Abstract
Despite over 50 years of clinical and basic studies, acute respiratory distress syndrome (ARDS) is still a critical challenge with high mortality worldwide. The severity of neutrophil activation was associated with disease severity. However, the detailed pathophysiology of the circulating polymorphonuclear neutrophil activation in ARDS remains unclear. To identify key pathways and genes in the ARDS-specific neutrophil phenotype distinct from sepsis, the datasets of blood polymorphonuclear neutrophils (PMNs) from patients with ARDS (GSE76293) and from sepsis patients (GSE49757) were chosen from the Gene Expression Omnibus (GEO) and analyzed using bioinformatics methods. A total of 220 differential expressed genes (DEGs) were overlapped between GSE49757 and GSE76293 in a Venn diagram. Pathway enrichment analysis results showed that DEGs in GSE76293 were mainly enriched in the MAPK signaling pathway, FoxO signaling pathway, and AMPK signaling pathway relative to GSE49757. We identified 30 hub genes in the protein-protein interaction network. By comparing with GSE49757, we speculated that GAPDH, MAPK8, PIK3CB, and MMP9 may play important roles in the progression of ARDS-specific circulating neutrophil activation. The findings may provide novel insights into the development of promising targets for the diagnosis and treatment of ARDS in the future.
1. Introduction
Acute respiratory distress syndrome (ARDS) is characterized by diffuse damage of the alveolar-capillary barrier, immune cell infiltration, protein-rich edema fluid in the alveoli, and severe gas-exchange abnormalities. Despite over 50 years of clinical and basic studies, ARDS is still a critical challenge with high mortality worldwide. ARDS increases healthcare costs and impairs quality of life seriously [1]. Therefore, getting a better understanding of the pathogenesis of ARDS is urgent and highly demanded.
Polymorphonuclear neutrophils (PMNs) are crucial for controlling infections as innate immune system cells [2]. Circulating PMNs become activated and penetrate the alveolar-capillary barrier into the airspaces in the progression of ARDS. PMNs in the alveoli inflammatory microenvironment become further activated to play an important role in phagocytosing pathogens, releasing reactive oxygen species and inducing neutrophil extracellular traps [3–6]. Subsequently, activated neutrophils lead to alveolar damage and further loss of lung function. However, the mechanisms of blood PMNs activation and infiltration in the development of ARDS remain poorly understood.
High-throughput gene profiling has been a powerful tool in revealing key pathways and genes of lung disease, such as ARDS and asthma [7, 8]. The transcriptomics mining of key pathways and genes offers a potential direction for future mechanism research. One of the major issues in previous high-throughput studies of ARDS was that samples were whole blood or total leukocytes rather than purified neutrophils. In the present study, we identified differential expressed genes (DEGs) in blood PMNs from patients with ARDS (GSE76293) and from sepsis patients (GSE49757) and used integrated bioinformatics methods to identify key pathways and genes in the ARDS-specific neutrophil phenotype distinct from sepsis. Our findings may provide novel potential targets for the diagnosis and treatment of ARDS.
2. Materials and Methods
2.1. Microarray Data
To investigate the ARDS-specific neutrophil phenotype distinct from sepsis, we searched expression profiles of ARDS blood PMNs and chose the datasets GSE76293 [9] and GSE49757 [10] from the Gene Expression Omnibus (GEO) [11]. In the current study, 12 ARDS blood PMNs samples and 12 HVT blood PMNs samples were used for analysis in the dataset GSE76293; 20 PMNs samples stimulated with severe sepsis plasma and 19 PMNs samples stimulated with HVT plasma were selected to verify the ARDS-specific neutrophil phenotype in the dataset GSE49757.
2.2. Identification of DEGs
DEGs were identified using GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r/), which is an online tool based on the GEOquery and Limma R packages [12]. The genes that met the cut-off criteria of an adjusted p value < 0.01 and a |log2 fold change| > 0.585 were considered DEGs. To indicate the intersection among DEGs between GSE49757 and GSE76293, a Venn diagram was produced by a Venn webtool (http://bioinformatics.psb.ugent.be/webtools/Venn/).
