Abstract
CircRNAs are a class of noncoding RNA species with a circular configuration that is formed by either typical spliceosome-mediated or lariat-type splicing. The expression of circRNAs is usually abnormal in many cancers. Several circRNAs have been demonstrated to play important roles in carcinogenesis. In this review, we will first provide an introduction of circRNAs biogenesis, especially the regulation of circRNA by RNA-binding proteins, then we will focus on the recent findings of circRNA molecular mechanisms and functions in cancer development. Finally, some open questions are also discussed.
Keywords: circRNAs, carcinogenesis, biogenesis, RNA-binding protein, flanking introns, genomic alternation
1. Introduction
Discovered more than three decades ago [1], circRNAs have attracted extensive attention in recent years [2,3,4]. As a member of the noncoding RNA family, circRNAs display a unique covalently closed circular form, which distinguishes it from its other noncoding RNA cousins such as microRNA and lncRNA. High-throughput RNA-seq studies have detected a large number of circRNAs with different lengths and types. Initial analyses of the sequencing data indicate they are specifically expressed during tissue/developmental stages and conserved between mice and humans [3,4]. Emerging evidence has demonstrated that some circRNAs have various biological functions and play potentially important roles in multiple diseases such as cancers [5,6,7].
Here, we summarize the expanding findings on circRNA and provide an up-to-date account of their biogenesis, regulatory mechanisms, and cellular functions in carcinogenesis.
2. The Biogenesis of circRNAs in Cancers
In eukaryotes, linear mRNA is formed by processing exons of pre-mRNAs through alternative splicing. Unlike linear RNAs, circRNAs were originally thought to be aberrant RNA splicing products [8]. Because of their covalent closed-loop structure, circRNAs are protected from degradation by RNases, so they are more stable than linear RNAs [3]. When analyzing 17 different cancer cohorts from the MiOncoCirc compendium, Vo et al. characterized circRNAs across >2000 cancer samples and observed a general decrease in total circRNA abundance compared to the adjacent normal tissues [9].
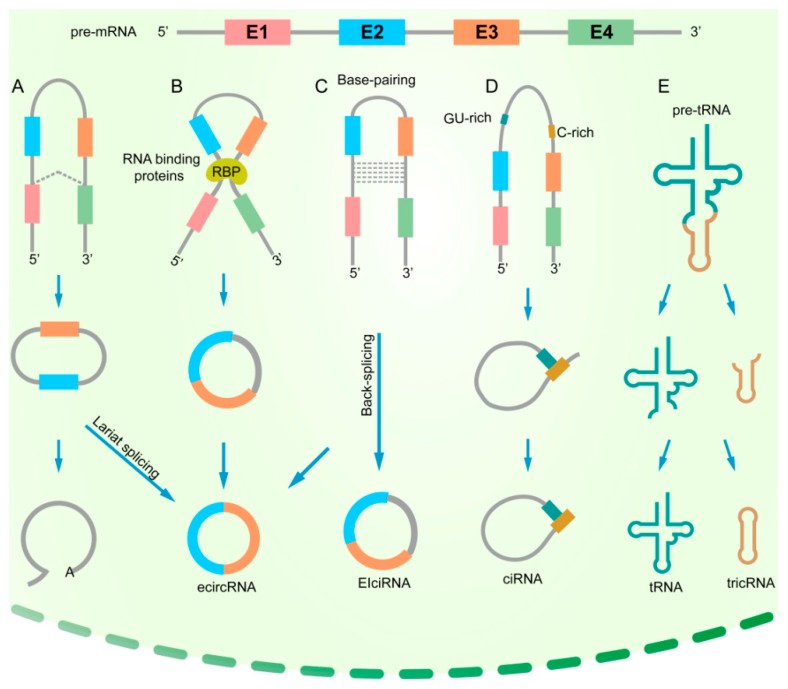
With the optimization of next-generation sequencing technology, several circRNA subtypes have been discovered in recent years. There are four main subtypes of circRNAs: exonic circRNAs (ecircRNAs), which are mainly derived from single or several exons; on the other side, circular intronic RNAs (ciRNAs) contain only introns; exonic-intronic circRNAs (EIciRNAs), which consist of both introns and exons; and tRNA intronic circRNAs (tricRNAs) are formed by splicing pre-tRNA intron (Figure 1). Currently, most of the identified circRNAs are exonic circRNAs.
Figure 1.
Biogenesis of circRNAs. (A) Lariat-driven circularization. When a pre-mRNA is spliced, the 3′ hydroxyl of the upstream exon interacts with the 5′ phosphate of the downstream exon to form a covalent linkage, producing a lariat that contains exons and introns. The 2′ hydroxyl of the 5′ intron reacts with the 5′ phosphate of the 3′-intron, followed by an interaction between the 3′ hydroxyl of the 3′ exon and the 5′ phosphate of the 5′ exon, through which an ecircRNA is formed. (B) RNA-binding protein (RBP)-driven circularization. RBPs can promote the interaction of the downstream intron and upstream intron, causing the formation of an ecircRNA. (C) Base-pairing-driven circularization. The downstream introns and upstream introns are paired based on inverse-repeating or complementary sequences. The introns are removed or retained to form ecircRNA or EIciRNA. (D) Biosynthesis of ciRNA. Formation of ciRNAs mainly depends on a 7-nt GU-rich element and an 11-nt C-rich element to escape debranching and exonucleolytic degradation. (E) Formation of tricRNA. tRNA splicing enzymes divide pre-tRNA into two parts: tricRNAs are generated by a 3′–5′ phosphodiester bond, and the other part generates tRNAs.
