Abstract
Epimedium wushanense (Berberidaceae) is recorded as the source plant of Epimedii Wushanensis Folium in the Chinese Pharmacopoeia. However, controversies exist on the classification of E. wushanense and its closely related species, namely, E. pseudowushanense, E. chlorandrum, E. mikinorii, E. ilicifolium, and E. borealiguizhouense. These species are often confused with one another because of their highly similar morphological characteristics. This confusion leads to misuse in the medicinal market threatening efficiency and safety. Here, we studied the plastid genomes of these Epimedium species. Results show that the plastid genomes of E. wushanense and its relative species are typical circular tetramerous structure, with lengths of 156,855–158,251 bp. A total of 112 genes were identified from the Epimedium plastid genomes, including 78 protein-coding, 30 tRNA, and 4 rRNA genes. A loss of rpl32 gene in E. chlorandrum was found for the first time in this study. The phylogenetic trees constructed indicated that E. wushanense can be distinguished from its closely related species. E. wushanense shows a closer relationship to species in ser. Dolichocerae. In conclusion, the use of plastid genomes contributes useful genetic information for identifying medicinally important species E. wushanense and provides new evidence for understanding phylogenetic relationships within the Epimedium genus.
Keywords: plastid genome, Epimedium, phylogenetic analysis, identification
1. Introduction
The classification and identification of the genus Epimedium L. (Berberidaceae) has always been a research hotspot. Stearn [1] proposed the most comprehensive classification system for this genus, which was supported by geographical distribution and C-banding of chromosomes. In Stearn’s monograph, genus Epimedium is divided into subgenera Rhizophyllum and Epimedium. Furthermore, subgenus Epimedium is divided into four sections, namely, Epimedium, Polyphyllon, Macroceras, and Diphyllon. The section Diphyllon, consisting of approximately 47 known species in China, is further subdivided into four series, namely, Campanulatae Stearn, Davidianae Stearn, Dolichocerae Stearn, and Brachycerae Stearn. However, controversies still exist in the classification and discrimination of Epimedium species. On the one hand, phylogenetic research based on molecular markers [2], flavonoid types [3], and amplified fragment length polymorphisms (AFLP) [4] could not support Stearn’s classification system. On the other hand, the majority of Epimedium species are easily confused with one another due to their highly similar morphology, thereby complicating the distinction between these species. E. wushanense T.S. Ying and its closely related species is a typical example of species complex. Ying et al. [5] listed the distribution of E. wushanense in Chongqing, Guizhou, Hubei, and Sichuan in the Flora of China. On the contrary, Guo et al. [6] concluded that E. wushanense from Guangxi and Guizhou should be recognized as a new species E. pseudowushanense B.L. Guo. Moreover, E. wushanense is easily confused with E. chlorandrum Stearn, E. mikinorii Stearn, E. ilicifolium Stearn, and E. borealiguizhouense S.Z. He & Y.K. Yang due to their highly similar morphological characteristics [7]. The dried leaves of E. wushanense are used as Epimedii Wushanensis Folium (Wushan Yinyanghuo) in the Chinese Pharmacopeia [8]. Considering differences in bioactive constituents among Epimedium species [9], incorrect identification of species is likely to result in potential risks in herbal efficacy and medical safety. However, only a few reports on the taxonomic analysis and authentication of E. wushanense and its related species are available [10,11]. Therefore, a systematic classification and comprehensive identification of E. wushanense and its closely related species is urgently needed.
The complete plastid genome, containing variant genetic information, has been extensively utilized in plant identification and phylogenetic studies. Most plastid genomes in angiosperms are circular DNA molecules ranging from 120 kb to 160 kb in length [12]. They have the advantages of maternal inheritance, single structure, and high conservation of gene content and genome structure [13,14]. Jose et al. [15] performed a comprehensive study on the evolution and variability of the genus Citrus to provide a good understanding of its phylogenetic relationships. Curci et al. [16] developed plastid resources for the genus Cynara and found that plastid genome can be used as a reliable and valuable molecular marker for Cynara identification. In addition, Pessoa-Filho et al. [17] investigated the phylogenetic relationships of four Urochloa species on the basis of their plastid genomes. Therefore, plastid genomes were expected to be an effective resource for discriminating E. wushanense and its related species.
Here, we studied the plastid genomes of these Epimedium species. We successfully discriminate them based on plastid genome sequence and provide useful taxonomic information for the genus Epimedium.