2.3. Gene Ontology (GO) and Pathway Enrichment Analyses
The Database for Annotation Visualization and Integrated Discovery (DAVID; http://david.ncifcrf.gov, version 6.7) [13, 14] was used to analyze the gene ontology and pathway of DEGs as previously described [15, 16]. p value < 0.05 was used as a threshold to define significantly enriched terms.
2.4. Protein-Protein Interactions (PPI) Network and Module Analysis
The Search Tool for the Retrieval of Interacting Genes (STRING; http://string-db.org, version 10.5) online database was used to predict interactions of DEGs [17]. A PPI network was drawn by Cytoscape (version 3.7.1). Furthermore, the plugins CytoNCA [18] and MCODE [19] of Cytoscape were used to identify the hub genes and modules in the PPI network.
2.5. Transcription Factor (TF) Regulatory Network Analysis
The iRegulon plugin in Cytoscape was used to predict TFs of the selected hub DEGs [20]. A normalized enrichment score (NES) >4 was considered the threshold value.
2.6. Relative mRNA Expression Level of Hub Genes
To verify the differences of relative mRNA expression level of hub genes between GSE49757 and GSE76293, we downloaded the matrix data of the two datasets from the GEO and analyzed the log2 normalized signal intensity of selected hub genes using GraphPad Prism 7.04.
2.7. Statistical Analysis
All statistical analyses in this study were performed using GraphPad Prism 7.04 (GraphPad Software, San Diego, CA, USA), and p < 0.05 was considered to be significant. Data are presented as mean ± SEM. Student's t‐test was used to compare difference.
3. Results
3.1. Identification of DEGs
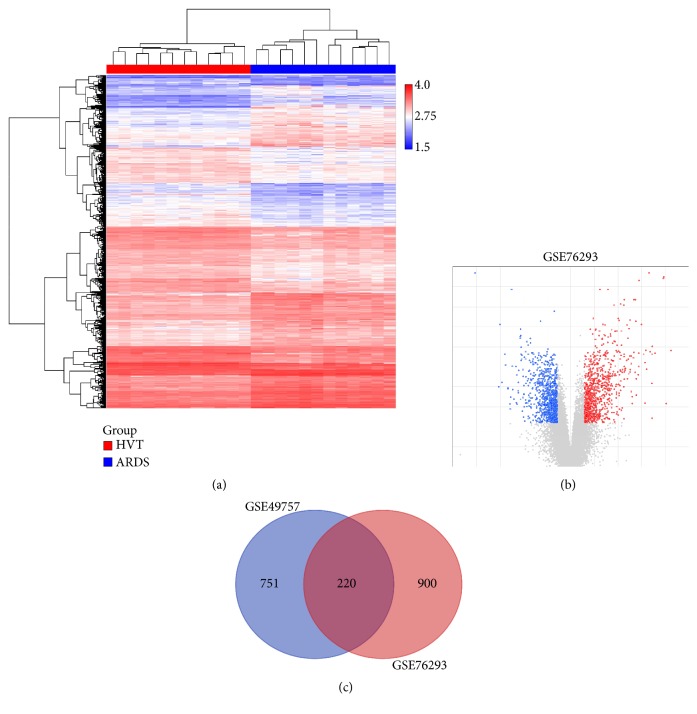
In total, 1120 mRNAs were significantly differentially expressed in ARDS blood PMNs compared with HVT blood PMNs, including 486 upregulated genes and 634 downregulated genes (Figures 1(a) and 1(b)). There were 971 DEGs in PMNs exposed to severe septic plasma compared with unstimulated controls, including 455 upregulated genes and 516 downregulated genes. A total of 220 genes were overlapped between GSE49757 and GSE76293 in the Venn diagram (Figure 1(c)).
Figure 1.
Hierarchical clustering, volcano plot, and Venn diagram of differentially expressed genes in GSE49757 and GSE76293. Hierarchical clustering indicates the gene expression profile of GSE76293 (a): red to blue colors refer to be high to low relative expression levels. Volcano plot of differentially expressed genes in GSE76293 (b): gray dots indicate no change; blue and red dots indicate downregulated and upregulated genes, respectively. Venn diagram indicates the intersection among DEGs between GSE49757 and GSE76293 (c).
