Abstract
The papilionoid legume genus Ormosia comprises approximately 130 species, which are distributed mostly in the Neotropics, with some species in eastern Asia and northeastern Australia. The taxonomy and evolutionary history remain unclear due to the lack of a robust species-level phylogeny. Chloroplast genomes can provide important information for phylogenetic and population genetic studies. In this study, we determined the complete chloroplast genome sequences of five Ormosia species by Illumina sequencing. The Ormosia chloroplast genomes displayed the typical quadripartite structure of angiosperms, which consisted of a pair of inverted regions separated by a large single-copy region and a small single-copy region. The location and distribution of repeat sequences and microsatellites were determined. Comparative analyses highlighted a wide spectrum of variation, with trnK-rbcL, atpE-trnS-rps4, trnC-petN, trnS-psbZ-trnG, trnP-psaJ-rpl33, and clpP intron being the most variable regions. Phylogenetic analysis revealed that Ormosia is in the Papilionoideae clade and is sister to the Lupinus clade. Overall, this study, which provides Ormosia chloroplast genomic resources and a comparative analysis of Ormosia chloroplast genomes, will be beneficial for the evolutionary study and phylogenetic reconstruction of the genus Ormosia and molecular barcoding in population genetics and will provide insight into the chloroplast genome evolution of legumes.
1. Introduction
The genus Ormosia Jacks. (Fabaceae, Papilionoideae) comprises approximately 130 species and has a disjunct distribution between the Neotropics and the eastern Asian and northeastern Australian Tropics, i.e., from southern India and southern China to northeastern Australia [1, 2]. Ormosia was defined as a segregate genus by the following combination of morphological characters: flowers with distinct, imbricate calyx lobes; an incurved style with a terminal or lateral (usually bilobed) stigma; and predominantly red, black, or bicolored seeds with a hard testa. Molecular phylogenetic analyses based on the chloroplast matK and trnL intron sequences in large-scale phylogenetic studies of papilionoid genera have consistently recovered a monophyletic Ormosia, yet sampling within the genus has been limited [1, 3]. Ormosia diverged as an early branch within the Genistoid clade, which is one of the fundamental lineages of papilionoid legumes [3]. The results also showed that matK and trnL intron sequences lacked variations among the species. However, only a few genomic resources have been explored in this genus. In GenBank, there are presently fewer than 500 sequences of Ormosia species.
In recent years, the chloroplast genome resources have been widely used in plant systematics and species identification [4–8]. The chloroplast genome is inherited in a maternal manner in the majority of plants and is smaller in size and has very low recombination compared with that of the nuclear and mitochondria genome [9]. Moreover, the chloroplast genome has a moderate rate of nucleotide evolution, which makes the chloroplast genome suitable for species identification and for phylogenetic studies at different taxonomic levels [10].
Most of the chloroplast genomes in angiosperms have a typical quadripartite structure, with two copies of inverted repeats (IRs) separating the large single-copy (LSC) and small single-copy regions (SSC) and the genome size ranging from 120 to 170 kb in length. A comparative analysis of the complete chloroplast genomes played an important role in understanding the chloroplast genome evolution. In this paper, we investigated the complete chloroplast genomes of five Ormosia species through next-generation sequencing (NGS). The objectives of this study were (i) to describe the structure of the Ormosia chloroplast genome; (ii) to identify highly divergent regions in the Ormosia chloroplast genome which suit DNA barcodes; and (iii) to calibrate the phylogenetic position of Ormosia based on phylogenomic analysis.
2. Materials and Methods
2.1. Plant Materials and DNA Extraction
The fresh and healthy leaves of five Ormosia species were collected from the Subtropical Forestry Experimental Center of Chinese Academy of forestry, Fenyi, Jiangxi (O. henryi and O. hosiei), Jiangmen, Guangdong (O. emarginata), Bawangling National Nature Reserve, Hainan (O. xylocarpa), and Longmen, Huizhou, Guangdong (O. semicastrata). Voucher specimens were deposited in the herbaria of the Institute of Botany (PE), China Academy of Sciences. Fresh leaves from each accession were immediately dried with silica gel prior to DNA extraction. The total genomic DNA was extracted following the method of Li et al. [11] and was purified using the Wizard DNA Clean-Up System (Promega, Madison, WI, USA). The DNA quality was assessed based on spectrophotometry and electrophoresis in 1% (w/v) agarose gel.
2.2. Illumina Sequencing, Assembly, and Annotation
The DNA was sheared to fragments of 400~600 bp using an ultrasonicator. Paired-end libraries were prepared with the NEBNext® Ultra™ DNA Library Prep Kit. The genome was then sequenced using the HiSeq X Ten platform (Illumina, Santiago, CA, USA).
The paired-end reads were qualitatively assessed and assembled with SPAdes 3.6.1 [12]. Chloroplast genome sequence contigs were selected from SPAdes software by performing a BLAST search using the Lupinus albus chloroplast genome sequence as a reference (GenBank accession number: KJ468099) and then were assembled with Sequencher 5.4.5 (Gene Codes, Ann Arbor, MI). To verify the assembly, four boundaries between the single-copy (SC) and the inverted repeat (IR) regions of the assembled sequences were confirmed by PCR amplification and Sanger sequencing using the primers by Dong et al. [13]. Chloroplast genome annotation was performed with Plann [14] using the Lupinus albus as reference sequence from GenBank. A chloroplast genome map was drawn using Genome Vx software [15].
