Abstract
We describe results from IMmotion150, a randomized phase 2 study of atezolizumab (anti-PD-L1) alone or combined with bevacizumab (anti-VEGF) versus sunitinib in 305 patients with treatment-naive metastatic renal cell carcinoma. Co-primary endpoints were progression-free survival (PFS) in intent-to-treat and PD-L1+ populations. Intent-to-treat PFS hazard ratios for atezolizumab + bevacizumab or atezolizumab monotherapy versus sunitinib were 1.0 (95% confidence interval (CI), 0.69-1.45) and 1.19 (95% CI, 0.82-1.71), respectively; PD-L1+ PFS hazard ratios were 0.64 (95% CI, 0.38-1.08) and 1.03 (95% CI, 0.63-1.67), respectively. Exploratory biomarker analyses indicated that tumor mutation and neoantigen burden were not associated with PFS. Angiogenesis, T-effector/IFN-γ response, and myeloid inflammatory gene expression signatures were strongly and differentially associated with PFS within and across the treatments. These molecular profiles suggest that prediction of outcomes with anti-VEGF and immunotherapy may be possible and offer mechanistic insights into how blocking VEGF may overcome resistance to immune checkpoint blockade.
Aberrant angiogenesis and antitumor immune suppression are hallmarks of many cancers. Clear-cell renal cell carcinoma (RCC) is associated with a hyperangiogenic state due to overproduction of vascular endothelial growth factor (VEGF) as a result of inactivation of the von Hippel–Lindau tumor-suppressor gene (VHL)1,2. Over the last decade, treatment for metastatic RCC (mRCC) has focused on targeting the VEGF signaling pathway with receptor tyrosine kinase inhibitors, such as sunitinib, or monoclonal antibodies that block VEGF, such as bevacizumab. Although VEGF pathway blockade is effective in many patients, it is associated with development of resistance3,4.
Expression of the immune checkpoint molecule programmed death-ligand 1 (PD-L1) on tumor cells and/or tumor-infiltrating immune cells (IC) has been reported to suppress antitumor immunity and is associated with poor prognosis in mRCC5–10. Ligation of PD-L1 to its receptor, programmed death-1 (PD-1), on T cells results in inhibition of proliferation and effector function of T cells11–13. Atezolizumab is a humanized engineered immunoglobulin G1 monoclonal antibody that selectively targets PD-L1 to block its interaction with PD-1 and the co-stimulatory molecule B7.1 to reinvigorate tumor-specific T-cell immunity. PD-L1/PD-1 inhibitors, including atezolizumab, have shown durable responses in patients with previously treated mRCC8,14.
In addition to its well-characterized role in angiogenesis, VEGF is also believed to play a role in cancer immune evasion15–18. Data from preclinical models and phase 1 studies suggest that anti-VEGF might enhance the antitumor activity of immune checkpoint blockade by improving T-cell infiltration, upregulating major histocompatibility complex class I expression, and reversing myeloid immunosuppression15,16,19–22. A phase 1 study combining atezolizumab and bevacizumab established the combination to be well-tolerated in RCC, with evidence of augmentation of antitumor immunity and encouraging antitumor activity compared with historical experience with either single agent alone21.
The phase 2 IMmotion150 study (), presented here, is the first randomized trial to investigate the clinical activity of atezolizumab with or without bevacizumab, against a standard-of-care anti-VEGF tyrosine kinase inhibitor, sunitinib, in patients with untreated mRCC. Although VEGF-targeted therapy and immune checkpoint inhibition are both established as standards of care for mRCC, biomarkers for response and resistance are lacking. Expression of a VEGF-inducible angiogenesis-associated gene signature has been previously associated with improved clinical outcomes for bevacizumab-containing therapy in patients with metastatic colorectal cancer23. PD-L1 expression by immunohistochemistry (IHC), gene expression signatures related to intratumoral CD8 T-effector cell presence and interferon γ (IFN-γ) activity, and somatic mutation load have previously been associated with immune checkpoint inhibitor activity in patients with melanoma, non-small-cell lung cancer (NSCLC), or bladder cancer24–28. Moreover, myeloid inflammation has been implicated in suppression of the antitumor immune response15–18. We therefore evaluated association of these components of the tumor microenvironment with clinical outcome in the present study. Here we describe the primary clinical results of IMmotion150 and the exploratory molecular analyses conducted to evaluate their potential predictive value and to inform development of personalized therapy in patients with mRCC.
Results
Efficacy.
The primary objective of this prospectively designed phase 2 trial was to estimate the efficacy, as measured by PFS, of atezolizumab + bevacizumab and atezolizumab monotherapy compared with sunitinib, rather than to seek for statistical significance of primary endpoints, in all randomized patients and in the PD-L1+ (PD-L1 on ≥ 1% of IC by IHC) population. Patients were enrolled in the study from 8 January 2014 to 16 March 2015. This report reflects the results of data with a clinical cutoff date of 17 October 2016 and a median survival follow-up of 20.7 months. All randomized patients were included in the intent-to-treat (ITT) population (N = 305) for all efficacy analyses. Patients in the safety analysis population (n = 304) received more than one dose of study drug. One patient in the sunitinib arm was excluded from the safety analysis due to withdrawal of consent before receiving study drug (Supplementary Fig. 1). Discontinuation of treatment was higher with atezolizumab (77.7%) and sunitinib (82.2%) than with atezolizumab + bevacizumab (68.3%), and disease progression was the most common reason for discontinuation among all treatment arms. Patient demographics were comparable across treatment arms for the safety and ITT populations. (Table 1 and see Methods).
Table 1|.
Baseline demographics and patient characteristics
| Sunitinib n = 101 | Atezolizumab + bevacizumab n = 101 | Atezolizumab n = 103 | |
|---|---|---|---|
| Age, median (range), in years | 61 (25-85) | 62 (32-88) | 61 (27-81) |
| Male, n (%) | 79 (78%) | 74 (73%) | 77 (75%) |
| KPS ≥ 80, n (%) | 94 (93%) | 99 (99%) | 101 (99%) |
| Predominant clear cell histology, n (%) | 96 (96%) | 97 (96%) | 95 (92%) |
| Sarcomatoid component, n (%) | 14 (14%) | 15 (15%) | 16 (15%) |
| Prior nephrectomy, n (%) | 88 (87%) | 88 (87%) | 89 (86%) |
| MSKCC risk category, n (%) | |||
| Favorable (0) | 21 (21%) | 30 (30%) | 26 (25%) |
| Intermediate (1 or 2) | 70 (69%) | 62 (61%) | 69 (67%) |
| Poor(≥3) | 10 (10%) | 9 (9%) | 8 (8%) |
| ≥ 1% PD-L1 expression on IC (PD-L1+), n (%) | 60 (59%) | 50 (50%) | 54 (52%) |
KPS, Karnofsky performance status; MSKCC, Memorial Sloan Kettering Cancer Center.
