606 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Mizoguchi A, Mizoguchi E, Animal models of IBD: linkage to human disease. Curr Opin Pharmacol 10, 578–587 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fang K, Zhang S, Glawe J, Grisham MB, Kevil CG, Temporal genome expression profile analysis during t-cell-mediated colitis: identification of novel targets and pathways. Inflamm Bowel Dis 18, 1411–1423 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu F, Dassopoulos T, Cope L, Maitra A, Brant SR, Harris ML, Bayless TM, Parmigiani G, Chakravarti S, Genome-wide gene expression differences in Crohn’s disease and ulcerative colitis from endoscopic pinch biopsies: insights into distinctive pathogenesis. Inflamm Bowel Dis 13, 807–821 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Cooney JM, Barnett MP, Brewster D, Knoch B, McNabb WC, Laing WA, Roy NC, Proteomic analysis of colon tissue from interleukin-10 gene-deficient mice fed polyunsaturated Fatty acids with comparison to transcriptomic analysis. J Prot Res 11, 1065–1077 (2012). [DOI] [PubMed] [Google Scholar]
- 8.M’Koma AE, Seeley EH, Washington MK, Schwartz DA, Muldoon RL, Herline AJ, Wise PE, Caprioli RM, Proteomic profiling of mucosal and submucosal colonic tissues yields protein signatures that differentiate the inflammatory colitides. Inflamm Bowel Dis 17, 875–883 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu K, Knutson CG, Wishnok JS, Fox JG, Tannenbaum SR, Serum metabolomics in a Helicobacter hepaticus mouse model of inflammatory bowel disease reveal important changes in the microbiome, serum peptides, and intermediary metabolism. J Prot Res 11, 4916–4926 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yau Y, Leong RW, Zeng M, Wasinger VC, Proteomics and metabolomics in inflammatory bowel disease. J Gastroenterol Hepatol 28, 1076–1086 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Huttenhower C, Kostic AD, Xavier RJ, Inflammatory bowel disease as a model for translating the microbiome. Immunity 40, 843–854 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kostic AD, Xavier RJ, Gevers D, The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology 146, 1489–1499 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ostanin DV, Bao J, Koboziev I, Gray L, Robinson-Jackson SA, Kosloski-Davidson M, Price VH, Grisham MB, T cell transfer model of chronic colitis: concepts, considerations, and tricks of the trade. Am J Physiol Gastrointest Liver Physiol 296, G135–146 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang S, Wang X, Igbal S, Wang Y, Asunkoya AO, Chen Z, Chen Z, Shin DM, Yuan H, Wang YA, Zhau HE, Chung LW, Ritenour C, Kucuk O, Wu D, Epidermal growth factor promotes protein degradation of epithelial protein lost in neoplasm (EPLIN), a putative metastasis suppressor, during epithelial-mesenchymal transition. J Biol Chem 288, 1469–1479 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui N, Hu M, Khalil RA, Biochemical and Biological Attributes of Matrix Metalloproteinases. Prog Mol Biol Transl Sci 147, 1–73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lattin JE, Schroder K, Su AI, Walker JR, Zhang J, Wiltshire T, Saijo K, Glass CK, Hume DA, Kellie S, Sweet MJ, Expression analysis of G Protein-Coupled Receptors in mouse macrophages. Immunome Res 4, 5 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lakschevitz FS, Visser MB, Sun C, Glogauer M, Neutrophil transcriptional profile changes during transit from bone marrow to sites of inflammation. Cell Mol Immunol 12, 53–65 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yip AM, Horvath S, Gene network interconnectedness and the generalized topological overlap measure. BMC Bioinformatics 8, 22–22 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang B, Horvath S, in Statistical Applications in Genetics and Molecular Biology. (2005), vol. 4. [Google Scholar]
- 21.Tibshirani R, Walther G, Hastie T, Estimating the number of clusters in a data set via the gap statistic. J R Stat Soc Series B StatMethodol 63, 411–423 (2001). [Google Scholar]
- 22.Nibbe RK, Koyuturk MR, Chance MR, An integrative -omics approach to identify functional sub-networks in human colorectal cancer. PLoS Comput Biol 6, e1000639 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maxwell S, Chance MR, Koyuturk M, Linearity of Network Proximity Measures: Implications for Set-based Queries and Significance Testing. Bioinformatics 33, 1354–1361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen JJ, von Mering C, STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43, D447–452 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, Buetow KH, PID: the Pathway Interaction Database. Nucleic Acids Res 37, D674–679 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cerami EG, Gross BE, Demir E, Rodchenkov I, Babur O, Anwar N, Schultz N, Bader GD, Sander C, Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res 39, D685–690 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, Caudy M, Garapati P, Gillespie M, Kamdar MR, Jassal B, Jupe S, Matthews L, May B, Palatnik S, Rothfels K, Shamovsky V, Song H, Williams M, Birney E, Hermjakob H, Stein L, D’Eustachio P, The Reactome pathway knowledgebase. Nucleic Acids Res 42, D472–477 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E, PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res 43, D512–520 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng YJ, Li YY, The role of p38 mitogen-activated protein kinase in the pathogenesis of inflammatory bowel disease. J Dig Dis 12, 327–332 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Chong C, Tan L, Lim L, Manser E, The mechanism of PAK activation. Autophosphorylation events in both regulatory and kinase domains control activity. J Biol Chem 276, 17347–17353 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Licciulli S, Maksimoska J, Zhou C, Troutman S, Kota S, Liu Q, Duron S, Campbell D, Chernoff J, Field J, Marmorstein R, Kissil JL, FRAX597, a small molecule