Abstract
Non-small-cell lung cancer, histologically classified into adenocarcinoma (AD) and squamous cell carcinoma, is one of the most deadly malignancies worldwide. Lung AD (LUAD) could benefit of a plethora of target therapies and, in the last few years, also of immunotherapies. Here we focused on a real-life cohort of LUAD and The Cancer Genome Atlas (TCGA)-LUAD dataset aiming to gain insights into the immune contexture of such a malignancy. We explored the mutational status of 41 genes and the expression of 94 genes, related to immune-checkpoint, inflammation, and stromal microenvironment. Surprisingly, we found that our cohort has a very low mutational burden if we consider our panel as its surrogate. Regarding gene expression data, we identified 31 genes significantly deregulated in tumor tissues compared with a pool of normal samples. Unsupervised hierarchical clustering of the deregulated genes is able to identify two clusters of tumor samples, differently enriched in alterations in actionable genes. In particular, we identified a cluster enriched in patients carrying KRAS alterations. In silico deconvolution, that is the inferring of tumor microenvironment composition by gene expression data, through TIMER algorithm has been performed to explore immune microenvironment. Estimation performed on our gene expression matrix showed that B cell infiltration is lower in the KRAS-mutated enriched cluster, as in the TCGA-LUAD dataset. Such a finding has been validated in situ through immunohistochemistry in an independent cohort. Moreover, cases in LUAD-TCGA with low B cell infiltration have a significantly worse overall survival than those with higher levels. In the real-life cohort we observed that cases belonging to cluster enriched in KRAS-mutated patients have a poor outcome. LUAD driven by KRAS mutation represents an unmet clinical need, being refractory to pharmacological inhibition. Our results link KRAS mutations to B cell infiltration. Thus, the present findings could be helpful in a better definition of immunotherapeutic approaches for KRAS mutated patients.
Keywords: LUAD, immunotherapy, tumor microenvironment, B cells, KRAS
1. Introduction
Lung cancer remains one of the most deadly malignancies worldwide. The majority of patients have non-small-cell lung cancer (NSCLC) and in an advanced stage [1]. They can be histologically classified into lung adenocarcinoma (LUAD) and squamous cell carcinoma, with the former being the most common subtype, comprising more than 50% of all NSCLC [1].
Historically, these patients were treated with platinum chemotherapy regardless of histological subtype. Despite an improvement in overall survival (OS) when compared with best supportive care, a therapeutic plateau has been reached with a response rate of about 20% and a median survival of 8–10 months [2]. Actionable genetic alterations (e.g., EGFR (growth factor receptor) mutations, ALK/ROS1 translocations) made it possible to treat patients in a targeted way, even if only a small percentage of them benefit from these approaches. In a different percentage of LUAD (20%–25%) the discovery of KRAS mutations has been associated with poorer overall survival (OS) than with KRAS wild-type tumors, particularly in the advanced-stage setting [3,4].
In order to improve the clinical outcome of these patients, a novel emerging approach is based on the immunotherapy that acts targeting specific immune regulatory checkpoints. In particular, Cytotoxic T-Lymphocyte Antigen 4, CTLA-4, and Programmed cell death protein 1/Programmed death-ligand1, PD-1/PD-L1, are the main checkpoints to be targeted [5]. Briefly, CTLA4 is involved in the early phase by reducing T-cell response to self-antigens; the axis PD-1/PD-L1 acts in the active stage of T-cell response in tumor microenvironment. Thus, targeting them lead to reduction of tumor growth through increasing the immune response. Biomarkers able to discriminate patients which benefits from immunotherapies are still under investigation. Tumor mutational burden, neoantigens, and PD-L1 expression are not sufficient probably because a deeper knowledge of tumor microenvironment (TME) is not taken into consideration. In particular, TME changes and the relationship with the biology of cancer cells (e.g., mutational status of peculiar genes or altered pathways) are needed. Indeed, conflicting results on the correlation among EGFR/KRAS (epidermal growth factor receptor/Kirsten Rat sarcoma virus) mutations and immune checkpoint inhibitors response have been evidenced. KRAS, which is considered as undruggable, has been demonstrated by several groups to be able to modulate the immune response, as in colorectal and pancreatic cancers [6,7,8].
Thus, to study how the mutational status of cancer cells could contribute to the enhancement of immunogenicity, should be able to indicate the sensitivity to immune checkpoint blockade [9]. It is well known that the interplay between immune microenvironment and cancer cells is highly dynamic in the different steps of cancer onset. Generally speaking, three important phases (elimination, equilibrium, escape) could resume the deep changes of the immune system in relation to carcinogenesis [10]. Such a process is called “immunoediting” and it is recognized as one of the hallmarks of cancer. Briefly, starting from the initial phases of immune surveillance and quiescence, respectively elimination and equilibrium, the immune system does not attack the tumor allowing its proliferation [11,12]. Several studies pointed out the different composition of TME of lung cancer, recently reviewed by Altorki et al. [13]. In the last few years, computational methods were developed to infer TME composition from bulk genomic data. These approaches, such as CIBERSORT [14] or TIMER [15] are deeply used to date to gain knowledge of TME.
In this study we explored the biological heterogeneity of the immune landscape of a real-life cohort of 20 patients affected by LUAD and of a cohort affected by LUAD and reported as TGCA dataset with the aim to identify potential biomarkers useful for a possible immune-therapy approach in these patients.
2. Results
2.1. Immune-Mutational Status
In the present study, we analyzed a cohort including 20 LUAD patients. The series comprises patients affected by advanced stage LUAD (stage IIIB-IV), carrying 7/20 (35%), 8/20 (40%), 8/20 (40%) EGFR, KRAS, and TP53 alterations, respectively.