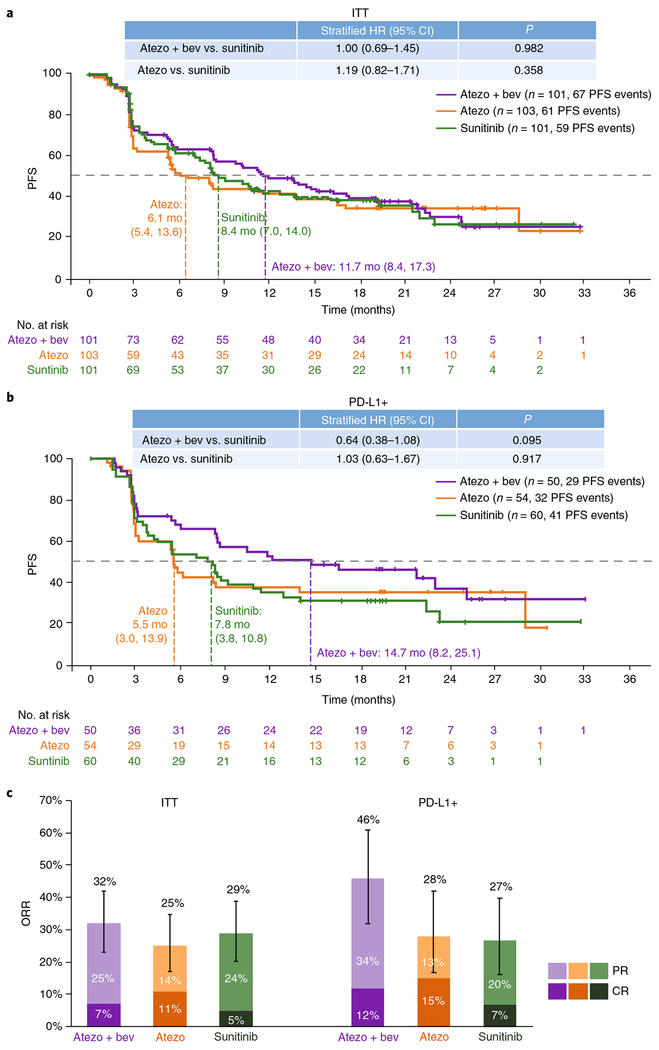
Independent review facility-assessed efficacy endpoints are summarized as follows. There were 67 PFS events among 101 randomized patients in the atezolizumab + bevacizumab arm, 61 PFS events among 103 randomized patients in atezolizumab monotherapy arm, and 59 PFS events among 101 randomized patients in sunitinib arm. Stratified analysis in the ITT population showed a median PFS of 11.7 months (95% CI, 8.4–17.3) with atezolizumab + bevacizumab versus 8.4 months (95% CI, 7.0–14.0) with sunitinib (hazard ratio (HR) 1.00; 95% CI, 0.69–1.45) and 6.1 months (95% CI, 5.4–13.6) with atezolizumab monotherapy (HR 1.19; 95% CI, 0.82–1.71 versus sunitinib; Fig. 1a). In the PD-L1+ population, there were 29 PFS events among 50 patients in the atezolizumab + bevacizumab arm, 32 PFS events among 54 patients in the atezolizumab monotherapy arm, and 41 PFS events among 60 patients in the sunitinib arm. The median PFS in the PD-L1+ population was 14.7 months (95% CI, 8.2–25.1) with atezolizumab + bevacizumab versus 7.8 months (95% CI, 3.8–10.8) with sunitinib (HR 0.64; 95% CI, 0.38–1.08) and 5.5 months (95% CI, 3.0–13.9) with atezolizumab monotherapy (HR 1.03; 95% CI, 0.63–1.67 versus sunitinib; Fig. 1b). The objective response rates (ORRs) were 32% (7% complete response (CR), 25% partial response (PR)), 25% (11% CR, 14% PR), and 29% (5% CR, 24% PR) with atezolizumab + bevacizumab, atezolizumab monotherapy, and sunitinib, respectively (Fig. 1c). In PD-L1+ patients, the ORRs were 46% (12% CR, 34% PR), 28% (15% CR, 13% PR), and 27% (7% CR, 20% PR) with atezolizumab + bevacizumab, atezolizumab monotherapy, and sunitinib, respectively (Fig. 1c). As of the clinical cutoff date of 17 October 2016, 148 investigator-assessed (INV) PFS events were observed in the atezolizumab + bevacizumab and sunitinib arms combined and 154 INV PFS events were observed in the atezolizumab monotherapy and sunitinib arms combined. The concordance between INV PFS (Supplementary Fig. 2) and independent review facility-assessed PFS (Fig. 1a,b) was 77% for all patients and was similar between study arms. PFS in key subgroups with atezolizumab + bevacizumab and atezolizumab monotherapy versus sunitinib is shown in Supplementary Fig. 3 and Supplementary Table 3. Of note, we observed a trend for improved efficacy (PFS) with higher expression of PD-L1 with atezolizumab + bevacizumab and for atezolizumab monotherapy versus sunitinib at a PD-L1 cutoff of ≥ 5% (unstratified analysis; Supplementary Fig. 3).
Fig. 1|. Positive independent review facility (IRF)-assessed efficacy associated with atezolizumab + bevacizumab in mRCC patients with PD-L1+ IC.
a,b, Kaplan-Meier curves depict IRF-assessed median PFS in the atezolizumab (atezo) + bevacizumab (bev), atezolizumab monotherapy, and sunitinib treatment arms in the (a) ITT population and (b) PD-L1+ (≥1% PD-L1 expression on IC by IHC) population across 33 months. Censored data are indicated by vertical tick marks in Kaplan-Meier curves. Sample numbers per group and timepoint indicated below each graph. HR calculated using stratified Cox proportional hazard regression models, and P values calculated using stratified log-rank test (for details, see Methods). P values are provided for descriptive purposes only and were not adjusted for multiple comparisons. Mo, months. c, ORRs as depicted by PR and CR for the ITT and PD-L1+ populations for each treatment arm (n = 101, 103, and 101 patients for atezolizumab + bevacizumab, atezolizumab monotherapy, and sunitinib treatment arms, respectively, in the ITT population; n = 50, 54, and 60 patients, respectively, in the PD-L1+ population). ORR values are indicated above each bar (with 95% CIs for ORR plotted as error bars). Values within the lighter and darker regions of the bars refer to the PR and CR rates, respectively
Safety.
Treatment-related adverse events (AEs) leading to discontinuation occurred in 9% of patients in the atezolizumab + bevacizumab arm, 3% in the atezolizumab monotherapy arm, and 9% in the sunitinib arm (Table 2). In the atezolizumab + bevacizumab group, proteinuria was the most common related AE leading to treatment discontinuation (5%). With sunitinib, the most common related AEs leading to treatment discontinuation were increased blood creatinine and palmar-plantar erythrodysesthesia syndrome (2% each). With atezolizumab monotherapy, nephritis, pancreatitis, and demyelination (1% each) were the treatment-related AEs leading to discontinuation of treatment. Supplementary Fig. 4 shows all-cause AEs occurring at frequency of ≥ 20% in the atezolizumab + bevacizumab and sunitinib arms or in the atezolizumab monotherapy and sunitinib arms, with a difference in incidence between the two arms of ≥ 5%; select AEs of special interest and all AEs occurring in ≥ 20% of patients in any of the three arms are also shown.
Table 2|.
Toxicity profile and safety summary
| Sunitinib n = 100 | Atezolizumab + bevacizumab n = 101 | Atezolizumab n = 103 | |
|---|---|---|---|
| Treatment duration, median (range), in months | 6.7 (0.1-33.1) | Atezolizumab: 11.8 (0.7-32.7)Bevacizumab: 10.3 (0.0-29.8) | 7.6 (0.0-33.1) |
| All-grade AEs, any cause, n (%) | 99 (99%) | 101 (100%) | 101 (98%) |
| Treatment-related AE | 96 (96%) | 91 (90%) | 86 (83%) |
| Grade 3/4 AEs, any cause, n (%) | 69 (69%) | 64 (63%) | 41 (40%) |
| Treatment-related grade 3/4 AEs | 57 (57%) | 40 (40%) | 17 (17%) |
| AEs leading to death, n (%)a | 2 (2%) | 3 (3%) | 2 (2%) |
| Treatment-related AEs leading to deatha | 2 (2%) | 1 (1%) | 0 |
| AEs leading to withdrawal from treatment, n (%) | 10 (10%) | 15 (15%) | 7 (7%) |
| Treatment-related AEs leading to withdrawal from treatment | 9 (9%) | 9 (9%) | 3 (3%) |
| AEs leading to dose modification or interruption, n (%) | 70 (70%) | 61 (60%) | 28 (27%) |
Sunitinib arm: sudden death (related), intestinal hemorrhage (related). Atezolizumab arm: hematophagic histiocytosis, lower respiratory tract infection. Atezolizumab + bevacizumab arm: intracranial hemorrhage (related), hemorrhage, pneumonia.
Molecular correlates of clinical outcome.
We conducted exploratory studies to evaluate molecular biomarkers, including gene expression and somatic mutations, relevant to the disease and tumor immune biology in RCC and their association with clinical outcomes within each treatment group and across treatment groups. Demographic and baseline characteristics in biomarker subgroups were generally consistent with those in the ITT population (Supplementary Table 1).
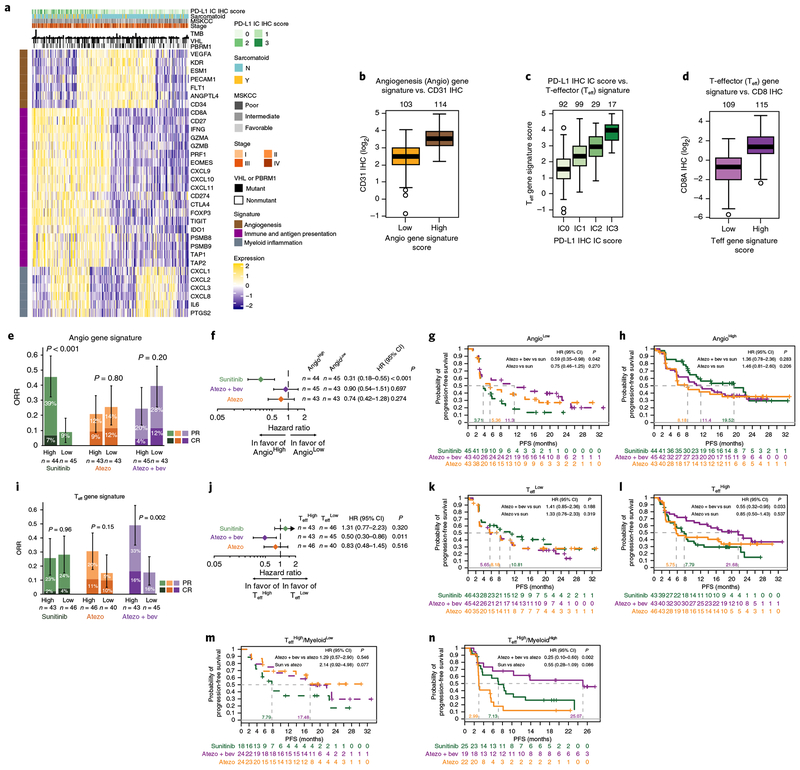
A heatmap of genes previously defined and representing angiogenesis23 and immune biology24,29–33 in 263 evaluable pretreatment tumors (Fig. 2a) showed distinct biological subgroups based on relative expression levels of angiogenesis (Angio), immune (including T-effector presence and function, IFN-γ response, checkpoint inhibitors, and antigen presentation), and myeloid inflammation-associated genes. The subgroup with high expression of the Angio gene signature (AngioHigh) was characterized by relatively higher vascular density as evaluated by CD31 IHC (Fig. 2b), whereas the subgroup with high expression of the T-effector (Teff) gene signature (TeffHigh) was positively associated with protein expression of PD-L1 on IC by IHC (Fig. 2c) and CD8 T-cell infiltration (Fig. 2d), indicative of pre-existing adaptive antitumor immunity. Additionally, differential expression of genes associated with myeloid inflammation within the TeffHigh and TeffLow subgroups was observed, suggesting further functional subcategories of these tumors (Fig. 2a). The associations of clinical outcomes in these biological subgroups within each treatment arm and across treatment arms are shown in Supplementary Table 2. The following comparisons represent a subset of the larger analysis. Biomarker associations with clinical outcomes are discussed when the 95% CI for HR did not cross 1 for PFS evaluation and when the 95% CI were non-overlapping for ORR comparisons.
