Abstract
Liquid biopsy (LB) is a non-invasive approach representing a promising tool for new precision medicine strategies for cancer treatment. However, a comprehensive analysis of its reliability for pancreatic cancer (PC) is lacking. To this aim, we performed the first meta-analysis on this topic. We calculated the pooled sensitivity, specificity, positive (LR+) and negative (LR−) likelihood ratio, and diagnostic odds ratio (DOR). A summary receiver operating characteristic curve (SROC) and area under curve (AUC) were used to evaluate the overall accuracy. We finally assessed the concordance rate of all mutations detected by multi-genes panels. Fourteen eligible studies involving 369 patients were included. The overall pooled sensitivity and specificity were 0.70 and 0.86, respectively. The LR+ was 3.85, the LR- was 0.34 and DOR was 15.84. The SROC curve with an AUC of 0.88 indicated a relatively high accuracy of LB for molecular characterization of PC. The concordance rate of all mutations detected by multi-genes panels was 31.9%. LB can serve as surrogate for tissue in the molecular profiling of PC, because of its relatively high sensitivity, specificity and accuracy. It represents a unique opportunity to be further explored towards its introduction in clinical practice and for developing new precision medicine approaches against PC.
Keywords: liquid biopsy, cfDNA, pancreatic cancer, precision medicine, circulating tumor cells (CTC)
1. Introduction
Pancreatic cancer is a lethal malignancy with an increasing incidence, and it is projected to become the second most common cause of cancer-related death in the world at the end of the next decade [1,2,3]. The most common type of pancreatic cancer is represented by ductal adenocarcinoma (PDAC), which is responsible for more than 95% of deaths from pancreatic cancer [3,4].
At the time of diagnosis, the vast majority of patients with PDAC present with locally advanced or metastatic disease and are thus not amenable to surgical resection with a curative purpose [3]. For such patients, the priority is to receive an undebatable diagnosis of cancer including useful information for addressing all potential therapeutic approaches. However, obtaining tissue for the diagnosis in this setting may be difficult and requires invasive procedures. The gold standard is (endoscopic-) ultrasound guided fine-needle aspiration (US-FNA), which provides a limited number of cells for cytological analysis, not always allowing complete molecular profiling [5,6]. Moreover, the abundance of tumor stroma in PDAC impairs the negative predictive value of US-FNA because of sampling error, sometimes requiring a repeat of the procedure with further risks for PDAC patients [6,7].
For all these reasons, there is an urgent need for new techniques or biomarkers to aid in diagnosis, staging and clinical-therapeutic decisions, overcoming contraindications associated with invasive procedures. To date, the unique widely available blood-based test for patients with PDAC is the determination of the carbohydrate antigen 19-9 (CA19-9); however, because of its limited specificity, it is used only as generic support for diagnosis and for surveillance of recurrence [8].
Notably, recent studies have explored the possibilities of obtaining from the blood useful information for cancer treatment analyzing circulating tumor cells (CTCs), circulating-free DNA (cfDNA) or RNA, exosomes and secretomes, all parameters that are comprised under the general definition of liquid biopsy (LB) [9,10,11]. This kind of non-invasive approach represents a promising tool for new precision medicine strategies, since LB may be used also for molecular profiling [12,13,14,15].
Although some studies have explored and highlighted the usefulness of LB for genetic analysis of patients with PDAC [16,17,18,19,20], a comprehensive analysis of its reliability for this tumor type is lacking. With this first systematic review and meta-analysis on this topic, we aimed to highlight the reliability of LB in assessing the molecular profile of PDAC in comparison with the mutational analysis on tissue specimen. In addition, a specific focus on KRAS mutational status is provided, as this gene is the most frequently mutated driver gene in PDAC.
2. Results
2.1. Search Results and Descriptive Findings
Among 387 potential eligible studies, 60 full text articles were retrieved. Of them, 14 studies were eligible for this meta-analysis (Figure S1, Table 1) [21,22,23,24,25,26,27,28,29,30,31,32,33,34].
Table 1.
Summarizing table of the most relevant features of the studies included in this meta-analysis.
| First Author of the Study, Year [21,22,23,24,25,26,27,28,29,30,31,32,33,34] | N. of Patients | Stage | Type of Tissue Specimen | Molecular Test for Tissue Specimen and Genes | Time Point of Tissue and Liquid Biopsy Test and Genes | Type of Liquid Biopsy | Molecular Test for Liquid Biopsy | TP | FP | TN | FN |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ako, 2017 [21] | 40 | I–II: 60%, III–IV: 40% | 16 SR and 24 EUS-FNA | PCR, KRAS | The same time | Plasma and serum, cfDNA | Droplet PCR, KRAS | 19 | 0 | 3 | 18 |
| Bernard, 2019a [22] | 34 | I–II: 68%, III–IV: 32% | 22 SR and 12 EUS-FNA | PCR, KRAS | The same time | Blood for cfDNA | Droplet digital PCR, KRAS | 20 | 1 | 11 | 2 |
| Brychta, 2016 [23] | 50 | I–II: 82%, III–IV: 18% | SR | Chip-based digital PCR, KRAS | The same time | Plasma (cfDNA) | Chip-based digital PCR, KRAS | 13 | 0 | 14 | 23 |
| Earl, 2015 [24] | 12 | NA | SR | PCR, KRAS | The same time | Plasma (cfDNA) | Droplet digital PCR, KRAS | 3 | 3 | 2 | 4 |
| Kinugasa, 2015 [25] | 75 | I–II: 3%, III–IV: 97% | EUS-FNA | PCR, KRAS | The same time | Serum (cfDNA) | Droplet digital PCR, KRAS | 43 | 4 | 15 | 13 |
| Kulemann, 2016 [26] | 11 | I–II: 91%, III–IV: 9% | NS | PCR, KRAS | Retrospective | Blood with isolation and analysis of CTCs | PCR, KRAS | 5 | 0 | 0 | 6 |
| Marchese, 2006 [27] | 30 | I–II: 83%,III–IV: 17% | 25 SR, 5 EUS-FNA | rflp-PCR KRAS | The same time | Serum (cfDNA) | rflp-PCR KRAS | 0 | 0 | 9 | 21 |
| Park, 2018a [28] | 17 | I–II: 18%,III–IV: 82% | EUS-FNA | PCR, KRAS | The same time | Plasma (cfDNA) | PCR, KRAS | 10 | 0 | 4 | 3 |
| Pishvaian, 2017a,* [29] | 16 | I–II: 0%,III–IV: 100% | EUS-FNA of pancreas or metastasis | 321 genes panel NGS | During treatment | cfDNA | 68 genes panel NGS | 6 | 1 | 0 | 9 |
| Sefrioui, 2017 [30] | 27 | NS | EUS-FNA/biopsy/SR | Digital PCR, KRAS | The same time | Plasma (cfDNA) | Digital PCR, KRAS | 14 | 3 | 5 | 5 |
| Shibata, 1998 [31] | 3 | I–II: 66.6%, III–IV: 33.3% | NS | nPCR, KRAS | The same time | Peripheral blood (CTCs separation) | nPCR, KRAS | 3 | 0 | 0 | 0 |
| Vietsch, 2018a,* [32] | 5 | I–II: 100%, III–IV: 0% | SR | 56 genes panel NGS | LB before surgery | cfDNA | 56 genes panel NGS | 0 | 0 | 0 | 5 |
| Wu, 2014 [33] | 36 | NS | NS | COLD-PCR, KRAS | The same time | Plasma (cfDNA) | COLD-PCR, KRAS | 26 | 0 | 10 | 0 |
| Zill, 2015 [34] | 13 | NS | EUS-FNA | NGS | The same time | Plasma (cfDNA) | 54 genes panel NGS | 12 | 0 | 0 | 1 |
| Total | 369 | I–II: 57%, III–IV: 43% | 11 studies: same time, 3 studies: other times | 12 studies: cfDNA from blood, 2 studies: CTCs separation | 174 | 12 | 73 | 110 |
Abbreviation: TP: true positive, FP: false positive, TN: true negative, FN: false negative; SR: surgical resected specimen; EUS-FNA: endoscopic ultrasound-guided fine-needle aspiration; cfDNA: circulating-free DNA; CTCs: circulating tumor cells; NS: not specified; rflp-PCR: restriction fragment length polymorphism-polymerase chain reaction; NGS: targeted next-generation sequencing for specific pancreatic cancer genes; nPCR: nested PCR; LB: liquid biopsy; COLD-PCR: co-amplification-at-lower-denaturation-temperature polymerase chain reaction; Note: Bernard, 2019a refers to the analysis of exoDNA in liquid biopsy; Park, 2018a refers to the analysis using PCR; Pishvaian, 2017a refers to a cohort of patients in which cfDNA has been analyzed; Vietsch, 2018a refers to patients whose liquid biopsy has been analyzed before surgical resection of pancreatic tumor; * in these studies, the rate of concordance of mutations between tumor tissue and liquid biopsy has been shown taking into account the four most important genes in pancreatic cancer (KRAS, TP53, SMAD4, and CDKN2A).
