Abstract
Cancer development requires a favorable tissue microenvironment. By deleting Myd88 in keratinocytes or specific bone marrow subpopulations in oncogenic RAS-mediated skin carcinogenesis, we show that IL17 from infiltrating T cells and IκBζ signaling in keratinocytes are essential to produce a permissive microenvironment and tumor formation. Both normal and RAS-transformed keratinocytes respond to tumor promoters by activating canonical NF-κB and IκBζ signaling, releasing specific cytokines and chemokines that attract Th17 cells through MyD88-dependent signaling in T cells. The release of IL17 into the microenvironment elevates IκBζ in normal and RAS-transformed keratinocytes. Activation of IκBζ signaling is required for the expression of specific promoting factors induced by IL17 in normal keratinocytes and constitutively expressed in RAS-initiated keratinocytes. Deletion of Nfkbiz in keratinocytes impairs RAS-mediated benign tumor formation. Transcriptional profiling and gene set enrichment analysis of IκBζ–deficient RAS-initiated keratinocytes indicate that IκBζ signaling is common for RAS transformation of multiple epithelial cancers. Probing The Cancer Genome Atlas datasets using this transcriptional profile indicates that reduction of IκBζ signaling during cancer progression associates with poor prognosis in RAS-driven human cancers.
Implications
The paradox that elevation of IκBζ and stimulation of IκBζ signaling through tumor extrinsic factors is required for RAS-mediated benign tumor formation while relative IκBζ expression is reduced in advanced cancers with poor prognosis implies that tumor cells switch from microenvironmental dependency early in carcinogenesis to cell-autonomous pathways during cancer progression.
Introduction
Emerging from an intense exploration of inflammation and cancer is the central role of NF-κB in both experimentally induced cancer in rodents and human cancers (1). As a nuclear transcription factor complex, NF-κB has both intrinsic action in promoting cell survival and extrinsic functions in regulating the expression of multiple released cytokines and chemokines. While the active nuclear NF-κB is commonly found in many cancers, mutations in the NF-κB family or its immediate regulators are rare and sporadic, suggesting that mutational mechanisms are unlikely to directly contribute to its activity in cancer. Furthermore, NF-κB has proved to be a difficult drug target (2). Thus, attention has turned to upstream regulators of this pathway. Among these is MyD88, an adaptor protein linked to toll-like receptor (TLR) and IL1 family receptors (IL1R, IL18R, and others) whose downstream effectors include the NF-κB complex. The discovery of frequent mutations in amino acid 265 of MyD88 leading to constitutive activation of NF-κB in B-cell lymphomas provided strong evidence for this pathway’s association with human cancer (3, 4). MyD88 protein is also overexpressed in the tumor epithelium of many human cancer types (5, 6). Modification of MyD88 in experimental studies repeatedly demonstrates the importance of its activity in cancer pathogenesis (reviewed in ref. 7).
Sorting out the contribution of cell-autonomous and microenvironmental influences on cancer pathogenesis is particularly suited for study in 7,12 dimethylbenz(a)anthracene (DMBA)/12-O-tetradecanoyl-phorbol-13-acetate (TPA; DMBA/TPA)-induced mouse skin squamous cancer because both Ras oncogene–driven transformation of target cells (DMBA initiation) and programmed changes in the skin microenvironment (TPA promotion) are distinct requirements for tumor formation. Oncogenic activation of Ras alleles is recognized as a cell-autonomous–initiating event for keratinocytes and an activated EGFR-IL1α-IL1R-MyD88-NF-κB axis is essential for creating the potential to form a tumor (8). However, RAS activation in the absence of a disturbance in tissue homeostasis is not sufficient for tumors to form in the intact skin. This distinguishes an intact tissue-based model of RAS oncogenesis from in vitro cell-autonomous acquisition of a transformed phenotype. The mouse skin model of tumor promotion then provided the first paradigm for a microenvironmental component in carcinogenesis that is now recognized as essential in human malignancies (9). In TPA-promoted mouse skin, knockout studies of TLR4 and MyD88 indicate that NF-κB signaling participate in tumor promotion (6, 8, 10–12). Furthermore, Myd88 ablation targeted to bone marrow progenitor cells reduces skin tumor induction, indicating intact extraepidermal bone marrow–derived cells are important in promotion (8). Successful tumor promotion also requires the presence of IL17 in the tumor microenvironment from an extra-epidermal source (10, 13, 14). The goal of the current studies was to explore the MyD88-dependent systemic factors that create a permissive microenvironment for the emergence of Ras oncogene–initiated keratinocytes in intact skin. The data revealed a MyD88-dependent roadmap through which IL17 is introduced into the skin microenvironment and influences both normal and initiated keratinocytes through IκBζ signaling, creating an enabling microenvironment for the selective outgrowth of RAS-transformed keratinocytes. While these observations were derived from a skin experimental model, the frequent association of IL17 and RAS oncogenesis suggests broader implications (15–20).
Materials and Methods
Tumor induction experiments
Mouse studies were performed under a protocol approved by the NCI and NIH Animal Care and Use Committee. The MyD88−/− colony was maintained on a C57BL/6NCr background. The dorsal skin of 7-week old mice was shaved with surgical clippers and subsequently checked for the lack of hair growth. Initiation was accomplished by a single topical application of 400 nmol of DMBA (Acros 40818-0050) in 0.2 mL acetone. Promoter treatments with 10 nmol of TPA (LC Laboratories P-1680) in 0.2 mL acetone, twice a week, were begun 1 week after initiation and continued for 20 weeks. Skin tumors induced by the initiation-promotion protocol were removed and both frozen and formalin fixed. The MyD88flox/flox mice (21) were crossed with CD4-Cre+, CD19-Cre+, Lyz2-Cre+, and CD11c-Cre+ (all from Jackson Laboratories) to obtain cell-specific deletion of MyD88. These mice were subjected to two-stage skin carcinogenesis as described above. For in vivo studies aimed at measuring signaling and gene expression analyses, mice were painted with TPA for 4.5 weeks (corresponding to eight or nine TPA applications) and full thickness skin biopsies were either snap frozen in liquid nitrogen or fixed in formalin three days after the last application of TPA. The IκBζ-deficient mice were provided by Dr. Shizuo Akira (Osaka University, Osaka, Japan; ref. 22). Primers used for the genotyping of the various mouse strains are listed in Supplementary Table S5.
Generation of Il1r1F/F mice
Two LoxP sites were introduced into introns 2 and 4 of the Il1r1 gene, such that Cre-mediated deletion results in excision of exons 3 and 4. The targeting construct was introduced into Bruce 4 C57Bl6 ES cells and homologous integration were used to generate Il1r1F/F mice. Selected clones were injected into C57BL/6-Albino blastocysts and transferred to pseudo-pregnant females. Chimeric offspring were bred with C57BL/6-Albino partners to generate F1 mice that were screened for integration by Southern analysis. The resulting mice were crossed with C57BL/6-FLPe mice to facilitate removal of the neomycin cassette and generate the Il1r1F allele.
Athymic nude mouse grafting
On day 3 in culture, Nfkbiz+/− or Nfkbiz−/− primary keratinocytes were infected with the v-rasHa retrovirus and on day 8 were trypsinized and used for grafting as described previously (23). Seven million keratinocytes were mixed with 4 million SENCAR mouse primary dermal fibroblasts (cultured for one week) and grafted onto the back of nude mice on a prepared skin graft site located in the midback region. Tumor dimensions were measured weekly using calipers, and approximate tumor volumes were determined by multiplying tumor height × length × width.
Cell culture and treatments
Primary mouse keratinocytes and hair follicle buds were isolated from newborn pups as described previously (23) and cultured in modified Eagle’s medium (S-MEM, Gibco 98-0216DJ), 7% Chelex-treated FCS (Gemini Bio Products), and 0.05 mmol/L calcium unless otherwise indicated. MyD88-, EGFR-, and IκBζ-deficient keratinocytes were isolated from pups obtained from respective heterozygous breeding pairs (24). LSL-HrasG12D mice were purchased from The Jackson Laboratory (stock #009046). TPA (Tocris) was diluted in DMSO. IL17A (R&D Systems, 421-ML-025), IL22 (R&D Systems, 582-ML-010), IL1ra (5 μg/mL, IL1ra or anakinra; Division of Veterinary Resources, NIH, Bethesda, MD) were diluted in culture medium and added to cell culture medium as indicated before cell harvesting. Neonatal human epidermal primary keratinocytes (Gibco, C-001-5C) were cultured in KSF media supplemented with human keratinocyte growth supplement (Gibco, S-001-5).
Retroviral and adenoviral constructs
The v-rasHa replication-defective ecotropic retrovirus was prepared using Psi2 producer cells. Retrovirus titers were routinely 1 × 107 virus/mL. Cultured primary keratinocytes were infected with v-rasHa retrovirus (here referred to as RAS or oncogenic RAS) on day 3 at a multiplicity of infection (MOI) of 1 in medium containing hexadimethrine bromide (4 μg/mL; Sigma, 107689). The IκBsr (IκB super repressor) adenovirus or Cre recombinase (Cre) were introduced into primary keratinocytes using an adenoviral construct driven by a cytomegalovirus promoter, and a similarly constructed adeno-GFP was used as control. Keratinocytes were adenovirus-infected for 30 minutes in serum-free medium with a MOI of 10 viral particles per cell and hexadimethrine bromide (4 μg/mL) to enhance uptake. Serum-containing medium was added to the cells for the next 48 hours after the infection. The cDNAs for human RAS oncogenes were purchased from Addgene: pBabe HRAS 12V (plasmid #12545), pBabe KRAS 12V (plasmid #12544). The promoter FerH (25) and RAS cDNAs were cloned into pLV-CE vector and high titer lentivirus (10e8 TU per mL) were produced (Cellomics Technology, LLC).
