To the Editor:
Heterozygous loss-of-function pathogenic variants in the TNF-α–induced protein 3 gene (TNFAIP3) cause autoinflammation due to haploinsufficiency of A20 protein (HA20). HA20 commonly manifests as severe orogenital ulceration and uveitis.1 Central nervous system (CNS) inflammation is rare but is reported in patients with HA20, and animal studies have shown that mice with mutant TNFAIP3 are prone to severe neuroinflammation.2, 3 Loss of A20 function causes spontaneous cerebral inflammation, as demonstrated by robust microglial activation, reactive astrogliosis, endothelial activation, increased oxidative/nitrosative stress, and expression of nuclear factor κ (NF-κB)–regulated proinflammatory soluble mediators, such as IL-1β, TNF, IL-6, and monocyte chemoattractant protein 1 (MCP-1) in the brain.3 CNS involvement as the sole clinical manifestation of heterozygous TNFAIP3 variants in human subjects has never been described.
We report a case of TNFAIP3-mediated autoinflammation manifesting as progressive neuroinflammation. We show that mutated A20 protein did not control interferon-dependent transcription, highlighting an entirely novel mechanism of autoinflammation in patients with HA20. Targeted treatment with the Janus kinase (JAK) inhibitor baricitinib led to marked clinical, radiologic, and immunologic improvement. Methods are provided in this Methods section in this article's Online Repository at www.jacionline.org.
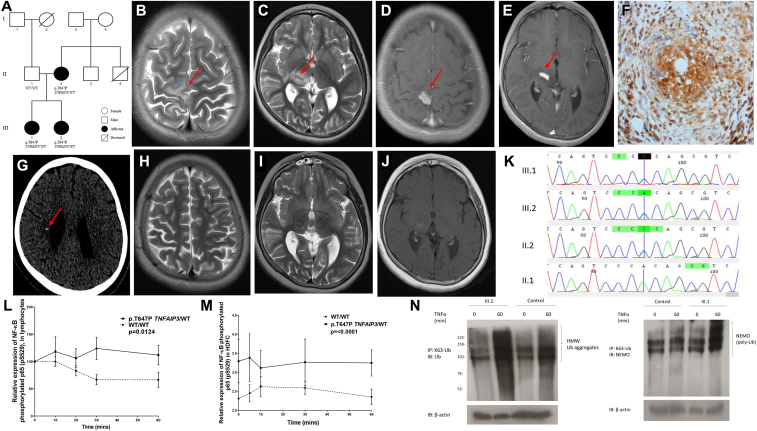
The proband (III.1) was an 8-year-old girl of nonconsanguineous Pakistani-Indian descent (Fig 1, A) who presented with left-sided focal seizures and hemiparesis, uveitis, and cognitive decline. She had choreoretinitis from the age of 6 months considered secondary to congenital infection; however, no pathogen had been identified. There was no history of systemic symptoms, and she had occasional mouth ulcers but no genital ulcers. Magnetic resonance imaging of her brain revealed contrast-enhancing T2-hypointense intracranial mass lesions affecting predominantly the grey matter of the paracentral lobule and the thalamus on the left, with surrounding edema (Fig 1, B-E). Laboratory tests included negative autoantibody screen results; and negative normal complement function study results; targeted NOD2 genetic testing; a nitroblue tetrazolium test; and immunoglobulin levels, lymphocyte subsets, and cerebrospinal fluid analysis (see Table E1 in this article's Online Repository at www.jacionline.org). She had a modestly increased erythrocyte sedimentation rate of 25 mm/h (normal range, <5 mm/h) and normal C-reactive protein levels (5 mg/L; normal range, <5 mg/L). Brain histology revealed granulomatous inflammation, focal necrosis, and calcification (Fig 1, F). Tuberculosis was still considered a possible differential diagnosis, and empiric anti-tuberculosis treatment was started (isoniazid/rifampicin/pyrazinamide/moxifloxacin). However, results of brain tissue culture, PCR tests for mycobacteria, and QuantiFERON tests were all negative. There was progression of the mass lesion, worsening of left-sided hemiplegia, and new-onset ataxia.
Fig 1.