The formation mechanisms of different types of circRNAs are also diversely regulated. Generation of exonic circRNAs can be achieved by both lariat-driven circularization and back splicing. When the upstream splice acceptor and downstream donor are close to forming a lariat containing the exons, the introns in the lariat are removed, and the exons are joined by a 5′–3′ phosphodiester bond (Figure 1A) [10]. Interactions between RNA-binding proteins (RBPs) form a ‘bridge’ within the upstream and downstream introns, followed by back splicing to form ecircRNAs (Figure 1B) [11,12]. During base-pairing-driven circularization, the downstream splicing donor is connected to the upstream splicing receptor depending on ALU complementary sequences. In the process of circRNA formation, the introns are removed or retained to form ecircRNA or EIciRNA(Figure 1C) [3]. The biosynthesis of ciRNAs mainly depends on a 7-nt GU-rich element and an 11-nt C-rich element to escape debranching and exonucleolytic degradation (Figure 1D) [13]. Unlike ecircRNAs, which are mainly located in the cytoplasm, ciRNAs and EIciRNAs are mainly distributed in the nucleus and play a crucial role in regulating parental gene transcription. A special intronic circRNA, tricRNA, has been found in Archaea and Drosophila [14,15,16]. The formation of tricRNA needs tRNA splicing enzymes to divide pre-tRNA into two parts: tricRNAs are generated by a 3′–5′ phosphodiester bond, and the other part generates tRNAs (Figure 1E) [15]. It is generally accepted that both the binding of RNA proteins and the presence of specific and repetitive sequences in the introns surrounding the circularizing exons determine the production of circRNAs.
2.1. Introns Flank circRNAs
Previously, analysis of the surrounding introns of circRNAs in human fibroblasts has indicated they are usually longer and harbor complementary ALU repeats [3]. A recent analysis of circRNAs in over 2000 cancer samples confirmed this finding [9]. These repetitive sequences are thought to facilitate circular RNA formation [17]. Besides, Ivanov et al. found that the reverse complementary sequences are conserved and highly enriched within introns that bracket circRNAs. They also successfully developed a computational system to identify novel circRNAs based on scoring the presence of reverse complementary sequences in human introns [18]. Furthermore, introns upstream or downstream circularized exons, are preferentially harbor more RNA editing events, which proved to affect the formation of circRNA [18,19].
2.2. Regulation of circRNA Biogenesis by RNA-Binding Proteins (RBPs)
RNA-binding proteins, which usually contain RNA-binding motifs, play a central role in transcriptional regulation of genes [20]. Because of the development of high-throughput methods, such as RNA sequencing and mass spectrometry, about ~1500 RBPs have been identified from the human genome until now [21,22]. Bioinformatics analyses of the alteration spectrum across thousands of human cancers samples identified ~9% of cancer-related RBPs, highlighting its potential roles in tumorigenesis [23]. RBPs can shorten the distance between the donor site and the receptor site by binding to the introns on the flank regions, thus promoting circularization of the exons in circRNA biogenesis.
QKI, KH domains containing RNA binding proteins, is the first identified RBP involved in circRNA formation during the epithelial to mesenchymal transition. It enhances circRNA formation by binding to its consensus target motif, single-stranded RNA (ssRNA), in introns that flank the circRNA-forming exons. Consequently, insertion of synthetic QKI binding sites into introns was sufficient to produce circRNAs [12]. Based on genome-wide siRNA screening and circRNA expression reporter assays, Li et al. identified that the immune factors nuclear factor 90 and its 110 isoform (NF90/NF110) could couple circRNA biogenesis and function during viral infection. They found the nuclear export of NF90/NF110 to the cytoplasm correlated with decreased circRNA expression, while NF90/NF110 bound to viral mRNAs to stimulate an antiviral immune response [24]. Epithelial splicing regulatory protein 1 (ESRP1) is an essential splicing factor during pluripotency [25]. A recent report found an elegant regulating network involved in circRNA and RNA-binding protein in human embryonic stem cells (hESCs). In this scenario, NANOG and OCT4 regulate ESRP1 expression in hESCs, which leads to the promotion of circBIRC6 generation. Consequently, circBIRC6 works as a ‘molecular reservoir’ of miR-34a and miR-145, collectively contributing to pluripotency maintenance [26].
Different RBPs may play different, or even opposite, roles in the back-splicing process. For example, all the above three RBPs can promote the production of circRNAs, whereas the RNA-editing enzyme (that edits adenosine to inosine) acts on RNA enzyme 1 (ADAR1) to inhibit circRNA formation [18]. ADAR1 binds double-stranded RNA to mediate adenosine-to-inosine (A-to-I) RNA editing [27]. Large scale analyses of multiple cancer samples from The Cancer Genome Atlas (TCGA) indicate RNA editing events in tumor samples correlate best with the ADAR1 expression level globally. Approximately 3.5% of detected RNA editing sites are associated with potential clinical relevance, many of which are in noncoding regions [19]. This is consistent with a report that A-to-I editing is also enriched with the base-paired or proximal regions of circularized exons [18]. A mechanistic model suggested RNA editing sites may disrupt the RNA–RNA interactions of introns to form larger structures [28]. This is supported by the evidence that ADAR1 depletion can up-regulate the formation of circRNAs [18]. The known instances of RBPs have been summarized in Table 1.
Table 1.
