Summary
Long-range (>10 μm) transport of electrons along networks of Geobacter sulfurreducens protein filaments, known as microbial nanowires, has been invoked to explain a wide range of globally important redox phenomena. These nanowires were previously thought to be type IV pili composed of PilA protein. Here we report a 3.7 Å resolution cryo-electron microscopy structure which surprisingly reveals that, rather than PilA, G. sulfurreducens nanowires are assembled by micrometer-long polymerization of the hexaheme cytochrome OmcS, with hemes packed within ~3.5–6 Å of each other. The inter-subunit interfaces show unique structural elements such as inter-subunit parallel-stacked hemes and axial coordination of heme by histidines from neighbouring subunits. Wild-type OmcS filaments show 100-fold greater conductivity than other filaments from an ΔomcS strain, highlighting the importance of OmcS to conductivity in these nanowires. This structure explains the remarkable capacity of soil bacteria to transport electrons to remote electron acceptors for respiration and energy sharing.
Introduction
Conductive filamentous appendages of common soil bacteria Geobacter (Reguera et al., 2005), referred to as microbial nanowires, play a critical role in long-range extracellular electron transfer for respiration (Malvankar et al., 2011) and interspecies electron exchange (Summers et al., 2010). These nanowires have been invoked to explain a wide range of globally important redox phenomena that influence carbon and mineral cycling in soils and sediments, bioremediation, corrosion, and anaerobic conversion of organic wastes to methane or electricity (Malvankar and Lovley, 2014; Malvankar et al., 2011). However, these filaments’ composition, structure and underlying conduction mechanism have remained uncertain because the filaments are difficult to solubilize for studies using traditional biochemical methods and X-ray crystallography. Geobacter sulfurreducens serves as a model organism for the broader phenomenon of extracellular electron transfer because it produces these conductive filaments (Tan et al., 2017) and has a fully sequenced genome and well-developed genetic system (Reguera et al., 2005). In contrast to other electron-transferring bacteria (Marsili et al., 2008), G. sulfurreducens does not use diffusing shuttle molecules but requires direct contact with an electron acceptor via conductive filaments for long-range extracellular electron transfer (Reguera et al., 2005).
G. sulfurreducens outer-surface c-type cytochromes, including the hexaheme cytochrome OmcS (Qian et al., 2011), are known to play a critical role in bacterial growth on insoluble electron acceptors, including Fe (III) oxide and electrodes (Holmes et al., 2006; Mehta et al., 2005). Nonetheless, previous studies proposed conductive Geobacter filaments were type IV pili composed of PilA protein for a number of reasons: i) electron-transferring cells showed high levels of messenger RNA for PilA (Childers et al., 2002); ii) the amino acid sequence of G. sulfurreducens PilA is similar to the N-terminal sequence of PilA from other type IV pili-producing bacteria (Reguera et al., 2005); iii) genomic organization of G. sulfurreducens pilus biosynthesis genes was also similar to other type IV pili-producing bacteria (Reguera et al., 2005); iv) a pilA deletion mutant strain lacked filaments and could not transfer electrons extracellularly (Reguera et al., 2005); v) point mutations in pilA caused cells to produce filaments with different conductivities than wild-type (WT) filaments (Adhikari et al., 2016; Tan et al., 2016a; Tan et al., 2017; Vargas et al., 2013). Because of these extensive literature data, previous studies, including some from authors of the present manuscript (S.E.Y. N.L.I., A.I.H and N.S.M.), presumed that conductive filaments were type IV PilA pili.
Despite these data, there has never been any direct evidence that conductive Geobacter extracellular filaments are composed of PilA. Instead, the filament composition was inferred from indirect evidence, including the presence of PilA in biochemical analyses (Tan et al., 2016b), or from low-resolution imaging by atomic force microscopy (AFM) and negative-staining transmission electron microscopy, which suggested filament dimensions similar to type IV pili (Reguera et al., 2005).
On the other hand, alternative functional roles have been suggested for PilA (Reguera et al., 2007). PilA is implicated in the secretion of OmcS to the outer surface (Liu et al., 2018; Richter et al., 2012), and overexpression of PilA is accompanied by overproduction of OmcS and extracellular filaments (Leang et al., 2013; Summers et al., 2010). Here, we show that the conductive extracellular appendages of wild-type G. sulfurreducens strain are primarily OmcS filaments. These appendages are polymerized filaments of OmcS with unique structural features that also provide a molecular basis for understanding long-range electronic transport in proteins. Gel electrophoresis, mass spectrometry, AFM, and conductivity measurements show that these nanowires are the same filaments that were previously thought to be type IV pili. Data from our measurements are consistent with previous studies, and our results establish previously unknown class of protein-based nanowires based on cytochrome polymerization.
Results and Discussion
The following results reconcile previous observations by directly identifying the composition and structure of conductive filaments using cryo-electron microscopy (Cryo-EM). We grew WT cultures using anodes of microbial fuel cells as the sole electron acceptors (O’Brien and Malvankar, 2017). These growth conditions promote production of conductive biofilms and filaments as well as overexpression of PilA in comparison with growth on soluble electron acceptors such as fumarate. (Malvankar et al., 2011). Consistent with previous studies (Tan et al., 2016b), we confirmed the presence of both PilA and OmcS with expected molecular weights of ~ 6.5 kDa and ~ 45 kDa, respectively, in our filament preparations using polyacrylamide gel electrophoresis (SDS-PAGE), peptide mass spectrometry and western immunoblotting (Fig. S1, methods).
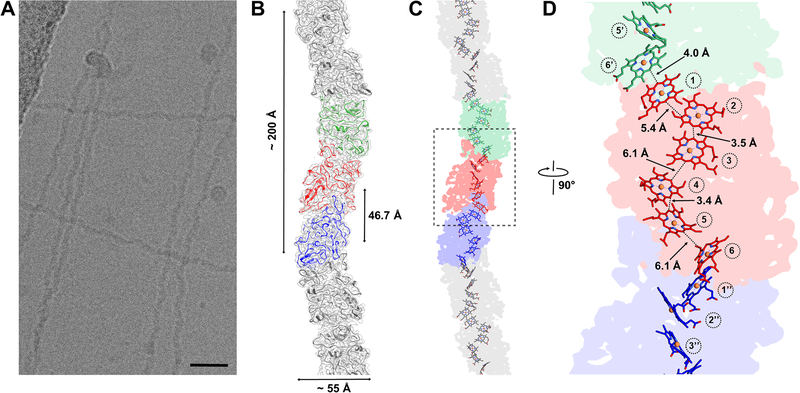
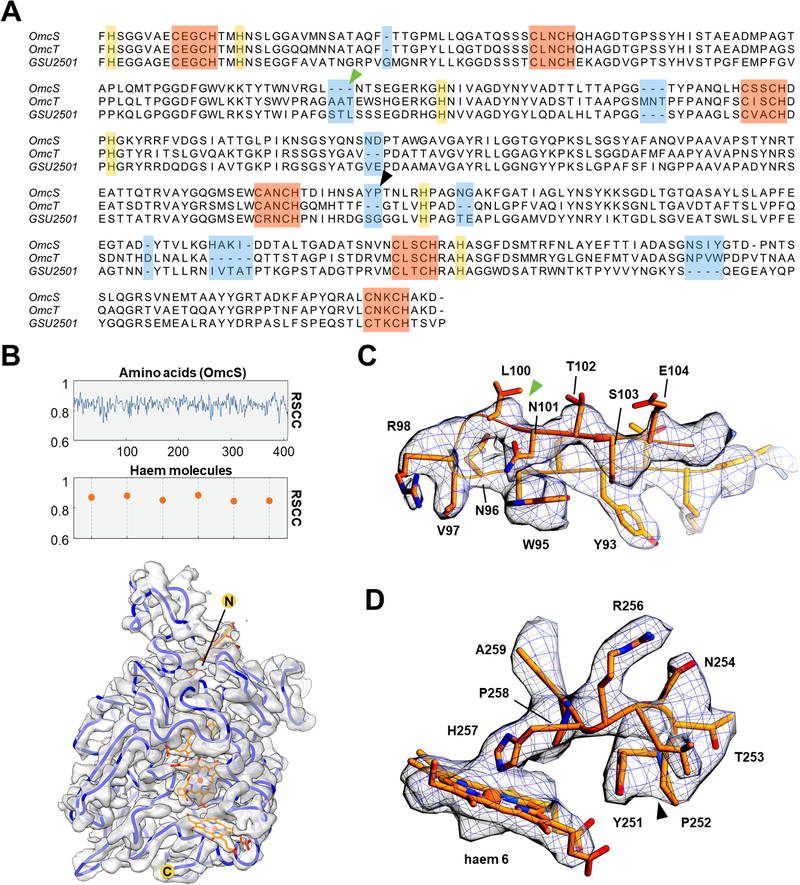
Cryo-EM images of filaments purified from the WT strain showed a sinusoidal morphology with a period of ~ 200 Å (Fig. 1A). Averaged power spectra from multiple filaments show a meridional layer line at ~ 1/(47 Å) (Fig. S4B), establishing that there are ~ 4.3 subunits per turn of a ~ 200 Å pitch 1-start helix (Fig. 1B). Using the iterative helical real space reconstruction (IHRSR) approach (Egelman, 2000), we were able to reach a resolution where the handedness of α-helices was clearly visible. The 1-start filament helix was left-handed, with a rise per subunit of 46.7 Å and a rotation of −83.1° (Fig. 1B), substantially different than type IV pili that typically show a rise of ~ 10 Å and a right-handed helix (Wang et al., 2017). The tracing of the Cα backbone of the protein subunit at this resolution revealed that the asymmetric unit contained at least 380 residues, which contrasts with the 61 residues present in PilA (Reardon and Mueller, 2013; Reguera et al., 2005). Further, there was no apparent internal symmetry that would arise in the asymmetric unit if it contained multiple copies of identical chains. The NMR structure of PilA (Reardon and Mueller, 2013) also failed to fit into the observed EM density map. Surprisingly, we found that there were six hemes per asymmetric unit, with the highest densities in the volume at the centres of these hemes, suggesting the presence of metal atoms. Given that the protein contains at least 380 residues and would therefore have a likely molecular weight between 40–50 kDa, we used SDS-PAGE and cut out the strongest band in the gel, which was at ~ 45 kDa (Fig. S1A), analyzing this band by mass spectrometry. Five proteins were identified, four of which had heme-binding motifs (CXXCH) and had masses between 45 and 49 kDa (Fig. S1B). Three of these proteins (OmcS, OmcT and GSU2501) had a very similar pattern of heme-binding motifs that approximately matched the initial Cα trace (Fig. 2B), and their sequences could be easily aligned (Fig. 2A). These three proteins had between 45–63% sequence identity with each other. Another protein found in mass spectrometry, OmcZ (Inoue et al., 2010), contained ~ 30 additional residues compared to the other three, and thus its sequence could not be aligned to the others, and furthermore the pattern of eight heme-binding motifs in OmcZ did not match that found in the map. The remaining protein, OmpJ, contains no heme (Afkar et al., 2005). Of the three possible candidates, only the OmcS sequence could be threaded through the map without any conflicts (Figure 2C,D, Fig. S2). There are nine regions of OmcT, GSU2501, or both which prevented their sequences from being fit into the map (Fig. 2A highlighted in blue, Fig. S2). In addition, the pattern of bulky amino acids clearly established that only the OmcS sequence was consistent with the map. For example, the elongated density from Arginine (R) 256 is clearly seen in the map (Fig. 2D). But in the sequences of OmcT and GSU2501, this residue would be a valine (Fig. 2A, highlighted in purple) and could not explain the map density. These studies show that OmcS is the only cytochrome found by mass spectrometry in our filament preparations in being consistent with the cryo-EM map.