Pedigree, magnetic resonance imaging features, and genetic sequencing results. A, Pedigree shows segregation of the p.T647P TNFAIP3 variant. B and C, Axial T2-weighted images at the level of the perirolandic cortex and basal ganglia show hypointense solid areas (arrows) surrounded by vasogenic edema in the right paracentral lobule (Fig 1, B) and thalamus (Fig 1, C). D and E, Axial T1-weighted image after contrast at the same levels showing intense enhancement of the solid lesions (arrows). F, Brain biopsy specimen showing necrotizing granulomatous inflammation with positive p65 nuclear stain. G, Computed tomographic brain scan demonstrating foci of calcifications (arrow) in the thalamic lesion and in the periventricular white matter within the area of vasogenic edema. H-J, Axial T2 and T1-weighted image after contrast showing almost complete resolution of the previously noted lesions. K, Sanger sequencing chromatogram of the TNFAIP3 gene aligned to reference sequence exon 8. The line indicates a heterozygous TNFAIP3 p.T647P variant in subjects III.1, III.2, and II.2. L and M, There was increased expression of phosphorylated p65 in patients' lymphocytes (P = .0124) and patients' HDFCs (P = .0001) compared with control cells. WT, Wild-type. N, TNF-stimulated dermal fibroblasts from subject III.1 showed increased abundance and molecular weight of Lys63-ubiquitinated NF-κB essential modulator (NEMO). HMW, High molecular weight; IB, immunoblotting; IP, immunoprecipitation.
At that point, the patient was considered to have an unclassified granulomatous neuroinflammatory disorder reminiscent of neurosarcoid. Based on this diagnosis, she received treatment with 2 mg/kg/d prednisolone, weaning to 0.5 mg/kg/d over 6 months and mycophenolate mofetil (1200 mg/m2/d). There was no response to this treatment, with a further increase in size of the brain lesion. Intravenous cyclophosphamide (6 doses, 500-750 mg/m2) was then given, and prednisolone continued at a higher dose (2 mg/kg/d). There was poor response to these treatments, and significant steroid-related side effects were noted (weight gain, Cushingoid appearance, and arterial hypertension).
A second brain biopsy again revealed necrotizing granulomatous inflammation and no evidence of infection. Whole-body positron emission tomography–computed tomography revealed no evidence of malignancy; brain computed tomography revealed intracerebral calcification (Fig 1, G). In view of the significant development of intracerebral calcification, she was considered to have an unclassified interferonopathy and was started on baricitinib (6 mg/d), an oral JAK1/JAK2 inhibitor that blocks interferon signaling (through a program sponsored by Eli Lilly and Company). This resulted in rapid clinical and radiologic improvement: 24 months later, she remains stable, with no further seizures and marked resolution of the intracerebral inflammatory lesion (Fig 1, H-J). Prednisolone was weaned off for the first time in 2 years. Her younger sister (subject III.2) had arthritis and a facial malar-type rash from the age of 6 months. Subject II.4 died at the age of 17 years from early-onset systemic lupus erythematosus. Subject II.2 had mild oral ulceration.
Whole-exome sequencing revealed a heterozygous c.A1939C (NM_001270508) p.T647P variant in TNFAIP3, which was confirmed by using Sanger sequencing in subject III.1 and also present in the symptomatic sibling (subject III.2) and mildly symptomatic mother (subject II.2; Fig 1, A and K). This variant resides in the fourth zinc finger domain of A20, an area that has E3 ubiquitin ligase activity and is involved in recruiting adaptor proteins, such as Tax-1–binding protein 1 and the A20-binding inhibitor of NF-κB, to enable A20 to exert its inhibitory function.4
PBMCs derived from heterozygotes for the p.T647P variant of TNFAIP3 showed increased expression of NF-κB phosphorylated p65 transcription factor compared with control cells (P = .0124; Fig 1, L). Similar differences were also observed in human dermal fibroblast cells (HDFCs) from subject III.1 compared with control HDFCs (P = .0001; Fig 1, M), suggesting that the heterozygous p.T647P TNFAIP3 variant impaired the ability of the A20 to regulate the canonical NF-κB pathway. TNF-stimulated HDFCs from subject III.1 also showed increased molecular weight of Lys63-ubiquitinated NF-κB essential modulator as a result of insufficient A20 deubiquitinase activity (Fig 1, N).