RNA-binding proteins that regulate circRNA biogenesis.
| Gene | Effect on the Formation of circRNA | Mechanisms | PMID |
|---|---|---|---|
| QKI | Promote | QKI enhances circRNA formation by binding to its consensus target single-stranded RNA (ssRNA) motif in introns that flank circRNA-forming exons. | 25768908 |
| TNRC6A | Promote | The RNA-binding protein TNRC6A binds to the flanking intron sequence of circHOMER1 and regulates the formation of circHOMER1. In the absence of TNRC6A, circHOMER1 cannot be effectively looped. | 29726904 |
| NF90/NF110 | Promote | NF90/NF110 stimulates circRNA formation by promoting the stability of intron complementary sequences | 28625552 |
| HNRNPL | Promote | Circular RNA formation regulated by HNRNPL back splicing. | 28611215 |
| MBL/MBNL1 | Promote | MBL/MBNL1 strongly and specifically binds with circMbl flanking introns, which contain conserved muscleblind binding sites and strongly affects circMbl biosynthesis | 25242144 |
| ESRP1 | Promote | The splicing factor ESRP1, which is controlled by the core pluripotency-associated factors, OCT4 and NANOG, can lead to the promotion of circBIRC6 generation. | 29074849 |
| FUS | Inhibit | FUS was found to regulate circRNA generation by binding introns that flank back-splicing junctions | 28358055 |
| DHX9 | Inhibit |
DHX9 can bind to IRAlus and possesses RNA helicase activity. It was speculated that DHX9 may inhibit circRNA expression by unwinding RNA pairs that flank circularized exons. |
28355180 |
| ADAR1 | Inhibit | ADAR1 binds double-stranded RNA to mediate adenosine-to-inosine (A-to-I) RNA editing to inhibit circRNA formation. | 25558066 |
2.3. Impacts of Genomic Alterations on the Formation of circRNAs
Oncogenic gene fusion is commonly found in cancer samples and plays a role in carcinogenesis. Guarnerio et al. hypothesized chromosome rearrangement will result in the juxtaposition at a close enough proximity to favor new events of back splicing, which would promote the generation of aberrant circRNAs [29]. To verify this hypothesis, they examined the most recurrent translocation fusion genes, such as PML-RARa and MLL/AF9 fusion genes, in leukemia. Circular forms of fusion circRNAs (f-circRNA) are indeed produced from transcribed exons of distinct genes and affected by the translocations. Furthermore, they also found circular EWSR1/FLI1 fusion in SK-NEP-1 sarcoma cells and EML4/ALK1 in H3122 lung cancer cells. However, a recent search of MiOncoCirc data, which included 17 cancer sequencing datasets, did not detect any f-circRNAs resulting from chromosomal translocations and deletions. Instead, they discovered read-through circRNA (rt-circRNA), a novel class of circular transcripts that span exons originating from two adjacent genes on the same strand. They found some of these rt-circRNA reads were commonly found across different cancer types, and they were even detected in normal tissues or in samples with a normal copy number of the parent genes, which indicated the prevalence of rt-circRNAs in cancer samples [9].
3. Functional Mechanisms of circRNAs
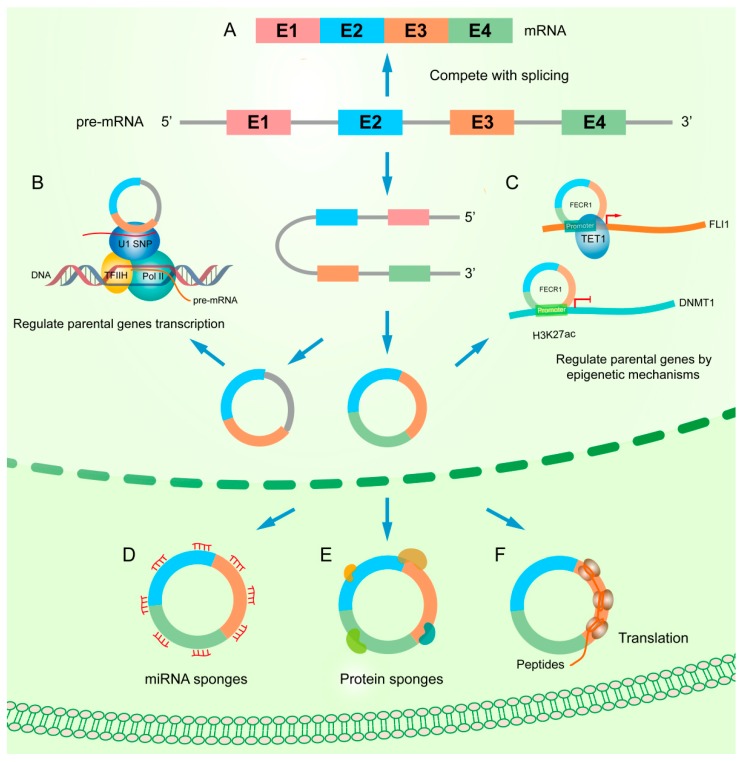
The functional mechanism of circRNAs are diverse in human cancer including acting as miRNA sponges, interacting with protein, regulating gene splicing or transcription, translating proteins, or peptide and epigenetic regulation (Figure 2). According to the targets of circRNAs, they can generally be classified into two categories: one is to regulate its hosting gene, the other is to target different ones. We have summarized the known instances in Table 2.
Figure 2.
The molecular mechanism of circRNAs (A) Competition with splicing. The formation of circRNA competes with the splicing of linear RNA. (B) Regulation of parental gene transcription. ciRNAs and EIciRNAs, with retained introns, can bind to U1 snRNP through RNA–RNA interactions and further interact with the Pol II transcription complex to enhance parental gene expression. (C) Circular RNA FECR1 from FLI1 gene interacts with FLI1 promoter, recruits TET1 demethylase, and induces extensive DNA demethylation in the CpG islands. In addition, FECR1 inhibits DNMT1, a critical enzyme that maintains DNA demethylation during DNA replication, by binding to its promoter region rich in H3K27ac. (D) miRNA sponges. CircRNA affects the expression of miRNA and downstream target genes by adsorbing miRNAs. (E) Protein sponges. CircRNA regulates target proteins by binding to them. (F) Translation. CircRNAs can be translated into peptides or proteins.
