Abstract
Transcriptional regulator KAISO plays a critical role in cell cycle arrest and apoptosis through modulation of p53 acetylation by histone acetyltransferase p300. KAISO potently stimulates apoptosis in cells expressing WT p53, but not in p53-mutant or p53-null cells. Here, we investigated how KAISO transcription is regulated by p53, finding four potential p53-binding sites (p53-responsive DNA elements; p53REs) located in a distal 5′-upstream regulatory element, intron 1, exon 2 coding sequence, and a 3′-UTR region. Transient transcription assays of pG5-p53RE-Luc constructs with various p53REs revealed that p53 activates KAISO (ZBTB33) transcription by acting on p53RE1 (−4326 to −4227) of the 5′-upstream region and on p53RE3 (+2929 to +2959) of the exon 2 coding region during early DNA damage responses (DDRs). ChIP and oligonucleotide pulldown assays further disclosed that p53 binds to the p53RE1 and p53RE3 sites. Moreover, ataxia telangiectasia mutated (ATM) or ATM-Rad3–related (ATR) kinase-mediated p53 phosphorylation at Ser-15 or Ser-37 residues activated KAISO transcription by binding its p53RE1 or p53RE3 sites during early DDR. p53RE1 uniquely contained three p53-binding half-sites, a structural feature important for transcriptional activation by phosphorylated p53 Ser-15·Ser-37. During the later DDR phase, a KAISO-mediated acetylated p53 form (represented by a p53QRQ acetyl-mimic) robustly activated transcription by acting on p53RE1 in which this structural feature is not significant, but it provided sufficient KAISO levels to confer a p53 “apoptotic code.” These results suggest that the critical apoptosis regulator KAISO is a p53 target gene that is differently regulated by phosphorylated p53 or acetylated p53, depending on DDR stage.
Keywords: p53, apoptosis, gene transcription, transcription factor, DNA damage, post-translational modification (PTM), DNA damage response, KAISO, mutant p53, p53QRQ, transcriptional regulation, DNA damage response (DDR), zinc finger and BTB domain-containing 33 (ZBTB33), stress response
Introduction
KAISO was originally isolated as a protein interacting with p120ctn, a structural analogue of β-catenin, a member of the Wnt signaling pathway (1). KAISO exhibits DNA-binding activity to KAISO-binding sites (CTGCNA) and methylated CpG dinucleotides of target genes associated with cancer and/or embryonic development (2–4). In that regard, KAISO interacts with transcriptional corepressors, such as NCoR1, Sin3A, and Groucho, to mediate DNA methylation-dependent transcriptional repression (5).
Previously, we demonstrated that KAISO is induced by DNA damage in p53-expressing cells and interacts with a p53/p300 complex to increase acetylation of p53 Lys-320 and Lys-382 residues while decreasing Lys-381 acetylation. Moreover, the p53 with this particular acetylation pattern potently induces cell cycle arrest and apoptosis by activating transcription of CDKN1A (cyclin-dependent kinase inhibitor 1) and various apoptosis effector genes. KAISO is a critical regulator of p53-mediated apoptosis, under genotoxic stress conditions, in mammalian cells (6). We also showed that APAF1, encoding the core molecule of the apoptosome, is transcriptionally activated by KAISO (7).
p53, normally short-lived and present at low levels, is a key mediator of cellular responses to a broad range of genotoxic stresses (8). In response to DNA damage and other cellular stresses, both the protein level and activities of p53 are greatly increased. The crucial regulators of the genotoxic stress response, ataxia telangiectasia mutated (ATM)2 and ATM-Rad3–related (ATR) protein kinases, phosphorylate many cellular substrates, including p53 (9–12). p53-dependent transcriptional outcomes can be determined by the presence of p53-responsive DNA elements (p53REs), levels of induced p53, p53 post-translational modifications, its interacting proteins, and the epigenetic landscape of p53 target gene promoters. In particular, post-translational modifications, including phosphorylation and acetylation, were shown to regulate p53 stability, site-specific DNA-binding activity, and transactivation potential. Protein kinases, including ATR, ATM, and DNA-PK, were shown to phosphorylate several serine and/or threonine residues of the N terminus of p53, effectively regulating its function. Phosphorylation of p53 Ser-15, in vivo, is stimulated by DNA-damaging agents, including ionizing radiation and UV (13). Ser-15 phosphorylation might stabilize p53 by preventing p53 nuclear export (14). In addition, phosphorylation of human p53 on serines 15 and 37 in vitro by DNA-PK was reported to inhibit p53 interaction with the its repressor, Mdm2 (15). Phosphorylation at Ser-37 is also important for the transcriptional activity of p53 (16).
Another important modification of p53, acetylation, also plays important roles in response to various types of DNA damage. Acetylation has been shown to increase p53 sequence-specific DNA-binding capacity, through the recruitment of coactivators, and to enhance its stabilization by inhibiting ubiquitination of p53 by MDM2 (17–20). Specifically, p53 is acetylated at Lys-370, Lys-372, Lys-381, and Lys-382 by p300/CBP and at Lys-320 by p300/CREB-binding protein–associated factor (PCAF) (21–24).
Phosphorylation at the p53 N terminus enhances its interaction with the acetyltransferase p300/CBP and acetylates p53. Phosphorylation at N-terminal serines, such as Ser-15, -33, and -37, has been reported to recruit p300/CBP and PCAF to induce p53 acetylation in response to DNA damage (25, 26). Phosphorylation of p53 at Ser-46 by UV-activated HIPK2 facilitates CBP-mediated acetylation of p53 at Lys-382 to promote p53-dependent gene expression (27). These reports suggest that phosphorylation and acetylation of p53 may selectively induce p53 target gene expression and exert p53 functions, such as apoptosis.
The gene TP53 is absent or mutated in a high proportion of human cancers (28, 29), often leading to expression of a full-length protein that is deficient in certain functions, such as specific DNA binding. Many p53-mutant forms exert a dominant-negative effect, serving to abrogate the ability of WT p53 to inhibit cellular transformation or mediate DNA repair (30). Disruption of p53 functions promotes checkpoint defects, cellular immortalization, genomic instability, and inappropriate survival, allowing the continued proliferation of abnormal cells (31, 32). Activated p53 binds to specific DNA sequences of its target genes after DNA damage, functioning as a homotetramer and binding to p53-response elements; however, mutants of p53, including each of the four “hot spots” frequently altered in human cancers, fail to bind to the p53-binding consensus dimer (33).
The consensus-binding site of p53 contains two copies of a 10-bp motif (5′-PuPuPuC(A/T)(T/A)GPyPyPy-3′), separated by a 0–21-bp spacer. One copy of the motif was insufficient for site-specific DNA binding (34). Genes activated by p53 include a negative cell cycle regulator, CDKN1A (p21), and pro-apoptotic genes, such as Bax and Puma, which trigger cell cycle arrest, DNA damage repair (DDR), cellular senescence, apoptosis, and other responses, to determine cell fate following genotoxic stress (35–37).
In this study, we found that KAISO expression is dramatically increased by DNA damage only in cells with WT p53 and not in cells with p53 hot spot mutations. Consequently, we investigated how KAISO gene expression is controlled by p53 through p53REs in its 5′-upstream regulatory and coding regions. During early phases of DDR, p53 phosphorylated at Ser-15 and Ser-37 up-regulates KAISO and early DDR genes to activate apoptosis during later phases of DDR. Phosphorylated p53 Ser-15·Ser-37 and “acetylation-coded” (“cell death code”) (6) p53 differentially regulate KAISO expression via time differences of their expression and differential reading of its structural features by p53RE1.
Results
KAISO gene expression is increased only in cells expressing WT p53 and not in the cells with hot spot p53 mutants or lacking p53
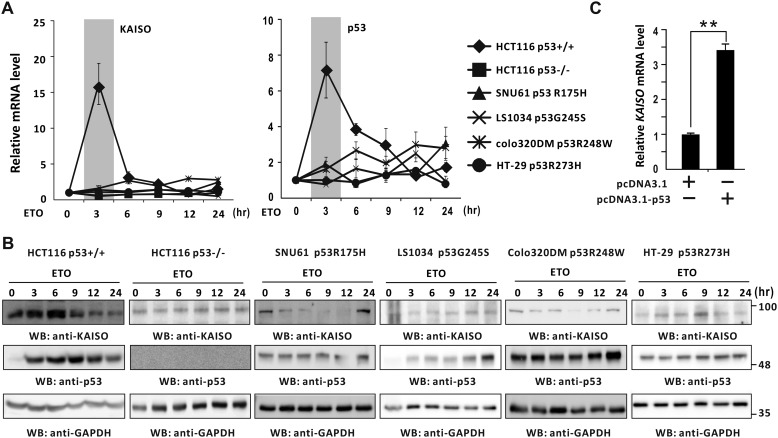
Previously, we showed that temporal expression patterns of KAISO and p53 are similar in cells treated with etoposide (6). We further investigated expression patterns of KAISO and p53 in various cell types expressing no p53, WT p53, or hot spot p53 mutations upon treatment with the DNA-damaging agent etoposide. KAISO and p53 mRNA levels were increased by 7–16-fold at 3 h of etoposide treatment in HCT116 colorectal cancer p53+/+ cells. However, in HCT116 p53−/−cells or in p53-mutated cancer cells, such as SNU61, Colo320DM, LS1034, and HT-29, neither KAISO nor TP53 mRNA levels were significantly increased by etoposide (Fig. 1A). We observed similar KAISO and p53 expression pattern at the protein level (Fig. 1B). In addition, ectopic p53 increased transcription of endogenous KAISO in H1299 lung cancer cells lacking endogenous p53 (Fig. 1C).
Figure 1.
Induction of KAISO expression by DNA damage is p53-dependent. A and B, RT-qPCR and Western blot analysis (WB) of KAISO and p53 expression. HCT116 p53+/+, HCT116 p53−/−, SNU61, Colo320DM, LS1034, and HT-29 cells were treated with etoposide (60 μm), harvested at the indicated times, and analyzed for mRNA by RT-qPCR and protein expression by Western blot analysis, respectively. GAPDH was used as control. C, RT-qPCR analysis. H1299 p53-null cells were transfected with p53 expression or control vector and analyzed for KAISO mRNA levels by RT-qPCR.
We also tested whether other types of DNA-damaging reagents (cisplatin, an alkylating reagent, and 5-fluorouracil, a pyrimidine analog) also induce KAISO transcription, via p53. Those assays showed that cisplatin induced KAISO transcription only in HCT116 cells expressing p53, in a fashion similar to that of etoposide (a topoisomerase inhibitor). In contrast, 5-fluorouracil induced KAISO transcription weakly and slowly in p53+/+ cells (Fig. S1). These results suggest that WT p53 is required for KAISO expression in cells treated with DNA-damaging agents.