2.3. Analysis of Tandem Repeats and Single Sequence Repeats
Five types of repeat sequences, including forward repeat, reverse repeat, complement repeat, palindromic repeat, and tandem repeat, were identified in the Ormosia chloroplast genomes. We used REPuter to identify forward repeat, reverse repeat, complement repeat, and palindromic repeat [16], in which the similarity percentage of the two repeat copies was at least 90%, the minimum repeat size was 30 bp, and the hamming distance was 3. Tandem repeats were identified using the web-based Tandem Repeats Finder (https://tandem.bu.edu/trf/trf.html), with 2, 7, and 7 set for the alignment parameters match, mismatch, and indel, respectively.
Single sequence repeats (SSRs) were identified by GMAT [17] with the parameters set at >10 for mononucleotide, >5 for dinucleotide, >4 for trinucleotide, and >3 for tetranucleotide, pentanucleotide, and hexanucleotide SSRs.
2.4. Comparison of Whole Chloroplast Genomes and Divergent Hotspot Identification
The mVISTA program (http://genome.lbl.gov/vista/mvista/submit.shtml) with Shuffle-LAGAN mode [18] was used to compare the Ormosia chloroplast genomes. The O. henryi chloroplast genome was used as a reference.
All five Ormosia sequenced chloroplast genomes were aligned using MAFFT v7 [19], assuming collinear genomes for the full alignment, and then were adjusted manually using Se-Al 2.0 [20]. A sliding window analysis was conducted to generate the nucleotide diversity of the chloroplast genome using the DnaSP v5.10 software [21]. The step size was set to 100 bp, with an 800-bp window length.
2.5. Phylogenetic Reconstruction
Eighty-one protein-coding sequences were present in 70 species from the family Fabaceae and one species from Moraceae as an outgroup were used for the phylogenetic reconstruction. The chloroplast genomes of these species were downloaded from GenBank (Table S1). Gene alignment was performed using MAFFT v7 [19]. Phylogenetic trees were constructed by the maximum likelihood (ML) and Bayesian inference (BI) analyses methods.
The program ModelFinder was used to find the optimal substitution mode [22], using both the Bayesian information criterion and the Akaike information criterion. Maximum likelihood (ML) analyses were performed using RAxML v.8.1.24. Statistical support for the branches (BS) was calculated by rapid bootstrap analyses with 1000 replicates.
Bayesian inference was conducted using MrBayes v3.2.2 [23] using the GTR+G+I model on the CIPRES Science Gateway. The default priors were utilized, along with the default heating scheme (one cold and three heated chains), and runs were conducted for 10 million generations with trees sampled every 1000 generations. The first 25% percent of trees from all runs were discarded as burn-in.
3. Results
3.1. Genome Sequencing and Assembly
Using the Illumina HiSeq X Ten system, the total DNA from five species of Ormosia was sequenced to produce 165,518,310–342,489,92 paired-end raw reads (150 bp average read length) per species. After screening, these paired-end reads through alignment with themselves, 209,546 to 813,022 chloroplast genome reads were extracted with 181 X to 702 X coverage (Table 1). The accuracy of inverted repeat junction regions in assembled sequences was further confirmed by PCR amplification and Sanger sequencing with specific primers. The finished, high-quality Ormosia chloroplast genome sequences that were thus obtained were used in the following analyses and were submitted to GenBank (accession numbers, MH571753, MH571754, and MK105448- MK105450).
Table 1.
Summary of the sequencing data for five Ormosia species.
| Species | Locality | Voucher | Raw data no. | Mapped reads no. | Mapped to reference genome (%) | Chloroplast genome coverage (X) |
|---|---|---|---|---|---|---|
| O. henryi | Fenyi, Jiangxi | BOP214710 | 34,248,992 | 390,755 | 1.14% | 337 |
| O. hosiei | Fenyi, Jiangxi | BOP214711 | 39,701,622 | 466,122 | 1.17% | 409 |
| O. emarginata | Jiangmen, Guangdong | BOP216254 | 16,518,310 | 813,022 | 4.92% | 702 |
| O. xylocarpa | Bawangling, Hainan | BOP216381 | 23,924,424 | 209,546 | 0.88% | 181 |
| O. semicastrata | Longmen, Guangdong | BOP217157 | 23,652,340 | 219,828 | 0.93% | 193 |
3.2. Chloroplast Genomes Features of Ormosia Species
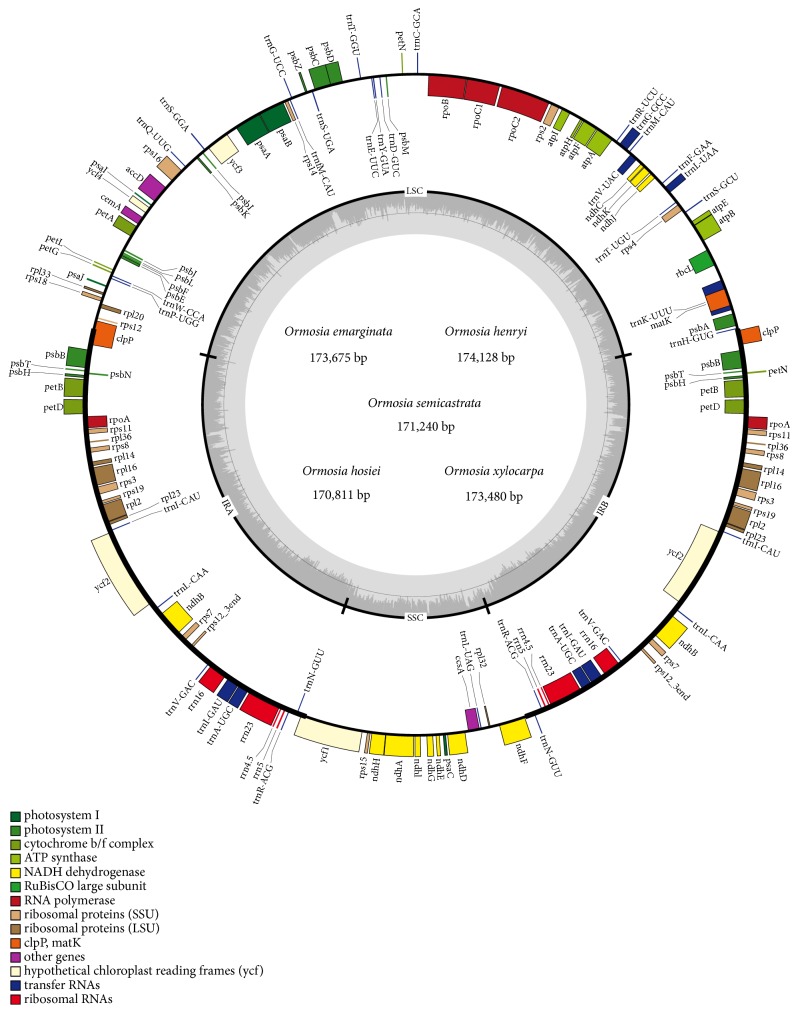
The five Ormosia chloroplast genomes ranged from 170,811 to 174,128 base pairs in length, with Ormosia henryi being the largest and Ormosia hosiei the smallest. All chloroplast genomes shared the common feature comprising two copies of IR (40,034–40,633 bp) separated by the LSC (71,728–74,231 bp) and SSC (18,295– 18,798 bp) regions (Figure 1, Table 2). The overall GC content was 35.7-36.0%, which indicated nearly identical levels among the five complete Ormosia chloroplast genomes.