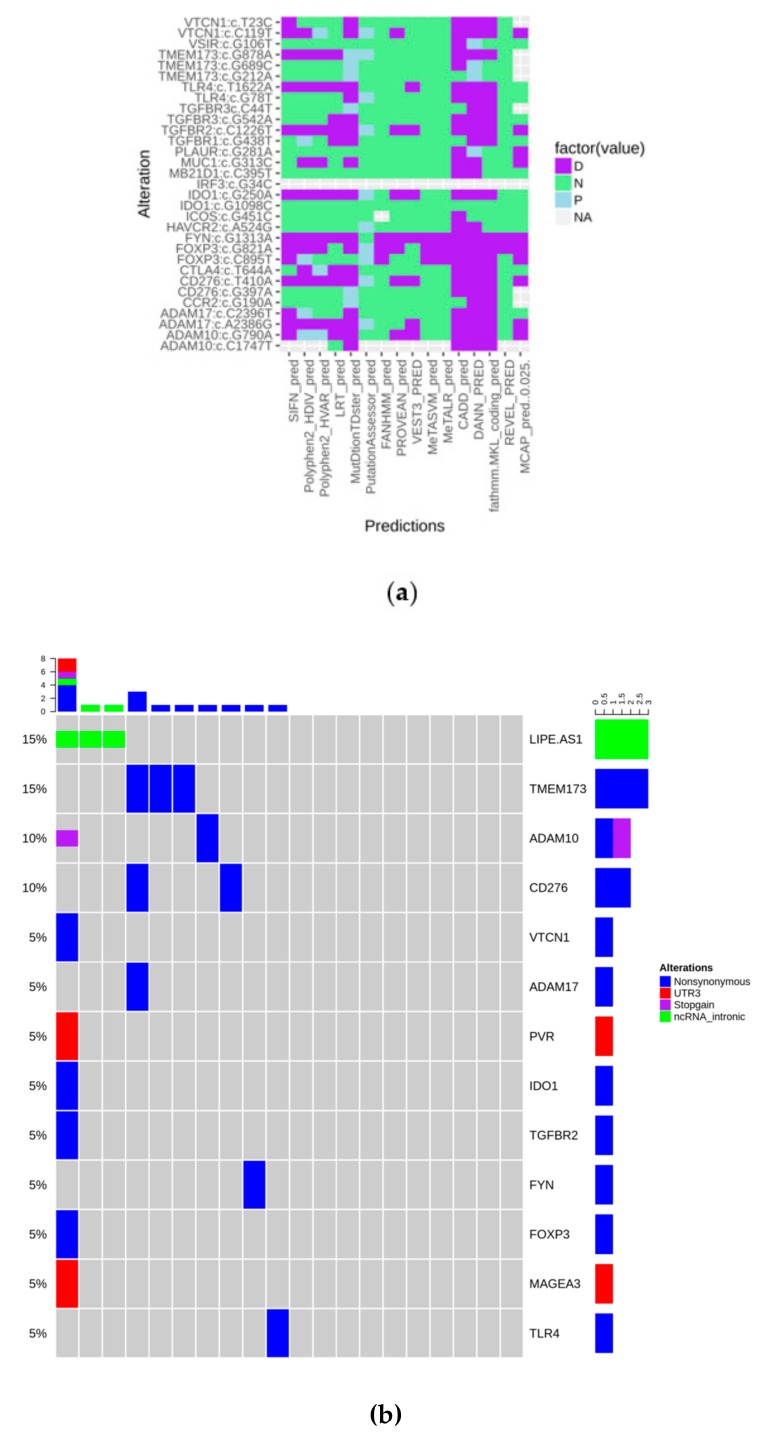
To explore the mutational status of genes involved in immunological/inflammatory response, we designed a targeted sequencing panel including 41 genes. After filtering, as described in the Materials and Methods section, variants were annotated and their deleteriousness was predicted through 16 tools, as shown in Figure 1a. They were considered as pathogenic when at least 9/16 algorithms agreed in their prediction. Alterations, that “survived” the filtering steps included, regards 13 genes over 41. PVR c.*27G > A, MAGEA3 c.*41C > T, and LIPE-AS1 were retained because of their probable regulating role.
Figure 1.
(a) Heatmap showing the predicted biological impact of detected alterations (D: Deleterious; N: Neutral; P: Predicted deleterious); (b) Oncoprint of the mutations that “survived” all the filtering steps.
The oncoprint plot in Figure 1b shows that alterations have no peculiar distribution in the analyzed cohort, indicating that the “immune mutational status” is substantially wild-type. Moreover, considering our panel as a surrogate to measure tumor mutational burden, it could be observed that it is very low in the present series.
2.2. Immune Gene Expression Results and Integration with Overall Molecular Data
Immune gene expression evaluation was performed through a custom designed targeted panel including 95 genes. To explore deregulation, we compared our tumor samples with nonpathological samples through the DeSeq2 algorithm. We detected 31/95 genes significantly deregulated (Table 1). Gene expression results were confirmed on LUAD-TCGA dataset (n = 574) through the web-browser of TIMER (Supplementary Material Figure S1). Only ITGAE, IL7, and HMGB1 expression observed in LUAD-TCGA have an opposite behavior with respect to what was found in our cohort.
Table 1.
Significantly deregulated genes in the tumor/normal comparison.
| log2FoldChange | Adjusted p-Value | |
|---|---|---|
| C10orf54 | −1.63 | 8.06 × 10−8 |
| TGFBR3 | −2.46 | 1.98 × 10−7 |
| IL6 | −3.39 | 2.43 × 10−7 |
| PLAU | 2.83 | 3.85 × 10−7 |
| MMP12 | 5.63 | 5.84 × 10−5 |
| TNFRSF9 | 2.12 | 0.0002 |
| ADORA2A | −1.38 | 0.0003 |
| IL2RA | 1.32 | 0.0011 |
| FOXP3 | 2.75 | 0.0013 |
