Abstract
Background
Pekin duck is an important animal model for its ability for fat synthesis and deposition. However, transcriptional dynamic regulation of adipose differentiation driven by complex signal cascades remains largely unexplored in this model. This study aimed to explore adipogenic transcriptional dynamics before (proliferation) and after (differentiation) initial preadipocyte differentiation in ducks.
Results
Exogenous oleic acid alone successfully induced duck subcutaneous preadipocyte differentiation. We explored 36 mRNA-seq libraries in order to study transcriptome dynamics during proliferation and differentiation processes at 6 time points. Using robust statistical analysis, we identified 845, 652, 359, 2401 and 1933 genes differentially expressed between -48 h and 0 h, 0 h and 12 h, 12 h and 24 h, 24 h and 48 h, 48 h and 72 h, respectively (FDR < 0.05, FC > 1.5). At the proliferation stage, proliferation related pathways and basic cellular and metabolic processes were inhibited, while regulatory factors that initiate differentiation enter the ready-to-activate state, which provides a precondition for initiating adipose differentiation. According to weighted gene co-expression network analysis, pathways positively related to adipogenic differentiation are significantly activated at the differentiation stage, while WNT, FOXO and other pathways that inhibit preadipocyte differentiation are negatively regulated. Moreover, we identified and classified more than 100 transcription factors that showed significant changes during differentiation, and found novel transcription factors that were not reported to be related to preadipoctye differentiation. Finally, we manually assembled a proposed regulation network model of subcutaneous preadipocyte differentiation base on the expression data, and suggested that E2F1 may serve as an important link between the processes of duck subcutaneous preadipocyte proliferation and differentiation.
Conclusions
For the first time we comprehensively analyzed the transcriptome dynamics of duck subcutaneous preadipocyte proliferation and differentiation. The current study provides a solid basis for understanding the synthesis and deposition of subcutaneous fat in ducks. Furthermore, the information generated will allow future investigations of specific genes involved in particular stages of duck adipogenesis.
Electronic supplementary material
The online version of this article (10.1186/s12864-019-6055-9) contains supplementary material, which is available to authorized users.
Keywords: Pekin duck, Subcutaneous fat, Preadipocyte differentiation, Transcriptome, Transcription factors, Regulation network model
Background
Adipose tissue has multiple roles in the regulation of insulin sensitivity [1], feed conversion ratio [2] and meat quality [3, 4] in animals. Certain amounts of intramuscular fat are required to meet consumer needs. Most investigations have focused on mammals, specifically the mouse and human. Adipocyte differentiation is a complex process regulated by multiple transcription factors (TFs), which affect expression level and activity of hundreds of proteins, resulting in dramatic changes in phenotypes [5–7]. Peroxisome Proliferator Activated Receptor Gamma (PPARγ) and CCAAT Enhancer Binding Protein Alpha (C/EBPα) are two master TFs supported by overwhelming evidence in vivo and in vitro [6],and many TFs and signaling pathways also participate in adipogenesis progression, playing either a positive or negative role [6, 8–10]. Gene networks, which integrate the mRNA and microRNA data, of brown adipose tissue have been created in the most recent study in the mouse model [11]. Avian species do not have brown adipose tissue (BAT), and lack Uncoupling Protein 1 (UCP1) [12]. Birds were thought to only share white adipose tissue [13, 14]. The subcutaneous white adipose tissue has important beneficial characteristics, including storage of lipid, secretion of adipokines, and anti-inflammatory roles [15]. Most studies on preadipocyte proliferation and adipogenic differentiation have been performed in vitro using human and murine cell lines [16, 17]. Recent studies showed that the process of preadipocyte differentiation in chicken has both similarities and differences with mammals [18]. Very limited studies based on candidate genes showed that several well-known TFs have similar expression patterns in chickens [19] and ducks [20] during adipocyte differentiation.
Duck is one of the most important meat sources in Asia, especially in China [21]. Roast Pekin duck requires a considerable skin-fat content, and Pekin duck has been used as a new model for studying behavior [22], meat quality [23], growth [4, 24] and fat synthesis and deposition [25]. Understanding the genetics of adipocyte differentiation is critical for controlling adipocyte deposition in ducks. However, no transcriptomic data have been reported during duck adipocyte differentiation. Elucidating the adipocyte differentiation process at the transcriptional level would be the foundation for further understanding of adipocyte biology in ducks.
This study aims to comprehensively analyze and compare gene expression profiles of 6 different time points during subcutaneous preadipocyte proliferation and differentiation. Our study explored 36 mRNA-seq libraries to obtain high quality differentially expressed gene sets across time points and constructed co-expression gene networks. We provide a reliable set of differentially expressed genes (DEGs) representing preadipocyte proliferation and differentiation. Furthermore, we have identified many known and novel TFs and signaling pathways associated with duck preadipocyte proliferation and differentiation. Finally, we provide a proposed regulation network model of subcutaneous preadipocyte differentiation.
Results
Duck subcutaneous preadipocyte differentiation
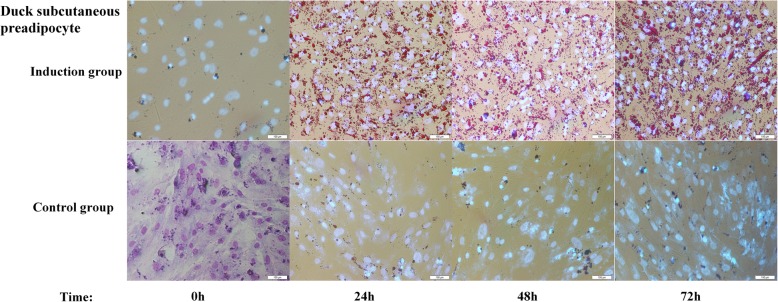
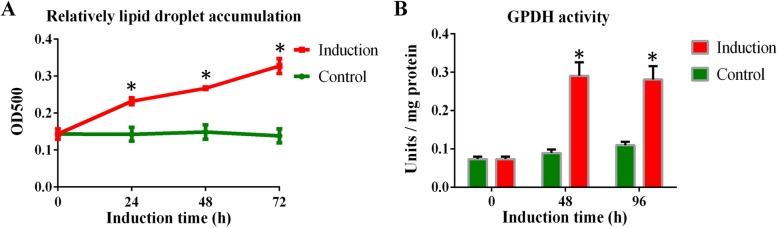
Cell morphological characteristics were measured at 0 h, 24 h, 48 h and 72 h during preadipocyte differentiation. Duck subcutaneous preadipocyte cultured in differentiation medium containing 300 μM oleic acid, showed a remarkable increase in lipid deposition compared with those cultured in the growth medium without oleic acid (Fig. 1). Lipid droplets in cells form as early as 24 h and showed a gradual increase in the percentage of cells with increased intracellular lipid content. Similarly, preadipocyte cultured in the induction medium showed significantly higher accumulation of lipid droplets compared with the control medium (Fig. 2a). GPDH enzyme activity increased significantly in comparison with control group at 48 h and 96 h of differentiation (Fig. 2b), with the addition of oleic acid. These results suggest that oleic acid alone can successfully induce differentiation in duck preadipocyte.
Fig. 1.
Morphological changes of duck subcutaneous preadipocyte cultured in differentiation medium (Induction group) or growth medium (Control group) at 200×. The picture in the lower left corner is the cell diagram after staining with Giemsa at 0 h. Bar, 100 μm
Fig. 2.
Intracellular lipid droplet accumulation and glycerol-3-phosphate dehydrogenase (GPDH) activity in duck preadipocytes cultured in differentiation medium (Induction) or growth medium (Control). (a) Relative quantification of lipid droplet accumulation within 72 h after induction. (b) GPDH activities were analyzed at 0 h, 48 h and 96 h post-induction. Bars indicate SD of the mean values (n = 3). *Statistically significant differences compared to the levels at 0 h (P < 0.05)
Transcriptome dynamics during preadipocyte proliferation and differentiation
Gene expression was studied over 120 h in a total of 36 samples (6 biological replicates at each point for each condition) using mRNA-seq. Each mRNA-seq library was sequenced to more than 20 million reads. Subsequently, clean reads were uniquely mapped to the duck reference genome (Anas_platyrhynchos.BGI_duck_1.0) with statistics for the mRNA-seq data referred to in Additional file 1: Table S1. All samples were hierarchically clustered based on Spearman’s correlation of gene expression. If the sample could not be clustered with other samples from the same time point, the sample was considered an outlier and was excluded from further analysis. To represent count data variability, standard error values were calculated per gene based on biological replicates FPKM (n = 5–6) at each time point, with the exception of the reference genes, where SE were calculated based on all samples and across all experimental groups (excluding the outlier sample -48 h-1 and 0 h-5, n = 34) (Additional file 2: Figure S1). The remaining 34 samples were used for subsequent analysis, and FPKM of obtained transcripts are provided in Additional file 3: Table S2.
Using a robust statistical analysis, we identified 845, 652, 359, 2401 and 1933 genes showing differential expression between -48 h and 0 h, 0 h and 12 h, 12 h and 24 h, 24 h and 48 h, 48 h and 72 h, respectively (FDR < 0.05, FC > 1.5) (Fig. 3). We found that gene expression patterns changed substantially between 24 h and 48 h in terms of the most DEGs at this stage relative to all other comparisons. A complete list of DEGs seen during the differentiation process is provided in Additional file 4: Table S3.
Fig. 3.