Table 2.
Different mechanism of circRNAs in human cancers.
| Function | CircRNA | Cancer Type | Expression | Targeting miRNA or Genes | Mechanisms | PMID |
|---|---|---|---|---|---|---|
| Acting as miRNA sponge | circAGFG1 | TNBC | Up | miR-195-5p/CCNE1 | CircAGFG1 can promote TNBC cell proliferation, mobility, and invasion as well as tumorigenesis and metastasis in vivo by acting as a ceRNA (competing endogenous RNA) of miR-195-5p to relieve the repressive effect of miR-195-5p on its target cyclin E1 (CCNE1). | 30621700 |
| circHIPK3 | CRC | Up | miR-7/FAK, IGF1R, YY1, EGFR | circHIPK3 promotes proliferation/migration | 29549306 | |
| circCDR1 | ESCC | Up | miR-7/HOXB13 | CirsCDR1 functions as the sponge of miR-7 and reactivates its downstream HOXB13-mediated NF-κB/p65 pathway. | 30082829 | |
| circHIPK3 | BCa | Down | miR-558/HPSE | circHIPK3 targets miR-558 to suppress the expression of HPSE and inhibits migration, invasion, and angiogenesis of bladder cancer cells in vitro and suppresses bladder cancer growth and metastasis in vivo. | 28794202 | |
| circTRIM33-12 | HCC | Down | miR-191/ TET1 | CircTRIM33-12 upregulate TET1 expression by sponging miR-191, resulting in significantly reduced 5-hydroxymethylcytosine (5hmC) levels in HCC cells. | 31153371 | |
| circTADA2As | TNBC | Down | miR-203a-3p/SOCS3 | circTADA2As suppresses cell proliferation, migration, invasion, and clonogenicity and possesses a tumor-suppressor capability. | 30787278 | |
| circLARP4 | GC | Down | miR-424-5p/ LATS1 | circLARP4 is mainly localized in the cytoplasm and inhibits biological behaviors of GC cells by sponging miR-424. | 28893265 | |
| circATP2B1 | ccRCC | Down | miR-204-3p/FN1 | CircATP2B1 is suppressed by ERβ, and then reduces miR-204-3p, which increases fibronectin 1 expression and enhances ccRCC cell invasion. | 29490945 | |
| circITCH | BCa | Down | miR-17, miR-224/p21, PTEN | circITCH promotes the aggressive biological behaviors of BCa via up-regulating the expression of p21 and PTEN through ‘sponging’ miR-17 and miR-224 | 29386015 | |
| circMTO1 | HCC | Down | miR-9/ p21 | circMTO1 can down-regulate p21 by acting as the sponge of oncogenic miR-9 to suppress hepatocellular carcinoma progression. | 28520103 | |
| circDB | HCC | Up | miR-34a/USP7 | CircDB promotes tumor growth and reduces DNA damage by suppressing miR-34a and activating the USP7/Cyclin A2 signaling pathway | 30546088 | |
| Binding to proteins | CircACC1 | CRC | Up | AMPK | CircACC1 functions to stabilize and promote the enzymatic activity of the AMPK holoenzyme by forming a ternary complex with the regulatory β and γ subunits. | 31155494 |
| circDNMT1 | BRCA | Up | P53, AUF1 | Ectopically expressed circDnmt1 promotes the nuclear translocation of both p53 and AUF1, p53 nuclear translocation induces cellular autophagy, while AUF1 nuclear translocation reduces Dnmt1 mRNA instability, resulting in increased Dnmt1 translation. | 29973691 | |
| circAGO2 | GC | Up | HuR | circAGO2 binds with HuR protein to promote its activation and enrichment on the 3′-untranslated region of target genes, which reduces AGO2 binding and repression of AGO2/miRNA-mediated gene silencing | 30341421 | |
| circPABPN1 | Hela cell | Up | HuR | The binding of CircPABPN1 to HuR inhibits HuR binding to PABPN1 mRNA and reduces PABPN1 translation. | 28080204 | |
| circFOXO3 | BRCA | Down | circFOXO3, p53 | CircFoxo3 promotes MDM2-induced p53 ubiquitination and subsequent degradation by binding to Foxo3 protein and p53, resulting in cell apoptosis. | 27886165 | |
| circCCNB1 | BRCA | Down | Ccnb1/Cdk1 | CircCcnb1 can interact with both Ccnb1 and Cdk1 proteins. Ectopic delivery of circCcnb1 inhibits tumor growth and extends mouse viability. | 31199987 | |
| Translating proteins or peptide | circCTNNB1 | HCC | Up | 370-amino acid β-catenin isoform | CircCTNNB1 produces a novel, 370 amino acid β-catenin isoform that uses the start codon as the linear β-catenin mRNA transcript, and translation is terminated at a new stop codon created by circularization. | 31027518 |
| CircPINT | GBM | Up | PINT87aa | lncRNA-PINT can be translated into a small peptide to suppress glioblastoma cell proliferation | 30367041 | |
| CircE7 | Derived from human papillomavirus and presented in CESC and HNSC | Up | E7 protein | Specific disruption of circE7 in CaSki cervical carcinoma cells reduces E7 protein levels and inhibits cancer cell growth both in vitro and in tumor xenografts. | 31127091 | |
| circSHPRH | GBM | Down | SHPRH-146aa | SHPRH-146aa is a tumor suppressor in human glioblastoma, which is translated by circ-SHPRH. | 29343848 | |
| circFBXW7 | GBM | Down | FBXW7-185aa | The spanning junction open reading frame in circ-FBXW7 driven by internal ribosome entry site encodes a novel 21 kDa protein (FBXW7-185aa). Upregulation of FBXW7-185aa in cancer cells inhibits proliferation and cell cycle acceleration. | 28903484 | |
| Regulating parental gene expression at multiple levels | circFECR1 | BRCA | Up | TET1, DNMT1. | CircFECR1 regulates DNA methylation and demethylation enzymes to control breast cancer tumor growth. | 30537986 |
| circYAP | BRCA | Up | eIF4G, PABP | CircYap can bind with Yap mRNA and the translation initiation associated proteins, eIF4G and PABP, which functionally leads to the suppression of Yap translation initiation. | 31092884 | |
| circEIF3J, circPAIP2 | Hela, HEK293 | Up | U1 snRNA | EIciRNAs predominantly localizes in the nucleus, interacts with U1 snRNP, and promotes transcription of their parental genes. | 25664725 |
BRCA: breast cancer; GC: Gastric cancer; BCa: bladder cancer; CRC: colorectal cancer; CESC: cervical squamous cell carcinoma and endocervical adenocarcinoma; HNSC: head and neck squamous cell carcinoma; GBM: primary glioblastomas; HCC: hepatocellular carcinoma; TNBC: Triple-negative breast cancer; ccRCC: Clear Cell Renal Cell Carcinoma; and ESCC: esophageal squamous cell carcinoma.