Immunoblotting
Cultured keratinocytes with or without v-rasHa transduction were lysed in RIPA lysis buffer (Cell Signaling Technology, 9806S) supplemented with Halt Protease and Phosphatase-Inhibitor cocktail (Thermo Fisher Scientific, 78442) and phenylmethylsulfonylfluoride (Cell Signaling Technology, 8553S). Proteins were quantified by the BCA method (Thermo Fisher Scientific, 23225) and separated by 4%–20% or 10% tris-HCl gels (Bio-Rad, 5671094). Nuclear extracts were prepared according to the NE-PER Kit Protocol (Pierce, 78835). To prepare lysates from skin biopsy samples, flash-frozen skin biopsies in liquid nitrogen were pulverized in RIPA buffer supplemented as above using ceramic beads (Bertin Instruments, CR-28) in reinforced tubes and a Precellys24 Homogenizer (Bertin Instruments). For analysis of keratinocyte differentiation in vitro, cultures were washed once with PBS (Ca2+- and Mg2+-free), and total cell lysates were prepared [5% SDS and 20% 2-mercaptoethanol in 0.25 M tris (pH 6.8)]. Total ERK1/2 (Cell Signaling Technology, 4695S), phosphorylated ERK1/2 (Cell Signaling Technologies, 9101S), Lamin B1 (Cell Signaling Technology, 13435), total NF-κB (Cell Signaling Technology, 8242S), phosphorylated NF-κB (Cell Signaling Technology, 3033S), total IκBα (Cell Signaling Technology, 9242S), and phosphorylated IκBα (Cell Signaling Technology, 2859S), were used at 1:1,000 dilution. Antibody to IκBζ was used at 1:500 (Invitrogen, 14-6801-82), antibody to HSP90 (Cell Signaling Technology, 4877S) was used at 1:2,000. Antibodies against K1 (BioLegend, Poly19056) and K10 (BioLegend, Poly19054) were used at 1:10,000, and antibody against β-Actin (Abcam, 6276) was used at 1:5,000 dilution. All antibodies were incubated overnight. ECL SuperSignal (Pierce) system was used for detection.
[3H] Thymidine incorporation assay
Keratinocytes were plated in 24-well plates, and three days after plating [3H] thymidine (PerkinElmer, NET027W001MC, 1 μCi per well) was added for 4 hours. Cultures were trypsinized, wells were transferred to glass fiber filters using a Brandel cell harvester, and incorporated counts read using a Wallac TriLux 1450 MicroBeta scintillation counter (PerkinElmer).
RT-PCR analysis
RNA was isolated from cultured cells using RLT RNeasy 96 Qiacube HT Kit (Qiagen, 74171) and on-column DNA digest (Qiagen, 79254) while tissue biopsies were first homogenized with TRIzol using the manufacturer’s protocol (Invitrogen, 15596018). Complementary DNA synthesis and real-time PCR analysis were conducted as described previously (26). Predesigned QuantiTect Primers (Qiagen) were used for all genes except for Gapdh, where the following primers were designed: 5′-CATGGCCTTCCGTGTTCCrA-3′ (forward) and 5′-GCGGCACGTCAGATCCA-3′ (reverse).
Measurement of cytokine/chemokines levels
Supernatant was collected, cleared of cells by centrifugation, and used to assay cytokine and chemokine levels. Cytokines and chemokines were measured using a Magnetic Luminex Assay (R&D Systems) according to the manufacturer’s instructions. Quantification and analysis were performed with the Bio-Plex MAGPIX Reader (Bio-Rad) using the Collaborative Protein Technology Resource core (Center for Cancer Research, NCI, Bethesda, MD).
Mouse tissue processing and phenotypic analysis
The method used was originally reported by Naik and colleagues (27). Dorsal skin was shaved with mini clippers (Wahl), adipose tissue was removed with a number 10 scalpel, and skin was cut in 1-cm by 1-cm pieces. Tissue samples were digested in RPMI containing 100 U/mL penicillin, 100 μg/mL streptomycin, 55 μmol/L β-mercaptoethanol, 20 μmol/L HEPES (HyClone), and 0.25 mg/mL Liberase-purified enzyme blend (Roche Diagnostic Corp., 05401020001) and incubated for 2 hours at 37°C and 5% CO2. Digested skin sheets were homogenized using 50-μm Medicon (BD Biosciences, 340591) and a Medimachine tissue homogenizer system (BD Biosciences). Single-cell suspensions were stained with either LIVE/DEAD Fixable Blue Dead Cell Stain Kit (Invitrogen, L23105) in Hank’s balanced salt solution (HBSS) to exclude dead cells. For detection of transcription factors, cells were stained using the Foxp3 staining set (eBioscience, 11-5773) according to the manufacturer’s protocol. For detection of intracellular cytokines, cells were fixed and permeabilized with BD Cytofix/Cytoperm and stained in BD Perm Wash buffer (BD Biosciences, 005-223-00). Cells were stained with the following antibodies purchased from either eBioscience or BD Biosciences: CD45.2 (eBio 104), TCRβ (eBioH57-597), IL17A (eBio17B7), IFNγ (eBioXMG1.2), Foxp3 (eBioFJK-16s), TCRγδ (eBioGL3, 25-5711). Staining was performed in the presence of Mouse BD Fc Block (BD Biosciences, 553142), 0.2 mg/mL purified rat IgG and 1 mg/mL of normal mouse serum (Jackson Immuno Research, 015-000-120).
T cell sorting for gene expression analysis
Twenty-four hours post last TPA application, animals were euthanized, and cells isolated from dorsal skin as described for phenotypic analysis. The cell pellet was resuspended in HBSS and stained with 4′, 6-diamidino-2-phenylindol (Sigma D9542) to exclude dead cells. Cells were preincubated with FcBlock (BD Biosciences, 553141), 0.2 mg/mL purified rat IgG and 1 mg/mL of normal mouse serum (Jackson Immuno Research, 015-000-120). Thereafter, the following antibodies were added: CD45 Pacific Blue (clone 30-F11, BD Biosciences), CD3 FITC (clone 145-2C11, e-Bioscience), and NK tetramer APC (kindly provided by Jeff Subleski). After staining, cells were washed and resuspended in sorting buffer (HBSS, 1% BSA, 55 μmol/L 2-mercaptoethanol, 20 mmol/L HEPES) and subjected to FACS sorting. Cells were gated on FSC/SSC/Alive/CD45+/CD3+/NK tetramer negative and collected in 200 μL of TRIzol LS reagent (Ambion, 10296010). Total RNA from sorted T cells was prepared using the Direct-zol RNA MiniPrep Kit (Zymo Research, R2060) according to the manufacturer’s instructions. Total RNA was amplified and subjected to microarray procedure using Affymetrix whole-genome mouse arrays 430 2.0. Data files were analyzed using BRBArrayTools (Biometric Research Branch, NCI).
Microarray and bioinformatic analysis
Total RNA was extracted from keratinocyte cultures with TRIzol (Invitrogen, 15596018) using the manufacturer’s protocol followed by purification of the clear phase on a Qiagen column including a DNA digest step. Four independent biological replicates were evaluated. Gene expression profiling was done using the Affymetrix Mouse Gene 1.0 ST array platform. RNA qualitytesting, microarray hybridization, and processing were done by the Laboratory of Molecular Technology in Frederick, MD. The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession GSE100836. Qlucore Omics Explorer software (version 3; Qlucore AB) was used to analyze the microarray data. A two-group comparison t test, a FDR ≤ 0.05 and a fold change > 1.5, were applied for selecting differentially expressed genes. Gene set enrichment analysis (GSEA) was carried out using Qlucore implementation and subsets of MSigDB collections, namely C2 (curated gene sets), C3 (motif gene sets), C4 (computational gene sets), C5 (GO gene sets), and C6 (oncogenic gene sets). Gene sets were considered significantly enriched when the FDR estimate did not exceed 5% in a collection. Furthermore, GSEA results were visualized as a network using the Enrichment Map Plugin version 2.1.1 (28) implemented in Cytoscape version 3.5.1 (29). The enrichment map was generated with top enriched gene sets (FDR < 2.5%), limited to core enrichment subsets (“leading edge”). Combined Jaccard and Overlap coefficients were used to construct the network with the default similarity cutoff of 0.375 and equal weight for each of the coefficients; in addition, only subgraphs containing at least three nodes were retained. The network was plotted with R ggnetwork (https://briatte.github.io/ggnetwork/) using the Kamada–Kawai algorithm to calculate the layout.
Survival profiles (Kaplan–Meier) of RAS-driven human cancer
From the RAS-transformed mouse keratinocyte Nfkbiz deficiency transcriptional profiles (Supplementary Table S3), we generated a list of differentially expressed genes with 2-fold change and FDR = 0.05.
For colon cancer patient prognosis analysis, we used The Cancer Genome Atlas (TCGA) Colon Adenocarcinoma (COAD) dataset and chose the 16 most upregulated genes and defined the sum of their expression as the index to divide the patients into low- and high-index groups by the median. Those patients with colon cancer with high index had significantly poor disease-free survival with HR = 3.6 (1.5–8.65) estimated by the Cox proportional hazards model after age adjustment and the log-rank P = 0.003.
For patients with pancreatic and lung cancer, we used the TCGA Pancreatic Ductal Adenocarcinoma (PAAD) dataset the TCGA Lung Adenocarcinoma (LUAD) dataset, respectively. The weighted sum of the 19 most upregulated genes (adding three additional genes to the 16 most upregulated) was used as the index to divide the patients into low and high groups. The weights used to define the index were the log2-fold changes obtained from the mouse Nfkbiz-deficiency profile. The TCGA Lung Squamous Cancer (LUSC) dataset was also explored but the high and low index groups had no significant difference in disease-free survival.