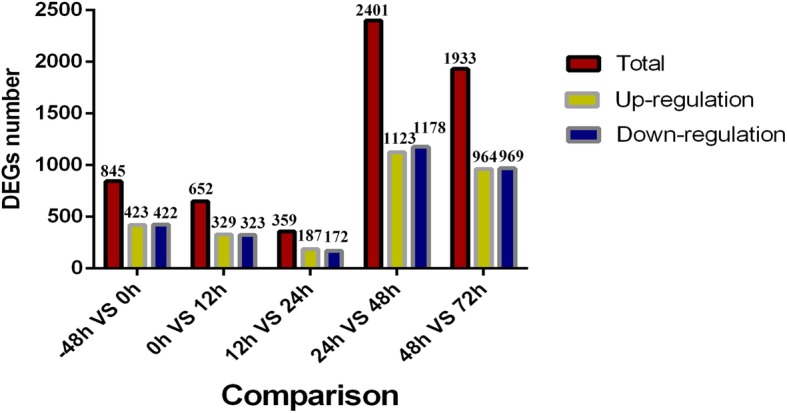
Histogram of the number of DEGs at different time points during preadipocyte differentiation stages
Functional analysis of differentially expressed genes during preadipocyte proliferation
Transcriptome expression analyses of duck preadipocyte proliferation stages showed that 423 genes were over-expressed at -48 h, and 422 genes were over-expressed at 0 h from 845 DEGs (− 48 h compared to 0 h). GO enrichment analysis showed genes over-expressed at -48 h compared to 0 h include those involved in mRNA catabolic process, mitotic cell cycle phase transition and cholesterol biosynthetic process. Genes over-expressed at 0 h compared to -48 h include those involved in actin cytoskeleton organization, calcium ion binding and collagen fibril organization. Surprisingly, 26 and 22 focal adhesion terms were significantly enriched at each time point (Table 1; Full list of enriched GO terms. Also refer to Additional file 5: Table S4). Moreover, KEGG pathway analysis showed genes over-expressed at -48 h compared to 0 h include those involved in ribosome, cell cycle and glycolysis. Genes over-expressed at 0 h compared to -48 h include those involved in regulation of ECM-receptor interaction, actin cytoskeleton and MAPK signaling pathway (Table 1; Full list of enriched pathways categories. Also refer to Additional file 6: Table S5).
Table 1.
Enriched GO terms and KEGG pathways of DEGs in the proliferation stage
| Proliferation stage | GO enrichment | Gene count |
Log (q-value) |
KEGG pathway | Gene count |
Log (q-value) |
|---|---|---|---|---|---|---|
| Up at -48 h | mRNA catabolic process | 44 | − 23.15 | Ribosome | 30 | −22.09 |
| Mitotic cell cycle phase transition | 45 | −16.92 | PI3K-Akt signaling pathway | 23 | −6.36 | |
| Ribosomal subunit | 34 | −22.87 | Cell cycle | 20 | −12.60 | |
| RNA splicing | 29 | −8.00 | Spliceosome | 14 | −6.20 | |
| Focal adhesion | 26 | −7.40 | DNA replication | 14 | −3.14 | |
| Cholesterol biosynthetic process | 16 | −11.48 | Glycolysis | 6 | −4.953 | |
| Up at 0 h | Actin cytoskeleton organization | 44 | −12.86 | Focal adhesion | 22 | −9.80 |
| Calcium ion binding | 42 | −10.43 | ECM-receptor interaction | 17 | −11.44 | |
| Cadherin binding | 29 | −11.76 | Regulation of actin cytoskeleton | 12 | −2.33 | |
| Focal adhesion | 25 | −5.88 | Calcium signaling pathway | 11 | −2.24 | |
| Collagen fibril organization | 13 | −9.30 | MAPK signaling pathway | 10 | −0.86 |
Co-expression network and module construction during preadipocyte differentiation
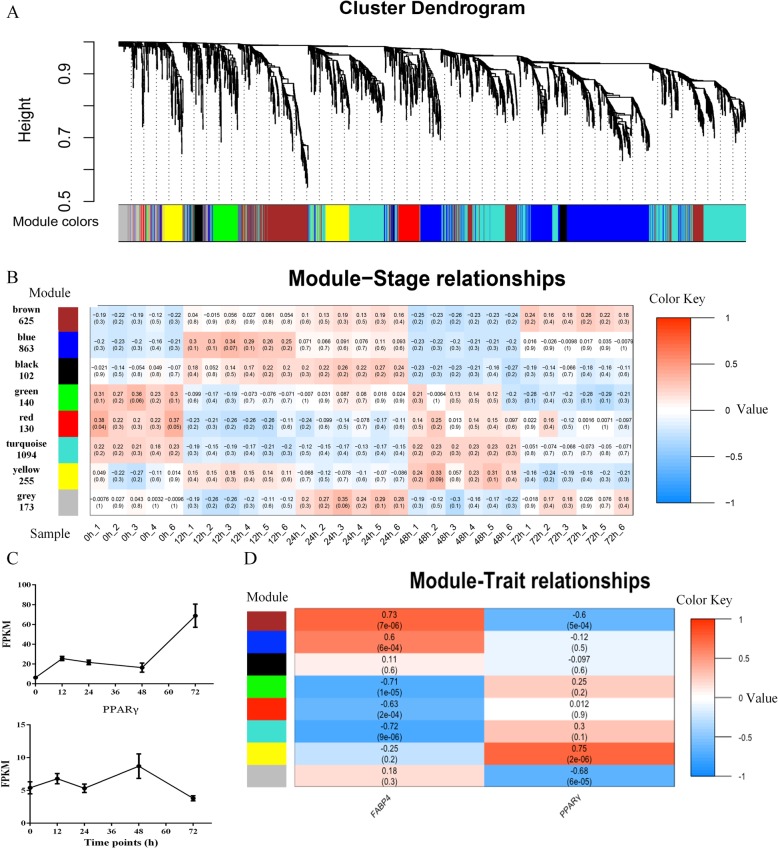
To gain insight into whole gene interaction networks of preadipocyte differentiation and lipid biosynthesis, we performed a Weighted Correlation Network Analysis (WGCNA) to identify groups of co-expressed genes using non-redundant DEGs (n = 3382) between any two adjacent time points during the differentiation stages. Modules associated with specific differentiation stage were identified based on the correlation between module eigengene and samples. As shown in the dendrogram (Fig. 4a), 8 consensus modules were identified in the analysis, labeled by different colors, with each containing at least 100 genes (Fig. 4b; Additional file 7: Table S6). MEblue, MEyellow and MEblack modules were highly and specifically accumulated at12h, 24 h and 48 h after differentiation, respectively (Figure 4b; Additional file 8: Figure S2), which indicated that these groups of genes might be responsible for positive regulation during differentiation. In contrast, MEturquoise, MEred and MEgreen modules were highly and specifically accumulated at 0 h and significantly decreased after differentiation (Fig. 4b), indicating that these groups of genes might be involved in maintaining pluripotency of preadipocyte or negative regulation of preadipocyte differentiation. Additionally, the MEbrown module was highly and specifically accumulated at 72 h after differentiation, which indicated that this group of genes might be responsible for lipid deposition of adipose cells at the end of differentiation. A further module-trait relationship analysis, using the expression level of PPARγ and Fatty acid binding protein 4 (FABP4) as the trait data, revealed that expression patterns of FABP4 were not only highly correlated with the MEbrown module, but also positively correlated with MEblue and MEyellow modules. PPARγ was only correlated with the MEblack module, which might relate to the slow rise of its expression after the beginning of differentiation (Fig. 4c, d).
Fig. 4.
Weighted gene co-expression network analyses (WGCNA) of DEGs identified from differentiation stages. (a) Hierarchical cluster tree showing 8 modules of co-expressed genes. Each of the 3382 DEGs are represented by a tree leaf and each of the modules by a major tree branch. The lower panel shows modules in designated colors. (b) Module–sample correlations and corresponding p-values (in parentheses). The left panel shows the 8 modules and the number of member genes. The color scale on the right shows module–sample correlations from − 1 (blue) to 1 (red). The labels on the bottom panel represent samples at different points in time. (c) mRNA-seq expression patterns of PPARγ and FABP4. (d) Module–trait correlations and corresponding p-values (in parentheses). The left panel shows the 8 modules and the colour scale on the right shows module–trait correlations from − 1 (blue) to 1 (red). The left panel “PPARγ” (bottom) represents the expression changes of PPARγ, which is the key transcription factor activating adipogenic differentiation. The right panel “FABP4” (bottom) represents the expression changes of FABP4, which is important for lipid hydrolysis and transportation of intracellular free fatty acid
Functional analysis of differentially expressed genes during preadipocyte differentiation
According to the above analysis, the expression level of genes belong to the MEblue, MEyellow and MEblack modules increased significantly after induction, but decreased rapidly in late differentiation, so they were considered as early positive response genes of differentiation. The genes in the MEturquosie, MEgreen and MEred modules were considered as early negative response genes, while the genes in the MEbrown module designated late response genes.
In the early positive response genes, most of the enriched GO terms related to the regulation of cellular protein localization, ribosome biogenesis and cellular response to lipid (Table 2; Additional file 5: Table S4). To obtain more detailed information, a pathway analysis was carried out using Metascape. Some primary adipose differentiation-related pathways, including those involved in cell cycle, MAPK, and PI3K-Akt signaling pathway were observed in the early positive response genes (Table 2; Additional file 6: Table S5). The GO enrichment analyses of the early negative response genes revealed distinct enrichment patterns. The three primary enriched terms were the actin filament-based process, response to growth factor, and regulation of system processes (Table 2; Additional file 5: Table S4). Pathway analysis of early negative response genes showed that apoptosis, WNT, FOXO and HIF signaling pathways (which inhibit adipose differentiation) were significantly enriched (Table 2; Additional file 6: Table S5). The functional annotations of the late response genes were linked to cell division, lipid biosynthetic process, steroid biosynthetic process, and fatty acid metabolic processes (Table 2; Additional file 5: Table S4). For pathway analysis, cell cycle, steroid biosynthesis and PPARγ signaling pathway were also significantly enriched (Table 2; Additional file 6: Table S5).
Table 2.
