Abstract
Living systems are capable of locomotion, reconfiguration, and replication. To perform these tasks, cells spatiotemporally coordinate the interactions of force-generating, “active” molecules that create and manipulate non-equilibrium structures and force fields that span up to millimeter length scales [1-3]. Experimental active matter systems of biological or synthetic molecules are capable of spontaneously organizing into structures [4, 5] and generating global flows [6-9]. However, these experimental systems lack the spatiotemporal control found in cells, limiting their utility for studying non-equilibrium phenomena and bioinspired engineering. Here, we uncover non-equilibrium phenomena and principles by optically controlling structures and fluid flow in an engineered system of active biomolecules. Our engineered system consists of purified microtubules and light-activatable motor proteins that crosslink and organize microtubules into distinct structures upon illumination. We develop basic operations, defined as sets of light patterns, to create, move, and merge microtubule structures. By composing these basic operations, we are able to create microtubule networks that span several hundred microns in length and contract at speeds up to an order of magnitude faster than the speed of an individual motor. We manipulate these contractile networks to generate and sculpt persistent fluid flows. The principles of boundary-mediated control we uncover may be used to study emergent cellular structures and forces and to develop programmable active matter devices.
Our scheme is based on a well-studied active system composed of stabilized microtubule filaments and kinesin motor proteins [4-8, 10-12]. In the original biochemical system, kinesin motors are linked together by practically irreversible biotin-streptavidin bonds. As linked motors pull on microtubules, a variety of phases and structures spontaneously emerge, such as asters, vortices, and networks. However, spatial and temporal control of these structures is limited [5, 13].
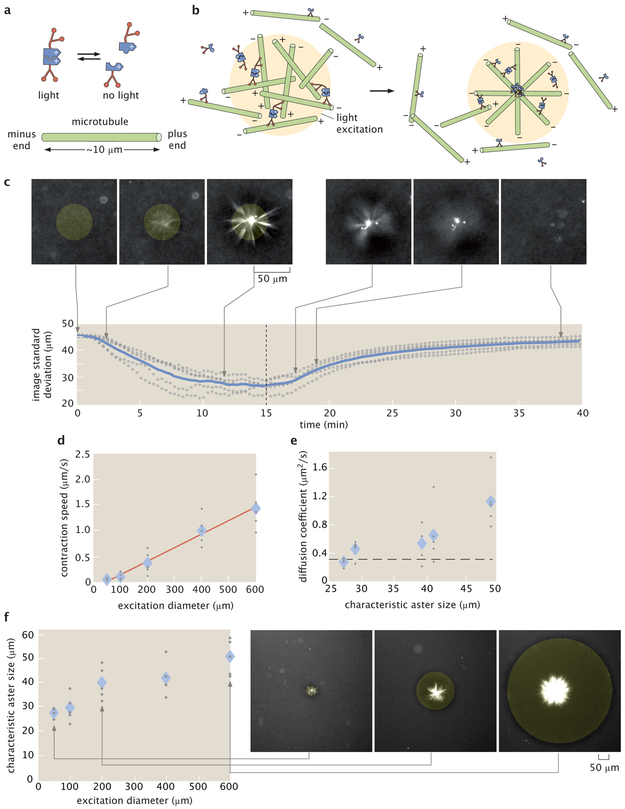
We engineered the system so that light activates reversible linking between motors (Fig. 1a) by fusing Kinesin I motors to optically-dimerizable iLID proteins [14]. Light patterns are projected into the sample throughout its depth and determine when and where motors link (see Supplementary Information for details). Outside of the light excitation volume, microtubules remain disordered, while inside the light volume, microtubules bundle and organize. The reversibility of the motor linkages allows structures to remodel as we change the light pattern. For a cylinder pattern of light excitation, microtubules contract into a 3D aster (Fig. 1b) (Supplementary Information 2.1, Video 1, Video 2). We use the projection of a cylinder of light as an operation for creating asters. We note that vortices, spirals, and extensile behavior are not observed under our conditions (Supplementary Information 2.2).