As reported in Table 1, the 14 studies eligible included 369 participants with pancreatic cancer, with a higher prevalence of stage I–II (57%) [21,22,23,24,25,26,27,28,29,30,31,32,33,34]. Eleven studies made the LB assessment at the same time as the tissue sample [21,22,23,24,25,27,28,30,31,33,34], and the remaining three at different time points [26,29,32]. The choice of tissue type for mutational analysis has been clearly specified in 11 studies: four studies used PDAC tissue from US-FNA [25,28,29,34], three from surgical resection [23,24,32] and four used PDAC tissue from US-FNA in case of metastatic or locally advanced PDAC and surgical resected tissue in case of early stage-PDAC [21,22,27,30]. The choice of the material as LB was represented in the vast majority of studies (12/14, 86%) by circulating-free DNA (cfDNA) from blood [21,22,23,24,25,27,28,29,30,32,33,34], whereas only two studies explored the use of circulating tumor cells (CTCs) for the same aim [26,31]. Eleven studies used polymerase chain reaction (PCR)-based techniques for the mutational analysis of KRAS status [21,22,23,24,25,26,27,28,30,31,33], whereas three studies performed next-generation sequencing (NGS) for a wider molecular analysis [29,32,34]. Overall, we identified 174 TP, 12 FP, 73 TN, 110 FN.
2.2. Quality of the Studies Included
As shown in Table S1, the risk of bias in the studies included is low, having only two studies reporting possible high risk of bias for flow and timing. However, it is unlikely that this affects our results since our stringent criteria for the selection of the studies ensure the high quality of the studies and of the specific procedures that have been performed.
2.3. Diagnostic Accuracy of the Liquid Biopsy
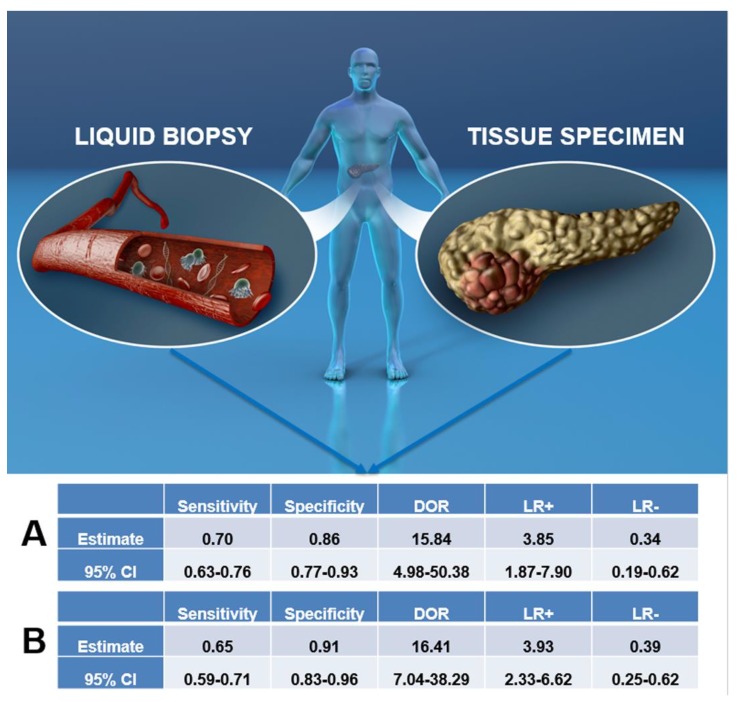
As reported in Figure 1A and Figure 2A, the overall pooled sensitivity and specificity were 0.70 (95% CI, 0.63–0.76) and 0.86 (95% CI, 0.77–0.93) respectively. It has to be clear that these summarizing values are referring to sensitivity and specificity with respect to concordance between LB and tissue molecular analyses and not with respect to cancer diagnosis more generally.
Figure 1.
This figure represents the core of this meta-analysis, in which we tested the reliability of liquid biopsy to serve as surrogate for tissue for molecular profiling of pancreatic cancer. The upper panel is a schematic representation of the differences between the materials used for liquid biopsy (above all from the blood, on the left) and for tissue specimen analysis (surgical resections or cytology, on the right). The section indicated with letter A shows the overall summary of diagnostic accuracy parameters for liquid biopsy compared to tissue specimen with this meta-analysis, whereas the section with letter B shows the same parameters, but obtained analyzing data regarding KRAS only. Abbreviations: CI: confidence interval; DOR: diagnostic odds ratio; LR+: positive likelihood ratio, LR−: negative likelihood ratio.