Fig. 1. Structure of microbial nanowires reveals closely-stacked hemes in an OmcS filament.
(A) Cryo-EM image of the purified wild-type electrically-conductive filaments showing a sinusoidal undulation with a pitch of ~ 200 Å shown in (B). Scale bar for (A), 200 Å. (B) The surface of the reconstruction (transparent gray) with superimposed ribbon models of the OmcS subunits with three subunits in the center in three different colors. (C) Each subunit contains six hemes closely stacked over the micrometer-lengths of the filaments. (D) A zoomed region of the box shown in (C) with the minimum observed edge-to-edge distances indicated between hemes numbered in circles. The distance between two hemes in adjacent subunits (heme 1 and heme 6′) is comparable to the distances between parallel stacked hemes within a subunit (heme 2:heme 3 and heme 4:heme 5).
Fig. 2. De novo atomic model building of OmcS filaments.
(A) Sequence-based alignment of OmcS and two other c-type cytochromes with similar molecular weight detected by mass spectroscopy (Fig. S1B). The six conserved CXXCH motifs responsible for heme binding are highlighted in red. The histidine residues paired with the CXXCH motifs in heme binding are highlighted in yellow. Regions with insertions or deletions compared to the OmcS sequence are highlighted in blue. (B) The per-residue real space correlation coefficient (RSCC) plot of the atomic model against the 3.7 Å cryo-EM map (top), with protein and ligand displayed separately. The protein Cα trace in blue with ligands (bottom), with N- and C-termini labeled. (C) and (D) Zoomed view of the regions indicated in (A) by green and black arrowheads, respectively, with the OmcS atomic model fit into the cryo-EM map. The green arrowhead in (C) indicates the location where the two other cytochromes (OmcT, GSU2501) show a three-residue insertion, not compatible with the map. The black arrowhead in (D) indicates a region where OmcT has a two-residue deletion, and GSU2501 has a serine and glycine rather than the tyrosine and proline found in OmcS. The map has extra density that could not be explained by a two-residue deletion or by serine and glycine.
We built an ab initio atomic model of OmcS, which was then used to refine a filament model. We directly estimated its resolution using a map:model comparison (Neumann et al., 2018; Subramaniam et al., 2016) which yielded an estimate of 3.7 Å (Fig. S3A). Strikingly, the “gold standard” measure of resolution employed by Relion (Scheres, 2012), which is based upon reproducibility, yielded an estimate of 3.2 Å. Comparing the Relion map (filtered to 3.2 Å) and our map generated in Spider (filtered to 3.6 Å) showed no significant differences (Fig. S3B,D) suggesting that the Relion estimate of resolution is overly optimistic. Overall, the fit of the model to the map (Fig. S3E,F), the good density for many sidechains, and the refinement statistics (Table 1) all validate our model of the OmcS filament.
Table 1:
Refinement statistics for the OmcS model
| OmcS model | |
|---|---|
| Helical symmetry | |
| Rise (Å) | 46.7 |
| Resolution (Å) | 3.7 |
| Model to map CC | 0.82 |
| Clash score, all atoms | 12.8 |
| Protein geometry | |
| Ramachandran favored (%) | 88.2 |
| Ramachandran outliers (%) | 0.4 |
| Rotamer outliers (%) | 0.0 |
| Cβ deviations > 0.25 Å | 0 |
| RMS deviations | |
| Bond (Å) | 0.01 |
| Angles (°) | 1.36 |
| PDB ID | 6EF8 |
| EMDB ID | EMD-9046 |
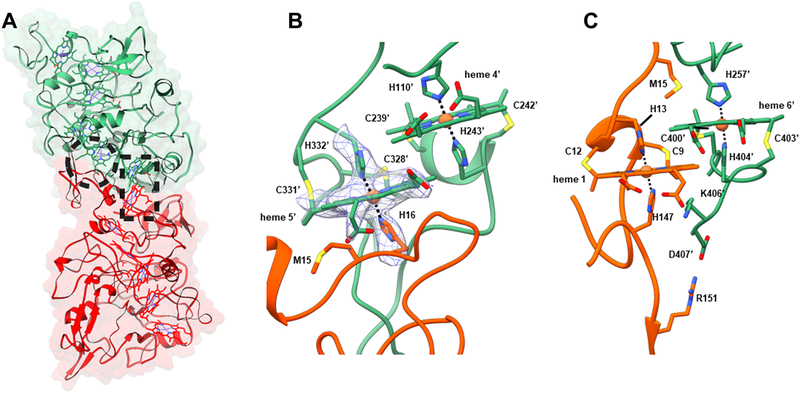
While it has been known for more than half a century that cytochromes can polymerize in ethanolic solutions (Margoliash and Lustgarten, 1962) and structures have been determined for aggregates up to tetramers (Hirota et al., 2010), natural polymerization of the type we observe here has not been previously described to our knowledge. Stacking arrangements of aromatic rings generally prefer parallel (offset face-to-face) or perpendicular (T-shaped) conformations (Janiak, 2000). The parallel stacking yields the highest electronic coupling, which maximizes electron transfer (Jiang et al., 2017), whereas the T-shape enhances structural stability (Janiak, 2000). Hemes in the OmcS nanowires form parallel-stacked pairs, with each pair perpendicular to the next, forming a continuous chain over the entire length of the filament (Fig. 1D). The minimum edge-to-edge distances between the parallel hemes is 3.4 to 4.1 Å, and 5.4 to 6.1 Å between the perpendicular stacked pairs. For all hemes in OmcS nanowires, two histidines axially coordinate iron at the centre of each heme and the vinyl groups of each heme form covalent thioether bonds with cysteines (Fig. 3). The bis-histidine axial ligation of the heme iron atoms are consistent with the coordination found in other c-type cytochromes (Clarke et al., 2011), and the cysteine linkages are consistent with the c-type hemes reported to occupy six heme-binding motifs of OmcS (Qian et al., 2011).
Fig. 3. Subunit interface interactions within OmcS filament.
(A) The large interface in the filament (~ 2,600 Å2 per subunit) is due to the complementarity between the upper portion of bottom subunit (red) and the lower portion of the top subunit (green). Residues in one subunit strongly interact via hemes shown in the dashed circle and rectangle and corresponding zoomed images are shown in (B) and (C) respectively. (B) Histidine 16 of the bottom subunit is coordinating the iron atom in heme 5′ of the top subunit. The cryo-EM densities corresponding to Histidine 16, Histidine 332’, and heme 5’ are shown in a mesh. (C) The stacking of heme 6′ from a top subunit with heme 1 from the subunit below.
