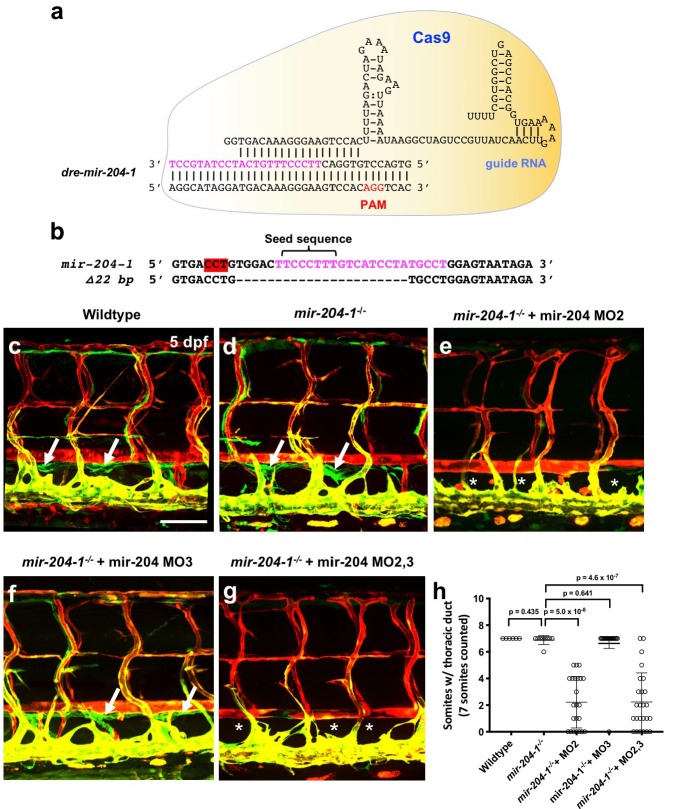
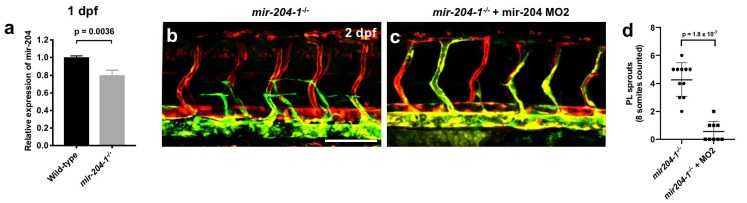
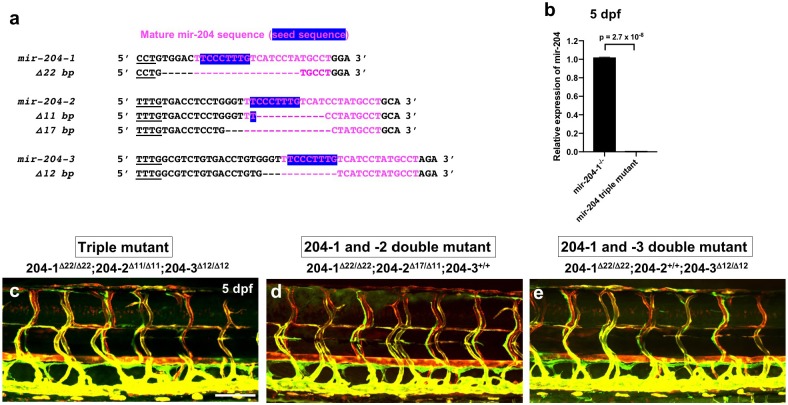
Figure 3. Mir-204 function is required for lymphatic development.
(a) Schematic of CRISPR/Cas9 and guide RNA targeting of mir-204–1. (b) Sequence alignment of wildtype and mir-204–1 mutant genomic DNA. The mature mir-204 sequence is noted in magenta, and the PAM sequence is highlighted in red (on the reverse strand). The mutant carries 22 bp deletion that removes 17 nucleotides of the mature mir-204 sequence. (c–g) Representative confocal images of the mid-trunk of 5 dpf wild type sibling (c), mir-204–1-/- mutant (d), MO2-injected mir-204–1-/- mutant (e), MO3-injected mir-204–1-/- mutant (f), and MO2 + MO3 co-injected mir-204–1-/- mutant (g) animals. Images are lateral views of Tg(mrc1a:eGFP)y251, Tg(kdrl:mCherry)y171 double-transgenic animals, rostral to the left. The thoracic duct is labeled with white arrows, and absence of the thoracic duct is noted with asterisks. (h) Quantification of thoracic duct formation in five dpf wild type (n = 6), mir-204–1-/- mutant (n = 9), MO2-injected mir-204–1-/- mutant (n = 23), MO2-injected mir-204–1-/- mutant (n = 19), and MO2 + MO3 co-injected mir-204–1-/- mutant animals (n = 25). The number of somitic segments with an intact thoracic duct was counted, with the same seven mid-trunk somites measured in each animal. Scale bar: 100 μm (c). All graphs are analyzed by t-test and the mean ± SD is shown.