Fig. 2|. Baseline tumor gene signature analyses.
a, Heatmap showing expression of genes of interest (rows) in 263 pretreatment tumors (columns). Normalized counts of genes related to angiogenesis (brown), immune and antigen presentation (purple), and myeloid inflammation (gray) were z-score transformed before visualization. Sample annotations include PD-L1 IHC status for tumor-infiltrating ICs, presence of sarcomatoid features, Memorial Sloan Kettering Cancer Center (MSKCC) score, tumor stage, number of mutations (TMB), and mutation status of VHL and PBRM1. b, Mean CD31 IHC staining intensity is higher in AngioHigh than in AngioLow (two-tailed t test, P=4.19×10−21). c,d, Teff signature scores are associated with (c) PD-L1 protein expression levels on IC by IHC (one-sided Wald test, P=3.26×10−20) and (d) intratumoral CD8A protein expression by IHC (two-tailed t test, P = 1.26×10−28). Box plot elements in b-d are defined in Methods. Sample numbers per group indicated above each graph. e, ORR= PR + CR in the AngioHigh and AngioLow populations for each treatment arm. Error bars represent 95% CI for ORR; P values calculated using a two-sided χ2 tests. f, Forest plots of PFS HRs and CIs for AngioHigh vs. AngioLow populations within each treatment arm. g,h, Kaplan-Meier curves showing the probability of PFS across treatment arms in the AngioLow (g) and AngioHigh (h) subgroups; HR calculated vs. sunitinib. i, ORR (PR + CR) in the TeffHigh and TeffLow populations for each treatment arm. Error bars represent 95% CIs for ORR; P values calculated using two-sided χ2 tests. j, Forest plots of PFS HRs and CIs for TeffHigh vs. TeffLow populations within each treatment arm. k,l, Kaplan-Meier curves showing probability of PFS across treatment arms in TeffLow (k) and TeffHigh (l) subgroups; HR calculated vs. sunitinib. m,n, Kaplan-Meier curves showing probability of PFS in TeffHighMyeloidLow (m) and TeffHighMyeloidHigh (n) subgroups; HR calculated vs. atezolizumab monotherapy. Censored data indicated by vertical tick marks in Kaplan-Meier curves. All HR and CI values for PFS were extracted from Cox proportional hazard regression models; median survival time per group is indicated. P values reported are for descriptive purposes only and were not adjusted for multiple comparisons. Sample numbers per group indicated below the graphs in e, g-i, and k-n, and within the graphs in f and j.
To determine whether highly angiogenic tumors were more responsive to antiangiogenic therapy, we investigated the association of the Angio gene signature with clinical outcome in each treatment arm. High expression of the Angio gene signature, based on median signature score, was associated with improved ORR (46% in AngioHigh versus 9% in AngioLow; Fig. 2e) and PFS (HR 0.31; 95% CI, 0.18–0.55; Fig. 2f) within the sunitinib treatment arm. When evaluated across treatment arms, no apparent difference in PFS was observed in the AngioHigh subgroup between the atezolizumab + bevacizumab and sunitinib arms or between the atezolizumab monotherapy and sunitinib arms (Fig. 2h). In the AngioLow subgroup, atezolizumab + bevacizumab demonstrated improved PFS versus sunitinib (HR 0.59; 95% CI, 0.35–0.98; Fig. 2g).
We next asked whether the presence of a pre-existing immune response, as identified by expression of the Teff gene signature, may be associated with clinical benefit to immunotherapy-containing regimens. High Teff gene signature expression, based on median signature score, was associated with improved ORR (49% in TeffHigh versus 16% in TeffLow; Fig. 2i) and PFS (HR 0.50; 95% CI, 0.30–0.86; Fig. 2j) versus low Teff gene signature expression within the atezolizumab + bevacizumab arm (Fig. 2j). When compared across treatment arms, high Teff gene signature expression was associated with improved PFS with atezolizumab + bevacizumab versus sunitinib (HR 0.55; 95% CI, 0.32–0.95; Fig. 2l).
Given that myeloid inflammation has been associated with suppression of the antitumor adaptive T-cell response18, we next investigated the contribution of the myeloid inflammation signature to clinical outcome. High myeloid inflammation gene signature expression (MyeloidHigh), based on median signature score, was associated with reduced PFS in the atezolizumab monotherapy arm (HR 2.98; 95% CI, 1.68–5.29) and, to a lesser extent, in the atezolizumab + bevacizumab arm (HR 1.71; 95% CI, 1.01–2.88), but not in the sunitinib arm (Supplementary Table 2). When compared across treatment arms, MyeloidHigh was associated with worse PFS with atezolizumab monotherapy versus sunitinib (HR 2.03; 95% CI, 1.21–3.40); however, this was not observed between atezolizumab + bevacizumab versus sunitinib (Supplementary Table 2).
In addition to evaluation of gene expression signatures that distinguish the clinical activity of atezolizumab + bevacizumab versus sunitinib, we investigated gene expression profiles that may differentiate the activity of atezolizumab + bevacizumab versus atezolizumab monotherapy. Teff, Angio, or myeloid inflammation gene expression signatures did not differentiate activity of atezolizumab + bevacizumab versus atezolizumab monotherapy when evaluated across the respective dichotomized expression subgroups (Supplementary Table 2). The heat map of the three gene signatures (Fig. 2a) showed a distinct population of MyeloidHigh tumors within the inflamed (TeffHigh) category of mRCC tumors. We asked whether the presence of myeloid inflammation within this subgroup of TeffHigh tumors impacted clinical outcome with the three therapies. Atezolizumab monotherapy had worse activity in the TeffHighMyeloidHigh tumors compared with the TeffHighMyeloidLow tumors (HR 3.82; 95% CI, 1.70–8.60; Supplementary Table 2). When compared across treatment arms, atezolizumab + bevacizumab showed improved PFS compared with atezolizumab monotherapy (HR 0.25; 95% CI, 0.10–0.60; Fig. 2n). No apparent difference in PFS was observed between atezolizumab + bevacizumab and atezolizumab monotherapy in the TeffHighMyeloidLow subgroup (Fig. 2m).
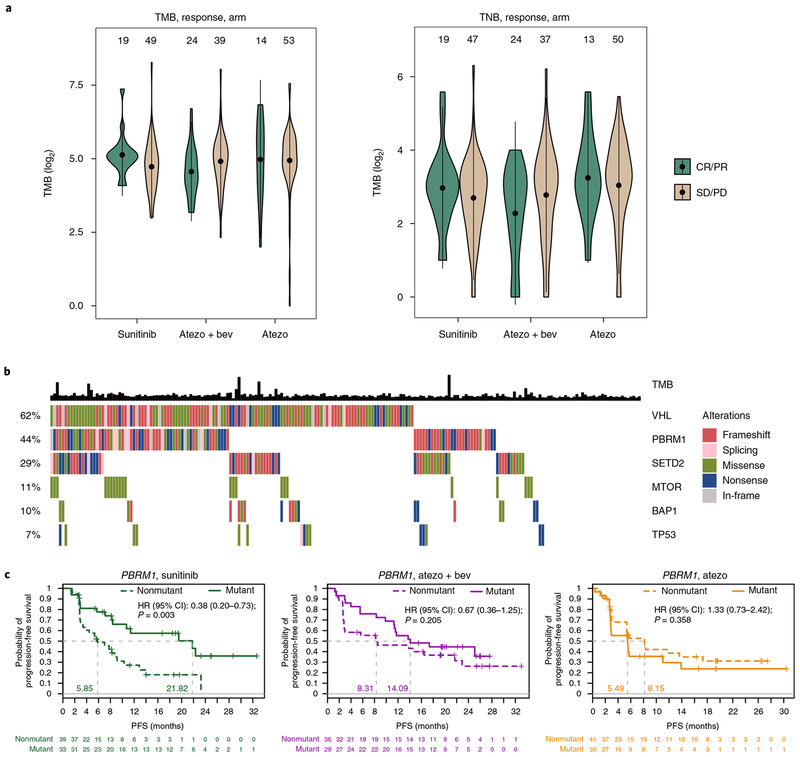
The presence of genomic instability, as identified by tumor mutation burden (TMB) and tumor neoantigen burden (TNB), has been associated with improved clinical outcome to checkpoint inhibitors25,34,35. In addition, small insertions and deletions (indels) and associated frameshift mutation burden (FMB) have been associated with increased immunogenicity in RCC36. However, to our knowledge, these markers of potential immunogenicity have not been previously studied in a randomized setting in mRCC. Whole-exome sequencing of 201 tumors and matched peripheral blood mononuclear cells showed that the median expressed TMB was 30 (range, 1–309; 95% CI, 27–32), the median TNB was 8 (range, 0–79; 95% CI, 7–9), the median indel load was 3 (range, 0–113; 95% CI, 2.4–3.9), and the median FMB was 2 (range, 0–55; 95% CI, 1.5–2.6). Notably, no apparent association was found between TMB, TNB, indels, or FMB and intratumoral Teff gene signature (Supplementary Fig. 5) or clinical benefit in any of the three treatment arms (Fig. 3a and Supplementary Fig. 6).