Statistical analysis
Unless otherwise specified, biochemical data were analyzed by Prism software, and significance values were assigned through Mann–Whitney U test, Student t test, or one-way ANOVA with Dunnett posttest. P < 0.05 was considered to be significant.
Results
Deletion of Myd88 in T cells reduces skin tumor formation
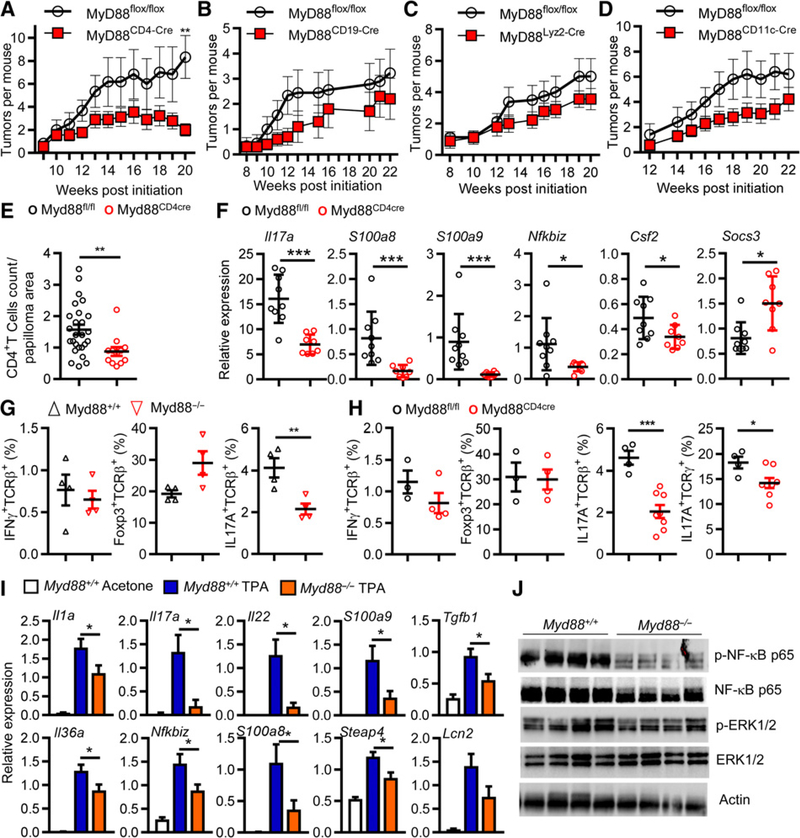
We reported that targeted deletion of Myd88 in bone marrow precursor cells largely prevented skin tumor formation that was reversed in mice reconstituted with wild-type bone marrow (8). To identify MyD88-dependent hematopoietic cells that contribute to skin carcinogenesis, we deleted Myd88 in selected subpopulations by Cre/Lox technology and induced skin tumors by DMBA/TPA. Deletion of Myd88 in T cells by crossing CD4-Cre mice with Myd88fl/fl mice substantially reduced the final skin tumor incidence by nearly 5-fold (Fig. 1A). In contrast, skin tumor incidence in mice with Myd88 deletion targeted to B cells (CD19-Cre), macrophages/granulocytes (Lyz2-Cre), and dendritic cells (CD11c-Cre) was less affected (Fig. 1B–D). Deletion of MyD88 in T cells substantially reduced the number of CD4+ T cells infiltrating the tumor microenvironment (Fig. 1E).
Figure 1.
Systemic or CD4+ T-cell-targeted deletion of MyD88 inhibits tumor development in chemically induced skin carcinogenesis and reduces the content of IL17A producing cells in TPA-treated skins. A-D, Panels represent the mean number of skin tumors per mouse (mean ± SEM). Mice were treated with DMBA/0.2 mL acetone at time 0 then with 10 nmol TPA/0.2 mL acetone twice a week for up to 20 weeks. MyD88fl/fl (n = 6) MyD88CD4-Cre (n = 8; A); MyD88fl/fl (n = 9) and MyD88CD19-Cre (n = 10; B); MyD88flfl (n = 8) and MyD88Lyz2-Cre (n = 9; C); and MyD88fl/fl (n = 5) and MyD88CD11c-Cre (n = 14; D). Significant differences in the number of papillomas that develop were calculated by Student t test (**, P < 0.001). E, Graphs represent the quantification of CD4+ T cells on papilloma sections from MyD88fl/fl and MyD88CD4-Cre. **, P < 0.01 versus MyD88fl/fl. F, qPCR analysis of differentially expressed genes in CD3+-sorted cell population isolated from MyD88fl/fl and MyD88CD4-Cre TPA-treated skins. G, Plots summarizing flow cytometric data of Foxp3, IL17A, IFNγ expression by live CD45+ TCRβ+ infiltrating the skin of MyD88fl/fl and MyD88−/− mice after 9 TPA applications. H, The same analysis was performed comparing MyD88fl/fl and MyD88CD4-Cre mice but IL17A+ TCRγ+ cells were also analyzed. Graphs show means ± SEM of 3 to 8 mice. F and G, **, P < 0.01 MyD88−/− and MyD88CD4-Cre, respectively, versus control group. I, Gene expression changes in skin biopsies collected 6 hours after the last TPA or acetone application (8 total) in WT and MyD88−/− mice. Expression levels were determined by semiquantitative PCR. *, P < 0.05 MyD88−/− versus control group. J, Immunoblot of total skin lysates from WT and MyD88−/− mice collected as described for I. p-, phosphorylated. Each lane represents a skin biopsy from an individual mouse. Data shown in A-I are representative of three independent experiments, while data shown in E and J are representative of two independent experiments. Data in A-J are means ± SEM.
IL17-producing T cells are reduced in TPA-treated skin of MyD88-deficient mice
RNA profiling of CD3+ cells extracted from skin of Myd88fl/fl and CD4-cre+/Myd88fl/fl mice after nine TPA treatments (Supplementary Table S1A) revealed that the expression of proinflammatory factors, including factors known to enhance tumor promotion (e.g., S100a8, S100a9, and Cxcl2), was reduced in skin infiltrating T cells with targeted Myd88 deficiency. The most highly modified ontology scored by the loss of Myd88 in T cells was that for Th17 cells (Supplementary Table S1B). qPCR analysis confirmed the reduced expression of tumor-promoting proinflammatory transcripts (e.g., S100a8 and S100a9) and a reduction of Il17a and Nfkbiz transcripts (Fig. 1F). After nine treatments with TPA, IL17A+ TCRβ+ T cells were significantly reduced in dissociated and FACS-sorted skin of both the Myd88-deficient mice and in the CD4-Cre/Myd88fl/f mice compared with controls, while other populations (IFNγ+ and Foxp3+) did not differ significantly (Fig. 1G and H). A substantial reduction in IL17+γδT cells (Fig. 1H) indicates that recruitment of resident T cells into the microenvironment may also be affected. These changes suggest that MyD88 is required to attract or polarize IL17-expressing T cells into the tumor-promoting microenvironment.
Myd88-deficient mice lack the cytokine/chemokine response to topical TPA
After multiple TPA treatments in vivo, a selective subset of promotion relevant cytokine and chemokine transcripts (30–32) was reduced in promoted whole skin from Myd88-deficient mice compared with similarly treated control mice (Fig. 1I). Notable in these assays, the absence of Myd88 reduces the cutaneous expression of members of an IL17 activation pathway (14, 33, 34) including transcripts for IL17A, Steap4, and IκBζ and a trending reduction in Lipocalin2. These data represent a composite of changes in keratinocytes, dermal cells, and infiltrating bone marrow–derived cells, and the reduction of IL17A transcripts and protein in TPA-treated Myd88 null skin is likely the result of reduced infiltration of IL17A-producing cells since IL17A is not expressed by keratinocytes (35). Also notable is the reduction of TGFβ that along with IL1 and IL6 contributes to the maturation of IL17-producing cells (36). Myd88 loss reduced the activation of cutaneous NF-κB by topical TPA but did not reduce ERK1/2 phosphorylation (Fig. 1J). The impairment of NF-κB activity in response to TPA likely underlies the reduction in the level of inflammatory chemokines and cytokines released by Myd88-deficient keratinocytes.
MyD88 is required for TPA responses in isolated mouse keratinocytes
Loss of Myd88 did not interfere with the activation of protein kinase C (PKC) by TPA in isolated keratinocytes as activation of PKCδ and ERK and downregulation of PKCα and PKCδ occurred equally in keratinocytes of both genotypes treated with TPA (Supplementary Fig. S1A), and the DNA synthesis response was identical in both genotypes to a range of TPA doses (Supplementary Fig. S1B)
However, cytokine and chemokine release by Myd88−/− keratinocytes after TPA treatment was markedly reduced compared with WT keratinocytes (Supplementary Fig. S1C) including reductions of CCL4 and IL1α that have been reported to recruit CD4+ T cells to the skin (37, 38). In fact, deletion of the IL1R in CD4+ T cells reduced the yield of carcinogen-induced skin tumors (Supplementary Fig. S1D). As suggested above, the absence of MyD88 impaired the activation of NF-κB (p65) and phosphorylation of IκBα in TPA-treated keratinocytes consistent with the in vivo data (Supplementary Fig. S1E).