3.2. GO and Pathway Enrichment Analyses
DEGs in GSE76293 were mainly associated with the following biological processes: apoptotic process, response to oxidative stress, response to lipopolysaccharide, response to tumor necrosis factor, and leukotriene signaling pathway (Figure 2(a)). The results also indicated that DEGs in GSE76293 were mainly enriched in the following pathways: MAPK signaling pathway, FoxO signaling pathway, AMPK signaling pathway, and TNF signaling pathway (Figure 2(b)).
Figure 2.
Biological process (BP) (a) and pathway enrichment (b) of differentially expressed genes in GSE49757 and GSE76293 (top 20, p < 0.05). Red to blue colors indicate low to high -log10 (p value) levels. Point size indicates the number of differentially expressed genes in the corresponding items.
DEGs in GSE49757 were mainly associated with the following biological processes: inflammatory response, response to lipopolysaccharide, apoptotic process, immune response, positive regulation of cytokine production, and positive regulation of NF-kappaB signaling (Figure 2(a)). The results also indicated that DEGs in GSE49757 were mainly enriched in the following pathways: NF-kappa B signaling pathway, cytokine-cytokine receptor interaction, NOD-like receptor signaling pathway, and TNF signaling pathway (Figure 2(b)).
3.3. PPI Network Analysis
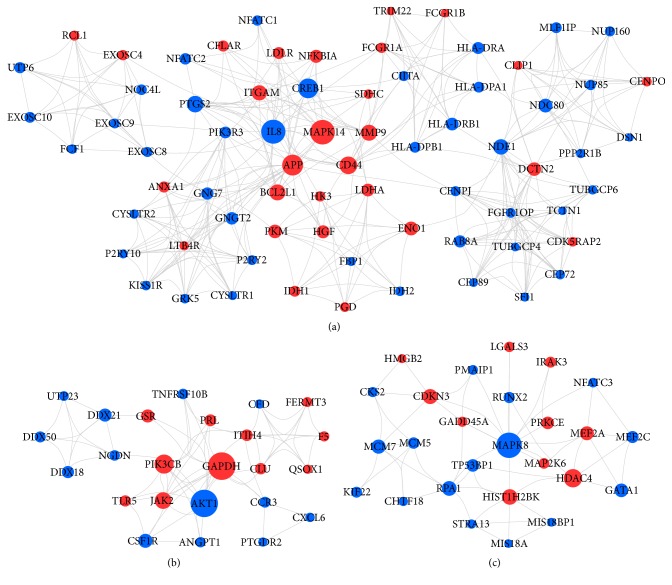
The PPI network of 899 nodes with 3757 protein interaction pairs was constructed to identify hub genes in ARDS circulating PMN activation in GSE76293. The top 30 hub genes with the higher degree were listed in Table 1. The three most significant modules of the PPI network were shown in Figure 3, in which GAPDH, AKT1, MAPK14, MAPK8, IL8, PIK3CB, and MMP9 were the top hub genes.
Table 1.
The top 30 hub genes of the PPI network in GSE76293. logFC: log2 fold change between two experimental conditions. adj.P.Val: p value after adjustment for multiple testing.