Fig. 3|. Association between tumor mutations and clinical outcome.
a, TMB and TNB are plotted by response group (CR and PR vs. SD (stable disease) and PD (progressive disease)) for each treatment arm. No apparent difference was observed between response groups in the sunitinib (two-tailed t test, P = 0.06), atezolizumab + bevacizumab (two-tailed t test, P = 0.14), and atezolizumab monotherapy arms (two-tailed t test, P = 0.93). The violin plots show the kernel probability density of the data. The point indicates the mean TMB or TNB in each group, while the bars represent the 95% CIs of the mean. Sample numbers per group indicated above the violin plots. b, Presence of loss-of-function mutations in genes of interest across 201 tumors. Based on functional prediction of mutation consequence, mutations were categorized as frameshift, splicing, missense, nonsense, or in-frame. The overall prevalence of mutations for each gene is shown on the left as percentages. In addition, TMB is shown. c, Patients were divided into two groups based on the presence of loss-of-function mutations (mutant vs. nonmutant). The probability of PFS for these two groups is shown for each treatment arm. HR and CIs were extracted from Cox proportional hazard regression models, comparing the mutant group with the nonmutant group. Censored data are indicated by vertical tick marks in Kaplan-Meier curves. Median survival time per group is indicated. P values reported are for descriptive purposes only and were not adjusted for multiple comparisons. Sample numbers per group and timepoint indicated below the graphs.
Evaluation of disease-associated biology showed that two of the most frequently mutated genes in this patient population were VHL (62%) and PBRM1 (44%; Fig. 3b). Mean Angio gene signature expression was higher in both VHL and PBRM1 mutants than in nonmutants (Supplementary Fig. 7). No apparent association was observed between VHL mutation status and PFS in any treatment group (Supplementary Fig. 8). Within-treatment-arm evaluation showed that PBRM1 mutations were associated with improved PFS in the sunitinib group (HR 0.38; 95% CI, 0.20–0.73; Fig. 3c). When compared across treatments, atezolizumab monotherapy was associated with worse PFS than sunitinib (HR 2.49; 95% CI, 1.26–4.91), and atezolizumab + bevacizumab showed improved PFS versus atezolizumab monotherapy (HR 0.42; 95% CI, 0.22–0.82) in the PBRM1 mutant subgroup.
Discussion
IMmotion150 is the first randomized study to evaluate the clinical activity of the combination of an antiangiogenesis agent and an immune checkpoint inhibitor in treatment-naive patients with mRCC. It is distinguished from other ongoing randomized trials investigating checkpoint inhibition in untreated mRCC by the inclusion of a PD-L1/PD-1 inhibitor monotherapy arm. The combination of atezolizumab + bevacizumab produced encouraging efficacy versus the most commonly applied kidney cancer therapy, sunitinib, in the subgroup of patients with tumors expressing PD-L1 on ≥ 1% of IC (54% of enrolled patients). This finding was further supported in a randomized phase 3 study (IMmotion151, )37. Atezolizumab also demonstrated antitumor activity when administered as a single agent and was well tolerated. Of note, the high response rate observed with atezolizumab monotherapy, including complete responses, support further investigation, including in the adjuvant setting for patients with resected high-risk RCC (IMmotion010, ). Safety in the atezolizumab + bevacizumab arm and the atezolizumab monotherapy arm was consistent with previous data for each drug alone, and AEs leading to treatment discontinuation were low.
A consistent trend of increasing efficacy with increasing levels of PD-L1 IC expression across both atezolizumab containing arms underscores the relevance of pre-existing immunity for differentiating the activity of atezolizumab and atezolizumab + bevacizumab from sunitinib in mRCC. This was particularly true in the atezolizumab + bevacizumab arm, in which bevacizumab appeared to enhance antitumor activity in immunogenic tumors. The predictive relevance of PD-L1 expression on IC is further supported by the strong correlation of PD-L1 IC as determined by IHC with the Teff immune gene signature. These findings are consistent with results from other recently reported studies in mRCC38,39. Specifically, in a phase 3 study evaluating ipilimumab + nivolumab versus sunitinib, improved clinical benefit was observed in patients expressing PD-L1 on tumor cells (24% of enrolled patients)39. These data support development of PD-L1 IHC and/or an immune gene expression signature based diagnostic or enrichment strategy in treatment-naive patients with mRCC.
To further identify determinants of differential activity across the three treatment groups, we interrogated three biological axes that we hypothesized play a role in the response to the treatment regimens studied: tumor angiogenesis, pre-existing immunity, and immunosuppressive myeloid inflammation. Sunitinib efficacy was enriched in highly angiogenic tumors (AngioHigh) and, in agreement with previous reports40, coincided with tumors exhibiting PBRM1 mutations. The combination of atezolizumab + bevacizumab improved clinical benefit compared with sunitinib in TeffHigh tumors. Atezolizumab monotherapy was effective in tumors with pre-existing immunity and a relatively lower expression of myeloid inflammation-associated genes (TeffHighMyeloidLow), but less so in immunogenic tumors with concomitantly high myeloid inflammation (TeffHighMyeloidHigh). Myeloid inflammation associated with high expression of IL-6, prostaglandins, and the CXCL8 family of chemokines has been implicated in accumulation of myeloid-derived suppressor cells in tumors and suppression of antitumor immunity41–46, and VEGF/VEGF receptor blockade has been shown to reduce myeloid-derived suppressor cells in tumors and blood in preclinical tumor models16,47 and human cancers22. The improved clinical outcome associated with atezolizumab + bevacizumab compared with atezolizumab monotherapy in the immunesuppressed TeffHighMyeloidHigh subgroup suggests that the addition of bevacizumab to atezolizumab may overcome innate inflammation-mediated resistance in these tumors. Although these findings require validation, the molecular subgroups identified in this study may have discriminatory characteristics with potential broad relevance for application of antiangiogenesis and checkpoint inhibitor-based therapies across a spectrum of cancers, in which VEGF expression may contribute to tumor immunosuppression. Notably, a recent study investigating chemotherapy in combination with atezolizumab + bevacizumab in NSCLC reported improved efficacy versus chemotherapy and bevacizumab alone48. The combination of atezolizumab + bevacizumab is also under evaluation in hepatocellular carcinoma, gastric, and ovarian cancer (, ).
The lack of association between TMB or TNB and efficacy in atezolizumab-containing treatment groups in RCC is in contrast to results for other malignancies, such as NSCLC24 and metastatic urothelial carcinoma25. Recently, it has been reported that indels and associated frameshift mutations may be an important source of high-affinity neoantigens and that their relatively higher frequency in RCC may drive its immunogenic phenotype despite relatively lower TMB levels36. Our analyses in a larger randomized cohort in RCC failed to confirm these findings. Thus, the underlying biological basis of immunogenicity in RCC remains to be identified, and other determinants of immunogenicity such as clonal indels36 and expression of endogenous retroviruses49 warrant further investigation.
Retrospective data from a heterogeneous population recently linked PBRM1 mutations with response to immune checkpoint inhibitors in renal cancer50. Our data does not support this finding. In agreement with other recent reports in first-line mRCC40,51, our data instead suggest association of PBRM1 mutations with improved outcome to sunitinib. Differences in the patient population and study design may account for these differences.