Activated NF-κB subunits are known to promote transcription of genes encoding proinflammatory cytokines.5 Levels of several proinflammatory cytokines were substantially greater in heterozygotes for p.T647P TNFAIP3 compared with those in healthy control subjects: IL-1β (P = .02), IL-6 (P = .02), IL-8 (P = .02), and TNF-α (P = .02).
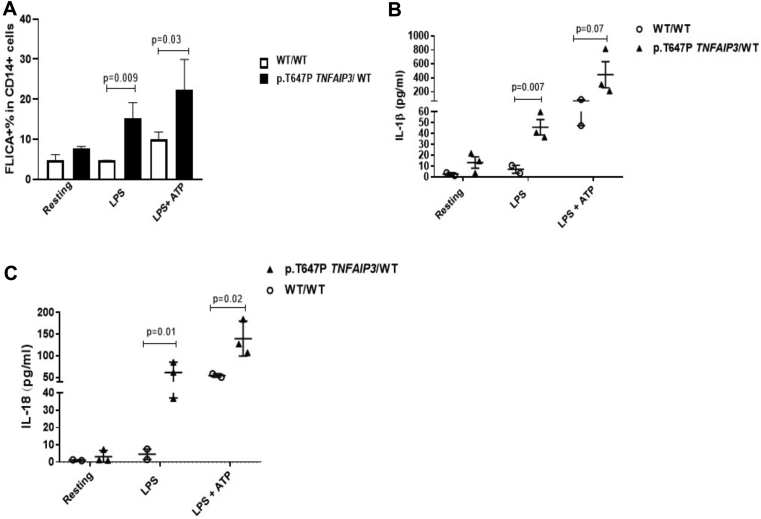
Recent studies suggest that A20 functions as a negative regulator of the NLR family pyrin domain containing 3 inflammasome independently of its role in NF-κB regulation.1 Consistent with these data, PBMCs from subject III.1 showed constitutive activation of the NLR family pyrin domain containing 3 inflammasome, which resulted in activation of caspase-1 (P = .009) and increased secretion of active IL-1β (P = .007) and IL-18 (P = .01, see Fig E1 in this article's Online Repository at www.jacionline.org).
Fig E1.
NLR family pyrin domain containing 3 (NLRP3) inflammasome activation associated with the p.T647P heterozygous variant in TNFAIP3. A, PBMCs from subject III.1 constitutively expressed higher levels of FLICA (caspase-1) in response to LPS (mean, 16.57; SEM, 2.533) compared with control subjects (mean, 4.667; SEM, 0.01; P = .009). B and C, There was increased release of IL-1β and IL-18 in supernatants from PBMCs derived from subject III.1 after LPS stimulation compared with that in healthy control subjects (P = .007 and P = .01, respectively). Differences in IL-1β, IL-18 secretion, and caspase-1 activation were also observed between PBMCs derived from subject III.1 and healthy control cells after ATP addition. Results are expressed as means and SEMs. P values of less than .05 determined by using the unpaired t test were considered significant. WT, Wild-type.
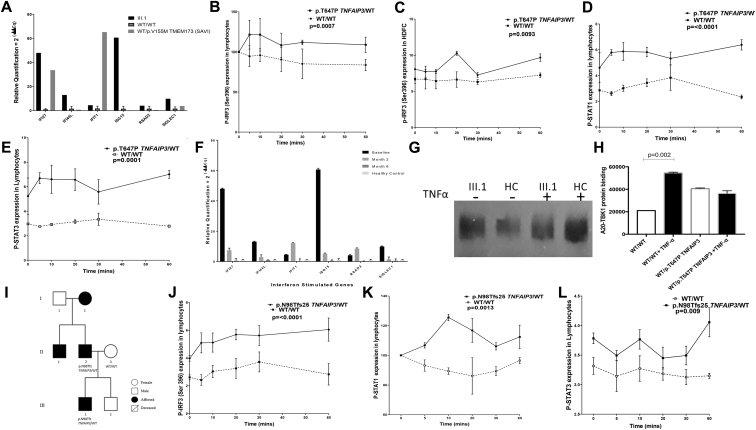
Activation of interferon regulatory factor 3 (IRF3), a critical transcription factor that regulates interferon immune responses, is negatively regulated by A20 through interaction with the NF-κB–activating kinase/TRAF family member–associated NF-κB activator–binding kinase 1 (TBK1).6 Thus we next examined whether interferon immune responses were impaired. Type 1 interferon-stimulated gene expression levels were upregulated in whole blood from subject III.1 (Fig 2, A); levels of serum IFN-α and IFN- β were also increased compared with those in control subjects (P = .04 and P = .049, respectively). We also observed enhanced expression of phosphorylated IRF3 in lymphocytes from all heterozygotes for p.T647P TNFAIP3 variants (P = .0007; Fig 2, B) and in HDFCs from subject III.1 compared with control subjects (P = .009; Fig 2, C). Lymphocytes from subject III.1 also demonstrated increased phosphorylation of signal transducer and activator of transcription (STAT) 1 and STAT3 compared with control subjects (P = .0001 and P = .0001, respectively); similar differences in STAT1 and STAT3 phosphorylation were observed in HDFCs (Fig 2, D and E, and see Fig E2, A and B, in this article's Online Repository at www.jacionline.org).