p53 activates KAISO transcription by acting on its p53RE1 (bp −4326 to −4227) of 5′-upstream regulatory regions and p53RE3 (bp +2929 to +2959) in its exon 2 coding sequence region
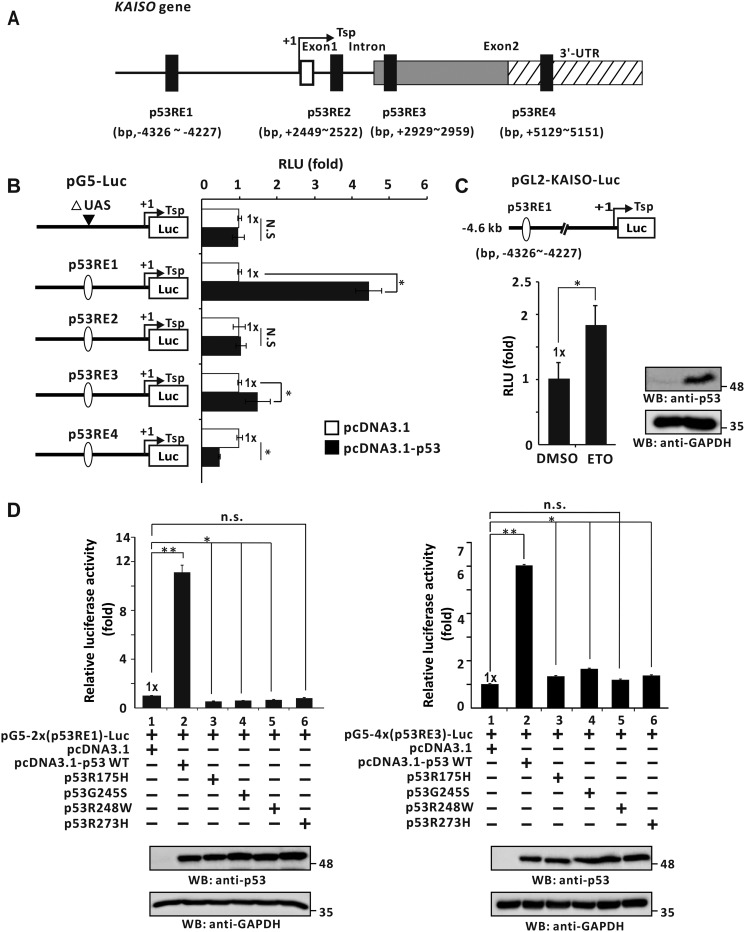
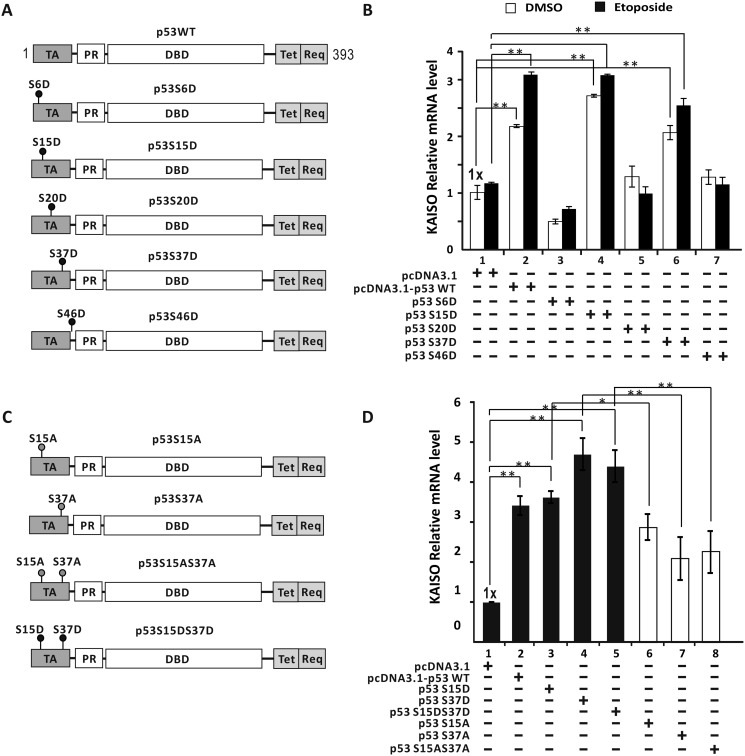
Because KAISO expression increased only in cells with WT p53, we investigated how p53 activates transcription of the KAISO gene. Computational analysis of the KAISO gene transcription unit (i.e. 5′-upstream regulatory region, promoter, exon, intron, and 3′-UTR) was conducted by MAC vector (version 7.2.3). This assessment revealed four potential p53REs: 5′-upstream regulatory region (bp −4326 to −4227), intron 1 (bp +2449 to +2522), exon 2 coding sequence region (bp +2929 to +2959), and 3′-UTR (bp +5129 to +5151) (Fig. 2A). We next prepared pG5-(p53RE)-Luc reporter constructs with the p53REs (one copy of all four distinct KAISO promoter p53REs) placed at the proximal promoter and tested which p53RE(s) is important for transcriptional activation by WT p53. In H1299 cells, ectopic p53 increased luciferase expression with p53RE1 in the KAISO 5′-upstream regulatory region and p53RE3 of the exon 2 coding sequence region, by 4.4- and 1.4-fold, respectively. These data suggest that p53RE1 may be more important for transcriptional activation of KAISO by p53 (Fig. 2B). In HCT116 p53+/+ cells transfected with pGL2-KAISO-Luc-4.6 kb construct with p53RE1, etoposide treatment increased reporter expression by 1.8-fold (Fig. 2C).
Figure 2.
p53 activates KAISO gene transcription by acting on the p53RE1 located in the 5′-upstream regulatory region. A, four potential p53REs identified in the KAISO gene: 5′-upstream regulatory region, intron 1, exon 2 coding sequence, and 3′-UTR. B, structure of the four pG5-Luc reporter plasmid constructs with the KAISO p53REs and transient transcription assays. The upstream activation sequence (UAS) (bp +1 to +97) was removed from pG5-Luc reporter plasmid. H1299 p53-null cells were transiently co-transfected with pG5–1x(p53RE)-Luc reporter plasmid and WT p53 expression vector. All assays were performed in triplicate. Error bars, S.D. C, HCT116 p53+/+ cells were transfected with pGL2-KAISO-Luc-4.6 kb containing the KAISO promoter (bp −4441 to +165). After treatment with etoposide for 24 h, luciferase activity was measured and normalized to total cellular protein. Data presented are the average of three independent assays. Error bars, S.D. *, p < 0.05. D, transient transcription analysis of the p53REs of the KAISO gene fusion reporter plasmid constructs. pG5–2x(p53RE1)-Luc or pG5–4x(p53RE3)-Luc and WT p53 expression vector or four hot spot p53 mutant expression vectors were transiently co-transfected in H1299 p53-null cells, and luciferase activities were measured. Luciferase activities were normalized to co-expressed β-gal activity, and data are presented as the average of three independent assays. Error bars, S.D. *, p < 0.05; **, p < 0.01; n.s., not significant. WB, Western blotting.
As described above, we successfully identified KAISO gene p53-response elements mediating transcriptional activation by WT p53 and depleted KAISO expression in cells expressing hot spot p53 mutants. Consequently, we next investigated whether hot spot p53 mutants could activate transcription of the reporter constructs with p53RE1 or -3 at the proximal promoter (pG5–2x(p53RE1)–Luc or pG5–4x(p53RE3)–Luc). Transient transcription assays in p53-null H1299 cells showed that WT p53 significantly increased reporter expression by acting on p53RE1 or p53RE3. However, hot spot p53 mutants could not increase reporter expression (Fig. 2D).
p53 directly binds p53RE1 and 3 of the KAISO gene
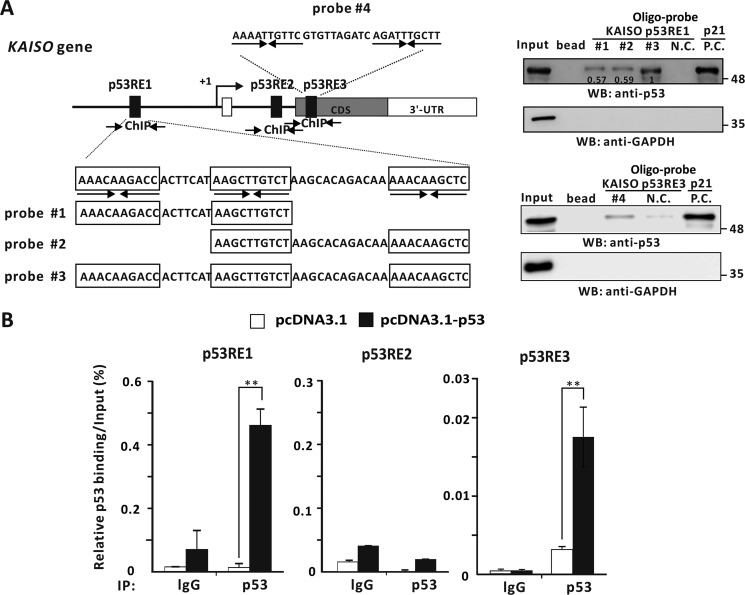
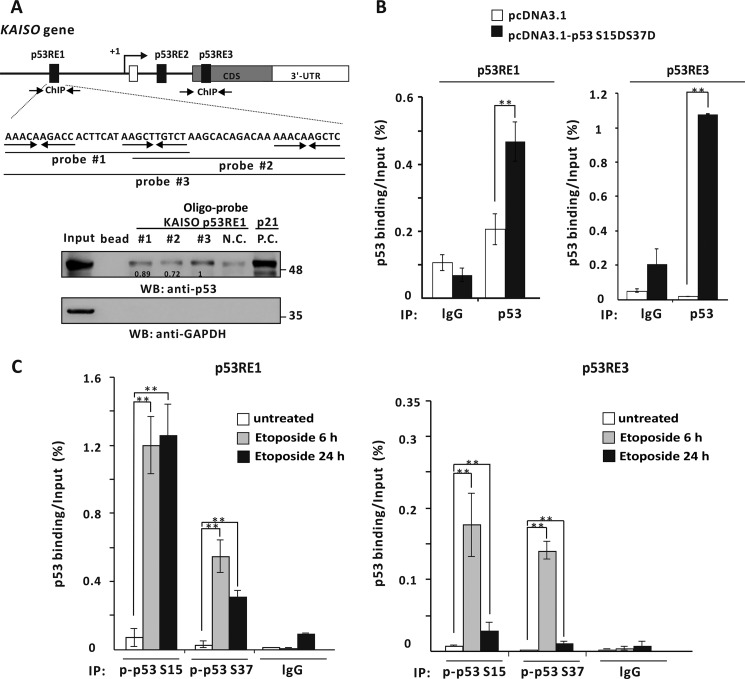
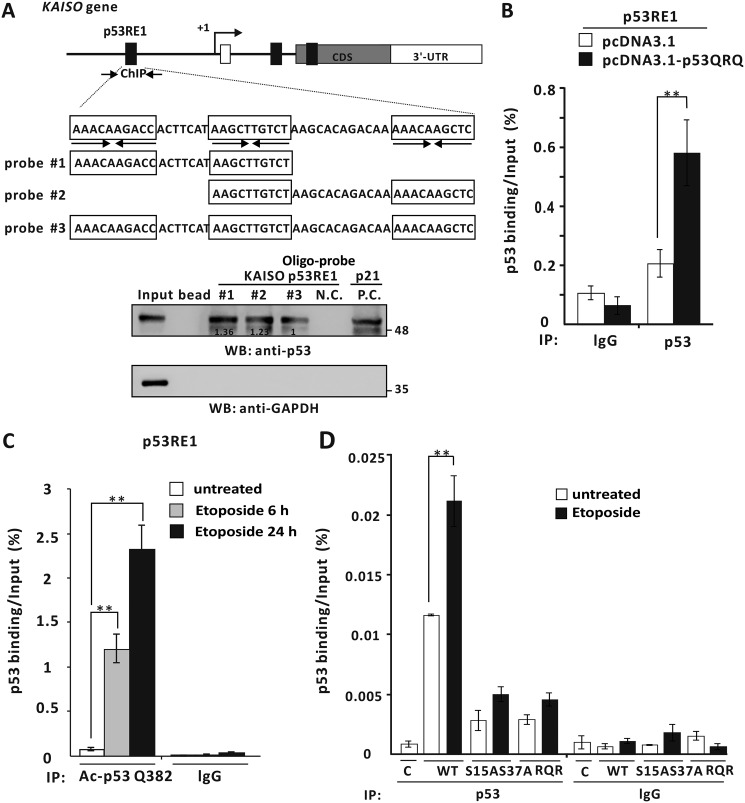
Because we showed that WT p53 enhances transcriptional activation of KAISO by acting on its p53-response elements of its 5′-upstream regulatory region and its exon 2 coding region, we investigated whether p53 actually binds to two KAISO p53-binding elements by oligonucleotide pulldown assays. Interestingly, p53RE of the 5′-upstream regulatory region contains three copies of the 10-bp p53-binding motif (5′-PuPuPuC(A/T)(T/A)GPyPyPy-3′). Consequently, we designed three probes, #1, #2, and #3, to cover two consensus p53 half-sites, with probe #3 containing three p53-binding half sites. Extracts from p53-null H1299 cells transfected with a p53 expression vector were incubated with the probes conjugated to agarose beads, showing that p53 bound to probes #1, #2, and #3 of p53RE1 and probe #4 of p53RE3. Interestingly, p53 bound more strongly to probe #3 (Fig. 3A). To further test whether p53 bound to p53REs of the KAISO gene, in vivo ChIP assays showed that p53 bound to p53RE1 and p53RE3 of KAISO but not to its intron 1 p53RE2 (Fig. 3B).
Figure 3.
p53 binds to the p53REs of the 5′-upstream regulatory region and coding region of KAISO. A, structure and oligonucleotide pulldown/Western blotting assay of p53REs of the KAISO gene. Endogenous p53REs of KAISO 5′-upstream regulatory region (ChIP#1, bp −4354 to −4254), intron 1 (ChIP#2, bp +2426 to +2556), and exon-coding sequence region (ChIP#3, bp +2885 to +2982) are shown above. Arrows, locations of the ChIP oligonucleotide primer sets spanning the p53-responsive elements (p53REs). Probe #1, #2, #3, and #4 represent the p53REs used for oligonucleotide pulldown assays. Shown is an oligonucleotide pulldown/Western blotting assay of p53 binding to the p53REs of KAISO 5′-upstream regulatory region and exon 2 coding sequence region. H1299 p53-null cell extracts with ectopic p53 were incubated with biotinylated double-stranded oligonucleotides. The mixtures were further incubated with streptavidin-agarose beads and precipitated by centrifugation. The precipitate was analyzed by a Western blotting assay (WB) using antibodies against p53 or GAPDH. B, ChIP assays of p53 binding to a potential p53RE of the endogenous KAISO 5′-upstream regulatory region and coding region. H1299 p53-null cells were transfected with a p53 expression vector and immunoprecipitated (IP) with anti-p53 antibody, followed by PCR amplification of the region flanking p53RE1, p53RE2, and p53RE3. **, p < 0.01. Error bars, S.D.