3.1. Interfering with Parental Gene Regulation
Accumulated reports observed that circRNA can, in a complicated way, regulate its parental gene in terms of epigenetic control, splicing, transcription, or translation. For example, Chen et al. reported that circFECR1, a FLI1 exonic circular RNA, interacts with the FLI1 promoter and recruits TET1 demethylase to induce DNA demethylation in the CpG islands. Concurrently, circFECR1 also binds to and downregulates DNMT1, the critical enzyme that maintains DNA demethylation during DNA replication. Thus, circFECR1 promotes tumor cell invasion by coordinately regulating TET1 and DNMT1 in breast cancers [30].
If the circRNA contains the same exon as the parental gene, the processing of circRNA will compete with the splicing of pre-mRNA. Previous studies have shown that back-splicing can compete with pre-mRNA splicing in circRNA biogenesis, resulting in low levels of linear mRNA that contain exon inclusions. In general, the more an exon is circularized, the less it presents in the processed mRNA [8,11].
CircRNA can also affect the transcription of its parental gene. For example, intron-retained circRNAs, such as EIciRNA are located in the nucleus. It is reported that EIciRNAs can bind to U1 snRNPs through RNA–RNA interactions, while U1 forms a complex with RNA polymerase II by binding to TFIIH. EIciRNAs regulate RNA polymerase II activity and promote parental gene transcription [28]. Similar to EIciRNA, ciRNA also can stimulate the transcription of its parental gene by up-regulating the activity of RNA polymerase II and enhance the expression of ankyrin repeat domain 52 protein (ANKRD52) [13].
Besides, some circRNAs have been demonstrated to regulate gene transcription via both RNA polymerase II complex and translation-related proteins. Recently, circYap was found to bind with Yap mRNA and the translation initiation associated proteins eIF4G and PABP. The complex containing overexpressed circYap abolishes the interaction of PABP on the poly (A) tail and eIF4G on the 5′-cap of the Yap mRNA, which functionally leads to the suppression of Yap translation initiation. Individually blocking the binding sites of circYap on Yap mRNA, or respectively mutating the binding sites for PABP and eIF4G, de-represses Yap translation [31].
3.2. Acting as miRNA Sponges
CircRNAs are stable in cells because they do not have a 5′end and a poly-A tail, which prevent ribonuclease degradation. Furthermore, there are multiple miRNA response elements on the circular sequences. Thus, circRNA is natural miRNA sponge. CDR1as consists of a single exon and 63 conserved binding sites for miR-7, can suppress miR-7’s activity, and can upregulate the expression of miR-7 targets such as SNCA, EGFR, and IRS2 [2]. CircRNAs can inhibit the expression of miRNAs by adsorption and, meanwhile, affect the molecular level of downstream target genes. Many studies have shown that the most general function of circRNAs is its action as a miRNA sponge to regulate target gene expression by inhibiting miRNA activity. One circRNA can regulate one or multiple miRNAs through multiple miRNA binding sites. CircRNAs regulate gene expression through binding to and releasing miRNAs from their downstream target genes. For example, we found circTADA2A was repressed in a large cohort of triple-negative breast cancer (TNBC) patients compared to adjacent normal tissues. Through bioinformatics analyses, miR-203a-3p and miR-302c-3p binding sites were identified in circTADA2A, which was confirmed in subsequent wet experiments. Functional analysis indicated circTADA2A-E6 binds with miR-203a-3p, thus affecting its downstream gene SOCS3 to suppress cell migration, invasion, and clonogenicity. Furthermore, their down-regulation was correlated with survival time in TNBC patients. We conclude that circTADA2A-E6 is a tumor-suppressor circRNA and could be utilized as a promising prognostic biomarkers and therapeutic target for TNBCs [32]. Interestingly, an oncogenic circRNA in TNBC was also discovered in a recent report. Yang et al. found that circAGFG1 was significantly up-regulated in TNBC, and it promoted TNBC progression through the circAGFG1/miR-195-5p/CCNE1 axis [33]. The same circRNA may even target different miRNAs to exert opposite functions in different cancers. For example, circHIPK3 targets miR-7 to promote colorectal cancer growth and metastasis, while it also targets miR-558 to suppress the expression of HPSE and inhibits migration, invasion, and angiogenesis of bladder cancer cells in vitro [34,35].