Fig 2.
Enhanced type 1 interferon signaling in immune cells derived from patients with the heterozygous p.T647P variant in TNFAIP3.A, Type 1 interferon–stimulated gene expression levels were upregulated in whole blood from subject III.1, with levels comparable with those seen in a patient with stimulator of interferon genes vasculopathy with onset in infancy (SAVI). Control data were derived from 13 subjects. B and C, There was increased expression of phosphorylated IRF3 in lymphocytes from all heterozygotes for p.T647P TNFAIP3 (P = .0007) and in HDFCs from subject III.1 (P = .009) compared with control cells. D and E, Lymphocytes from p.T647P TNFAIP3 heterozygotes demonstrated increased STAT1 and STAT3 expression (P = .0001). F, Treatment with an oral JAK1/2 inhibitor resulted in a significant decrease in type I interferon gene expression. G and H, In fibroblasts derived from subject III.1, there was impaired ability of the p.T647P A20 protein to bind to TBK1 in response to TNF-α. I, Pedigree for a family of patients with heterozygous p.N98Tfs25 TNFAIP3–associated autoinflammation. J-L, There was also increased expression of phosphorylated IRF3 (P = .0001), STAT1 (P = .0013), and STAT3 (P = .009) in lymphocytes derived from the p.N98Tfs25 TNFAIP3 heterozygote compared with control cells.
Fig E2.
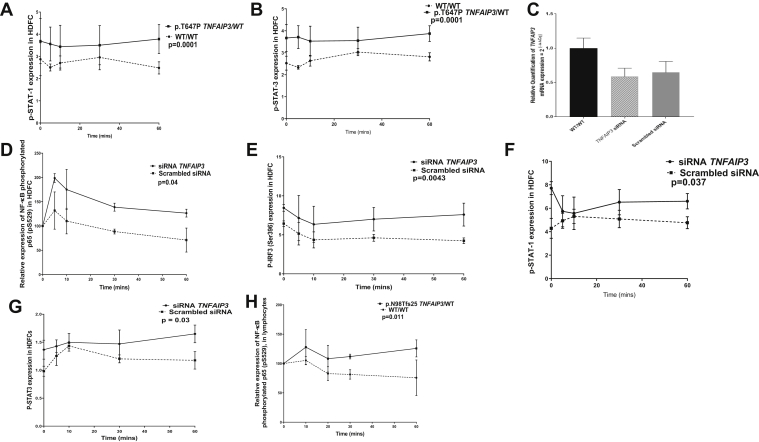
siRNA-mediated silencing of TNFAIP3 enhances type I interferon signaling. A and B, HDFCs from subject III.1 also demonstrated increased phosphorylation of STAT1 and STAT3 compared with healthy control cells (P ≤ .0001 and P ≤ .0001, respectively). C-G, siRNA-mediated silencing of TNFAIP3 in HDFCs resulted in enhanced expression of phosphorylated p65 (P = .04), IRF3 (P = .004), STAT1 (P = .03), and STAT3 (P = .03) compared with scrambled siRNA control cells. G and H, There was increased expression of phosphorylated p65 in lymphocytes from a patient with the p.N98Tfs25 TNFAIP3 variant in comparison with healthy control cells (P = .011). Experiments were performed in triplicate, and results were expressed as means and SEMs. P values of less than .05 determined by using the unpaired t test and ANOVA were considered significant.