Figure 1.
Gene map of Ormosia chloroplast genome. The genes inside and outside the circle are transcribed in the clockwise and counterclockwise directions, respectively. Genes in different functional groups are shown in different colors. The thick lines indicate the extent of the inverted repeats (IRa and IRb) that separate the genomes into small single-copy (SSC) and large single-copy (LSC) regions.
Table 2.
Summary statistics for assembly of five Ormosia species chloroplast genomes.
| Species | O. henryi | O. hosiei | O. emarginata | O. xylocarpa | O. semicastrata |
|---|---|---|---|---|---|
| Length (bp) | 174,128 | 170,811 | 173,675 | 173,480 | 171,240 |
| LSC (bp) | 74,231 | 72,448 | 73,727 | 73,537 | 71,728 |
| IR (bp) | 40,588 | 40,034 | 40,575 | 40,633 | 40,448 |
| SSC (bp) | 18,721 | 18,295 | 18,798 | 18,677 | 18,616 |
| Gene number | 110 | 110 | 110 | 110 | 110 |
| Protein coding genes | 76 | 76 | 76 | 76 | 76 |
| tRNA | 30 | 30 | 30 | 30 | 30 |
| rRNA | 4 | 4 | 4 | 4 | 4 |
| GC content (%) | 35.7 | 36 | 35.8 | 35.9 | 35.9 |
| Accession number | MH571754 | MH571753 | MK105448 | MK105449 | MK105450 |
Ormosia chloroplast genomes all have 112 different genes arranged in the same order, including 78 protein-coding genes, 30 tRNAs, and 4 rRNAs. Among these genes, twelve of the protein-coding genes and six of the tRNA genes contained introns; 16 genes harbored a single intron and two genes (ycf3 and clpP) harbored two introns. 42 genes were duplicated in the IR region, including 31 protein-coding genes, 7 tRNA genes, and 4 rRNA genes (rrn5, rrn4.5, rrn23, and rrn16). The trnK-UUU had the largest intron, which contained the matK gene. The 5′-end exon of the rps12 gene was located in the LSC region, and the intron and two copies of 3′-end exon were located in the IR regions.
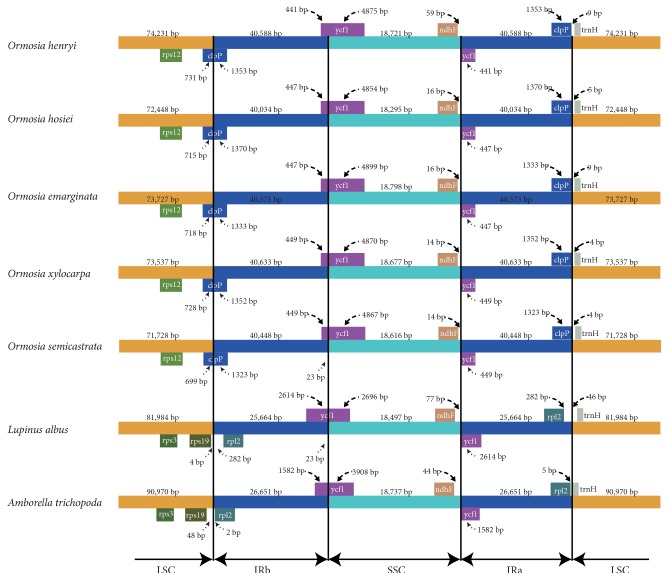
3.3. IR Expansion and Contraction
The IR boundary regions of five Ormosia species and Lupinus albus (Fabaceae) and Amborella trichopoda were compared, and the results showed that the border of the Ormosia chloroplast genomes was slightly different from that of other genomes (Figure 2). In Ormosia, the boundary of IRb/LSC occurred within the gene clpP, resulting in the duplication of a portion of this gene (1,323-1,370 bp) in the IR region. The boundary of IRb/LSC in Lupinus albus and Amborella trichopoda occurred between rps19 and rpl2 and between rpl2 and trnH-GUG on the IRa/LSC side, with 0 and 282 noncoding nucleotides between these two genes. The IRa/SSC border extended into ycf1, resulting in a pseudogene in the five Ormosia species. The length of the ycf1 pseudogene was 4,854-4,899 bp in Ormosia, 2,696 bp in Lupinus albus, and 3,908 bp in Amborella trichopoda. Furthermore, ndhF deviated from the IRb/SSC in Ormosia by 14-59 bp. There were 4-9 bp of noncoding sequence between IRa/LSC border and the 3'-end of gene trnH-GUG in the LSC region. Taken together, the IR in Ormosia had a 15 kb expansion compared with other lineages, and the IR boundary regions varied slightly within the Ormosia chloroplast genomes.