| Gene symbol | Degree | logFC | adj.P.Val |
|---|---|---|---|
| GAPDH | 106 | 1.28 | 2.69E-06 |
| AKT1 | 105 | -1.23 | 6.93E-05 |
| RIPK4 | 102 | -1.62 | 3.42E-04 |
| TOP2A | 87 | 0.97 | 3.44E-03 |
| MAPK14 | 69 | 1.50 | 1.74E-07 |
| MAPK8 | 68 | -0.97 | 2.15E-03 |
| CXCL8 | 67 | -2.12 | 1.97E-03 |
| SMARCA4 | 66 | -0.62 | 7.63E-03 |
| PIK3CB | 60 | 0.65 | 3.56E-03 |
| APP | 55 | 1.37 | 4.14E-03 |
| CREB1 | 51 | -0.68 | 6.18E-03 |
| JAK2 | 47 | 0.95 | 5.29E-05 |
| CD44 | 43 | 1.82 | 2.85E-06 |
| HDAC4 | 42 | 0.97 | 1.66E-05 |
| PTGS2 | 39 | -1.47 | 5.15E-03 |
| H2AFV | 39 | -0.68 | 2.36E-03 |
| MMP9 | 37 | 0.69 | 2.75E-04 |
| BCL2L1 | 37 | 0.84 | 3.26E-03 |
| SKP2 | 36 | -0.69 | 5.53E-03 |
| POLR2I | 35 | -0.69 | 2.77E-03 |
| NDE1 | 35 | -0.72 | 8.75E-04 |
| ITGAM | 34 | 0.79 | 5.92E-05 |
| SNU13 | 34 | -0.78 | 3.66E-03 |
| CSF1R | 33 | -2.10 | 1.08E-05 |
| HIST1H2BK | 32 | 0.60 | 1.05E-04 |
| MIB2 | 31 | -0.73 | 5.11E-04 |
| CDKN3 | 30 | 0.89 | 6.48E-04 |
| UBE2D4 | 29 | -0.59 | 1.51E-03 |
| NDC80 | 29 | -1.13 | 6.52E-04 |
Figure 3.
The three most significant modules of the PPI network in GSE76293. Red and blue circles indicate upregulated and downregulated differentially expressed genes, respectively. Circle size indicates the node degree.
3.4. TF Regulatory Network Analysis
The TFs which regulated the top 50 hub genes in the PPI network were predicted. With a threshold of an NES > 4, a total of six TFs (E2F1, NFKB1, NFYA, PBX3, EGR1, and RELA) were revealed to be associated with 30 target hub genes in Figure 4.
Figure 4.
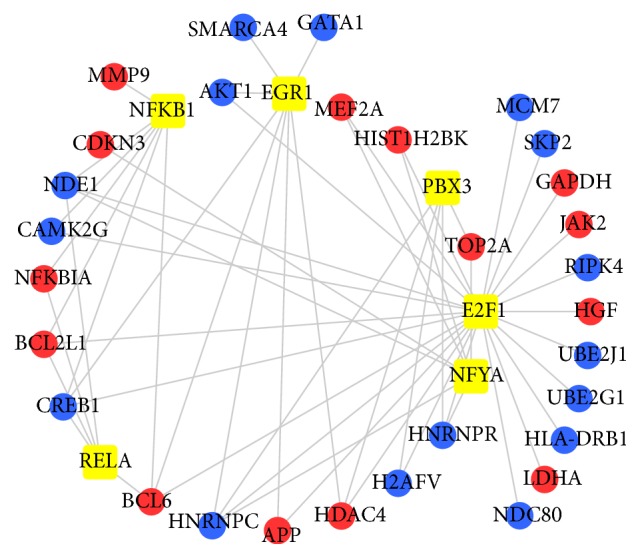
TF regulatory network of the top 50 hub genes in the PPI network in GSE76293. Yellow squares indicate TFs. Red and blue circles indicate upregulated and downregulated differentially expressed genes, respectively.
3.5. Relative mRNA Expression Level of Hub Genes
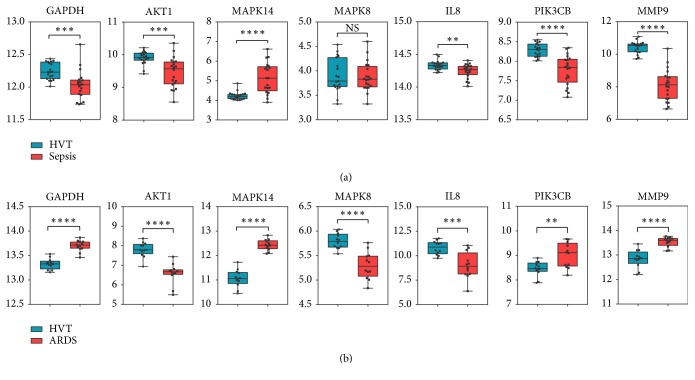
To identify the hub genes of ARDS-specific neutrophil phenotype distinct from sepsis, we compared the relative mRNA expression levels of selected hub genes in both GSE49757 and GSE76293. We found that AKT1 and IL8 were downregulated, while MAPK14 was upregulated in both GSE49757 and GSE76293 (Figure 5). At the same time, the relative expression trends of GAPDH, MAPK8, PIK3CB, and MMP9 were different between GSE49757 and GSE76293 (Figure 5).