3.3. Binding to Proteins
Some circRNAs that harbor binding sites for RNA-binding proteins may serve as protein sponges or decoys in addition to acting as miRNA sponges. For instance, the circular RNA Ccnb1 could bind with H2AX in p53 mutant cells and suppress mutant p53 in tumor progression. This study found that circ-Ccnb1 could interact with both Ccnb1 and Cdk1 proteins, thus counteracting the effects of p53 mutations in breast cancer [36]. In colorectal cancer tissues, increasing AMPK activation is usually associated with upregulated expression of circACC1 [37]. CircACC1 functions to stabilize and promote the enzymatic activity of the AMPK holoenzyme by binding with the regulatory β and γ subunits. Argonaute 2 (AGO2) is a core component of the miRNA-induced silencing complex. Recently, Chen et al. identified that circAGO2, one intronic circRNA generated from the AGO2 gene, binds with the HuR protein to facilitate HuR-repressed functions of AGO2–miRNA complexes in gastric cancer [38]. CircRNA can also form functional complexes with proteins. CircFOXO3, which is highly expressed in noncancer cells, could arrest the function of CDK2 and block cell cycle progression via formation of a circFOXO3–p21–CDK2 ternary complex [39]. Undoubtedly, to coordinate the different cellular processes, some of the proteins that bind with circRNA may also be an essential regulator of the biogenesis of circRNA [24].
3.4. Translating Proteins or Peptide
circRNAs were initially defined as a distinct class of endogenous noncoding RNA that could not translate proteins. Recently, strong evidence from several research groups has shown that circRNAs can encode proteins. Liang et al. reported that circCTNNB1 produces a novel, 370 amino acid CTNNB1 (i.e., β-catenin) isoform that uses the start codon as the linear β-catenin mRNA transcript, and translation is terminated at a new stop codon created by circularization. They found that this novel isoform can stabilize full-length β-catenin by antagonizing GSK3β-induced β-catenin phosphorylation and degradation, leading to activation of the Wnt pathway [40]. In addition, Zhang et al. reported that circular lncRNA-PINT can be translated into a small peptide to suppress glioblastoma cell proliferation; this action is mediated by trapping PAF1c to inhibit translational elongation of oncogenes [41]. However, the molecular mechanism of translation remains largely unknown. Generally speaking, linear mRNA translation normally requires a 5′end 7-methylguanosine (m7G) cap structure and a 3′ poly-A tail. CircRNA may be translated into peptides and proteins in other novel ways because circRNAs lack both caps and poly(A) tails. One report indicated that circRNAs can be translated from an artificial internal ribosomal entry site (IRES) to generate a functional green fluorescent protein (GFP) [42]. And another mechanism is that circular RNA containing an infinite open reading frame (ORF) can be efficiently translated to produce proteins [43]. Based on ribosome footprinting assays of fly head extracts, Pamudurti et al. identified that a group of circRNAs is translated in a cap-independent manner. They share the start codon with the hosting RNA, encoding specific domains from hosting protein [44].
4. Perspectives
In recent years, considerable interest has focused on circRNA in cancers. In this review, we outline current understandings on its biogenesis, molecular mechanism, and function, but there are also some open questions. Firstly, controls of circRNA and its parental gene are thought to be intensely coupled together [11,30,31]. However, Smid et al. developed a new circRNA prediction method, which did not rely on unmapped reads or known splice junctions. When analyzing random-primed cDNA libraries from a large primary breast cancer cohort, they found only a small part of circRNAs could be explained by known splicing events [45]. Although it cannot completely rule out the technical disparities between this study and previous investigations, it raises the possibility that specific regulatory processes and functions for circRNAs exist. Future studies are needed to systematically compare the datasets and analysis methods to clarify the relationship between circRNA and its host gene.
Besides, genetic or epigenetic factors contributing to circRNA regulation have not been fully explored in cancers. For example, genomic aberrations include both chromosome translocation and short nucleotide variants [46]. Canonical short variants such as inversions, insertions, translocations, deletions, and duplications may introduce or interrupt elements in flanking regions; thus, they should contribute to the formation of circRNAs. Although short repeats affect circRNA production, as found in a previous reporter assay, it should be proven in clinical samples that some of the well-established cancer-associated short variants correlate with cancers via circRNA [17]. Thus, a high-throughput bioinformatics pipeline should be developed to analyze the large cancer sequencing datasets available in the public repository. In a recent analysis of sequencing data from the dorsolateral prefrontal cortex, Liu et al. combined the expression levels of circRNAs with genetic cis-acting SNPs and found that partial circQTL SNPs might influence circRNA formation by altering the canonical splicing site or the reverse complementary sequence match [47]. It would be interesting to conduct a similar approach in cancer samples and to see if some of these circQTL SNPs are highly linked to genome-wide association study signals in different cancer types.
Finally, circRNAs possess several characteristics that are well-suitable to serve as biomarkers in cancers. Some circRNAs have tissue-specific expressions and correlate with clinical indicators [4,9,32]. They are protected from endonuclease degradation and are stable in formalin-fixed, paraffin-embedded (FFPE) tissues [48]. Notably, recent reports have shown that circRNAs can be secreted in exosomes, saliva, and urine [9,49,50]. Therefore, circRNAs could be further explored to be utilized as a surrogate for cancer diagnostics.
Acknowledgments
We sincerely thank Shantou University Medical College of China and Li Ka-Shing foundation for their support of this research.