2. Results and Discussion
2.1. Plastid Genome Structures of E. wushanense and Its Closely Related Species
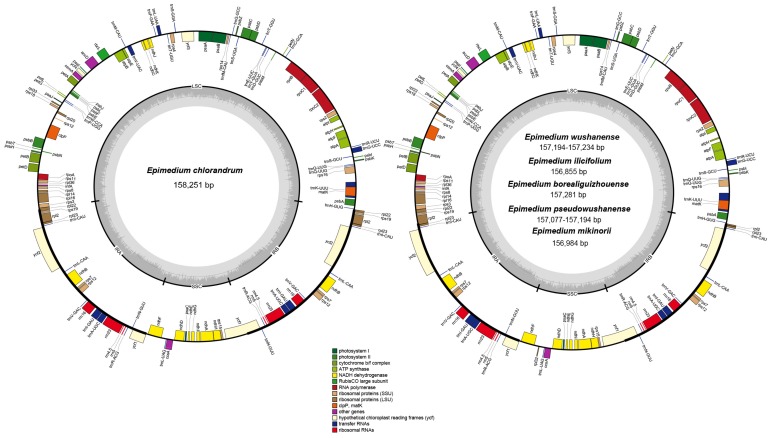
The plastid genome length of the six Epimedium species (E. wushanense, E. pseudowushanense, E. chlorandrum, E. mikinorii, E. ilicifolium, and E. borealiguizhouense) ranges from 156,855 bp to 158,251 bp. All the nine plastid genomes displayed a typical quadripartite structure, which consists of a pair of inverted repeats (IRs, 25,775–27,693 bp) separated by a large single-copy (LSC) region (86,627–88,603 bp) and a small single-copy (SSC) region (16,238–17,091 bp) (Figure 1, Table 1). The average GC content of the nine plastid genomes was 38.80%. Congruent with previous research, a total of 112 genes were identified from the nine plastid genomes, including 78 protein-coding, 30 tRNA, and 4 rRNA genes [18] (Figure 1). Similar with other angiosperms [19,20], a total of 18 genes containing introns were found. Among them, three genes, namely, clpP, rps12, and ycf3, contain two introns. The other 15 genes (atpF, rpoC1, rpl2, rpl16, petB, petD, rps16, ndhA, ndhB, trnA-UGC, trnG-UCC, trnL-UAA, trnK-UUU, trnI-GAU, and trnV-UAC) contain one intron. The loss of plastid rpl32 gene in Populus and Thalictrum species has been found, and the corresponding rpl32 gene has been transferred in the nuclear genome [21,22,23]. E. chlorandrum was treated as a synonym of E. acuminatum in the research of Zhang et al. [24]. Compared with E. acuminatum, an 859 bp deletion (including ndhF-rpl32 intergenic spacer, partial sequence; rpl32 gene, complete sequence; and rpl32-trnL intergenic spacer, partial sequence) in E. chlorandrum was found for the first time in our study. The deletion was verified through PCR amplification and Sanger sequencing with specific primers rpl32_1F/rpl32_1R (Table S1). The special plastid genome structure of E. chlorandrum suggests that it is different from E. acuminatum.
Figure 1.
Gene map of the complete plastid genomes of E. wushanense and its closely related species. Genes drawn inside the circle are transcribed clockwise, whereas those outside are transcribed counterclockwise. The darker gray in the inner circle corresponds to the GC content, whereas the lighter gray corresponds to the AT content.
Table 1.
Plastid genome characteristics of E. wushanense and its closely related species.
| Latin Name | Voucher No. | Accession No. | LSC Length/bp | SSC Length/bp | IR Length/bp | Genome Size/bp | GC Content/% |
|---|---|---|---|---|---|---|---|
| E. wushanense | HBXS | MK408753 | 88,527 | 17,091 | 25,788 | 157,194 | 38.80 |
| E. wushanense | HBXS-2 | MK992920 | 88578 | 17,090 | 25,783 | 157,234 | 38.80 |
| E. pseudowushanense | GZJH | MK408750 | 88,449 | 17,070 | 25,779 | 157,077 | 38.78 |
| E. pseudowushanense | GZNW | MK992919 | 88,538 | 17,079 | 25,788 | 157,193 | 38.78 |
| E. mikinorii | HBES | MK408752 | 88,360 | 17,074 | 25,775 | 156,984 | 38.81 |
| E. mikinorii | GZLS | MK992918 | 88,522 | 16,833 | 25,783 | 156,921 | 38.80 |
| E. chlorandrum | SCMP | MK408754 | 86,627 | 16,238 | 27,693 | 158,251 | 38.89 |
| E. ilicifolium | SXBX | MK992921 | 88,217 | 17,038 | 25,800 | 156,855 | 38.77 |
| E. borealiguizhouense | GZQB | MK408751 | 88,603 | 17,040 | 25,819 | 157,281 | 38.78 |
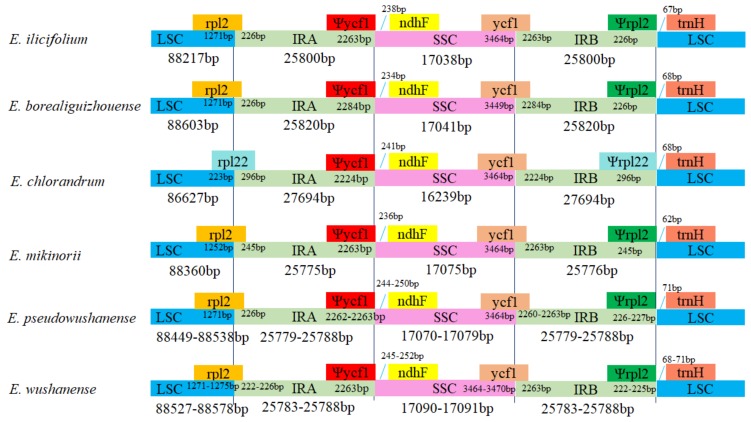
The contraction and expansion at the boundary of the IR regions were considered the main reasons for the length variation of the plastid genomes [25]. In this study, the gene ycf1 crossed the SSC/IR region, and the pseudogene fragment ycf1 was located at the IRA region with 2224–2284 bp in the plastid genomes of the six Epimedium species. Meanwhile, evident variations at the IR and LSC boundary regions were observed among the plastid genomes of E. chlorandrum and the other five species (E. wushanense, E. pseudowushanense, E. borealiguizhouense, E. ilicifolium, and E. mikinorii). The gene rpl22 crossed the LSC/IRA region, and Ψrpl22 with 296 bp was located at the IRB region in the plastid genome of E. chlorandrum, which was congruent with E. acuminatum. Otherwise, the gene rpl2 crossed the LSC/IRA region in the other five species, and Ψrpl2 with 222–245 bp was located in the IRB region. This finding is congruent with the E. dolichostemon, E. lishischenii, and E. pseudowushanense investigated by Zhang et al. [18]. At the junction of the IRA/SSC region, the distance between ψycf1 and ndhF ranged from 234 to 252b p. In addition, the distance between ψrpl22 and trnH in E. chlorandrum was 68 bp, whereas that between ψrpl2 and trnH in the other four species ranged from 62 to 71 bp at the junction of the IRB/LSC region (Figure 2).