Figure 5.
The relative mRNA expression level of the seven hub genes in GSE49757 (a) and GSE76293 (b). All data are means ± SEM, Student's t-test. ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; NS, not significant.
4. Discussion
Acute respiratory distress syndrome (ARDS) is characterized by diffuse damage of the alveolar-capillary barrier, immune cell infiltration, protein-rich edema fluid in the alveoli, and severe gas-exchange abnormalities. The severity of neutrophil activation and infiltration was associated with disease severity [21, 22]. However, the detailed pathophysiology of the ARDS-specific neutrophil activation remains unclear, especially the complicated molecular mechanisms.
The rapid development of high-throughput detection technology in recent decades has provided technical support for genome-wide analyses of changes in gene expression related to ARDS. However, one of the issues in previous high-throughput studies of ARDS was that samples were whole blood or total leukocytes rather than purified neutrophils. In the present study, we chose the dataset GSE76293 including 12 ARDS blood PMNs samples and 12 HVT blood PMNs samples. There were 1120 DEGs in ARDS blood PMNs compared with HVT controls, including 486 upregulated mRNAs and 634 downregulated mRNAs.
Because not all patients with severe sepsis develop ARDS, a comparison of circulating neutrophils from patients with ARDS with neutrophils from severe septic patients may allow us to find the ARDS-specific neutrophil phenotype distinct from sepsis. Therefore, we searched sepsis blood neutrophils expression profile in the GEO and chose the dataset GSE49757, in which 20 PMNs samples stimulated with severe septic plasma and 19 PMNs samples stimulated with HVT plasma samples were used for analysis. There were 971 DEGs between severe septic plasma samples and controls, including 455 upregulated genes and 516 downregulated genes. The overlap between GSE49757 and GSE76293 contained 220 genes in the Venn diagram.
DEGs in GSE76293 were mainly enriched in the following biological processes: response to oxidative stress, leukotriene signaling pathway, and response to tumor necrosis factor compared with GSE49757. KEGG enrichment analysis results showed that DEGs in GSE76293 were mainly involved in the MAPK signaling pathway, FoxO signaling pathway, and AMPK signaling pathway relative to GSE49757. These findings, while preliminary, suggested that these biological processes and pathways played important roles in the ARDS-specific neutrophil phenotype distinct from sepsis.
To identify hub genes involved in the activation of circulating PMNs in ARDS, we constructed the PPI network and predicted its key modules. We found that GAPDH, AKT1, MAPK14, MAPK8, IL8, PIK3CB, and MMP9 were the top hub genes in the three most significant modules. We speculated that the seven hub genes may affect the circulating PMN activation in ARDS. To further identify whether the seven hub genes were involved in the ARDS-specific neutrophil phenotype distinct from sepsis, we verified the relative mRNA expression of the seven genes in GSE49757. We found that AKT1 and IL8 were downregulated; MAPK14 was upregulated in both GSE49757 and GSE76293. However, the relative expression trends of GAPDH, MAPK8, PIK3CB, and MMP9 were different between GSE49757 and GSE76293. We speculated that GAPDH, MAPK8, PIK3CB, and MMP9 may play important roles in the ARDS-specific neutrophil phenotype distinct from sepsis.
Phosphoinositide 3-kinases (PI3Ks) participate in most pathophysiology processes in almost all human tissues. PIK3CB played an important role in neutrophil survival, priming, activation, and ROS production [23–26]. Previous reports have shown that AKT was one of the important effectors of PI3K signaling [27]. PIK3CB resulted in the phosphorylation of AKT; in turn, AKT activation participated in the activation of downstream effectors of the PI3K pathway. Our results showed that PIK3CB was upregulated, whereas AKT1 was significantly downregulated in ARDS blood PMNs compared with HVT blood PMNs. We speculated that one possible explanation for these results may be that the PI3K pathway interacts with other signaling pathways and forms complex interaction networks, which cause the specific cellular response.