Figure 2.
Comparison of the border positions of the LSC, SSC, and IR regions.
3.4. Analysis of Repeat Elements
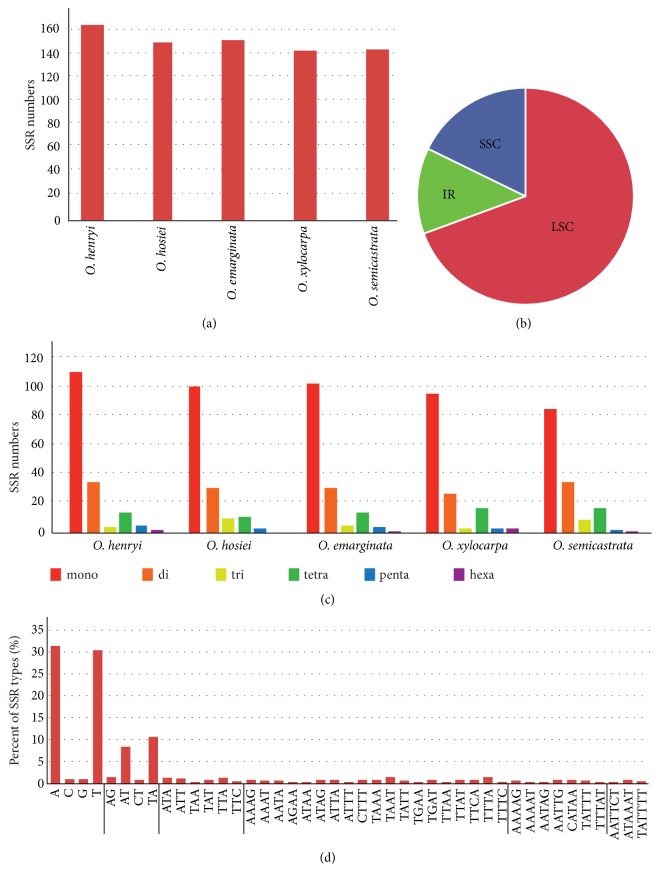
Each Ormosia chloroplast genomes contained 147 to 169 SSRs (Figure 3(a)). Among these SSRs, most were located in the LSC/SSC regions (85.2-88.1%, Figure 3(b)). The average of mono-, di-, tri-, and tetranucleotide SSRs were 62.92%, 20.54%, 4.01%, and 9.43%, respectively. Hexanucleotide SSRs were very rare across the chloroplast genomes (Figure 3(c)). SSRs in Ormosia chloroplast genomes were especially rich in AT and rarely contained CG (Figure 3(d)). Almost all SSRs (61.37%) were mononucleotide A/T repeats; C/G mononucleotide SSRs were rarely present (1.55%). AT/TA repeats were the most common (90.57%) among dinucleotide SSRs.
Figure 3.
Analysis of perfect simple sequence repeats (SSRs) in five Ormosia chloroplast genomes. (a) Number of SSRs detected in five chloroplast genomes. (b) Frequency of identified SSRs in LSC, IR, and SSC regions. (c) Number of SSR types detected in five chloroplast genomes. (d) Frequency of identified SSR motifs in different repeat class types.
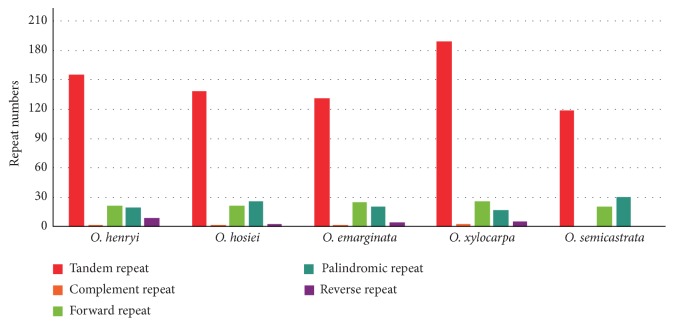
In addition to the SSRs, we employed REPuter and the Tandem Repeats Finder to analyze the repeat sequences of the five Ormosia chloroplast genomes (Figure 4). We classified sequence repeat motifs into five categories: forward, reverse, complement, palindromic, and tandem repeats. Ormosia contained 21-26 forward repeats, 0-9 reverse repeats, 0-2 complement repeats, 17-30 palindromic repeats, and 118-188 tandem repeats.
Figure 4.
Analysis of repeated sequences in five Ormosia chloroplast genomes.