Figure 2.
Comparison of the borders of the LSC, SSC and IR regions among plastid genomes of E. wushanense and its closely related species.
2.2. Codon Usage
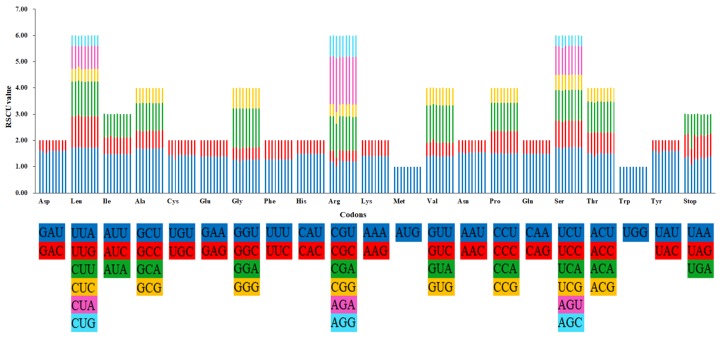
All the protein-coding genes consist of 26,058 (E. borealiguizhouense) to 26,333 (E. chlorandrum) codons in the nine Epimedium plastid genomes. These codons encode 20 amino acids (Table S2, Figure 3). The most and least amino acids in Epimedium species are respectively leucine (9.3–10.5%) and cysteine (1.1–1.5%). Apart from methionine and tryptophan, other amino acid codons all have preferences. The results showed a strong bias in codon usage for A and U at the third codon position (Figure 3). Similar results for codon usage were also observed in the plastid genomes of Taxillus and Panax species [19,25]. Interestingly, the usage of codons in E. borealiguizhouense showed preference for higher G/C content at the 3rd codon position, as well as AGA and stop codon UGA, comparing with the other five species (Figure 3).
Figure 3.
Codon content of 20 amino acids and stop codons in all protein-coding genes of the plastid genomes of E. wushanense and its closely related species. The histogram from the left to right side represents plastid genomes of E. wushanense (HBXS, HBXS-2), E. borealiguizhouense, E. chlorandrum, E. mikinorii (HBES, GZLS), E. pseudowushanense (GZJH, GZNW) and E. ilicifolium, respectively.
2.3. SSRs and Repeat Structure Analyses
SSRs, consisting of tandem repeated DNA sequences in the plastid genome, present high levels of polymorphism [26]. SSRs have been used as molecular markers for species identification and phylogenetic investigations [26,27]. In this study, 71–79 SSRs were detected in the nine Epimedium plastid genomes using the MISA tool (Table S3). The majority of SSRs in all species were mononucleotides repeats (88.1–92.4%), and the A/T mononucleotides repeats were the most common, with a proportion of 84.6–89.9% (Table 2). Pentanucleotide SSRs were detected in all the species sequenced in our study except for E. borealiguizhouense. Aside from population GZNW of E. pseudowushanense, no hexanucleotide SSR existed in the other five species. Congruent with previous research [18], most of these SSR loci were located in the LSC region, followed by the SSC and IR regions.
Table 2.
Types and amounts of SSRs in the E. wushanense and its closely related species.
| SSR Type | Repeat Unit | Amount/Ratio (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| E. borealiguizhouense | E. mikinorii | E. chlorandrum | E. pseudowushanense | E. wushanense | E. ilicifolium | |||||
| GZQB | HBES | GZLS | SCMP | GZJH | GZNW | HBXS | HBXS-2 | SXBX | ||
| Mono | A/T | 64/88.9 | 61/85.9 | 67/88.2 | 66/84.6 | 68/88.3 | 67/87.0 | 69/88.5 | 71/89.9 | 65/85.5 |
| C/G | 2/2.8 | 2/2.8 | 2/2.6 | 3/3.8 | 2/2.6 | 2/2.6 | 2/2.6 | 2/2.5 | 2/2.6 | |
| Di | AT/AT | 1/1.4 | 2/2.8 | 2/2.6 | 1/1.3 | 2/2.6 | 2/2.6 | 2/2.6 | 3/3.8 | 2/2.6 |
| Tri | AAT/ATT | 1/1.4 | 1/1.4 | 1/1.3 | 2/2.6 | 1/1.3 | 1/1.3 | 1/1.3 | 0/0 | 1/1.3 |
| AGG/CCT | 3/4.2 | 3/4.2 | 3/3.9 | 3/3.8 | 3/3.9 | 3/3.9 | 3/3.8 | 3/3.8 | 3/3.9 | |
| AAG/CTT | 1/1.4 | 1/1.4 | 0/0 | 1/1.3 | 0/0 | 0/0 | 0/0 | 0/0 | 1/1.3 | |
| Tetra | AAAT/ATTT | 0/0 | 0/0 | 0/0 | 1/1.3 | 0/0 | 0/0 | 0/0 | 0/0 | 1/1.3 |
| Penta | AAGAT/ATCTT | 0/0 | 1/1.4 | 1/1.3 | 1/1.3 | 1/1.3 | 1/1.3 | 1/1.3 | 0/0 | 0/0 |