Our OmcS model filament has a low percentage of α- and 310-helices (~ 13%) as well as β-strands (~ 6%), leaving ~ 81% of the model as turns and coil - consistent with previous secondary structure studies of OmcS monomers (Qian et al., 2011). We compared the OmcS protomer within the filament with a group of three crystallographic structures of other multi-heme c-type cytochromes (PDB IDs: 1ofw, 3ucp, and 3ov0). These structures showed 45, 49, and 60% turns and coils, respectively, with uniformly hydrophobic cores surrounding the hemes and heme-binding residues. Cores in our model of OmcS also included buried charges (Arginine at locations 333, 344 and 375) that lack proximal compensating charges, as well as buried sidechain hydroxyls (Tyrosine at locations 186, 231, 385). In addition, our model of OmcS has a salt bridge between protein chains, Aspartate 407 to Arginine 151, which, along with cysteines in heme binding motifs, are highly conserved amino acids in OmcS (Ashkenazy et al., 2016). The lack of structural homology seen with these other c-type cytochromes is consistent with the observation that there is no conserved fold for this family of proteins (Bertini et al., 2006).
The model shows that each OmcS subunit contacts only one subunit on either side, so that all connectivity in the filament is along the left-handed 1-start helix (Fig. 1B). The interface between adjacent subunits is extensive, with ~ 2,600 Å2 of surface area buried per subunit (Fig. 3A). In addition to the buried surface area, interactions between adjacent subunits incorporate additional stabilizing elements unique to the filament structure. Histidine 16 in each subunit coordinates the iron in heme 5 of an adjacent subunit (Fig. 3B). Furthermore, heme pairs at the interface are parallel rather than T-shaped with ~ 4 Å edge-to-edge distance (Figures 1D, 3B,C). This parallel stacking and inter-subunit coordination of heme may contribute substantially to the stability of the protein-protein interface. In addition, the presence of parallel-stacked hemes at the interface suggests facile transport of electrons between monomers.
There is no precedent for such seamless micrometer-long polymerization of hundreds of cytochromes to our knowledge. Based on previous studies that have shown that cytochromes could form tetramers, the OmcS polymerization could be due to successive domain swapping, where the c-terminal helix can be displaced from its original position in the monomer and histidine-heme coordination can be perturbed significantly (Hirota et al., 2010). We have determined the structure for the OmcS protomer within the filament, but the structure of isolated OmcS monomer is needed to provide insight into this surprising polymerization process.
The atomic structure was solved with filaments from conductive biofilms of electrode-grown cells that require long-distance electron transport. However, fumarate-grown cells can also produce conductive filaments (Ing et al., 2017; Leang et al., 2013; Leang et al., 2010; Malvankar et al., 2011; Reguera et al., 2005). Filaments purified with fumarate-grown cells showed similar structure to filaments purified from electrode-grown cells (Figures S4A,B). Moreover, previously published images of intact G. sulfurreducens filaments attached to cells (Leang et al., 2013) showed structural features similar to purified filaments such as an identical helical rise of 47 Å (Fig. S4C). These results showed that purified OmcS filaments are similar in dimensions and structure to cell-attached filaments that were previously thought to be type IV pili (Leang et al., 2013). Importantly, these studies also indicate that the formation of OmcS filaments is a natural process and not due to artificial preparation or pH conditions that can cause cytochrome c to form filamentous structures under extremely denaturing conditions (Haldar et al., 2014).
The cryo-EM images showed another filamentous structure that was thinner than the OmcS filament (Fig. S5). The averaged power spectrum of these filaments showed similar layer lines to the OmcS filament, but with a slightly different axial rise of ~ 57 Å and rotation of ~ 160 degrees, suggesting that this thinner filament is also not a type IV pilus and could potentially be another cytochrome filament. Due to much lower abundance of this filament, it was not possible to build an atomic model or to determine its composition. No filaments with power spectra consistent with type IV pili were observed in purified filaments preparations or in previously published images of intact, cell-attached filaments (Leang et al., 2013).
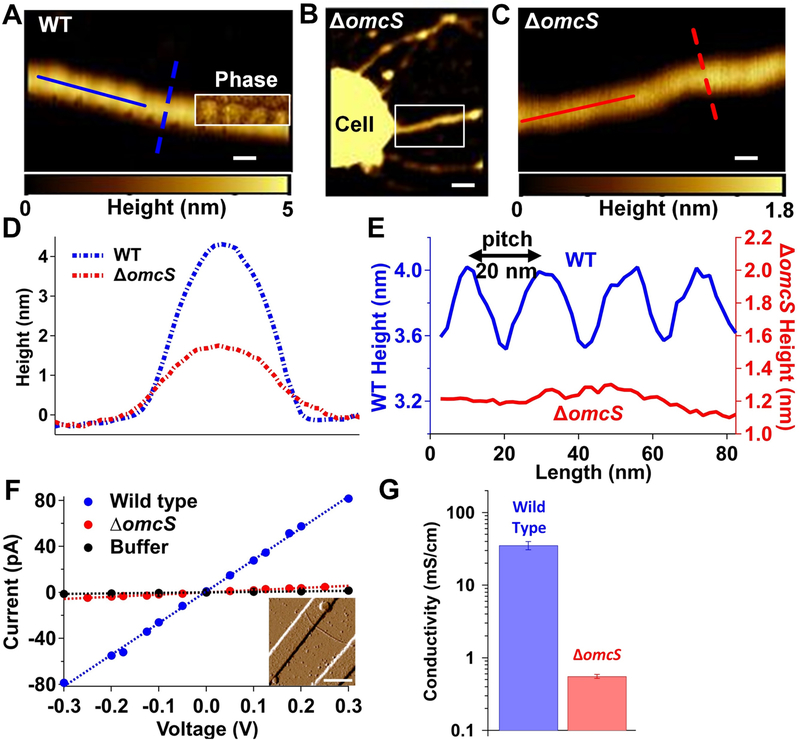
To evaluate the contribution of OmcS to the conductivity of these filaments, the direct current (DC) conductivity of individual OmcS filaments of WT strain were compared with sparse filaments produced by a ΔomcS strain. G. sulfurreducens forms a variety of filaments in response to genetic mutations (Klimes et al., 2010). Therefore, it is not surprising that the ΔomcS strain also forms filaments that were previously thought to be conductive type IV pili (Leang et al., 2010). Atomic force microscopy (AFM; Fig. 4A–E) revealed distinct structural features for WT G. sulfurreducens filaments versus the type IV pili of other species (Wang et al., 2017) and filaments from the ΔomcS strain (Leang et al., 2010). In contrast to the linear and smooth-surfaced structure of ΔomcS filaments, WT filaments exhibited an axial height periodicity with a 20-nm pitch (Fig. 4E), consistent with the helical pitch determined by cryo-EM (Fig. 1B). Filaments of the ΔomcS strain showed no apparent axial periodicity under AFM. Moreover, ΔomcS filament thickness (~1.7 nm height measured by AFM) was half that of WT filaments (~ 4 nm) (Fig. 4D,E). This substantial thickness difference and distinct axial periodicity observed for OmcS filaments was used to confirm that filaments studied for electrical measurements are the same OmcS nanowires characterized by cryo-EM.
Fig. 4. Electrical measurements show that OmcS is required for filament conductivity.
AFM height image of filaments from (A) WT strain (B) ΔomcS strain and (C) zoomed image of region shown in B. Inset in A is the phase image overlaid on height image that shows the repeating pattern. Scale bars: A, C, 20 nm; B, 100 nm. (D) The height profile for filaments of WT and ΔomcS strains using lateral cross section [dashed lines in (A) and (C)]. (E) Longitudinal height profile for filaments of WT and ΔomcS strain [solid lines in (A) and (C)]. (F) Current-voltage profile for individual filaments of WT and ΔomcS strain compared to buffer alone. Inset: AFM images for filaments by ΔomcS strain across gold electrodes. Scale bar, 500 nm. (G) Comparison of DC conductivity (left) and carrier density (right) for filaments of WT and ΔomcS strains. Error bars represent S.E.M of three biological replicates.
AFM was further used to locate an individual OmcS filament bridging two gold electrodes (Fig. 4F, inset). Our DC conductivity measurements of individual OmcS filaments fully hydrated in buffer yielded values comparable to previous measurements of WT filaments (Adhikari et al., 2016), further suggesting that OmcS filaments are identical to the WT nanowires discussed in previous studies as Type IV pili. DC conductivity measurements of individual ΔomcS filaments showed a very low conductivity that was more than 100-fold lower than OmcS filaments (Fig. 4F,G). Our conductivity measurements thus show that OmcS is required for high conductivity of these ~4 nm-thick filaments.
Our finding that G. sulfurreducens nanowires are OmcS filaments is consistent with previous physiological studies. These studies highlight the importance of OmcS in extracellular electron transfer (Holmes et al., 2006; Leang et al., 2013; Leang et al., 2010; Mehta et al., 2005; Summers et al., 2010). OmcS is one of the most abundant cytochromes found in the proteome for electricity-producing G. sulfurreducens and is required only during extracellular electron transfer to insoluble electron acceptors such as Fe (III) oxide (Holmes et al., 2006; Mehta et al., 2005). It is also critical for direct interspecies electron transfer between syntrophic Geobacter co-cultures as evolved co-culture overexpressed OmcS, and deletion of the omcS gene inhibited bacterial ability to exchange electrons (Summers et al., 2010).
OmcS also plays a critical role in long-range electron transport to electrodes in current-producing biofilms. Both microarray and quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis have demonstrated that cells show higher transcript levels for OmcS than for any other protein during the early stages of growth on electrodes (Holmes et al., 2006). Furthermore, immunogold localization has shown that OmcS is distributed throughout conductive G. sulfurreducens biofilms (Leang et al., 2013) and that deletion of the omcS gene inhibits electricity production (Holmes et al., 2006). However, the role of OmcS in conductivity was overlooked because ΔomcS biofilms were conductive and produced high current densities (Malvankar et al., 2011). These results might be due to a reciprocal relationship (Park and Kim, 2011) between the expression of OmcS and that of OmcZ, a cytochrome essential for current production (Nevin et al., 2009). The ΔomcS biofilms may compensate for the loss of OmcS by increasing the production of OmcZ.