Small interfering RNA (siRNA)–mediated silencing of TNFAIP3 in HDFCs resulted in upregulation of phosphorylated p65 (P = .04), phosphorylated IRF3 (P = .0043), phosphorylated STAT1 (P = .037), and phosphorylated STAT3 (P = .03) expression compared with that seen in scrambled siRNA control cells (see Fig E2, C-G).
We documented almost complete clinical and radiologic resolution of neuroinflammation in subject III.1 (Fig 1, H-J) and normalization of interferon-induced gene expression in whole blood (Fig 2, F) after treatment with baricitinib.
To establish whether loss of A20-negative regulatory control over IRF3 activation was secondary to the reduced binding capacity of the A20 protein to TBK1, we next used a coimmunoprecipitation assay. In HDFCs from subject III.1, there was no significant upregulation in binding of TBK1 to the A20 protein in response to TNF-α in contrast to significantly increased binding of A20/TBK1 observed in control cells (Fig 2, G and H).
We next examined whether interferon-mediated immune responses might be generally dysregulated in patients with HA20, irrespective of neurological involvement, in a 4-year-old nonconsanguineous white male with HA20 without any neurological involvement (Fig 2, I). The patient presented with recurrent oral inflammation and penile ulceration and was heterozygous for the p.N98Tfs25 TNFAIP3 variant, as was his symptomatic father who had similar clinical features from early childhood. We confirmed significant upregulation of p65 (P = .011) phosphorylated IRF3 (P = .0001; Fig 2, J), and STAT1 (P = .0013) and STAT3 expression (P = .009; Fig 2, K and L) in lymphocytes from this boy compared with levels seen in healthy control subjects.
We expand the spectrum of clinical presentation associated with HA20 caused by variants in TNFAIP3, which now includes progressive neuroinflammation. We provide insights into the mechanism for the observed immunophenotype through loss of A20-mediated negative regulatory control of IRF3 activation and subsequent dysregulated interferon pathway signaling. TNFAIP3-mediated autoinflammation should now be considered in the differential diagnosis of neuroinflammation, particularly in the presence of intracerebral calcification and uveitis. JAK inhibition might represent a novel therapeutic approach for autoinflammation and neuroinflammation associated with heterozygous TNFAIP3 variants.
Neuroinflammation is rare but has been previously reported in patients with HA20. In a recent case series CNS vasculitis was reported in 2 (13%) of 16 patients with HA20. However, the true frequency of CNS involvement in patients with HA20 might be underappreciated because not all patients were systematically assessed for presence of neurological involvement.2 Therefore we suggest that clinicians should consider screening for neuroinflammation in all patients with suspected HA20. Future collaborative studies might facilitate more detailed phenotype-genotype correlation and could help identify which patients with HA20 might be more at risk of neuroinflammation, but currently, these data do not exist. Notably, animal studies have previously suggested that heterozygous variants in TNFAIP3 cause milder neuroinflammatory changes compared with the severe neuroinflammation observed in complete A20 knockout mice.3 Therefore the heterozygous state in our patients might contribute to the less severe phenotype observed in some of our patients. Of note, immune cells from subjects II.2 and III.2, as well as heterozygotes for the p.T647P TNFAIP3 variant, exhibited enhanced NF-κB activity and IRF3 activation, but these subjects currently have a much milder phenotype, further emphasizing the previously described clinical heterogeneity of HA20, even within the same kindred.1, 2 Additional modifying alleles and genetic and/or environmental risk factors (eg, intercurrent infection or other triggers) might play a role in modifying the phenotype and influence susceptibility to or disease severity of patients with HA20.
Footnotes
All research at Great Ormond Street Hospital NHS Foundation Trust and UCL Great Ormond Street Institute of Child Health is made possible by the National Institute for Health Research (NIHR) Great Ormond Street Hospital Biomedical Research Centre. The views expressed are those of the author(s) and not necessarily those of the National Health Service, the NIHR, or the Department of Health.