| NT5E | 2.64 | 0.0013 |
| MUC1 | 2.48 | 0.0015 |
| TNFRSF18 | 2.05 | 0.0039 |
| TNFSF18 | 2.87 | 0.0039 |
| MMP9 | 2.17 | 0.0045 |
| HPSE | 1.26 | 0.0051 |
| IL12B | 5.57 | 0.0051 |
| CSPG4 | −1.59 | 0.0079 |
| CEACAM1 | 1.75 | 0.010 |
| TGFBR2 | −0.89 | 0.010 |
| ITGAE | 1.21 | 0.014 |
| BTLA | 1.61 | 0.018 |
| TLR4 | −0.89 | 0.026 |
| TBK1 | 0.98 | 0.026 |
| IL7 | 0.88 | 0.028 |
| FAS | −0.73 | 0.029 |
| CXCL10 | 1.19 | 0.034 |
| MS4A1 | 1.79 | 0.034 |
| HMGB1 | 0.72 | 0.040 |
| MB21D1 | 0.97 | 0.042 |
| SERPINE1 | −1.23 | 0.042 |
| CCL13 | 1.72 | 0.048 |
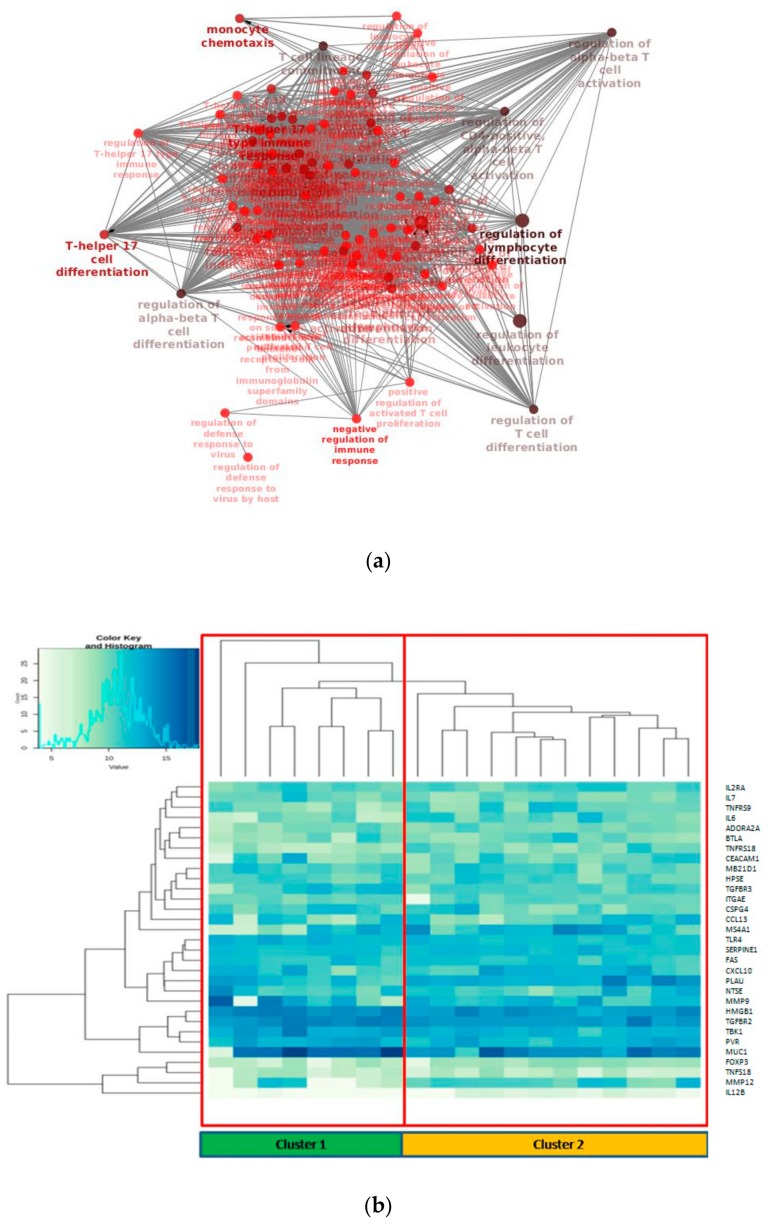
GO/KEGG enrichment of significantly deregulated genes has been depicted through Cluepedia/Cluego network (Figure 2a). In particular, it could be observed the enrichment of terms related to T cell differentiation, as could be expected, but more interestingly terms regarding T helper 17 lymphocytes (Th17) were found.
Figure 2.
(a) Biological network of the enriched GO (Gene Ontology) terms related to differentially expressed genes in the tumor/normal setting; (b) Unsupervised hierarchical clustering of the 31 deregulated genes.
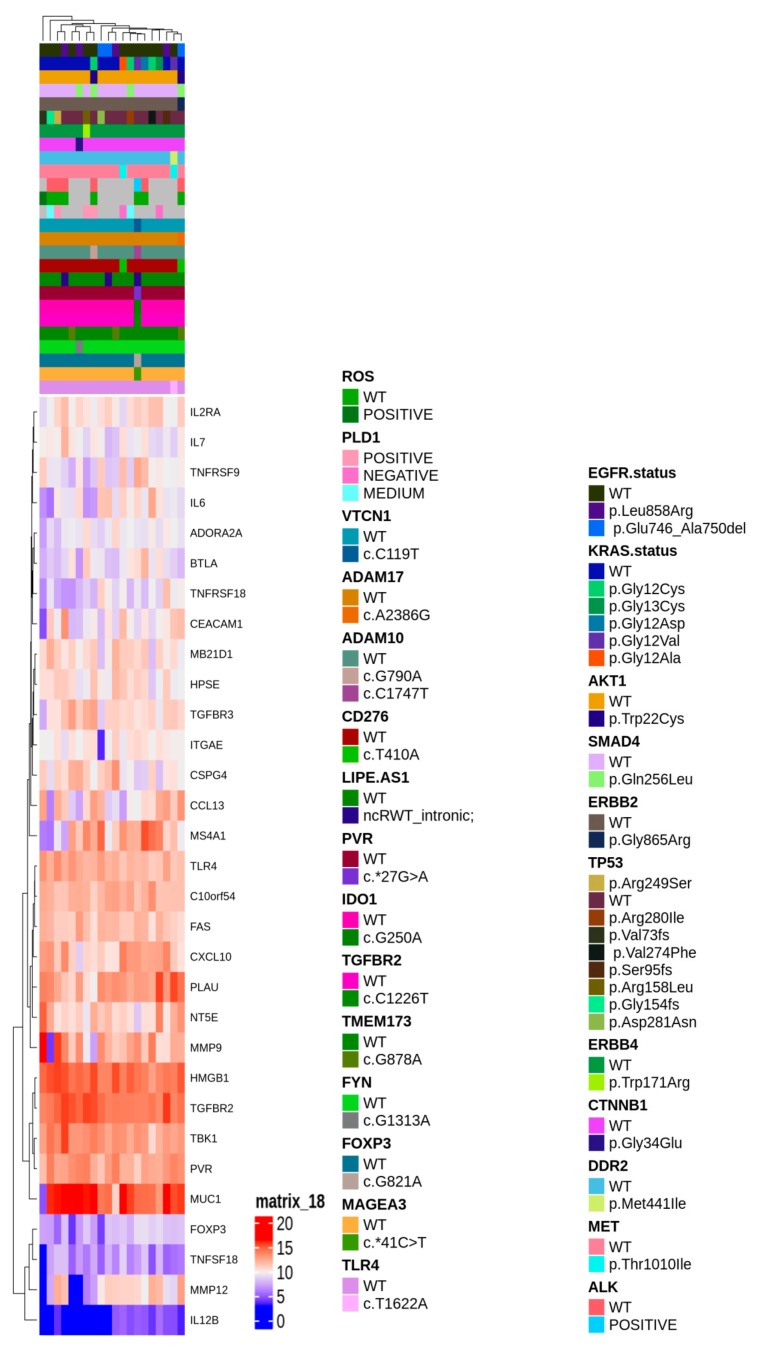
To gain insight of such deregulated genes, unsupervised hierarchical clustering of LUAD tumor samples was performed, which splitted the cohort into 2 clusters (Figure 2b). Thus, we aimed to identify a molecular feature able to distinguish the two identified clusters. A complex heatmap (Figure 3) including all molecular data available for our cohort (colon and lung panel results, ALK/ROS1 translocations and PD-L1 staining when available) was drawn. It could be seen that almost all KRAS mutated patients are grouped in cluster 2 (7/8 mutated cases in cluster 2) (Figure 3). In particular, alterations in KRAS are p.G12X and p.G13X, where X stands for the mutated amino acid.
Figure 3.
Complex heatmap annotated with all the genetic alterations detected in the real-life cohort.
2.3. Immune Infiltration Estimation and Overall Survival
The identification of two clusters differently enriched in KRAS mutation carrying patients and the identification of pathway/GO terms of the deregulated genes are not sufficient to infer the composition of microenvironment.