Using immunogold localization, previous studies found that OmcS is associated with filaments (Leang et al., 2013; Leang et al., 2010; Summers et al., 2010). AFM images of filaments (Malvankar et al., 2012) as well as these antibody-labelling results (Leang et al., 2013; Leang et al., 2010; Summers et al., 2010) were interpreted as showing isolated OmcS monomers binding to the surfaces of the PilA filaments rather than showing antibodies directly binding to OmcS filaments. In light of the result presented here, a reinterpretation of these previous studies suggests that the antibodies may have been directly binding to the subunits of the OmcS filaments.
While no evidence of PilA was found in the structure of any of the purified filaments, non-filamentous PilA was present in our samples in lower abundance compared to OmcS (Fig. S1A), consistent with prior studies (Tan et al., 2016b). Multiple studies have shown that PilA is required for secretion of OmcS to the extracellular environment, as pilA deletion eliminated the presence of OmcS in outer-surface preparations (Liu et al., 2018; Richter et al., 2012). Overexpression of PilA is often accompanied by overproduction of OmcS and filaments (Leang et al., 2013; Summers et al., 2010), further suggesting that PilA is involved in secretion of OmcS filaments and explaining previous correlations found between PilA and biofilm conductivity (Malvankar et al., 2011). The requirement of PilA for the synthesis of OmcS filaments thus also explains the inability of ΔpilA cells to grow on insoluble electron acceptors such as Fe (III) oxides (Reguera et al., 2005) and electrodes (Reguera et al., 2006). A number of other bacteria have also been shown to require non-filamentous type IV pilins for the secretion of extracellular proteins (Hager et al., 2006). One possibility is that PilA is a pseudopilin as a part of a Type 2 Secretion System (T2SS) (Nivaskumar and Francetic, 2014), and OmcS is exported by this T2SS. Previous studies have shown that T2SS pseudopili can be secreted outside the cell (Nivaskumar and Francetic, 2014; Vignon et al., 2003). A similar mechanism could explain the presence of non-filamentous PilA in our filament preparations.
This potential ability of PilA to regulate the secretion and assembly of the OmcS nanowires, as well as that of other multi-heme cytochromes (Liu et al., 2018; Richter et al., 2012), could explain how point mutations in pilA caused cells to produce filaments with different conductivities than wild-type (WT) filaments. Filaments produced by cells with point mutations in pilA showed very different morphology than WT filaments (Tan et al., 2016a). For example, substitution of two residues in PilA with tryptophan yielded mutant cells with filaments which surprisingly had half the diameter of WT filaments (Tan et al., 2016a). Based on our finding that conductive filaments are composed of cytochromes, this very large structural change in the filaments of mutant strains suggests that the observed change in conductivity may be due to filaments of different cytochromes or of different conformations of OmcS. Further structural studies on the filaments produced by these pilA mutants are needed to fully reconcile these studies with our finding of OmcS filaments functioning as nanowires.
In summary, our findings show that conductive G. sulfurreducens filaments are polymerized chains of OmcS. The filament structure has hemes closely stacked along the micrometer-length of the filament, establishing the molecular basis for electronic conductivity in these nanowires. Functional characterization of conductivity in individual filaments shows that OmcS is required for the filament conductivity. The structure presented here provides insights into supramolecular protein nanowires, explaining the remarkable capacity of soil bacteria to transport electrons to extracellular electron acceptors for respiration and for sharing of energy and nutrients (Malvankar et al., 2011) with syntrophic partners (Summers et al., 2010) that are hundreds of micrometers away. The advances in understanding of the structural basis for conductivity in microbial nanowires presented here can provide design principles for development of future bioelectronic interfaces between living cells and devices.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Nikhil S. Malvankar at Nikhil.Malvankar@yale.edu
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Bacterial strains and growth conditions.
The Geobacter sulfurreducens wild-type (WT) strain DL1 (ATCC 51573, DSMZ 12127) (Coppi et al., 2001) and the omcS deletion mutant strain (Mehta et al., 2005) (designated ΔomcS) were obtained from our laboratory culture collection. The cultures were maintained at 30 °C or at 25 °C under strictly anaerobic conditions in growth medium supplemented with acetate (10 mM) as the electron donor and fumarate (40 mM) as the electron acceptor in sterilized and degassed NBAF medium (O’Brien and Malvankar, 2017). 1L NBAF medium contained the following: 0.04 g/L calcium chloride dihydrate, 0.1 g/L, magnesium sulfate heptahydrate, 1.8 g/L sodium bicarbonate, 0.5 g/L sodium carbonate, 0.42 g/L potassium phosphate monobasic, 0.22 g/L potassium phosphate dibasic, 0.2 g/L ammonium chloride, 0.38 g/L potassium chloride, 0.36 g/L sodium chloride, vitamins and minerals as listed in (O’Brien and Malvankar, 2017). Resazurin was omitted and 1 mM cysteine was added as an electron scavenger. All chemicals obtained from Fisher Scientific unless otherwise noted. For filament samples used to build the atomic model, the wild-type strain was grown on electrodes under electron acceptor-limiting conditions that induce filament expression (O’Brien and Malvankar, 2017) (Fig. 1A).
METHODS DETAILS
G. sulfurreducens filament preparation and biochemical characterization.
Filaments were separated from bacteria and extracted via centrifugation (Tan et al., 2016a) and maintained in 150 mM ethanolamine buffer at pH 10.5 in a manner similar to structural studies on bacterial filaments (Wang et al., 2017). Cells were gently scraped from the surface using a plastic spatula and isotonic wash buffer (20.02 × 10−3 M morpholinepropanesulfonic acid, 4.35 × 10−3 M NaH2 PO4 ·H2O, 1.34 ×10 −3 M KCl, 85.56 ×10−3 M NaCl, 1.22 × 10−3 M MgSO4 ·7H2O, and 0.07 × 10 −3 M CaCl2 ·2H2O), then collected by centrifugation and re-suspended in 150 × 10−3 M ethanolamine (pH 10.5). Filaments were mechanically sheared from the cell surface using a Waring Commercial Blender (Cat. No. 7011S) at low speed for 1 min, and then cells were removed by centrifugation at 13,000 g before collecting filaments with an overnight 10% ammonium sulfate precipitation and subsequent centrifugation at 13,000 g. Collected filaments were re-suspended in ethanolamine buffer then cleaned by centrifugation at 23,000 g to remove debris and a second 10% ammonium sulfate precipitation with centrifugation at 13,000 g (Tan et al., 2016a). The final filament preparation was re-suspended in 200 μl ethanolamine buffer. Filament preparations were further passed through 0.2 μm filters to remove any residual cells and stored at 4 °C. Cell-free filament preparations were imaged first with transmission electron microscopy to ensure sample quality (Fig. 1A). Dilute 5 μl solutions containing filaments were placed on gold electrodes to achieve individual filaments across two gold electrodes (Fig. 4F, Inset). Prior to all measurements, filaments were imaged with AFM and height measurements were performed to confirm the presence of individual filaments. For all conductivity measurements, samples were maintained under hydrated buffer environments (150 mM ethanolamine) and the pH of the buffer was equilibrated to pH 7 using HCl (Malvankar et al., 2011). Transmission electron microscopy imaging was used to confirm that filaments maintain their structure and remain morphologically similar at all pH conditions.
For polyacrylamide gel electrophoretic separation (SDS-PAGE) (Fig. S1A), all filament samples were prepared in ethanolamine buffer and dried in a speedvac. Samples were then resuspended in 500 μl ultrapure deionized water and centrifuged at 18,000 × g at 4°C for 1 hour. The pellets were dried again in the speedvac, resuspended in water and dried repetitively 2 times. The final dried samples were resuspended in 5–8 μl 2.5% sodium-dodecyl sulphate (SDS) to disassemble filaments into their constituent monomers. The samples were incubated at room temperature for at least 4 hours, then diluted 3-fold with deionized water. The denatured samples were boiled in 1X SDS sample buffer that included b-mercaptoethanol for 12 min. The samples were run on a 16% Tricine protein gel (ThermoFisher Scientific, Carlsbad, CA) initially at constant voltage of 30 V for 18 minutes before changing to 190 V for 12 minutes. Precision Plus Protein Prestained molecular weight standards (BioRad, Hercules, CA) and Low Range Protein Ladder (Thermo Scientific) were used to compare the molecular weight of PilA and cytochromes in the filament preparations. Gels were immediately washed at least 3 times with ultra-pure deionized water over a 1-hour period, stained with Coomassie R-250 stain (Thermo Scientific, Rockford, IL), and destained overnight.
Custom monospecific anti-PilA antibody was synthesized by Pacific Immunology (Ramona, CA) by immunizing two rabbits with synthetic peptide sequence containing targeted epitope on the native protein, PilA, and then affinity purifying the serum against that peptide sequence. Specificity of antibody in the serum was confirmed by ELISA after 1st stage of immune response and then verified again with purified antibody (1:125,000 titer) at the final stage. Synthetic peptide sequence contained following 21 amino acids from C-terminus of PilA, Cys-RNLKTALESAFADDQTYPPES, to which a cysteine was added to N-terminus and then was conjugated with Keyhole Limpet Hemocyanin prior to immunization. The molecular weight of the synthetic peptide was verified by HPLC and ESI-MS.