3.5. Sequence Divergence and Divergence Hotspot Regions
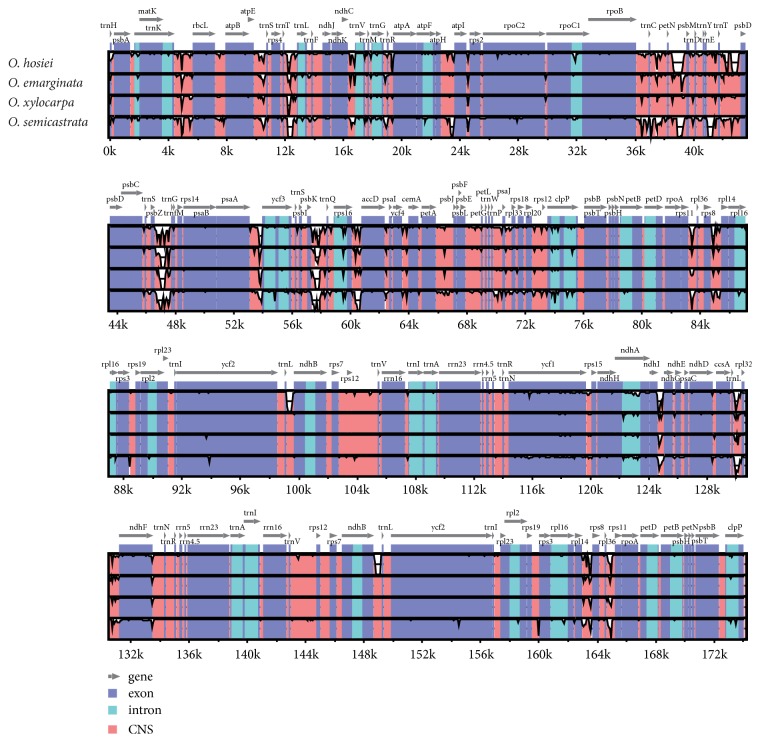
A comparative analysis based on mVISTA was performed among the five chloroplast genomes of Ormosia to investigate the levels of sequence divergence (Figure 5). VISTA-based similarity graphical information portrays sequence identity among the five Ormosia chloroplast genomes with a reference to the O. henryi chloroplast genomes. The organization of the chloroplast genome among Ormosia was essentially colinear and gene order conservation. The results also showed that the IR regions and coding region were more conserved than SC region and noncoding regions.
Figure 5.
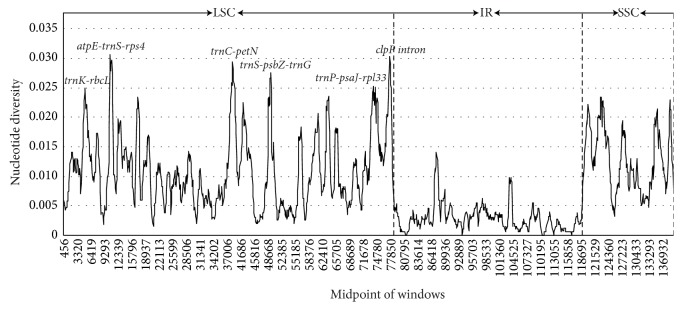
Sliding window analysis of the Ormosia chloroplast genomes (window length: 800 bp; step size: 100 bp). x-axis: position of the midpoint of a window; y-axis: nucleotide diversity of each window.
To identify the sequence divergence hotspots, the nucleotide diversity (pi, π) value within 800 bp was calculated (Figure 6) with DnaSP 5.0 software. In the Ormosia chloroplast genomes, the pi values varied from 0 to 0.03063. The IR region was more conserved than that of the LSC and SSC regions among the five genomes. Six hypervariable regions (Pi > 0.025) were uncovered among the Ormosia chloroplast genomes. They were trnK-rbcL, atpE-trnS-rps4, trnC-petN, trnS-psbZ-trnG, trnP-psaJ-rpl33, and clpP intron. All six regions were located in the LSC region.
Figure 6.
Visualization of genome alignment of the chloroplast genomes of five Ormosia species using O. henryi as reference using mVISTA. The y-scale axis represents the percent identity within 50%–100%. Dashed rectangles indicate highly divergent regions among Ormosia.
3.6. Phylogenomic Analysis
Chloroplast phylogenomics has been proved to be effective in resolving complex relationships at the order level, such as Saxifragales [6]; family level, such as Nelumbonaceae [5]; and the lower taxonomic level, such as Juglans [4] and Forsythieae [8]. In this study, we used 81 protein-coding genes to calibrate the phylogenetic position of Ormosia in the Fabaceae.
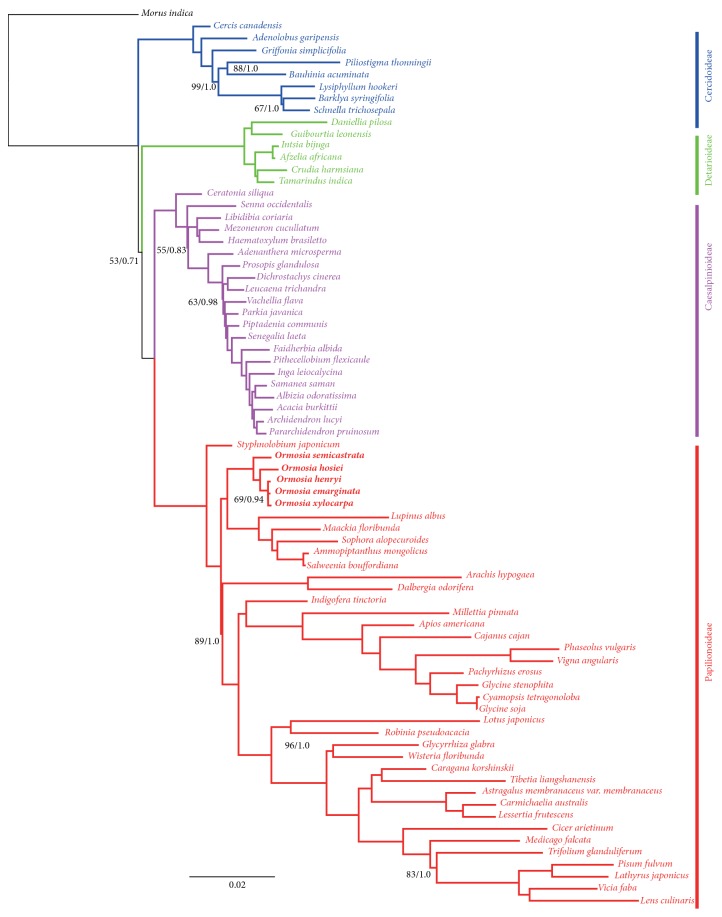
ML and Bayesian analyses based on 81 protein-coding genes produced identical tree topologies, with 100% bootstrap support (BS) form ML and 1.0 Bayesian posterior probabilities (PP) at nearly every node (Figure 7, Figure S1). The phylogeny was congruent with the published matK gene-based phylogenies and showed Cercidoideae as a basal, Caesalpinioideae and Papilionoideae forming sister groups [1]. The result showed that Dialioideae was sister to Caesalpinioideae + Papilionoideae, though with lower bootstrap support and posterior probability values (53 BS/0.71 PP). There were two reasons that might explain the tree topology with lower bootstrap support. Firstly, the inferred phylogenetic trees combine short and long internodes branches, indicating rapid radiation [6, 24]. Secondly, incomplete lineage sorting is proposed as a potential explanation for incongruence among characters [25, 26]. Ormosia was in the Papilionoideae clade and was sister to the Lupinus clade.