| AATAT/ATATT | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 1/1.3 | |
| Hexa | AACGAC/CGTTGT | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 1/1.3 | 0/0 | 0/0 | 0/0 |
| Total | 72/100 | 71/100 | 76/100 | 78/100 | 77/100 | 77/100 | 78/100 | 79/100 | 76/100 | |
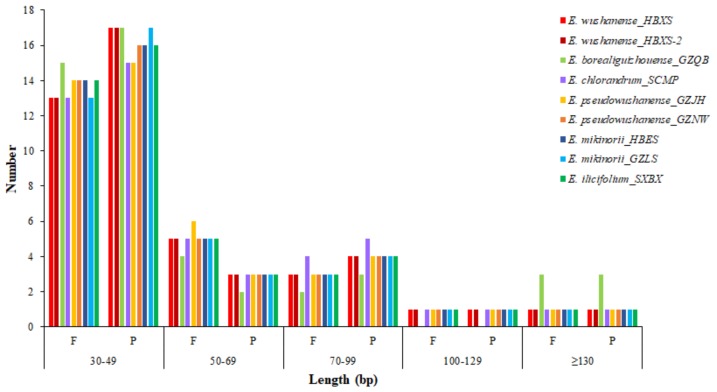
Repetitive structures in plastid genomes is closely related to the plastid genome rearrangement and variation, which is of great importance for phylogenetic and population genetics studies [28]. A total of 49 repeats were identified in each of the nine Epimedium plastid genomes by REPuter (Table S4). E. chlorandrum, E. borealiguizhouense, and E. ilicifolium all possess 24 forward and 25 palindromic repeats. Both of the two populations of E. wushanense consist of 23 forward and 26 palindromic repeats; whereas the two populations of E. pseudowushanense and E. mikinorii comprise different number of forward and palindromic repeats (Figure 4). The lengths of repeats in the plastid genome of E. borealiguizhouense ranged from 32 bp to 189 bp, whereas those in the other eight plastid genomes ranged from 32 bp to 131 bp (Figure 4, Table S4). The special repetitive structure of E. borealiguizhouense could distinguish it from E. wushanense. The copy lengths with 30–49 bp are the most common (57.1–65.3%), whereas those with lengths longer than 100 bp are minimal (8.2–12.2%) (Figure 4). These repeats were located in the LSC and IR regions, and no repeat was found in the SSC region. Interestingly, the intergenic spacer (IGS) and protein coding (pCD) regions are the most common distribution places for repeats. Previous studies showed that repeated sequences in many angiosperm lineages are mainly distributed in the IGS and intron regions of plastid genomes [19,20,29]. However, a larger number of repeats were found in the pCD regions than those in the noncoding regions in the present study, which is similar with the report of Zhang et al. [18] (Table S4).
Figure 4.
Repeat sequences in the plastid genomes of E. wushanense and its closely related species. F, forward; P, palindrome.
2.4. Genome Sequence Divergence
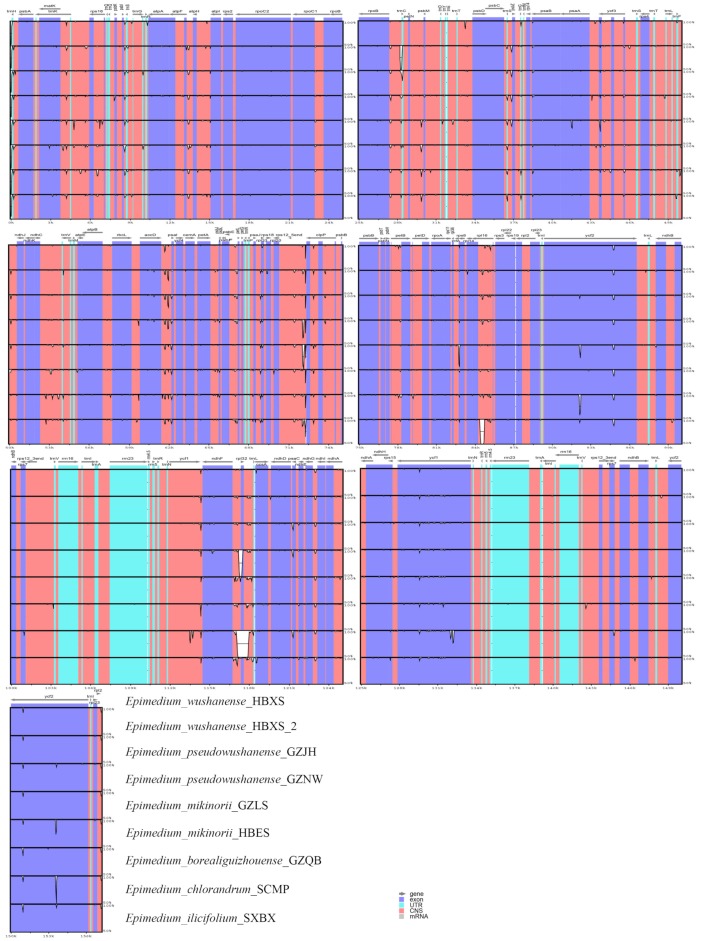
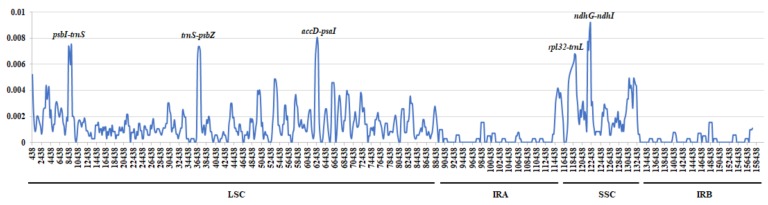
The nine Epimedium plastid genomes obtained from the present study were compared and plotted using mVISTA with E. wushanense (HXBX) as a reference (Figure 5). The plots illustrate the high similarity of the nine Epimedium plastid genome sequences. The sequence similarity ranged from 99.4% (E. chlorandrum) to 99.8% (E. wushanense, HXBX-2) among these samples. Meanwhile, the coding region exhibited a lower divergence than the non-coding region, and sequence similarity was close to 100% in the coding region. The highly variable regions in the nine Epimedium plastid genomes were identified by the sliding window analysis using DnaSP. The average Pi value for nine Epimedium plastid genomes was 0.00121. The IR regions exhibited lower nucleotide diversity than the LSC and SSC regions. Meanwhile, the results showed that the most divergent regions were localized in the IGS region. A total of five regions showed higher Pi values than other regions (Pi > 0.006), namely, psbI-trnS, trnS-psbZ, accD-psaI, rpl32-trnL, and ndhG-ndhI (Figure 6). These divergent regions, which comprise abundant variation information, can be used to develop molecular markers for authenticating E. wushanense and its closely related species.