For LC-MS/MS analysis of filaments (Fig. S1B), the PilA band (~ 6.5 kDa) and OmcS bands (~ 50 kDa and 30 kDa) were extracted from the protein gel and treated with trypsin to digest the protein (Fig. S1). Proteomic analysis of the cleaved peptides from filaments of electrode-grown cells) was performed by the Proteomics Mass Spectrometry Facilities at Yale University Results gave unique amino acid sequence matches with the OmcS and c-terminal domain of PilA (Fig. S1) whereas the hydrophobicity of the N-terminal domain may have interfered with the sequence detection procedure (Ing et al., 2017).
Filament preparation from fumarate-grown cells (Fig. S4A).
G. sulfurreducens was grown and used for filament purification (Ing et al., 2017). Briefly, G. sulfurreducens (DSMZ strain 12127) was grown anaerobically in 1 L cultures of sterilized and degassed NBAF medium (0.04 g/L calcium chloride dihydrate, 0.1 g/L magnesium sulphate heptahydrate, 1.8 g/L sodium bicarbonate, 0.5 g/L sodium carbonate, 0.42 g/L potassium phosphate monobasic, 0.22 g/L potassium phosphate dibasic, 0.2 g/L ammonium chloride, 0.38 g/L potassium chloride, 0.36 g/L sodium chloride, vitamins and minerals, using 20 mM acetate as the electron donor and 40 mM fumarate as the electron acceptor). Resazurin was omitted and 1mM cysteine was added as an electron scavenger. All chemicals were obtained from Fisher Scientific unless otherwise noted.
Upon reaching stationary phase, cells were pelleted by centrifugation at 5000 × g for 20 min at 4° C. Cell pellets were removed and extracellular filaments were purified from the supernatant through dropwise additions of 1.1 M aqueous MgCl2, which was added to a final ratio of 1:10 for aqueous MgCl2:supernatant. Supernatants were stored at 4° C overnight and centrifuged at 50,000 × g for 1 h at 4° C to precipitate extracellular filaments. Pellets of extracellular filaments were resuspended with ultrapure water adjusted to pH 4.3 with HCl and passed through 0.2 μm filters. Additional contaminants were removed from the resuspended pellets using 50 kDa dialysis tubing. After two 1 L exchanges of pH 4.3 water, the dialyzed sample was removed from dialysis tubing and stored at 4° C.
For analysis of filaments from fumarate-grown cells (Fig. S4), the purified filament samples were analysed for protein content by 12.5% PhastGel (GE Healthcare) and stained with Coomassie. There were two prominent protein bands around 47 kDa, and these were cut from the gel, digested by trypsin overnight, and then analysed by mass spec at the University of Virginia core facility. The LC-MS system consisted of a Thermo Electron Q Exactive HFX mass spectrometer system with an Easy Spray ion source connected to a Thermo 75 μm × 15 cm C18 Easy Spray column. 5 μL of the extract was injected and the peptides eluted from the column by an acetonitrile/0.1 M formic acid gradient at a flow rate of 0.3 μL/min over 2.0 hours. The nanospray ion source was operated at 1.9 kV. The digest was analysed using the rapid switching capability of the instrument acquiring a full scan mass spectrum to determine peptide molecular weights followed by product ion spectra (10 HCD) to determine amino acid sequence in sequential scans. This mode of analysis produces approximately 60,000 MS/MS spectra of ions ranging in abundance over several orders of magnitude.
Prior to use, purified filaments were characterized by MALDI-TOF. For MALDI-TOF analysis, filaments were denatured with 10% OG overnight and then diluted with ultrapure water to a final OG concentration of 2% prior to the addition of matrix solution (α-Cyano-4-hydroxycinnamic acid dissolved in a 2:1 solution of ultrapure water: acetonitrile and 0.2% trifluoroacetic acid). The final sample for MALDI was a 12:7:5 ratio of matrix solution to ultrapure water to diluted and denatured protein. Mass spectra were collected in positive ion mode.
Cryo-EM sample preparation conditions.
For samples used to build the atomic model, holey carbon coated Quantifoil grids (R 2μm/2μm) were used. Prior to use, the grids were floated in 0.05% Triton 100X solution (Cheung et al., 2013). Cryo-EM specimens were prepared with a FEI Vitrobot Mark IV at 22 °C with 100% humidity. 3 μL of sample solution containing WT filaments were dropped on the grids and spread gently by pipette tip before loading to the Vitrobot, blotted for 5.5 s and plunged freezing in liquid ethane.
Cryo-EM data collection conditions.
Micrographs were acquired by FEI Titan Krios electron microscopy performed at 300kV equipped with a Gatan K2 summit camera. Quantum energy filter with a slit width at 20 eV was applied to remove inelastically scattered electrons. Movies were collected using super-resolution imaging mode with a physical pixel size of 1.05 Å and an exposure rate of 6.8 electrons per pixel per second. A total exposure time of 9.75 seconds was fractioned into 30 frames and 1447 movies were generated using serial EM auto-collection. The data collection and processing parameters are summarized below:
| Magnification | 130,000 X |
| Voltage (kV) | 300 |
| Electron exposure (e−/Å2) | 63 |
| Energy filter slit width | 20 eV |
| Physical Pixel Size (Å) | 1.05 |
| Defocus range (μm) | 0.5 – 3.0 |
For analysis of filaments from fumarate-grown cells (Fig. S4), sample (4 μL, ~1 μg/μl) was applied to discharged lacey carbon grids and plunged frozen using a Vitrobot Mark IV (FEI, Inc.). Grids were imaged in a Titan Krios at 300 keV and recorded with a Falcon III direct electron detector at 1.4 Å per pixel. Micrographs were collected using a defocus range of 1.25–2.25 μm, with a total exposure of 2 s (amounting to ~ 60 electrons/Å2) distributed into 24 fractions. All the micrographs were first motion corrected (ignoring the first fraction) using MotionCorr (Li et al., 2013) version 2 and then used for CTF estimation by the CTFFIND3 program (Mindell and Grigorieff, 2003).
Cryo-EM image analysis.
For filaments of fumarate-grown cells (Fig. S4), images were extracted using the e2helixboxer program within EMAN2 (Tang et al., 2007) from the dose-weighted fractions 2–10 (amounting to ~ 20 electrons/Å2), after the images were corrected for the CTF through multiplication by the theoretical CTF (a Wiener filter in the limit of a very poor SNR). A total of 17,800 overlapping 384-px long segments (with a shift of 55 pixels, ~ 1.5 times the axial rise per subunit) were generated. The determination of the helical symmetry was unambiguous given the 1/(47 Å) meridional layer-line and the 1-start helix of ~ 200 Å pitch. A reconstruction was generated using the IHRSR method implemented in Spider (Egelman, 2000), and this reconstruction was subsequently filtered to 20 Å for the starting reference in the reconstruction of the second dataset. To build the atomic model of filaments from electrode-grown cells, a total of 573 images were selected, motion-corrected, dose weighted (amounting to ~ 20 electrons/Å2) and CTF-corrected in the same manner as for filaments of electrode-grown cells. About 2,000 long filaments were extracted using e2helixboxer and then 384-pixel long 28,293 overlapping segments with a shift of 60 pixels were generated. The final volume from the IHRSR reconstruction was estimated to have a resolution of ~ 3.7 Å based on the model:map FSC (Fig. S3). It was filtered to 3.6 Å and sharpened with a negative B-factor of 100. Micrographs with all fractions and boxing coordinates were also imported into Relion for 3D reconstruction. A similar final reconstruction to the one from SPIDER was generated in Relion using the same helical symmetry, starting with the SPIDER volume filtered to 10 Å. Interestingly this reconstruction was estimated to have a resolution of 3.2 Å resolution based on a “gold standard” map:map FSC.
Model building of OmcS filaments.
First, about 400 protein Cα atoms and six heme cofactors corresponding to a single subunit within the filaments were built manually in Coot (Emsley and Cowtan, 2004). Then the density corresponding to a single subunit was segmented from the experimental filament density using Chimera (Pettersen et al., 2004). Using the proteins identified in the MS/MS results and real space fitting to the cryo-EM density, the sequence of the filament was unambiguously determined to be cytochrome OmcS. The full length OmcS protein as well as six heme molecules were then built manually in Coot. Then the OmcS/heme was rebuilt with the RosettaCM protocol (Wang et al., 2015) to remove bad geometries. A total of 3,000 full-length models were generated and the top 20 models were selected based on Rosetta’s energy function, Ramachandran plots and overall fit to the map. These 20 models were then combined into one model by manual editing in Coot using the criteria of the local fit to the density map and the geometry statistics of the model. To better refine heme interacting areas at this resolution, bond/angle restraints for the heme molecule itself, His-Fe, and Cys-heme thioester bonds were created based on high resolution cytochrome c3 crystal structures NrfB (PDB 2P0B) and NrfHA (PDB 2J7A). Then a filament model was generated and further refined using Phenix with additional heme area restraints, and MolProbity (Williams et al., 2018) was used to evaluate the quality of the filament model. The refinement statistics are given in Table 1.
Atomic force microscopy (AFM).