Temporal expression of mRNA and protein of KAISO, p53, phosphorylated p53, acetylated p53, and p53 target genes
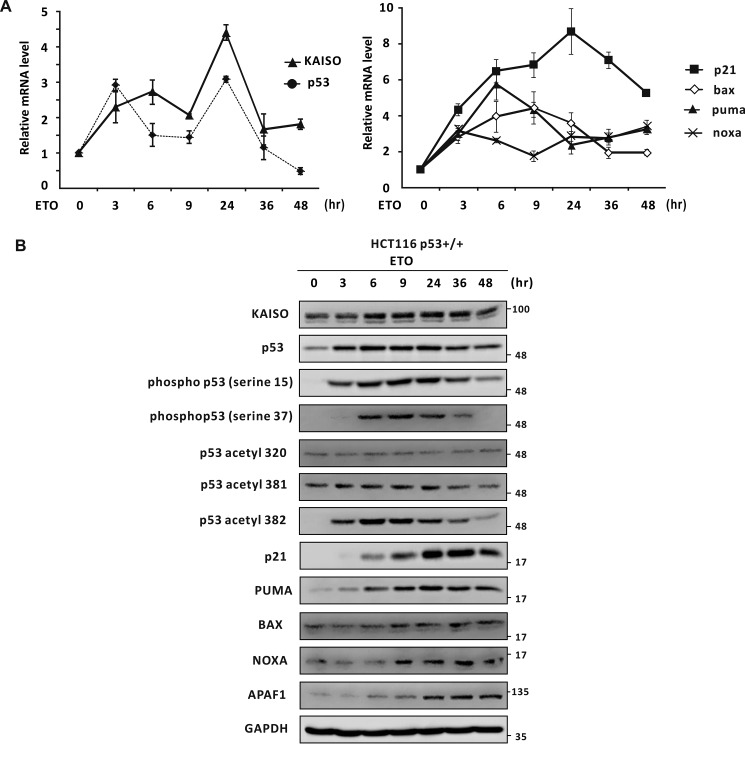
In humans, a number of serine residues at the p53 N terminus, comprising its transactivation domain, become phosphorylated by various kinases, including ATM and ATR, during early DNA damage responses (9–16). Because KAISO is an early response p53 target gene induced by DNA damage and is a critical regulator of apoptosis, we suspected that phosphorylated p53 may play a role in KAISO transcription activation, and a particular combination of phosphorylation at serine or threonine residues of p53 may be important in specific KAISO promoter binding and subsequent transcription activation. First, we investigated the expression patterns of KAISO, p53, p53 target genes (regulating cell cycle arrest and apoptosis (p21, Bax, Puma, and Noxa)), and p53 post-transcriptional modifications (PTMs) in HCT116 p53+/+ cells treated with etoposide. Upon etoposide treatment, KAISO mRNA levels gradually increased up to 6 h, reactivated at 9 h, and peaked at 24 h. In contrast, p53 mRNA levels increased by 3 h, reactivated at 9 h, peaked at 24 h, and decreased thereafter. Moreover, expression of the p53 target genes p21, BAX, and PUMA increased by 3 h of etoposide treatment and increased steadily until 6 h. Among these, p21 expression decreased after peaking at 24 h, similar to the expression pattern of KAISO. Conversely, BAX and PUMA expression levels increased by 6 or 9 h and decreased thereafter, whereas NOXA transcription increased by 3 h and reactivated at 9 h, resembling the expression pattern of p21 (Fig. 4A).
Figure 4.
Temporal expression of mRNA and protein of KAISO, p53, p53 phosphorylated at Ser-15 or Ser-37, p53 acetylated at Lys-320, Lys-381, or Lys-382, and a p53 target gene. A and B, temporal expression of mRNA and protein of KAISO, p53, p53 phosphorylated at Ser-15 or Ser-37, or p53 acetylated at Lys-320, Lys-381, or Lys-382, and p53 target genes in HCT116 p53+/+ cells treated with etoposide (ETO). The HCT116 p53+/+ cells treated with etoposide (60 μm) were harvested at the indicated times and analyzed for mRNA by RT-qPCR and protein expression by Western blot analysis, respectively. GAPDH was used as control. Error bars, S.D.
We also analyzed protein expression levels of KAISO, p53, p53 target genes, and p53 PTMs to explore their regulatory relationships, using HCT116 p53+/+ cells. Upon etoposide treatment, KAISO expression increased steadily from 3 to 36 h, whereas p53 expression increased from 3 to 24 h, peaking at 24 h and decreasing thereafter. Specifically, phosphorylation of p53 Ser-15 and Ser-37 increased at 3 h of etoposide treatment, and peaked at 9 or 24 h and then decreased thereafter, reminiscent of the expression pattern of p53. In contrast, acetylation of p53 showed different patterns, depending on the location of the acetylated lysine. For example, lysine 320 acetylation was low and remained relatively unchanged, whereas lysine 381 acetylation remained unchanged but decreased weakly after 36 h. Moreover, acetylation of p53 lysine 382 showed a pattern similar to that of phosphorylation of p53 Ser-15 or Ser-37. This pattern of p53 modifications may be involved in the regulation of p53 target gene expression (Fig. 4B).
It was intriguing that this pattern (“code”) of p53 Ser-15·Ser-37 phosphorylation and Lys-381/382 acetylation preceded induction of p21, PUMA, BAX, NOXA, and APAF1 (key factors regulating cell cycle arrest and apoptosis), suggesting that these particular modifications are important for expression of these genes. Overall, the protein expression patterns of KAISO and p53 were similar (Fig. 4B), suggesting that this p53 modification code may be important for KAISO expression during DNA damage responses.
p53 phosphorylated at Ser-15 and/or Ser-37 activates KAISO gene transcription
We next investigated whether p53 phosphorylation is important for KAISO transcriptional activation and which of five p53 serine residues, known to be phosphorylated upon treatment with etoposide, were critical for KAISO gene transcription (Fig. 5A). Consequently, we transfected WT p53 or phosphomimic p53 mutants (p53-S6D, S15D, S20D, S37D, and S46D) into p53-null H1299 cells, with or without etoposide treatment, and analyzed endogenous KAISO mRNA expression. Those results showed that the p53-S15D and -S37D mutants up-regulated KAISO, similar to WT p53. However, whereas transcriptional activation of KAISO by WT p53, p53-S15D, and p53-S37D was slightly higher in cells treated with etoposide, the difference was statistically insignificant, suggesting that Ser-15 or Ser-37 phosphorylation is sufficient for KAISO transcription activation during early DNA damage responses in p53-null, etoposide-treated H1299 cells (Fig. 5B). Furthermore, we prepared additional p53 expression constructs, including nonphosphorylatable p53 mutants (p53-S15A, p53-S37A, and p53-S15A·S37A) and dual p53 phosphomimic (p53-S15D·S37D) (Fig. 5C). We then transfected these plasmid expression vectors into p53-null H1299 cells and analyzed KAISO gene expression without etoposide treatment. KAISO expression is induced by WT p53, but nonphosphorylatable p53 mutants (p53-S15A, S37A, and S15A·S37A) gave relatively weaker transcription activation than WT p53. Considering that p53-S15A, S37A, and S15A·S37A similarly activated KAISO transcription, single phosphorylation at either Ser-15 or Ser-37 appeared to be sufficient for transactivation (Fig. 5D). Moreover, transcriptional activation potential of phosphorylation of p53 Ser-15 was not affected by phosphorylation of Ser-37, and vice versa.
Figure 5.
Phosphorylated p53 at Ser-15 and/or Ser-37 residues activates KAISO transcription. A, WT p53 and mimics of phosphorylated p53 at serine residues. Filled circles, phosphomimics, p53 serine residue X; TA, transactivation domain (residues 1–42); PR, proline-rich domain (residues 40–92); DBD, DNA-binding domain (residues 101–306); Tet, tetramerization domain (residues 307–355); Reg, regulatory domain (residues 356–393). B, RT-qPCR analysis of KAISO mRNA expression. H1299 p53-null cells were transfected with WT p53 expression or p53 phosphomimic mutant expression vector and analyzed for KAISO mRNA, with or without etoposide (60 μm) treatment for 3 h. C, structures of p53, p53-S15D and/or -S37D, and p53-S15A and/or -S37A. p53-S15A/S37A is a dominant-negative form of p53-S15D/S37D. Filled circles, phosphorylation target p53 serine residue. Filled red circles, nonphosphorylated p53 serine residue. D, RT-qPCR analysis. H1299 p53-null cells were transfected with WT p53, p53 phosphomimic mutant (Ser → Asp), or p53 nonphosphorylatable Ser-15/37 → Ala-15/37 mutant expression vector and analyzed for KAISO mRNA for 3 h. Error bars, S.D.; *, p < 0.05; **, p < 0.01.
Alternatively, we tested whether the p53 phosphomimics (p53-S15D, p53-S37D, and p53-S15D·S37D) could activate KAISO gene transcription. All three p53 mutants activated KAISO transcription similarly, suggesting that p53 Ser-15 and/or Ser-37 residues may be phosphorylated, because p53 S15D and/or S37D could up-regulate KAISO (Fig. 5D). As described above (Fig. 5B), simultaneous modification of Ser-15 and Ser-37 did not provide additional transcriptional activation.
Identification of unique structural features of the p53RE1 of KAISO involved in transcription activation by phosphorylated p53 Ser-15 and/or Ser-37
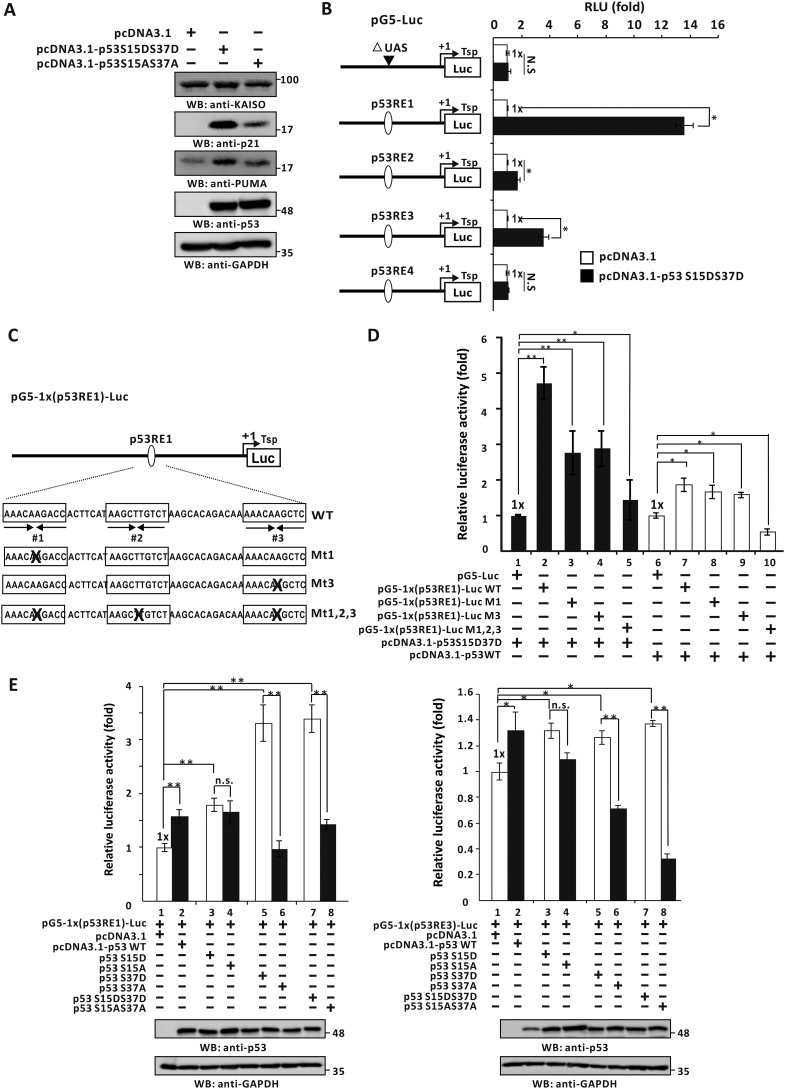
Because p53-S15D and/or -S37D could up-regulate KAISO mRNA expression, we tested whether KAISO protein expression could be rescued by p53-S15D·S37D in p53-null H1299 cells by Western blotting. We transfected H1299 cells with a p53 phosphomimic expression vector (p53-S15D·S37D) or a nonphosphorylatable p53 mutant (p53-S15A·S37A) and analyzed expression patterns of KAISO, p53, and p53 target gene (p21, PUMA) proteins. Expression levels of the p53 mutants were comparable. KAISO expression was increased by the p53 phosphomimic p53-S15D·S37D but not noticeably increased by the nonphosphorylatable p53-S15A·S37A, which only weakly induced the p53 target genes p21 and PUMA. Together, these data demonstrate that functional rescue of KAISO expression is only possible with WT p53 (phosphorylated p53 Ser-15 and Ser-37) in p53-null H1299 cells (Fig. 6A).