Figure 1:
Light-switchable active matter system enables optical control over aster formation, decay and size. a, Schematic of light-dimerizable motors. b, Schematic of light-controlled reorganization of microtubules into an aster. c, Images of labeled microtubules during aster assembly and decay and corresponding image spatial standard deviation versus time. The blue line is the mean of 5 experiments and the gray dots represent individual experiments. The dashed line is when the activation light is removed, transitioning from creation to decay. d, Max contraction speed versus excitation diameter. The red line is a linear fit. e, Diffusion coefficients versus characteristic aster size. The characteristic size is the image spatial standard deviation at the 15 minute time point shown in (c). The dashed line represents the diffusion coefficient of a 7 μm microtubule (Supplementary Information 2.11). f, Aster characteristic size versus excitation diameter with representative images. In (d, e, f) the diamonds represent the mean of 5 experiments and the gray dots represent individual experiments. In (c, f), the yellow shaded disks represent the light pattern.
Our temporal control over aster formation allows us to study the dynamics of their creation and decay (Fig. 1c) (Video 3) through time lapse imaging (Supplementary Information 2.3). We characterize these dynamics by measuring the spatial width of the distribution of fluorescently-labeled microtubules using image standard deviation (Supplementary Information 2.4). During aster formation, the distribution of microtubules within a cylinder pattern contracts. After 10–15 min, the distribution reaches a steady state, indicating that the aster is fully formed. To quantify a characteristic aster size (Supplementary Information 2.5), we measure the image standard deviation at 15 min (Supplementary Information 2.6). Once the excitation light is removed, asters begin to decay into free microtubules. The spatial distribution of microtubules widens over time, returning to the initial uniform distribution. Further, aster decay is reversible (Supplementary Information 2.7).
To understand scaling behavior, we investigate how the dynamics of aster formation and decay depend on excitation volume. During formation, microtubule distributions contract. The contraction speed (Supplementary Information 2.8) grows with the diameter of the excitation cylinder (Fig. 1d). Similar scaling of contraction speed has been observed for actomyosin systems [15] (Supplementary Information 2.9) and modeled for generic networks [16]. Alternatively, contraction can be measured by a characteristic contraction timescale [17] (see Supplementary Information 2.8). During decay, microtubule distributions spread in a manner consistent with diffusion (Supplementary Information 2.10). The effective diffusion coefficient is independent of characteristic aster size (Fig. 1e) and is consistent with what is expected for free microtubules (Supplementary Information 2.11). Further, we manipulate aster size through the diameter of the excitation volume (Fig. 1f) and find a scaling dependence (Supplementary Information 2.12) that shows similarities to the dependence of spindle size on confining volumes [18].
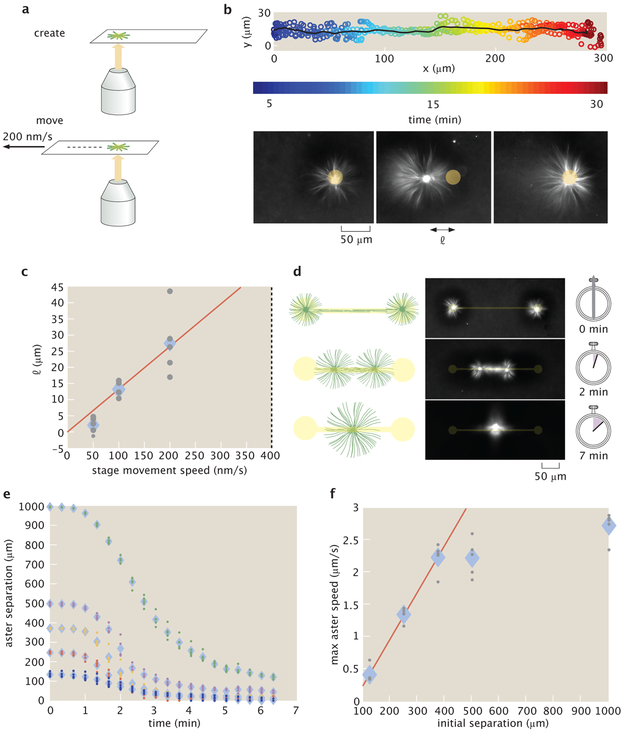
Moving activation patterns are responsible for dynamically re-positioning structures and forces within a cell [19]. We are able to similarly move asters by re-positioning light patterns relative to the sample slide by moving the slide stage (Fig. 2a). We are also able to move asters by directly moving the light pattern, however, moving the stage allows for a greater range of travel. As the stage moves, the asters track with the light pattern, traveling up to hundreds of microns relative to the slide (Fig. 2b) (Video 4) (Supplementary Information 2.13). The aster maintains a steady state distance l between itself and the light pattern (Fig. 2c). We find that asters are always able to track the pattern for stage speeds up to 200 nm/s. At 400 nm/s asters are not able to stay with the pattern, setting an “escape velocity” that is comparable to the motor speeds measured in gliding assays (Supplementary Information 2.16). When the stage stops moving, the aster returns to the center of the light pattern, indicating that the aster is experiencing a restoring force. We can characterize aster movement as caused by an effective potential (Supplementary Information 2.14), and observe mesoscopic phenomena that may inform the underlying mechanisms of aster motion (Supplementary Information 2.15).