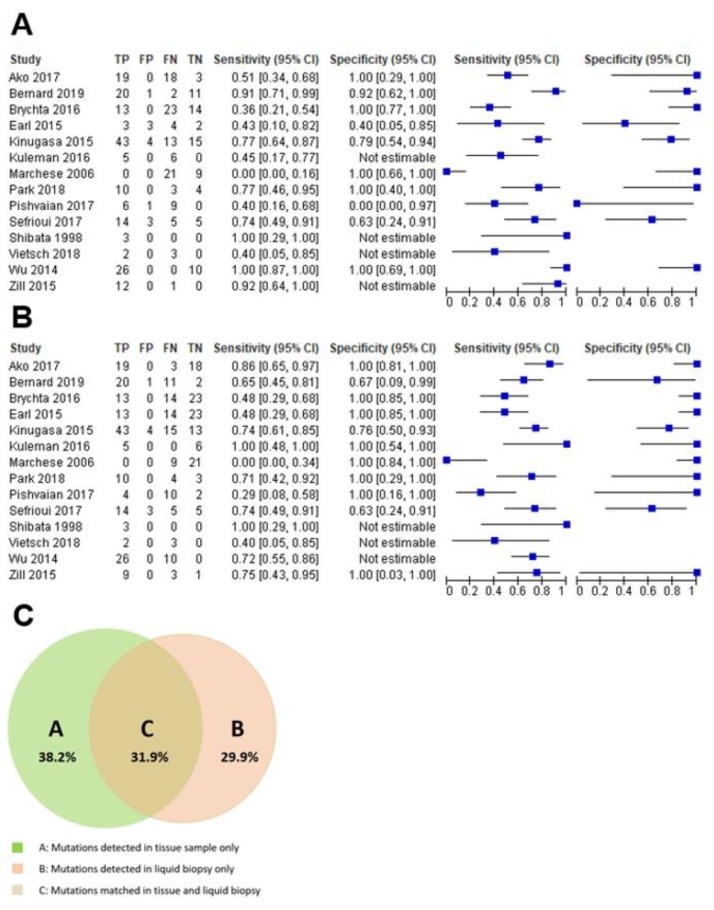
Figure 2.
(A) shows the sensitivity and specificity meta-analysis plot for the appropriateness of liquid biopsy compared to tissue specimen, whereas (B) shows the same plot, but obtained analyzing data regarding KRAS mutational status only. (C) is a Venn diagram showing, for studies based on next-generation sequencing, the concordance rate of all mutations detected by the analysis on tissue specimens and by the analysis on liquid biopsy. The area on the left (green area) indicates mutations detected only in tissue samples, the area on the right (red area) indicates mutations detected only in liquid biopsy, whereas the middle area indicates mutations detected in both tissue samples and liquid biopsy.
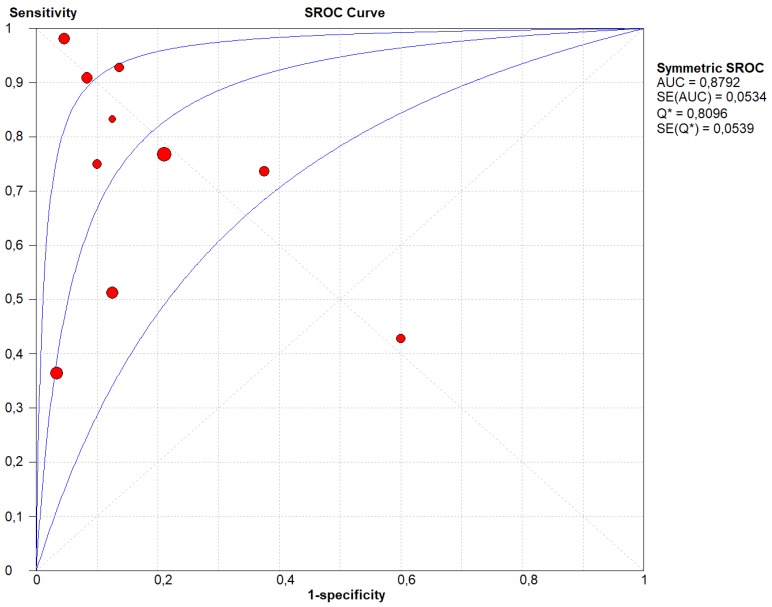
Our results showed that LR+ was 3.85 (95% CI, 1.87–7.90), LR- was 0.34 (95% CI, 0.19–0.62) and DOR was 15.84 (95% CI, 4.98–50.38) (Figure 1A). Between-study heterogeneity was significant in the sensitivity, specificity, and the DOR (I-square estimated to respectively 84.2%, 54.6%, and 50.3%), but the stage of the pancreatic cancer (I–II vs. III–IV) did not explain any of these high I-squares (beta = 0.28, p = 0.28 for sensitivity; beta = 0.89, p = 0.45 for specificity; beta = 0.51, p = 0.28 for DOR). Similar results were evident for the type of molecular analysis of both tissue samples and LB (divided in PCR and NGS) (beta = −4.24, p = 0.17 for sensitivity; beta = 2.81, p = 0.32 for specificity; beta = −3–24, p = 0.68 for DOR). We did not find any evidence of threshold effect (Spearman correlation coefficient: 0.157; p = 0.756). Figure 3 shows the corresponding SROC curve with AUC of 0.880 indicating that LB is capable of identifying the mutations present in pancreatic cancer with a relatively high accuracy.
Figure 3.
Summary Receiver Operating Curve (SROC) of the appropriateness liquid biopsy, taking tissue specimen as the outcome. In this figure, the blue lines represent the AUC (central line) with its 95% CI (external lines) calculated with a meta-analytic approach, while red dots represent the sensitivity and specificity data for each study.
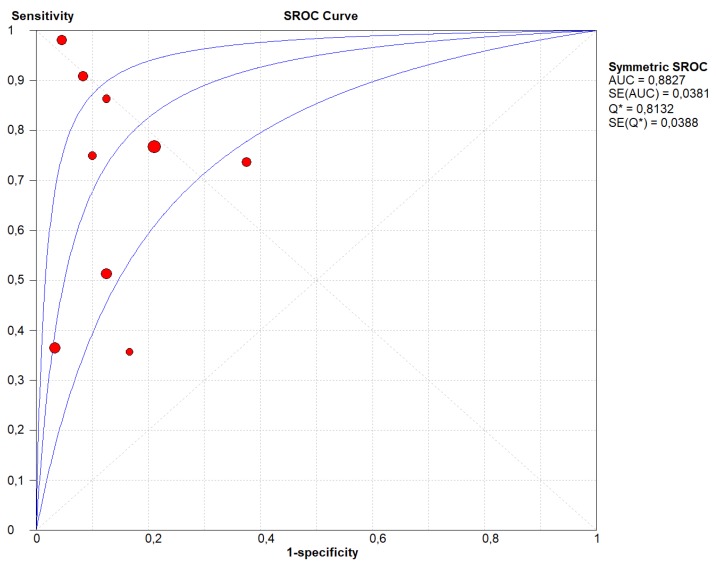
Figure 1B and Figure 2B show the same analyses, taking KRAS mutation as the outcome. As summarized in Table S2, the TP were 169, the FP 11, TN 78 and the FN 111. The overall pooled sensitivity and specificity were 0.65 (95% CI, 0.59–0.71) and 0.91 (95% CI, 0.83–0.96) respectively. Our results showed that LR+ was 3.93 (95% CI, 2.33–6.62), LR- was 0.39 (95% CI, 0.25–0.62) and DOR was 16.41 (95% CI, 7.04–38.29) (Figure 1B). We observed a significant heterogeneity for sensitivity (I-square = 87.0%), but not for specificity and the DOR (I-square = 46.0% and 0%, respectively). Again, the stage of the cancer did not explain the heterogeneity found (beta = 0.89, p = 0.26), as well as the type of molecular analysis (PCR vs. NGS, beta = −2.82, p = 0.46). We did not find any evidence of threshold effect (Spearman correlation coefficient: 0.148; p = 0.622). Figure 4 shows the SROC curve of KRAS only, with AUC of 0.882 indicating a relatively high accuracy also for LB investigating KRAS only.