Figure 6.
Identification of p53-response element and transcriptional regulation of KAISO by p53 and a p53-S15D·S37D, a mimic of phosphorylated p53 Ser-15 and Ser-37. A, Western blot analysis (WB) of KAISO, p53 target genes (p21, PUMA), and p53 expression of the cell extracts prepared from the H1299 p53-null cells transfected with control vector, p53-S15D·S37D, or p53-S15A·S37A. N.S., not significant. B, structure of the four pG5-Luc reporter plasmid constructs with the KAISO p53REs and transient transcription assays. H1299 p53-null cells were transiently co-transfected with pG5–1x(p53RE)-Luc reporter plasmid and p53-S15D·S37D expression vector. All assays were performed in triplicate. Error bars, S.D. C and D, structure of the pG5–1x(p53RE1)-Luc reporter plasmid constructs with WT p53RE1 and mutations at p53RE half-sites (Mt1, Mt2, Mt1,2,3) and transient transcription assays. H1299 p53-null cells were transfected with reporter plasmids, WT p53, and a mimic of phosphorylated p53-S15D·S37D expression vector and were analyzed for luciferase activity. Reporter activities were normalized to co-expressed β-gal activity. Data presented are the average of three independent assays. Error bars, S.D. *, p < 0.05; **, p < 0.01. E, pG5-Luc reporter plasmid constructs with p53RE1 or p53RE3 of the KAISO gene and transient transcription assays. H1299 p53-null cells were transfected with reporter plasmids and WT p53 or a phosphomimic mutant p53-S15D and/or S37D or p53 nonphosphorylatable p53-S15A or S37A expression vector and were analyzed for luciferase activity. Reporter activities were normalized to co-expressed β-gal activity. Data presented are the average of three independent assays. Error bars, S.D. *, p < 0.05; **, p < 0.01.
As shown in Fig. 2, WT p53 activated reporter gene transcription by acting on the KAISO promoter p53REs-1 and/or -3. We also investigated how p53-phospho-S15D·S37D activates KAISO transcription, showing that ectopic p53-S15D·S37D increased expression of p53RE1 and -RE3 luciferase fusion vectors by 13.6- and 3.5-fold, respectively, in p53-null H1299 cells. Transcription activation by p53-S15D·S37D was 3 times stronger than that by WT p53 (Fig. 6B).
Interestingly, unlike many other p53-response elements with two half binding elements (33), p53RE1 contains three half p53-binding elements. Consequently, we investigated whether this unique structural feature was recognized by WT p53 or the p53 dual phosphomimic p53-S15D·S37D and was functional in transcriptional regulation.
To investigate which two copies of p53 half-sites could mediate transcriptional activation, we prepared four KAISO promoter-luciferase gene reporter constructs: pG5–1x(p53RE1)-Luc WT, pG5–1x(p53RE1)-Luc MT1, pG5–1x(p53RE1)-Luc MT3, and pG5–1x(p53RE1)-Luc MT1, -2, and -3 with mutations introduced at p53-binding half-sites (Fig. 6C). Transcription assays showed that ectopic WT p53 activated transcription of the pG5–1x(p53RE#1)-Luc WT reporter construct by 1.8-fold. Mutations of the p53RE1 half-site at MT1 or MT3 weakly decreased p53 reporter expression, and mutation of all three p53-binding sites effectively nullified reporter expression (Fig. 6D, lanes 6–10).
Interestingly, the p53 dual phosphomimic, p53-S15D·S37D, strongly activated KAISO expression by 4.7-fold, whereas the p53RE1-MT1 or MT3 mutations down-regulated KAISO transcription by 50%, and mutation of all three sites completely abolished KAISO transcriptional activation by p53 S15D·S37D (Fig. 6D, lanes 1–5). These results suggest that the unique structural feature of p53RE1 is functional and important for KAISO transcriptional activation by p53-S15D·S37D or possibly by p53 endogenously phosphorylated during early DNA damage responses.
We next investigated whether phosphorylated p53 Ser-15 and/or Ser-37 could activate transcription of the reporter constructs with p53RE1 or -3 within the KAISO proximal promoter (pG5–1x(p53RE1)–Luc or pG5–1x(p53RE3)–Luc). Transient transcription assays in p53-null H1299 cells showed that the p53 phosphomimic mutants, p53-S37D and p53 S15D·S37D, increased reporter expression by acting on p53RE1 or p53RE3, whereas nonphosphorylatable p53-S37A and p53 S15A·S37A could not activate reporter expression. However, the p53 phosphomimic, p53-S15D, increased reporter expression by acting on p53RE1 or p53RE3, whereas nonphosphorylatable p53-S15A did not decrease reporter expression significantly. These findings indicate that phosphorylated p53-Ser-37·Ser-15 and/or Ser-37 alone are necessary for KAISO gene expression via p53RE1 and/or p53RE3 (Fig. 6E).
We then wished to determine whether the p53 phosphomimic p53-S15D·S37D physically interacts with KAISO p53RE1, using oligonucleotide pulldown assays. To that end, extracts from p53-null H1299 cells transfected with a p53-S15D·S37D expression vector were incubated with agarose beads conjugated to the above-described probes #1, #2, and #3 of p53RE1. The p53-S15D·S37D bound more strongly to the probe #3, in accord with the above data (Fig. 7A). We further tested whether this particular form of p53 bound to p53-binding elements in the endogenous KAISO gene promoter, via ChIP. Those assays clearly showed that p53-S15D·S37D strongly bound p53RE1 and -RE3 (Fig. 7B).
Figure 7.
The endogenous phosphorylated p53 at Ser-15 or Ser-37 and p53-S15D·S37D, a mimic of phosphorylated p53 Ser-15 and Ser-37, bind to the p53RE1 and -RE3 of the KAISO gene. A, oligonucleotide pulldown/Western blotting assay of p53 binding to the p53RE1 of KAISO. Mimics of phosphorylated p53-S15D·S37D bind to the p53RE1 in vitro. Shown is the structure of p53RE1 and three probes used in oligonucleotide pulldown assays of p53 binding. H1299 p53-null cell extracts with ectopic p53-S15D·S37D expression were incubated with biotinylated double-stranded oligonucleotides, precipitated, and analyzed by a Western blotting assay (WB) using antibodies against p53 or GAPDH. B, ChIP assays of p53-S15D·S37D binding to the p53RE1 and -RE3 of endogenous KAISO. H1299 p53-null cells were transfected with a p53-S15D·S37D expression vector and immunoprecipitated (IP) with an anti-p53 antibody, followed by PCR amplification of the region flanking p53RE1 and -RE3. Shown is the average of three independent assays. Error bars, S.D. **, p < 0.05. C, ChIP assays of endogenously phosphorylated p53 Ser-15 or Ser-37 binding to p53RE1 and -RE3 of the endogenous KAISO gene. HCT116 p53+/+ cells treated with etoposide (60 μm) were harvested at 6 or 24 h, immunoprecipitated with a specific anti-phosphorylated p53 Ser-15 or Ser-37 antibody, and amplified by PCR of the region flanking p53RE1 and -RE3. Shown is the average of three independent assays. Error bars, S.D. **, p < 0.05.
We next examined whether phosphorylated p53-Ser15 or Ser-37 binds to the p53 REs of KAISO and influences KAISO transcription at 6 and 24 h post-etoposide treatment (coinciding with up-regulation of p53 and KAISO genes; Fig. 4A). ChIP assays, using an antibody specific to phosphorylated p53 Ser-15 or Ser-37 showed that the phosphorylated p53 bound well to p53RE1 at 6 and 24 h. Interestingly, phosphorylated p53 Ser-15 or Ser-37 bound well to p53RE3 at 6 h but decreased significantly by 24 h. These interesting binding patterns, revealed by ChIP, revealed differential DNA binding patterns of phosphorylated p53 Ser-15 and/or Ser-37, providing information on how KAISO may be regulated.
The revealed DNA–p53 binding interaction patterns matched very well with the Western blotting and RT-qPCR results. For example, p53 Ser-15 phosphorylation increased over 3–24 h, whereas p53 Ser-37 phosphorylation increased over 6–9 h and decreased by 24 h. These expression patterns were also reflected in DNA–p53 binding at p53RE1 and -RE3. Overall, our data showed that p53 RE1 is the primary element interacting with phosphorylated and acetylated p53 and that p53RE3 is only mediating transcriptional activation by phosphorylated p53 (Fig. 7C).
p53QRQ, a mimic of KAISO-mediated acetylated p53, activates KAISO transcription via p53RE1
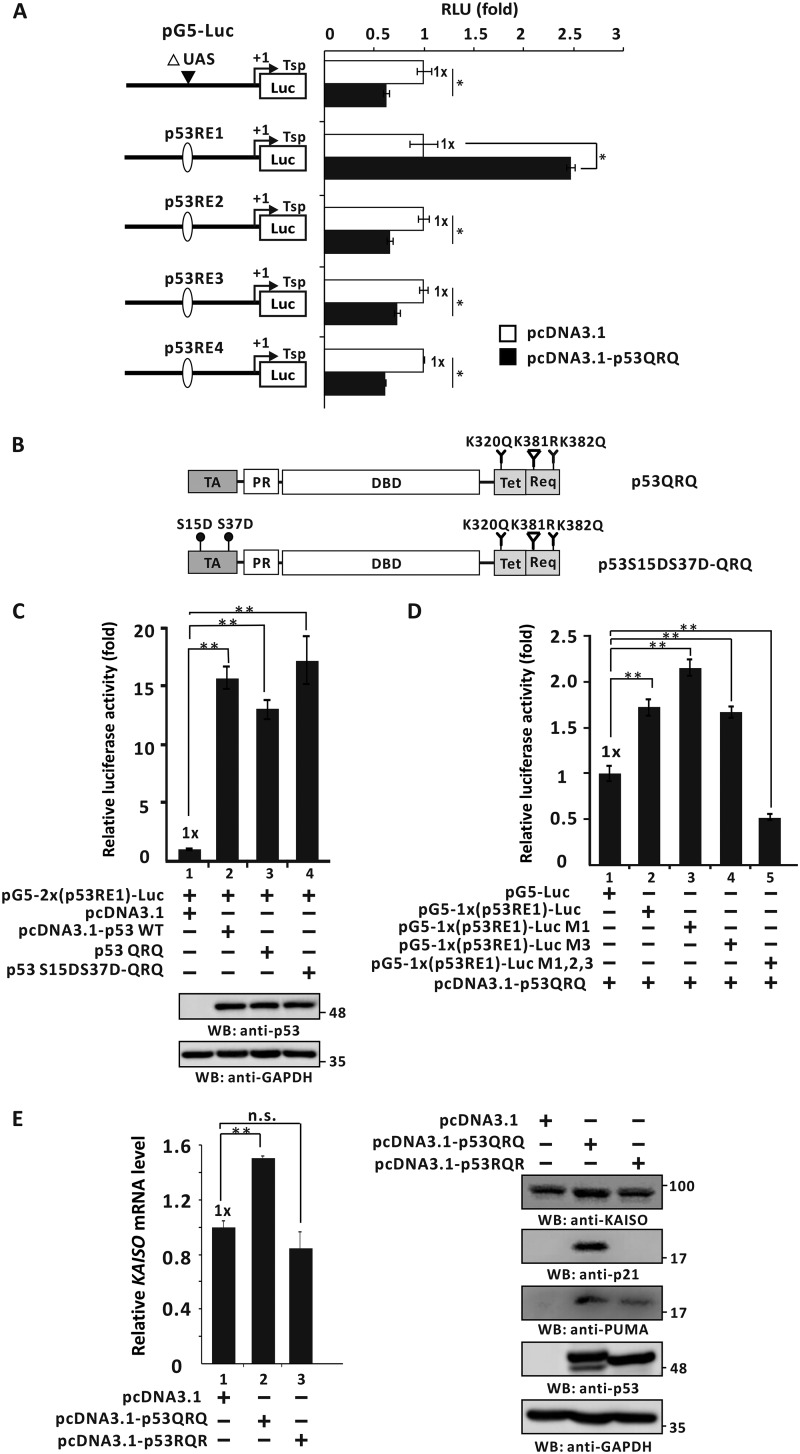
As noted above, KAISO expression is induced during early DDRs and up-regulated again, at later time periods, when cells are exposed to etoposide. Moreover, a KAISO–p53–p300 complex formation increases acetylation of p53 Lys-320 and Lys-381 residues but decreases acetylation of Lys-381 by p300 (5), and these events are important to increase p53 DNA binding and transcriptional activation of p53 target apoptotic genes at later time periods. Moreover, p53QRQ (K320Q, K381R, and K382Q), a p53 triacetyl mimic, represents a KAISO-mediated p53 acetylation pattern or code that might regulate KAISO transcription. Therefore, we tested which p53RE(s) might be important for transcriptional activation of KAISO by the phosphomimic p53QRQ, which may be functionally important in later stages of DDRs.