IL17 elicits tumor-promoting responses in normal keratinocytes
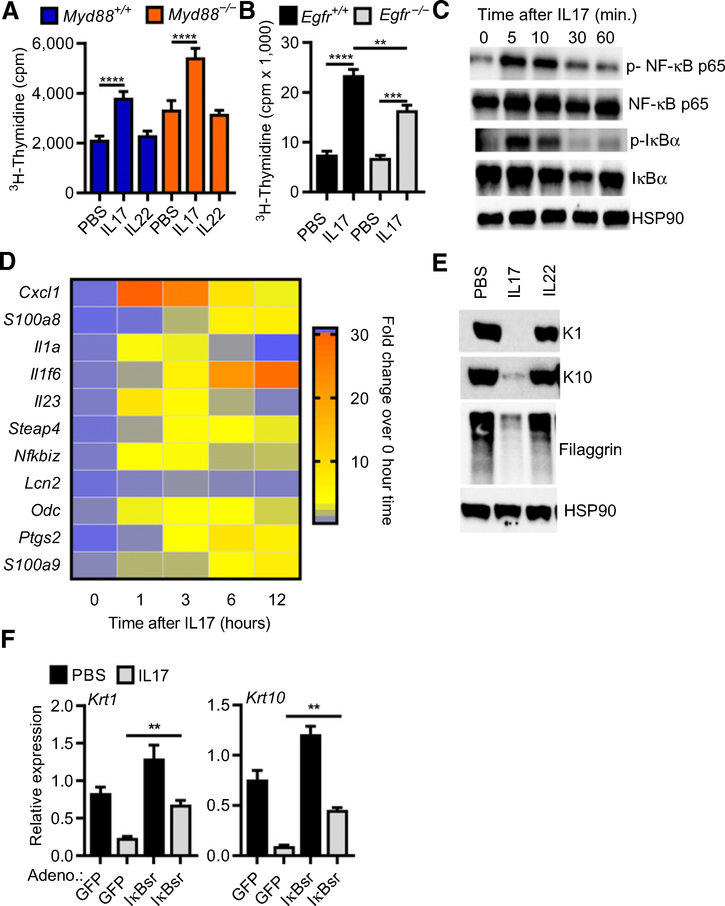
The infiltration of CD4+-IL17 cells into the tumor microenvironment to enhance tumor formation suggests that IL17 has unique effects on normal and initiated keratinocytes. Recombinant IL17, but not IL22, stimulated thymidine incorporation in normal keratinocytes through a mechanism largely independent of EGFR and MyD88 (Fig. 2A and B). In addition to proliferation, IL17 treatment activated canonical NF-κB through phosphorylation and loss of IκBα (Fig. 2C) and increased expression of multiple tumor-promoting factors (S100a8, S100a9, Odc, Cxcl1, Il1a, Il1f6, and Ptgs2; Fig. 2D). IL17 treatment also prevented keratinocyte terminal differentiation as indicated by the inhibition of the expression of transcripts and protein for the suprabasal keratins K1 and K10 and the late differentiation marker filaggrin (Fig. 2E). Like other modifiers of keratinocyte differentiation (8), IL17 uses the canonical NF-κB pathway to modify keratinocyte differentiation because its effect is partially reversed by preventing IκBα degradation using an adenoviral IκBα super-repressor (Fig. 2F).
Figure 2.
IL17 stimulates proliferation, upregulates cytokine expression, and inhibits differentiation of normal keratinocytes. Tritiated thymidine incorporation was measured in WT and MyD88−/− (A) or WT and EGFR−/− (B) keratinocyte cultures treated with IL17 or IL22 for 24 hours, bars represent the mean ± SEM value of four replicates. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. C, Immunoblotting of total cell extracts from primary keratinocytes treated with IL17 for the indicated period. HSP90, heatshock protein 90; p-, phosphorylated. D, Heatmap representation of gene expression analysis performed by real-time PCR quantification from keratinocytes treated for 1, 3, 6, and 12 hours with IL17. E, Total SDS cell extracts from PBS, IL17- or IL22-treated keratinocytes were immunoblotted with specific antibodies recognizing early (K1 and K10) and late (filaggrin) markers of differentiation from cultures maintained under differentiating conditions (0.12 mmol/L Ca++) for 24 hours. F, Real-time PCR quantification of K1 and K10 mRNAs in keratinocytes infected with GFP (control) or degradation-resistant IκBα (IκBsr Ad) adenovirus to block NF-κB activity and treated with IL17 under differentiating conditions (**, P < 0.01; ***, P < 0.001).
IL17 elicits responses in oncogenic RAS-initiated keratinocytes relevant for tumor formation
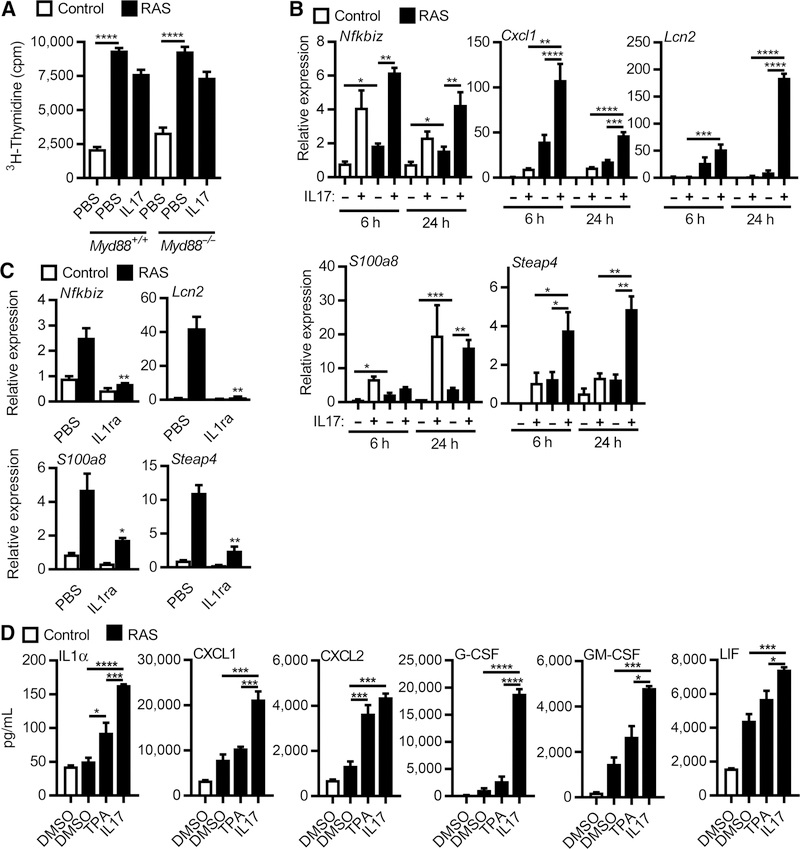
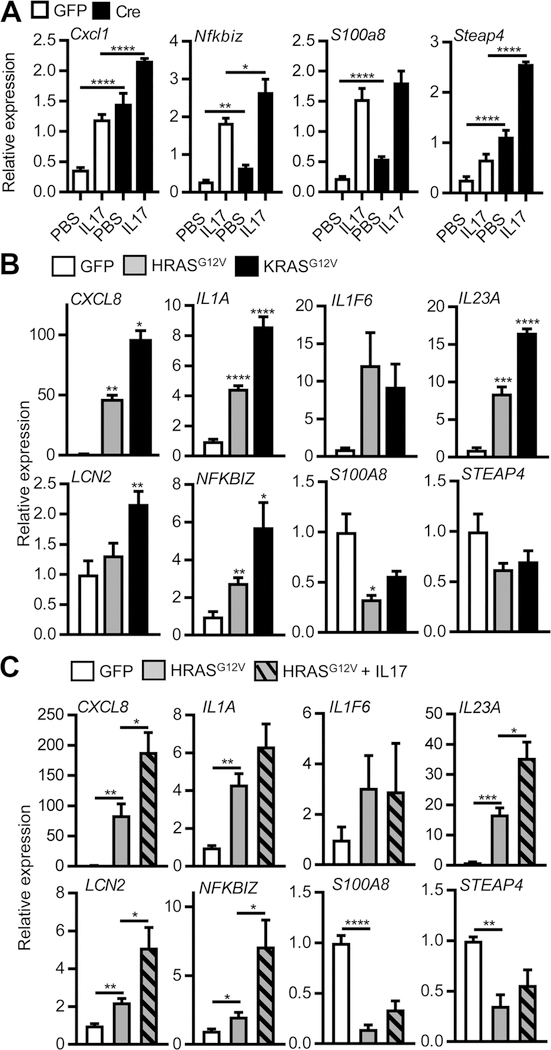
The high proliferation rate of RAS-transformed initiated keratinocytes is independent of MyD88 (Fig. 3A) but unlike normal keratinocytes, IL17 did not further enhance proliferation as measured by thymidine incorporation (Fig. 3A). Global gene expression analysis of RAS keratinocytes identified 65 genes differentially expressed upon treatment with IL17 (Supplementary Table S2). Most of these transcripts were upregulated and included typical IL17 response genes (Lcn2, Steap4, and Nfkbiz) and genes associated with oncogene-induced inflammation (S100a8, S100a9, and Cxcl1). The differential expression of these genes was validated by qPCR (Fig. 3B). Interestingly, upregulation of these transcripts in RAS-transformed keratinocytes depended on intact IL1 signaling as it was blocked by IL1ra (Fig. 3C). IL17 was more potent than TPA at enhancing the release of inflammatory mediators from RAS oncogene–transformed keratinocytes (Fig. 3D) implicating that IL17 signaling is through a pathway more powerful on initiated keratinocytes than a classic tumor promoter. These responses are not dependent on high retroviral RAS expression as expression of these cytokines was also elevated and enhanced by IL17 in keratinocytes expressing only a single oncogenic Hras allele from the endogenous Hras promoter, as would be the case for DMBA initiation in vivo (Fig. 4A). Transformation of human keratinocytes with oncogenic H- and K-RAS lentiviruses also upregulates transcripts for NFKBIZ and proinflammatory factors, a response that was further enhanced by IL17 (Fig. 4B and C). The constitutive elevation of Nfkbiz transcripts in RAS-transformed mouse and human keratinocytes and the further induction by IL17 raised the issue of what role the IκBζ signaling pathway may play in skin carcinogenesis.