Figure 4.
Summary Receiver Operating Curve (SROC) of the appropriateness liquid biopsy, taking KRAS mutational status as the outcome. In this figure, the blue lines represent the AUC (central line) with its 95% CI (external lines) calculated with a meta-analytic approach, while red dots represent the sensitivity and specificity data for each study.
For NGS studies, the concordance rate of all mutations detected by multi-genes panels analyzing tissue specimens vs. LB was 31.9%, while 38.2% of mutations were detected on tissue specimens only and 29.9% on LB only (Figure 2C).
3. Discussion
In the present meta-analysis, 14 different studies containing a total of 369 patients, with matched molecular analysis of PDAC tissue specimens and LB, were analyzed. The overall pooled sensitivity and specificity of LB compared to the mutational analysis on tumor tissue were 0.70 (95% CI, 0.63–0.76) and 0.86 (95% CI, 0.77–0.93), respectively. Considering those studies that analyzed KRAS mutations only, the sensitivity was slightly lower (0.65; 95% CI, 0.59–0.71), but the specificity was higher (0.91; 95% CI, 0.83–0.96). Notably, the SROC curves indicate that, compared to tissue specimens, LB demonstrated a high accuracy in determining the mutational asset of PDAC (AUC of 0.880 for all studies and of 0.882 for those regarding KRAS only).
A recent study indicated that a large body of literature has been produced in recent years related to LB in cancer, but less than 1% regarded PDAC [35]. Despite this restricted number of studies on PDAC compared to other cancer types (e.g., lung and breast cancers), LB represents also for this cancer a very promising tool in the era of precision medicine. The values obtained by the quantitative synthesis of this meta-analysis, indeed, does indicate a potential role of LB in the management of PDAC patients.
The major advantage of LB is that it does not require an invasive procedure, highlighting its very promising role firstly in the management of old and/or low-performance status patients [33]. Indeed, with a reliable radiological background, the detection of a multigene PDAC-specific molecular profiling could support the diagnosis of pancreatic cancer without the need of further and hazardous analyses. Notably, in case of the use of multigene panels for molecular analysis, LB could highlight with a reasonable certainty the presence of druggable mutations, with potential important implications for targeted therapies. Thus, also based on this meta-analysis, LB should be encouraged in all cases presenting significant contraindications to invasive or mini-invasive procedures.
Notably, higher concentrations of cfDNA and/or higher numbers of CTCs have been demonstrated in case of late-stage diseases, particularly in case of metastatic or locally-advanced PDAC [20,36,37,38,39,40,41,42]. Although in our meta-analysis a sub-stratification by stage was not possible due to statistical reasons (i.e., the rate of false positive in early stages was zero for all the studies included), the use of LB in late-stage PDAC should guarantee higher sensitivity and specificity compared to those obtained in a localized disease. This advantage may also be translated into the use of LB for the follow-up of metastatic patients during therapy, which is another potentially useful application of LB for the management of PDAC patients [12,22,32,43].
Another potential advantage of LB may be represented by the possibility of overcoming the issue linked to tumor heterogeneity, potentially showing mutations not present in a restricted area of tumor tissue. Although some studies have shown that driver gene mutations in PDAC are usually maintained during clonal evolution [44,45], this aspect may be even more evident for metastatic PDAC, in which a biopsy of the primary tumor or of the metastatic tissue cannot always represent such a complex molecular landscape [46,47,48]. In our meta-analysis, four studies presented data derived from NGS [28,29,32,34]. About one third of all mutations were detected simultaneously by sequencing on tissue specimens and LB, but 38.2% and 29.9% of all mutations were detected respectively on tissue specimens and on LB only. These significant differences could be partly explained by intra-tumor heterogeneity and by clonal evolution of cancer cells, which are well-recognized important biological issues influencing tumor representativeness by tissue specimens. Notably, Vietsch and colleagues demonstrated that cancer heterogeneity within each patient was much more evident in the mutational landscape of cfDNA than in tumor tissue DNA [32]. In their paper, indeed, they demonstrated that the majority (78%) of mutations in the cfDNA were not detected in the primary tumor tissue; this finding demonstrated that a tissue section of a given tumor could fail to represent the molecular makeup of the entire cancer. About the differences in mutation detection using PDAC tissue vs. LB, the lack of standardized and well-established procedures for the molecular analysis of LB could also have partly affected the reliability of the molecular profiling. This point has been further complicated by recent evidence that has described the detection of hotspot PDAC mutations in patients without cancer, an issue that is still under investigation [25].
Regarding the slightly higher specificity, we have found for KRAS compared with all genes, it is important to acknowledge that the KRAS mutation is not highly specific for PDAC, even if matched with histology [49]. Indeed, it can occur frequently in precursor lesions as well as in cancers of other organs, such as colorectal and lung malignancies [50,51,52]. In this regard, it is important to stretch that the specificity regarding KRAS that has emerged in the current meta-analysis reflects the correlation existing between the primary tumor tissue and the LB, and not the overall specificity of KRAS mutational status for PDAC diagnosis.
Despite of these important limitations, LB may become a fundamental tool in the routine clinical management of PDAC patients because of its several advantages, also highlighted by the results of this meta-analysis. However, many aspects can be better addressed in the future to improve LB sensitivity, specificity, and accuracy, allowing it to be introduced as a new and decisive tool for the development of precision medicine in the clinical care of PDAC patients [53]. As the American Society of Clinical Oncology and the College of American Pathologists have highlighted in a recommendation paper, indeed, further studies with standardized technologies are needed to introduce LB in clinical practice [54]. Of note, the introduction of the analysis with LB of epigenetic markers [55,56], of exosome-derived DNA [22,36], and of gene expression profiling/circulating proteins [35,57,58] may represent important steps in improving this kind of non-invasive approach for the molecular characterization of PDAC. Particularly, about epigenetic alterations, DNA methylation may represent a very promising horizon in this changing scenario. Recent studies had indeed shown that DNA methylation signatures were highly consistent between cfDNA and the genomic DNA from its tissue origins also in cancer models [59,60]. Furthermore, if in terms of somatic mutations a high inter-tumor heterogeneity may be found, in the same tumor category the epigenetic profiles are quite similar, and can be of great help, for example, in identifying the actual tumor origin also with LB and in metastatic settings [60]. Gai and colleagues suggested that tissue-specific methylation biomarkers are reasonably consistent across cancer patients for plasma DNA-based cancer testing [60]. Notably, Chan and colleagues reported that, using whole genome bisulfite sequencing of plasma cfDNA, the methylation profile could serve as a general approach for the diagnosis of multiple types of cancers [61]. It is also of importance to report that, besides DNA methylation, recent studies have also highlighted the promising potential of cfDNA fragmentation patterns as an emerging research direction in cancer liquid biopsy [62]. Indeed, cfDNA molecules were not randomly fragmented, but they showed strong size patterns that are associated with the nucleosome footprint, indicating that cfDNA fragmentation patterns may have certain biological and/or clinical potential in liquid biopsy. However, current knowledge on cfDNA fragmentation patterns, either from a biological basis or clinical utilities, is considerably still preliminary and without direct implications for clinical practice [62]. Another important aspect, which should be further elucidated and validated by future research, is the enhanced detection of cfDNA by specific DNA fragment size analysis (90–150 base-pairs), recently uncovered by Mouliere and colleagues [63]. Indeed, with the analysis of size-selected cfDNA, they identified clinically actionable mutations and copy number alterations that were otherwise not detected.