The combination of ipilimumab + nivolumab recently showed improved overall survival in all randomized patients, compared with sunitinib in patients with previously untreated mRCC, while no PFS benefit was observed in all randomized patients. Enhanced clinical benefit was demonstrated for both endpoints in patients with intermediate and poor risk determined by the International Metastatic Renal Cell Carcinoma Consortium (IMDC) score. However, patients with favorable prognostic risk showed superior PFS with sunitinib39. Of note, results presented here suggest that atezolizumab + bevacizumab has clinical activity across prognostic risk groups, including favorable risk patients, as identified by the Memorial Sloan Kettering Cancer Center score52. Ad hoc analysis of the IMDC prognostic risk groups, derived from clinical data reported at screening, revealed a concordance rate of 78% overall between the two prognostic risk models in the atezolizumab + bevacizumab versus sunitinib arms and corroborated antitumor activity of the combination therapy as measured by PFS across risk groups.
The work presented here is limited by the size and hypothesis-generating nature of the study, requiring further evaluation of the clinical and biomarker findings in larger randomized trials. Ongoing phase 3 trials investigating the combination of VEGF and PD-L1/PD-1 inhibition will provide further insights into the role of biomarkers and prognostic risk scores for cancer immunotherapy in mRCC (IMmotion151 (), , , and ).
Overall, data from IMmotion150 suggest that atezolizumab + bevacizumab may particularly enhance PFS benefit in patients with pre-existing antitumor immunity (as determined by a high Teff score and PD-L1 IC expression) compared with sunitinib. In addition, we report comprehensive biomarker analyses that expand our understanding of the biology of kidney cancer and may identify patient populations that derive benefit from sunitinib, atezolizumab, or the combination of atezolizumab + bevacizumab to enable personalized therapy in patients with mRCC. Furthermore, our findings identify myeloid inflammation as a potential mechanism of innate resistance to atezolizumab monotherapy in mRCC that may be overcome by the addition of bevacizumab. The findings from this study will be evaluated in the phase 3 study IMmotion151, which will examine atezolizumab + bevacizumab versus sunitinib in first-line mRCC, and the phase 3 IMmotion010 trial, which will examine atezolizumab in the adjuvant setting.
Methods
Study design and outcomes.
IMmotion150 () is a prospectively designed, phase 2, multicenter, randomized, open-label study conducted in 96 institutions that was designed to evaluate the safety and provide preliminary evidence of activity of atezolizumab + bevacizumab vs. sunitinib, and atezolizumab monotherapy vs. sunitinib, as well as to inform the study design of the phase 3 trial (IMmotion151; ). The sample size was ~100 patients per arm, and a 70% event rate was deemed adequate for estimation of effect size (including median PFS and HR) in the ITT and PD-L1+ subgroups by the trial steering committee. While this study is large for a randomized phase 2, its size allowed for subset analysis of biomarkers, thus increasing the study’s relevance.
The original primary endpoint was PFS per Response Evaluation Criteria in Solid Tumors version 1.1 (RECIST v1.1) via IRF in the ITT population. Although patients were stratified by ≥ 5% PD-L1 expression on IC, the definition of PD-L1 positivity was revised from ≥ 5% to ≥ 1% PD-L1 expression on IC, based on phase 1a data, and the study protocol was amended to create PD-L1 expression on IC as a co-primary endpoint of IRF-assessed PFS. This amendment likely contributed to the slight imbalance in the numbers of PD-L1+ patients between treatment arms (sunitinib, 59%; atezolizumab, 52%; atezolizumab + bevacizumab, 50%; Table 1). Secondary endpoints included investigator (INV)-assessed PFS, ORR, and duration of response (DOR) per RECIST v1.1, overall survival (OS), patient-reported outcomes, and safety. Key exploratory objectives included evaluation of the relationship between the expression of predictive and prognostic exploratory biomarkers and their association with disease status and efficacy, as defined by ORR and PFS. All data are reported per IRF assessment, unless otherwise stated.
This study was conducted in full conformance with the International Conference on Harmonization E6 guidelines for Good Clinical Practice and the principles of the Declaration of Helsinki, or the laws and regulations of the country in which the research was conducted, whichever afforded the greater protection to the individual.
Participants.
Eligible patients were ≥ 18 years of age, had a Karnofsky performance score ≥ 70, and had unresectable advanced or mRCC with a component of clear cell histology and/or sarcomatoid histology not previously treated with any systemic agents for RCC. Patients were required to have adequate hematologic and end-organ function. Patients were excluded if they had known active brain or spinal cord metastases, uncontrolled pleural/pericardial effusion or ascites, or uncontrolled hypercalcemia.
Randomization and masking.
After written informed consent was obtained and eligibility determined, the study site obtained each patient’s identification number and treatment assignment from the interactive voice/web response system (IxRS). Stratification factors at the time of randomization included Memorial Sloan Kettering Cancer Center (MSKCC) risk category52 (low, intermediate, or high risk), prior nephrectomy (yes or no), and PD-L1 status (≥ 5% or < 5% PD-L1 expression on IC) as determined by IHC staining using the SP142 assay. Stratified permuted block randomization was used to assign patients in a 1:1:1 ratio to one of three treatment arms: atezolizumab + bevacizumab, atezolizumab alone, or sunitinib. The study was open-label and allocation was unmasked.
Procedures.
Study treatment consisted of atezolizumab 1,200 mg fixed intravenous dose + bevacizumab 15 mg/kg every three weeks, atezolizumab 1,200 mg fixed intravenous dose every three weeks, or sunitinib 50 mg/d orally for four weeks followed by two weeks of rest. On disease progression (as assessed by the investigator per RECIST v1.1), patients randomized to atezolizumab monotherapy or sunitinib had the option to cross over and receive the atezolizumab + bevacizumab combination in some regions (option was not available in Europe). Crossover and OS data are immature and not presented here but will be the subject of a future manuscript. In the absence of unacceptable toxicity, treatment with the combination continued until evidence of PD. Where permitted, patients in the atezolizumab-containing arms could continue to be treated beyond disease progression per RECIST v1.1 until lack of clinical benefit; those in non-European nations could cross over to atezolizumab + bevacizumab therapy at any time after disease progression per RECIST v1.1, provided all eligibility criteria were met.
During the study, each cycle was 6 weeks (42 d) in duration, and data on tumor measurement and survival status were collected for evaluation of PFS, milestone PFS (at 24, 52, and 76 weeks), OS, and ORR per RECIST v1.1. Tumor assessments occurred at baseline and every 12 weeks ± 5 business days after Cycle 1, Day 1, or more frequently if clinically indicated. Patients who discontinued first-line treatment or crossover treatment were followed up for survival approximately every 3 months until death, withdrawal of consent, loss to follow-up, or study termination. Patients who discontinued study treatment for reasons other than disease progression (e.g., toxicity) continued to undergo scheduled tumor assessments (every 12 weeks) until death, disease progression per RECIST v1.1, withdrawal of consent, or study termination, whichever occurred first.
Statistical analysis.
Kaplan–Meier methodology was used to estimate the median PFS for each treatment arm, and Kaplan–Meier curves were produced. PFS was defined as the time from the date of randomization to first occurrence of PD or death from any cause, whichever occurred first. Patients without PD or death were censored at the last assessment date. If no assessment was performed, data was censored at the randomization date. The primary analysis was triggered when 140 INV PFS events occurred among patients treated with atezolizumab + bevacizumab and with sunitinib or among patients treated with atezolizumab alone and with sunitinib, whichever occurred later. The HR estimates and their 95% CIs were determined by using the stratified Cox proportional hazards model. The stratification factors included prior nephrectomy, tumor PD-L1 status, and MSKCC score, and were determined based on data from the electronic case report form; if such data were missing, data collected by the IxRS at the time of randomization were used. Descriptive statistics include means, medians, ranges, and s.d., as appropriate. Interim analyses occurred before the analysis presented here, and endpoints were modified during the trial to help inform the randomized phase 3 study. The analysis, therefore, was exploratory. The patient-reported outcomes of symptom severity and interference with daily functioning were collected using the MD Anderson Symptom Inventory and will be the subject of a future manuscript.
Tumor specimens from patients acquired < 12 months before study treatment were required for enrollment in the study. Tumor specimens were prospectively tested for PD-L1 expression on ICs by a central laboratory using the SP142 IHC assay (Ventana, Tucson, AZ). IC staining was defined as follows: any discernible PD-L1 staining of any intensity in IC covering < 1% or absent (IC 0), ≥ 1% to < 5% (IC 1), ≥ 5% to < 10% (IC 2), or ≥ 10% (IC3) of tumor area occupied by tumor cells and by associated intratumoral and contiguous peritumoral desmoplastic stroma. Statistical analysis of the biomarker component of this study focused on comparisons of the two study arms with the control arm. Analysis was also performed within study arms comparing high and low expression (Supplementary Table 2). A final component of this analysis explored outcomes specifically in the stromal high subpopulation, which appeared resistant to immune therapy. This analysis was ad hoc in nature.
Gene expression analyses.
Whole-transcriptome profiles were generated for 263 patients using TruSeq RNA Access technology (Illumina). RNA-seq reads were first aligned to ribosomal RNA sequences to remove ribosomal reads. The remaining reads were aligned to the human reference genome (NCBI Build 38) using GSNAP53,54 version 2013-10-10, allowing a maximum of two mismatches per 75 base sequence (parameters: ‘-M 2 -n 10 -B 2 -i 1 -N 1 -w 200000 -E 1 –pairmax-rna = 200000 -clip-overlap). To quantify gene expression levels, the number of reads mapped to the exons of each RefSeq gene was calculated using the functionality provided by the R/Bioconductor package GenomicAlignments55.