Mitogen‐activated protein kinases (MAPKs) are involved in most of the cellular responses to harmful stimuli (like infection, oxidative stress, etc.) [28–31]. One interesting finding was that MAPK14 was significantly upregulated while MAPK8 was downregulated in ARDS blood PMNs compared with HVT blood PMNs. It is difficult to explain this result, but it might be related to the functional differences in apoptosis between MAPK14 and MAPK8. Prior studies have noted that circulating neutrophils from patients with ARDS exhibited delayed apoptosis. Phosphorylation of MAPK14 led to the inhibition of neutrophil apoptosis [32], while MAPK8 induced apoptosis or growth inhibition [33, 34]. We speculated that the differential expression of MAPK14 and MAPK8 may contribute to the delayed apoptosis of circulating neutrophils.
In patients with ARDS, immunocytes such as macrophages in the regions of pulmonary injury secrete chemokines, of which IL-8 is the typical neutrophil chemokine [5, 35]. Previous studies have shown that IL-8 was elevated in bronchoalveolar lavage fluid PMNs from patients with ARDS. Here, we noticed that IL-8 was downregulated in ARDS blood PMNs compared with HVT controls in GSE76293. In addition, IL-8 expression was also downregulated in PMNs exposed to severe septic plasma compared to unstimulated controls in GSE49757. The reason for this is not clear, but one possible explanation for these results may be that the downregulation of IL-8 contributes to circulating neutrophil recruitment into the regions rich in IL-8.
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a key enzyme in the process of glycolysis. GAPDH is often considered a housekeeping gene and a control for western bolt and qPCR because of the stable and high expression in most cells and tissues. However, increasing studies have reported that GAPDH was associated with many physiological functions, such as inflammatory and immune responses [36–38]. Piszczatowski RT et al. found that myeloid zinc finger-1 (MZF-1) regulated the translation of GAPDH [39]. Here, we found that GAPDH was upregulated in ARDS blood PMNs compared with HVT blood PMNs in GSE76293 and might be regulated by the E2F1 transcription factor [40]. In addition, the present study showed that GAPDH was downregulated in PMNs exposed to severe septic plasma relative to unstimulated controls in GSE49757. Therefore, it needs to be considered with caution when GAPDH is used as a control for qPCR in future studies.
Despite the results obtained above, there were some limitations in this study. Because of the relatively small sample size and the heterogeneity of ARDS, the findings need to be interpreted with caution. The results need to be further validated in a large number of samples, and the mechanisms need to be investigated both in vitro and vivo in the future.
5. Conclusions
In conclusion, we identified key pathways and genes involved in the ARDS-specific neutrophil phenotype distinct from sepsis. We speculated that GAPDH, MAPK8, PIK3CB, and MMP9 may play important roles in the progression of ARDS-specific circulating neutrophil activation. The findings may provide novel insights into the development of promising targets for the diagnosis and treatment of ARDS in the future.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 81770076, No. 81570074, and No. 81600054) and Shandong Taishan Scholars and Young Experts Program.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
References
- 1.Williams A. E., Jose R. J., Mercer P. F., et al. Evidence for chemokine synergy during neutrophil migration in ARDS. Thorax. 2017;72(1):66–73. doi: 10.1136/thoraxjnl-2016-208597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brazil J. C., Parkos C. A. Pathobiology of neutrophil-epithelial interactions. Immunological Reviews. 2016;273:94–111. doi: 10.1111/imr.12446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zemans R. L., Colgan S. P., Downey G. P. Transepithelial migration of neutrophils. American Journal of Respiratory Cell and Molecular Biology. 2009;40(5):519–535. doi: 10.1165/rcmb.2008-0348TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Summers C., Singh N. R., White J. F., et al. Pulmonary retention of primed neutrophils: a novel protective host response, which is impaired in the acute respiratory distress syndrome. Thorax. 