inhibitor of the p21-activated kinases, inhibits tumorigenesis of neurofibromatosis type 2 (NF2)-associated Schwannomas. J Biol Chem 288, 29105–29114 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laubitz D, Harrison CA, Midura-Kiela MT, Ramalingam R, Larmonier CB, Chase JH, Caporaso JG, Besselsen DG, Ghishan FK, Kiela PR, Reduced Epithelial Na+/H+ Exchange Drives Gut Microbial Dysbiosis and Promotes Inflammatory Response in T Cell-Mediated Murine Colitis. PLoS One 11, e0152044 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Itakura A, Aslan JE, Kusanto BT, Phillips KG, Porter JE, Newton PK, Nan X, Insall RH, Chernoff J, McCarty OJ, p21-Activated kinase (PAK) regulates cytoskeletal reorganization and directional migration in human neutrophils. PLoS One 8, e73063 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bär F, Bochmann W, Widok A, von Medem K, Pagel R, Hirose M, Yu X, Kalies K, König P, Böhm R, Herdegen T, Reinicke AT, Büning J, Lehnert H, Fellermann K, Ibrahim S, Sina C, Mitochondrial gene polymorphisms that protect mice from colitis. Gastroenterology 145, 1055–1063 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Linares PM, Gisbert JP, Role of growth factors in the development of lymphangiogenesis driven by inflammatory bowel disease: a review. Inflamm Bowel Dis 17, 1814–1821 (2011). [DOI] [PubMed] [Google Scholar]
- 36.D’Alessio S, Correale C, Tacconi C, Gandelli A, Pietrogrande G, Vetrano S, Genua M, Arena V, Spinelli A, Peyrin-Biroulet L, Fiocchi C, Danese S, VEGF-C-dependent stimulation of lymphatic function ameliorates experimental inflammatory bowel disease. J Clin Invest 124, 3863–3878 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cromer WE, Ganta CV, Patel M, Traylor J, Kevil CG, Alexander JS, Mathis JM, VEGF-A isoform modulation in an preclinical TNBS model of ulcerative colitis: protective effects of a VEGF164b therapy. J Transl Med 11, 207 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drake JM, Graham NA, Stoyanova T, Sedghi A, Goldstein AS, Cai H, Smith DA, Zhang H, Komisopoulou E, Huang J, Graeber TG, Witte ON, Oncogene-specific activation of tyrosine kinase networks during prostate cancer progression. Proc Natl Acad Sci USA 109, 1643–1648 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martyn KD, Kim M-J, Quinn MT, Dinauer MC, Knaus UG, p21-activated kinase (Pak) regulates NADPH oxidase activation in human neutrophils. Blood 106, 3962–3969 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khare V, Dammann K, Asboth M, Krnjic A, Jambrich M, Gasche C, Overexpression of PAK1 promotes cell survival in inflammatory bowel diseases and colitis-associated cancer. Inflamm Bowel Dis 21, 287–296 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ostanin DV, Bao J, Koboziev I, Gray L, Robinson-Jackson SA, Kosloski-Davidson M, Price VH, Grisham MB, T cell transfer model of chronic colitis: concepts, considerations, and tricks of the trade. Am J Physiol Gastrointest Liver Physiol 296, G135–146 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ting L, Rad R, Gygi SP, Haas W, MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat Methods 8, 937–940 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wessel D, Flugge UI, A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem 138, 141–143 (1984). [DOI] [PubMed] [Google Scholar]
- 44.Chen Y, Zhu J, Lum PY, Yang X, Pinto S, MacNeil DJ, Zhang C, Lamb J, Edwards S, Sieberts SK, Leonardson A, Castellini LW, Wang S, Champy MF, Zhang B, Emilsson V, Doss S, Ghazalpour A, Horvath S, Drake TA, Lusis AJ, Schadt EE, Variations in DNA elucidate molecular networks that cause disease. Nature 452, 429–435 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McAlister GC, Nusinow DP, Jedrychowski MP, Wühr M, Huttlin EL, Erickson BK, Rad R, Haas W, Gygi SP, MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal Chem 86, 7150–7158 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eng JK, McCormack AL, Yates JR, An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 5, 976–989 (1994). [DOI] [PubMed] [Google Scholar]
- 47.Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villén J, Haas W, Sowa ME, Gygi SP, A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edwards A, Haas W, Multiplexed Quantitative Proteomics for High-Throughput Comprehensive Proteome Comparisons of Human Cell Lines. Methods Mol Biol 1394, 1–13 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Van Rechem C, Black JC, Boukhali M, Aryee MJ, Gräslund S, Haas W, Benes CH, Whetstine JR, Lysine demethylase KDM4A associates with translation machinery and regulates protein synthesis. Cancer Discov 5, 255–263 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kodani T, Rodriguez-Palacios A, Corridoni D, Lopetuso L, Di Martino L, Marks B, Pizarro J, Pizarro T, Chak A, Cominelli F, Flexible colonoscopy in mice to evaluate the severity of colitis and colorectal tumors using a validated endoscopic scoring system. J Vis Exp, e50843 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T, Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27, 431–432 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Colonic histology from animals analyzed in the study.
Table S5. Pathway analysis for each data set.
Table S6. Modules from trans-omic co-expression network analysis.
Table S7. Statistics for trans-omic co-expression network analysis.
Table S8. Phospho site lists for each kinase used in GSEA.
Table S9. Statistics for GSEA-based kinase enrichment.
Fig. S2. Schematic of experimental design.
Fig. S3. Functional annotation of phosphorylation sites for mammalian proteins.
Fig. S4. Human Pak1 and Pak2 signaling network neighborhood.
Fig. S5. Pak activation during acute colitis.
Table S1. Affymetrix microarray quantification of gene expression in individual samples.
Table S2. MS-based quantification of proteins in individual samples.
Table S3. MS-based quantification of phospho-peptides in individual samples.
Table S4. Differential expression analysis for each data set.