The limitations of our study are the overall small sample size, but this is predominantly owing to the recent development of the application of LB in PDAC studies, and the different molecular analysis that we have quantitatively summarized. Finally, the heterogeneity of our outcomes can be considered as another limitation of our study. This high heterogeneity and the differences that have emerged in both sensitivity and specificity may derive from the varying technologies applied in the studies, as well as in the different timings of LB analysis or also in differences of sequencing depths in NGS studies. Although in our meta-regression analysis we did not find significant moderators to explain this aspect, this point suggests that further studies with well-standardized techniques are needed before the introduction of LB in routine diagnostic activity.
4. Materials and Methods
This systematic review adhered to the MOOSE guidelines [64] and PRISMA statement [65], following a predetermined but unpublished protocol.
4.1. Inclusion and Exclusion Criteria
Studies were eligible if they met the following criteria: (1) A prospective cohort or retrospective study design; (2) contained a clear comparison of mutational analysis using cancer tissue samples vs. LB in patients with pancreatic cancer; (3) contained histological or cytological diagnosis of pancreatic cancer (pancreatic ductal adenocarcinoma); and (4) were published in a peer review journal or published abstract.
Exclusion criteria were: (1) Comparison of patients with pancreatic cancer with patients without cancer (healthy volunteers, chronic pancreatitis); (2) no presence of invasive cancer (e.g., intraductal papillary mucinous neoplasm, so called IPMN); (3) no data regarding molecular analysis of tissue samples and LB in the title/abstract; (4) no data on mutational profiling; (5) diagnosis of non-epithelial malignancies (i.e., lymphomas) or of other histology other than PDAC (e.g., acinar cell carcinoma), and (6) case reports, in vitro or animal studies. We considered articles in any language.
4.2. Data Sources and Literature Search Strategy
Two investigators (C.L., N.V.) independently searched PubMed, SCOPUS, and Embase until 31 January 2019. The search terms used in PubMed included combinations of the following keywords: (“pancreatic” OR “pancreas” OR “pancrea*”) AND (“cell free DNA” OR “cfDNA” OR “circulating DNA” OR “circulating tumor cells” OR “circulating tumour cell” OR “CTC” OR “CTCs”) AND (“diagnosis” OR “sensitivity” OR “specificity” OR “accuracy”). A similar search was carried out in SCOPUS and Embase. We considered the reference lists of all included articles and of previous related reviews.
4.3. Study Selection
Following the searches as outlined above, after removal of duplicates, two independent reviewers (C.L., N.V.) screened titles and abstracts of all potentially eligible articles. The two authors applied the eligibility criteria, considered the full texts, and a final list of included articles was reached through consensus with a third author (A.N.).
4.4. Data Extraction
Two authors were involved in data extraction in a standardized Microsoft Excel database. Specifically, one author (C.L.) extracted data from the included articles and a second independent author (N.V.) validated the data. For each article, we extracted information about authors, year of publication, number of patients, tumor stage, type of tissue samples, type of LB, type of molecular analysis of both tissue samples and LB (divided in PCR and NGS), number of true positive, true negative, false positive and false negative results, using the molecular analysis on tissue samples as reference.
4.5. Outcomes
The primary outcomes were sensitivity, specificity, positive and negative likelihood ratios, diagnostic odds ratio (DOR), and the area under the curve (AUC) of the molecular analysis with LB. For those studies which used multigene-panels of next-generation sequencing for molecular analysis, the concordance rate between the molecular profiles of tissue samples and LB was also reported. A similar (secondary) analysis of concordance was also performed considering the mutational status of the KRAS gene only.
4.6. Assessment of Study Quality
Based on the revised quality assessment of diagnosis, accuracy studies-2 (QUADAS-2) criteria, the included articles were evaluated as at high risk (−) or low risk (+) by four key domains: Patient selection, index test, reference standard, and flow and timing [66].
4.7. Data Synthesis and Statistical Analysis
We used RevMan Manager 5.3 and Meta-Disc software 5.1.32 to conduct this meta-analysis. All the studies reported the data regarding true positive (TP), true negative (TN), false positive (FP) and false negative (FN). Therefore, we were able to calculate the pooled sensitivity (TP/TP + FN), specificity (SPE) (TN/TN + FP), negative likelihood ratio (LR−), positive likelihood ratios (LR+) and DOR with their 95% confidence intervals. At the same time, we constructed the summary receiver operator characteristic (SROC) curve and calculated the area under the SROC curve based on the sensitivity and specificity of each study. In case of high heterogeneity (as indicated by an I2 > 50%), we performed a meta-regression analysis, taking the stage of the tumor (I–II vs. III–IV) and the type of molecular analysis of both tissue samples and LB (divided in PCR and NGs) as potential moderators.
For those studies that used multi-genes panel for DNA sequencing, we also compared the concordance rate of mutations detected by the analysis on tissue specimens and the analysis on LB, designed a summarizing Venn diagram.
5. Conclusions
To conclude, although further studies are needed to improve the standardization and the applicability of LB, we have demonstrated in this meta-analysis that this non-invasive approach can serve as surrogate to tissue in the molecular profiling of PDAC, because of its relatively high sensitivity, specificity, and accuracy. Thus, LB could be introduced in clinical practice and should be further explored towards the development of new precision medicine approaches against pancreatic cancer.
Acknowledgments
The Authors thank Cristina Girimondo for the assistance in retrieving manuscripts from electronic and libraries’ archives and Davide Gamberini of “Grafica Biomedica” for the graphical support.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/11/8/1152/s1, Figure S1: PRISMA checklist for this meta-analysis, Table S1: QUADAS-2 tool: Risk of bias and applicability judgments of the present meta-analysis, Table S2: Data considering the mutational status of KRAS gene only.
Author Contributions
C.L., N.V., A.N., A.S.: study conception and design; C.L., N.V., A.N., A.S.: data extraction and elaboration; all authors: data interpretation; all authors: discussion of the findings; C.L., N.V., A.N., A.S.: paper writing; all authors: final editing and approval of the manuscript in its present form.