Figure 3.
IL17 enhances transcriptional activity and cytokine release but not proliferation of RAS keratinocytes. A, Tritiated thymidine incorporation was measured in control or RAS-transduced WT and Myd88−/− keratinocyte cultures treated with IL17 for 24 hours. Bars represent the mean ± SEM value of four replicates. B, Real-time PCR quantification of mRNAs from control and RAS-transduced keratinocytes stimulated with IL17 for 6 or 24 hours. Bars represent the mean ± SEM value of four replicates. C, Real-time PCR analysis of mRNA expression in control or RAS-transduced keratinocytes treated with PBS or IL1R antagonist (IL1ra, anakinra). D, Cytokine and chemokine concentrations in culture supernatants from control and RAS-transduced keratinocytes cultures collected 24 hours after TPA or IL17 treatments and assayed by multiplex ELISA. For A-E, **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Figure 4.
IL17 enhances transcription of downstream effectors in mouse keratinocytes with endogenous mutant Ras activation and human keratinocytes transformed by human mutant RAS. A, Real-time PCR quantification of mRNAs from GFP and Cre adenovirus-transduced LSL-HrasG12D mouse keratinocytes stimulated with IL17. B, Real-time PCR quantification of mRNAs from GFP, mutant HRASG12V, and KRASG12V lentivirus-transduced human keratinocytes. C, Real-time PCR quantification of mRNAs from GFP and mutant HRASG12V lentivirus-transduced human keratinocytes treated with human IL17A. For A-C, *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
IκBζ signaling is essential for keratinocyte response to IL17 and tumor formation by oncogenic RAS
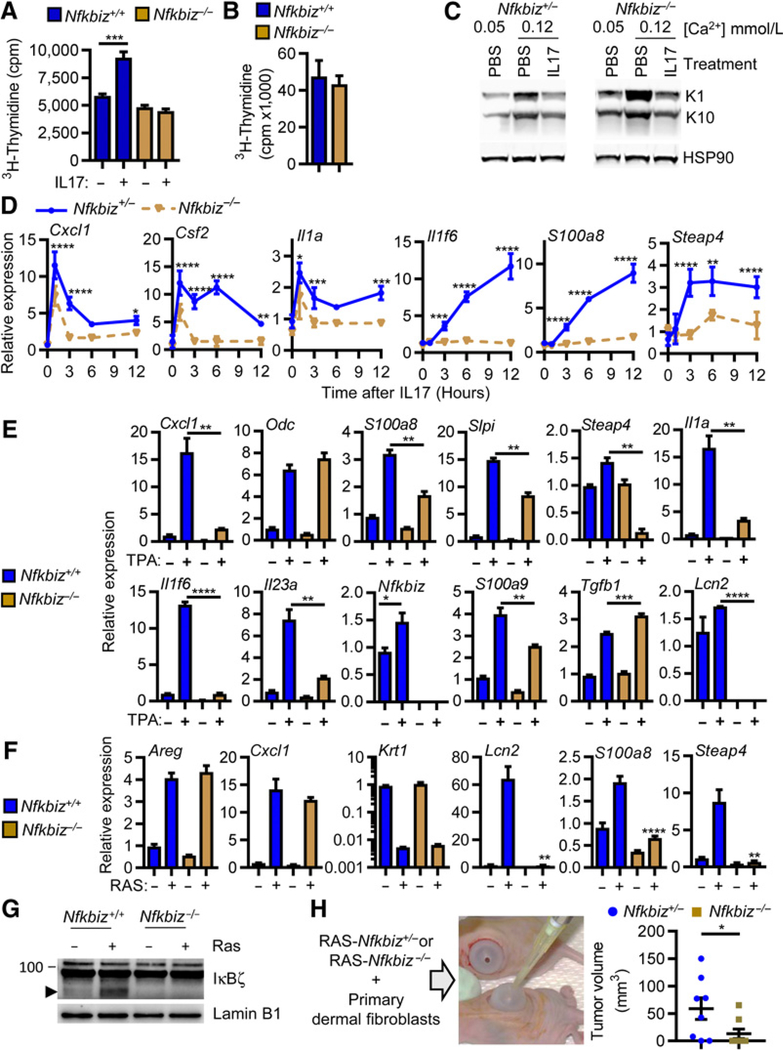
IL17 is known to regulate an alternate NF-κB transcription pathway through the upregulation of the NF-κB p50–binding cofactor IκBζ, thereby resulting in selective activation or inhibition of NF-κB–dependent genes (39–41). The importance of the IL17-IκBζ pathway in skin has been clearly shown in a mouse model of psoriasis (42). We discovered that in response to IL17, normal keratinocytes genetically ablated for Nfkbiz were not stimulated to proliferate (Fig. 5A) while proliferation was unaffected by loss of IκBζ in RAS-transformed keratinocytes (Fig. 5B). Likewise, loss of IκBζ did not prevent IL17 inhibition of normal keratinocyte differentiation (Fig. 5C), but ablation of Nfkbiz reduced the IL17- (Fig. 5D), TPA- (Fig. 5E), and oncogenic RAS-mediated expression of transcripts for proinflammatory cytokines and chemokines important for tumor formation (Fig. 5F). It should be noted that TPA also increased the expression of Nfkbiz in keratinocytes as has been reported previously (43). Thus, the tumor-promoting effects of both TPA and IL17 merge through IκBζ signaling, and IκBζ is both a target and effector of PKC signaling in the tumor promotion phase of skin carcinogenesis. The upregulation of IκBζ is also prominently detected in the nucleus of keratinocytes transduced with oncogenic RAS (Fig. 5G). To test the relevance of the IκBζ pathway for RASmediated tumor formation, Nfkbiz heterozygous and null keratinocytes (22) were transduced with oncogenic RAS and grafted to an orthotopic site on a prepared nude mouse skin bed (Fig. 5H). While papillomas developed in most recipients of RAS-transduced Nfkbiz heterozygous keratinocytes, mice receiving RAS-transduced Nfkbiz-null keratinocytes either did not form papillomas or the few tumors that formed were significantly smaller when measured at 20 days (Fig. 5G). Because Nfkbiz deficiency did not reduce the proliferation of RAS-transformed cells in vitro, the reduced tumor yield and smaller tumor size were likely due to diminished responsiveness of Nfkbiz-deficient RAS-initiated keratinocytes to internal signaling and to factors that modify the tumor microenvironment to promote tumor growth or survival.
Figure 5.
TPA, IL17, and the RAS oncogene are dependent on IkBz signaling for induction of tumor-promoting transcripts and tumor formation in vivo. Tritiated thymidine incorporation was measured in WT or Nfkbiz−/− normal keratinocytes treated with IL17 for 24 hours (A) or RAS-transduced keratinocytes (B). Bars represent the mean ± SEM value of four replicates. C, Total SDS cell extracts from Nfkbiz+/− and Nfkbiz−/− keratinocytes were immunoblotted with specific antibodies recognizing K1 and K10 from cultures under basal (0.05 mmol/L Ca++) conditions or treated with IL17 under differentiating conditions (0.12 mmol/L Ca++). Realtime PCR quantification of mRNAs from Nfkbiz+/− and Nfkbiz−/− keratinocytes treated for 1, 3, 6, and 12 hours with IL17 (D) or from WT or Nfkbiz−/− normal keratinocyte treated with TPA (E) or from control and RAS-transduced WT and Nfkbiz−/− keratinocytes (F). Symbols represent the mean ± SEM value of three replicates. G, Immunoblotting of nuclear extracts from control and RAS-transduced WT and Nfkbiz−/− keratinocytes. Arrow head denotes specific band. H, Primary keratinocytes Nfkbiz+/− or Nfkbiz−/− were transduced with a retrovirus expressing oncogenic RAS and grafted together with WT dermal fibroblasts onto the backs of athymic mice. Each dot represents mean volume of individual tumors at day 20 postgrafting. Data shown represent two independent experiments and are reported as mean ± SEM. For all panels, *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Multiple downstream effectors of RAS transformation are regulated through IκBζ signaling
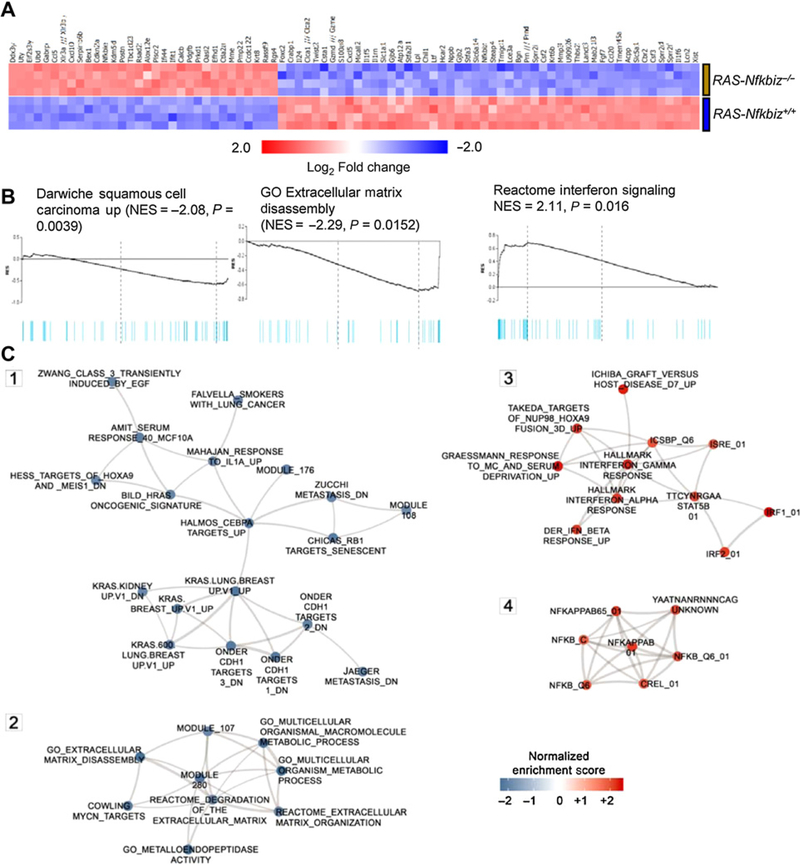
The contribution of IκBζ to the gene expression signature of RAS-mediated tumor initiation was explored by profiling the transcriptome of keratinocytes transformed by oncogenic RAS in the presence or absence of an IκBζ signaling pathway. The expression of 296 genes was significantly affected (by at least 1.5-fold change and FDR < 5%) by IκBζ deficiency (Supplementary Table S3; Fig. 6A), thereby confirming a selective impact of IκBζ signaling on the RAS-transformed phenotype. The global gene expression data from Nfkbiz-deficient and -sufficient RAS–initiated keratinocytes were analyzed by GSEA (Fig. 6B) using Qlucore implementation and subsets of MSigDB collections, namely C2 (curated gene sets), C3 (motif gene sets), C4 (computational gene sets), C5 (GO gene sets), and C6 (oncogenic gene sets). Gene sets were considered significantly enriched when the FDR estimate did not exceed 5% in a collection. Furthermore, GSEA results were visualized as a functional overlap network (subgraphs in Fig. 6C and described in Materials and Methods). This analysis revealed multiple IκBζ-regulated pathways that are highly associated with the transcriptome from RAS-transformed keratinocytes sufficient or deficient in IκBζ (Fig. 6B and C). For example, expression of specific proinflammatory factors that contribute to carcinogenesis are common to multiple experimental and human cancers associated with RAS transformation from diverse organ sites (Fig. 6C, subgraph 1; Supplementary Table S4). We now show that the upregulation of these factors is commonly dependent on IκBζ signaling (Supplementary Table S4). Similarly, Subgraph 2 (Fig. 6C; Supplementary Table S4) describes common groups of genes that are rich in components that modulate the extracellular matrix, particularly through the expression of membrane bound and secreted proteases. These genes are highly expressed in keratinocytes after RAS transformation, as we reported previously (44). Enrichment of these genes has IκBζ dependency in common. Subgraphs 3 and 4 (Fig. 6C; Supplementary Table S4) reveal common transcripts that may be suppressed by the action of IκBζ signaling because they increase in its absence in RAS-transformed keratinocytes. These include components of the NF-κB pathway (subgraph 4) and their downstream effectors and IFN pathway transcripts (subgraph 3; Fig. 6C; Supplementary Table S4).