Figure 2:
Moving and merging operations of asters with dynamic light patterns. a, Asters are moved relative to the slide by repositioning the microscope stage. b, Overlay of five individual trajectories of aster movement relative to slide moving at 200 nm/s. The line represents the mean trajectory. Time lapse images show the position of the aster relative to the light pattern. l is the displacement of the aster from center of the light pattern. c, l versus stage speed. The dotted line at 400 nm/s represents the escape velocity. The red line is a linear fit. d, Illustration of the aster merge operation by a connected excitation pattern and the corresponding time series of images. e, Distance between merging asters over time for different initial separations. f, Maximum speeds of asters as measured from (e). The red line is a linear fit to the first three data points. In (c, e, f) the diamonds represent the mean of 5 experiments and the dots represent individual experiments.
Intriguingly, we find that asters formed near each other interact by spontaneously merging. To study this interaction, we construct an aster merger operation, where asters are connected with light (Fig. 2d) (Video 5). At the beginning of the merging process, a network of bundled microtubules forms, which connects the asters. The connecting network begins to contract and the asters move towards each other (Fig. 2e). The speed at which asters merge (Supplementary Information 2.8) increases as a function of linking distance up to a speed of roughly 2.5 μm/s (Fig. 2f). The scaling of aster merger speed as a function of distance is similar to the observed relationship of contraction speed as a function of the excitation cylinder size discussed above. We note that the maximum observed merger speed is about an order of magnitude higher than the speeds observed during gliding assays (Supplementary Information 2.16), which is analogous to how cell migration speeds can exceed single motor speeds [20]. Our ability to move and merge microtubule asters reveals that they are not steady state structures as previously observed [5], but are dynamic and constantly remodeling.
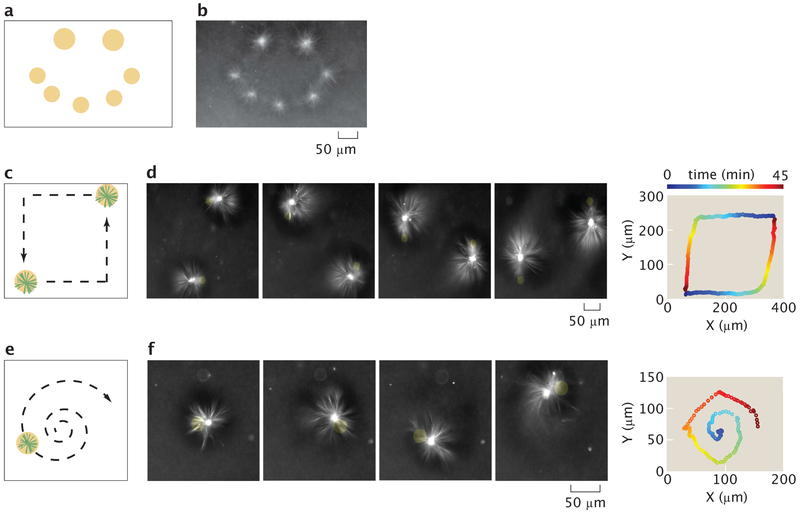
The capability to perform successive operations remains a fundamental step towards engineering with active matter. Our ability to form dynamic light-defined compartments of active molecules enables us to execute multiple aster operations. By composing aster creation operations, we are able to form asters of differing sizes and place them at prescribed positions in parallel (Fig. 3a, b) (Video 6). Once asters are created, they can be simultaneously moved by using multiple dynamic light patterns (Fig. 3c, d) (Video 7). Further, aster trajectories are not limited to rectilinear motion but can be moved along complex trajectories (Fig. 3e, f) (Video 8). During movement, there are inflows of microtubule bundles created in the light pattern, which feed into the aster. There are also outflows of microtubules, which appear as comet-tail streams following the asters (Fig. 3d, f). These mass flows illustrate some of the complex non-equilibrium dynamics that are introduced by moving boundaries of molecular activity. The new capability to simultaneously generate and manipulate asters provides a basis for “programming” complex systems of interacting non-equilibrium structures.