To visualize individual filaments, 5 μl of buffer solution containing filaments were deposited on mica or on a silicon wafer insulated by a 100 nm silicon dioxide dielectric layer with gold electrodes patterned by nanoimprint lithography (Tan et al., 2016a). For nanoimprinting, the substrate was cleaned with a Piranha solution (H2SO4:H2O2 = 3:1) and a diluted Hydrofluoric acid solution before patterning. Two layers of resists (50-nm-thick poly methyl methacrylate and 60-nm-thick UV-curable resist) were then sequentially spin-coated onto the cleaned substrate. Circuit patterns including nanoelectrodes separated by nano-sized gaps, microscale fan outs, and contact pads were transferred from a quartz mold to the UV resist with nanoimprint lithography in a homemade imprint chamber. The residual UV-resist layer and the poly methyl methacrylate underlayer were removed by reactive ion etching with fluorine based (CHF3/O2) and oxygen-based gases respectively. The excess buffer was absorbed with filter paper. The sample was air-dried and was mounted on a metal puck (Oxford Instrument, Cypher ES). AFM experiments were performed using soft cantilevers (ASYELEC-01, Oxford Instrument Co.) with a nominal force constant of 2 N/m and resonance frequencies of 70 KHz. The free-air amplitude of the tip was calibrated with the Asylum Research software and the spring constant was captured by the thermal vibration method. The sample was imaged with a Cypher ES scanner using intermittent tapping (AC-air topography) mode. AFM showed that gold electrodes were bridged with individual filaments to facilitate conductivity measurements (Fig. 4F, inset). To visualize helical features of filaments, higher-resolution AFM was performed in attractive force imaging mode.
Direct current (DC) conductivity measurements.
All DC conductivity measurements of filaments were performed under fully hydrated buffer conditions in a 2-electrode configuration inside a triaxially shielded dark box using a probe station (MPI Corp.) connected to a semiconductor parameter analyser (Keithley 4200A-SCS) equipped with preamplifiers, allowing 0.1 fA current resolution and 0.5 μV voltage resolution. A DC voltage, typically in the range of −0.5V to +0.5V, was applied between the two electrodes and the current was measured over a minimum period of 120 seconds until the steady state was reached. The linearity of the I-V characteristics was maintained by applying an appropriate low voltage and the slope of the I-V curve was used to determine the conductance (G). Measurements were performed at low voltages (< 0.5 V) and over longer times (> 100 seconds) to ensure a lack of electrochemical leakage currents or faradic currents as evidenced by the absence of significant DC conductivity in buffer or filaments of ΔomcS strain that were maintained under identical buffer conditions as filaments of the WT strain (Fig. 4F,G). All analysis was performed using IGOR Pro (WaveMetrics Inc.).
Conductivity calculations.
The conductivity (σ) of filaments was calculated using the relation (Malvankar et al., 2011) σ = G∙(L/A) where G is the conductance, L is the length of the filament, and A = πr2 is the area of cross section of filament with 2r as the height of the filament measured by AFM (Fig. 4F,G).
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification and statistical analyses employed in this publication pertain to the analysis on electron microscopy data and the determination of structures by electron microscopy, which are integral parts of existing algorithms and software used.
DATA AND SOFTWARE AVAILABILITY
All data for OmcS filament were deposited in EMDB and PDB with the following entry codes: EMDB: EMD-9046 and PDB: 6EF8.
Supplementary Material
Fig. S1. Biochemical characterizations of filament preparations show PilA and OmcS proteins, Related to Figures 1 and 2 (A) SDS-PAGE gel of filaments showing OmcS and PilA at expected molecular weights of ~ 45 kDa and ~ 6.5 kDa. LC-MS/MS analysis of metalloproteins in purified G. sulfurreducens filaments from (B) the ~ 45 kDa band and (C) the ~ 30 kDa band. EMPAI: Exponentially modified protein abundance index. LC-MS/MS analysis showing matched peptides in (D) OmcS in ~ 45 kDa band and (E) PilA in the ~ 6.5 kDa band. (F) Western immunoblotting using anti-PilA antibody showing PilA in purified filament preparations (left) and cell lysate (right).
Fig. S2. De novo atomic model building of OmcS filaments, Related to Figures 1 and 2 (A) Sequence-based alignment of OmcS and two other c-type cytochromes detected by mass spectrometry, with the same highlighted colors used in Fig 2A. Regions indicated by red, orange, yellow, green, cyan, blue and purple arrowheads are shown in (B)-(G), respectively, with OmcS atomic model fit into the cryo-EM map. Both OmcT and GSU2501 have deletions or insertions at those regions and cannot fit into the cryo-EM map.
Fig. S3. Illustration of cryo-EM map quality, Related to Figures 1 and 2 (A) The model:map FSC (Fourier Shell Correlation) calculation using a 0.38 criterion, which is sqrt (0.143), estimates the Spider map to have a resolution of 3.7 Å. (B) A view of amino acids 325–350 fit into the Spider map used in (A) after filtering the map to 3.6 Å. (C) The Relion map:map “gold standard” FSC using the 0.143 criterion estimates the Relion map to have a resolution of 3.2 Å. (D) A view of the same region as (B) but with the Relion map which has been filtered to 3.2 Å, shows that the Relion estimate of resolution is considerably over-optimistic. (E) Fitting of six heme co-factors into the corresponding cryo-EM densities. (F) The Histidine 51 interaction with heme 2 shown along with surrounding residues fit into the cryo-EM density.
Fig. S4. Power spectra of purified filaments from fumarate-grown strain and cell-attached filaments are similar to power spectra from filaments of the electrode-grown strain used to build the atomic model, Related to Figure 1 and 2 (A) OmcS filaments (black arrow) from the fumarate-grown strain. Some flagellar filaments (white arrow) are present in this strain. Scale bar, 500 Å. (B) The averaged power spectrum of the OmcS filaments from fumarate-grown cells (left) shows similar layer lines and helical symmetry as that from filaments of electrode-grown cells (right). (C) Power spectra generated from previously published images (Leang et al., 2013) of intact filaments attached to G. sulfurreducens cells. Left, the power spectrum of a bundle of filaments showing the meridional layer line (blue arrow) at 1/(47 Å). Because of the bundling, a strong sampling is seen on the near-equatorial layer line. Right, averaged power spectra from 13 isolated segments to avoid the sampling. These power spectra show that the cell-attached filaments have symmetry similar to that as observed in cryo-EM images of purified OmcS filaments, with a rotation of 84 degrees and a rise of 47 Å per subunit.
Fig. S5. The thinner filament observed in cryo-EM imaging, Related to Figure 1 (A) The cryo-EM images frequently showed a thinner filament (white arrow) compared to OmcS filaments (black arrow). Scale bar, 200 Å. (B) The averaged power spectrum of the thinner filaments shows layer lines similar to those from the OmcS filament, but with a somewhat different axial rise of ~ 57 Å and rotation of ~ 160°. (C) A helical reconstruction of the thinner filament filtered to 20 Å. (D) The projection of the reconstruction in (C) with a low threshold (left) and a high threshold (right). The high threshold shows strong peaks separated by approximately 10 Å, labelled by yellow dots, which could be hemes.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monospecific antibody anti-PilA | This paper | N/A |
| Bacterial and Virus Strains | ||
| Geobacter sulfurreducens wild-type strain DL1 | Coppi et al., 2001 | ATCC 51573 |
| Geobacter sulfurreducens wild-type strain PCA | Ing et al., 2017 | DSMZ 12 127 |
| Geobacter sulfurreducens omcS deletion mutant strain (designated ΔomcS) | Mehta et al., 2005 | N/A |
| Biological Samples | ||
| Purified OmcS filaments from electrode-grown cells | This paper | N/A |
| Purified OmcS filaments from fumarate-grown cells | Ing et al., 2017 | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Common lab reagents | N/A | N/A |
| NBAF growth Medium | O’Brien and Malvankar, 2017 | N/A |
| Trypsin | Sigma-Aldrich | Cat #: T7409 |
| Ethanolamine | Sigma-Aldrich | Cat #: E9508 |
| Sodium-dodecyl sulphate | Ambion, Inc | Cat #: AM9822 |
| Deposited Data | ||
| OmcS filament Cryo-EM map | This paper | EMDB: EMD-9046 |
| OmcS filament structure | This paper | PDB: 6EF8 |
| Software and Algorithms | ||
| Spider package | Egelman, 2000 | http://spider.wadsworth.org/spider_doc/spider/docs/spider.html |
| CTFFIND3 | Mindell and Grigorieff, 2003 | http://grigoriefflab.janelia.org/ctf |
| EMAN2 | Tang et al., 2007 | http://blake.bcm.tmc.edu/emanwiki/EMAN2 |
| IHRSR | Egelman, 2000 | N/A |
| COOT | Emsley and Cowtan, 2004 | http://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| CHIMERA | Pettersen et al., 2004 | http://www.rbvi.ucsf.edu/chimera/ |
| MolProbity | Williams et al., 2018 | http://molprobity.biochem.duke.edu/ |
| Relion | Scheres, 2012 | https://www2.mrc-lmb.cam.ac.uk/relion |
| RosettaCM | Wang et al., 2015 | https://www.rosettacommons.org/docs/latest/application_documentation/structure_prediction/RosettaCM |
| Other | ||
| Cypher ES Atomic Force Microscope | Oxford Instrument | N/A |
| Probe station with triaxially shielded dark box | MPI Corp. | N/A |
| semiconductor parameter analyser | Keithley | 4200A-SCS |
Acknowledgments
Y.G. and N.S.M. thank Derek Lovley for providing strains, Eric Martz, Ruchi Jain, Danial Shapiro, Atanu Acharya and Yong Xiong for helpful discussions, and Shenping Wu as well as Marc Llaguno for help with cryo-EM. We also thank Hao Jiang and Qiangfei Xia for providing nanoelectrodes and Olivera Francetic for helpful discussions about the T2SS.This research was partially supported by the Career Award at the Scientific Interfaces from Burroughs Welcome Fund (to N.S.M.), the National Institutes of Health Director’s New Innovator award (1DP2AI138259–01 to N.S.M.), NIH R35GM122510 (to E.H.E.), the NSF CAREER award no. 1749662 (to N.S.M.). Research was sponsored by the Defense Advanced Research Project Agency (DARPA) Army Research Office (ARO) and was accomplished under Cooperative Agreement Number W911NF-18-2-0100 (with N.S.M.). This research was supported NSF Graduate Research Fellowship awards 2017224445 (to J.P.O.) and DGE-1321846 (to N.L.I.), the Air Force Office of Scientific Research, grant FA9550-14-1-0350 (to A.I.H.). Research in the Malvankar laboratory is also supported by the Charles H. Hood Foundation Child Health Research Award, and the Hartwell Foundation Individual Biomedical Research Award. The cryo-EM imaging of filaments from fumarate-grown cells, conducted at the Molecular Electron Microscopy Core facility at the University of Virginia, was supported by the School of Medicine and built with NIH grant G20-RR31199. The Titan Krios and Falcon II direct electron detector were obtained with NIH S10-RR025067 and S10-OD018149, respectively. The cryo-EM imaging of filaments used to solve the atomic structure was conducted at the Yale University’s Cryo-EM resource facility.