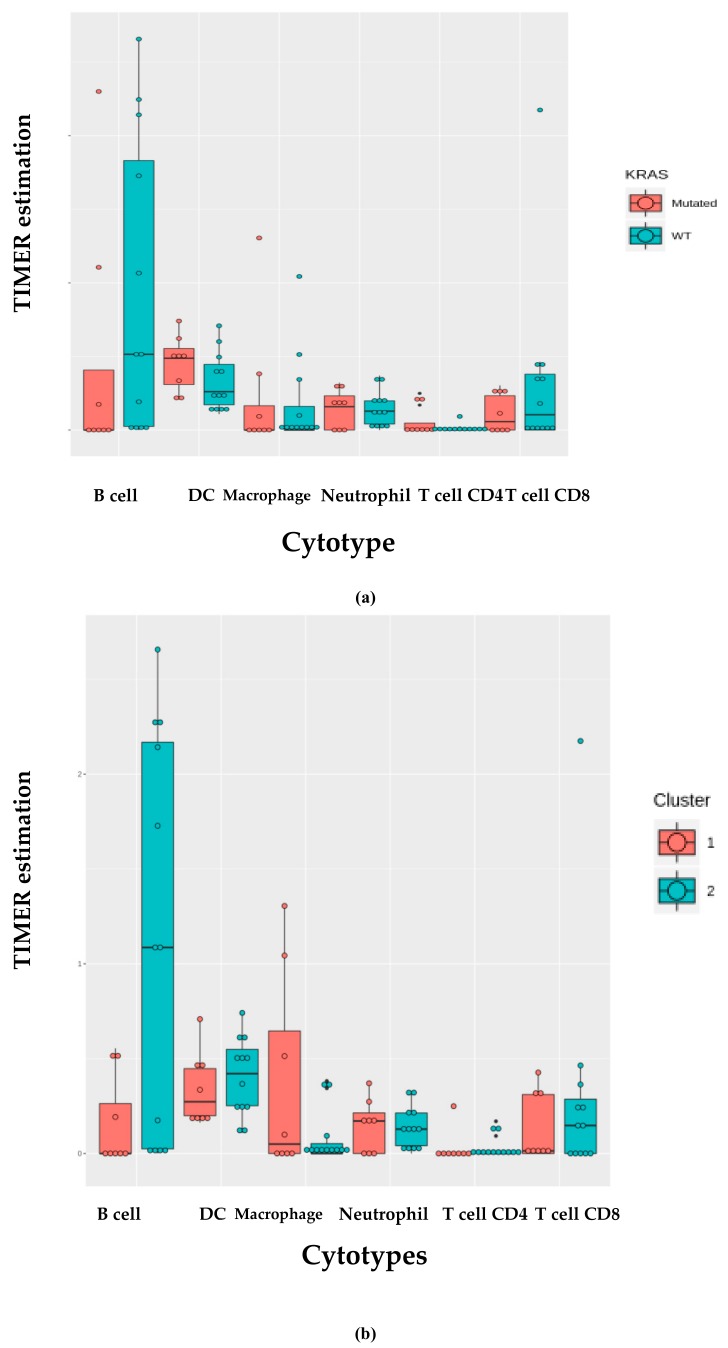
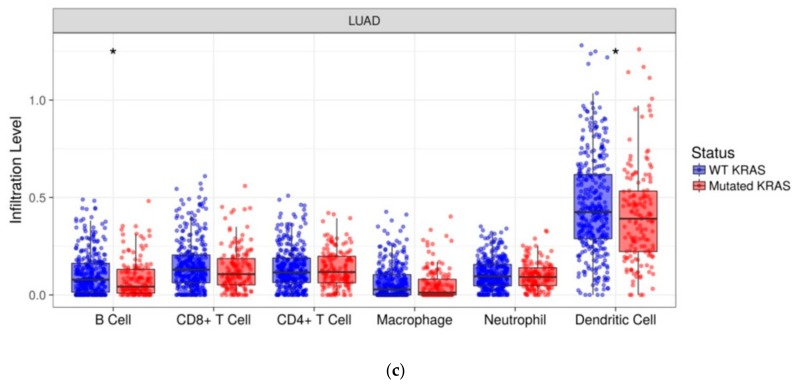
We investigated the immune infiltration through TIMER deconvolution approach, which is more reliable than other methods (e.g., CIBERSORT that suffers from collinearity bias). Estimation performed on our gene expression matrix showed that, after stratification based both on cluster and KRAS mutational status, B cell infiltration is lower in both cluster 2 subsets and KRAS mutated patients than in cases belonging to cluster 1and in KRAS WT (p = 0.06) (Figure 4a,b). Notably, also in the LUAD-TCGA dataset, B cell infiltration is significantly low in KRAS mutated patients (Figure 4c). Dendritic cell infiltration, which is significantly lower in KRAS mutated cases than wild-type ones in LUAD-TCGA, has an opposite behavior in the real-life cohort we analyzed.
Figure 4.
TIMER deconvolution from 95-gene expression matrix of real-life cohort stratifying for (a) KRAS mutational status and (b) cluster assignment; (c) TIMER estimation of TCGA-LUAD dataset.
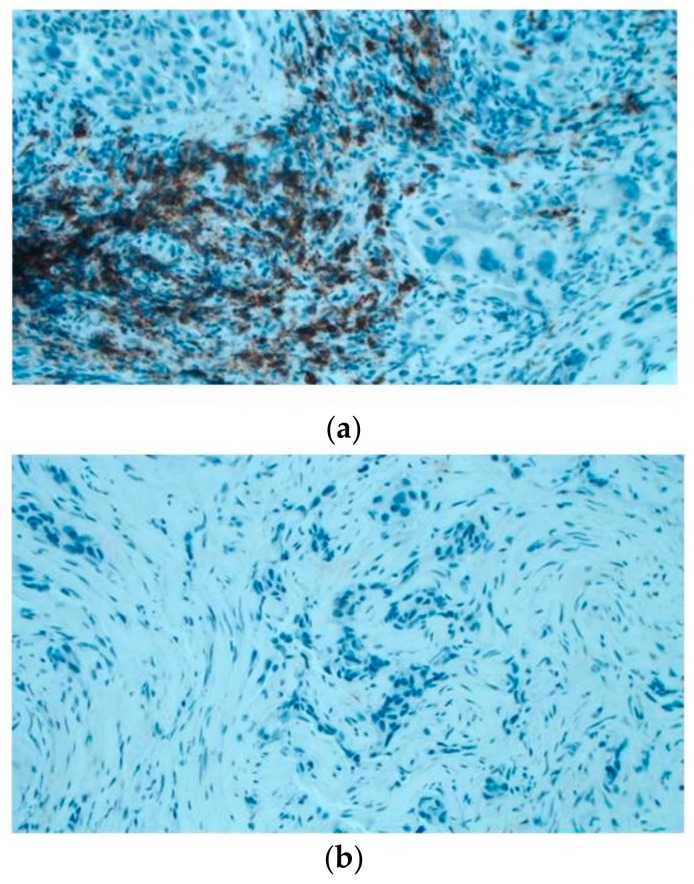
Then, the amount of B lymphocyte infiltration has been evaluated in situ through immunohistochemistry in an independent cohort including 15 cases. It was observed that 5/6 (83%) of KRAS WT cases have B cell infiltration (20%–70% of total infiltration); whilst 5/9 KRAS (55%) mutated showed no B cell infiltration (Figure 5).