Figure 7.
Phylogenetic tree obtained from maximum likelihood and Bayesian inference methods of 81 genes for 82 taxa. Numbers above nodes indicate ML bootstrap support value (ML-BP)/Bayesian posterior probability (BI-PP). Nodes with 100 ML-BP/1.0 BI-PP/100 MP-BP are not marked.
Phylogenetic analysis based on 81 protein-coding genes (Figure 7) successfully resolved relationships among the sampled species of Ormosia. O. semicastrata occupied the most basal position, which was sister to the rest of the Ormosia species. O. hosiei was sister to O. henryi, O. emarginata, and O. xylocarpa, which formed a clade.
4. Discussion
4.1. Chloroplast Genome Evolution of Ormosia
In this study, using the next-generation sequencing method, we sequenced five new chloroplast genomes of Ormosia. The complete chloroplast genomes ranged from 170,811 to 174,128 bp, which is longer compared to that of the other angiosperms. The chloroplast genomes of Ormosia species were structurally conserved, and no rearrangement events were detected in this study. Meanwhile, the genome divergence was low. mVISTA results revealed high similarities among chloroplast genomes, which suggested that the Ormosia cpDNAs were rather conserved. The Ormosia chloroplast genome was structurally similar to that of most angiosperms chloroplast genomes and the IR region showed lower sequence divergence than SSC and LSC regions possibly due to copy corrections between the IR sequences by gene conversion [27].
The organization of the Ormosia chloroplast genomes was similar to that of the angiosperm genome, except for the IR expansion. The boundaries of repeat/single copy represent highly variable regions and often influence the genome size of the chloroplast genome. The information of the IR expansion and contraction can be used to study the genome evolution among plant lineages. In this study, by comparing the inverted repeat/single-copy (IR/SC) boundaries, we detected a 15 kb IR expansion in Ormosia. The position of all four IR/SC junctions can vary even among closely related species in angiosperm chloroplast genomes. The shifts are small among the Ormosia species, involving up to several hundred bp (Figure 2). Larger IR expansions occur less frequently and outnumber large contractions [28]. For example, there is a 10 kb IR contraction in Schisandraceae [29]. In Petroselinum, the IR contracted ~1.5 kb at the IRB-LSC boundary compared with other Apiales species [30].
The IRb and LSC boundaries typically occur between rpl2 and trnH-GUG in most angiosperms [31]. Several elegant models have been proposed to explain the diversification of the IR boundary regions sequences. Goulding et al. [32] and Wang et al. [31] proposed a model that the double-strand break in the IR and LSC boundaries followed by strand invasion and recombination to explain the larger IR expansion. The second model is the recombination between the short repeats or poly(A) of tRNA genes, which may affect the position of the IR boundary [30]. The third model is the indels, which caused a mismatch that resulted in the upstream sequence becoming a single copy [33].
Variation in the chloroplast genome size and gene order within groups is relatively rare. However, the Fabaceae chloroplast genome exhibited significant size variation, chloroplast genome rearrangements, and gene and intron losses [28, 34]. There were also many IR boundary shifts in the legumes [28, 35]. Therefore, further research with expanded sampling is urgently needed to determine the IR boundary shifts and genome rearrangements in the Fabaceae.
4.2. Highly Variable Chloroplast Markers for Evaluating Ormosia Phylogeny and DNA Barcoding
Because of the more than 120 species, great morphological diversity, disjunct distribution of the genus Ormosia, its DNA barcoding and species phylogenetic relationships are still difficult to unravel. Only a few studies focused on the phylogeny and taxonomy of Ormosia by molecular phylogeny. The chloroplast genome markers, the rbcL, matK, trnH-psbA, trnL-F, ndhF, rpoB, and ycf1 genes, have been used widely to investigate taxonomy and DNA barcoding [10, 36, 37]. Nevertheless, increasingly more studies showed that those markers had low discriminatory power and insufficient information for phylogenetic analysis [38, 39].
The indel and single nucleotide substitute mutation events were not random but were clustered as “hotspots” in the chloroplast genome. Those highly variable regions that evolve very rapidly and meet the criteria required to be a DNA barcode. The strategy of searching the potential DNA barcodes has been successfully applied to Diospyros [40], Yam [41], Oryza [42], and Lagerstroemia [43]. Based on the five compared Ormosia chloroplast genomes, six highly variable regions (trnK-rbcL, atpE-trnS-rps4, trnC-petN, trnS-psbZ-trnG, trnP-psaJ-rpl33, and clpP intron) were identified. The regions trnS-psbZ-trnG and clpP intron have been the focus of previous studies to assess the DNA barcodes in angiosperms [10]. Therefore, further work on investigating whether these markers could be recommended as effective, specific barcodes for Ormosia species is necessary.