Figure 6.
The RAS transcriptome is dependent on IκBz. A, Heatmap visualization of relative expression of the top 82 genes whose expression is altered by at least 2-fold when comparing RAS-keratinocytes Nfkbiz+/+ with RAS keratinocytes Nfkbiz−/−. Genes are ordered by fold change. B, GSEA enrichment plots are shown for top-enriched or top-depleted gene sets in RAS-transduced Nfkbiz−/− keratinocytes. The black line is the running enrichment score calculated along the ranked gene list; the vertical light blue bars in the plot indicate the position of the genes from the respective gene set. Supplementary Table S4 lists all GSEA estimates for these gene sets, including “leading edge” genes. C, Network visualization of top-enriched functional gene sets from MSigDB collections (GSEA FDR < 5.0%) in RAS-transduced Nfkbiz−/− keratinocytes. Network nodes represent subsets of the enriched gene sets including GSEA core enriched genes (“leading edge”)and are shown in blue and red for gene sets underexpressed and overexpressed in the RAS-transduced Nfkbiz−/− keratinocytes, respectively. Edges in the network represent mutual overlap of genes between the gene sets and the thickness of an edge is proportional to the combined similarity coefficient, ranging between 0.375 to 1.0. Supplementary Table S4 contains the underlying network data.
IκBζ-dependent RAS gene signature predicts disease free survival in COAD, LUAD, and PAAD
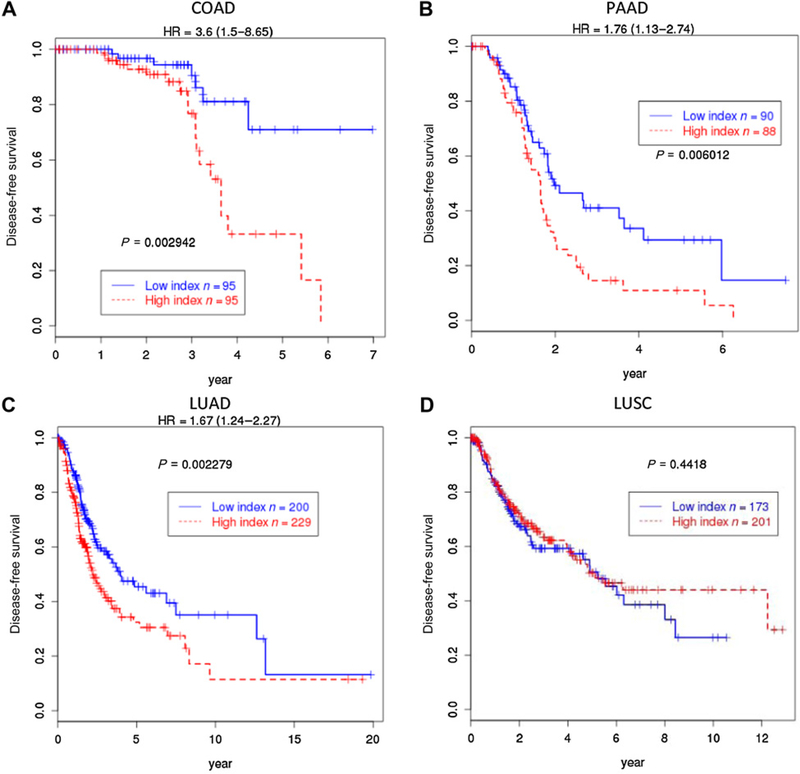
Using a subset of genes from the transcriptional profile developed for IκBζ dependency in RAS-transformed keratinocytes, we interrogated TCGA data from human cancers where oncogenic RAS is a key driver. We chose the 16 most upregulated genes upon loss of Nfkbiz in RAS keratinocytes to probe the TCGA COAD dataset and defined the sum of their expression as the index to divide the patients into low and high index groups by the median. Those patients with colon cancer with high index had significantly poor survival with HR = 3.6 and P = 0.003 (Fig. 7A). Similarly, for patients with pancreatic cancer, we used the TCGA PAAD dataset and selected 19 most upregulated genes by adding three additional genes to the 16 genes and define the weighted sum of the 19 genes as the index to divide the patients into low and high groups. The weights used to define the index were the log2-fold changes obtained from the mouse Nfkbiz deficiency experiment (Supplementary Table S3). The patients with pancreatic cancer with high index had significantly worse prognosis with HR = 1.76 and P = 0.006 (Fig. 7B). A similar relation of high index with poor prognosis was detected in the TCGA LUAD (Fig. 7C) dataset with HR = 1.67 and P = 0.002, but the relationship with TCGA LUSC was not significant (Fig. 7D). This difference could imply specificity for RAS mutations or RAS dependence for oncogenesis. Because high index represents higher expression of genes normally repressed by IκBζ in RAS keratinocytes, it implies that those patient tumors with poor prognosis have lower IκBζ activity. One possible cause for the lower IκBζ activity is a significant reduction of NFKBIZ expression with increasing tumor stage in COAD (Supplementary Fig. S2A). For human skin cancer, although based on a small population size, the trend to cancer progression and reduced NFKBIZ expression is also detected in two independent studies of human cutaneous cancers progressing through normal, actinic keratosis, and squamous cell cancer (Supplementary Fig. S2B).
Figure 7.
IkBz-dependent RAS gene signature predicts disease free survival in COAD, PAAD, and LUAD. A, A 16-gene sum index from the most overexpressed genes in RAS keratinocytes depleted of Nfkbiz was used to divide patients with COAD with high- and low-index groups to plot Kaplan-Meier curves. The patients with high index had worse prognosis with HR = 3.6 (1.5–8.65) and log-rank P = 0.003. A 19-gene weighted sum index from the same profile was used to divide the patients with PAAD (B), LUAD (C), and LUSC (D) with high- and low-index groups. The 16-gene set is a subset of the 19 genes and the weights used to compute the index were log2-fold changes derived from Nfkbiz-dependent gene in mouse RAS keratinocytes (Supplementary Table S3). The patients with PAAD with high index had worse prognosis with HR = 1.76 (1.13–2.74) and the log-rank P = 0.006. The patients with LUAD with high index had worse prognosis with HR = 1.67 (1.24–2.27) and the log-rank P = 0.002. But for the patients with LUSC, the high- and low-index groups had no significant difference in survival.