Gene signatures were defined as follows: Angio23: VEGFA, KDR, ESM1, PECAM1, ANGPTL4, and CD34; Teff24: CD8A, EOMES, PRF1, IFNG, and CD274; myeloid inflammation29–33: IL-6, CXCL1, CXCL2, CXCL3, CXCL8, and PTGS2. These three gene expression signatures were defined based on previously published associations with their respective biology.
To calculate scores for each of these signatures, counts were first normalized using edgeR’s normalization factors56, followed by filtering out genes with low coverage (i.e., not reaching 0.25 CPM (counts per million) in at least one-tenth of available samples) and log2-transformation using limma’s voom57. Then for each sample, the average expression of all genes in a given signature was computed, and is reported as the sample’s signature score. For each gene signature, patients were divided into two groups based on the median gene signature score of all tumors: high gene signature expression was defined as expression at or above median levels, and low gene signature expression was defined as expression below the median.
For the heatmap (Fig. 2a), each patient was placed into high or low groups for all three gene expression signatures: Angio, Teff, and myeloid inflammation (based on median expression, as described above). Subsequently, patients were sorted by the combination of these three groups: first TeffHighAngioLow patients are shown, sorted by myeloid inflammation low/high; then TeffHighAngioHigh patients are shown, sorted by myeloid inflammation high/low; then TeffLowAngioHigh patients are shown, sorted by myeloid inflammation low/high; finally, TeffLowAngioLow patients are shown, sorted by myeloid inflammation high/low. Also, the ordering of the genes was predetermined, based on biological function. Z-score-transformed normalized counts are shown.
Whole-exome sequencing and variant calling.
Whole-exome sequencing (WES) data was generated for 208 patients, sequencing DNA extracted from both tumor as well as peripheral blood mononuclear cells using the Agilent SureSelect v5 (51 MB) kit on a HiSeq 2500 (Illumina) sequencer. FASTQ reads were aligned to the human reference genome (NCBI Build 38) using GSNAP53,54 v. 2013-10-10 (parameters: ‘-M 2 -n 10 -B 2 -i 1 –pairmax-dna = 1000 –terminal-threshold = 1000 –gmap-mode = none –clip-overlap’). Duplicate reads in the resulting BAM file were marked using PicardTools, and indels realigned using the GATK IndelRealigner tool.
Somatic variants were called using a union of Lofreq v2.1.258 and Strelka59 calls. Indel qualities were assigned to the alignments using ‘lofreq indelqual –dindel’, and somatic mutations were called using ‘lofreq somatic’ with the ‘–call-indels’ option. Strelka-based somatic mutations were called using the Strelka-provided configuration file strelka_config_bwa_default.ini, with the only modification being the setting ‘isSkipDepthFilters = 1’ instead of ‘isSkipDepthFilters = 0’. Somatic mutations were annotated for effects on transcripts using Ensembl Variant Effect Predictor60 on RefSeq-based gene models. To identify nonsynonymous mutations, mutations were retained when the most severe consequence was one of frameshift_variant, stop_lost, stop_gained, start_lost, initiator_codon_variant, inframe_insertion, inframe_deletion, missense_variant, coding_sequence_variant, or protein_altering_variant. For single-gene mutation status, allelic frequency of a variant had to be at least 5% for categorization of the patient sample as mutant.
Indel calling.
Only Strelka59 variant calls were considered for identifying indels. Predicted indels were further categorized into frameshift indels (fs_indels), inframe indels (inframe_indels), or other indels (other_indels). Frameshift indels included stop_lost, as there is novel sequence downstream of the indel, which from the point of view of neoantigen generation is equally effective as frameshift indels, although the functional impact is distinct as there is no truncation of sequence. Indels whose most severe effect was other than frameshift or in-frame indels were classified as other indels. All nonindel mutations were categorized as substitutions. A given genomic locus was categorized as one of the above classes, with no repetition, i.e., for the purpose of quantifying genomic mutation load in each of the above categories, one genomic mutation was counted only once, based on the most severe effect across all transcripts corresponding to that locus.
Neoantigen prediction.
Expressed mutations were identified by tallying RNA-seq alignments for identified mutations in the exome data using the tally Variants function from the R package VariantTools61. The neoantigen potential of each mutation was predicted after identifying HLA genotypes of the subjects and assigning the optimal HLA-neoepitope pair across all HLA alleles and 8- to 11-mer peptides containing the mutation, based on minimum IC50 value predicted by NetMHCcons62. HLA genotyping was completed on whole-exome data from PBMCs, using Polysolver63.
Biomarker association analyses.
For testing associations between a continuous variable and a binary trait (Fig. 2b,d and Supplementary Fig. 7), two-tailed t tests were used. Otherwise, for testing associations between a continuous variable and a categorical variable with more than two levels (Fig. 2c), likelihood-ratio test P values were calculated using ANOVA. For testing associations between two continuous variables, Pearson correlation coefficients were calculated (Supplementary Fig. 5).
For testing associations of signature scores or mutational status with PFS, Cox proportional hazard regression models were fit, using survival as response and mutational status or signature score as terms, comparing high- to low-signature score tumors or mutant to nonmutant. For testing associations of TMB, TNB, indels, and FMB with PFS, patients were split into subgroups based on TMB/TNB quartiles. Unstratified Cox proportional hazard regression models were fit and likelihood-ratio test P values extracted. To achieve a normal distribution of the data, TMB and TNB were log2-transformed. We excluded 8, 32 and 41 patients with 0 predicted neoantigens, indels, or frameshift mutations, respectively, from mutation load analyses.
In bar graphs representing ORR and violin plots representing the distribution of TMB and TNB, respectively, the error bars indicate the 95% CIs. For ORR, these were computed using the Clopper–Pearson method64. For box plots, the actual box represents the middle 50% of the data, with the line in the middle indicating the median. The box edges (hinges) are the 25 th and 75 th percentiles. The lines (whiskers) show the largest or smallest observation that falls within a distance of 1.5 × the box size from the nearest hinge; if any observations fall farther away, they are considered outliers and are shown separately.
All P values reported are presented for descriptive purposes only and not adjusted for multiple testing. Biomarker associations with clinical outcomes are further discussed in the main text when the 95% CIs for HR did not cross 1 for PFS evaluation and when the 95% CIs were non-overlapping for ORR comparisons.
Role of the funding source.
The funder of the study (F. Hoffmann-La Roche, AG.) provided study drugs and collaborated with academic authors on study design, data collection, analysis, and interpretation. All authors verify that IMmotion150 was conducted per protocol, which was approved by each site’s independent ethics committee. Those committees are as follows: Asan Medical Center Ethics Committee, Asan Medical Center, IRB; Austin Health HREC, Research Ethics Unit; Austin Health, Austin Health Human Research Ethics Committee; Bellberry Human Research Ethics Committee; CEI de Clinica Bajio CLINBA, Comite de Etica en Investigacion; CEP – ISCMPOA; CEP – PUCRS; CEP para Analise de Projetos de Pesquisa do HCFMUSP e da FMUSP, Hospital da Universidade de Sao Paulo; Chang Gung Med Found, Institutional Review Board; Chungnam National University Hospital, IRB; Cleveland Clinic Florida, Cleveland Clinic Institutional Review Board; Comitato Etico A.O.S. Camillo – Forlanini; Comitato Etico Area Vasta Sud Est; Comitato Etico dell’ Azienda Osped. A. Cardarelli; Comitato Etico Dell’irccs San Matteo Di Pavia; Comitato Etico Di Area Vasta Romagna E Irst; Comitato Etico Irccs Ospedale San Raffaele; Comitato Etico Provinciale Modena; Comitato Etico-Milano Area C C/O A. O. Ospedale Niguarda Ca’Granda; Comitê de Ética em Pesquisa; Universidade Regional do Noroeste do Estado do Rio Grande do Sul UNIJUÍ; Concord Repatriation General Hospital HREC; Copernicus Group IRB; Dana-Farber Cancer Institute Institutional Review Board; E.C. of Altai Oncological Center; EC at FSI MSROI n.a. Hertsen of Rosmedtechnology; EC of FSBI Privolzhsky Federal Medical Research Centre; EC of Moscow City Oncol. Hospital #62; Ethic Committee, University Clinical Center Sarajevo; Ethical Clearance Committee on Human Rights; Ethics Committee University Clinical Centre of the Republic of Srpska; Institutional Review Board, Faculty of Medicine; Lakeridge Health Research Ethics Board; London - Central Research Ethics Committee; Macquarie University Human Research Ethics Committee; McGill University, Sir Mortimer B Davis Jewish General Hospital, Ethics Board; MedStar Health Research Institute-Georgetown Univ. Oncology IRB; Memorial Sloan Kettering Cancer Center; Institutional Review Board; National Cancer Center Institutional Review Board; Nippon Medical School Hospital Institutional Review Board; Nova Scotia Health Authority Research Ethics Board; Ontario Cancer Research Ethics Board; Parkway Independent Ethics Committee; Research Ethics Committee, Nat. Taiwan Univ. Hosp.; Research Ethics Committee. Fac. Med. Chiang Mai University; Seoul National University Bundang Hospital IRB; Seoul National University Hospital; IRB; Severance Hospital- Yonsei University; IRB; SingHealth Centralised IRB, Review Board B; Siriraj Institutional Review Board; Songklanagarind Ethics Committee; St John of God Health Care Ethics Committee; The IRB, Taichung Veterans General Hospital; The University of Chicago IRB; UC Irvine office of Research Administration; US Oncology, Inc. Institutional Review Board; Vanderbilt University Institutional Review Board; Videnskabsetiske Komité Region Midt; Sundhedssekr; WIRB. All authors had access to the study data and confirm data accuracy and completeness. Manuscript medical writing assistance was provided by a sponsor-funded professional medical writer. The corresponding author had final responsibility for the decision to submit for publication.