Acknowledgments
This research was funded by the “cooperation project between CAF and the Province (2019sy03).” The authors would like to acknowledge and thank Dr. Yanlei Liu and Dr. Chao Xu at IBCAS (Institute of Botany, Chinese Academy of Sciences) for technical support with the Illumina sequencing.
Data Availability
The five Ormosia chloroplast genomes are available in GenBank database (accession numbers: MH571753, MH571754, and MK105448- MK105450).
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Authors' Contributions
Bin Li designed the study. Hongshan Liu, Zhihai Su, Shuiqing Yu, Jialin Liu, Xiaojuan Yin, Guowei Zhang, and Wei Liu collected samples and performed the experiment. Bin Li analyzed the data and wrote the manuscript. All of the authors discussed the results and approved the final manuscript. Hongshan Liu and Zhihai Su have contributed equally to this work.
Supplementary Materials
Table S1. A list of the 72 taxa sampled from GenBank in this study for phylogeny analysis. Figure S1. Phylogenetic tree reconstruction of 82 taxa using Bayesian inference methods based on 81 genes chloroplast genome sequences.
References
- 1.Azani N., Babineau M., Bailey C. D., et al. A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny. Taxon. 2017;66(1):44–77. [Google Scholar]
- 2.Lewis G. P. Legumes of the World: Royal Botanic Gardens Kew. 2005. [Google Scholar]
- 3.Cardoso D., de Queiroz L. P., Pennington R. T., et al. Revisiting the phylogeny of papilionoid legumes: new insights from comprehensively sampled early-branching lineages. American Journal of Botany. 2012;99(12):1991–2013. doi: 10.3732/ajb.1200380. [DOI] [PubMed] [Google Scholar]
- 4.Dong W., Xu C., Li W., et al. Phylogenetic resolution in juglans based on complete chloroplast genomes and nuclear DNA sequences. Frontiers in Plant Science. 2017;8, article 1148 doi: 10.3389/fpls.2017.01148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xue J.-H., Dong W.-P., Cheng T., Zhou S.-L. Nelumbonaceae: systematic position and species diversification revealed by the complete chloroplast genome. Journal of Systematics and Evolution. 2012;50(6):477–487. doi: 10.1111/j.1759-6831.2012.00224.x. [DOI] [Google Scholar]
- 6.Dong W., Xu C., Wu P., et al. Resolving the systematic positions of enigmatic taxa: manipulating the chloroplast genome data of Saxifragales. Molecular Phylogenetics and Evolution. 2018;126:321–330. doi: 10.1016/j.ympev.2018.04.033. [DOI] [PubMed] [Google Scholar]
- 7.Chen Z., Grover C. E., Li P., et al. Molecular evolution of the plastid genome during diversification of the cotton genus. Molecular Phylogenetics and Evolution. 2017;112:268–276. doi: 10.1016/j.ympev.2017.04.014. [DOI] [PubMed] [Google Scholar]
- 8.Ha Y.-H., Kim C., Choi K., Kim J.-H. Molecular phylogeny and dating of forsythieae (Oleaceae) provide insight into the miocene history of eurasian temperate shrubs. Frontiers in Plant Science. 2018;9, article 99 doi: 10.3389/fpls.2018.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Twyford A. D., Ness R. W. Strategies for complete plastid genome sequencing. Molecular Ecology Resources. 2017;17(5):858–868. doi: 10.1111/1755-0998.12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong W., Liu J., Yu J., Wang L., Zhou S., Moustafa A. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS ONE. 2012;7(4, article e35071) doi: 10.1371/journal.pone.0035071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J., Wang S., Jing Y., Wang L., Zhou. A modified CTAB protocol for plant DNA extraction. Chinese Bulletin of Botany. 2013;48(1):72–78. [Google Scholar]
- 12.Bankevich A., Nurk S., Antipov D. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong W., Xu C., Cheng T., Lin K., Zhou S. Sequencing angiosperm plastid genomes made easy: a complete set of universal primers and a case study on the phylogeny of saxifragales. Genome Biology and Evolution. 2013;5(5):989–997. doi: 10.1093/gbe/evt063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang D. I., Cronk Q. C. B. Plann: a command-line application for annotating plastome sequences. Applications in Plant Sciences. 2015;3(8, article 1500026) doi: 10.3732/apps.1500026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conant G. C., Wolfe K. H. GenomeVx: simple web-based creation of editable circular chromosome maps. Bioinformatics. 2008;24(6):861–862. doi: 10.1093/bioinformatics/btm598. [DOI] [PubMed] [Google Scholar]
- 16.Kurtz S., Choudhuri J. V., Ohlebusch E., Schleiermacher C., Stoye J., Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Research. 2001;29(22):4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X., Wang L. GMATA: an integrated software package for genome-scale SSR mining, marker development and viewing. Frontiers in Plant Science. 2016;7, article 1350 doi: 10.3389/fpls.2016.01350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frazer K. A., Pachter L., Poliakov A., Rubin E. M., Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Research. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katoh K., Standley D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.1996. Se-Al: sequence alignment editor, version 2.0. [Google Scholar]
- 21.Librado P., Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 22.Kalyaanamoorthy S., Minh B. Q., Wong T. K., von Haeseler A., Jermiin L. S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods. 2017;14(6):587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ronquist F., Teslenko M., van der Mark P., et al. Mrbayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Systematic Biology. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma P., Zhang Y., Zeng C., Guo Z., Li D. Chloroplast phylogenomic analyses resolve deep-level relationships of an intractable bamboo tribe arundinarieae (Poaceae) Systematic Biology. 2014;63(6):933–950. doi: 10.1093/sysbio/syu054. [DOI] [PubMed] [Google Scholar]