Figure 5.
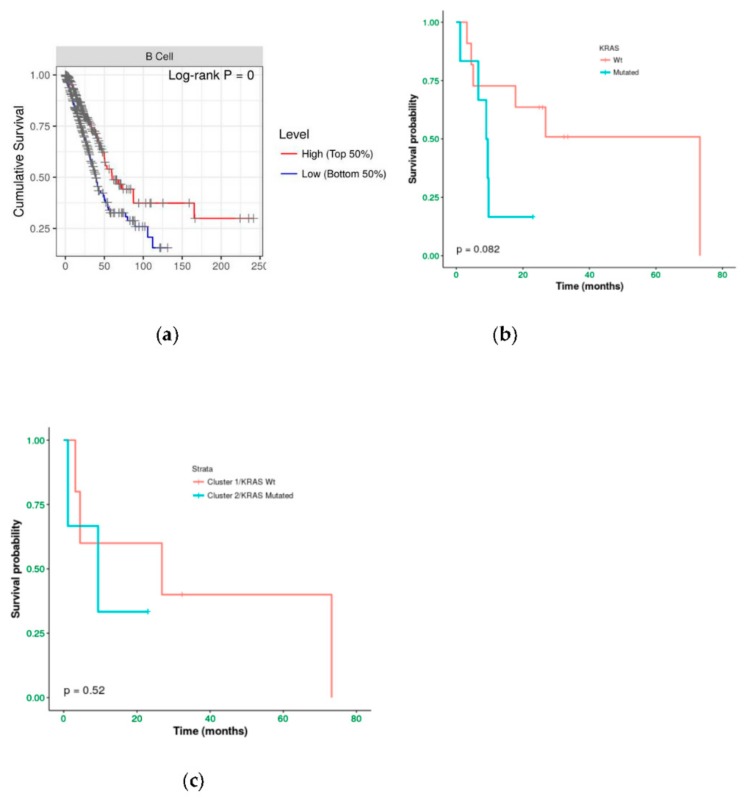
Immunoistochemical evaluation of B cell infiltration in (a) KRAS WT (60% of infiltration) and (b) mutated cases (0% infiltration), respectively (magnification: 20×) Moreover, cases in LUAD-TCGA with low B cell infiltration have a significantly worse overall survival than those with higher levels (Figure 6a). Interestingly, in the real-life cohort we observed the same results stratifying for presence/absence of KRAS mutation, with mutated patients having a poor outcome (Figure 6b). Moreover, mutated patients in cluster 2 have a worse overall survival than cluster 1 KRAS WT cases (Figure 6c).
Of note, immune infiltration estimation on LUAD-TCGA data has been performed on RNA-Seq data whilst we performed estimation in our cohort using a matrix including 95 genes. Thus, our custom RNA panel has a good performance in TIMER estimation, being able to confirm the result related to B cell infiltration.
3. Discussion
The advent of immunotherapies posed a step forward in the treatment of many solid tumors, in particular LUAD. In the present study, both the analysis of real-life cohorts and TCGA-LUAD data allowed to gain insights into the biology of such a disease. Through the set-up of two NGS custom panels, both for DNA and RNA, including genes related to major immunity checkpoints and pathways, it was possible to dissect different aspects of LUAD. 41-gene DNA panel evidenced no mutational enrichment. This is an important finding because mutational status of genes involved in immunological/inflammatory pathways could be less important than their fine regulation. Tumor mutational burden (TMB) is a potential biomarker, also in NSCLC, for response to immunotherapy targeting PD-1/PD-L1 [16,17]. If our panel is considered as a surrogate of TMB, thus our result confirmed previous findings highlighting the lower mutational burden of LUAD with respect to squamous cell carcinoma [18,19]. Moreover, the role of TMB as a predictive biomarker is still controversial, both if it is considered solely or in combination with other ones, such as PD-L1 expression. Regarding 95-gene panel expression analysis, 31 genes were detected as significantly deregulated comparing tumor/normal samples. Unsupervised hierarchical clustering of tumor samples evidenced two clusters, which were found to be differently enriched in KRAS-mutant samples. KRAS mutations are detected almost exclusively in LUAD [20,21] and they are associated to poor survival, in particular in advanced stages [22,23]. Moreover, KRAS mutant is biologically heterogeneous in terms of type of alteration and concomitant mutations. The type of KRAS mutation has been dissected with respect to patient survival. No specific distribution has been observed of the nine types of mutations at codons 12, 13 e 61 in NSCLC in the three co-occurring mutation clusters [24]. However, in vitro studies highlighted different behaviors on the downstream pathways, such a PI3K-AKT affinity for KRAS-G12C or KRAS-G12V [25]. In NSCLC patients KRAS-G12C has been correlated to poorer overall survival than KRAS-G12D mutants or KRAS-G12A [26]. Nevertheless, the association of specific KRAS mutations to survival needs more data and appropriate study design. The cohort of the present study carries only alterations at codon 12 and 13. In our cohort, patients in KRAS-mutant enriched cluster, 33% of samples carry a mutation in TP53, but only the half of them are KRAS/TP53 co-mutant. Of note, TP53 mutants in the other cluster are all KRAS wild-type. Thus, cluster 2 is enriched both in KRAS mutation carriers and KRAS/TP53 double mutants. Skoulidis et al. identified three subgroups of KRAS-driven LUAD based on co-occurring mutations. In particular, the so-called KP cluster, including KRAS/TP53 mutants, has been demonstrated to be featured by immunoediting, T-cell inflammation, and resistance to adaptive immune response and also JAK-STAT pathway activation and interferon signaling [24].