Disclosure of potential conflict of interest: C. M. Mulhern acknowledges support from Great Ormond Street Hospital Children's Charity. Y. Hong is funded by an Arthritis Research UK project grant (21791). E. Omoyinmi received support from Rosetrees Trust and SOBI. P. A. Brogan acknowledge support from Great Ormond Street Hospital Children's Charity and received institutional grants from SOBI, Novartis, Roche, Novimmune, and Chemocentryx and consultancy fees from Roche, Novartis, UCB, and SOBI. D. Eleftheriou was supported by Arthritis Research UK (grant 20164 and 21593) and has received institutional grants from Roche, Pfizer and Lilly, and SOBI. The rest of the authors declare that they have no relevant conflicts of interest.
Methods
Patients
This study was approved by the Bloomsbury ethics committee (ethics no. 08H071382). We obtained written informed consent from all the patients, family members, and healthy control subjects (ethics no. 11/LO/0330) who participated in the study. Baricitinib was made available through the following program sponsored by Eli Lilly and Company (Indianapolis, Ind): “Compassionate use treatment protocol I4V-MC-JAGA: treatment of conditions expected to benefit from JAK 1/2 inhibition: CANDLE, CANDLE-related conditions, SAVI and severe juvenile dermatomyositis” (NCT01724580).
Genetic sequencing
Whole-exome sequencing was performed with the Illumina HiSeq 300 platform (Illumina, San Diego, Calif). Data were analyzed in the Galaxy platform (usegalaxy.org), and single nucleotide polymorphism were annotated by using wANNOVAR. The additional case of HA20 studied as disease control was sequenced on the Vasculitis and Inflammation Panel.E1
The TNFAIP3 variant was confirmed, and familial segregation was ascertained by using PCR and Sanger sequencing with the BigDye v3.1 kit (Applied Biosystems, Foster City, Calif) and primers to amplify and sequence exon 8 (forward: ATCTCTGTATCGGTGGGGTG and reverse: TTGTCACTGTCGGTAGAAAACG).
PBMC isolation and stimulation
PBMCs were isolated from freshly drawn heparinized blood by using gradient density centrifugation with Lymphoprep (STEMCELL Technologies, Vancouver, British Columbia, Canada). Blood was diluted with the equal volume of RPM1 640 medium and overlaid on an equal volume of Ficoll-Paque reagent and then centrifuged at 800g for 10 minutes. The PBMC layer was taken, washed, and resuspended in warm RPMI with 10% FCS. Cells were seeded at 2 × 106 cells/well in a 6-well plate and processed for protein extraction or stimulated with 100 ng/mL TNF-α for up to 60 minutes to induce NF-κB activation.
Immunoprecipitation and immunoblotting
Whole-cell lysates were prepared with RIPA buffer (Thermo Fisher Scientific, Waltham, Mass) and 1% proteinase inhibitor from isolated PBMCs or dermal fibroblasts. For immunoprecipitation, lysates were mixed with antibodies (1:100) and incubated with 50 μL of protein A Dynabeads (Thermo Fisher Scientific) overnight at 4 °C. Immunoprecipitates were boiled in 30 μL of 2× Laemmli buffer with 10% β-mercaptoethanol and washed with PBS. For immunoblotting, samples were subjected to SDS-PAGE analysis and electrotransferred onto polyvinylidene difluoride membranes (Millipore, Temecula, Calif). Membranes were blocked with milk and probed with primary and secondary antibodies and visualized with the enhanced chemiluminescence detection system (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom). The following antibodies were used: A20/TNFAIP3 (#5630 CST), TBK1 (D1B4; Cell Signaling Technology, Danvers, Mass), ACTN (MAB 1501R; Merck Millipore, Burlington, Mass), RIP1 (#3493, Cell Signaling Technology), K65-Ub antibody (05-1308; Merck), Ub (sc-271289), and NF-κB essential modulator (sc-56919).