Further assessments showed that in p53-null H1299 cells, ectopic p53QRQ increased luciferase expression of reporter constructs only via p53RE1 in the KAISO 5′-upstream regulatory region and not via p53RE3 of the exon 2 coding sequence region and was clearly different from p53-S15A and/or p53-S37A (Fig. 8A). To investigate whether p53 PTMs (i.e. phosphorylated p53 form, p53QRQ, and potentially transient phosphorylated p53QRQ form) affect p53's transcriptional activation of KAISO via p53RE1, p53-null H1299 cells were transfected with the pG5–2x(p53RE1)-Luc construct and expression vectors for p53, p53QRQ, and p53-S15D·S37D-QRQ, showing that transcription of the KAISO reporter construct was increased by WT p53 and similarly by p53-S15D·S37D-QRQ and p53QRQ (Fig. 8, B and C).
Figure 8.
p53QRQ, a mimic of p53 with KAISO-medicated apoptotic code (acetylation at 320 and 382, no acetylation at p53 Lys-381) activates the transcription of a KAISO reporter via the p53RE1 of the 5′-upstream regulatory region of KAISO gene. A, structure of the four pG5-Luc reporter plasmid constructs with the p53REs of the KAISO gene and transient transcription assays. Identification of the p53RE critical for transcriptional activation of KAISO by p53QRQ. H1299 p53-null cells transfected with pG5-Luc reporter plasmid with p53RE and p53QRQ expression vectors were analyzed for luciferase activity, normalized to co-expressed β-gal activity. Data presented are the average of three independent assays. Error bars, S.D. *, p < 0.05; **, p < 0.01. B and C, structures of p53QRQ and p53-S15D·S37D-QRQ, and transient transcription assays. Shown below are the structures of p53QRQ and p53-S15D·S37D-QRQ. Transient transcription analysis of the p53REs of KAISO gene fusion reporter plasmid constructs is shown. pG5–2x(p53RE1)-Luc and WT p53 or p53QRQ or p53-S15D·S37D-QRQ expression vectors were transiently co-transfected in H1299 p53-null cells, and luciferase activities were measured. Luciferase activities were normalized to co-expressed β-gal activity, and data presented are the average of three independent assays. D, transient transcription assays. Mutations were introduced to test the function of each p53-binding half-sites in transcriptional activation of the reporter gene by p53QRQ. H1299 p53-null cells were transiently co-transfected with pG5–1x(p53RE1)–Luc reporter plasmid and p53QRQ expression vector. All assays were performed in triplicate. Error bars, S.D. E, RT-qPCR analysis and Western blot analysis (WB). H1299 p53-null cells were transfected with control vector, p53QRQ, or p53RQR expression and analyzed for KAISO, p53 target genes (p21, PUMA), and p53 expression. Data presented are the average of three independent assays. Error bars, S.D.; *, p < 0.05; **, p < 0.01; n.s., not significant.
The p53 phosphomimic p53-S15D and/or -S37D can activate early transcription of KAISO, a transcriptional regulator that is critical for activation of apoptotic gene expression. The actual player in apoptosis is a post-translationally modified p53 form having a particular functional code, as mimicked by p53QRQ. This particular form of p53 selectively binds apoptotic gene promoters to activate their transcription and later activates KAISO gene transcription to provide KAISO required for the generation of sufficiently PTM-coded p53 for apoptosis. Transient transcription assays showed that p53QRQ activated the KAISO promoter by 1.8-fold, whereas mutations of p53-binding half-site 1 or 3 showed little effect on transcriptional activation, although mutations of all three sites abolished transcriptional activation by p53QRQ (Fig. 8D). This mode of regulation is quite different from those of p53-S15D and/or -S37D (with regard to -fold activation and binding behavior to the p53RE1 and/or -RE3), suggesting a specific role of phosphorylated p53 during early phases of DDR transient and p53 with its KAISO-mediated “acetylation code” in late phases of DDR. Furthermore, we investigated whether ectopic p53QRQ and/or p53 functionally negative forms of transient p53QRQ, such as p53RQR (K320R, K381Q, and K382R), can modulate endogenous KAISO gene expression after 24 h of H1299 cell culture. Those assays showed that p53QRQ activated KAISO gene expression at both the mRNA and protein levels, but p53RQR could not. Likewise, p53 target gene expression was activated by p53QRQ, but only very weakly by p53RQR, although p53QRQ and -RQR were comparably expressed (Fig. 8E).
p53QRQ binds to the p53RE1 of the KAISO gene, and its binding is enhanced at a late stage of DNA damage responses
We also investigated whether transcriptional regulatory behaviors, like DNA binding, were different for p53QRQ. Oligonucleotide pulldown assays showed that p53QRQ binds to the p53RE1 of the KAISO 5′-upstream regulatory region, and ChIP assays showed that p53 binds to KAISO's p53RE1. However, unlike the phosphomimic p53-S15D and/or -S37D, p53QRQ showed similar binding activity to all three probes (e.g. p53QRQ bound to probes #1 and #2 similarly). However, the presence of all three intact elements did not increase DNA-binding activity of p53QRQ (Fig. 9, A and B).
Figure 9.
The mimic of a KAISO-mediated acetylated p53, p53QRQ, binds to the p53RE1 of the 5′-upstream regulatory region of KAISO. A, structure and oligonucleotide pulldown/Western blotting assay (WB) of p53RE1 of the KAISO gene. p53RE1 contains three p53-binding half-sites. The locations of oligonucleotide primer–binding sites of the ChIP assay of p53QRQ binding are indicated by arrows. Probes #1 to #3 used in oligonucleotide pulldown assays are indicated below. Oligonucleotide pulldown and Western blotting assays of p53QRQ binding to p53RE1 probes are shown. H1299 p53-null cells extracts with ectopic p53QRQ were incubated with biotinylated double-stranded oligonucleotides. The mixtures were further incubated with streptavidin-agarose beads and precipitated by centrifugation. The precipitate was analyzed by a Western blotting assay using antibodies against p53 (DO-1) or GAPDH. p53QRQ bound similarly to all three probes. 3′-UTR was used as negative control. p53RE1 of the p21/CDKN1A gene was positive control. B, ChIP assay of p53QRQ binding to the p53RE1 of the endogenous KAISO gene in the cells transfected with pcDNA3.1-p53QRQ. IP, immunoprecipitation. C, ChIP assays of endogenously acetylated p53-K382 binding to p53RE1 of endogenous KAISO. HCT116 p53+/+ cells treated with etoposide (60 μm) were harvested at 6 or 24 h, immunoprecipitated with a specific anti-acetylated p53 Lys-382 antibody, and amplified by PCR of the region flanking p53RE1. Shown is the average of three independent assays. Error bars, S.D. **, p < 0.05. D, PTMs of p53, including phosphorylation of Ser-15 and Ser-37 and acetylation of Lys-382, PTMs critical for binding the KAISO p53RE1. Dominant-negative forms of phosphorylated p53 at Ser-15 and/or Ser-37 (p53-S15A·S37A) or the KAISO-mediated acetylated form (p53RQR) were expressed in H1299 cells treated with etoposide (60 μm) and analyzed for p53 binding. The mutations strongly attenuated p53 binding.
We next examined whether acetylated p53 Lys-382 binds to the p53REs of KAISO and was involved in KAISO transcription at the time points of 6 and 24 h post-etoposide treatment, times at which p53 and KAISO were both activated. Interestingly, acetylated p53 lysine 382 bound well to p53RE1 at 6 h, and intriguingly, binding was increased further at 24 h. These binding patterns, revealed by ChIP, are interesting and show the temporal differential DNA-binding patterns of acetylated p53 Lys-382 (representing p53 acetylated Lys-320 and Lys-382 and nonacetylated Lys-381 or p53QRQ) (Fig. 9C).
We also investigated the effect of p53 phosphorylation on p53 acetylation and binding to p53RE1 and, thereby, KAISO expression. ChIP assays showed that whereas WT p53 bound well to p53RE1, neither p53-S15A and/or pS37A nor p53RQR (two dominant negative forms of p53-S15D and/or p53-S37D and p53QRQ) bound p53RE1, suggesting again the importance of phosphorylation or acetylation at Ser-15, Ser-37, Lys-320, Lys-381, and Lys-382 in binding to the p53RE1 during the induction of KAISO in early or late DNA damage responses (Fig. 9D).
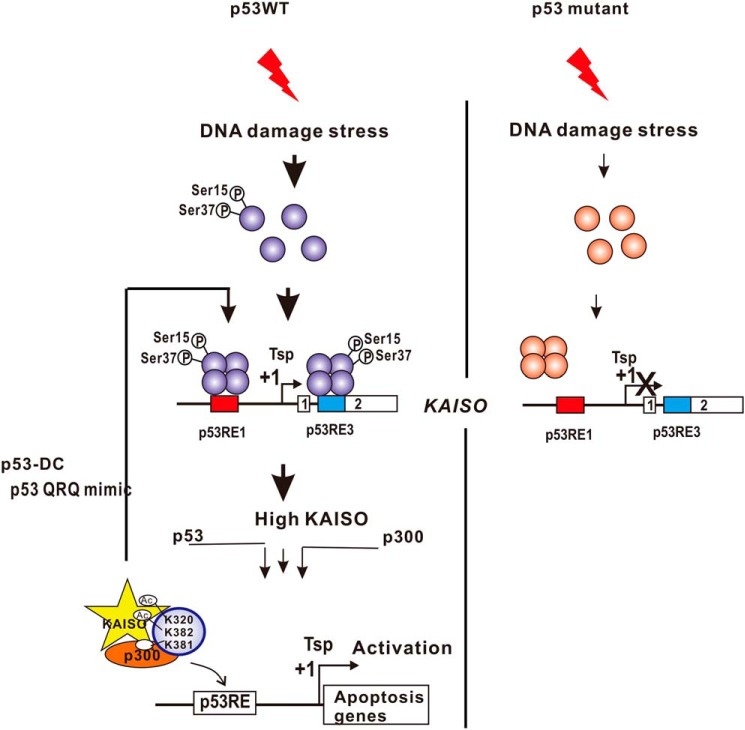
The results of our studies highlight the importance of phosphorylated p53 (particularly phosphorylation at Ser-15 and Ser-37) in the initial early-phase expression of KAISO and acetylated p53 (p53 Ac-K281, -K320, and -K382) in the induction of KAISO for apoptotic gene expression. Although p53RE1 located in the 5′-upstream regulatory element plays a major role, p53RE3 is also important for up-regulation of initial early-phase KAISO induction, and structural features of p53RE1 are critical for promoter element recognition by phosphorylated p53 Ser-15 and/or Ser-37. Moreover, this structural feature is not recognized by p53, with KAISO-mediated acetylation code for late-period KAISO induction, in the DNA damage response (Fig. 10).
Figure 10.
Hypothetical model of KAISO gene transcriptional activation by p53 during DNA damage responses. In cells undergoing DNA damage responses (e.g. by treatment with etoposide, cisplatin, etc.), KAISO is transcribed as an early DNA damage response gene, activated by p53, and robustly activated again at later periods of the DNA damage response, which is critical for apoptosis. KAISO gene and post-translational modification of p53, either by phosphorylation or acetylation, and KAISO promoter structure (particularly p53RE1 and p53RE3) play important roles. DNA-damaging stress induces phosphorylation of Ser-15 and/or Ser-37 of p53 by ATM or ATR. This particular form of p53 binds the two elements but preferentially to the p53RE1, with three unique p53-binding half-sites. The initial pulse of KAISO expression is critical in the generation of a KAISO-mediated acetylated form of p53 (Ac-K320, deacetylated Lys-381, Ac-382; mimicked by p53QRQ). In turn, acetylated p53 (called the p53 “death code,” p53DC) robustly activates KAISO gene transcription by acting on p53RE1, thus generating sufficient KAISO and thereby apoptotic p53 required for the induction of apoptotic gene expression.