Footnotes
Declaration of Interests
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information includes five figures and can be found with this article online at http://dx.doi.org/XXXX/j.cell.2019.XXX
References
- Adhikari R, Malvankar N, Tuominen M, and Lovley D (2016). Conductivity of individual Geobacter pili. RSC Advances 6, 8354–8357. [Google Scholar]
- Afkar E, Reguera G, Schiffer M, and Lovley DR (2005). A novel Geobacteraceae-specific outer membrane protein J (OmpJ) is essential for electron transport to Fe (III) and Mn (IV) oxides in Geobacter sulfurreducens. BMC microbiology 5, 41–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, and Ben-Tal N (2016). ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Research 44, W344–W350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertini I, Cavallaro G, and Rosato A (2006). Cytochrome c: occurrence and functions. Chemical Reviews 106, 90–115. [DOI] [PubMed] [Google Scholar]
- Cheung M, Kajimura N, Makino F, Ashihara M, Miyata T, Kato T, Namba K, and Blocker AJ (2013). A method to achieve homogeneous dispersion of large transmembrane complexes within the holes of carbon films for electron cryomicroscopy. Journal of Structural Biology 182, 51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childers SE, Ciufo S, and Lovley DR (2002). Geobacter metallireducens accesses insoluble Fe(iii) oxide by chemotaxis. Nature 416, 767–769. [DOI] [PubMed] [Google Scholar]
- Clarke TA, Edwards MJ, Gates AJ, Hall A, White GF, Bradley J, Reardon CL, Shi L, Beliaev AS, Marshall MJ, et al. (2011). Structure of a bacterial cell surface decaheme electron conduit. Proceedings of the National Academy of Sciences 108, 9384–9389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppi MV, Leang C, Sandler SJ, and Lovley DR (2001). Development of a genetic system for Geobacter sulfurreducens. Applied and Environmental Microbiology 67, 3180–3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelman EH (2000). A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy 85, 225–234. [DOI] [PubMed] [Google Scholar]
- Emsley P, and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta Crystallographica Section D 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Hager AJ, Bolton DL, Pelletier MR, Brittnacher MJ, Gallagher LA, Kaul R, Skerrett SJ, Miller SI, and Guina T (2006). Type IV pili-mediated secretion modulates Francisella virulence. Molecular Microbiology 62, 227–237. [DOI] [PubMed] [Google Scholar]
- Haldar S, Sil P, Thangamuniyandi M, and Chattopadhyay K (2014). Conversion of amyloid fibrils of cytochrome c to mature nanorods through a honeycomb morphology. Langmuir 31, 4213–4223. [DOI] [PubMed] [Google Scholar]
- Hirota S, Hattori Y, Nagao S, Taketa M, Komori H, Kamikubo H, Wang Z, Takahashi I, Negi S, and Sugiura Y (2010). Cytochrome c polymerization by successive domain swapping at the C-terminal helix. Proceedings of the National Academy of Sciences 107, 12854–12859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes DE, Chaudhuri SK, Nevin KP, Mehta T, Methe BA, Liu A, Ward JE, Woodard TL, Webster J, and Lovley DR (2006). Microarray and genetic analysis of electron transfer to electrodes in Geobacter sulfurreducens. Environmental Microbiology 8, 1805–1815. [DOI] [PubMed] [Google Scholar]
- Ing NL, Nusca TD, and Hochbaum AI (2017). Geobacter sulfurreducens pili support ohmic electronic conduction in aqueous solution. Physical Chemistry Chemical Physics 19, 21791–21799. [DOI] [PubMed] [Google Scholar]
- Inoue K, Qian X, Morgado L, Kim B, Mester T, Izallalen M, Salgueiro C, and Lovley D (2010). Purification and characterization of omcZ, an outer-surface, octaheme c-type cytochrome essential for optimal current production by Geobacter sulfurreducens. Applied and Environmental Microbiology 76, 3999–4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janiak C (2000). A critical account on π-π stacking in metal complexes with aromatic nitrogen-containing ligands. Journal of the Chemical Society, Dalton Transactions, 3885–3896. [Google Scholar]
- Jiang X, Futera Z, Ali ME, Gajdos F, von Rudorff GF, Carof A, Breuer M, and Blumberger J (2017). Cysteine linkages accelerate electron flow through tetra-heme protein STC. Journal of the American Chemical Society 139, 17237–17240. [DOI] [PubMed] [Google Scholar]
- Klimes A, Franks AE, Glaven RH, Tran H, Barrett CL, Qiu Y, Zengler K, and Lovley DR (2010). Production of pilus-like filaments in Geobacter sulfurreducens in the absence of the type IV pilin protein PilA. FEMS Microbiology Letters 310, 62–68. [DOI] [PubMed] [Google Scholar]
- Leang C, Malvankar NS, Franks AE, Nevin KP, and Lovley DR (2013). Engineering Geobacter sulfurreducens to produce a highly cohesive conductive matrix with enhanced capacity for current production. Energy & Environmental Science 6, 1901–1908. [Google Scholar]
- Leang C, Qian XL, Mester T, and Lovley DR (2010). Alignment of the c-type cytochrome OmcS along pili of Geobacter sulfurreducens. Applied and Environmental Microbiology 76, 4080–4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, and Cheng Y (2013). Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nature Methods 10, 584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhuo S, Rensing C, and Zhou S (2018). Syntrophic growth with direct interspecies electron transfer between pili-free Geobacter species. The ISME Journal 12, 2142–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvankar NS, and Lovley DR (2014). Microbial nanowires for bioenergy applications. Current Opinion in Biotechnology 27, 88–95. [DOI] [PubMed] [Google Scholar]
- Malvankar NS, Tuominen MT, and Lovley DR (2012). Lack of cytochrome involvement in long-range electron transport through conductive biofilms and nanowires of Geobacter sulfurreducens. Energy & Environmental Science 5, 8651–8659. [Google Scholar]
- Malvankar NS, Vargas M, Nevin KP, Franks AE, Leang C, Kim BC, Inoue K, Mester T, Covalla SF, Johnson JP, et al. (2011). Tunable metallic-like conductivity in microbial nanowire networks. Nature Nanotechnology 6, 573–579. [DOI] [PubMed] [Google Scholar]
- Margoliash E, and Lustgarten J (1962). Interconversion of horse heart cytochrome c monomer and polymers. Journal of Biological Chemistry 237, 3397–3405. [PubMed] [Google Scholar]
- Marsili E, Baron DB, Shikhare ID, Coursolle D, Gralnick JA, and Bond DR (2008). Shewanella secretes flavins that mediate extracellular electron transfer. Proceedings of the National Academy of Sciences 105, 3968–3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta T, Coppi MV, Childers SE, and Lovley DR (2005). Outer membrane c-type cytochromes required for Fe (III) and Mn (IV) oxide reduction in Geobacter sulfurreducens. Applied and Environmental Microbiology 71, 8634–8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell JA, and Grigorieff N (2003). Accurate determination of local defocus and specimen tilt in electron microscopy. Journal of Structural Biology 142, 334–347. [DOI] [PubMed] [Google Scholar]
- Neumann P, Dickmanns A, and Ficner R (2018). Validating resolution revolution. Structure 26, 785–795. e784. [DOI] [PubMed] [Google Scholar]
- Nevin KP, Kim BC, Glaven RH, Johnson JP, Woodard TL, Methé BA, DiDonato RJ Jr, Covalla SF, Franks AE, Liu A, et al. (2009). Anode biofilm transcriptomics reveals outer surface components essential for high density current production in Geobacter sulfurreducens fuel cells. PLoS ONE 4, e5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nivaskumar M, and Francetic O (2014). Type II secretion system: A magic beanstalk or a protein escalator. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1843, 1568–1577. [DOI] [PubMed] [Google Scholar]
- O’Brien JP, and Malvankar NS (2017). A simple and low-cost procedure for growing Geobacter sulfurreducens cell cultures and biofilms in bioelectrochemical systems. Current Protocols in Microbiology 43, A.4K.1–A.4K.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I, and Kim B-C (2011). Homologous overexpression of omcZ, a gene for an outer surface c-type cytochrome of Geobacter sulfurreducens by single-step gene replacement. Biotechnology Letters 33, 2043. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera—a visualization system for exploratory research and analysis. Journal of Computational Chemistry 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Qian X, Mester T, Morgado L, Arakawa T, Sharma ML, Inoue K, Joseph C, Salgueiro CA, Maroney MJ, and Lovley DR (2011). Biochemical characterization of purified OmcS, a c-type cytochrome required for insoluble Fe(III) reduction in Geobacter sulfurreducens. Biochimica et Biophysica Acta (BBA) - Bioenergetics 1807, 404–412. [DOI] [PubMed] [Google Scholar]