Abbreviations
| circRNA | Circular RNA |
| lncRNA | Long noncoding RNA |
| ciRNAs | Circular intronic RNAs |
| EIciRNAs | Exonic-intronic circRNAs |
| TricRNA | tRNA intronic circular RNA |
| RBPs | RNA-binding proteins |
| siRNA | Small interfering RNA |
| hESCs | Human embryonic stem cells |
| TCGA | The Cancer Genome Atlas |
| PMID | PubMed Unique Identifier |
| SNPs | Single nucleotide polymorphisms |
Funding
This work has been supported in part by the National Natural Science Foundation of China (No. 81673037).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Hsu M.T., Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339–340. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 2.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 3.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M., et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 5.Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B., Luo Y., Lyu D., Li Y., Shi G., et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maass P.G., Glazar P., Memczak S., Dittmar G., Hollfinger I., Schreyer L., Sauer A.V., Toka O., Aiuti A., Luft F.C., et al. A map of human circular RNAs in clinically relevant tissues. J. Mol. Med. 2017;95:1179–1189. doi: 10.1007/s00109-017-1582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piwecka M., Glazar P., Hernandez-Miranda L.R., Memczak S., Wolf S.A., Rybak-Wolf A., Filipchyk A., Klironomos F., Cerda Jara C.A., Fenske P., et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017:357. doi: 10.1126/science.aam8526. [DOI] [PubMed] [Google Scholar]
- 8.Cocquerelle C., Mascrez B., Hetuin D., Bailleul B. Mis-splicing yields circular RNA molecules. Faseb J. 1993;7:155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 9.Vo J.N., Cieslik M., Zhang Y., Shukla S., Xiao L., Zhang Y., Wu Y.M., Dhanasekaran S.M., Engelke C.G., Cao X., et al. The Landscape of Circular RNA in Cancer. Cell. 2019;176:869–881. doi: 10.1016/j.cell.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zaphiropoulos P.G. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc. Natl. Acad. Sci. USA. 1996;93:6536–6541. doi: 10.1073/pnas.93.13.6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 12.Conn S.J., Pillman K.A., Toubia J., Conn V.M., Salmanidis M., Phillips C.A., Roslan S., Schreiber A.W., Gregory P.A., Goodall G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y., Zhang X.O., Chen T., Xiang J.F., Yin Q.F., Xing Y.H., Zhu S., Yang L., Chen L.L. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 14.Abelson J., Trotta C.R., Li H. tRNA splicing. J. Biol. Chem. 1998;273:12685–12688. doi: 10.1074/jbc.273.21.12685. [DOI] [PubMed] [Google Scholar]
- 15.Salgia S.R., Singh S.K., Gurha P., Gupta R. Two reactions of Haloferax volcanii RNA splicing enzymes: joining of exons and circularization of introns. Rna. 2003;9:319–330. doi: 10.1261/rna.2118203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu Z., Filonov G.S., Noto J.J., Schmidt C.A., Hatkevich T.L., Wen Y., Jaffrey S.R., Matera A.G. Metazoan tRNA introns generate stable circular RNAs in vivo. Rna. 2015;21:1554–1565. doi: 10.1261/rna.052944.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang D., Wilusz J.E. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ivanov A., Memczak S., Wyler E., Torti F., Porath H.T., Orejuela M.R., Piechotta M., Levanon E.Y., Landthaler M., Dieterich C., et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170–177. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 19.Han L., Diao L., Yu S., Xu X., Li J., Zhang R., Yang Y., Werner H.M.J., Eterovic A.K., Yuan Y., et al. The Genomic Landscape and Clinical Relevance of A-to-I RNA Editing in Human Cancers. Cancer Cell. 2015;28:515–528. doi: 10.1016/j.ccell.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Licatalosi D.D., Darnell R.B. RNA processing and its regulation: global insights into biological networks. Nat. Rev. Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerstberger S., Hafner M., Tuschl T. A census of human RNA-binding proteins. Nat. Rev. Genet. 2014;15:829–845. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neelamraju Y., Hashemikhabir S., Janga S.C. The human RBPome: from genes and proteins to human disease. J. Proteom. 2015;127:61–70. doi: 10.1016/j.jprot.2015.04.031. [DOI] [PubMed] [Google Scholar]
- 23.Wang Z.L., Li B., Luo Y.X., Lin Q., Liu S.R., Zhang X.Q., Zhou H., Yang J.H., Qu L.H. Comprehensive Genomic Characterization of RNA-Binding Proteins across Human Cancers. Cell Rep. 2018;22:286–298. doi: 10.1016/j.celrep.2017.12.035. [DOI] [PubMed] [Google Scholar]
- 24.Li X., Liu C.X., Xue W., Zhang Y., Jiang S., Yin Q.F., Wei J., Yao R.W., Yang L., Chen L.L. Coordinated circRNA Biogenesis and Function with NF90/NF110 in Viral Infection. Mol. Cell. 2017;67:214–227. doi: 10.1016/j.molcel.2017.05.023. [DOI] [PubMed] [Google Scholar]