Figure 5.
Sequence identity plots comparing nine Epimedium plastid genomes with E. wushanense as a reference.
Figure 6.
Nucleotide diversity graphs of nine Epimedium plastid genomes. The x-axis represents position of the midpoint of a window, and the y-axis represents Pi value.
2.5. Phylogenetic and Taxonomic Implications for the Genus Epimedium
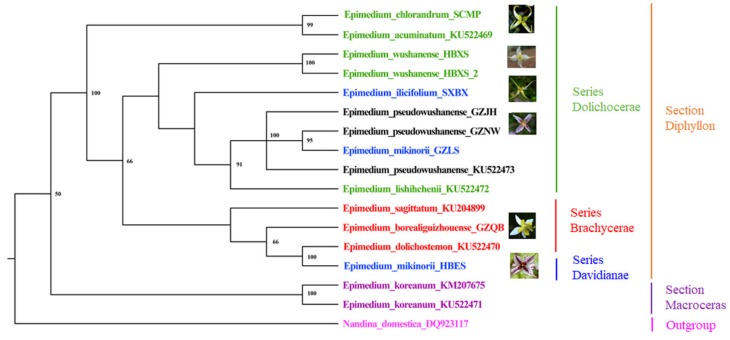
The classification and identification of the genus Epimedium has always been controversial. The plastid genomes contained much variation, which can be used to discriminate genetically close species [30]. In this study, the results showed that E. wushanense can be distinguished from its relative species (Figure 7). Phylogenetic analysis based on karyomorphology [31] and molecular markers [4,32] has been done to confirm the evolution of this genus. However, the further subdivision of the Chinese sect. Diphyllon was poorly resolved. In this study, protein-coding genes of 17 plastid genomes from 11 Epimedium species and one outgroup species (Nandina domestica) were used to construct the phylogenetic trees using maximum likelihood (ML) method. The phylogenetic tree generated by the ML, including ML bootstrap values, is shown in Figure 7. Within Epimedium, the 11 species were divided into four clades. E. koreanum was first separated from the other Epimedium species, which was congruent with Stearn’s classification system. However, the relationship of the other species belonging to sect. Diphyllon did not completely support Stearn’s classification. E. wushanense, E. lishihchenii, E. chlorandrum, and E. acuminatum were divided into ser. Dolichocerae by Stearn [1]. Three specimens of E. pseudowushanense and two specimens of E. wushanense clustered as single branch respectively, supporting E. pseudowushanens as a new species in-dependent of E. wushanense [6]. Based upon the floral characters, E. pseudowushanense was classified into ser. Dolichocerae by Guo et al. [6]. Zhang et al. [10] proposed that E. pseudowushanense should be grouped into ser. Davidianae and that E. wushanense should be moved from ser. Dolichocerae to ser. Davidianae. In our study, E. pseudowushanense was clustered with E. lishihchenii and E. wushanense that were grouped into ser. Dolichocerae, supporting the taxonomic views of Stearn [1] and Guo et al. [6]. Moreover, E. ilicifolium was clustered with species belonging to ser. Dolichocerae into one branch, which supports the opinion of moving it from ser. Davidianae to ser. Dolichocerae [10]. In previous research, Guo et al. [3] grouped E. mikinorii from ser. Davidianae [1] to ser. Dolichocerae according to its petal characteristics. However, E. mikinorii was first clustered with E. dolichostemon in one branch and then clustered with E. borealiguizhouense in one clade. Hence, E. mikinorii was inferred to have a close relationship with species belonging to ser. Brachycerae. The population GZLS, a transitional species between E. pseudowushanense and E. mikinorii, was clustered with E. pseudowushanense into one branch, demonstrating that it may have a closer evolutionary relationship to E. pseudowushanense. The present study provides valuable information for identifying Epimedium species and resolving their phylogenetic relationships. To further explore the phylogeny of the whole genus, more plastid genomes of Epimedium species are needed. At present, the plastid genomes of plants have been applied as a useful tool for exploring the phylogenetic relationships in many genera [15,16,17,30]. Some studies report the conflicts in phylogenetic trees based on plastid and nuclear genome data [33,34]. In the future work, we will thoroughly analyze the phylogenetic relationships of the genus Epimedium derived from both plastid and nuclear genome data, thus comprehensively understanding the evolutionary relationship of this genus.
Figure 7.
Phylogenetic relationships of 11 Epimedium species constructed based on the 87 protein-coding genes with maximum likelihood (ML). Numbers at the nodes represent ML bootstrap values. Stearn’s classification for each species is represented by different color, and species in the same series are represented by the same color.
3. Materials and Methods
3.1. Taxon Sampling, DNA Extraction, and Sequencing
Totally 16 specimens representing 11 Epimedium species were collected. Among them, nine samples from six Epimedium species were sampled from Guizhou, Hubei, Shaanxi, and Sichuan provinces of China (Table S5). These samples were all identified by taxonomist Prof. Yanqin Xu at Jiangxi University of Traditional Chinese Medicine and taxonomist Prof. Shunzhi He at Guiyang College of Traditional Chinese Medicine. All corresponding voucher samples were deposited in the Herbarium of the Institute of Medicinal Plant Development, Chinese Academy of Medicinal Sciences, Beijing, China. Before DNA extraction, the fresh leaves were frozen at −20 °C Total genomic DNA was extracted and purified using the Plant Genomic DNA Rapid Extraction kit (Bioteke Corporation, Beijing, China) in accordance with the manufacturer’s instructions. The quantified DNA was then used to construct shotgun libraries with an average insert size of 500 bp and sequenced using Illumina Hiseq X in accordance with the manufacturer’s manual. Approximately 5 GB of raw data from each sample were produced with 150 bp pair-end read lengths. Moreover, seven plastid genomes from six Epimedium species [18,35,36] were downloaded from GenBank (Table S5).