To gain a deeper knowledge of the microenvironment, TCGA-LUAD cohort has been analyzed through TIMER, a deconvolution algorithm, which highlighted a significant low infiltration level of B-cells and dentritic cells in KRAS mutants. B-cell result has been confirmed using the 95-gene panel expression matrix both stratifying by KRAS mutational status and clustering information and by IHC on an independent cohort. The function in anti-tumor immunity of B cells has emerged in lung cancer in several studies. Indeed, it has been shown that B lymphocytes are able to promote T cell activation and expansion. B cells, which are responsible for humoral immunity, are able in lung to differentiate into plasma cells producing antibodies against tumor antigens (e.g., TP53, NY-ESO-1) and the high concentration of tumor-antigen antibodies has been associated to a high proportion of follicular B cells [27]. Plasma cells, follicular B cells [28], and co-localization of CD8+ and CD4+ T cells and B lymphocytes are associated to long overall survival in NSCLC patients [29,30]. Such a result has been confirmed also in the TCGA-LUAD dataset. However, the association of B cell infiltration and KRAS alteration is the most interesting finding of the present study. The role of KRAS in the modulation of the tumor microenvironment has been demonstrated not only for the immune component but also the stromal one. GO/KEGG analysis evidenced a significant enrichment of terms related to Th17 lymphocytes and in the KRAS-mutants enriched cluster the upregulation of IL6 and MMP12 has been detected, confirming the crosstalk between tumor and Th17 cells [31,32]. To date, few data have demonstrated the link between the mutational status of KRAS and B cell. The role of KRAS in the B lymphopoiesis has been shown by Chen et al. [33] that demonstrated how the lack of KRAS in murine model impairs B cell development and maturation. Nevertheless, models to study the crosstalk between KRAS mutant cells and B lymphocytes are mandatory because this relationship needs to be studied to improve the (immune)therapeutic setting of LUAD patients. Indeed, recently Meng et al. [34] demonstrated the ability to target tumor neoantigens, as KRAS mutants, eliciting the humoral immune response through infiltrating B cells in pancreatic cancer. Such a mechanism is promising because it is similar to the NY-ESO-1 approach in melanoma [35,36]. Regarding NSCLC, it has been shown that in vitro cultivated infiltrating B cells can present antigens and activate CD4+ T cells [37]. It is a very interesting study on the role of immune regulation in TME of NSCLC. However, a major effort is needed to link mutations harbored by tumor to the capacity to present antigens of B cells.
In particular, KRAS mutation/low B cell infiltration association with respect to overall survival highlighted the possibility to set-up novel immunotherapies, such as the B lymphocytes-based approach. Indeed, recent clinical studies testing allogeneic B cells from PBMCs (peripheral blood mononuclear cells) of donors in renal cell tumor and melanoma [38] and adoptive B-cell transfer approach [39] showed promising results.
In conclusion, the present study, exploiting through genomic data the biology of LUAD, confirmed the importance of the mutational status regarding the relationship between the tumor cell and microenvironment. Intriguingly, the association of KRAS alterations to B cell infiltration and survival shed a light to the set-up of new therapeutic approaches. Moreover, the results coming from deconvolution on the expression matrix of the custom targeted panel demonstrated that it could be a useful tool to explore the TME composition, as demonstrated through the TCGA-LUAD dataset molecular data, and in situ validations.
4. Materials and Methods
4.1. Sample Cohort
Twenty advanced-stage (IIIB and IV) NSCLC patients were enrolled by the Department of Thoracic Oncology of the Giovanni Paolo II Institute of Bari between November 2016 and May 2017.Evaluation of survival was performed according to RECIST 1.1 criteria with a median time OS time of 9.67 months. Moreover 8 patients who underwent surgery with lung tumor free diagnosis were enrolled by the Department of Surgery of San Paolo Hospital, Bari. All cases were diagnosed according to the current WHO guidelines. A cohort including 15 LUAD cases, whose KRAS mutational status was known, was used to validate deconvolution results by the in situ approach.
The study was approved by the local Ethics Committee of the IRCCS “Giovanni Paolo II” of Bari (CE prot. n. 540/2015) and was performed in accordance with the international standards of good clinical practice. All patients signed informed consent.
Blood samples from healthy patients were collected and stored as germline biological material.
4.2. DNA and RNA Extraction
Between three to six FFPE tissue sections (6 μm thick) with adequate tumor cellularity selected by a pathologist (>50%), were macro dissected and subjected to DNA and RNA isolation using QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany) and RNeasy FFPE Kit(Qiagen), respectively, according to manufacturers’ protocols. DNA was also isolated from blood samples using QIAamp DNA Blood Midi Kit (Qiagen). DNA and RNA concentration was determined using Qubit 2.0 fluorometer (Thermo Fisher Scientific, Waltham, MA, USA).
4.3. Ion Torrent PGM Sequencing
Sequencing was performed as previously reported in [40]. Briefly, two custom panels were designed through the Ion Ampliseq designer tool, one including the coding region of 41 genes to detect DNA alterations and one to study the gene expression of 95 genes. Both of them include genes involved in immune regulation and inflammation. Moreover, also CE-IVD Oncomine Solid Tumor (Thermo Fischer Scientific) was used to assess the mutational status of actionable genes.
All generated reads were aligned to human genome hg19 using the Torrent Suite Server.
4.4. Variant Calling and Filtering
Variant calling and filtering was described in [40]. Briefly, the callsetwas generated merging results from the Somatic High-Stringency Variant Caller plugin of Torrent Suite and Vardict [41] algorithm. Germline variants were filtered out using a pool of healthy controls. Annovar [42] was used to functionally annotate variants. Regarding results of the CE-IVD Oncomine Solid Tumor panel, an optimized Ion Reporter workflow was used and only variants annotated in COSMIC (Catalog Of Somatic Mutations in Cancer) database (https://cancer.sanger.ac.uk/cosmic).
The OncoPrint plot was designed with the ComplexHeatmap R package [43].
4.5. Gene Expression Detection
Gene expression was evaluated, as stated above, through a custom panel including 95 genes. Ten (10)ng of input RNA were reverse transcribed for target enrichment by the Ion AmpliSeq Library Kit 2.0 (Thermo Fisher Scientific). Libraries were then quantified and pooled in an equimolar ratio. Then libraries were pooled and templated using the Ion OneTouch 2 system. Sequencing of the Ion 318 chip was carried out on the Personal Genome Machine (PGM, Thermo Fisher Scientific) according to the Ion PGM Sequencing 200 Kit (Thermo Fisher Scientific). Read count matrix was downloaded from the Torrent Server and analyzed with DeSeq2 R package [44]. Deconvolution was performed through the web interface of TIMER (https://cistrome.shinyapps.io/timer/) both to the analyzed TCGA-LUAD dataset and real-life cohort [10].
4.6. Immunohistochemistry and Immunohistochemical Assessment
Paraffin-embedded tissue sections of patients fixed in formalin were cut into 4µm, the slides were processed and stained by the indirect immunoperoxidase method using the Dako Omnis Platform (Dako, Santa Clara, CA, USA). The slides underwent deparaffinization, followed by antigen retrieval with EnVision FLEX Target Retrieval Solution, Low pH for PD-L1, and High pH for CD20. The sections were incubated with primary antibodies Monoclonal Mouse Anti-Human PD-L1 (Clone 22C3; Dako Omnis; diluited at 1:30) and Monoclonal Mouse Anti-Human CD20cy (Clone L26; Dako Omnis; ready-to-Use). After endogenous peroxidase activity blocking with EnVision FLEX Peroxidase-Blocking Reagent, the products of the antigen–antibody reactions were visualized by EnVision FLEX DAB + Chromogen and EnVsion FLEX Substrate Buffer. Cell nuclei were stained with Hematoxylin (8 min). Tonsil FFPE sections were used as positive controls. For negative control, the primary antibody was omitted.