Enriched GO terms and KEGG pathways of different response gene sets at differentiation stages
| Differentiation stage | GO enrichment (biological process) | Gene count |
Log (q-value) |
KEGG pathway | Gene count |
Log (q-value) |
|---|---|---|---|---|---|---|
| Early positive response gene set | Muscle structure development | 59 | −6.72 | PI3K-Akt signaling pathway | 31 | −2.40 |
| ncRNA metabolic process | 52 | −6.68 | Regulation of actin cytoskeleton | 19 | −1.39 | |
| Cellular protein localization | 46 | −5.57 | MAPK signaling pathway | 17 | −0.43 | |
| Ribosome biogenesis | 34 | −6.12 | DNA replication | 14 | −4.43 | |
| Early negative response gene set | Actin filament-based process | 63 | −6.70 | WNT signaling pathway | 27 | −7.21 |
| Response to growth factor | 57 | −5.08 | Apoptosis | 18 | −2.86 | |
| Muscle structure development | 52 | −4.71 | FOXO signaling pathway | 17 | −2.74 | |
| Regulation of system process | 50 | −5.51 | HIF-1 signaling pathway | 14 | −2.47 | |
| Late response gene set | Cell division | 65 | −28.52 | Cell cycle | 18 | −7.29 |
| Lipid biosynthetic process | 31 | −3.473 | Steroid biosynthesis | 8 | −6.08 | |
| Steroid biosynthetic process | 21 | −8.212 | p53 signaling pathway | 8 | −2.12 | |
| Fatty acid metabolic process | 14 | −0.713 | PPARγ signaling pathway | 7 | −1.38 |
Expression pattern analysis of differentially expressed TFs during differentiation
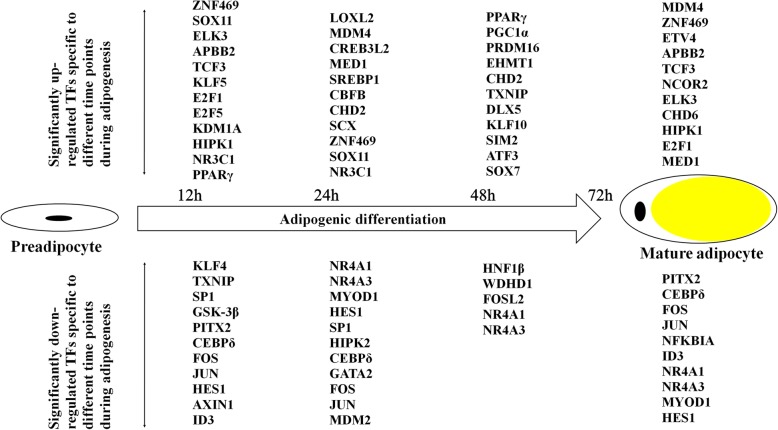
Activation and repression of defined transcription factors are essential for the commitment of progenitors to a specific differentiation lineage, setting the stage for a gene expression pattern characteristic of each mature cell type. Adipocyte differentiation is regulated by multiple TFs, and cooperative interactions among these transcription factors drive the expression of downstream target genes that are necessary for the generation and maintenance of adipocyte characteristics such as lipid accumulation and insulin sensitivity. A total of 164 differentially expressed TFs were obtained by alignment of DEGs with ITFP and TRANSFAC databases (Additional file 9: Table S7). A map of TF signature patterns at different time points during the differentiation process is shown in Fig. 5. Some TFs, such as E2F Transcription Factor 1 (E2F1), E2F Transcription Factor 5 (E2F5), Nuclear Receptor Subfamily 3 Group C Member 1 (NR3C1) and Krüppel Like Factor 5 (KLF5), which directly induce PPARγ expression and initial preadipocyte differentiation, were immediately up-regulated in the early differentiation stage (Fig. 5; Additional file 10: Figure S3). Similarly, some TFs involved in the inhibition of adipocyte differentiation are also enriched in the early negative response gene set, including GATA Binding Protein 2 (GATA2), GATA Binding Protein 3 (GATA3), HES Family BHLH Transcription Factor 1 (HES1) and Myogenic Differentiation 1 (MYOD1) (Fig. 5; Additional file 10: Figure S3). A considerable number of TFs that were not reported to be involved in regulation networks of adipocyte differentiation included Zinc Finger Protein 469 (ZNF469), SRY-Box 11 (SOX11) and Transcription Factor 3 (TCF3) (Fig. 5; Additional file 10: Figure S3).
Fig. 5.
TFs expression at different time points during preadipocyte differentiation stages. Significantly up-regulated TFs expressed at the different time points (12 h, 24 h, 48 h and 72 h) are shown in the top half of the panel, while significantly down-regulated transcription factors at the same time points are shown in the lower half of the panel
Validation of DEGs by RT-qPCR
Three samples from 0 h, 12 h, 24 h and 48 h were randomly selected for RT-qPCR to validate some key factors involved in adipose differentiation. These included E2F1, E2F5, PPARγ, GSK3β, CCND1, AXIN1, SOX11 and ZNF469. The same cell samples used in mRNA-seq were used for RT-qPCR validation. The results showed that the expression patterns of these genes were highly consistent with the mRNA-seq results (Additional files 11, 12: Table S8, Figure S4).
Discussion
This study is the first report to investigate global transcriptome changes during preadipocyte differentiation in ducks. Here, we not only obtained a relatively complete dynamic transcriptome map of subcutaneous fat differentiation in ducks, but also found many known or unknown TFs and signaling pathways associated with preadipocyte proliferation and differentiation, which is beneficial for determining optimal breeding for subcutaneous fat deposition in Pekin duck.
Regulation events of preadipocyte proliferation
A total of 845 DEGs were obtained between -48 h and 0 h by pair wise comparison. The premise of inducing preadipocyte differentiation is that the proliferating preadipocyte become growth-arrested by contact inhibition. This process is accompanied by many regulatory events, ultimately providing a special microenvironment for initiating differentiation. Thus, GO enrichment and pathway analysis were performed to explore the functions of DEGs during proliferation. As expected, consistent with growth arrested, the cell cycle and related pathways were significantly down-regulated; for example cell cycle, DNA replication, glycolysis and PI3K-Akt signaling pathway during different proliferation stages. Meanwhile, proliferation associated with basic cellular and metabolic processes (transcription, ribosome biogenesis, translation and protein folding) were also down-regulated. The PI3K-Akt signaling pathway is a major mediator of cellular proliferation, survival and differentiation [26]. Phosphatase And Tensin Homolog (PTEN), a primary and classical inhibitor of the PI3K-Akt pathway [27], was significantly up-regulated at 0 h (Fig. 6a). Some previous tumor research has reported that the PI3K-Akt pathway, glycolysis and DNA Methyltransferase 1 (DNMT1) cooperate to activate cell proliferation and cross-regulate each other in a positive feedback loop to provide the sufficient amount of ATP and metabolic intermediates required for rapid proliferation [28, 29]. We further analyzed changes in the transcriptional levels of DNMT1 and several genes associated with glycolysis. Interesting, the expression of these genes showed a very high agreement with the PI3K pathway, which was significantly down-regulated at 0 h (Fig. 6B). In addition, DNMT1 is a major DNA methyltransferase responsible for maintaining self-renewal and the undifferentiated state in mesenchymal stem cells [30], whereas its knockout can accelerate preadipocyte differentiation [31], and the expression patterns of PPARγ and DNMT1 showed opposite trends in some cell lines [32].
Fig. 6.
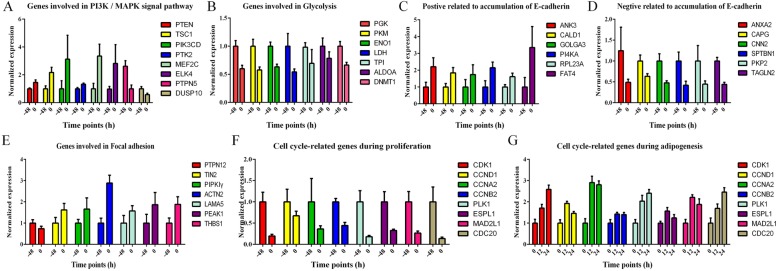
mRNA-seq expression patterns of some key regulatory or functional genes responsible for regulating pathways during proliferation or differentiation stages. (a) Genes involved in PI3K / MAPK signal pathway. (b) Genes involved in glycolysis. (c) Positive related to accumulation of E-cadherin. (d) Negative related to accumulation of E-cadherin. (e) Genes involved in focal adhesion. (f) Cell cycle related genes during proliferation stage. (g) Cell cycle related genes during differentiation stages
On the other hand, cadherin binding, calcium ion binding, focal adhesion and MAPK signaling pathway were up-regulated at 0 h. Cadherin is a calcium-dependent cell adhesion molecule that is important for the formation of adherens junctions to bind cells to each other, and the loss of its function can directly promote cell proliferation and tumor progression [33]. Moreover, mitotic cell cycle phase transition is inhibited by over-expression of cadherin in cells, which is down-regulated at 0 h [34, 35] (Fig. 6c, d). 26 and 22 focal adhesion terms were also significantly enriched at -48 h and 0 h respectively. Focal adhesion is the primary site of cell adhesion to the substrate, which links the extracellular matrix, via membrane-bound receptors, to the cell’s cytoskeleton, and plays a critical role in many fundamental processes such as embryonic morphogenesis, angiogenesis and inflammation [36, 37]. We observed that the expression of Protein Tyrosine Phosphatase Non-Receptor Type 12 (PTPN12), an important phosphatase which enables increased focal adhesions as well as inhibits tumor growth [38], was significantly down-regulated at 0 h. Similarly, both Talin2 (TIN2) and phosphatidylinositol phosphate kinase type I Gamma (PIPKIγ) play a role in focal adhesion formation [39], and their expression increased at 0 h (Fig. 6e). According to the above analysis, both of the cadherin binding terms and the focal adhesion terms play a negative role in cell cycle transition and mitosis of duck preadipocyte (Fig. 7).
Fig. 7.
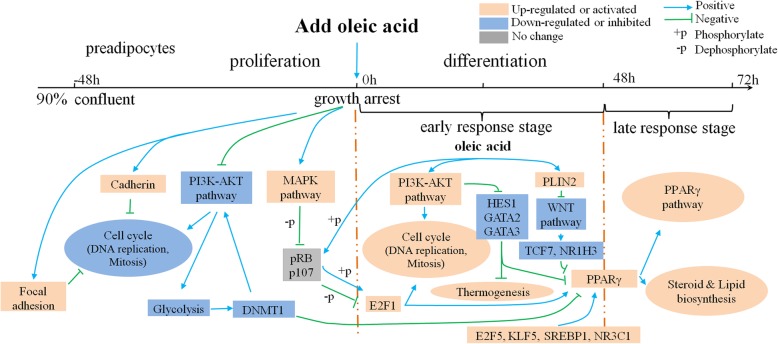
Regulation network models in duck subcutaneous preadipocyte proliferation and differentiation stage. The pink or blue boxes represent the genes or pathways, which were identified by our study, while the gray boxes represent genes from published literature
The MAPK signaling pathway relies on a series of phosphatase cascades to sustain activation of p38α, ultimately inhibiting cell proliferation [40, 41]. Dual Specificity MAP Kinase Phosphatase 10 (DUSP10) or Protein Tyrosine Phosphatase, Non-Receptor Type 5 (PTPN5) which act as upstream inhibitors and dephosphorylase p38α, were significantly decreased, whereas Myocyte Enhancer Factor 2C (MEF2C) and ETS Domain Containing Protein 4 (ELK4), which act downstream of p38α [40, 42, 43], were increased at 0 h (Fig. 6a). In fact, the activated p38 will further dephosphorylate the Retinoblastoma (pRB) and Cyclin Dependent Kinase Inhibitor 1B (p27Kip1) in confluent cultures [41]. Furthermore, the dephosphorylated pRB binds to the E2F binding site on the PPARγ promoter, preventing E2F1 triggering the expression of PPARγ during the early stages of adipogenesis [44, 45]. All these results suggested that there may also be such a positive feedback loop in duck subcutaneous preadipocyte that is directly or indirectly involved in the regulation of cell proliferation and differentiation (Fig. 7).