Reporting Summary.
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data availability.
The data that support the findings of this study, including anonymized genetic data from 163 patients who granted informed consent to share such data, are made available at the European Genome-Phenome Archive (EGA) under accession number EGAS00001002928. Furthermore, we provide qualified researchers access to additional individual clinical patient level data through the clinical study data request platform (https://clinicalstudydatarequest.com/). Details of Roche’s Data Sharing Policy are available here (https://clinicalstudydatarequest.com/Study-Sponsors/Study-Sponsors-Roche.aspx).
Supplementary Material
Acknowledgements
The authors thank A. Bailey for her contributions to development of the protocol and Z. Boyd for his contributions to the development of the PD-L1 IHC assay and its implementation in this study. Support for third-party writing assistance for this manuscript—by P.S. Davies of Health Interactions, Inc.—was provided by F. Hoffmann-La Roche, AG. This study was sponsored by F. Hoffmann-La Roche, AG. Authors were funded by NCI grants P50 CA101942-13 to D.F.M, M.B.A., and T.K.C.; P30 CA008748 to R.J.M.; and P30 CA14599 to W.M.S.
Competing interests
D.F.M. reports a consulting/advisory role for Bristol-Myers Squibb, Merck, Roche/Genentech, Pfizer, Exelixis, Novartis, Eisai, X4 Pharmaceuticals, and Array BioPharma; and reports that his home institution receives research funding from Prometheus Laboratories. M.B.A. has been a paid consultant to Roche/Genentech, Bristol-Myers Squibb, Merck, Pfizer, Novartis, Exelixis, and Eisai. R.J.M. reports consulting fees from Roche/Genentech, Novartis, Pfizer, Eisai, and Exelixis, and research funds from Roche/Genentech, Bristol Myers Squibb, Pfizer, Novartis, Eisai, and Exelixis to the hospital for which he is employed. B.I.R. reports research funding to his institution from Roche/Genentech during the conduct of the study and grants/fees from Pfizer and Merck outside the submitted work. B.E. reports honoraria and research funding from Bristol-Myers Squibb, Novartis, Pfizer, and Ipsen; honoraria from Eusa Pharma, Roche, and Eisai; and research funding from Aveo. L.F. reports research funding to his institution from Roche/Genentech outside the submitted work. R.W.J. reports a consulting/advisory role with Bristol-Myers Squibb, Nektar, Genoptix, Eisai, Novartis, and Exilixis and research funding from Merck and Bristol-Myers Squibb; his home institution is in a consulting/advisory role with Merck and receives research funding from Roche/Genentech, X4 Pharmaceuticals, and Amgen. S.K.P. reports honoraria and a consulting/advisory role with Novartis, Astellas Pharma, Pfizer, Aveo, Myriad Pharmaceuticals, Roche/Genentech, Exelixis, Bristol-Myers Squibb, Ipsen, and Eisai and honoraria and research funding from Medivation. M.S. reports stock option interest in Amphivena Therapeutics, Intensity Therapeutics, and Adaptive Biotechnologies; a consulting role with Bristol-Myers Squibb, Roche/Genentech, AstraZeneca/MedImmune, Pfizer, Nektar, Lilly, Merck, Alexion Pharmaceuticals, Theravance, Baxalta/Shire, Seattle Genetics, Ignyta, Pierre Fabre, Incyte, Newlink Genetics, Celldex, Gritstone, and Innate Pharma; and an advisory role with Symphogen, Adaptimmune, Omniox, Lycera, and Molecular Partners. W.K.R. reports research funding to her home institution from Pfizer, Novartis, Tracon Pharmaceuticals, Bristol-Myers Squibb, Calithera Biosciences, and Peloton Therapeutics and research funding to an immediate family member from Incyte and Merck. W.M.S. reports honoraria and a consulting/advisory role with CVS Caremark, AstraZeneca, Bristol-Myers Squibb, Roche/Genentech, and Pfizer; research funding to his home institution from AstraZeneca, Bayer, Bristol-Myers Squibb, Boehringer Ingelheim, Exelixis, Novartis, Roche/Genentech, Pfizer, Merck, Janssen, and X4 Pharmaceuticals; and other relationships with UpToDate and American Cancer Society. M.E.G. acknowledges NHS funding to the NIHR Biomedical Research Centre at the Royal Marsden Hospital and Institute of Cancer Research, London UK. A.R. reports honoraria, accommodations, and a consulting/advisory role with Pfizer, Novartis, and Bristol-Myers Squibb; a consulting/advisory role with Ipsen and Roche; and research funding to his home institution from Pfizer and Novartis. S.B. reports personal fees and nonfinancial support for advisory boards from Pfizer, Astellas, Bristol-Myers Squibb, and Novartis; nonfinancial support for advisory boards from Bayer and Roche/Genentech; and nonfinancial support from Exelixis. V.G. reports grants from Bristol-Myers Squibb, Merck, Pfizer, and AstraZeneca, personal fees and nonfinancial support from Bristol-Myers Squibb, Merck, Roche, Novartis, Ipsen, Pfizer, AstraZeneca, Eisai, Eusa Pharma, and Cerulean outside the submitted work. T.K.C. reports consulting/advisory fees from AstraZeneca, Bayer, Bristol-Myers Squibb, Cerulean, Eisai, Foundation Medicine, Exelixis, Roche/Genentech, GlaxoSmithKline, Merck, Novartis, Peloton, Pfizer, Prometheus Laboratories, and Corvus; and research funding to his home institution from AstraZeneca, Bristol-Myers Squibb, Exelixis, Genentech, GlaxoSmithKline, Merck, Novartis, Peloton, Pfizer, Roche, Tracon, and Eisai. C. Schiff and P.S.H. report employment, including stock, with Genentech, Inc. G.D.F. reports employment, including stock, with Genentech, Inc. and stock with Foundation Medicine. T.P. reports honoraria and a consulting/advisory role with Roche/Genentech, Bristol-Myers Squibb, and Merck; a consulting/advisory role with AstraZeneca and Novartis; research funding from AstraZeneca/MedImmune and Roche/Genentech; and other relationships with Ipsen and Bristol-Myers Squibb (ASCO). M.A.H, D.N., S.J., E.P-L., J.Q., and D.S.C. are employees of Genentech, Inc. A.T. is an employee of Roche Products Ltd. J.A.R., J.H., T.H., C. Suárez, and R.D. have nothing to disclose.
Footnotes
Supplementary information is available for this paper at https://doi.org/10.1038/s41591-018-0053-3.