3.2. Genome Assembly and Genome Annotation
Low-quality reads including a mixture data of nuclear and organelle genomes were trimmed with the software Trimmomatic [37]. The clean reads were then extracted and mapped to the reference database. Such database is constructed with all plastid genomes of plants from the National Center for Biotechnology Information on the basis of their coverage and similarity. The extracted reads were assembled to contigs with SOAPdenovo (v2, BGI HK Research Institute, Hong Kong, China) [38]. Complete plastid genome sequences were obtained with the combination and extension of the resulting contigs. Gaps in the assemblies were bridged by Sanger sequencing with specific primers designed for PCR based on their flanking sequences. To ensure assembly accuracy, the four junctions between the inverted repeats (IRs) and single-copy (SC) regions were verified through PCR amplification and Sanger sequencing with specific primers. All the specific primers are listed in Table S1.
The plastid genome annotation was performed using the software CPGAVAS [39], with manual corrections using the Apollo genome editor [40]. The plastid genome maps were generated using the Organellar-Genome DRAW (v1.2, Max Planck Institute of Molecular Plant Physiology, Potsdam, Germany) [41] with default settings and checked manually. The complete and correct plastid genome sequences of the six species were deposited in GenBank under accession numbers of MK408750-MK408754 and MK992918-MK992921 (Table S5). Raw sequences are available in the Sequence Read Archive of the NCBI under the accession numbers SAMN12430115-SAMN12430123 (Table S5).
3.3. Genome Structure Analyses and Genome Comparison
The GC content was analyzed using the software Mega 6.0 [42]. Simple sequence repeats (SSRs) were identified using the MISA Perl script (http://pgrc.ipk-gatersleben.de/misa/). Microsatellites were detected, including mono-, di-, tri-, tetra-, penta-, and hexanucleotide repeats, with thresholds of 10 repeat units for mono-, six repeat units for di-, four repeat units for tri- and tetra-, and three repeat units for penta- and hexanucleotide SSRs. The REPuter program [43] was run to identify the location and size of forward, palindromic, reverse, and complement repeats in the plastid genomes under the following criterion: Cutoff n ≥ 30 bp and 90% sequence identities (Hamming distance of 3). The alignment for the plastid genomes was performed and plotted using the mVISTA program [44]. The distribution of codon usage for all protein-coding genes was investigated using MEGA 6.0 [42] with the relative synonymous codon usage (RSCU) ratio [45]. To calculate the nucleotide diversity (Pi) and detect highly variable sites among Epimedium plastid genomes, DNA polymorphism analysis was performed using DnaSP (DNA Sequence Polymorphism) v6 [46] with 200 bp step size and 800 bp window length.
3.4. Phylogenetic Analysis
The 87 protein-coding genes were extracted from plastid genomes generated from the present research and downloaded from GenBank. The sequences were concatenated and aligned using MAFFT v7 [47], and Nandina domestica (DQ923117) was used as an outgroup. Appropriate nucleotide substitution models and parameters for the ML was conducted by running Likelihood Ratio Tests in PAUP*4.0 b10 [48] and using MrModeltest [49]. Model testing showed that the general time reversible (GTR) including rate variation among sites (+G) and invariable sites (+I) (= GTR + G + I) was the best fit model to the data sets under the Akaike Information Criterion (AIC). The ML analysis was conducted using the best-fitting model with 1000 bootstrap replicates.
4. Conclusions
This work reports nine new complete plastid genomes from E. wushanense and its closely related species, thereby enriching the plastid genome data of the Epimedium genus. A loss of rpl32 gene in E. chlorandrum, which is a unique feature to distinguish it from other species, was found. The phylogenetic tree constructed demonstrated that E. wushanense can be separated from its relative species. Moreover, the plastid genome data provide new evidence for classifying Epimedium species, which will contribute valuable information for further exploring the phylogenetic relationships of the genus Epimedium.
Acknowledgments
We would like to acknowledge Shunzhi He at Guiyang College of Traditional Chinese Medicine for identifying experimental samples.
Supplementary Materials
Supplementary Materials can be found at https://www.mdpi.com/1422-0067/20/16/4003/s1. Table S1: Primers used for genome sequence validation, Table S2: Codon usage within the plastid genomes of E. wushanense and its closely related species, Table S3: Distribution of SSR loci in the plastid genomes of E. wushanense and its closely related species, Table S4: Repeated sequences identified in the plastid genomes of E. wushanense and its closely related species, Table S5: Voucher information and GenBank accession numbers for the Epimedium samples.
Author Contributions
Conceptualization, X.P.; data curation, M.G., L.R., B.L. and Y.L.; formal analysis, M.G., L.R. and B.L.; funding acquisition, X.P.; investigation, M.G., L.R. and Y.X.; resources, Y.X. and B.G.; supervision, X.P.; visualization, M.G. and L.R.; writing—original draft, M.G., L.R. and X.P.; writing—review & editing, J.S., N.M., S.C. and X.P.