Discussion
KAISO, a critical regulator of apoptosis, is induced by etoposide and inhibits cell proliferation by inducing apoptosis in p53-expressing cells (6, 7). However, little is known of how KAISO transcription is regulated. Here, we demonstrate that KAISO is a target gene of p53, which can activate KAISO expression via p53RE1 (bp −4326 to −4227) and p53RE3 (bp +2929 to +2959) elements in colon cancer cells (Figs. 1 and 2B).
p53 is a multifunctional tumor suppressor protein and is considered the “guardian of the genome” (38). Stress-induced p53 activation involves p53 post-translational modifications on multiple sites by phosphorylation, acetylation, and other modifications (22, 40–42). Phosphorylation has been studied most intensively and has been proposed to play a critical role in the stabilization and activation of p53. We proposed that an early PTM of p53, phosphorylation at Ser-15 and Ser-37 residues, increased KAISO expression during early cellular DNA damage responses. Multiple serine/threonine kinases have been implicated in p53 phosphorylation. Saito et al. (43) suggested that ATM mediates phosphorylation of p53 at multiple sites in response to ionizing radiation. In particular, Ser-15 phosphorylation has been shown to enhance the interaction of p53 with the transcriptional co-activators CBP and PCAF, which subsequently acetylates and activates transactivational activity by p53. ATM mainly phosphorylates Ser-15, in response to ionizing radiation and chemotherapeutic drugs, whereas ATR phosphorylates both Ser-15 and Ser-37 when cells are exposed to UV and inhibitors of replication (10, 11, 21, 44).
Our study revealed that p53 is phosphorylated at Ser-15 and Ser-37 residues during early-phase DNA damage responses, a time point at which KAISO is also induced, in HCT116 cells expressing WT p53 (Fig. 4B). Our data further suggest that p53 phosphorylated at Ser-15 and Ser-37 may play a role in KAISO gene transcription by acting on p53RE1 and p53RE3 (Fig. 6, B–E).
p53 acetylation also plays an important role in transcriptional activation of p53 target genes. Previous studies have shown that p53 acetylation enhances its ability to bind DNA and promote transcription of downstream targets after DNA damage. p53 can be acetylated by various histone acetyltransferases (e.g. p300/CBP, Tip60, and PCAF) and deacetylated by histone deacetylases (HDAC1, -2, -5, and -6 and sirtuin (SIRT)-1, -2, and -3), respectively (17, 45–50). Phosphorylation of p53 at Ser-15, Ser-33, and Ser-37 was reported to recruit p300/CBP and PCAF to acetylate p53 in response to DNA damage (24, 25). Moreover, Wang et al. (39) showed that p53 phosphorylation at Ser-15 leads to acetylation of p53 at Lys-320 and Lys-373/Lys-382 by p300 upon treatment with the DNA-hypomethylating agent 5-aza-2′-deoxycytidine. Upon treatment of the cells with etoposide, KAISO forms a complex with p53 and p300 to modulate acetylation at three key lysine residues of p53 by acetyltransferase p300, increasing acetylation at Lys-320 and Lys-382 while decreasing acetylation of Lys-381. Through such p53 post-translational modifications, KAISO up-regulates various apoptotic genes to induce cell death (6).
We also observed that KAISO transcription is activated twice upon treatment with etoposide, both in the early phase and later phase of the DDR process. Therefore, we hypothesized that phosphorylated p53 activates transcription of KAISO in early DDR, and acetylated p53 activates KAISO in late DDR. We found that the phosphorylated p53 at Ser-15 and/or Ser-37 can activate early-phase KAISO, required to invoke the p53 “death code,” which is required to activate KAISO robustly and to induce transcription of apoptotic genes. Intriguingly, both phosphorylated and acetylated p53 showed differences in selection of p53-binding response elements (p53REs) and in the magnitude of transcription activation. Specifically, p53 phosphorylated at Ser-15 and/or Ser-37 selectively bound p53RE1 and -RE3 but preferred p53RE1 with the unique arrangement of three p53-binding half-sites. In contrast, acetylated p53, generated by interaction with KAISO and p300, is a potent transactivator of KAISO, acting only on p53RE1, but not particularly affected by the unique structure of p53RE1. Our study indicates that the mode of KAISO transcriptional activation by acetylated p53 during late DDR may be different from KAISO activation mediated by phosphorylated p53 Ser-15 and Ser-37 in early DDR time periods.
Both phosphorylated p53 and acetylated p53 may be expressed and coexist, and they are thought to regulate different sets of genes, depending on p53 PTM codes. For example, KAISO expression is regulated by mixed PTMs of p53, with transcriptional activation by phosphorylated p53 as well as acetylated p53 and the KAISO-mediated acetylation code (i.e. p53QRQ) during DDR responses. In that event, p53QRQ continuously increases p53 target genes, such as p21, PUMA, and BAX, as well as KAISO. In HCT116 p53+/+ cells, KAISO mRNA levels peaked at 6 h and then increased again to peak at 24 h (Fig. 4A). KAISO can also be regulated by the different modes. As described above, our studies highlight the importance of post-translationally modified forms of p53 in the activation of KAISO gene transcription and apoptosis during early and late stages of genotoxic stress. Our studies also implicate that mutation in any of the p53 codons for Ser-15, Ser-37, Lys-320, Lys-381, and Lys-382 likely causes defective KAISO and apoptotic gene induction.
We also tested whether KAISO can induce apoptosis in WT p53 or hot spot mutant–expressing cells by flow cytometry. The SNU61, Colo320DM, LS1034, and HT-29 cells expressing endogenous p53 hot spot mutant were transfected with KAISO expression vector and were analyzed for apoptosis. Ectopic KAISO expression in these cells did not stimulate apoptosis. In the H1299-null cells transfected with KAISO, WT p53, p53-R175H, p53-G245S, p53-R248W, and p53-R273H expression vector, only the cells transfected with WT p53 and KAISO showed strong apoptosis (Fig. S2), suggesting the critical roles of functional p53 and KAISO apoptotic cell death.
In conclusion, we herein investigated the molecular mechanism of KAISO gene transcription by post-translationally modified p53 during DNA damage responses, successfully identifying two p53-responsive DNA regulatory elements (p53RE1 and p53RE3) critical for the activation of the KAISO promoter. Moreover, p53 phosphorylation at Ser-15 and/or Ser-37 and p53 with the KAISO-mediated acetylation code differentially recognize p53RE1 or p53RE3. p53RE1 has unique structural features preferably recognized by phosphorylated p53-Ser-15·Ser-37, which are important in early induction of KAISO gene transcription. Complex formation of p53, KAISO, and p300 resulted in p53 coded with a particular acetylation pattern that only recognizes p53RE1 may contribute to KAISO gene transcription during late DNA damage responses. Such findings have implications for transactivational activity by p53 and other tumor-suppressing signal pathways.
Experimental procedures
Plasmids, antibodies, and reagents
To prepare KAISO promoter-Luc gene fusion reporter plasmids, the KAISO promoter was PCR-amplified from genomic DNA isolated from HEK293 cells and cloned into pGL2-Luc vector to generate a reporter plasmid (pGL2-KAISO-Luc-4.6 kb). The following oligonucleotide PCR primers were used for pGL2-KAISO-Luc-4.6 kb plasmid. The primers were designed to amplify a 1606-bp DNA fragment (bp −4441 to −2835) and 3000-bp DNA fragment (bp −2835 to +165), respectively. The following primer pairs were used: for the 1606-bp DNA fragment region (bp −4441 to −2835), 5′-GATCGGTACCGATATCCACAAACTGACCTCGC-3′ (forward) and 5′-GATCCTCGAGCTCACGGTTTCCAGAATATGCC-3′ (reverse); for the 3000-bp DNA fragment region (bp −2835 to +165), 5′-GATCCTCGAGCAGTTTGACACTATGCATAGTT-3′ (forward) and 5′-GATCAAGCTTCAAGAAGAAGCCGCACTCCCCA-3′ (reverse).
The pG5-(p53RE)–Luc reporter plasmid was prepared by cloning a p53RE double-stranded oligonucleotide into pG5-Luc. Short oligonucleotides were heated at 95 °C for 5 min and cooled slowly to room temperature. Oligonucleotides were phosphorylated at their 5′-ends by T4 polynucleotide kinase (Takara, CA) and Klenow enzyme (Roche, Mannheim, Germany) by incubating at 37 °C for 30 min. Oligonucleotide sequences of the p53REs of the KAISO gene of the four pG5-Luc reporter plasmid constructs were as follows: p53RE1 (55-mer) of the 5′-upstream regulatory region, 5′-CTAGCAAACAAGACCACTTCATAAGCTTGTCTAAGCACAGACAAAAACAAGCTCC-3′ (forward) and 5′-TCGAGGAGCTTGTTTTTGTCTGTGCTTAGACAAGCTTATGAAGTGGTCTTGTTTG-3′ (reverse); intron region p53RE2 (79-mer), 5′-CTAGCAAACTTGCTCAACTCAGGAGCAAGCCATGAAATTGGACACTTGTTCCAAAAGCCAACCTGTATGAACAATTTCC-3′ (forward) and 5′-TCGAGGAAATTGTTCATACAGGTTGGCTTTTGGAACAAGTGTCCAATTTCATGGCTTGCTCCTGAGTTGAGCAAGTTTG-3′ (reverse); p53RE3 of the exon 2 coding region (36-mer), 5′-CTAGCAAAATTGTTCGTGTTAGATCAGATTTGCTTC-3′ (forward) and 5′-TCGAGAAGCAAATCTGATCTAACACGAACAATTTTG-3′ (reverse); p53RE4 in 3′-UTR (28-mer), 5′-CTAGCAAATAAGTTTTAGAGTTGTTCTC-3′ (forward) and 5′-TCGAGAGAACAACTCTAAAACTTATTTG-3′ (reverse). Oligonucleotide primers were purchased from Macrogen (Seoul, Korea). Preparation of pcDNA3.1, pcDNA3.1-KAISO, pcDNA3.1-p53, pcDNA3.1-p53QRQ, pcDNA3.1-p53-R175H, pcDNA3.1-p53-G245S, pcDNA3.1-p53-R248W, and pcDNA3.1-p53-R273H plasmids used was reported elsewhere (24). All plasmid constructs were verified by sequencing.
Antibodies against p53 (DO1; sc-126), GAPDH (6C5; sc-32233), p21 (H-51; sc-397), BAX (B-9; sc-7480), and NOXA (FL-54; sc-30209) were purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX). Antibody against PUMA (NBP1–76639) was purchased from Novus Biologicals (Centennial, CO). Antibodies against p53 Ser-15 (9284S), p53 Ser-37 (9289S), p53 (9282S), and APAF1 (8969S) were purchased from Cell Signaling Technology (Boston, MA). Antibody against p53 acetyl-Lys-381 (ab61241), p53 acetyl-Lys-382 (ab75754), and KAISO (ab12723) were purchased from Abcam (Cambridge, UK). Antibody against p53 acetyl-Lys-320 (02-1283) was purchased from Sigma-Aldrich. To obtain a polyclonal antibody against KAISO, a white rabbit was immunized subcutaneously with recombinant HIS-KAISO(ΔZF) (amino acids 1–499) protein six times at 2-week intervals. Blood was collected, incubated at 37 °C for 90 min, and centrifuged. Following centrifugation, the supernatant was incubated with Affi-Gel 10 gel beads (Bio-Rad) cross-linked to a recombinant KAISO(ΔZF) protein. The precipitated beads were then collected and washed with PBS, and the antibody was eluted in 1 m Tris (pH 7.6). Most of the chemical reagents (etoposide, DMSO) were purchased from Sigma.
Cell cultures
HEK293A, HCT116 p53+/+, HCT116 p53−/−, and HT-29 cells were cultured in Dulbecco's modified Eagle's medium (Gibco-BRL), and H1299, SNU61, Colo320DM, and LS1034 cells were cultured in RPMI medium 1640 supplemented with 10% fetal bovine serum (Gibco-BRL) and 1% penicillin-streptomycin. Cultures were maintained in humidified incubators at 37 °C in an atmosphere of 5% CO2 and 95% air. SNU61, Colo320DM, and LS-1034 cells were obtained from the Korean Cell Line Bank (Seoul, Korea).
qPCR analysis of KAISO, p53, and GAPDH mRNA expression in cells
Total RNA was isolated from the HCT116 p53+/+, HCT116 p53−/−, H1299, SNU61, Colo320DM, LS1034, and HT-29 cells using TRIzol reagent (Invitrogen). cDNAs were synthesized using 2 μg of total RNA, oligo(dT) primer, and Superscript reverse transcriptase II (200 units/μl) (Invitrogen). RT-qPCR was performed using SYBR Green Master Mix (Applied Biosystems, Foster City, CA). Reactions were subjected to quantification using the ABI PRISM 7300 RT-PCR system (Applied Biosystems). All reactions were performed in triplicate. GAPDH mRNA or 18S RNA was used as control. The oligonucleotide primers used for the qPCR assays are as follows: KAISO, 5′-CCGAGATTCTGCCCACAAA-3′ (forward) and 5′-GGGCGAGTTATTGCTAGCACTAG-3′ (reverse); 18S RNA, 5′-AGTCCCTGCCCTTTGTACACA-3′ (forward) and 5′-GATCCGAGGGCCTCACTAAAC-3′ (reverse); p53, 5′-CCTGAGGTTGGCTCTGACTGTA-3′ (forward) and 5′-AAAGCTGTTCCGTCCCAGTAGA-3′ (reverse); GAPDH, 5′-ACCACAGTCCATGCCATCAC-3′ (forward) and 5′-TCCACCACCCTGTTGCTGTA-3′ (reverse). All qPCR primers were purchased from Macrogen.