Regulation of preadipocyte differentiation
Differentiation itself is characterized by changes in cell morphology and regulated by complex molecular events that are initiated by adipogenic hormonal stimulus. Based on co-expression network analysis, we divided the DEGs obtained at different stages of differentiation into three response gene sets: early positive response, early negative response and late response gene sets.
Regulation of actin cytoskeleton, DNA replication, PI3K-Akt signaling pathway and other functional pathways were significantly enriched in the early positive response gene set, with most of them having been reported to be involved in the early stage of the differentiation [46–48]. DNA replication is one of the key events taking place in early adipogenesis, and inhibition of DNA synthesis at this stage blocks differentiation [49]. A hall mark of differentiation is a pronounced change in cell shape, which is determined by dynamics of the actin cytoskeleton [50]. The consequent rapid increase in actin leads to interaction of actin with other adipogenic inhibitors, and allows expression of PPARγ followed by adipogenic differentiation [51]. Interesting, many cell cycle-related genes and the PI3K-Akt signaling pathway, which were down-regulated during the proliferation stage, increased significantly during the differentiation stage (Fig. 6F, G). This is not surprising as those contact inhibited preadipocyte re-enter the cell cycle after hormonal induction, arrest proliferation and, again, finally undergo terminal differentiation [6]. More importantly, the inducing agent, oleic acid, has been reported to stimulate the proliferation of various cells by activating the PI3K-Akt pathway [52, 53]. Phosphorylated pRB combined with highly expressed CCND1 releases activated E2F1, ultimately initiating PPARγ transcription [44]. Moreover, E2F1 global knockout mice have a limited ability to accumulate adipose tissue in response to high-fat feeding [44]. Corroborating this, some mice enable developing adipose depot act on E2F1 to stimulate PPARγ without the need for other early adipogenesis transcription factors, such as CCAAT Enhancer Binding protein β or δ (C/EBPβ, C/EBPδ) [8, 54]. These reports and our transcriptomic profiling together suggests that, oleic acid- PI3K-Akt pathway- pRB- E2F1 -mediated activation cascade mechanism could facilitate the expression of PPARγ during the early stage of differentiation (Fig. 7).
As expected, WNT, HIF, FOXO and other genes known to be involved in the inhibition of the preadipocyte differentiation signaling pathway, were significantly enriched in the early negative response gene set. The WNT signaling pathway maintains preadipocyte in an undifferentiated state through inhibition of C/EBPα and PPARγ. The canonical WNT signaling pathway inhibits the kinase activity of complexes containing glycogen synthase kinase 3 β (GSK3β), Axis Inhibitor 1 (Axin1), β-catenin and other proteins [55]. This complex targets β-catenin for rapid degradation through phosphorylation [56]. Thus, once hypophosphorylated due to WNT signaling, β-catenin is stabilized and translocates to the nucleus where it binds the TCF/LEF family of transcription factors to negatively regulate PPARγ transcription [56, 57]. Consistent with this, expression of PLIN2, GSK-3β and AXIN1 increased rapidly at 12 h, while expression of TCF/LEF family transcription factor, T Cell Specific Transcription Factor 7 (TCF7) and Nuclear Receptor Subfamily 1 Group H Member 3 (NR1H3) decreased significantly. On the other hand, oleic acid has been reported to stimulate the expression of Perilipin2 (PLIN2) in 3 T3-L1 cells [58]. Meanwhile, PLIN2 has been shown to activate AXIN1 and GSK3β, thereby inhibiting the WNT signaling pathway [59]. Consistent with these reports, our data predict that an oleic acid- PLIN2- WNT pathway- β-catenin- TCF7 mediated negative regulatory cascade mechanism could further enhance the expression of PPARγ during the early stage of differentiation (Fig. 7).
In addition, we found more differentially expressed TFs at the early response stage (0 h–48 h) compared to late response stage (48 h–72 h). NR3C1 [60], KLF5 [61] and Sterol Regulatory Element Binding Transcription Factor 1 (SREBP1) [62], which directly or indirectly facilitate the expression of PPARγ, were significantly up-regulated within 24 h (Additional file 10: Figure S3). At the same time, some TFs known to inhibit adipocyte differentiation also decreased significantly, including GATA2 [63], GATA3 [63, 64], HES1 [65] and MYOD1 [66] (Additional file 10: Figure S3), with most of them having been reported to be inhibited by the activated PI3K-AKT signaling pathway. Although the brown adipose differentiation pathway associated with thermogenicity has been lost in the avian lineage [67], we also found several brown adipose determination TFs increased significantly at the early response stage. These include PPARG Coactivator 1 Alpha (PGC1α) [68], Euchromatic Histone Lysine Methyltransferase 1 (EHMT1) [69] and PR/SET Domain 16 (PRDM16) [70] (Additional file 10: Figure S3). In fact, the formation of brown adipose tissue shares many common differentiation regulatory nodes with white adipose tissue [71], so their rise may be related to the decline of differentiation inhibitors, such as GATA2, GATA3 and HES1 (Fig. 7). However the role of these genes in duck subcutaneous preadipocyte differentiation requires further investigation.
The lipid biosynthetic process, steroid biosynthetic process, PPARγ and p53 signaling pathways implicated in the regulation of lipid and lipoprotein metabolism [72, 73], were enriched in the late response stage. At the same time, some of the markers related to fat metabolism and nutrient transport, which were also highly expressed in the late stage of preadipocyte differentiation, also increased significantly, with the expression of particular genes even increasing by more than 50 fold (i.e., FABP4, PLIN2). This is consistent with previous studies which show that the preadipocyte have been transformed into adipocyte after 72 h of induction [19].
Finally, we also identified TFs (i.e., ZNF469 and SOX11) that have not been reported previously to be involved in the regulation of adipose differentiation, which were up-regulated during the differentiation stage. ZNF469 has been proposed as a candidate gene for keratoconus, and its mutation is associated with brittle cornea syndrome [74]. Previous study on the evolution of adipose tissue has shown that the ZNF469 gene in Pekin duck is highly variable compared with its wild ancestor, which may be one of the factors causing the excessive deposition of adipose tissue in Pekin duck [75]. SOX11 was reported to inhibit osteogenic differentiation of preadipocyte, but the relationship with adipocyte differentiation has not been reported [76]. In fact, preadipocyte are delicately balanced for their differentiation direction - numerous in vitro investigations have demonstrated that adipose-induction factors inhibit osteogenesis, and conversely, bone-induction factors hinder adipogenesis [66]. Taken together, we speculate that the ZNF469 and SOX11 have a positive effect on Pekin duck subcutaneous preadipocyte differentiation.
Genes with high expression across all stages
Dynamic changes in gene expression reflect intrinsic mechanisms of an organism’s response to developmental and environmental signals. Although genes with high and constant expression levels in all stages may also exhibit the characteristics of the cell itself. In the present study, 1000 genes with FPKM over 30 (Additional file 13: Table S9) were selected to carry out functional enrichment analysis. As expected, transcription, ribosome biogenesis, translation and protein folding were identified, indicating active growth and metabolism in adipose cells and tissues [77]. Extracellular matrix (ECM)-receptor interaction was also significantly enriched in these genes (Additional file 14: Table S10). The ECM of adipose tissues undergoes constant remodeling to allow adipocytes and their precursor cells (preadipocytes) to change cell shape and function in adaptation to nutritional cues by interacting with the receptor on the cell surface [78]. Moreover, as belonging to the ‘receptor’ term integrins subunit beta 1 (ITGB1) and CD44 were positively correlated with insulin resistance and glycemic control in human subjects [79, 80]. In line with this, HFD-fed CD44 knockout mice remained considerably more insulin-sensitive and glucose- tolerant than HFD-fed wild-type control mice and exhibited lower blood insulin levels [81]. Furthermore, domestic poultry adipose tissue is considered to be fairly insensitive to insulin (insulin resistance) with lipolysis being under glucagon control, due to intensive genetic selection for rapid growth [82]. In this sense, these highly expressed ECM receptors may contribute to insulin resistance in poultry.
Conclusions
This study is the first report exploring transcriptome changes during the differentiation of preadipocyte into adipocyte in ducks. In total, 845 and 3382 DEGs were identified in the preadipocyte proliferation and differentiation stages. We not only found many known and novel TFs and signaling pathways associated with duck preadipocyte proliferation and differentiation, but also provide a proposed regulation network model of subcutaneous preadipocyte differentiation. Our study provides a solid transcriptional analysis with which to facilitate functional studies on preadipocyte differentiation in ducks.
Methods
Duck subcutaneous preadipocytes isolation
Pekin ducks were provided by Beijing Golden Star Ltd. All ducks in this study were given continuous access to a standard commercial feed ration and water as described in our previous study [4, 21]. In order to reduce the suffering of animals, three ducks were moved to the laboratory which provided isolation, thereby minimizing noise and distractions. Ducks were sacrificed under deep anesthesia with sodium pentobarbital (Sigma). The subcutaneous adipose tissues were collected for the primary culture of subcutaneous preadipocytes. The experimental procedure was in accordance with the guidelines of the China agricultural University Animal Care Committee. Subcutaneous preadipocytes from three ducks were prepared by the method as described before [19], with some modifications. Briefly, subcutaneous adipose tissue was collected under sterile conditions from a 16-day-old female duck and washed with PBS. The clean adipose tissue was minced into fine sections and digested with 15 mL of digestion Solution [DMEM/F12 (Dulbecco’s modified Eagle’s medium/Ham’s nutrient mixture F-12), 100 mM HEPES, 4% BSA, 2 mg/mL collagenase I (Invitrogen), pH 7. 4] for 65 min at 37 °C in a water bath shaker. After incubation, growth medium (DMEM/F12, 10% FBS, 100 U/mL penicillin and streptomycin) was added to stop digestion. The mixture was filtered through nylon screens with 70 μm mesh openings to remove undigested tissue and large cell aggregates. The filtered suspensions were centrifuged at 300×g for 10 min to separate floating adipocytes from preadipocytes. The harvested preadipocytes were then re-suspended with 10 mL of Blood Cell Lysis Buffer (Invitrogen), and incubated at room temperature for 10 min. Finally, the obtained preadipocytes were seeded into T25 flasks at a suitable density and cultured in a humidified atmosphere of 95% air and 5% CO2 at 37 °C until 90% confluency. The preadipocytes were then serially subcultured at a 1:2 split ratio until differentiation experiments began.