Caspase-1 activity
PBMCs were seeded in a 96-well plate at a density of 1.6 × 105 cells/well (8.0 × 105 cells/mL). Relevant wells were primed with 100 ng/mL LPS for 4 hours and, if required, then stimulated with 10 mmol/L ATP for 30 minutes. Caspase-1 activity was measured by using FLICA (ImmunoChemistry Technologies, Bloomington, Minn), a cell-permeable fluorescent probe (FAM-YVAD-FMK) that binds active caspase-1. Cells were incubated for 1 hour with FLICA at 37°C and stained with phycoerythrin (PE)–conjugated anti-CD14 (BD, Franklin Lakes, NJ) to identify monocytes. Cells were subsequently analyzed for the frequency of CD14+FLICA+ cells.
Interferon-stimulated gene RNA expression from whole blood
Whole blood was collected into PAXgene tubes and RNA extracted by using PreAnalytix RNA isolation. RNA concentrations were assessed with a NanoDrop (FLUOstar Omega; BMG Labtech, Offenburg, Germany). We performed quantitative reverse transcription PCR analysis with the iTaq Universal SYBR Green Supermix (172-5121; Bio-Rad Laboratories, Hercules, Calif) and cDNA derived from 400 ng total RNA. Using Qiagen QuantiTect primers (Qiagen, Hilden, Germany) for IFI27 (NM_001130080), IFI44L (NM_006820), IFIT1 (NM_001001887), ISG15 (NM_005101), RSAD2 (NM_080657), and SIGLEC1 (NM_023068), the relative abundance of each of these target transcripts were normalized to expression level of HPRT1 (NM_000194, HS_HPRT1_1_SG), as assessed by using CFX Maestro software (Bio-Rad Laboratories).
IRF3, STAT1, STAT3, and p65 phosphorylation assays
PBMCs were treated with 100 ng/mL TNF-α for up to 60 minutes. Cells were fixed with BD Cell Fix Buffer (10 minutes at 37°C) and then permeabilized with BD Perm Buffer III (30 minutes at 4°C) before staining with PE–anti–NF-κB p65 pS529 (catalog no. 565447; BD Biosciences, San Jose, Calif), PE–anti-IRF3 (catalog no. 612564; BD Biosciences), PE–anti-STAT1 (catalog no. 61256; BD Biosciences), or Alexa Fluor 488–STAT3 (catalog no. 557814; BD Biosciences) antibodies. Flow cytometric analysis was performed on a BD Calibur flow cytometer. Results were analyzed with FlowJo software (version 10.4.2, TreeStar, Ashland, Ore).
Dermal fibroblasts and siRNA silencing of TNFAIP3 experiments
Dermal fibroblasts were explanted from subject III.1 and from control subjects without any identifiable inflammatory condition and maintained in DMEM/F12 (DMEM/Nutrient Mixture F-12) supplemented with 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin (Life Technologies, Grand Island, NY) at 37°C in 5% CO2 in a humidified incubator. siRNA knockdown was performed in these fibroblasts through transient transfection with 100 nmol/L of siRNA targeting TNFAIP3 (SC-37007; Santa Cruz Biotechnology, Dallas, Tex) or a negative control. siRNAs were transfected through complexation with Lipofectamine RNAiMAX (Thermo Fisher Scientific), according to the manufacturer's instructions. At 48 hours after transfection, knockdown efficiency was evaluated by using quantitative real-time PCR. QuantiTect TNFAIP3 primers were used (QT00070301; Qiagen).
Cytokine measurement
Multiple cytokines were quantified at different time points in the sera of affected patients (subjects III.1, III.2, and II.2), family members (subject II.1), and healthy control subjects by using an electrochemiluminescence immunoassay with Meso Scale Discovery plates (MSD, Rockville, Md), according to the manufacturer's instructions. Data were analyzed with Discovery Workbench 4.0. Control sera were obtained from healthy pediatric control subjects (n = 15; 9 female subjects; median age, 14 years; age range, 12-18 years) with local ethics approval (REC 11/LO/0330).
Statistical analyses
All experiments were complicated in triplicate, unless otherwise specified. Results are expressed as means and SEMs or medians and ranges, where appropriate. All statistics (ANOVA, unpaired Student t test, or Mann-Whitney U test) and graphs were produced by using Prism software (version 7; GraphPad Software, La Jolla, Calif). P values of less than .05 were considered significant.
Table E1.