2014;69(7):623–629. doi: 10.1136/thoraxjnl-2013-204742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zemans R. L., Matthay M. A. What drives neutrophils to the alveoli in ARDS? Thorax. 2017;72(1):1–3. doi: 10.1136/thoraxjnl-2016-209170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grommes J., Soehnlein O. Contribution of neutrophils to acute lung injury. Molecular Medicine. 2011;17(3-4):293–307. doi: 10.2119/molmed.2010.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desai A. A., Hysi P., Garcia J. G. Integrating genomic and clinical medicine: searching for susceptibility genes in complex lung diseases. Translational Research. 2008;151(4):181–193. doi: 10.1016/j.trsl.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kan M., Shumyatcher M., Himes B. E. Using omics approaches to understand pulmonary diseases. Respiratory Research. 2017;18(1) doi: 10.1186/s12931-017-0631-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juss J. K., House D., Amour A., et al. Acute respiratory distress syndrome neutrophils have a distinct phenotype and are resistant to phosphoinositide 3-kinase inhibition. American Journal of Respiratory and Critical Care Medicine. 2016;194(8):961–973. doi: 10.1164/rccm.201509-1818OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khaenam P., Rinchai D., Altman M. C., et al. A transcriptomic reporter assay employing neutrophils to measure immunogenic activity of septic patients' plasma. Journal of Translational Medicine. 2014;12(1, article no. 65) doi: 10.1186/1479-5876-12-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barrett T., Wilhite S. E., Ledoux P., et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Research. 2013;41(1):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sean D., Meltzer P. S. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and bioconductor. Bioinformatics. 2007;23(14):1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 13.Huang D. W., Sherman B. T., Lempicki R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang D. W., Sherman B. T., Lempicki R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 15.Liu T., Zhang T., Zhou F., et al. Identification of genes and pathways potentially related to PHF20 by gene expression profile analysis of glioblastoma U87 cell line. Cancer Cell International. 2017;17, article 87 doi: 10.1186/s12935-017-0459-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan Z., Li L., Fang Q., et al. Integrated bioinformatics analysis of master regulators in anaplastic thyroid carcinoma. BioMed Research International. 2019;2019:13. doi: 10.1155/2019/9734576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franceschini A., Szklarczyk D., Frankild S., et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Research. 2013;41(1):D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang Y., Li M., Wang J., Pan Y., Wu F.-X. CytoNCA: a cytoscape plugin for centrality analysis and evaluation of protein interaction networks. BioSystems. 2015;127:67–72. doi: 10.1016/j.biosystems.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 19.Bandettini W. P., Kellman P., Mancini C., et al. MultiContrast Delayed Enhancement (MCODE) improves detection of subendocardial myocardial infarction by late gadolinium enhancement cardiovascular magnetic resonance: a clinical validation study. Journal of Cardiovascular Magnetic Resonance. 2012;14(1, article no. 83) doi: 10.1186/1532-429X-14-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janky R., Verfaillie A., Imrichova H., et al. iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Computational Biology. 2014;10(7) doi: 10.1371/journal.pcbi.1003731.e1003731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abraham E. Neutrophils and acute lung injury. Critical Care Medicine. 2003;31(4, supplement):S195–S199. doi: 10.1097/01.CCM.0000057843.47705.E8. [DOI] [PubMed] [Google Scholar]
- 22.Fujishima S. Pathophysiology and biomarkers of acute respiratory distress syndrome. Journal of Intensive Care. 2014;2, article 32(1) doi: 10.1186/2052-0492-2-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kulkarni S., Sitaru C., Jakus Z., et al. PI3Kbeta plays a critical role in neutrophil activation by immune complexes. Science Signaling. 2011;4(168, article no. ra23) doi: 10.1126/scisignal.2001617. [DOI] [PubMed] [Google Scholar]