- Reardon PN, and Mueller KT (2013). Structure of the type IVa major pilin from the electrically conductive bacterial nanowires of Geobacter sulfurreducens. Journal of Biological Chemistry 288, 29260–29266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguera G, McCarthy KD, Mehta T, Nicoll JS, Tuominen MT, and Lovley DR (2005). Extracellular electron transfer via microbial nanowires. Nature 435, 1098–1101. [DOI] [PubMed] [Google Scholar]
- Reguera G, Nevin KP, Nicoll JS, Covalla SF, Woodard TL, and Lovley DR (2006). Biofilm and nanowire production leads to increased current in Geobacter sulfurreducens fuel cells. Applied and environmental microbiology 72, 7345–7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguera G, Pollina RB, Nicoll JS, and Lovley DR (2007). Possible nonconductive role of Geobacter sulfurreducens pilus nanowires in biofilm formation. Journal of Bacteriology 189, 2125–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter LV, Sandler SJ, and Weis RM (2012). Two isoforms of Geobacter sulfurreducens PilA have distinct roles in pilus biogenesis, cytochrome localization, extracellular electron transfer, and biofilm formation. Journal of Bacteriology 194, 2551–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH (2012). RELION: implementation of a Bayesian approach to cryo-EM structure determination. Journal of Structural Biology 180, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam S, Earl LA, Falconieri V, Milne JL, and Egelman EH (2016). Resolution advances in cryo-EM enable application to drug discovery. Current Opinion in Structural Biology 41, 194–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers ZM, Fogarty HE, Leang C, Franks AE, Malvankar NS, and Lovley DR (2010). Direct exchange of electrons within aggregates of an evolved syntrophic coculture of anaerobic bacteria. Science 330, 1413–1415. [DOI] [PubMed] [Google Scholar]
- Tan Y, Adhikari RY, Malvankar NS, Pi S, Ward JE, Woodard TL, Nevin KP, Xia Q, Tuominen MT, and Lovley DR (2016a). Synthetic biological protein nanowires with high conductivity. Small 12, 4481–4485. [DOI] [PubMed] [Google Scholar]
- Tan Y, Adhikari RY, Malvankar NS, Ward JE, Nevin KP, Woodard TL, Smith JA, Snoeyenbos-West OL, Franks AE, Tuominen MT, et al. (2016b). The low conductivity of Geobacter uraniireducens pili suggests a diversity of extracellular electron transfer mechanisms in the genus Geobacter. Frontiers in Microbiology 7, 980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y, Adhikari RY, Malvankar NS, Ward JE, Woodard TL, Nevin KP, and Lovley DR (2017). Expressing the Geobacter metallireducens PilA in Geobacter sulfurreducens yields pili with exceptional conductivity. mBio 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, and Ludtke SJ (2007). EMAN2: An extensible image processing suite for electron microscopy. Journal of Structural Biology 157, 38–46. [DOI] [PubMed] [Google Scholar]
- Vargas M, Malvankar NS, Tremblay PL, Leang C, Smith J, Patel P, Synoeyenbos-West O, Nevin KP, and Lovley DR (2013). Aromatic amino acids required for pili conductivity and long-range extracellular electron transport in Geobacter sulfurreducens. mBio 4, e00105–00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignon G, Köhler R, Larquet E, Giroux S, Prévost M-C, Roux P, and Pugsley AP (2003). Type IV-Like Pili Formed by the Type II Secreton: Specificity, Composition, Bundling, Polar Localization, and Surface Presentation of Peptides. Journal of Bacteriology 185, 3416–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Coureuil M, Osinski T, Orlova A, Altindal T, Gesbert G, Nassif X, Egelman EH, and Craig L (2017). Cryoelectron microscopy reconstructions of the Pseudomonas aeruginosa and Neisseria gonorrhoeae type IV pili at sub-nanometer resolution. Structure 25, 1423–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RY-R, Kudryashev M, Li X, Egelman EH, Basler M, Cheng Y, Baker D, and DiMaio F (2015). De novo protein structure determination from near-atomic-resolution cryo-EM maps. Nature Methods 12, 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CJ, Headd JJ, Moriarty NW, Prisant MG, Videau LL, Deis LN, Verma V, Keedy DA, Hintze BJ, Chen VB, et al. (2018). MolProbity: More and better reference data for improved all-atom structure validation. Protein Science 27, 293–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Biochemical characterizations of filament preparations show PilA and OmcS proteins, Related to Figures 1 and 2 (A) SDS-PAGE gel of filaments showing OmcS and PilA at expected molecular weights of ~ 45 kDa and ~ 6.5 kDa. LC-MS/MS analysis of metalloproteins in purified G. sulfurreducens filaments from (B) the ~ 45 kDa band and (C) the ~ 30 kDa band. EMPAI: Exponentially modified protein abundance index. LC-MS/MS analysis showing matched peptides in (D) OmcS in ~ 45 kDa band and (E) PilA in the ~ 6.5 kDa band. (F) Western immunoblotting using anti-PilA antibody showing PilA in purified filament preparations (left) and cell lysate (right).
Fig. S2. De novo atomic model building of OmcS filaments, Related to Figures 1 and 2 (A) Sequence-based alignment of OmcS and two other c-type cytochromes detected by mass spectrometry, with the same highlighted colors used in Fig 2A. Regions indicated by red, orange, yellow, green, cyan, blue and purple arrowheads are shown in (B)-(G), respectively, with OmcS atomic model fit into the cryo-EM map. Both OmcT and GSU2501 have deletions or insertions at those regions and cannot fit into the cryo-EM map.
Fig. S3. Illustration of cryo-EM map quality, Related to Figures 1 and 2 (A) The model:map FSC (Fourier Shell Correlation) calculation using a 0.38 criterion, which is sqrt (0.143), estimates the Spider map to have a resolution of 3.7 Å. (B) A view of amino acids 325–350 fit into the Spider map used in (A) after filtering the map to 3.6 Å. (C) The Relion map:map “gold standard” FSC using the 0.143 criterion estimates the Relion map to have a resolution of 3.2 Å. (D) A view of the same region as (B) but with the Relion map which has been filtered to 3.2 Å, shows that the Relion estimate of resolution is considerably over-optimistic. (E) Fitting of six heme co-factors into the corresponding cryo-EM densities. (F) The Histidine 51 interaction with heme 2 shown along with surrounding residues fit into the cryo-EM density.
Fig. S4. Power spectra of purified filaments from fumarate-grown strain and cell-attached filaments are similar to power spectra from filaments of the electrode-grown strain used to build the atomic model, Related to Figure 1 and 2 (A) OmcS filaments (black arrow) from the fumarate-grown strain. Some flagellar filaments (white arrow) are present in this strain. Scale bar, 500 Å. (B) The averaged power spectrum of the OmcS filaments from fumarate-grown cells (left) shows similar layer lines and helical symmetry as that from filaments of electrode-grown cells (right). (C) Power spectra generated from previously published images (Leang et al., 2013) of intact filaments attached to G. sulfurreducens cells. Left, the power spectrum of a bundle of filaments showing the meridional layer line (blue arrow) at 1/(47 Å). Because of the bundling, a strong sampling is seen on the near-equatorial layer line. Right, averaged power spectra from 13 isolated segments to avoid the sampling. These power spectra show that the cell-attached filaments have symmetry similar to that as observed in cryo-EM images of purified OmcS filaments, with a rotation of 84 degrees and a rise of 47 Å per subunit.
Fig. S5. The thinner filament observed in cryo-EM imaging, Related to Figure 1 (A) The cryo-EM images frequently showed a thinner filament (white arrow) compared to OmcS filaments (black arrow). Scale bar, 200 Å. (B) The averaged power spectrum of the thinner filaments shows layer lines similar to those from the OmcS filament, but with a somewhat different axial rise of ~ 57 Å and rotation of ~ 160°. (C) A helical reconstruction of the thinner filament filtered to 20 Å. (D) The projection of the reconstruction in (C) with a low threshold (left) and a high threshold (right). The high threshold shows strong peaks separated by approximately 10 Å, labelled by yellow dots, which could be hemes.
Data Availability Statement
All data for OmcS filament were deposited in EMDB and PDB with the following entry codes: EMDB: EMD-9046 and PDB: 6EF8.