Induction of duck preadipocytes differentiation
Prepared duck preadipocytes were seeded into 6-well plates at a density of 1 × 105 cells per well and cultured with growth medium until achieving 90% confluence. After 2 days, the growth medium was removed and replaced with differentiation medium (growth medium supplemented with 300 μM oleic acid) and medium was changed every 2 days until day 3 of differentiation, which was similar to the procedure used with chicken preadipocytes [83]. The design and sampling strategy are described in Additional file 15: Figure S5. Cells were collected for mRNA-seq at -48 h, 0 h, 12 h, 24 h, 48 h and 72 h. Each interval included six biological replicates (n = 6), with 36 samples collected for mRNA-seq in total.
Oil red O staining and measurement of lipid droplet accumulation
Lipid droplets were stained with oil red O (Sigma) according to Shang Z et al. [83]. Briefly, the cells were washed three times with PBS and fixed with 10% (v/v) paraformaldehyde for 30 min at room temperature. Then cells were washed with PBS and stained with 1% Oil Red O working solution [Oil Red O dye in 60% (v/v) isopropyl alcohol] for 40 min. The cells were counterstained with Hoechst 33342 after removing the residual Oil Red O and repeatedly washed using distilled water. Staining work at each time point included three biological replicates (n = 3). Finally, observation and photographing of cell phenotypes were conducted under an inverted fluorescent microscope (Leica) at 200X magnification.
Lipid droplet accumulation was measured by oil red O extraction assay. First, oil red O stained cells were prepared by the above method. Then, oil red O was extracted by adding 1 mL of 100% (v/v) isopropyl alcohol, and measured at 500 nm using an ultraviolet spectrophotometer (Pharmacia). Adjacent plate wells with identical treatment were trypsinized, diluted and counted with a hemocytometer to normalize the extraction results [83, 84].
Glycerol-3-phosphate dehydrogenase (GPDH) assay
GPDH is a rate-limiting enzyme for fatty acyl-CoA biosynthesis and its enzyme activity will rise significantly in the late stages of differentiation. The differentiated preadipocytes were collected at 0 h, 48 h, and 96 h. GPDH assay was conducted using a GPDH Activity Colorimetric assay kit (Sigma). Each time point included three biological replicates for GPDH analysis (n = 3). Protein concentrations of cell culture homogenates were determined by BCA protein assay kit (Sigma) using bovine serum albumin as the standard. GPDH activity was reported as nmol/min/mL [19].
RNA extraction and cDNA library preparation
The different stages of clean preadipocytes were homogenized in TRIzol (Invitrogen) and processed following the manufacturer’s protocol. The quantity and quality of RNA were assessed via Nanodrop. All RNA samples had an RNA integrity number value > 8.0, and an optical density 260:280 ratio > 1.9. Approximately 5 μg of total RNA was then used for mRNA-seq using the Illumina sequencing platform. Briefly, the mRNA was enriched using magnetic beads with oligo (dT) primer, and then randomly fragmented using Fragmentation buffer. The first-strand and the second-strand cDNA were synthesized using First Strand Enzyme Mix and Second Strand/End Repair Enzyme Mix (Vazyme Biotech). The products were purified by AMPure XP beads (Beckman Coulter) and the end of the double strand was then repaired and A-tailed. Suitably sized fragments were selected using AMPure XP beads (Beckman Coulter) to construct the cDNA library by PCR. Following construction, double-stranded cDNA libraries were sequenced on an Illumina HiSeq X-10 with PE150 mode at the Novogene Inc.
Assembly-guided transcript discovery
The raw reads with adaptors removed were filtered according to the following criteria: 1) reads with unknown nucleotides (N) larger than 5%; 2) reads containing more than 30% bases with Q-value < 20. The clean reads were used for further analysis.
The mRNA-seq guide-assembly was performed using the HISAT2 and StringTie pipeline [85]. The paired-end reads of adipose samples were aligned to the duck reference genome individually using the hierarchical indexing for spliced alignment of transcripts program HISAT2 (−v2.0.5). For this purpose, we built an index file for the duck reference genome (Anas_platyrhynchos.BGI_duck_1.0) using HISAT2-build. StringTie was used to assemble each sample based on alignment file for each sample and merged all predicted transcripts into a unified transcript model. We compared reference-guided transcripts with the known annotations to assess the quality of transcript predictions.
The novel transcripts from assembled transcripts were extracted using gffread and annotated with known protein sequences database from Uniprot (www.uniprot.org) using the Blastx algorithm with a cutoff e-value of 10− 5. Ultimately, we merged all known transcripts and new annotated transcripts into a non-redundant gene set for quantification and differential expression analysis.
Differentially expressed gene identification
Transcripts were quantified via the Salmon (−v0.8.2) software using the transcriptome-based quasi-mapping mode, and clean reads of samples were mapped to the gene set individually. Once expression level for each transcript in each sample (− 48 h, 0 h, 12 h, 24 h, 48 h and 72 h) was quantified, the data were summarized to a gene-level. First, we calculated sample-to-sample distances to assess the data quality using DESeq2 (version1.16.1) [86]. Differential expression analysis at gene-level between 6 time points of adipose samples was performed using DESeq2. Significance for differential expression was accepted at the Benjamini-Hochberg adjusted P (FDR) < 0.05 level, and fold change (FC) > 1.5. Finally, we used Metascape (http://metascape.org) to get the enriched GO terms and KEGG pathways of differentially expressed genes (DEGs). In order to avoid poorly expressed genes, genes with FPKM < 0.5 were filtered before conducting DEG analysis.
Construction and visualization of co-expression network
The weighted correlation network analysis (WGCNA) relies on the hypothesis that strongly correlated expression levels of a group of genes, referred to as “modules”, may work cooperatively in related pathways, contributing together to the phenotype [87]. We found clusters (modules) of highly correlated DEGs, for summarizing such clusters using the module eigengene or an intramodular hub gene, for relating modules to one another (using eigengene network methodology), and for calculating module membership measures using WGCNA. In order to analyze the influence of power value on the scale independence and mean connectivity, we used the function connectivity from package WGCNA, with the “randomly selected genes” parameter set at 4000, other parameters set as default, and the power parameter pre-calculated by the pickSoft Threshold function of WGCNA. We next summarized the expression values using the function collapse Rows implemented in the R package WGCNA. The interactions (correlations) of each module were analyzed and visualized by heat map. Further, the co-expression network of highly coordinated genes among most of the modules was visualized and analyzed by Cytoscape (version 2.8.3).
GO and KEGG pathway enrichment analyses
To investigate genes from one gene ontology GO term (http://metascape.org/gp/index.html), a hypergeometric p-value was calculated and adjusted as a q-value, where the background was set to be genes in the whole genome. GO terms with q < 0.05 were considered significantly enriched, and GO enrichment analysis elucidated the biological functions of the DEGs. The log10 value (p-value) denotes enrichment scores that represent the significance of GO term enrichment among DEGs. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was also performed to predict the molecular interactions and reaction networks associated with DEGs. Using the same method as that used for GO enrichment analysis, significantly enriched KEGG pathways were identified.
Identification of transcription factors from DEGs
ITFP (http://itfp.biosino.org/itfp) and TRANSFAC (http://www.gene-re gulation.com/pub/databases.html) provide data on eukaryotic transcription factors. Data relating to human transcription factors, and their binding site motifs were downloaded from ITFP and TRANSFAC. Based on the downloaded data, DEGs were used for screening transcription factors. Furthermore, we assembled a time-specific map of the expression of transcription factors after induction based on DEGs obtained during the differentiation stage. The selected and displayed differentially expressed TFs from different time points were identified using a t-test at a fold change of ≥1.5 and probability (P < 0.05) compared with 0 h.
Validation of mRNA-seq data using quantitative real-time PCR
We randomly selected three samples from 0 h, 12 h, 24 h and 48 h, which were consistent with the library preparation sample, for RT-qPCR and calculated their correlation with the corresponding FPKM in mRNA-seq data. First strand cDNA was synthesized using the PrimeScript RT Master Mix kit according to the supplier’s protocol (Takara Bio Inc). Pairs of primers for each gene were designed from the CDs sequence of the target gene from the National Centre for Biotechnology Information (NCBI) (Additional file 11: Table S8). Quantitative real-time (RT-qPCR) was performed in duplicate reactions including SYBR Premix ExTaq II (Takara Bio Inc), specific forward and reverse primer, diluted cDNA and RNase free water. Quantification of selected gene expression was performed using the comparative threshold cycle (2-ΔΔCT) method by normalizing the expression of the target genes to a reference gene (GAPDH). The RT-qPCR results for all genes were statistically tested using the Student’s t-test.
Generation of gene network
We conducted further analysis of the DEGs obtained in our study and manually assembled a proposed regulation network model of preadipocyte proliferation and differentiation based on published literature (Fig. 7). Previous study reported that Cadherin, focal adhesion, DNA methylation and PI3K-AKT signaling pathway are mainly involved in regulation of cell cycle, mitosis and cancer. Similarly, several studies have revealed the importance of some TFs and pathways in regulating adipogenesis both in vitro and in vivo. These include PPARγ [6], E2F1 [44], GATA2 [63], PI3K-AKT [26, 53] and WNT signaling pathway [57]. Also, in our main network analysis, these TFs and pathways, were affected by growth arrest and oleic acid, consistent with their role in proliferation and differentiation, and were therefore incorporated from regulation network analysis of duck subcutaneous preadipocyte differentiation.