Subject III.1's routine clinical laboratory investigations
| Laboratory investigations | Subject III.1 (reference range) |
|---|---|
| Autoantibodies persistent >3 mo | Absent∗ |
| Hemoglobin | 10 g/L (120-160 g/L) |
| Platelet count | 182 × 109/L (150-450 × 109/L) |
| WBC count | 6.25 × 109/L (4.0-11 × 109/L) |
| Lymphocyte subsets | Normal |
| IgG | 19.9 g/L (3.1-13.8 g/L) |
| IgA | 1.94 g/L (0.4-0.7 g/L) |
| IgM | 2.29 g/L (0.5-2.2 g/L) |
| IgD | 7 kU/L (2-100 kU/L) |
| Adenovirus, CMV, EBV, HSV, VZV, parechovirus PCR | Negative |
| Toxoplasma species PCR | Negative |
| Mycoplasma species antibodies | Negative |
| QuantiFERON | Negative |
| Nitroblue tetrazolium test | Normal |
| Brucella species serology | Negative |
| Toxocara species serology | Normal |
| 16s PCR and 18s PCR CSF | Negative |
| JC and BK virus | Negative |
| Complement C3 | 1.77 g/L (0.75-1.65 g/L) |
| Complement C4 | 0.24 g/L (0.14-0.54 g/L) |
| Liver enzymes | ALT: 15 U/L (10-25 U/L) ALP: 96 U/L (150-380 U/L) |
| CSF white cell count | <1 × 106 |
| CSF cytospin | Negative |
| CSF oligoclonal bands | Negative |
CSF, Cerebrospinal fluid.
Autoantibodies tested were as follows: antinuclear antibodies, anti-neutrophil cytoplasm antibodies, rheumatoid factor, anti-tissue transglutaminase antibodies, anti-thyroid peroxidase antibodies, anti-myelin oligodendrocyte antibodies, anti-yo, anti-hu, anti-ri antibodies, N-methyl-D-aspartate receptor antibodies, rheumatoid factor antibodies, celiac screen antibodies, β2-glycoprotein, and anti-cardiolipin antibodies.
References
- 1.Zhou Q., Wang H.Y., Schwartz D.M., Stoffels M., Park Y.H., Zhang Y. Loss-of-function mutations in TNFAIP3 leading to A20 haploinsufficiency cause an early-onset autoinflammatory disease. Nat Genet. 2016;48:67. doi: 10.1038/ng.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aeschlimann F.A., Batu E.D., Canna S.W., Go E., Gul A., Hoffmann P. A20 haploinsufficiency (HA20): clinical phenotypes and disease course of patients with a newly recognised NF-kB-mediated autoinflammatory disease. Ann Rheum Dis. 2018;77:728–735. doi: 10.1136/annrheumdis-2017-212403. [DOI] [PubMed] [Google Scholar]
- 3.Guedes R.P., Csizmadia E., Moll H.P., Ma A., Ferran C., da Silva C.G. A20 deficiency causes spontaneous neuroinflammation in mice. J Neuroinflammation. 2014;11:122. doi: 10.1186/1742-2094-11-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao L.L., Coope H., Grant S., Ma A., Ley S.C., Harhaj E.W. ABIN1 protein cooperates with TAX1BP1 and A20 proteins to inhibit antiviral signaling. J Biol Chem. 2011;286:36592–36602. doi: 10.1074/jbc.M111.283762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee Y., Lee E., Zilbernnan-Rudenko J., Chen Y., Gucek M., Siegel R.M. A20 regulates NF-kappa B activation through K48 linked polyubiquitination of NEMO. J Immunol. 2017:198. [Google Scholar]
- 6.Saitoh T., Yamamoto M., Miyagishi M., Taira K., Nakanishi M., Fujita T. A20 is a negative regulator of IFN regulatory factor 3 signaling. J Immunol. 2005;174:1507–1512. doi: 10.4049/jimmunol.174.3.1507. [DOI] [PubMed] [Google Scholar]
Reference
- Omoyinmi E., Standing A., Keylock A., Price-Kuehne F., Gomes S.M., Rowczenio D. Clinical impact of a targeted next-generation sequencing gene panel for autoinflammation and vasculitis. PLoS One. 2017;12:e0181874. doi: 10.1371/journal.pone.0181874. [DOI] [PMC free article] [PubMed] [Google Scholar]