Figure 3:
Operations for creating and moving asters are composed to make different desired patterns and trajectories. a, Sketch for using excitation cylinders to simultaneously pattern asters of different sizes. b, Resultant pattern of asters corresponding to (a). c, Illustration of simultaneous control of two different aster trajectories, as indicated by the dashed arrows. d, Time lapse and the 2D trace of the aster trajectories corresponding to (c). The trajectory trace is color-coded to represent progression in time. e, Dynamically projected spiral to illustrate curvilinear motion. f, time lapse and the 2D trace of the aster trajectory. Time is color coded as in (d).
In our aster merging, moving, and trajectory experiments, we observe fluid flow of the buffer, as inferred by the advection of microtubules and small fluorescent aggregates. Similar cytoskeletaldriven flow is critical for the development and morphogenesis of various unicellular and multicellular organisms [21-27].
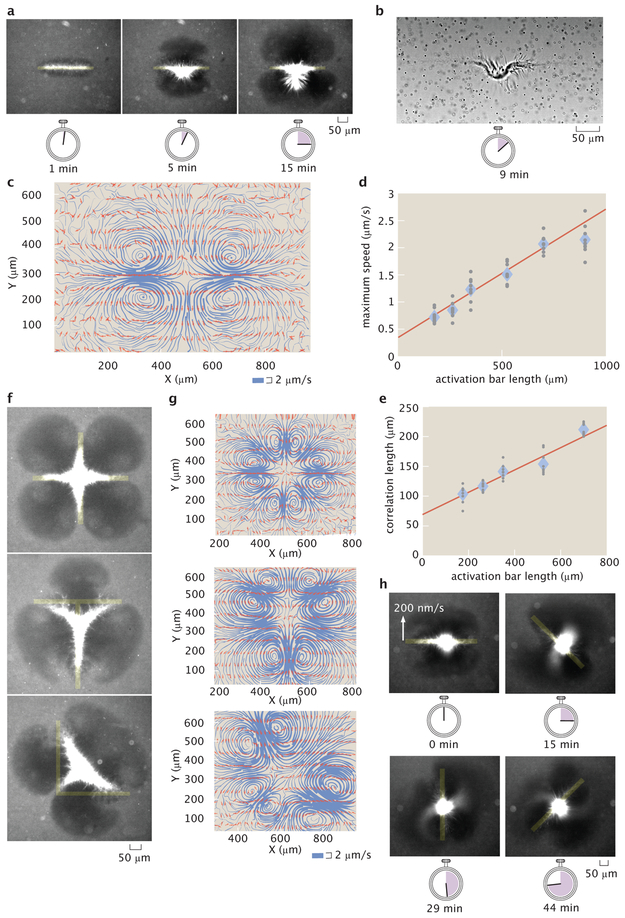
Based on these observations, we seek to generate and tune flows in our engineered system with light, which may also provide insight into the mechanics of cellular fluid flow. Recent work has used light to thermally induce cytoplasmic flows [28]. Here, we can generate fluid flows with light by activating contractile microtubule networks with the rectangular bar pattern used during aster merging (Fig. 4a) (Video 9). Brightfield images reveal a structurally changing microtubule network (Fig. 4b) (Video 10), which appears to drive the fluid flow. We observe there are minimum size and angle limits for these microtubule structures, as well as for asters (Supplementary Information 2.17).
Figure 4:
Advective fluid flow is created and controlled with patterned light. a, Microtubule organization created by an activation bar that is a 350 μm × 20 μm rectangular light pattern. Time series demonstrate continuous contraction of microtubules towards the pattern center along the major axis. b, Brightfield image of (a) shows a contracting microtubule network and tracer particles used to measure fluid flow. c, Streamline plots of background buffer flow from (a). The streamline thickness represents the flow speed. The arrows indicate the flow direction. d, Averaged maximum flow speed versus activation bar length. e, Averaged correlation length (size) of flow field versus activation bar length. f, Superposition of activation bars generate different patterns of contractile microtubules. g, Corresponding streamline plots. h, Time lapse of a light pattern rotating with an edge speed of 200 nm/s. In (d, e) the diamonds represents the mean of 9 experiments and the gray dots represent individual experiments. The red line is a linear fit to the data.
We measure the flow fields with tracer particles (Supplementary Information 2.18). The pattern of the flow is 2D (Supplementary Information 2.19) and stable throughout the experiment (Supplementary Information 2.20), consisting of inflows and outflows of microtubules, as illustrated by streamline plots (Fig. 4c)(Supplementary Information 2.21). The competition of these flows ensures that microtubules do not continuously accumulate in the illuminated region and that the surrounding medium is not completely depleted of microtubules.