Additional files
Table S1. Statistics relating to the mRNA-seq data. (XLSX 12 kb)
Figure S1. Heatmap of the differentiation of biological replicates of duck subcutaneous preadipocyte. The colors ranging from white to blue represent Pearson correlation coefficients ranging from 0 to 1, indicating low to high correlations, respectively. (PDF 498 kb)
Table S2. List of genes expressed during the entire stage. (XLSX 8936 kb)
Table S3. List of all DEGs during complete duck subcutaneous preadipocyte differentiation. (XLSX 929 kb)
Table S4. GO term analysis of DEGs at different stages. (XLSX 248 kb)
Table S5. KEGG pathway analysis of DEGs at different stages. (XLSX 51 kb)
Table S6. Genes involved in eight modules. (XLSX 147 kb)
Figure S2. Classification of DEGs throughout the entire differentiation process and visualization of gene expression levels of significant modules. (PDF 1241 kb)
Table S7. List of differentially expressed TFs obtained in differentiation stage. (XLSX 30 kb)
Figure S3. mRNA-seq expression patterns of some key functional genes or TFs during differentiation stage. (PDF 629 kb)
Table S8. Validation of mRNA-seq using RT-qPCR. (XLSX 28 kb)
Figure S4. Validation of some key regulatory factors during adipocyte differentiation stage (0 h, 12 h, 24 h and 48 h) using RT-qPCR. Transcript abundance is presented as fold change± S.E.M (n = 3) using GAPDH as a reference gene (delta-delta method). (PDF 387 kb)
Table S9. List of selected genes with FPKM >30 and stably expressed during the entire stage. (XLSX 582 kb)
Table S10. GO terms and KEGG pathway analysis of selected genes with FPKM >30 and stably expressed during the entire stage. (XLSX 80 kb)
Figure S5. Design and sampling strategy of the differentiation process of duck subcutaneous preadipocyte. (PDF 1095 kb)
Acknowledgments
We thank Beijing Golden Star Inc. for providing the Pekin ducks. We also thank Dr. Jacqueline Smith for help in revising the paper.
Abbreviations
- AXIN1
Axis Inhibitor 1
- BAT
Brown adipose tissue
- C/EBPα
CCAAT Enhancer Binding Protein Alpha
- C/EBPδ
CCAAT Enhancer Binding Protein Delta
- CCND1
G1/S Specific CyclinD1
- DEGs
Differentially expressed genes
- DNMT1
DNA Methyltransferase 1
- DUSP10
Dual Specificity MAP Kinase Phosphatase 10
- E2F1
E2F Transcription Factor 1
- E2F5
E2F Transcription Factor 5
- EHMT1
Euchromatic Histone Lysine Methyltransferase 1
- ELK4
ETS Domain Containing Protein 4
- FABP4
Fatty Acid Binding Protein 4
- GATA2
GATA Binding Protein 2
- GATA3
GATA Binding Protein 3
- GO
Gene ontology
- GPDH
Glycerol‑3‑phosphate dehydrogenase
- GSK3β
Glycogen Synthase Kinase 3 Beta
- HES1
Hes Family BHLH Transcription Factor 1
- ITGB1
Integrins Subunit Beta 1
- KEGG
Kyoto encyclopedia of genes and genomes
- KLF5
Kruppel Like Factor 5
- MEF2C
Myocyte Enhancer Factor 2C
- MYOD1
Myogenic Differentiation 1
- NCBI
National Center for Biotechnology Information
- NR1H3
Nuclear Receptor Subfamily 1 Group H Member 3
- NR3C1
Nuclear Receptor Subfamily 3 Group C Member 1
- p27Kip1
Cyclin Dependent Kinase Inhibitor 1B
- PGC1α
Phosphatidylinositol phosphate kinase type I Gamma
- PIPKIγ
PPARG Coactivator 1 Alpha
- PLIN2
Perilipin2
- PPARγ
Peroxisome Proliferator Activated Receptor Gamma
- PRDM16
PR/SET Domain 16
- PTEN
Phosphatase And Tensin Homolog
- PTPN12
Protein Tyrosine Phosphatase Non-Receptor Type 1
- PTPN5
Protein Tyrosine Phosphatase, Non-Receptor Type 5
- RB
Retinoblastoma
- RT-qPCR
Quantitative real-time Polymerase Chain Reaction
- SCD1
Stearoyl CoA Desaturase 1
- SOX11
SRY-Box 11
- SREBP1
Sterol Regulatory Element Binding Transcription Factor 1
- TCF3
Transcription Factor 3
- TCF4
T Cell Specific Transcription Factor 4
- TCF7
T Cell Specific Transcription Factor 7
- TFs
Transcription factors
- TIN
Talin
- UCP1
Uncoupling Protein 1
- ZNF469
Zinc Finger Protein 469
Authors’ contributions
ZCH conceived and designed the experimental plan. ZW, XQL and SRC collected samples and measurement data. ZW, ZTY and FZ participated in bioinformatics analyses. ZW, NY, TEP, and ZCH drafted and revised this manuscript. All authors read and approved the final manuscript.
Funding
The work was supported by the National Waterfowl-Industry Technology Research System (CARS-42-09), the National Scientific Supporting Projects of China (2015BAD03B06), the National Natural Science Foundation of China (31572388), Primary Research & Development Plan of Jiangsu Province (BE2017349) and the Program for Changjiang Scholar and Innovation Research Team in University (IRT1191). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The data supporting the conclusions of this article (raw mRNA-seq reads) are available in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under accession number SRX4646736 (https://www.ncbi.nlm.nih.gov/sra/SRX4646736).
Ethics approval and consent to participate
Housing and care of Pekin ducks and collection of adipose samples for use in the described experiments was conducted following methods approved by the Animal Care and Use Committee of China Agricultural University (permit number: SYXK 2007–0023).
Consent for publication
Not Applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zheng Wang, Email: 936608496@qq.com.
Zhong-Tao Yin, Email: 479816697@qq.com.
Fan Zhang, Email: 13580484516@163.com.
Xiao-Qin Li, Email: 291032861@qq.com.
Si-Rui Chen, Email: csr@cau.edu.cn.
Ning Yang, Email: nyang@cau.edu.cn.
Tom E. Porter, Email: teporter@umd.edu
Zhuocheng Hou, Email: zchou@cau.edu.cn.
References
- 1.Stenvers DJ, Scheer F, Schrauwen P, la Fleur SE, Kalsbeek A. Circadian clocks and insulin resistance. Nat Rev Endocrinol. 2019;15(2):75–89. doi: 10.1038/s41574-018-0122-1. [DOI] [PubMed] [Google Scholar]
- 2.Saintilan R, Sellier P, Billon Y, Gilbert H. Genetic correlations between males, females and castrates for residual feed intake, feed conversion ratio, growth rate and carcass composition traits in large White growing pigs. J Anim Breed Genet. 2012;129(2):103–106. doi: 10.1111/j.1439-0388.2011.00972.x. [DOI] [PubMed] [Google Scholar]
- 3.Hausman GJ, Dodson MV, Ajuwon K, Azain M, Barnes KM, Guan LL, Jiang Z, Poulos SP, Sainz RD, Smith S, et al. Board-invited review: the biology and regulation of preadipocytes and adipocytes in meat animals. J Anim Sci. 2009;87(4):1218–1246. doi: 10.2527/jas.2008-1427. [DOI] [PubMed] [Google Scholar]
- 4.Zhu F, Yuan JM, Zhang ZH, Hao JP, Yang YZ, Hu SQ, Yang FX, Qu LJ, Hou ZC. De novo transcriptome assembly and identification of genes associated with feed conversion ratio and breast muscle yield in domestic ducks. Anim Genet. 2015;46(6):636–645. doi: 10.1111/age.12361. [DOI] [PubMed] [Google Scholar]
- 5.Cerk IK, Wechselberger L, Oberer M. Adipose triglyceride lipase regulation: an overview. Curr Protein Pept Sci. 2018;19(2):221–233. doi: 10.2174/1389203718666170918160110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mota DSP, Richard AJ, Hang H, Stephens JM. Transcriptional regulation of adipogenesis. Compr Physiol. 2017;7(2):635–674. doi: 10.1002/cphy.c160022. [DOI] [PubMed] [Google Scholar]
- 7.Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, Tavernier G, Langin D, Spiegelman BM. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007;6(1):38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farmer SR. Transcriptional control of adipocyte formation. Cell Metab. 2006;4(4):263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White UA, Stephens JM. Transcriptional factors that promote formation of white adipose tissue. Mol Cell Endocrinol. 2010;318(1–2):10–14. doi: 10.1016/j.mce.2009.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gubelmann C, Schwalie PC, Raghav SK, Roder E, Delessa T, Kiehlmann E, Waszak SM, Corsinotti A, Udin G, Holcombe W, et al. Identification of the transcription factor ZEB1 as a central component of the adipogenic gene regulatory network. Elife. 2014;3:e3346. doi: 10.7554/eLife.03346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pahlavani M, Wijayatunga NN, Kalupahana NS, Ramalingam L, Gunaratne PH, Coarfa C, Rajapakshe K, Kottapalli P, Moustaid-Moussa N. Transcriptomic and microRNA analyses of gene networks regulated by eicosapentaenoic acid in brown adipose tissue of diet-induced obese mice. BBA Mol Cell Biol L. 2018;1863(12):1523–1531. doi: 10.1016/j.bbalip.2018.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mezentseva NV, Kumaratilake JS, Newman SA. The brown adipocyte differentiation pathway in birds: an evolutionary road not taken. BMC Biol. 2008;6(17):1–13. doi: 10.1186/1741-7007-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes J, Criscuolo F. Evolutionary history of the UCP gene family: gene duplication and selection. BMC Evol Biol. 2008;8:306. doi: 10.1186/1471-2148-8-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newman SA, Mezentseva NV, Badyaev AV. Gene loss, thermogenesis, and the origin of birds. Ann N Y Acad Sci. 2013;1289:36–47. doi: 10.1111/nyas.12090. [DOI] [PubMed] [Google Scholar]
- 15.Ma X, Lee P, Chisholm DJ, James DE. Control of adipocyte differentiation in different fat depots; implications for pathophysiology or therapy. Front Endocrinol. 2015;6(1). [DOI] [PMC free article] [PubMed]