5. Conclusions
The present paper, for the first time, links KRAS mutational status, which is responsible for a poor survival in LUAD patients, and B cell infiltration, which, in turn, has been indicated as a good prognostic marker. Notwithstanding the small size of the real-life cohort, the in silico exploration of the TCGA-LUAD dataset and the immunohistochemistry validation of B cell infiltration on an independent cohort indicate that the results could be further explored in a more extensive way. The aforementioned relationship between KRAS and B lymphocytes, moreover, will be explored in a larger cohort, including also LUAD patients treated with standard immunotherapies. To gain functional insights into the response to immune checkpoint inhibitors and the link between KRAS and B cells,anin vistrostudy will be planned.
Figure 6.
Kaplan–Meier curves of (a) TCGA-LUAD cohort stratifying for B cell infiltration and real-life cohort stratified for (b) KRAS mutational status and (c) cluster assignment.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/11/8/1145/s1, Figure S1: Expression of the deregulated genes in the real life cohort in TCGA-LUAD dataset.
Author Contributions
Investigation, P.R., P.D., L.R., B.M.A., D.B.G., Z.A.; writing—review and editing, M.M., C.A., P.P., Z.F.A.; validation, B.M.A.; funding acquisition, T.S.; supervision, T.S.; formal analysis, visualization, writing—original draft preparation, D.S.S.
Funding
This research was funded by Italian Ministry of Health (RC-2018).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Kumarakulasinghe N.B., Zanwijk N., Soo R.A. Molecular targeted therapy in the treatment of advanced stage non-small cell lung cancer (NSCLC) Respirology. 2015;20:370–378. doi: 10.1111/resp.12490. [DOI] [PubMed] [Google Scholar]
- 2.Schiller J.H., Harrington D., Belani C.P., Langer C., Sandler A., Krook J., Zhu J., Johnson D.H. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N. Engl. J. Med. 2002;346:92–98. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 3.Liu K., Guo J., Liu K., Fan P., Zeng Y., Xu C., Zhong J., Li Q., Zhou Y. Integrative analysis reveals distinct subtypes with therapeutic implications in KRAS-mutant lung adenocarcinoma. EBioMedicine. 2018;36:196–208. doi: 10.1016/j.ebiom.2018.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen N., Fang W., Lin Z., Peng P., Wang J., Zhan J., Hong S., Huang J., Liu L., Sheng J., et al. KRAS mutation-induced upregulation of PD-L1 mediates immune escape in human lung adenocarcinoma. Cancer Immunol. Immunother. 2017;66:1175–1187. doi: 10.1007/s00262-017-2005-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lesterhuis W.J., Haanen J.B., Punt C.J. Cancer immunotherapy–Revisited. Nat. Rev. Drug Discov. 2011;10:591–600. doi: 10.1038/nrd3500. [DOI] [PubMed] [Google Scholar]
- 6.Tran E., Robbins P.F., Lu Y.C., Prickett T.D., Gartner J.J., Jia L., Pasetto A., Zheng Z., Ray S., Groh E.M., et al. T-Cell Transfer Therapy Targeting Mutant KRAS in Cancer. N. Engl. J. Med. 2016;375:2255–2262. doi: 10.1056/NEJMoa1609279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y.T., Scanlan M.J., Sahin U., Tureci O., Gure A.O., Tsang S., Williamson B., Stockert E., Pfreundschuh M., Old L.J., et al. A testicular antigen aberrantly expressed in human cancers detected by autologous antibody screening. Proc. Natl. Acad. Sci. USA. 1997;94:1914–1918. doi: 10.1073/pnas.94.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neumann F., Wagner C., Kubuschok B., Stevanovic S., Rammensee H.G., Pfreundschuh M. Identification of an antigenic peptide derived from the cancer-testis antigen NY-ESO-1 binding to a broad range of HLA-DR subtypes. Cancer Immunol. Immunother. 2004;53:589–599. doi: 10.1007/s00262-003-0492-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miura Y., Sunaga N. Role of Immunotherapy for Oncogene-Driven Non-Small Cell Lung Cancer. Cancers. 2018;10:245. doi: 10.3390/cancers10080245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCoach C.E., Bivona T.G. The evolving understanding of immunoediting and the clinical impact of immune escape. J. Thorac. Dis. 2018;10:1248–1252. doi: 10.21037/jtd.2018.03.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dempke W.C.M., Fenchel K., Dale S.P. Programmed cell death ligand-1 (PD-L1) as a biomarker for non-small cell lung cancer (NSCLC) treatment-are we barking up the wrong tree? Transl. Lung Cancer Res. 2018;7:S275–S279. doi: 10.21037/tlcr.2018.04.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diggs L.P., Hsueh E.C. Utility of PD-L1 immunohistochemistry assays for predicting PD-1/PD-L1 inhibitor response. Biomark. Res. 2017;5:12. doi: 10.1186/s40364-017-0093-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altorki N.K., Markowitz G.J., Gao D., Port J.L., Saxena A., Stiles B., McGraw T., Mittal V. The lung microenvironment: An important regulator of tumour growth and metastasis. Nat. Rev. Cancer. 2019;19:9–31. doi: 10.1038/s41568-018-0081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y., Hoang C.D., Diehn M., Alizadeh A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li T., Fan J., Wang B., Traugh N., Chen Q., Liu J.S., Li B., Liu X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017;77:e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yarchoan M., Hopkins A., Jaffee E.M. Tumor Mutational Burden and Response Rate to PD-1 Inhibition. N. Engl. J. Med. 2017;377:2500–2501. doi: 10.1056/NEJMc1713444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yarchoan M., Johnson B.A., 3rd, Lutz E.R., Laheru D.A., Jaffee E.M. Targeting neoantigens to augment antitumour immunity. Nat. Rev. Cancer. 2017;17:569. doi: 10.1038/nrc.2017.74. [DOI] [PubMed] [Google Scholar]
- 18.Kleppe M., Levine R.L. Tumor heterogeneity confounds and illuminates: Assessing the implications. Nat. Med. 2014;20:342–344. doi: 10.1038/nm.3522. [DOI] [PubMed] [Google Scholar]