Site-directed mutagenesis
The p53 mutants were prepared by site-directed mutagenesis using the pcDNA3.1-p53 expression plasmid as a template. Conditions of PCR were as follows: denaturation step at 95 °C for 5 min, followed by 18 cycles at 95 °C for 30 s, 55 °C for 1 min, and an elongation step at 68 °C for 10 min. The oligonucleotides used for generation of the mutants were as follows: p53-R175H, 5′-GACGGAGGTTGTGAGGCACTGCCCCCACCATGAGC-3′ (forward) and p53-R175H 5′-GCTCATGGTGGGGGCAGTGCCTCACAACCTCCGTC-3′ (reverse); p53-G245S, 5′-CAGTTCCTGCATGGGCAGCATGAACCGGAGGCCCA-3′ (forward) and p53-G245S 5′-TGGGCCTCCGGTTCATGCTGCCCATGCAGGAACTG-3′ (reverse); p53-R248W, 5′-CATGGGCGGCATGAACTGGAGGCCCATCCTCACCA-3′ (forward) and 5′-TGGTGAGGATGGGCCTCCAGTTCATGCCGCCCATG-3′ (reverse); p53-R273H, 5′-GAACAGCTTTGAGGTGCATGTTTGTGCCTGTCCTG-3′ (forward) and 5′-CAGGACAGGCACAAACATGCACCTCAAAGCTGTTC-3′ (reverse); p53-S15A, 5′-CGTCGAGCCCCCTCTGGCTCAGGAAACATTTTCAG-3′ (forward) and 5′-CTGAAAATGTTTCCTGAGCCAGAGGGGGCTCGACG-3′ (reverse); p53-S15D, 5′-CGTCGAGCCCCCTCTGGATCAGGAAACATTTTCAG-3′ (forward) and 5′-CTGAAAATGTTTCCTGATCCAGAGGGGGCTCGACG-3′ (reverse); p53-S37A, 5′-CTGTCCCCCTTGCCGGCCCAAGCAATGGATGATT-3′ (forward) and 5′-AATCATCCATTGCTTGGGCCGGCAAGGGGGACAG-3′ (reverse); p53-S37D, 5′-CTGTCCCCCTTGCCGGACCAAGCAATGGATGATT-3′ (forward) and 5′-AATCATCCATTGCTTGGTCCGGCAAGGGGGACAG-3′ (reverse).
Transcriptional analysis of reporter plasmid with p53-responsive element of KAISO gene
Various pG5-(p53RE)s-Luc reporter plasmids and pcDNA3.1-p53, pcDNA3.1-p53-S15D, pcDNA3.1-p53-S15A, pcDNA3.1-p53-S37D, pcDNA3.1-p53-S37A, pcDNA3.1-p53-S15D·S37D, pcDNA3.1-p53-S15A·S37A, pcDNA3.1-p53QRQ (K320Q, K381R, K382Q), or pcDNA3.1-p53-S15D·S37D-QRQ expression plasmids were transiently transfected into H1299 cells using Lipofectamine Plus (Invitrogen). After 24–36 h of culture, the cells were harvested and analyzed for luciferase activity using a Microplate LB 96V luminometer (EG&G Berthold, Gaithersburg, MD). Reporter activity was normalized to the activity of co-expressed β-gal to normalize the transfection efficiency and is shown as the average of three independent assays.
Western blot analysis of protein expression
Transfected cells were harvested and lysed in radioimmune precipitation buffer (50 mm Tris-HCl (pH 8.0), 1% Nonidet P-40, 0.25% sodium deoxycholic acid, 150 mm NaCl, 1 mm EGTA, and complete Mini-Protease mixture). The cell lysates (50 μg) were separated using 8 or 10% SDS-polyacrylamide gels. The proteins were transferred onto Immun-BlotTM PVD membranes (Bio-Rad), and the membranes were blocked with 5% skim milk (BD Biosciences) or 3% BSA. Membrane blots were then incubated with antibodies followed by incubation with anti-mouse or rabbit secondary antibodies conjugated to horseradish peroxidase (Vector Laboratories, Burlingame, CA). Protein bands were visualized with ECL solution (PerkinElmer Life Sciences).
FACS analysis
H1299, SNU61, Colo320DM, LS1034, and HT-29 cells were transfected with the KAISO expression vector or control vectors and washed, stained with propidium diodide (1 μg/ml; BD Biosciences) and annexin V-FITC (1:25; BD Biosciences) in annexin V binding buffer (10 mm HEPES (pH 7.4), 140 mm NaCl, 5 mm CaCl2), and incubated for 15 min at room temperature in the dark. Apoptotic cells were analyzed using a FACSCalibur flow cytometer (BD Biosciences) at detection wavelengths of 488 nm (excitation) and 575 nm (peak emission). The obtained data were analyzed using ModFit LT 2.0 (Verity Software House, Inc.) and Wind MDI 2.8 (Joseph Trotter, Scripps Research Institute). For FACS analysis of apoptosis, cells were transfected with the KAISO, WT p53, or mutant p53 expression vectors and stained using propidium iodide and an annexin V apoptosis detection kit (BD Biosciences).
Oligonucleotide pulldown assays
Cells were lysed in HKMG buffer (10 mm HEPES (pH 7.9), 100 mm KCl, 5 mm MgCl2, 10% glycerol, 1 mm DTT, and 0.5% Nonidet P-40). Oligonucleotide probes were heated at 95 °C for 5 min and cooled slowly to room temperature to allow annealing. The cellular extracts were incubated with 1-μg biotinylated, double-stranded oligonucleotides of various potential p53-response elements of the KAISO gene (p53RE#1, p53RE#2, p53RE#3, and p53RE#4) for 16 h. To collect the DNA-bound proteins, the mixtures were incubated with streptavidin-agarose beads for 4 h, washed five times with HKMG buffer, and precipitated by centrifugation. The precipitates were analyzed by Western blotting using antibodies against KAISO and p53, as described above. Oligonucleotide sequences were as follows: KAISO 5′-upstream regulatory region p53RE#1 (37-mer), 5′-AGACAAAACAAGACCACTTCATAAGCTTGTCTAAGCA-3′ (forward) and 5′-TGCTTAGACAAGCTTATGAAGTGGTCTTGTTTTGTCT-3′ (reverse); p53RE#2 (42-mer), 5′-TCATAAGCTTGTCTAAG CACAGACAAAAACAAGCTCACTGT-3′ (forward) and 5′-ACAGTGAGCTTGTTTTTGTCTGTGCTTAGACAAGCTTATGAA-3′ (reverse); p53RE#3 (59-mer), 5′-AGACAAAACAAGACCACTTCATAAGCTTGTCTAAGCACAGACAAAAACAAGCTCACTGT-3′ (forward) and 5′-ACAGTGAGCTTGTTTTTGTCTGTGCTTAGACAAGCTTATGAAGTGGTCTTGTTTTGTCT-3′ (reverse); exon 2 KAISO-coding region p53RE#4, 5′-GTTCTAAAATTGTTCGTGTTAGATCAGATTTGCTTGATGA-3′ (forward) and 5′-TCATCAAGCAAATCTGATCTAACACGAACAATTTTAGAAC-3′ (reverse); KAISO 3′-UTR p53RE4 (negative control), 5′-GGCTACCACATAGTAGAGAATGGAATGAAG-3′ (forward) and 5′-CTTCATTCCATTCTCTACTATGTGGTAGCC-3′ (reverse); p21 promoter p53RE (positive control), 5′-GTCAGGAACATGTCCCAACATGTTGAGCTC-3′ (forward) and 5′-GAGCTCAACATGTTGGGACATGTTCCTGAC-3′ (reverse).
Quantitative ChIP and qPCR analysis (ChIP-qPCR)
To test the interaction between p53 and the KAISO promoter elements, H1299 cells were transfected with 5 μg of expression vector of pcDNA3.1-p53, pcDNA3.1-p53QRQ, pcDNA3.1-p53RQR, pcDNA3.1-p53-S15D·S37D, or pcDNA3.1-p53-S15A·S37A. HCT116 p53+/+ cells were treated with etoposide for 6 h or 24 h. Cells were fixed with 1% formaldehyde final concentration, for 20 min at room temperature, to cross-link protein–DNA complexes. Cells were harvested, washed three times in 1× PBS, and lysed in a 1% SDS, 50 mm Tris-HCl (pH 8.0), 10 mm EDTA. The lysate was sonicated to shear genomic DNA into fragments ranging from 500 to 1000 bp. The sonicated supernatant was then diluted 10-fold with ChIP dilution buffer (1% SDS, 1% Triton X-100, 16.7 mm Tris-HCl (pH 8.0), 167 mm NaCl, 1.2 mm EDTA) and precleared with salmon sperm DNA/protein A–agarose-50% slurry for 1 h at 4 °C. Precleared supernatant was incubated with antibody overnight at 4 °C with rotation. To collect DNA–protein–antibody complex, salmon sperm DNA/protein A–agarose-50% slurry was added to the mixture. The mixture was incubated for 2 h at 4 °C, with rotation, and then pelleted by centrifugation. Beads were washed three times in 1× PBS, including a protease inhibitor, incubated with 300 μl of elution buffer (1% SDS, 0.1 m Na2CO3), and rotated for 15 min to remove excess agarose. Eluted supernatants, as well as input DNA samples, were then decross-linked by incubating at 65 °C for 4 h. The supernatant was extracted with phenol/chloroform/isoamyl alcohol, precipitated with ethanol to recover DNA, and incubated with oligonucleotide primer sets designed to amplify regions flanking the p53-binding sites of the KAISO gene. Primer sets are as follows: p53RE#1 of KAISO 5′-upstream regulatory region, 5′-ATCACTGGGAGCCTGATCAAG-3′ (forward) and 5′-TGGTGTTTTCTGGGTGTCACA-3′ (reverse); p53RE#2 of the KAISO first intron region, 5′-GTTCAAATCTCTGTAACAATG-3′ (forward) and 5′-AACCAAAGTTCAGCATAATTT-3′ (reverse); p53RE#3 of the KAISO exon 2 coding region, 5′-CTGGGCAAGTTGTTGAACTG-3′ (forward) and 5′-CTAATAACTGCCCTGATTTAA-3′ (reverse); p53RE#4 of the KAISO 3′-UTR, 5′-GTGTGTTTTGTCTTGTATATG-3′ (forward) and 5′-AGTACTGACTTATTCTGACAT-3′ (reverse).
Statistical analysis
Student's t test was used for the statistical analyses. p values of <0.05 were considered statistically significant.
Author contributions
S.-H. C. and M.-W. H. conceptualization; S.-H. C., D.-I. K., and M.-W. H. data curation; S.-H. C., D.-I. K., S.-Y. C., and M.-W. H. formal analysis; S.-H. C. and M.-W. H. validation; S.-H. C., D.-I. K., S.-Y. C., M.-K. K., and M.-W. H. investigation; S.-H. C. and M.-W. H. writing-original draft; S.-H. C. writing-review and editing; K.-S. K. methodology; M.-W. H. resources; M.-W. H. software; M.-W. H. supervision; M.-W. H. funding acquisition; M.-W. H. visualization; M.-W. H. project administration.
Supplementary Material
This work was supported by JeonRack-HooSok Grant 2016R1E1A1A02921938 (to M.-W. H.), Do-Yak Research Grant 2011-0028817 (to M.-W. H.), and Medical Research Center Grant 2011-0030086 (to M.-W. H.) from the National Research Foundation of Korea (NRF) of the Korean Government (MSIP). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1 and S2.
- ATM
- ataxia telangiectasia mutated
- ATR
- ATM-Rad3–related
- qPCR
- quantitative real-time PCR
- DNA-PK
- DNA-dependent protein kinase
- CBP
- CREB-binding protein
- PCAF
- p300/CBP-associated factor
- DDR
- DNA damage response
- p53RE
- p53-responsive DNA elements
- PTM
- post-translational modification
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase.