- 16.Green CR, Wallace M, Divakaruni AS, Phillips SA, Murphy AN, Ciaraldi TP, Metallo CM. Branched-chain amino acid catabolism fuels adipocyte differentiation and lipogenesis. Nat Chem Biol. 2016;12(1):15–21. doi: 10.1038/nchembio.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruiz-Ojeda Francisco, Rupérez Azahara, Gomez-Llorente Carolina, Gil Angel, Aguilera Concepción. Cell Models and Their Application for Studying Adipogenic Differentiation in Relation to Obesity: A Review. International Journal of Molecular Sciences. 2016;17(7):1040. doi: 10.3390/ijms17071040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang T, Zhang X, Han K, Zhang G, Wang J, Xie K, Xue Q. Genome-Wide Analysis of lncRNA and mRNA Expression During Differentiation of Abdominal Preadipocytes in the Chicken. G3 (Bethesda) 2017;7(3):953–966. doi: 10.1534/g3.116.037069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsubara Y, Sato K, Ishii H, Akiba Y. Changes in mRNA expression of regulatory factors involved in adipocyte differentiation during fatty acid induced adipogenesis in chicken. Comp Biochem Physiol A Mol Integr Physiol. 2005;141(1):108–115. doi: 10.1016/j.cbpb.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 20.Wang S, Zhang Y, Xu Q, Yuan X, Dai W, Shen X, Wang Z, Chang G, Wang Z, Chen G. The differentiation of preadipocytes and gene expression related to adipogenesis in ducks (Anas platyrhynchos) PLoS One. 2018;13(5):e196371. doi: 10.1371/journal.pone.0196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin F, Zhu F, Hao J, Yang F, Hou Z. In vivo prediction of the carcass fatness using live body measurements in Pekin ducks. Poult Sci. 2018;97(7):2365–2371. doi: 10.3382/ps/pey079. [DOI] [PubMed] [Google Scholar]
- 22.Zhu F, Gao Y, Lin F, Hao J, Yang F, Hou Z. Systematic analysis of feeding behaviors and their effects on feed efficiency in Pekin ducks. J Anim Sci Biotechnol. 2017;8:81. doi: 10.1186/s40104-017-0212-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng MT, Zhu F, Yang YZ, Yang FX, Hao JP, Chen SR, Hou ZC. Genome-wide association study reveals novel loci associated with body size and carcass yields in Pekin ducks. BMC Genomics. 2019;20(1):1. doi: 10.1186/s12864-018-5379-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Z, Li M, Cheng H, Fan W, Yuan Z, Gao Q, Xu Y, Guo Z, Zhang Y, Hu J, et al. An intercross population study reveals genes associated with body size and plumage color in ducks. Nat Commun. 2018;9(1):2648. doi: 10.1038/s41467-018-04868-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fouad AM, El-Senousey HK. Nutritional factors affecting abdominal fat deposition in poultrY. A review. Asian-Australas J Anim Sci. 2014;27(7):1057–1068. doi: 10.5713/ajas.2013.13702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sai J, Owens P, Novitskiy SV, Hawkins OE, Vilgelm AE, Yang J, Sobolik T, Lavender N, Johnson AC, McClain C, et al. PI3K inhibition reduces mammary tumor growth and facilitates antitumor immunity and anti-PD1 responses. Clin Cancer Res. 2017;23(13):3371–3384. doi: 10.1158/1078-0432.CCR-16-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yanagisawa S, Baker JR, Vuppusetty C, Fenwick P, Donnelly LE, Ito K, Barnes PJ. Decreased phosphatase PTEN amplifies PI3K signaling and enhances proinflammatory cytokine release in COPD. Am J Phys Lung Cell Mol Phys. 2017;313(2):L230–L239. doi: 10.1152/ajplung.00382.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7(1):11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 29.Luo X, Hong L, Cheng C, Li N, Zhao X, Shi F, Liu J, Fan J, Zhou J, Bode AM, et al. DNMT1 mediates metabolic reprogramming induced by Epstein-Barr virus latent membrane protein 1 and reversed by grifolin in nasopharyngeal carcinoma. Cell Death Dis. 2018;9(6):619. doi: 10.1038/s41419-018-0662-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsai CC, Su PF, Huang YF, Yew TL, Hung Hung SC. Oct4 and Nanog directly regulate Dnmt1 to maintain self-renewal and undifferentiated state in mesenchymal stem cells. Mol Cell. 2012;47(2):169–182. doi: 10.1016/j.molcel.2012.06.020. [DOI] [PubMed] [Google Scholar]
- 31.Londono GT, Lu C, Lodato PM, Tse S, Olejniczak SH, Witze ES, Thompson CB, Wellen KE. DNMT1 is regulated by ATP-citrate lyase and maintains methylation patterns during adipocyte differentiation. Mol Cell Biol. 2013;33(19):3864–3878. doi: 10.1128/MCB.01495-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pazienza V, Tavano F, Francavilla M, Fontana A, Pellegrini F, Benegiamo G, Corbo V, di Mola FF, Di Sebastiano P, Andriulli A, et al. Time-qualified patterns of variation of PPARgamma, DNMT1, and DNMT3B expression in pancreatic cancer cell lines. PPAR Res. 2012;2012:890875. doi: 10.1155/2012/890875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christofori G. Changing neighbours, changing behavior: cell adhesion molecule-mediated signalling during tumour progression. EMBO J. 2003;22(10):2318–2323. doi: 10.1093/emboj/cdg228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim JH, Kushiro K, Graham NA, Asthagiri AR. Tunable interplay between epidermal growth factor and cell-cell contact governs the spatial dynamics of epithelial growth. Proc Natl Acad Sci. 2009;106(27):11149–11153. doi: 10.1073/pnas.0812651106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kimura K, Teranishi S, Kawamoto K, Nishida T. Protection of human corneal epithelial cells from hypoxia-induced disruption of barrier function by hepatocyte growth factor. Exp Eye Res. 2010;90(2):337–343. doi: 10.1016/j.exer.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 36.Provenzano PP, Keely PJ. Mechanical signaling through the cytoskeleton regulates cell proliferation by coordinated focal adhesion and rho GTPase signaling. J Cell Sci. 2011;124(Pt 8):1195–1205. doi: 10.1242/jcs.067009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balaban NQ, Schwarz US, Riveline D, Goichberg P, Tzur G, Sabanay I, Mahalu D, Safran S, Bershadsky A, Addadi L, et al. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat Cell Biol. 2001;3(5):466–472. doi: 10.1038/35074532. [DOI] [PubMed] [Google Scholar]
- 38.Sun T, Aceto N, Meerbrey KL, Kessler JD, Zhou C, Migliaccio I, Nguyen DX, Pavlova NN, Botero M, Huang J, et al. Activation of multiple proto-oncogenic tyrosine kinases in breast cancer via loss of the PTPN12 phosphatase. Cell. 2011;144(5):703–718. doi: 10.1016/j.cell.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Pereda JM, Wegener KL, Santelli E, Bate N, Ginsberg MH, Critchley DR, Campbell ID, Liddington RC. Structural basis for phosphatidylinositol phosphate kinase type Igamma binding to Talin at focal adhesions. J Biol Chem. 2005;280(9):8381–8386. doi: 10.1074/jbc.M413180200. [DOI] [PubMed] [Google Scholar]
- 40.Kumar GS, Zettl H, Page R, Peti W. Structural basis for the regulation of the mitogen-activated protein (MAP) kinase p38alpha by the dual specificity phosphatase 16 MAP kinase binding domain in solution. J Biol Chem. 2013;288(39):28347–28356. doi: 10.1074/jbc.M113.499178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Faust D, Dolado I, Cuadrado A, Oesch F, Weiss C, Nebreda AR, Dietrich C. p38alpha MAPK is required for contact inhibition. Oncogene. 2005;24(53):7941–7945. doi: 10.1038/sj.onc.1208948. [DOI] [PubMed] [Google Scholar]
- 42.Nordle AK, Rios P, Gaulton A, Pulido R, Attwood TK, Tabernero L. Functional assignment of MAPK phosphatase domains. Proteins. 2007;69(1):19–31. doi: 10.1002/prot.21477. [DOI] [PubMed] [Google Scholar]
- 43.Francis DM, Kumar GS, Koveal D, Tortajada A, Page R, Peti W. The differential regulation of p38alpha by the neuronal kinase interaction motif protein tyrosine phosphatases, a detailed molecular study. Structure. 2013;21(9):1612–1623. doi: 10.1016/j.str.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fajas L, Landsberg RL, Huss-Garcia Y, Sardet C, Lees JA, Auwerx J. E2Fs regulate adipocyte differentiation. Dev Cell. 2002;3(1):39–49. doi: 10.1016/S1534-5807(02)00190-9. [DOI] [PubMed] [Google Scholar]
- 45.Komori T. Regulation of Rb family proteins by Cdk6/Ccnd1 in growth plates. Cell Cycle. 2013;12(14):2161–2162. doi: 10.4161/cc.25515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marquez MP, Alencastro F, Madrigal A, Jimenez JL, Blanco G, Gureghian A, Keagy L, Lee C, Liu R, Tan L, et al. The role of cellular proliferation in adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells. Stem Cells Dev. 2017;26(21):1578–1595. doi: 10.1089/scd.2017.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jung AR, Kim RY, Kim HW, Shrestha KR, Jeon SH, Cha KJ, Park YH, Kim DS, Lee JY. Nanoengineered polystyrene surfaces with nanopore array pattern alters cytoskeleton organization and enhances induction of neural differentiation of human adipose-derived stem cells. Tissue Eng Part A. 2015;21(13–14):2115–2124. doi: 10.1089/ten.tea.2014.0346. [DOI] [PubMed] [Google Scholar]
- 48.Estes BT, Diekman BO, Guilak F. Monolayer cell expansion conditions affect the chondrogenic potential of adipose-derived stem cells. Biotechnol Bioeng. 2008;99(4):986–995. doi: 10.1002/bit.21662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fajas L. Adipogenesis: a cross-talk between cell proliferation and cell differentiation. Ann Med. 2003;35(2):79–85. doi: 10.1080/07853890310009999. [DOI] [PubMed] [Google Scholar]
- 50.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 51.Nobusue H, Onishi N, Shimizu T, Sugihara E, Oki Y, Sumikawa Y, Chiyoda T, Akashi K, Saya H, Kano K. Regulation of MKL1 via actin cytoskeleton dynamics drives adipocyte differentiation. Nat Commun. 2014;5:3368. doi: 10.1038/ncomms4368. [DOI] [PubMed] [Google Scholar]
- 52.Hardy S, Langelier Y, Prentki M. Oleate activates phosphatidylinositol 3-kinase and promotes proliferation and reduces apoptosis of MDA-MB-231 breast cancer cells, whereas palmitate has opposite effects. Cancer Res. 2000;60(22):6353–6358. [PubMed] [Google Scholar]
- 53.Arous C, Naimi M, Van Obberghen E. Oleate-mediated activation of phospholipase D and mammalian target of rapamycin (mTOR) regulates proliferation and rapamycin sensitivity of hepatocarcinoma cells. Diabetologia. 2011;54(4):954–964. doi: 10.1007/s00125-010-2032-1. [DOI] [PubMed] [Google Scholar]