- 19.Chalmers Z.R., Connelly C.F., Fabrizio D., Gay L., Ali S.M., Ennis R., Schrock A., Campbell B., Shlien A., Chmielecki J., et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9:34. doi: 10.1186/s13073-017-0424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Project, The Clinical Lung Cancer Genome; Network Genomic Medicine NGM. A genomics-based classification of human lung tumors. Sci. Transl. Med. 2013;5:209ra153. doi: 10.1126/scitranslmed.3006802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cancer Genome Atlas Research Network Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mascaux C., Iannino N., Martin B., Paesmans M., Berghmans T., Dusart M., Haller A., Lothaire P., Meert A.P., Noel S., et al. The role of RAS oncogene in survival of patients with lung cancer: A systematic review of the literature with meta-analysis. Br. J. Cancer. 2005;92:131–139. doi: 10.1038/sj.bjc.6602258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marabese M., Ganzinelli M., Garassino M.C., Shepherd F.A., Piva S., Caiola E., Macerelli M., Bettini A., Lauricella C., Floriani I., et al. KRAS mutations affect prognosis of non-small-cell lung cancer patients treated with first-line platinum containing chemotherapy. Oncotarget. 2015;6:34014–34022. doi: 10.18632/oncotarget.5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skoulidis F., Byers L.A., Diao L., Papadimitrakopoulou V.A., Tong P., Izzo J., Behrens C., Kadara H., Parra E.R., Canales J.R., et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 2015;5:860–877. doi: 10.1158/2159-8290.CD-14-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ihle N.T., Byers L.A., Kim E.S., Saintigny P., Lee J.J., Blumenschein G.R., Tsao A., Liu S., Larsen J.E., Wang J., et al. Effect of KRAS oncogene substitutions on protein behavior: Implications for signaling and clinical outcome. J. Natl. Cancer Inst. 2012;104:228–239. doi: 10.1093/jnci/djr523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nadal E., Chen G., Prensner J.R., Shiratsuchi H., Sam C., Zhao L., Kalemkerian G.P., Brenner D., Lin J., Reddy R.M., et al. KRAS-G12C mutation is associated with poor outcome in surgically resected lung adenocarcinoma. J. Thorac. Oncol. 2014;9:1513–1522. doi: 10.1097/JTO.0000000000000305. [DOI] [PubMed] [Google Scholar]
- 27.Germain C., Gnjatic S., Tamzalit F., Knockaert S., Remark R., Goc J., Lepelley A., Becht E., Katsahian S., Bizouard G., et al. Presence of B cells in tertiary lymphoid structures is associated with a protective immunity in patients with lung cancer. Am. J. Respir. Crit. Care Med. 2014;189:832–844. doi: 10.1164/rccm.201309-1611OC. [DOI] [PubMed] [Google Scholar]
- 28.Lohr M., Edlund K., Botling J., Hammad S., Hellwig B., Othman A., Berglund A., Lambe M., Holmberg L., Ekman S., et al. The prognostic relevance of tumour-infiltrating plasma cells and immunoglobulin kappa C indicates an important role of the humoral immune response in non-small cell lung cancer. Cancer Lett. 2013;333:222–228. doi: 10.1016/j.canlet.2013.01.036. [DOI] [PubMed] [Google Scholar]
- 29.Schalper K.A., Brown J., Carvajal-Hausdorf D., McLaughlin J., Velcheti V., Syrigos K.N., Herbst R.S., Rimm D.L. Objective measurement and clinical significance of TILs in non-small cell lung cancer. J. Natl. Cancer Inst. 2015;107 doi: 10.1093/jnci/dju435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kinoshita T., Muramatsu R., Fujita T., Nagumo H., Sakurai T., Noji S., Takahata E., Yaguchi T., Tsukamoto N., Kudo-Saito C., et al. Prognostic value of tumor-infiltrating lymphocytes differs depending on histological type and smoking habit in completely resected non-small-cell lung cancer. Ann. Oncol. 2016;27:2117–2123. doi: 10.1093/annonc/mdw319. [DOI] [PubMed] [Google Scholar]
- 31.Chang S.H. Tumorigenic Th17 cells in oncogenic Kras-driven and inflammation-accelerated lung cancer. Oncoimmunology. 2015;4:e955704. doi: 10.4161/21624011.2014.955704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McAllister F., Bailey J.M., Alsina J., Nirschl C.J., Sharma R., Fan H., Rattigan Y., Roeser J.C., Lankapalli R.H., Zhang H., et al. Oncogenic Kras activates a hematopoietic-to-epithelial IL-17 signaling axis in preinvasive pancreatic neoplasia. Cancer Cell. 2014;25:621–637. doi: 10.1016/j.ccr.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y., Zheng Y., You X., Yu M., Fu G., Su X., Zhou F., Zhu W., Wu Z., Zhang J., et al. Kras Is Critical for B Cell Lymphopoiesis. J. Immunol. 2016;196:1678–1685. doi: 10.4049/jimmunol.1502112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meng Q., Valentini D., Rao M., Maeurer M. KRAS RENAISSANCE(S) in Tumor Infiltrating B Cells in Pancreatic Cancer. Front. Oncol. 2018;8:384. doi: 10.3389/fonc.2018.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jager E., Chen Y.T., Drijfhout J.W., Karbach J., Ringhoffer M., Jager D., Arand M., Wada H., Noguchi Y., Stockert E., et al. Simultaneous humoral and cellular immune response against cancer-testis antigen NY-ESO-1: Definition of human histocompatibility leukocyte antigen (HLA)-A2-binding peptide epitopes. J. Exp. Med. 1998;187:265–270. doi: 10.1084/jem.187.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jager E., Nagata Y., Gnjatic S., Wada H., Stockert E., Karbach J., Dunbar P.R., Lee S.Y., Jungbluth A., Jager D., et al. Monitoring CD8 T cell responses to NY-ESO-1: Correlation of humoral and cellular immune responses. Proc. Natl. Acad. Sci. USA. 2000;97:4760–4765. doi: 10.1073/pnas.97.9.4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bruno T.C., Ebner P.J., Moore B.L., Squalls O.G., Waugh K.A., Eruslanov E.B., Singhal S., Mitchell J.D., Franklin W.A., Merrick D.T., et al. Antigen-Presenting Intratumoral B Cells Affect CD4(+) TIL Phenotypes in Non-Small Cell Lung Cancer Patients. Cancer Immunol. Res. 2017;5:898–907. doi: 10.1158/2326-6066.CIR-17-0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeng M., Kikuchi H., Pino M.S., Chung D.C. Hypoxia activates the K-ras proto-oncogene to stimulate angiogenesis and inhibit apoptosis in colon cancer cells. PLoS ONE. 2010;5:e10966. doi: 10.1371/journal.pone.0010966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sparmann A., Bar-Sagi D. Ras-induced interleukin-8 expression plays a critical role in tumor growth and angiogenesis. Cancer Cell. 2004;6:447–458. doi: 10.1016/j.ccr.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 40.Silvestris N., Brunetti O., Pinto R., Petriella D., Argentiero A., Fucci L., Tommasi S., Danza K., De Summa S. Immunological mutational signature in adenosquamous cancer of pancreas: An exploratory study of potentially therapeutic targets. Expert Opin. Ther. Targets. 2018;22:453–461. doi: 10.1080/14728222.2018.1456530. [DOI] [PubMed] [Google Scholar]
- 41.Lai Z., Markovets A., Ahdesmaki M., Chapman B., Hofmann O., McEwen R., Johnson J., Dougherty B., Barrett J.C., Dry J.R., et al. VarDict: A novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 2016;44:e108. doi: 10.1093/nar/gkw227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang H., Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat. Protoc. 2015;10:1556–1566. doi: 10.1038/nprot.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 44.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.