Discussion
A pathway to tumor promotion unveiled
The studies reported here propose a model by which a tissue-restrained oncogenic RAS-initiated epithelial cell establishes and responds to a microenvironment conducive to selective clonal outgrowth of transformed cells (Supplementary Fig. S3). Although presented in the context of skin, it is likely that tissue-constrained–initiated cells in multiple internal epithelia require altered tissue homeostasis via inflammation, hormones, injury to proliferate unimpeded by neighboring cells, or tissue architecture. All these stimuli are tumor-promoting factors. The emerging importance of IL17 as a mediator of tumor growth at multiple organ sites (15–20) supports the broader implications of these results. This analysis has also provided a roadmap for the events that follow the introduction of IL17-producing cells into the tumor microenvironment through the chemo-attractive consequences of RAS oncogenic activation of NF-κB and IκBζ signaling and the consequences of applying tumor-promoting agents to initiated skin. IL17, through IκBζ signaling, produces selective transcriptional changes in normal and incipient tumor cells, amplifying the signals coming from the promoting agent and encouraging the clonal expansion and outgrowth of RAS-initiated cells. Because RAS-initiated keratinocytes, unlike normal keratinocytes, are inherently resistant to terminal differentiation (45), they have the advantage of vertical growth to form visible papillomas. Of relevance to these observations, papillomas at high risk for malignant conversion that emerge first from promoted initiated skin, expressed higher levels of IL17 in transcriptional profiles than papillomas at low risk for malignant conversion (46).
Signaling through MyD88/NF-κB is essential for tumor promotion
A requirement for MyD88 signaling in experimental cutaneous squamous carcinogenesis has been previously documented (6, 8, 12) with a focus on tumor cell–autonomous functions. We now show a requirement for intact MyD88 signaling in the host, at both the target tissue independent of incipient tumor cells and in hematopoietic-derived cells remote from the site of tumor formation. Locally, the MyD88-dependent release of IL1α and CCL4 are good candidates to attract Th17 cells to the tumor microenvironment. Chung and colleagues (47) have documented a requirement for IL1α to induce the polarization of Th17 cells in the T-cell population, to maintain that phenotype and in combination with TGFβ and IL6 to elevate the production of IL17 by these cells. Deletion of the IL1R from T cells reduces tumor formation in skin supporting a crucial distant function for IL1/IL1R signaling in skin carcinogenesis (Supplementary Fig. S1). This connection of MyD88, IL1α and IL17 in cutaneous biology was previously seen in autoimmune cutaneous inflammation induced by deletion of Foxp3, where elevated cutaneous IL17, IL1α and skin inflammation are corrected by crossing Foxp3-null mice with Myd88-null mice (48). Similarly, deletion of IL1R in T cells prevented the protumorigenic cytokines IL17A and IL22 expression in a colorectal cancer mouse model (49).
Recruitment of Th17 cells and release of IL17 in the skin microenvironment are essential for tumor formation
In our study, the targeted deletion of MyD88 in T cells reduced the infiltration of CD4+ cells in TPA-treated skin, indicating that signals from skin were activating NF-κB in T cells to stimulate their migration, and transcriptional profiling alerted us to the specific deficiency in IL17 signaling. Previous studies have documented that directly deleting Il17 or the Il17r gene encoding its receptor from the mouse genome or targeted deletion of epidermal Traf3ip2 (also known as Act1 or CISK), the adapter required for IL17 signaling, reduced DMBA/TPA-induced skin tumors (10, 13, 14). Supporting these preclinical findings in mice, the tumor microenvironment surrounding human skin squamous and basal cell carcinomas is abundant in CD4+IL17+ T cells (50). Several previous studies have also reported that CD4+ T cells contribute to skin carcinogenesis (37, 51, 52). Those studies focused on T-cell extrinsic pathways to explore the basis for the infiltration. By targeting the NF-κB pathway directly in CD4+ T cells, we demonstrate the intrinsic requirements in the target hematopoietic population that respond to the chemotactic signals emanating from the tissue targeted for tumor formation. This, then, would appear to be the basic connection between the host and the target tissue permissive for tumor development. Little previous work on IL17 in the skin has focused on the differential response of normal and incipient tumor cells other than to show that recombinant IL17 can induce hyperplasia when injected intradermally (14). We now demonstrate the powerful IL17 responses involved to produce hyperplasia (stimulated proliferation and blocked differentiation of normal keratinocytes) and the upregulation of multiple cytokines and chemokines (normal and initiated keratinocytes) relevant for tumor eruption. Unexpectedly, we now discover that some of these IL17 effects are mediated through IκBζ signaling, and the consequences of RAS transformation in keratinocytes, including tumor formation, are strongly dependent on IκBζ, independent of IL17. In fact, exposure to IL17 elevates IκBζ expression in normal and RAS-initiated keratinocytes possibly creating a feed-forward loop. Multiple reports indicate that IκBζ has both gene-activating and gene-suppressing effects in combination with components of NF-κB.
IκBζ contributes to multiple stages in tumor formation
Nfkbiz genetic deficiency in mice indirectly produces an inflammatory skin phenotype mimicking atopic dermatitis by inducing skin dysbiosis and over-representation of the pathobiont Staphylococcus xylosus (53, 54). GSEA from RNA profiling of RAS-transformed IκBζ heterozygous- and homozygous-deficient keratinocytes confirms a diverse pattern of gene expression changes. For example, the loss of key inflammatory mediators (e.g., S100 proteins) and modifiers of the extracellular matrix (e.g., MMPs and TMPRSS) in RAS-initiated keratinocytes deficient in IκBζ could contribute to the reduction of tumor formation. These factors released from initiated cells enhance a receptive microenvironment for tumor formation in the skin. While these enrichment studies were performed in mouse keratinocytes, the high association with datasets from multiple gene expression studies in other organ sites (MSigDB) suggest that IκBζ is an active participant in RAS-related carcinogenesis in general. GSEA also revealed that IκBζ-mediated transcriptional regulation suppresses IFN-responsive genes in RAS-transformed keratinocytes, a function that could influence tumor growth through effects on immune modulation. While both tumor cell–autonomous and microenvironmental IκBζ signaling are important for the early stages of RAS-mediated tumor formation in mouse skin, the transcriptional signature associated with the loss of IκBζ signaling in RAS transformation marks a poor prognosis for human patients with cancer. This implies that during cancer progression, RAS-transformed cells switch from a microenvironmental-driven IκBζ dependency to more cell autonomy and alternative drivers. The discovery of IκBζ signaling as an integral component of cancer pathogenesis, with particular relevance to cancers driven by oncogenic RAS, warrants further study both as a therapeutic target for early neoplastic lesions and a prognostic indicator of cancer progression.
Supplementary Material
Acknowledgments
The authors thank Steve Jay for tumor volume measurements, Marta Custer and Jennifer Waters for the mouse colony care, Roberta Mattai and Kathleen Noer for cell sorting prior to microarray analysis, Jeff Subleski for reagents and advice on cell sorting, Professor Shizuo Akira from Osaka University (Osaka, Japan) for Nfkbiz−/− mice. This work was supported by funds from Intramural Research Program of the NIH, NCI, Center for Cancer Research, Project ZIA BC 004504.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary data for this article are available at Molecular Cancer Research Online (http://mcr.aacrjournals.org/).
References
- 1.DiDonato JA, Mercurio F, Karin M. NF-kappaB and the link between inflammation and cancer. Immunol Rev 2012;246:379–400. [DOI] [PubMed] [Google Scholar]
- 2.Tafani M, Pucci B, Russo A, Schito L, Pellegrini L, Perrone GA, et al. Modulators of HIF1alpha and NFkB in cancer treatment: is it a rational approach for controlling malignant progression? Front Pharmacol 2013;4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ngo VN, Young RM, Schmitz R, Jhavar S, Xiao W, Lim KH, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature 2011; 470:115–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Treon SP, Xu L, Yang G, Zhou Y, Liu X, Cao Y, et al. MYD88 L265P somatic mutation in Waldenstrom’s macroglobulinemia. N Engl J Med 2012;367: 826–33. [DOI] [PubMed] [Google Scholar]
- 5.Je EM, Kim SS, Yoo NJ, Lee SH. Mutational and expressional analyses of MYD88 gene in common solid cancers. Tumori 2012;98:663–9. [DOI] [PubMed] [Google Scholar]