- 24.Hawkins P. T., Stephens L. R. PI3K signalling in inflammation. Biochimica et Biophysica Acta. 2015;1851:882–897. doi: 10.1016/j.bbalip.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 25.Boyle K. B., Gyori D., Sindrilaru A., et al. Class IA phosphoinositide 3-kinase beta and delta regulate neutrophil oxidase activation in response to Aspergillus fumigatus hyphae. The Journal of Immunology. 2011;186(5):2978–2989. doi: 10.4049/jimmunol.1002268. [DOI] [PubMed] [Google Scholar]
- 26.Fruman D. A., Chiu H., Hopkins B. D., Bagrodia S., Cantley L. C., Abraham R. T. The PI3K pathway in human disease. Cell. 2017;170(4):605–635. doi: 10.1016/j.cell.2017.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manning B. D., Toker A. AKT/PKB signaling: navigating the network. Cell. 2017;169(3):381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta J., Nebreda A. R. Roles of p38α mitogen-activated protein kinase in mouse models of inflammatory diseases and cancer. FEBS Journal. 2015;282(10):1841–1857. doi: 10.1111/febs.13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi J., Sun S. Tumor necrosis factor receptor-associated factor regulation of nuclear factor κb and mitogen-activated protein kinase pathways. Frontiers in Immunology. 2018;9, article 1849 doi: 10.3389/fimmu.2018.01849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arthur J. S., Ley S. C. Mitogen-activated protein kinases in innate immunity. Nature Reviews Immunology. 2013;13(9):679–692. doi: 10.1038/nri3495. [DOI] [PubMed] [Google Scholar]
- 31.Zhou Y. Y., Li Y., Jiang W. Q., Zhou L. F. MAPK/JNK signalling: a potential autophagy regulation pathway. Bioscience Reports. 2015;35(3) doi: 10.1042/bsr20140141.e00199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin W.-C., Lin C.-F., Chen C.-L., Chen C.-W., Lin Y.-S. Inhibition of neutrophil apoptosis via sphingolipid signaling in acute lung injury. The Journal of Pharmacology and Experimental Therapeutics. 2011;339(1):45–53. doi: 10.1124/jpet.111.181560. [DOI] [PubMed] [Google Scholar]
- 33.Khan M. A., Farahvash A., Douda D. N., et al. JNK activation turns on LPS- and gram-negative bacteria-induced NADPH oxidase-dependent suicidal NETosis. Scientific Reports. 2017;7, article 3409(1) doi: 10.1038/s41598-017-03257-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kato T., Noma H., Kitagawa M., Takahashi T., Oshitani N., Kitagawa S. Distinct role of c-Jun N-terminal kinase isoforms in human neutrophil apoptosis regulated by tumor necrosis factor-α and granulocyte-macrophage colony-stimulating factor. Journal of Interferon & Cytokine Research. 2008;28(4):235–243. doi: 10.1089/jir.2007.0075. [DOI] [PubMed] [Google Scholar]
- 35.Williams A. E., Chambers R. C. The mercurial nature of neutrophils: still an enigma in ARDS? American Journal of Physiology-Lung Cellular and Molecular Physiology. 2014;306(3):L217–L230. doi: 10.1152/ajplung.00311.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mookherjee N., Lippert D. N., Hamill P., et al. Intracellular receptor for human host defense peptide LL-37 in monocytes. The Journal of Immunology. 2009;183:2688–2696. doi: 10.4049/jimmunol.0802586. [DOI] [PubMed] [Google Scholar]
- 37.Zhou Y., Yi X., Stoffer J. B., et al. The multifunctional protein glyceraldehyde-3-phosphate dehydrogenase is both regulated and controls colony-stimulating factor-1 messenger rna stability in ovarian cancer. Molecular Cancer Research. 2008;6(8):1375–1384. doi: 10.1158/1541-7786.MCR-07-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takaoka Y., Goto S., Nakano T., et al. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) prevents lipopolysaccharide (LPS)-induced, sepsis-related severe acute lung injury in mice. Scientific Reports. 2014;4, article no. 5204 doi: 10.1038/srep05204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Piszczatowski R. T., Rafferty B. J., Rozado A., Tobak S., Lents N. H. The glyceraldehyde 3-phosphate dehydrogenase gene (GAPDH) is regulated by myeloid zinc finger 1 (MZF-1) and is induced by calcitriol. Biochemical and Biophysical Research Communications. 2014;451(1):137–141. doi: 10.1016/j.bbrc.2014.07.082. [DOI] [PubMed] [Google Scholar]
- 40.Ertosun M. G., Hapil F. Z., Osman Nidai O. E2F1 transcription factor and its impact on growth factor and cytokine signaling. Cytokine & Growth Factor Reviews. 2016;31:17–25. doi: 10.1016/j.cytogfr.2016.02.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.