Funding
This work has been supported by University of Verona [personal funds for research] (C.L.), University of Verona Internalization Program “Cooperint” (C.L., L.A.A.B.), Associazione Italiana Ricerca Cancro (AIRC) grant n. 12182 (A.S.), European Community Grant Transcan Bio-PaC (A.S.), FP7 European Community Grant Cam-Pac [no: 602783] (A.S.) and Italian Health Ministry [RF-2013-02359692] (M.G.D. and A.S.). The funding agencies had no role in the collection, analysis and interpretation of data, and in the writing of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Kamisawa T., Wood L.D., Itoi T., Takaori K. Pancreatic cancer. Lancet. 2016;388:73–85. doi: 10.1016/S0140-6736(16)00141-0. [DOI] [PubMed] [Google Scholar]
- 3.Rahib L., Smith B.D., Aizenberg R., Rosenzweig A.B., Fleshman J.M., Matrisian L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 4.Luchini C., Capelli P., Scarpa A. Pancreatic ductal adenocarcinoma and its variants. Surg. Pathol. Clin. 2016;9:547–560. doi: 10.1016/j.path.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 5.Kitano M., Yoshida T., Itonaga M., Tamura T., Hatamaru K., Yamashita Y. Impact of endoscopic ultrasonography on diagnosis of pancreatic cancer. J. Gastroenterol. 2019;54:19–32. doi: 10.1007/s00535-018-1519-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DiPardo B.J., Winograd P., Court C.M., Tomlinson J.S. Pancreatic cancer circulating tumor cells: Applications for personalized oncology. Expert Rev. Mol. Diagn. 2018;18:809–820. doi: 10.1080/14737159.2018.1511429. [DOI] [PubMed] [Google Scholar]
- 7.Baek H.W., Park M.J., Rhee Y.Y., Lee K.B., Kim M.A., Park I.A. Diagnostic accuracy of endoscopic ultrasound-guided fine needle aspiration cytology of pancreatic lesions. J. Pathol. Transl. Med. 2015;49:52–60. doi: 10.4132/jptm.2014.10.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh S., Tang S.J., Sreenarasimhaiah J., Lara L.F., Siddiqui A. The clinical utility and limitations of serum carbohydrate antigen (CA19-9) as a diagnostic tool for pancreatic cancer and cholangiocarcinoma. Dig. Dis. Sci. 2011;56:2491–2496. doi: 10.1007/s10620-011-1709-8. [DOI] [PubMed] [Google Scholar]
- 9.Lewis A.R., Valle J.W., McNamara M.G. Pancreatic cancer: Are “liquid biopsies” ready for prime-time? World J. Gastroenterol. 2016;22:7175–7185. doi: 10.3748/wjg.v22.i32.7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crowley E., Di Nicolantonio F., Loupakis F., Bardelli A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013;10:472–484. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- 11.Siravegna G., Marsoni S., Siena S., Bardelli A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017;14:531–548. doi: 10.1038/nrclinonc.2017.14. [DOI] [PubMed] [Google Scholar]
- 12.Bettegowda C., Sausen M., Leary R.J., Kinde I., Wang Y., Agrawal N., Bartlett B.R., Wang H., Luber B., Alani R.M., et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gale D., Lawson A.R.J., Howarth K., Madi M., Durham B., Smalley S., Calaway J., Blais S., Jones G., Clark J., et al. Development of a highly sensitive liquid biopsy platform to detect clinically-relevant cancer mutations at low allele fractions in cell-free DNA. PLoS ONE. 2018;13:e0194630. doi: 10.1371/journal.pone.0194630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lambros M.B., Seed G., Sumanasuriya S., Gil V., Crespo M., Fontes M., Chandler R., Mehra N., Fowler G., Ebbs B., et al. Single-Cell Analyses of Prostate Cancer Liquid Biopsies Acquired by Apheresis. Clin. Cancer Res. 2018;24:5635–5644. doi: 10.1158/1078-0432.CCR-18-0862. [DOI] [PubMed] [Google Scholar]
- 15.Gorges T.M., Penkalla N., Schalk T., Joosse S.A., Riethdorf S., Tucholski J., Lücke K., Wikman H., Jackson S., Brychta N., et al. Enumeration and Molecular Characterization of Tumor Cells in Lung Cancer Patients Using a Novel In Vivo Device for Capturing Circulating Tumor Cells. Clin. Cancer Res. 2016;22:2197–2206. doi: 10.1158/1078-0432.CCR-15-1416. [DOI] [PubMed] [Google Scholar]
- 16.Le Calvez-Kelm F., Foll M., Wozniak M.B., Delhomme T.M., Durand G., Chopard P., Pertesi M., Fabianova E., Adamcakova Z., Holcatova I., et al. KRAS mutations in blood circulating cell-free DNA: A pancreatic cancer case-control. Oncotarget. 2016;7:78827–78840. doi: 10.18632/oncotarget.12386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takai E., Yachida S. Circulating tumor DNA as a liquid biopsy target for detection of pancreatic cancer. World J. Gastroenterol. 2016;22:8480–8488. doi: 10.3748/wjg.v22.i38.8480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moravec R., Divi R., Verma M. Detecting circulating tumor material and digital pathology imaging during pancreatic cancer progression. World J. Gastrointest. Oncol. 2017;9:235–250. doi: 10.4251/wjgo.v9.i6.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maire F., Micard S., Hammel P., Voitot H., Lévy P., Cugnenc P.H., Ruszniewski P., Puig P.L. Differential diagnosis between chronic pancreatitis and pancreatic cancer: Value of the detection of KRAS2 mutations in circulating DNA. Br. J. Cancer. 2002;87:551–554. doi: 10.1038/sj.bjc.6600475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sikora K., Bedin C., Vicentini C., Malpeli G., D’Angelo E., Sperandio N., Lawlor R.T., Bassi C., Tortora G., Nitti D., et al. Evaluation of cell-free DNA as a biomarker for pancreatic malignancies. Int. J. Biol. Markers. 2015;30:e136–e141. doi: 10.5301/jbm.5000088. [DOI] [PubMed] [Google Scholar]
- 21.Ako S., Nouso K., Kinugasa H., Dohi C., Matushita H., Mizukawa S., Muro S., Akimoto Y., Uchida D., Tomoda T., et al. Utility of serum DNA as a marker for KRAS mutations in pancreatic cancer tissue. Pancreatology. 2017;17:285–290. doi: 10.1016/j.pan.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 22.Bernard V., Kim D.U., San Lucas F.A., Castillo J., Allenson K., Mulu F.C., Stephens B.M., Huang J., Semaan A., Guerrero P.A., et al. Circulating Nucleic Acids Are Associated with Outcomes of Patients With Pancreatic Cancer. Gastroenterology. 2019;156:108–118.e4. doi: 10.1053/j.gastro.2018.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brychta N., Krahn T., von Ahsen O. Detection of KRAS Mutations in Circulating Tumor DNA by Digital PCR in Early Stages of Pancreatic Cancer. Clin. Chem. 2016;62:1482–1491. doi: 10.1373/clinchem.2016.257469. [DOI] [PubMed] [Google Scholar]