- 54.Tanaka T, Yoshida N, Kishimoto T, Akira S. Defective adipocyte differentiation in mice lacking the C/EBPbeta and/or C/EBPdelta gene. EMBO J. 1997;16(24):7432–7443. doi: 10.1093/emboj/16.24.7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li VS, Ng SS, Boersema PJ, Low TY, Karthaus WR, Gerlach JP, Mohammed S, Heck AJ, Maurice MM, Mahmoudi T, et al. Wnt signaling through inhibition of beta-catenin degradation in an intact Axin1 complex. Cell. 2012;149(6):1245–1256. doi: 10.1016/j.cell.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 56.Verheyen EM, Gottardi CJ. Regulation of Wnt/beta-catenin signaling by protein kinases. Dev Dyn. 2010;239(1):34–44. doi: 10.1002/dvdy.22019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Christodoulides C, Lagathu C, Sethi JK, Vidal-Puig A. Adipogenesis and WNT signalling. Trends Endocrinol Metab. 2009;20(1):16–24. doi: 10.1016/j.tem.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takahashi Y, Shinoda A, Kamada H, Shimizu M, Inoue J, Sato R. Perilipin2 plays a positive role in adipocytes during lipolysis by escaping proteasomal degradation. Sci Rep. 2016;6:20975. doi: 10.1038/srep20975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu X, Lu X, Song K, Blackman MR. Natural functions of PLIN2 mediating Wnt/LiCl signaling and glycogen synthase kinase 3 (GSK3)/GSK3 substrate-related effects are modulated by lipid. Mol Cell Biol. 2016;36(3):421–437. doi: 10.1128/MCB.00510-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang S, Wei H, Song T, Yang Y, Peng J, Jiang S. Transcriptome comparison between porcine subcutaneous and intramuscular stromal vascular cells during adipogenic differentiation. PLoS One. 2013;8(10):e77094. doi: 10.1371/journal.pone.0077094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oishi Y, Manabe I, Tobe K, Tsushima K, Shindo T, Fujiu K, Nishimura G, Maemura K, Yamauchi T, Kubota N, et al. Kruppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell Metab. 2005;1(1):27–39. doi: 10.1016/j.cmet.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 62.Kim JB, Wright HM, Wright M, Spiegelman BM. ADD1/SREBP1 activates PPARgamma through the production of endogenous ligand. Proc Natl Acad Sci. 1998;95(8):4333–4337. doi: 10.1073/pnas.95.8.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rui W, Jin Z, Zhe G, Song H. The methylation of C/EBP beta gene promoter and regulated by GATA-2 protein. Mol Biol Rep. 2013;40(2):797–801. doi: 10.1007/s11033-012-2117-7. [DOI] [PubMed] [Google Scholar]
- 64.Tong Q, Dalgin G, Xu H, Ting CN, Leiden JM, Hotamisligil GS. Function of GATA transcription factors in preadipocyte-adipocyte transition. Science. 2000;290(5489):134–138. doi: 10.1126/science.290.5489.134. [DOI] [PubMed] [Google Scholar]
- 65.Ross DA, Hannenhalli S, Tobias JW, Cooch N, Shiekhattar R, Kadesch T. Functional analysis of Hes-1 in preadipocytes. Mol Endocrinol. 2006;20(3):698–705. doi: 10.1210/me.2005-0325. [DOI] [PubMed] [Google Scholar]
- 66.Chen Q, Shou P, Zheng C, Jiang M, Cao G, Yang Q, Cao J, Xie N, Velletri T, Zhang X, et al. Fate decision of mesenchymal stem cells: adipocytes or osteoblasts? Cell Death Differ. 2016;23(7):1128–1139. doi: 10.1038/cdd.2015.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mezentseva NV, Kumaratilake JS, Newman SA. The brown adipocyte differentiation pathway in birds: an evolutionary road not taken. BMC Biol. 2008;6:17. doi: 10.1186/1741-7007-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Desjardins EM, Steinberg GR. Emerging role of AMPK in Brown and Beige adipose tissue (BAT): implications for obesity, insulin resistance, and type 2 diabetes. Curr Diab Rep. 2018;18(10):80. doi: 10.1007/s11892-018-1049-6. [DOI] [PubMed] [Google Scholar]
- 69.Ohno H, Shinoda K, Ohyama K, Sharp LZ, Kajimura S. EHMT1 controls brown adipose cell fate and thermogenesis through the PRDM16 complex. Nature. 2013;504(7478):163–167. doi: 10.1038/nature12652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Seale P, Conroe HM, Estall J, Kajimura S, Frontini A, Ishibashi J, Cohen P, Cinti S, Spiegelman BM. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J Clin Invest. 2011;121(1):96–105. doi: 10.1172/JCI44271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shao M, Gupta RK. Transcriptional brakes on the road to adipocyte thermogenesis. BBA Mol Cell Biol Lipids. 2019;1864(1):20–28. doi: 10.1016/j.bbalip.2018.05.010. [DOI] [PubMed] [Google Scholar]
- 72.Krstic Jelena, Reinisch Isabel, Schupp Michael, Schulz Tim, Prokesch Andreas. p53 Functions in Adipose Tissue Metabolism and Homeostasis. International Journal of Molecular Sciences. 2018;19(9):2622. doi: 10.3390/ijms19092622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Booth A. D., Magnuson A. M., Cox-York K. A., Wei Y., Wang D., Pagliassotti M. J., Foster M. T. Inhibition of adipose tissue PPARγ prevents increased adipocyte expansion after lipectomy and exacerbates a glucose-intolerant phenotype. Cell Proliferation. 2016;50(2):e12325. doi: 10.1111/cpr.12325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yildiz E, Bardak H, Gunay M, Bardak Y, Imamoglu S, Ozbas H, Bagci O. Novel zinc finger protein gene 469 (ZNF469) variants in advanced keratoconus. Curr Eye Res. 2017;42(10):1396–1400. doi: 10.1080/02713683.2017.1325910. [DOI] [PubMed] [Google Scholar]
- 75.Chen L, Luo J, Li JX, Li JJ, Wang DQ, Tian Y, Lu LZ. Transcriptome analysis of adiposity in domestic ducks by transcriptomic comparison with their wild counterparts. Anim Genet. 2015;46(3):299–307. doi: 10.1111/age.12294. [DOI] [PubMed] [Google Scholar]
- 76.Choi MK, Seong I, Kang SA, Kim J. Down-regulation of Sox11 is required for efficient osteogenic differentiation of adipose-derived stem cells. Mol Cell. 2014;37(4):337–344. doi: 10.14348/molcells.2014.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lempiainen H, Shore D. Growth control and ribosome biogenesis. Curr Opin Cell Biol. 2009;21(6):855–863. doi: 10.1016/j.ceb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 78.Lin, Chun TH, Kang L. Adipose extracellular matrix remodelling in obesity and insulin resistance. Biochem Pharmacol. 2016;119:8–16. doi: 10.1016/j.bcp.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kang L, Ayala JE, Lee-Young RS, Zhang Z, James FD, Neufer PD, Pozzi A, Zutter MM, Wasserman DH. Diet-induced muscle insulin resistance is associated with extracellular matrix remodeling and interaction with integrin alpha2beta1 in mice. Diabetes. 2011;60(2):416–426. doi: 10.2337/db10-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kodama K, Horikoshi M, Toda K, Yamada S, Hara K, Irie J, Sirota M, Morgan AA, Chen R, Ohtsu H, et al. Expression-based genome-wide association study links the receptor CD44 in adipose tissue with type 2 diabetes. Proc Natl Acad Sci. 2012;109(18):7049–7054. doi: 10.1073/pnas.1114513109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kang HS, Liao G, DeGraff LM, Gerrish K, Bortner CD, Garantziotis S, Jetten AM. CD44 plays a critical role in regulating diet-induced adipose inflammation, hepatic steatosis, and insulin resistance. PLoS One. 2013;8(3):e58417. doi: 10.1371/journal.pone.0058417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ji B, Ernest B, Gooding JR, Das S, Saxton AM, Simon J, Dupont J, Metayer-Coustard S, Campagna SR, Voy BH. Transcriptomic and metabolomic profiling of chicken adipose tissue in response to insulin neutralization and fasting. BMC Genomics. 2012;13:441. doi: 10.1186/1471-2164-13-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shang Zhouchun, Guo Lin, Wang Ning, Shi Hui, Wang Yuxiang, Li Hui. Oleate promotes differentiation of chicken primary preadipocytesin vitro. Bioscience Reports. 2014;34(1):51–57. doi: 10.1042/BSR20130120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ramirez-Zacarias JL, Castro-Munozledo F, Kuri-Harcuch W. Quantitation of adipose conversion and triglycerides by staining intracytoplasmic lipids with oil red O. Histochemistry. 1992;97(6):493–7. [DOI] [PubMed]
- 85.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11(9):1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genomic Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Langfelder P, Horvath S. WGCNA an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Statistics relating to the mRNA-seq data. (XLSX 12 kb)
Figure S1. Heatmap of the differentiation of biological replicates of duck subcutaneous preadipocyte. The colors ranging from white to blue represent Pearson correlation coefficients ranging from 0 to 1, indicating low to high correlations, respectively. (PDF 498 kb)
Table S2. List of genes expressed during the entire stage. (XLSX 8936 kb)
Table S3. List of all DEGs during complete duck subcutaneous preadipocyte differentiation. (XLSX 929 kb)
Table S4. GO term analysis of DEGs at different stages. (XLSX 248 kb)
Table S5. KEGG pathway analysis of DEGs at different stages. (XLSX 51 kb)
Table S6. Genes involved in eight modules. (XLSX 147 kb)
Figure S2. Classification of DEGs throughout the entire differentiation process and visualization of gene expression levels of significant modules. (PDF 1241 kb)
Table S7. List of differentially expressed TFs obtained in differentiation stage. (XLSX 30 kb)
Figure S3. mRNA-seq expression patterns of some key functional genes or TFs during differentiation stage. (PDF 629 kb)
Table S8. Validation of mRNA-seq using RT-qPCR. (XLSX 28 kb)
Figure S4. Validation of some key regulatory factors during adipocyte differentiation stage (0 h, 12 h, 24 h and 48 h) using RT-qPCR. Transcript abundance is presented as fold change± S.E.M (n = 3) using GAPDH as a reference gene (delta-delta method). (PDF 387 kb)
Table S9. List of selected genes with FPKM >30 and stably expressed during the entire stage. (XLSX 582 kb)
Table S10. GO terms and KEGG pathway analysis of selected genes with FPKM >30 and stably expressed during the entire stage. (XLSX 80 kb)
Figure S5. Design and sampling strategy of the differentiation process of duck subcutaneous preadipocyte. (PDF 1095 kb)
Data Availability Statement
The data supporting the conclusions of this article (raw mRNA-seq reads) are available in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under accession number SRX4646736 (https://www.ncbi.nlm.nih.gov/sra/SRX4646736).