- 6.Coste I, Le Corf K, Kfoury A, Hmitou I, Druillennec S, Hainaut P, et al. Dual function of MyD88 in RAS signaling and inflammation, leading to mouse and human cell transformation. J Clin Invest 2010;120:3663–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salcedo R, Cataisson C, Hasan U, Yuspa SH, Trinchieri G. MyD88 and its divergent toll in carcinogenesis. Trends Immunol 2013;34:379–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cataisson C, Salcedo R, Hakim S, Moffitt BA, Wright L, Yi M, et al. IL-1R-MyD88 signaling in keratinocyte transformation and carcinogenesis. J Exp Med 2012;209:1689–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valkenburg KC, de Groot AE, Pienta KJ. Targeting the tumour stroma to improve cancer therapy. Nat Rev Clin Oncol 2018;15:366–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L, Yi T, Zhang W, Pardoll DM, Yu H. IL-17 enhances tumor development in carcinogen-induced skin cancer. Cancer Res 2010;70: 10112–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mittal D, Saccheri F, Venereau E, Pusterla T, Bianchi ME, Rescigno M. TLR4-mediated skin carcinogenesis is dependent on immune and radioresistant cells. EMBO J 2010;29:2242–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swann JB, Vesely MD, Silva A, Sharkey J, Akira S, Schreiber RD, et al. Demonstration of inflammation-induced cancer and cancer immunoediting during primary tumorigenesis. Proc Natl Acad Sci U S A 2008;105: 652–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He D, Li H, Yusuf N, Elmets CA, Athar M, Katiyar SK, et al. IL-17 mediated inflammation promotes tumor growth and progression in the skin. PLoS One 2012;7:e32126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu L, Chen X, Zhao J, Martin B, Zepp JA, Ko JS, et al. A novel IL-17 signaling pathway controlling keratinocyte proliferation and tumorigenesis via the TRAF4-ERK5 axis. J Exp Med 2015;212:1571–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le Gouvello S, Bastuji-Garin S, Aloulou N, Mansour H, Chaumette MT, Berrehar F, et al. High prevalence of Foxp3 and IL17 in MMR-proficient colorectal carcinomas. Gut 2008;57:772–9. [DOI] [PubMed] [Google Scholar]
- 16.Carmi Y, Rinott G, Dotan S, Elkabets M, Rider P, Voronov E, et al. Microenvironment-derived IL-1 and IL-17 interact in the control of lung metastasis. J Immunol 2011;186:3462–71. [DOI] [PubMed] [Google Scholar]
- 17.McAllister F, Bailey JM, Alsina J, Nirschl CJ, Sharma R, Fan H, et al. Oncogenic Kras activates a hematopoietic-to-epithelial IL-17 signaling axis in preinvasive pancreatic neoplasia. Cancer Cell 2014;25: 621–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loncle C, Bonjoch L, Folch-Puy E, Lopez-Millan MB, Lac S, Molejon MI, et al. IL17 functions through the novel REG3beta-JAK2-STAT3 inflammatory pathway to promote the transition from chronic pancreatitis to pancreatic cancer. Cancer Res 2015;75:4852–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Housseau F, Wu S, Wick EC, Fan H, Wu X, Llosa NJ, et al. Redundant innate and adaptive sources of IL17 production drive colon tumorigenesis. Cancer Res 2016;76:2115–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walch-Ruckheim B, Mavrova R, Henning M, Vicinus B, Kim YJ, Bohle RM, et al. Stromal fibroblasts induce CCL20 through IL6/C/EBPbeta to support the recruitment of Th17 cells during cervical cancer progression. Cancer Res 2015;75:5248–59. [DOI] [PubMed] [Google Scholar]
- 21.Kleinridders A, Schenten D, Konner AC, Belgardt BF, Mauer J, Okamura T, et al. MyD88 signaling in the CNS is required for development of fatty acid-induced leptin resistance and diet-induced obesity. Cell Metab 2009;10: 249–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamamoto M, Yamazaki S, Uematsu S, Sato S, Hemmi H, Hoshino K, et al. Regulation of Toll/IL-1-receptor-mediated gene expression by the inducible nuclear protein IkappaBzeta. Nature 2004;430:218–22. [DOI] [PubMed] [Google Scholar]
- 23.Lichti U, Anders J, Yuspa SH. Isolation and short-term culture of primary keratinocytes, hair follicle populations and dermal cells from newborn mice and keratinocytes from adult mice for in vitro analysis and for grafting to immunodeficient mice. Nat Protoc 2008;3: 799–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Threadgill DW, Dlugosz AA, Hansen LA, Tennenbaum T, Lichti U, Yee D, et al. Targeted disruption of mouse EGF receptor: effect of genetic background on mutant phenotype. Science 1995;269:230–4. [DOI] [PubMed] [Google Scholar]
- 25.Day CP, Carter J, Bonomi C, Esposito D, Crise B, Ortiz-Conde B, et al. Lentivirus-mediated bifunctional cell labeling for in vivo melanoma study. Pigment Cell Melanoma Res 2009;22:283–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cataisson C, Pearson AJ, Tsien MZ, Mascia F, Gao JL, Pastore S, et al. CXCR2 ligands and G-CSF mediate PKCalpha-induced intraepidermal inflammation. J Clin Invest 2006;116:2757–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, et al. Compartmentalized control of skin immunity by resident commensals. Science 2012;337:1115–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 2010;5:e13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gebhardt C, Nemeth J, Angel P, Hess J. S100A8 and S100A9 in inflammation and cancer. Biochem Pharmacol 2006;72:1622–31. [DOI] [PubMed] [Google Scholar]
- 31.Rundhaug JE, Fischer SM. Molecular mechanisms of mouse skin tumor promotion. Cancers 2010;2:436–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen J, Abel EL, Riggs PK, Repass J, Hensley SC, Schroeder LJ, et al. Proteomic and pathway analyses reveal a network of inflammatory genes associated with differences in skin tumor promotion susceptibility in DBA/2 and C57BL/6 mice. Carcinogenesis 2012;33:2208–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiricozzi A, Guttman-Yassky E, Suarez-Farinas M, Nograles KE, Tian S, Cardinale I, et al. Integrative responses to IL-17 and TNF-alpha in human keratinocytes account for key inflammatory pathogenic circuits in psoriasis. J Invest Dermatol 2011;131:677–87. [DOI] [PubMed] [Google Scholar]
- 34.Shen F, Hu Z, Goswami J, Gaffen SL. Identification of common transcriptional regulatory elements in interleukin-17 target genes. J Biol Chem 2006;281:24138–48. [DOI] [PubMed] [Google Scholar]
- 35.Amatya N, Garg AV, Gaffen SL. IL-17 Signaling: the Yin and the Yang. Trends Immunol 2017;38:310–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mangan PR, Harrington LE, O’Quinn DB, Helms WS, Bullard DC, Elson CO, et al. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature 2006;441:231–4. [DOI] [PubMed] [Google Scholar]
- 37.Ortiz ML, Kumar V, Martner A, Mony S, Donthireddy L, Condamine T, et al. Immature myeloid cells directly contribute to skin tumor development by recruiting IL-17-producing CD4+ T cells. J Exp Med 2015; 212:351–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koenders MI, Devesa I, Marijnissen RJ, Abdollahi-Roodsaz S, Boots AM, Walgreen B, et al. Interleukin-1 drives pathogenic Th17 cells during spontaneous arthritis in interleukin-1 receptor antagonist-deficient mice. Arthritis Rheum 2008;58:3461–70. [DOI] [PubMed] [Google Scholar]
- 39.Yamazaki S, Muta T, Takeshige K. A novel IkappaB protein, IkappaB-zeta, induced by proinflammatory stimuli, negatively regulates nuclear factor-kappaB in the nuclei. J Biol Chem 2001;276:27657–62. [DOI] [PubMed] [Google Scholar]
- 40.Kao CY, Kim C, Huang F, Wu R. Requirements for two proximal NF-kappaB binding sites and IkappaB-zeta in IL-17A-induced human beta-defensin 2 expression by conducting airway epithelium. J Biol Chem 2008;283: 15309–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karlsen JR, Borregaard N, Cowland JB. Induction of neutrophil gelatinase-associated lipocalin expression by co-stimulation with interleukin-17 and tumor necrosis factor-alpha is controlled by IkappaB-zeta but neither by C/ EBP-beta nor C/EBP-delta. J Biol Chem 2010;285:14088–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johansen C, Mose M, Ommen P, Bertelsen T, Vinter H, Hailfinger S, et al. IkappaBzeta is a key driver in the development of psoriasis. Proc Natl Acad Sci U S A 2015;112:E5825–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin-Oliva D, Aguilar-Quesada R, O’Valle F, Munoz-Gamez JA, Martinez-Romero R, Garcia Del Moral R, et al. Inhibition of poly(ADP-ribose) polymerase modulates tumor-related gene expression, including hypoxia-inducible factor-1 activation, during skin carcinogenesis. Cancer Res 2006; 66:5744–56. [DOI] [PubMed] [Google Scholar]
- 44.Cataisson C, Michalowski AM, Shibuya K, Ryscavage A, Klosterman M, Wright L, et al. MET signaling in keratinocytes activates EGFR and initiates squamous carcinogenesis. Sci Signal 2016;9:ra62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuspa SH, Morgan DL. Mouse skin cells resistant to terminal differentiation associated with initiation of carcinogenesis. Nature 1981;293:72–4. [DOI] [PubMed] [Google Scholar]
- 46.Glick A, Ryscavage A, Perez-Lorenzo R, Hennings H, Yuspa S, Darwiche N. The high-risk benign tumor: evidence from the two-stage skin cancer model and relevance for human cancer. Mol Carcinog 2007;46:605–10. [DOI] [PubMed] [Google Scholar]
- 47.Chung Y, Chang SH, Martinez GJ, Yang XO, Nurieva R, Kang HS, et al. Critical regulation of early Th17 cell differentiation by interleukin-1 signaling. Immunity 2009;30:576–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rivas MN, Koh YT, Chen A, Nguyen A, Lee YH, Lawson G, et al. MyD88 is critically involved in immune tolerance breakdown at environmental interfaces of Foxp3-deficient mice. J Clin Invest 2012;122:1933–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dmitrieva-Posocco O, Dzutsev A, Posocco DF, Hou V, Yuan W, Thovarai V, et al. Cell-type-specific responses to interleukin-1 control microbial invasion and tumor-elicited inflammation in colorectal cancer. Immunity 2019;50:166–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nardinocchi L, Sonego G, Passarelli F, Avitabile S, Scarponi C, Failla CM, et al. Interleukin-17 and interleukin-22 promote tumor progression in human nonmelanoma skin cancer. Eur J Immunol 2015;45:922–31. [DOI] [PubMed] [Google Scholar]
- 51.Daniel D, Meyer-Morse N, Bergsland EK, Dehne K, Coussens LM, Hanahan D. Immune enhancement of skin carcinogenesis by CD4+ T cells. J Exp Med 2003;197:1017–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yusuf N, Nasti TH, Katiyar SK, Jacobs MK, Seibert MD, Ginsburg AC, et al. Antagonistic roles of CD4+ and CD8+ T-cells in 7,12-dimethylbenz(a)anthracene cutaneous carcinogenesis. Cancer Res 2008;68: 3924–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim Y, Lee YS, Yang JY, Lee SH, Park YY, Kweon MN. The resident pathobiont Staphylococcus xylosus in Nfkbiz-deficient skin accelerates spontaneous skin inflammation. Sci Rep 2017;7:6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shiina T, Konno A, Oonuma T, Kitamura H, Imaoka K, Takeda N, et al. Targeted disruption of MAIL, a nuclear IkappaB protein, leads to severe atopic dermatitis-like disease. J Biol Chem 2004;279:55493–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.