- 24.Earl J., Garcia-Nieto S., Martinez-Avila J.C., Montans J., Sanjuanbenito A., Rodríguez-Garrote M., Lisa E., Mendía E., Lobo E., Malats N., et al. Circulating tumor cells (Ctc) and kras mutant circulating free Dna (cfdna) detection in peripheral blood as biomarkers in patients diagnosed with exocrine pancreatic cancer. BMC Cancer. 2015;15:797. doi: 10.1186/s12885-015-1779-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kinugasa H., Nouso K., Miyahara K., Morimoto Y., Dohi C., Tsutsumi K., Kato H., Matsubara T., Okada H., Yamamoto K. Detection of K-ras gene mutation by liquid biopsy in patients with pancreatic cancer. Cancer. 2015;121:2271–2280. doi: 10.1002/cncr.29364. [DOI] [PubMed] [Google Scholar]
- 26.Kulemann B., Liss A.S., Warshaw A.L., Seifert S., Bronsert P., Glatz T., Pitman M.B., Hoeppner J. KRAS mutations in pancreatic circulating tumor cells: A pilot study. Tumour Biol. 2016;37:7547–7554. doi: 10.1007/s13277-015-4589-2. [DOI] [PubMed] [Google Scholar]
- 27.Marchese R., Muleti A., Pasqualetti P., Bucci B., Stigliano A., Brunetti E., De Angelis M., Mazzoni G., Tocchi A., Brozzetti S. Low correspondence between K-ras mutations in pancreatic cancer tissue and detection of K-ras mutations in circulating DNA. Pancreas. 2006;32:171–177. doi: 10.1097/01.mpa.0000202938.63084.e3. [DOI] [PubMed] [Google Scholar]
- 28.Park G., Park J.K., Son D.S., Shin S.H., Kim Y.J., Jeon H.J., Lee J., Park W.Y., Lee K.H., Park D. Utility of targeted deep sequencing for detecting circulating tumor DNA in pancreatic cancer patients. Sci. Rep. 2018;8:11631. doi: 10.1038/s41598-018-30100-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pishvaian M.J., Joseph Bender R., Matrisian L.M., Rahib L., Hendifar A., Hoos W.A., Mikhail S., Chung V., Picozzi V., Heartwell C., et al. A pilot study evaluating concordance between blood-based and patient-matched tumor molecular testing within pancreatic cancer patients participating in the Know Your Tumor (KYT) initiative. Oncotarget. 2016;8:83446–83456. doi: 10.18632/oncotarget.13225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sefrioui D., Blanchard F., Toure E., Rahib L., Hendifar A., Hoos W.A., Mikhail S., Chung V., Picozzi V., Heartwell C., et al. Diagnostic value of CA19.9, circulating tumour DNA and circulating tumour cells in patients with solid pancreatic tumours. Br. J. Cancer. 2017;117:1017–1025. doi: 10.1038/bjc.2017.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shibata K., Mori M., Kitano S., Akiyoshi T. Detection of ras gene mutations in peripheral blood of carcinoma patients using CD45 immunomagnetic separation and nested mutant allele specific amplification. Int. J. Oncol. 1998;12:1333–1338. doi: 10.3892/ijo.12.6.1333. [DOI] [PubMed] [Google Scholar]
- 32.Vietsch E.E., Graham G.T., McCutcheon J.N., Javaid A., Giaccone G., Marshall J.L., Wellstein A. Circulating cell-free DNA mutation patterns in early and late stage colon and pancreatic cancer. Cancer Genet. 2017;218–219:39–50. doi: 10.1016/j.cancergen.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu J., Zhou Y., Zhang C.Y., Song B.B., Wang B.L., Pan B.S., Lou W.H., Guo W. Co-amplification at lower denaturation-temperature PCR combined with unlabled-probe high-resolution melting to detect KRAS codon 12 and 13 mutations in plasma-circulating DNA of pancreatic adenocarcinoma cases. Asian Pac. J. Cancer Prev. 2014;15:10647–10652. doi: 10.7314/APJCP.2014.15.24.10647. [DOI] [PubMed] [Google Scholar]
- 34.Zill O.A., Greene C., Sebisanovic D., Siew L.M., Leng J., Vu M., Hendifar A.E., Wang Z., Atreya C.E., Kelley R.K., et al. Cell-Free DNA Next-Generation Sequencing in Pancreatobiliary Carcinomas. Cancer Discov. 2015;5:1040–1048. doi: 10.1158/2159-8290.CD-15-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otandault A., Anker P., Al Amir Dache Z., Guillaumon V., Meddeb R., Pastor B., Pisareva E., Sanchez C., Tanos R., Tousch G., et al. Recent advances in circulating nucleic acids in oncology. Ann. Oncol. 2019;30:374–384. doi: 10.1093/annonc/mdz031. [DOI] [PubMed] [Google Scholar]
- 36.Allenson K., Castillo J., San Lucas F.A., Scelo G., Kim D.U., Bernard V., Davis G., Kumar T., Katz M., Overman M.J., et al. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann. Oncol. 2017;28:741–747. doi: 10.1093/annonc/mdx004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ankeny J.S., Court C.M., Hou S., Li Q., Song M., Wu D., Chen J.F., Lee T., Lin M., Sho S., et al. Circulating tumour cells as a biomarker for diagnosis and staging in pancreatic cancer. Br. J. Cancer. 2016;114:1367–1375. doi: 10.1038/bjc.2016.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen J.D., Javed A.A., Thoburn C., Wong F., Tie J., Gibbs P., Schmidt C.M., Yip-Schneider M.T., Allen P.J., Schattner M., et al. Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers. Proc. Natl. Acad. Sci. USA. 2017;114:10202–10207. doi: 10.1073/pnas.1704961114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Premasekharan G., Gilbert E., Okimoto R.A., Hamirani A., Lindquist K.J., Ngo V.T., Roy R., Hough J., Edwards M., Paz R., et al. An improved CTC isolation scheme for pairing with downstream genomics: Demonstrating clinical utility in metastatic prostate, lung and pancreatic cancer. Cancer Lett. 2016;380:144–152. doi: 10.1016/j.canlet.2016.06.017. [DOI] [PubMed] [Google Scholar]
- 40.Takai E., Totoki Y., Nakamura H., Kato M., Shibata T., Yachida S. Clinical Utility of Circulating Tumor DNA for Molecular Assessment and Precision Medicine in Pancreatic Cancer. Adv. Exp. Med. Biol. 2016;924:13–17. doi: 10.1007/978-3-319-42044-8_3. [DOI] [PubMed] [Google Scholar]
- 41.Bidard F.C., Huguet F., Louvet C., Mineur L., Bouché O., Chibaudel B., Artru P., Desseigne F., Bachet J.B., Mathiot C., et al. Circulating tumor cells in locally advanced pancreatic adenocarcinoma: The ancillary CirCe 07 study to the LAP 07 trial. Ann. Oncol. 2013;24:2057–2061. doi: 10.1093/annonc/mdt176. [DOI] [PubMed] [Google Scholar]
- 42.Gemenetzis G., Groot V.P., Yu J., Ding D., Teinor J.A., Javed A.A., Wood L.D., Burkhart R.A., Cameron J.L., Makary M.A., et al. Circulating Tumor Cells Dynamics in Pancreatic Adenocarcinoma Correlate with Disease Status: Results of the Prospective CLUSTER Study. Ann. Surg. 2018;268:408–420. doi: 10.1097/SLA.0000000000002925. [DOI] [PubMed] [Google Scholar]
- 43.Oellerich M., Schütz E., Beck J., Kanzow P., Plowman P.N., Weiss G.J., Walson P.D. Using circulating cell-free DNA to monitor personalized cancer therapy. Crit. Rev. Clin. Lab. Sci. 2017;54:205–218. doi: 10.1080/10408363.2017.1299683. [DOI] [PubMed] [Google Scholar]