- 25.Jones G., Sagitov S., Oxelman B. Statistical inference of allopolyploid species networks in the presence of incomplete lineage sorting. Systematic Biology. 2013;62(3):467–478. doi: 10.1093/sysbio/syt012. [DOI] [PubMed] [Google Scholar]
- 26.Mirarab S., Bayzid M. S., Warnow T. Evaluating summary methods for multilocus species tree estimation in the presence of incomplete lineage sorting. Systematic Biology. 2014 doi: 10.1093/sysbio/syu063. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y., Iaffaldano B. J., Zhuang X., Cardina J., Cornish K. Chloroplast genome resources and molecular markers differentiate rubber dandelion species from weedy relatives. BMC Plant Biology. 2017;17(1):p. 34. doi: 10.1186/s12870-016-0967-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y.-H., Wicke S., Wang H., et al. Plastid genome evolution in the early-diverging legume subfamily cercidoideae (Fabaceae) Frontiers in Plant Science. 2018;9, article 138 doi: 10.3389/fpls.2018.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li B., Zheng Y. Dynamic evolution and phylogenomic analysis of the chloroplast genome in Schisandraceae. Scientific Reports. 2018;8(1, article 9285) doi: 10.1038/s41598-018-27453-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Downie S. R., Jansen R. K. A comparative analysis of whole plastid genomes from the apiales: expansion and contraction of the inverted repeat, mitochondrial to plastid transfer of DNA, and identification of highly divergent noncoding regions. Systematic Botany. 2015;40(1):336–351. doi: 10.1600/036364415X686620. [DOI] [Google Scholar]
- 31.Wang R., Cheng C., Chang C., Wu C., Su T., Chaw S. Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evolutionary Biology. 2008;8(1, article 36) doi: 10.1186/1471-2148-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goulding S. E., Wolfe K. H., Olmstead R. G., Morden C. W. Ebb and flow of the chloroplast inverted repeat. MGG Molecular & General Genetics. 1996;252(1-2):195–206. doi: 10.1007/BF02173220. [DOI] [PubMed] [Google Scholar]
- 33.Hansen D. R., Dastidar S. G., Cai Z., et al. Phylogenetic and evolutionary implications of complete chloroplast genome sequences of four early-diverging angiosperms: buxus (Buxaceae), Chloranthus (Chloranthaceae), Dioscorea (Dioscoreaceae), and Illicium (Schisandraceae) Molecular Phylogenetics and Evolution. 2007;45(2):547–563. doi: 10.1016/j.ympev.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 34.Cai Z., Guisinger M., Kim H., et al. Extensive reorganization of the plastid genome of Trifolium subterraneum (Fabaceae) Is associated with numerous repeated sequences and novel DNA insertions. Journal of Molecular Evolution. 2008;67(6):696–704. doi: 10.1007/s00239-008-9180-7. [DOI] [PubMed] [Google Scholar]
- 35.Schwarz E. N., Ruhlman T. A., Sabir J. S., et al. Plastid genome sequences of legumes reveal parallel inversions and multiple losses of rps16 in papilionoids. Journal of Systematics and Evolution. 2015;53(5):458–468. doi: 10.1111/jse.12179. [DOI] [Google Scholar]
- 36.Dong W., Cheng T., Li C., et al. Discriminating plants using the DNA barcode rbcLb: an appraisal based on a large dataset. Molecular Ecology Resources. 2014;14(2):336–343. doi: 10.1111/1755-0998.12185. [DOI] [PubMed] [Google Scholar]
- 37.Dong W., Xu C., Li C., et al. ycf1, the most promising plastid DNA barcode of land plants. Scientific Reports. 2015;5, article 8348 doi: 10.1038/srep08348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gernandt D. S., Geada López G., Ortiz García S., Liston A. Phylogeny and classification of Pinus. Taxon. 2005;54(1):29–42. doi: 10.2307/25065300. [DOI] [Google Scholar]
- 39.Yang J., Vázquez L., Chen X., et al. Development of chloroplast and nuclear DNA markers for Chinese oaks (Quercus subgenus Quercus) and assessment of their utility as DNA barcodes. Frontiers in Plant Science. 2017;8, article 816 doi: 10.3389/fpls.2017.00816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li W., Liu Y., Yang Y., et al. Interspecific chloroplast genome sequence diversity and genomic resources in Diospyros. BMC Plant Biology. 2018;18(1, article 210) doi: 10.1186/s12870-018-1421-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao J., Jiang D., Zhao Z., et al. Development of chloroplast genomic resources in Chinese yam (Dioscorea polystachya) BioMed Research International. 2018;2018:11. doi: 10.1155/2018/6293847.6293847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song Y., Chen Y., Lv J., et al. Development of chloroplast genomic resources for oryza species discrimination. Frontiers in Plant Science. 2017;8, article 1854 doi: 10.3389/fpls.2017.01854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu C., Dong W., Li W., et al. Comparative analysis of six lagerstroemia complete chloroplast genomes. Frontiers in Plant Science. 2017;8(15, article 15) doi: 10.3389/fpls.2017.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. A list of the 72 taxa sampled from GenBank in this study for phylogeny analysis. Figure S1. Phylogenetic tree reconstruction of 82 taxa using Bayesian inference methods based on 81 genes chloroplast genome sequences.
Data Availability Statement
The five Ormosia chloroplast genomes are available in GenBank database (accession numbers: MH571753, MH571754, and MK105448- MK105450).