Funding
This research was funded by grants from the National Natural Science Foundation of China (No. 81573541), CAMS Innovation Fund for Medical Sciences (CIFMS) (No. 2017-I2M-1-013), and the State Scholarship Fund of China (No. 201808110108).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Stearn W.T. The Genus Epimedium and Other Herbaceous Berberidaceae. Timber Press; Portland, OR, USA: 2002. [Google Scholar]
- 2.Zhang M.L., Uhink C.H., Kadereit J.W. Phylogeny and biogeography of Epimedium/Vancouveria (Berberidaceae): Western North American-East Asian disjunctions, the origin of European mountain plant taxa, and East Asian species diversity. Syst. Bot. 2007;32:81–92. doi: 10.1600/036364407780360265. [DOI] [Google Scholar]
- 3.Guo B.L., Pei L.K., Xiao P.G. Further research on taxonomic significance of flavonoids in Epimedium (Berberidaceae) J. Syst. Evol. 2008;46:874–885. [Google Scholar]
- 4.De Smet Y., Goetghebeur P., Wanke S., Asselman P., Samain M.S. Additional evidence for recent divergence of Chinese Epimedium (Berberidaceae) derived from AFLP, chloroplast and nuclear data supplemented with characterisation of leaflet pubescence. Plant Ecol. Evol. 2012;145:73–87. doi: 10.5091/plecevo.2012.646. [DOI] [Google Scholar]
- 5.Ying T.S., Boufford D.E., Brach A.R. Flora of China. Volume 19. Science Press; St. Louis, MO, USA: Botanical Garden Press; Beijing, China: 2011. Epimedium Linnaeus; pp. 787–799. [Google Scholar]
- 6.Guo B.L., He S.Z., Zhong G.Y., Xiao P.G. Two new species of Epimedium (Berberidaceae) from China. Acta Phytotaxon. Sin. 2007;45:813–821. doi: 10.1360/aps06138. [DOI] [Google Scholar]
- 7.He S.Z. The Genus Epimedium of China in Colour. Guizhou Publishing Group; Guizhou, China: 2014. [Google Scholar]
- 8.Chinese Pharmacopoeia Commission . Pharmacopoeia of the People’s Republic of China. Volume I Chemical Industry Press; Beijing, China: 2015. [Google Scholar]
- 9.Ma H., He X., Yang Y., Li M., Hao D., Jia Z. The genus Epimedium: An ethnopharmacological and phytochemical review. J. Ethnopharmacol. 2011;134:519–541. doi: 10.1016/j.jep.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y., Dang H., Li J., Wang Y. The Epimedium wushanense (Berberidaceae) species complex, with one new species from Sichuan, China. Phytotaxa. 2014;172:39. doi: 10.11646/phytotaxa.172.1.5. [DOI] [Google Scholar]
- 11.Guo M.Y., Xu Y.Q., Ren L., He S.Z., Pang X.H. A systematic study on DNA barcoding of medicinally important genus Epimedium L. (Berberidaceae) Genes Basel. 2018;9:637. doi: 10.3390/genes9120637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wicke S., Schneeweiss G.M., dePamphilis C.W., Muller K.F., Quandt D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011;76:273–297. doi: 10.1007/s11103-011-9762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palmer J.D., Jansen R.K., Michaels H.J., Chase M.W., Manhart J.R. Chloroplast DNA Variation and Plant Phylogeny. Ann. Mo. Bot. Gard. 1988;75:1180–1206. doi: 10.2307/2399279. [DOI] [Google Scholar]
- 14.Wu F.H., Chan M.T., Liao D.C., Hsu C.T., Lee Y.W., Daniell H., Duvall M.R., Lin C.S. Complete chloroplast genome of Oncidium Gower Ramsey and evaluation of molecular markers for identification and breeding in Oncidiinae. BMC Plant Bio. 2010;10:68. doi: 10.1186/1471-2229-10-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carbonell-Caballero J., Alonso R., Ibanez V., Terol J., Talon M., Dopazo J. A Phylogenetic Analysis of 34 Chloroplast Genomes Elucidates the Relationships between Wild and Domestic Species within the Genus Citrus. Mol. Biol. Evol. 2015;32:2015–2035. doi: 10.1093/molbev/msv082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Curci P.L., De Paola D., Sonnante G. Development of chloroplast genomic resources for Cynara. Mol. Ecol. Resour. 2016;16:562–573. doi: 10.1111/1755-0998.12457. [DOI] [PubMed] [Google Scholar]
- 17.Pessoa-Filho M., Martins A.M., Ferreira M.E. Molecular dating of phylogenetic divergence between Urochloa species based on complete chloroplast genomes. BMC Genom. 2017;18:516. doi: 10.1186/s12864-017-3904-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y., Du L., Liu A., Chen J., Wu L., Hu W., Zhang W., Kim K., Lee S., Yang T., et al. The Complete Chloroplast Genome Sequences of Five Epimedium Species: Lights into Phylogenetic and Taxonomic Analyses. Front. Plant Sci. 2016;7:306. doi: 10.3389/fpls.2016.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou J., Chen X., Cui Y., Sun W., Li Y., Wang Y., Song J., Yao H. Molecular Structure and Phylogenetic Analyses of Complete Chloroplast Genomes of Two Aristolochia Medicinal Species. Int. J. Mol. Sci. 2017;18:1839. doi: 10.3390/ijms18091839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X., Zhou J., Cui Y., Wang Y., Duan B., Yao H. Identification of Ligularia Herbs Using the Complete Chloroplast Genome as a Super-Barcode. Front. Pharmacol. 2018;9:695. doi: 10.3389/fphar.2018.00695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cusack B.P., Wolfe K.H. When gene marriages don’t work out: Divorce by subfunctionalization. Trends Genet. 2007;23:270–272. doi: 10.1016/j.tig.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 22.Ueda M., Fujimoto M., Arimura S., Murata J., Tsutsumi N., Kadowaki K. Loss of the rpl32 gene from the chloroplast genome and subsequent acquisition of a preexisting transit peptide within the nuclear gene in Populus. Gene. 2007;402:51–56. doi: 10.1016/j.gene.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 23.Park S., Jansen R.K., Park S. Complete plastome sequence of Thalictrum coreanum (Ranunculaceae) and transfer of the rpl32 gene to the nucleus in the ancestor of the subfamily Thalictroideae. BMC Plant Biol. 2015;15:40. doi: 10.1186/s12870-015-0432-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y., Dang H., Li S., Li J., Wang Y. Five new synonyms in Epimedium (Berberidaceae) from China. PhytoKeys. 2015;49:1–12. doi: 10.3897/phytokeys.49.8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim K.J., Lee H.L. Complete Chloroplast Genome Sequences from Korean Ginseng (Panax schinseng Nees) and Comparative Analysis of Sequence Evolution among 17 Vascular Plants. DNA Res. 2004;11:247–261. doi: 10.1093/dnares/11.4.247. [DOI] [PubMed] [Google Scholar]