- 25.Cieply B., Park J.W., Nakauka-Ddamba A., Bebee T.W., Guo Y., Shang X., Lengner C.J., Xing Y., Carstens R.P. Multiphasic and Dynamic Changes in Alternative Splicing during Induction of Pluripotency Are Coordinated by Numerous RNA-Binding Proteins. Cell Rep. 2016;15:247–255. doi: 10.1016/j.celrep.2016.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu C.Y., Li T.C., Wu Y.Y., Yeh C.H., Chiang W., Chuang C.Y., Kuo H.C. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat. Commun. 2017;8:1149. doi: 10.1038/s41467-017-01216-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bass B.L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L., et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 29.Guarnerio J., Bezzi M., Jeong J.C., Paffenholz S.V., Berry K., Naldini M.M., Lo-Coco F., Tay Y., Beck A.H., Pandolfi P.P. Oncogenic Role of Fusion-circRNAs Derived from Cancer-Associated Chromosomal Translocations. Cell. 2016;165:289–302. doi: 10.1016/j.cell.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 30.Chen N., Zhao G., Yan X., Lv Z., Yin H., Zhang S., Song W., Li X., Li L., Du Z., et al. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol. 2018;19:218. doi: 10.1186/s13059-018-1594-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu N., Yuan Z., Du K.Y., Fang L., Lyu J., Zhang C., He A., Eshaghi E., Zeng K., Ma J., et al. Translation of yes-associated protein (YAP) was antagonized by its circular RNA via suppressing the assembly of the translation initiation machinery. Cell Death Differ. 2019 doi: 10.1038/s41418-019-0337-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu J.Z., Shao C.C., Wang X.J., Zhao X., Chen J.Q., Ouyang Y.X., Feng J., Zhang F., Huang W.H., Ying Q., et al. circTADA2As suppress breast cancer progression and metastasis via targeting miR-203a-3p/SOCS3 axis. Cell Death Dis. 2019;10:175. doi: 10.1038/s41419-019-1382-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang R., Xing L., Zheng X., Sun Y., Wang X., Chen J. The circRNA circAGFG1 acts as a sponge of miR-195-5p to promote triple-negative breast cancer progression through regulating CCNE1 expression. Mol. Cancer. 2019;18:4. doi: 10.1186/s12943-018-0933-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Zeng K., Chen X., Xu M., Liu X., Hu X., Xu T., Sun H., Pan Y., He B., Wang S. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9:417. doi: 10.1038/s41419-018-0454-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Li Y., Zheng F., Xiao X., Xie F., Tao D., Huang C., Liu D., Wang M., Wang L., Zeng F., et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. Embo Rep. 2017;18:1646–1659. doi: 10.15252/embr.201643581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fang L., Du W.W., Awan F.M., Dong J., Yang B.B. The circular RNA circ-Ccnb1 dissociates Ccnb1/Cdk1 complex suppressing cell invasion and tumorigenesis. Cancer Lett. 2019;459:216–226. doi: 10.1016/j.canlet.2019.05.036. [DOI] [PubMed] [Google Scholar]
- 37.Li Q., Wang Y., Wu S., Zhou Z., Ding X., Shi R., Thorne R.F., Zhang X.D., Hu W., Wu M. CircACC1 Regulates Assembly and Activation of AMPK Complex under Metabolic Stress. Cell Metab. 2019;30:157–173. doi: 10.1016/j.cmet.2019.05.009. [DOI] [PubMed] [Google Scholar]
- 38.Chen Y., Yang F., Fang E., Xiao W., Mei H., Li H., Li D., Song H., Wang J., Hong M., et al. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ. 2019;26:1346–1364. doi: 10.1038/s41418-018-0220-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du W.W., Yang W., Liu E., Yang Z., Dhaliwal P., Yang B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang W.C., Wong C.W., Liang P.P., Shi M., Cao Y., Rao S.T., Tsui S.K., Waye M.M., Zhang Q., Fu W.M., et al. Translation of the circular RNA circbeta-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 2019;20:84. doi: 10.1186/s13059-019-1685-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang M., Zhao K., Xu X., Yang Y., Yan S., Wei P., Liu H., Xu J., Xiao F., Zhou H., et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat. Commun. 2018;9:4475. doi: 10.1038/s41467-018-06862-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y., Wang Z. Efficient backsplicing produces translatable circular mRNAs. Rna. 2015;21:172–179. doi: 10.1261/rna.048272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abe N., Matsumoto K., Nishihara M., Nakano Y., Shibata A., Maruyama H., Shuto S., Matsuda A., Yoshida M., Ito Y., et al. Rolling Circle Translation of Circular RNA in Living Human Cells. Sci. Rep. 2015;5:16435. doi: 10.1038/srep16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pamudurti N.R., Bartok O., Jens M., Ashwal-Fluss R., Stottmeister C., Ruhe L., Hanan M., Wyler E., Perez-Hernandez D., Ramberger E., et al. Translation of CircRNAs. Mol. Cell. 2017;66:9–21. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smid M., Wilting S.M., Uhr K., Rodriguez-Gonzalez F.G., de Weerd V., Prager-Van der Smissen W.J.C., van der Vlugt-Daane M., van Galen A., Nik-Zainal S., Butler A., et al. The circular RNome of primary breast cancer. Genome Res. 2019;29:356–366. doi: 10.1101/gr.238121.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sudmant P.H., Rausch T., Gardner E.J., Handsaker R.E., Abyzov A., Huddleston J., Zhang Y., Ye K., Jun G., Fritz M.H., et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Z., Ran Y., Tao C., Li S., Chen J., Yang E. Detection of circular RNA expression and related quantitative trait loci in the human dorsolateral prefrontal cortex. Genome Biol. 2019;20:99. doi: 10.1186/s13059-019-1701-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang F., Zhao X., Dong H., Xu J. circRNA expression analysis in lung adenocarcinoma: comparison of paired fresh frozen and formalin-fixed paraffin-embedded specimens. Biochem. Biophys. Res. Commun. 2018;500:738–743. doi: 10.1016/j.bbrc.2018.04.145. [DOI] [PubMed] [Google Scholar]
- 49.Li Y., Zheng Q., Bao C., Li S., Guo W., Zhao J., Chen D., Gu J., He X., Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bahn J.H., Zhang Q., Li F., Chan T.M., Lin X., Kim Y., Wong D.T., Xiao X. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin. Chem. 2015 doi: 10.1373/clinchem.2014.230433. [DOI] [PMC free article] [PubMed] [Google Scholar]