References
- 1. Daniel J. M., and Reynolds A. B. (1999) The catenin p120(ctn) interacts with Kaiso, a novel BTB/POZ domain zinc finger transcription factor. Mol. Cell. Biol. 19, 3614–3623 10.1128/MCB.19.5.3614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Daniel J. M., Spring C. M., Crawford H. C., Reynolds A. B., and Baig A. (2002) The p120(ctn)-binding partner Kaiso is a bi-modal DNA-binding protein that recognizes both a sequence-specific consensus and methylated CpG dinucleotides. Nucleic Acids Res. 30, 2911–2919 10.1093/nar/gkf398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lopes E. C., Valls E., Figueroa M. E., Mazur A., Meng F. G., Chiosis G., Laird P. W., Schreiber-Agus N., Greally J. M., Prokhortchouk E., and Melnick A. (2008) Kaiso contributes to DNA methylation-dependent silencing of tumor suppressor genes in colon cancer cell lines. Cancer Res. 68, 7258–7263 10.1158/0008-5472.CAN-08-0344 [DOI] [PubMed] [Google Scholar]
- 4. Park J. I., Kim S. W., Lyons J. P., Ji H., Nguyen T. T., Cho K., Barton M. C., Deroo T., Vleminckx K., Moon R. T., and McCrea P. D. (2005) Kaiso/p120-catenin and TCF/β-catenin complexes coordinately regulate canonical Wnt gene targets. Dev. Cell 8, 843–854 10.1016/j.devcel.2005.04.010 [DOI] [PubMed] [Google Scholar]
- 5. Yoon H. G., Chan D. W., Reynolds A. B., Qin J., and Wong J. (2003) N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Mol. Cell 12, 723–734 10.1016/j.molcel.2003.08.008 [DOI] [PubMed] [Google Scholar]
- 6. Koh D. I., Han D., Ryu H., Choi W. I., Jeon B. N., Kim M. K., Kim Y., Kim J. Y., Parry L., Clarke A. R., Reynolds A. B., and Hur M. W. (2014) KAISO, a critical regulator of p53-mediated transcription of CDKN1A and apoptotic genes. Proc. Natl. Acad. Sci. U.S.A. 111, 15078–15083 10.1073/pnas.1318780111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Koh D. I., An H., Kim M. Y., Jeon B. N., Choi S. H., Hur S. S., and Hur M. W. (2015) Transcriptional activation of APAF1 by KAISO (ZBTB33) and p53 is attenuated by RelA/p65. Biochim. Biophys. Acta 1849, 1170–1178 10.1016/j.bbagrm.2015.07.008 [DOI] [PubMed] [Google Scholar]
- 8. Vousden K. H., and Lane D. P. (2007) p53 in health and disease. Nat. Rev. Mol. Cell Biol. 8, 275–283 10.1038/nrm2147 [DOI] [PubMed] [Google Scholar]
- 9. Abraham R. T. (2001) Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15, 2177–2196 10.1101/gad.914401 [DOI] [PubMed] [Google Scholar]
- 10. Tibbetts R. S., Brumbaugh K. M., Williams J. M., Sarkaria J. N., Cliby W. A., Shieh S. Y., Taya Y., Prives C., and Abraham R. T. (1999) A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev. 13, 152–157 10.1101/gad.13.2.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Banin S., Moyal L., Shieh S., Taya Y., Anderson C. W., Chessa L., Smorodinsky N. I., Prives C., Reiss Y., Shiloh Y., and Ziv Y. (1998) Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 281, 1674–1677 10.1126/science.281.5383.1674 [DOI] [PubMed] [Google Scholar]
- 12. Canman C. E., Lim D. S., Cimprich K. A., Taya Y., Tamai K., Sakaguchi K., Appella E., Kastan M. B., and Siliciano J. D. (1998) Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281, 1677–1779 10.1126/science.281.5383.1677 [DOI] [PubMed] [Google Scholar]
- 13. Siliciano J. D., Canman C. E., Taya Y., Sakaguchi K., Appella E., and Kastan M. B. (1997) DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev. 11, 3471–3481 10.1101/gad.11.24.3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang Y., and Xiong Y. (2001) A p53 amino-terminal nuclear export signal inhibited by DNA damage-induced phosphorylation. Science 292, 1910–1915 10.1126/science.1058637 [DOI] [PubMed] [Google Scholar]
- 15. Shieh S. Y., Ikeda M., Taya Y., and Prives C. (1997) DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 91, 325–334 10.1016/S0092-8674(00)80416-X [DOI] [PubMed] [Google Scholar]
- 16. Dohoney K. M., Guillerm C., Whiteford C., Elbi C., Lambert P. F., Hager G. L., and Brady J. N. (2004) Phosphorylation of p53 at serine 37 is important for transcriptional activity and regulation in response to DNA damage. Oncogene 23, 49–57 10.1038/sj.onc.1207005 [DOI] [PubMed] [Google Scholar]
- 17. Liu L., Scolnick D. M., Trievel R. C., Zhang H. B., Marmorstein R., Halazonetis T. D., and Berger S. L. (1999) p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol. 19, 1202–1209 10.1128/MCB.19.2.1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhao Y., Lu S., Wu L., Chai G., Wang H., Chen Y., Sun J., Yu Y., Zhou W., Zheng Q., Wu M., Otterson G. A., and Zhu W. G. (2006) Acetylation of p53 at lysine 373/382 by the histone deacetylase inhibitor depsipeptide induces expression of p21(Waf1/Cip1). Mol. Cell. Biol. 26, 2782–2790 10.1128/MCB.26.7.2782-2790.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li M., Luo J., Brooks C. L., and Gu W. (2002) Acetylation of p53 inhibits its ubiquitination by Mdm2. J. Biol. Chem. 277, 50607–50611 10.1074/jbc.C200578200 [DOI] [PubMed] [Google Scholar]
- 20. Ito A., Kawaguchi Y., Lai C. H., Kovacs J. J., Higashimoto Y., Appella E., and Yao T. P. (2002) MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 21, 6236–6245 10.1093/emboj/cdf616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Appella E., and Anderson C. W. (2001) Post-translational modifications and activation of p53 by genotoxic stresses. Eur. J. Biochem. 268, 2764–2772 10.1046/j.1432-1327.2001.02225.x [DOI] [PubMed] [Google Scholar]
- 22. Bode A. M., and Dong Z. (2004) Post-translational modification of p53 in tumorigenesis. Nat. Rev. Cancer 4, 793–805 10.1038/nrc1455 [DOI] [PubMed] [Google Scholar]
- 23. Gu W., and Roeder R. G. (1997) Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90, 595–606 10.1016/S0092-8674(00)80521-8 [DOI] [PubMed] [Google Scholar]
- 24. Lill N. L., Grossman S. R., Ginsberg D., DeCaprio J., and Livingston D. M. (1997) Binding and modulation of p53 by p300/CBP coactivators. Nature 387, 823–827 10.1038/42981 [DOI] [PubMed] [Google Scholar]
- 25. Sakaguchi K., Herrera J. E., Saito S., Miki T., Bustin M., Vassilev A., Anderson C. W., and Appella E. (1998) DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 12, 2831–2841 10.1101/gad.12.18.2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lambert P. F., Kashanchi F., Radonovich M. F., Shiekhattar R., and Brady J. N. (1998) Phosphorylation of p53 serine 15 increases interaction with CBP. J. Biol. Chem. 273, 33048–33053 10.1074/jbc.273.49.33048 [DOI] [PubMed] [Google Scholar]
- 27. Hofmann T. G., Möller A., Sirma H., Zentgraf H., Taya Y., Dröge W., Will H., and Schmitz M. L. (2002) Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat. Cell Biol. 4, 1–10 10.1038/ncb715 [DOI] [PubMed] [Google Scholar]
- 28. Hollstein M., Sidransky D., Vogelstein B., and Harris C. C. (1991) p53 mutations in human cancers. Science 253, 49–53 10.1126/science.1905840 [DOI] [PubMed] [Google Scholar]
- 29. Berglind H., Pawitan Y., Kato S., Ishioka C., and Soussi T. (2008) Analysis of p53 mutation status in human cancer cell lines: a paradigm for cell line cross-contamination. Cancer Biol. Ther. 7, 699–708 10.4161/cbt.7.5.5712 [DOI] [PubMed] [Google Scholar]
- 30. Zambetti G. P., and Levine A. J. (1993) A comparison of the biological activities of wild-type and mutant p53. FASEB J. 7, 855–865 10.1096/fasebj.7.10.8344485 [DOI] [PubMed] [Google Scholar]
- 31. Vousden K. H., and Lu X. (2002) Live or let die: the cell's response to p53. Nat. Rev. Cancer 2, 594–604 10.1038/nrc864 [DOI] [PubMed] [Google Scholar]
- 32. Vogelstein B., Lane D., and Levine A. J. (2000) Surfing the p53 network. Nature 408, 307–310 10.1038/35042675 [DOI] [PubMed] [Google Scholar]
- 33. Freed-Pastor W. A., and Prives C. (2012) Mutant p53: one name, many proteins. Genes Dev. 26, 1268–1286 10.1101/gad.190678.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. el-Deiry W. S., Kern S. E., Pietenpol J. A., Kinzler K. W., and Vogelstein B. (1992) Definition of a consensus binding site for p53. Nat. Genet. 1, 45–49 10.1038/ng0492-45 [DOI] [PubMed] [Google Scholar]
- 35. Vousden K. H., and Prives C. (2009) Blinded by the light: the growing complexity of p53. Cell 137, 413–431 10.1016/j.cell.2009.04.037 [DOI] [PubMed] [Google Scholar]
- 36. Kruse J. P., and Gu W. (2009) Modes of p53 regulation. Cell 137, 609–622 10.1016/j.cell.2009.04.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rozan L. M., and El-Deiry W. S. (2007) p53 downstream target genes and tumor suppression: a classical view in evolution. Cell Death Differ. 14, 3–9 10.1038/sj.cdd.4402058 [DOI] [PubMed] [Google Scholar]
- 38. Lane D. P. (1992) Cancer: p53, guardian of the genome. Nature 358, 15–16 10.1038/358015a0 [DOI] [PubMed] [Google Scholar]
- 39. Wang H., Zhao Y., Li L., McNutt M. A., Wu L., Lu S., Yu Y., Zhou W., Feng J., Chai G., Yang Y., and Zhu W. G. (2008) An ATM- and Rad3-related (ATR) signaling pathway and a phosphorylation-acetylation cascade are involved in activation of p53/p21Waf1/Cip1 in response to 5-aza-2′-deoxycytidine treatment. J. Biol. Chem. 283, 2564–2574 10.1074/jbc.M702454200 [DOI] [PubMed] [Google Scholar]
- 40. Meek D. W., and Anderson C. W. (2009) Posttranslational modification of p53: cooperative integrators of function. Cold Spring Harb. Perspect. Biol. 1, a000950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Olsson A., Manzl C., Strasser A., and Villunger A. (2007) How important are posttranslational modifications in p53 for selectivity in target-gene transcription and tumour suppression? Cell Death Differ. 14, 1561–1575 10.1038/sj.cdd.4402196 [DOI] [PubMed] [Google Scholar]
- 42. Brooks C. L., and Gu W. (2003) Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr. Opin. Cell Biol. 15, 164–171 10.1016/S0955-0674(03)00003-6 [DOI] [PubMed] [Google Scholar]
- 43. Saito S., Goodarzi A. A., Higashimoto Y., Noda Y., Lees-Miller S. P., Appella E., and Anderson C. W. (2002) ATM mediates phosphorylation at multiple p53 sites, including Ser46, in response to ionizing radiation. J. Biol. Chem. 277, 12491–12494 10.1074/jbc.C200093200 [DOI] [PubMed] [Google Scholar]
- 44. Canman C. E., Lim D. S., Cimprich K. A., Taya Y., Tamai K., Sakaguchi K., Appella E., Kastan M. B., and Siliciano J. D. (1998) Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281, 1677–1679 10.1126/science.281.5383.1677 [DOI] [PubMed] [Google Scholar]
- 45. Tang Y., Luo J., Zhang W., and Gu W. (2006) Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol. Cell. 24, 827–839 10.1016/j.molcel.2006.11.021 [DOI] [PubMed] [Google Scholar]
- 46. Luo J., Su F., Chen D., Shiloh A., and Gu W. (2000) Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature 408, 377–381 10.1038/35042612 [DOI] [PubMed] [Google Scholar]
- 47. Juan L. J., Shia W. J., Chen M. H., Yang W. M., Seto E., Lin Y. S., and Wu C. W. (2000) Histone deacetylases specifically down-regulate p53-dependent gene activation. J. Biol. Chem. 275, 20436–20443 10.1074/jbc.M000202200 [DOI] [PubMed] [Google Scholar]
- 48. Solomon J. M., Pasupuleti R., Xu L., McDonagh T., Curtis R., DiStefano P. S., and Huber L. J. (2006) Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Mol. Cell. Biol. 26, 28–38 10.1128/MCB.26.1.28-38.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Peck B., Chen C. Y., Ho K. K., Di Fruscia P., Myatt S. S., Coombes R. C., Fuchter M. J., Hsiao C. D., and Lam E. W. (2010) SIRT inhibitors induce cell death and p53 acetylation through targeting both SIRT1 and SIRT2. Mol. Cancer Ther. 9, 844–855 10.1158/1535-7163.MCT-09-0971 [DOI] [PubMed] [Google Scholar]
- 50. Chen J., Wang A., and Chen Q. (2017) SirT3 and p53 deacetylation in aging and cancer. J. Cell. Physiol. 232, 2308–2311 10.1002/jcp.25669 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.