- 26.Powell W., Morgante M., Mcdevitt R., Vendramin G.G., Rafalski J.A. Polymorphic simple sequence repeat regions in chloroplast genomes: Applications to the population genetics of pines. Proc. Natl. Acad. Sci. USA. 1995;92:7759–7763. doi: 10.1073/pnas.92.17.7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiao Y., Jia H., Li X., Chai M., Jia H., Chen Z., Wang G., Chai C., van de Weg E., Gao Z. Development of simple sequence repeat (SSR) markers from a genome survey of Chinese bayberry (Myrica rubra) BMC Genom. 2012;13:201. doi: 10.1186/1471-2164-13-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cavalier-Smith T. Chloroplast Evolution: Secondary Symbiogenesis and Multiple Losses. Curr. Biol. 2002;12:R62–R64. doi: 10.1016/S0960-9822(01)00675-3. [DOI] [PubMed] [Google Scholar]
- 29.Yang J.B., Yang S.X., Li H.T., Yang J., Li D.Z. Comparative chloroplast genomes of Camellia species. PLoS ONE. 2013;8:e73053. doi: 10.1371/journal.pone.0073053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kane N., Sveinsson S., Dempewolf H., Yang J.Y., Zhang D., Engels J.M., Cronk Q. Ultra-barcoding in cacao (Theobroma spp.; Malvaceae) using whole chloroplast genomes and nuclear ribosomal DNA. Am. J. Bot. 2012;99:320–329. doi: 10.3732/ajb.1100570. [DOI] [PubMed] [Google Scholar]
- 31.Sheng M.Y., Wang L.J., Tian X.J. Karyomorphology of eighteen species of genus Epimedium (Berberidaceae) and its phylogenetic implications. Genet. Resour. Crop Evol. 2010;57:1165–1176. doi: 10.1007/s10722-010-9556-6. [DOI] [Google Scholar]
- 32.Zhang Y., Yang L., Chen J., Sun W., Wang Y. Taxonomic and phylogenetic analysis of Epimedium L. based on amplified fragment length polymorphisms. Sci. Hortic. Amst. 2014;170:284–292. doi: 10.1016/j.scienta.2014.02.025. [DOI] [Google Scholar]
- 33.Olmstead R.G., Bedoya A.M. Whole genomes: The holy grail. A commentary on: ‘Molecular phylogenomics of the tribe Shoreeae (Dipterocarpaceae) using whole plastidgenomes’. Ann. Bot. 2019;123:iv–v. doi: 10.1093/aob/mcz055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee-Yaw J.A., Grassa C.J., Joly S., Andrew R.L., Rieseberg L.H. An evaluation of alternative explanations for widespread cytonuclear discordance in annual sunflowers (Helianthus) New Phytol. 2019;221:515–526. doi: 10.1111/nph.15386. [DOI] [PubMed] [Google Scholar]
- 35.Lee J.H., Kim K., Kim N.R., Lee S.C., Yang T.J., Kim Y.D. The complete chloroplast genome of a medicinal plant Epimedium koreanum Nakai (Berberidaceae) Mitochondrial DNA A. 2016;27:4342–4343. doi: 10.3109/19401736.2015.1089492. [DOI] [PubMed] [Google Scholar]
- 36.Sun Y., Moore M.J., Zhang S., Soltis P.S., Soltis D.E., Zhao T., Meng A., Li X., Li J., Wang H. Phylogenomic and structural analyses of 18 complete plastomes across nearly all families of early-diverging eudicots, including an angiosperm-wide analysis of IR gene content evolution. Mol. Phylogenet. Evol. 2016;96:93–101. doi: 10.1016/j.ympev.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 37.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J., He G., Chen Y., Pan Q., Liu Y., et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu C., Shi L., Zhu Y., Chen H., Zhang J., Lin X., Guan X. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genom. 2012;13:715. doi: 10.1186/1471-2164-13-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee E., Helt G.A., Reese J.T., Munoz-Torres M.C., Childers C.P., Buels R.M., Stein L., Holmes I.H., Elsik C.G., Lewis S.E. Web Apollo: A web-based genomic annotation editing platform. Genome Biol. 2013;14:R93. doi: 10.1186/gb-2013-14-8-r93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lohse M., Drechsel O., Bock R. OrganellarGenomeDRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007;52:267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- 42.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kurtz S., Choudhuri J.V., Ohlebusch E., Schleiermacher C., Stoye J., Giegerich R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frazer K.A., Pachter L., Poliakov A., Rubin E.M., Dubchak I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharp P.M., Li W.H. The codon adaptation index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rozas J., Ferrer-Mata A., Sánchez-DelBarrio J.C., Guirao-Rico S., Librado P., Ramos-Onsins S.E., Sánchez-Gracia A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017;34:3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- 47.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swofford D.L. PAUP Ver.# 4b10. Sinauer Associates; Sunderland, MA, USA: 2002. [Google Scholar]
- 49.Nylander J.A.A. MrModeltest v2. Program Distributed by the Author. Evolutionary Biology Centre, Uppsala University; Uppsala, Sweden: 2004. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.