We manipulate the properties of the flow field through the geometry of the activation volume. The size (Supplementary Information 2.22) and speed of the flow field depend linearly on the length of the activation bar (Fig. 4d, e). The scaling of the flow speed is similar to the relationships for both the formation rate versus activation diameter and the aster merging speed versus separation. The positioning and number of inflows, outflows, and vortices are determined by the extrema of the light pattern geometry (Fig. 4f, g) (Video 11, Video 12, Video 13). A model that uses a series of point forces following the observed microtubule networks is able to recreate similar inflows and outflows (Supplementary Information 2.23), suggesting that forces from microtubule bundles drive the flow.
Furthermore, the shape of the flow field has a temporal dependence on the light pattern. We modulate the flow field to create an “active stir bar” by applying a rotating light pattern (Fig. 4h) (Video 14). While simplified active matter systems are able to spontaneously generate global flows [6, 8], in vivo cytoskeletal-driven fluid flows can be controlled and highly structured [21, 22, 26]. Our results demonstrate the creation and dynamic manipulation of localized, structured fluid flow in an engineered active matter system for the first time.
In this work, we uncover active matter phenomena through the creation and manipulation of nonequilibrium structures and resultant fluid flows. Our ability to define boundaries of protein activity with light enables unprecedented control of an active matter system’s organization (Supplemental Information 2.25). We find scaling rules of contractile networks, movement of non-equilibrium structures, and modulation of flow fields. This framework may be built upon to create active matter devices that control fluid flow. Future work will explore spatiotemporal limits of non-equilibrium structures, the interplay of mass flows and structural changes, and develop new theories of nonequilibrium mechanics and dynamics. Our approach of understanding through construction creates a path towards a generalizable theory of non-equilibrium systems, engineering with active matter, and understanding biological phenomena.
Supplementary Material
Acknowledgements
The authors would like to thank Maya Anjur-Dietrich, John Brady, Jehoshua Bruck, Vahe Galstyan, Soichi Hirokawa, Christina Hueschen, Yuri Lazebnik, Wendell Lim, Wallace Marshall, Dyche Mullins, Dan Needleman, Paul Rothemund, and Erik Winfree for influential scientific discussions. We thank Lukasz Bugaj, Zvonimir Dogic, Adam Frost, Walter Huynh, Rustem Ismagilov, Linnea Metcalf, Henry Nguyen, and Ron Vale for advice and assistance during development of the experimental system. Koen van den Dries for assistance with 3D visualization of asters. Paul Sternberg for use of a microscopy system for initial light activation experiments. We are grateful to Nigel Orme for assistance with figures and illustrations. The authors would like to acknowledge support from the NIH through grants 1R35 GM118043–01 (RP) and NIH DP5 OD012194 (MT); the NSF through NSF 1330864 (MT); the John Templeton Foundation as part of the Boundaries of Life Initiative Grants 51250 & 60973 (RP); The Foundational Questions Institute and Fetzer Franklin Fund through FQXi 1816 (RP, MT); and the UCSF Center for Systems and Synthetic Biology NIGMS P50 GM081879 (MT).
Footnotes
Data Availability The data that support the findings of this study are available from the Caltech Research Data Repository: https://data.caltech.edu/records/1160. All plasmids used in this study are available on Addgene. All other reagents and source code used for this study are available from the corresponding author upon reasonable request.
Competing Interests The authors declare no competing interests.