- 44.Makohon-Moore A.P., Zhang M., Reiter J.G., Bozic I., Allen B., Kundu D., Chatterjee K., Wong F., Jiao Y., Kohutek Z.A., et al. Limited heterogeneity of known driver gene mutations among the metastases of individual patients with pancreatic cancer. Nat. Genet. 2017;49:358–366. doi: 10.1038/ng.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Connor A.A., Denroche R.E., Jang G.H., Lemire M., Zhang A., Chan-Seng-Yue M., Wilson G., Grant R.C., Merico D., Lungu I., et al. Integration of Genomic and Transcriptional Features in Pancreatic Cancer Reveals Increased Cell Cycle Progression in Metastases. Cancer Cell. 2019;35:267–282.e7. doi: 10.1016/j.ccell.2018.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Molinari C., Marisi G., Passardi A., Matteucci L., De Maio G., Ulivi P. Heterogeneity in Colorectal Cancer: A Challenge for Personalized Medicine? Int. J. Mol. Sci. 2018;19:3733. doi: 10.3390/ijms19123733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Franczak C., Filhine-Tressarieu P., Broséus J., Gilson P., Merlin J.L., Harlé A. Clinical Interest of Circulating Tumor DNA in Oncology. Arch. Med. Res. 2018;49:297–305. doi: 10.1016/j.arcmed.2018.10.007. [DOI] [PubMed] [Google Scholar]
- 48.Sumbal S., Javed A., Afroze B., Zulfiqar H.F., Javed F., Noreen S., Ijaz B. Circulating tumor DNA in blood: Future genomic biomarkers for cancer detection. Exp. Hematol. 2018;65:17–28. doi: 10.1016/j.exphem.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 49.Luchini C., Pea A., Yu J., He J., Salvia R., Riva G., Weiss M.J., Bassi C., Cameron J.L., Hruban R.H., et al. Pancreatic cancer arising in the remnant pancreas is not always a relapse of the preceding primary. Mod. Pathol. 2019;32:659–665. doi: 10.1038/s41379-018-0183-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riva G., Pea A., Pilati C., Fiadone G., Lawlor R.T., Scarpa A., Luchini C. Histo-molecular oncogenesis of pancreatic cancer: From precancerous lesions to invasive ductal adenocarcinoma. World J. Gastrointest. Oncol. 2018;10:317–327. doi: 10.4251/wjgo.v10.i10.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ferrer I., Zugazagoitia J., Herbertz S., John W., Paz-Ares L., Schmid-Bindert G. KRAS-Mutant non-small cell lung cancer: From biology to therapy. Lung Cancer. 2018;124:53–64. doi: 10.1016/j.lungcan.2018.07.013. [DOI] [PubMed] [Google Scholar]
- 52.Huang D., Sun W., Zhou Y., Li P., Chen F., Chen H., Xia D., Xu E., Lai M., Wu Y., et al. Mutations of key driver genes in colorectal cancer progression and metastasis. Cancer Metastasis Rev. 2018;37:173–187. doi: 10.1007/s10555-017-9726-5. [DOI] [PubMed] [Google Scholar]
- 53.Pea A., Cheng L., Luchini C. Dissecting the molecular landscape of pancreatic cancer: Towards a precision medicine approach. Exp. Rev. Prec. Med. Drug Develop. 2019;4:113–119. doi: 10.1080/23808993.2019.1604135. [DOI] [Google Scholar]
- 54.Merker J.D., Oxnard G.R., Compton C., Diehn M., Hurley P., Lazar A.J., Lindeman N., Lockwood C.M., Rai A.J., Schilsky R.L., et al. Circulating Tumor DNA Analysis in Patients With Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J. Clin. Oncol. 2018;36:1631–1641. doi: 10.1200/JCO.2017.76.8671. [DOI] [PubMed] [Google Scholar]
- 55.Shen S.Y., Singhania R., Fehringer G., Chakravarthy A., Roehrl M.H.A., Chadwick D., Zuzarte P.C., Borgida A., Wang T.T., Li T., et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature. 2018;563:579–583. doi: 10.1038/s41586-018-0703-0. [DOI] [PubMed] [Google Scholar]
- 56.Li W., Zhang X., Lu X., You L., Song Y., Luo Z., Zhang J., Nie J., Zheng W., Xu D., et al. 5-Hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic biomarkers for human cancers. Cell. Res. 2017;27:1243–1257. doi: 10.1038/cr.2017.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Görner K., Bachmann J., Holzhauer C., Kirchner R., Raba K., Fischer J.C., Martignoni M.E., Schiemann M., Alunni-Fabbroni M. Genetic analysis of circulating tumor cells in pancreatic cancer patients: A pilot study. Genomics. 2015;106:7–14. doi: 10.1016/j.ygeno.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 58.Cohen J.D., Li L., Wang Y., Thoburn C., Afsari B., Danilova L., Douville C., Javed A.A., Wong F., Mattox A., et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926–930. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gai W., Ji L., Lam W.K.J., Sun K., Jiang P., Chan A.W.H., Wong J., Lai P.B.S., Ng S.S.M., Ma B.B.Y., et al. Liver- and colon-specific DNA methylation markers in plasma for investigation of colorectal cancers with or without liver metastases. Clin. Chem. 2018;64:1239–1249. doi: 10.1373/clinchem.2018.290304. [DOI] [PubMed] [Google Scholar]
- 60.Gai W., Sun K. Epigenetic Biomarkers in Cell-Free DNA and Applications in Liquid Biopsy. Genes (Basel). 2019;10:32. doi: 10.3390/genes10010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chan K.C.A., Jiang P., Chan C.W., Sun K., Wong J., Hui E.P., Chan S.L., Chan W.C., Hui D.S., Ng S.S., et al. Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc. Natl. Acad. Sci. USA. 2013;110:18761–18768. doi: 10.1073/pnas.1313995110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun K., Jiang P., Wong A.I.C., Cheng Y.K.Y., Cheng S.H., Zhang H., Chan K.C.A., Leung T.Y., Chiu R.W.K., Lo Y.M.D. Size-tagged preferred ends in maternal plasma DNA shed light on the production mechanism and show utility in noninvasive prenatal testing. Proc. Natl. Acad. Sci. USA. 2018;115:E5106–E5114. doi: 10.1073/pnas.1804134115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mouliere F., Chandrananda D., Piskorz A.M., Moore E.K., Morris J., Ahlborn L.B., Mair R., Goranova T., Marass F., Heider K., et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci. Transl. Med. 2018;10 doi: 10.1126/scitranslmed.aat4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stroup D.F., Berlin J.A., Morton S.C., Olkin I., Williamson G.D., Rennie D., Moher D., Becker B.J., Sipe T.A., Thacker S.B. Meta-analysis of observational studies in epidemiology: A proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA. 2000;283:2008–2012. doi: 10.1001/jama.283.15.2008. [DOI] [PubMed] [Google Scholar]
- 65.Liberati A., Altman D.G., Tetzlaff J., Mulrow C., Gøtzsche P.C., Ioannidis J.P., Clarke M., Devereaux P.J., Kleijnen J., Moher D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: Explanation and elaboration. BMJ. 2009;339:b2700. doi: 10.1136/bmj.b2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Whiting P.F., Rutjes A.W., Westwood M.E., Mallett S., Deeks J.J., Reitsma J.B., Leeflang M.M., Sterne J.A., Bossuyt P.M., QUADAS-2 Group QUADAS-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011;155:529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.