References
- 1.Kaelin WG Jr. The von Hippel-Lindau gene, kidney cancer, and oxygen sensing. J. Am. Soc. Nephrol 14, 2703–2711 (2003). [DOI] [PubMed] [Google Scholar]
- 2.George DJ & Kaelin WG Jr. The von Hippel-Lindau protein, vascular endothelial growth factor, and kidney cancer. N. Engl. J. Med 349, 419–421 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Motzer RJ et al. Tivozanib versus sorafenib as initial targeted therapy for patients with metastatic renal cell carcinoma: results from a phase III trial. J. Clin. Oncol 31, 3791–3799 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark JI et al. Impact of sequencing targeted therapies with high-dose interleukin-2 immunotherapy: an analysis of outcome and survival of patients with metastatic renal cell carcinoma from an on-going observational Il-2 clinical trial: PROCLAIMSM. Clin. Genitourin. Cancer 15, 31–41.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herbst RS et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 515, 563–567 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choueiri TK et al. PD-L1 expression in nonclear-cell renal cell carcinoma. Ann. Oncol 25, 2178–2184 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson RH et al. PD-1 is expressed by tumor-infiltrating immune cells and is associated with poor outcome for patients with renal cell carcinoma. Clin. Cancer Res 13, 1757–1761 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Motzer RJ et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N. Engl. J. Med 373, 1803–1813 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson RH et al. Costimulatory B7-H1 in renal cell carcinoma patients: indicator of tumor aggressiveness and potential therapeutic target. Proc. Natl. Acad. Sci. USA 101, 17174–17179 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson RH et al. Tumor B7-H1 is associated with poor prognosis in renal cell carcinoma patients with long-term follow-up. Cancer Res. 66, 3381–3385 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Keir ME, Butte MJ, Freeman GJ & Sharpe AH PD-1 and its ligands in tolerance and immunity. Annu. Rev. Immunol 26, 677–704 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Latchman YE et al. PD-L1-deficient mice show that PD-L1 on T cells, antigen-presenting cells, and host tissues negatively regulates T cells. Proc. Natl. Acad. Sci. USA 101, 10691–10696 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang J et al. The novel costimulatory programmed death ligand 1/B7.1 pathway is functional in inhibiting alloimmune responses in vivo. J. Immunol 187, 1113–1119 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McDermott DF et al. Atezolizumab, an anti-programmed death-ligand 1 antibody, in metastatic renal cell carcinoma: long-term safety, clinical activity, and immune correlates from a phase Ia study. J. Clin. Oncol 34, 833–842 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Elamin YY, Rafee S, Toomey S & Hennessy BT Immune effects of bevacizumab: killing two birds with one stone. Cancer Microenviron. 8, 15–21 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kusmartsev S et al. Oxidative stress regulates expression of VEGFR1 in myeloid cells: link to tumor-induced immune suppression in renal cell carcinoma. J. Immunol 181, 346–353 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Roland CL et al. Inhibition of vascular endothelial growth factor reduces angiogenesis and modulates immune cell infiltration of orthotopic breast cancer xenografts. Mol. Cancer Ther 8, 1761–1771 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Gabrilovich DI & Nagaraj S Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol 9, 162–174 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hodi FS et al. Bevacizumab plus ipilimumab in patients with metastatic melanoma. Cancer Immunol. Res 2, 632–642 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roland CL et al. Cytokine levels correlate with immune cell infiltration after anti-VEGF therapy in preclinical mouse models of breast cancer. PLoS One 4, e7669 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wallin JJ et al. Atezolizumab in combination with bevacizumab enhances antigen-specific T-cell migration in metastatic renal cell carcinoma. Nat. Commun 7, 12624 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osada T et al. The effect of anti-VEGF therapy on immature myeloid cell and dendritic cells in cancer patients. Cancer Immunol. Immunother 57, 1115–1124 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brauer MJ et al. Identification and analysis of in vivo VEGF downstream markers link VEGF pathway activity with efficacy of anti-VEGF therapies. Clin. Cancer Res 19, 3681–3692 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Fehrenbacher L et al. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet 387, 1837–1846 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Rosenberg JE et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet 387, 1909–1920 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snyder A et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med 371, 2189–2199 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rizvi NA et al. Activity and safety of nivolumab, an anti-PD-1 immune checkpoint inhibitor, for patients with advanced, refractory squamous non-small-cell lung cancer (CheckMate 063): a phase 2, single-arm trial. Lancet Oncol. 16, 257–265 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balar AV et al. Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: a single-arm, multicentre, phase 2 trial. Lancet 389, 67–76 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scheller J, Chalaris A, Schmidt-Arras D & Rose-John S The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochim. Biophys. Acta 1813, 878–888 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Russo RC, Garcia CC, Teixeira MM & Amaral FA The CXCL8/IL-8 chemokine family and its receptors in inflammatory diseases. Expert Rev. Clin. Immunol 10, 593–619 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Ha H, Debnath B & Neamati N Role of the CXCL8-CXCR1/2 axis in cancer and inflammatory diseases. Theranostics 7, 1543–1588 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zelenay S et al. Cyclooxygenase-dependent tumor growth through evasion of immunity. Cell 162, 1257–1270 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Powles T et al. Immune biomarkers associated with clinical benefit from atezolizumab (MPDL3280a; anti-PD-L1) in advanced urothelial bladder cancer (UBC). J. Immunother. Cancer 3, 83 (2015). [Google Scholar]
- 34.Rizvi NA et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carbone DP et al. First-line nivolumab in stage IV or recurrent non-small-cell lung cancer. N. Engl. J. Med 376, 2415–2426 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turajlic S et al. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. Lancet Oncol. 18, 1009–1021 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Motzer RJ et al. IMmotion151: a randomized phase III study of atezolizumab plus bevacizumab vs. sunitinib in untreated metastatic renal cell carcinoma (mRCC). J. Clin. Oncol 36, abstr, 578 (2018). [Google Scholar]
- 38.Choueiri TK et al. First-line avelumab + axitinib therapy in patients (pts) with advanced renal cell carcinoma (aRCC): Results from a phase Ib trial. J. Clin. Oncol 35, 4504 (2017). [Google Scholar]
- 39.Motzer R Nivolumab plus ipilimumab versus aunitinib in advanced renal-cell carcinoma. N. Engl. J. Med 378, 1277–1290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Voss MH et al. Integrated biomarker analysis for 412 renal cell cancer (RCC) patients (pts) treated on the phase 3 COMPARZ trial: correlating common mutation events in PBRM1 and BAP1 with angiogenesis expression signatures and outcomes on tyrosine kinase inhibitor (TKI) therapy. J. Clin. Oncol 35, 4523 (2017). [Google Scholar]
- 41.Prima V, Kaliberova LN, Kaliberov S, Curiel DT & Kusmartsev S COX2/mPGES1/PGE2 pathway regulates PD-L1 expression in tumor-associated macrophages and myeloid-derived suppressor cells. Proc. Natl. Acad. Sci. USA 114, 1117–1122 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coussens LM & Werb Z Inflammation and cancer. Nature 420, 860–867 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Q et al. The CXCL8-CXCR1/2 pathways in cancer. Cytokine Growth Factor Rev. 31, 61–71 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuan M et al. Tumor-derived CXCL1 promotes lung cancer growth via recruitment of tumor-associated neutrophils. J. Immunol. Res 10.1155/2016/6530410 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sumida K et al. Anti-IL-6 receptor mAb eliminates myeloid-derived suppressor cells and inhibits tumor growth by enhancing T-cell responses. Eur. J. Immunol 42, 2060–2072 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Najjar YG et al. Myeloid-derived suppressor cell subset accumulation in renal cell carcinoma parenchyma is associated with intratumoral expression of IL1β, IL8, CXCL5, and Mip-1α. Clin. Cancer Res 23, 2346–2355 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Draghiciu O, Nijman HW, Hoogeboom BN, Meijerhof T & Daemen T Sunitinib depletes myeloid-derived suppressor cells and synergizes with a cancer vaccine to enhance antigen-specific immune responses and tumor eradication. OncoImmunology 4, e989764 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reck M et al. Primary PFS and safety analyses of a randomized phase III study of carboplatin + paclitaxel +/− bevacizumab, with or without atezolizumab in 1 L non-squamous metastatic NSCLC (IMpower150). Ann. Oncol 10.1093/annonc/mdx760.002 (2017). [DOI] [Google Scholar]
- 49.Rooney MS, Shukla SA, Wu CJ, Getz G & Hacohen N Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160, 48–61 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miao D et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science 359, 801–806 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hsieh JJ et al. Genomic biomarkers of a randomized trial comparing first-line everolimus and sunitinib in patients with metastatic renal cell carcinoma. Eur. Urol 71, 405–414 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Motzer RJ et al. Survival and prognostic stratification of 670 patients with advanced renal cell carcinoma. J. Clin. Oncol 17, 2530–2540 (1999). [DOI] [PubMed] [Google Scholar]
- 53.Wu TD & Nacu S Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26, 873–881 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu TD, Reeder J, Lawrence M, Becker G & Brauer MJ GMAP and GSNAP for genomic sequence alignment: enhancements to speed, accuracy, and functionality. Methods Mol. Biol 1418, 283–334 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Lawrence M et al. Software for computing and annotating genomic ranges. PLOS Comput. Biol 9, e1003118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ritchie ME et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilm A et al. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 40, 11189–11201 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saunders CT et al. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 28, 1811–1817 (2012). [DOI] [PubMed] [Google Scholar]
- 60.McLaren W et al. The Ensembl variant effect predictor. Genome Biol. 17, 122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lawrence M, Degenhardt J & Gentleman R VariantTools: tools for working with genetic variants. version 1.12.0 Bioconductor https://bioconductor.org/packages/release/bioc/html/VariantTools.html (2018).
- 62.Karosiene E, Lundegaard C, Lund O & Nielsen M NetMHCcons: a consensus method for the major histocompatibility complex class I predictions. Immunogenetics 64, 177–186 (2012). [DOI] [PubMed] [Google Scholar]
- 63.Shukla SA et al. Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nat. Biotechnol 33, 1152–1158 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Clopper CJ & Pearson ES The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika 26, 404–413 (1934). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study, including anonymized genetic data from 163 patients who granted informed consent to share such data, are made available at the European Genome-Phenome Archive (EGA) under accession number EGAS00001002928. Furthermore, we provide qualified researchers access to additional individual clinical patient level data through the clinical study data request platform (https://clinicalstudydatarequest.com/). Details of Roche’s Data Sharing Policy are available here (https://clinicalstudydatarequest.com/Study-Sponsors/Study-Sponsors-Roche.aspx).