References
- 1.Marchetti MC et al. Hydrodynamics of soft active matter. Rev. Mod. Phys 85, 1143–1189 (2013). [Google Scholar]
- 2.Dumont S & Prakash M Emergent mechanics of biological structures. Molecular Biology of the Cell 25, 3461–3465. issn: 1059–1524, 1939–4586 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Needleman D & Dogic Z Active matter at the interface between materials science and cell biology. Nature Reviews Materials 2, 17048 issn: 2058–8437 (2017). [Google Scholar]
- 4.Nédélec FJ, Surrey T, Maggs AC & Leibler S Self-organization of microtubules and motors. Nature 389, 305 issn: 1476–4687 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Surrey T, Nédélec F, Leibler S & Karsenti E Physical Properties Determining SelfOrganization of Motors and Microtubules. Science 292, 1167–1171. issn: 0036–8075, 1095–9203 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Sanchez T, Chen DTN, DeCamp SJ, Heymann M & Dogic Z Spontaneous motion in hierarchically assembled active matter. Nature 491, 431 issn: 1476–4687 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeCamp SJ, Redner GS, Baskaran A, Hagan MF & Dogic Z Orientational order of motile defects in active nematics. Nature Materials 14, 1110 issn: 1476–4660 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu K-T et al. Transition from turbulent to coherent flows in confined three-dimensional active fluids. Science 355, eaal1979 issn: 0036–8075, 1095–9203 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Bricard A, Caussin J-B, Desreumaux N, Dauchot O & Bartolo D Emergence of macroscopic directed motion in populations of motile colloids. Nature 503, 95–98. issn: 1476–4687 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Nédélec F, Surrey T & Maggs AC Dynamic Concentration of Motors in Microtubule Arrays. Physical Review Letters 86, 3192–3195 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Lee HY & Kardar M Macroscopic equations for pattern formation in mixtures of microtubules and molecular motors. Physical Review. E, Statistical, Nonlinear, and Soft Matter Physics 64, 056113 issn: 1539–3755 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Keber FC et al. Topology and dynamics of active nematic vesicles. Science 345, 1135–1139. issn: 0036–8075, 1095–9203 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aoyama S, Shimoike M & Hiratsuka Y Self-organized optical device driven by motor proteins. Proceedings of the National Academy of Sciences 110, 16408–16413. issn: 0027–8424, 1091–6490 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guntas G et al. Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proceedings of the National Academy of Sciences of the United States of America 112, 112–117. issn: 1091–6490 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schuppler M, Keber FC, Kröger M & Bausch AR Boundaries steer the contraction of active gels. Nature Communications 7, 13120 issn: 2041–1723 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belmonte JM, Leptin M & Nédélec F A theory that predicts behaviors of disordered cytoskeletal networks. Molecular Systems Biology 13, 941 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foster PJ, Fürthauer S, Shelley MJ & Needleman DJ Active contraction of microtubule networks. Elife 4, e10837 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Good MC, Vahey MD, Skandarajah A, Fletcher DA & Heald R Cytoplasmic Volume Modulates Spindle Size During Embryogenesis. Science 342, 856–860. issn: 0036–8075, 10959203 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weiner OD, Marganski WA, Wu LF, Altschuler SJ & Kirschner MW An ActinBased Wave Generator Organizes Cell Motility. PLOS Biology 5, 1–11 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gardel ML, Schneider IC, Aratyn-Schaus Y & Waterman CM Mechanical Integration of Actin and Adhesion Dynamics in Cell Migration. Annual Review of Cell and Developmental Biology 26, 315–333 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Theurkauf WE Premature microtubule-dependent cytoplasmic streaming in cappuccino and spire mutant oocytes. Science 265, 2093–2096 (1994). [DOI] [PubMed] [Google Scholar]
- 22.Ganguly S, Williams LS, Palacios IM & Goldstein RE Cytoplasmic streaming in Drosophila oocytes varies with kinesin activity and correlates with the microtubule cytoskeleton architecture. Proceedings of the National Academy of Sciences 109, 15109–15114 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldstein RE, Tuval I & van de Meent J-W Microfluidics of cytoplasmic streaming and its implications for intracellular transport. Proceedings of the National Academy of Sciences 105, 3663–3667 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drescher K, Dunkel J, Cisneros LH, Ganguly S & Goldstein RE Fluid dynamics and noise in bacterial cell–cell and cell–surface scattering. Proceedings of the National Academy of Sciences 108, 10940–10945 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drescher K, Goldstein RE, Michel N, Polin M & Tuval I Direct Measurement of the Flow Field around Swimming Microorganisms. Phys. Rev. Lett. 105, 168101 (2010). [DOI] [PubMed] [Google Scholar]
- 26.He B, Doubrovinski K, Polyakov O & Wieschaus E Apical constriction drives tissue-scale hydrodynamic flow to mediate cell elongation. Nature 508, 392 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shinar T, Mana M, Piano F & Shelley MJ A model of cytoplasmically driven microtubulebased motion in the single-celled Caenorhabditis elegans embryo. Proceedings of the National Academy of Sciences 108, 10508–10513. issn: 0027–8424 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mittasch M et al. Non-invasive perturbations of intracellular flow reveal physical principles of cell organization. Nature Cell Biology 20, 344–351. issn: 1476–4679 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.