Abstract
The nickel-dependent carbon monoxide dehydrogenase (CODH) employs a unique heterometallic nickel–iron–sulfur cluster, termed the C-cluster, to catalyze the interconversion of CO and CO2. Like other complex metalloenzymes, CODH requires dedicated assembly machinery to form the fully intact and functional C-cluster. In particular, nickel incorporation into the C-cluster depends on the maturation factor CooC; however, the mechanism of nickel insertion remains poorly understood. Here, we compare X-ray structures (1.50–2.48 Å resolution) of CODH from Desulfovibrio vulgaris (DvCODH) heterologously expressed in either the absence (DvCODH−CooC) or presence (DvCODH+CooC) of co-expressed CooC. We find that the C-cluster of DvCODH−CooC is fully loaded with iron but does not contain any nickel. Interestingly, the so-called unique iron ion (Feu) occupies both its canonical site (80% occupancy) and the nickel site (20% occupancy), with addition of reductant causing further mismetallation of the nickel site (60% iron occupancy). We also demonstrate that a DvCODH variant that lacks a surface-accessible iron–sulfur cluster (the D-cluster) has a C-cluster that is also replete in iron but lacks nickel, despite co-expression with CooC. In this variant, all Feu is in its canonical location, and the nickel site is empty. This D-cluster–deficient CODH is inactive despite attempts to reconstitute it with nickel. Taken together, these results suggest that an empty nickel site is not sufficient for nickel incorporation. Based on our findings, we propose a model for C-cluster assembly that requires both CooC and a functioning D-cluster, involves precise redox-state control, and includes a two-step nickel-binding process.
Keywords: nickel, iron–sulfur protein, metalloenzyme, one-carbon metabolism, oxidation-reduction (redox), X-ray crystallography, bioremediation catalyst, carbon monoxide dehydrogenase (CODH), heterometallic cofactor
Introduction
Anerobic carbon monoxide dehydrogenases (CODHs)4 catalyze the reversible oxidation of CO to CO2, enabling certain microbes, such as Rhodospirillum rubrum and Carboxydothermus hydrogenoformans, to live on CO as a sole source of carbon and energy (1, 2). This microbial activity accounts for the removal of an estimated 108 tons of CO from the lower atmosphere each year, making CODHs an important part of the global carbon cycle (3). Given that CO is a toxic pollutant and a component of fossil fuel emissions, CODH has attracted attention as a possible bioremediation catalyst. Similarly, CODHs also have potential applications in the capture and removal of CO2 via the Wood–Ljungdahl pathway of carbon fixation, in which the CO that is generated is incorporated into the acetyl group of acetyl-CoA (4). Use of CODH in such capacities would benefit from an ability to produce large amounts of active enzyme. In particular, CODH activity requires a complex heterometallic nickel–iron–sulfur cofactor (termed the C-cluster), the biogenesis of which is poorly understood.
The CODH C-cluster is housed within a homodimeric protein scaffold that contains two additional iron–sulfur clusters, termed the B- and D-clusters, that are used for electron transfer during catalysis (Fig. 1A). The D-cluster, depending on the bacterial species, is either a [4Fe–4S] or [2Fe–2S] cluster that resides at the CODH dimer interface and serves as an electron conduit to external redox partners, such as ferredoxins, whereas the B-cluster is a [4Fe–4S] cluster that mediates electron transfer between the C- and D-clusters (5–9). The C-cluster is a structurally unique metallocluster composed of a distorted [Ni–3Fe–4S] cubane linked through a sulfide ion (SL) to a mononuclear iron site (Feu) (Fig. 1B) (5, 6). This canonical C-cluster architecture is essential for catalysis as it allows for binding of CO at the nickel ion of the cubane, activating it for nucleophilic attack by a water molecule ligated in immediate proximity at Feu (10–13). To access this chemistry, organisms require dedicated cellular machinery for C-cluster assembly, similar to the requirements of other complex metalloclusters (14). Our understanding of the C-cluster assembly process, however, remains incomplete. It is still unknown what the biosynthetic origin of the iron–sulfur scaffold of the cluster is, how the nickel–iron–sulfur cluster is assembled, and what roles individual accessory proteins play in this assembly process.
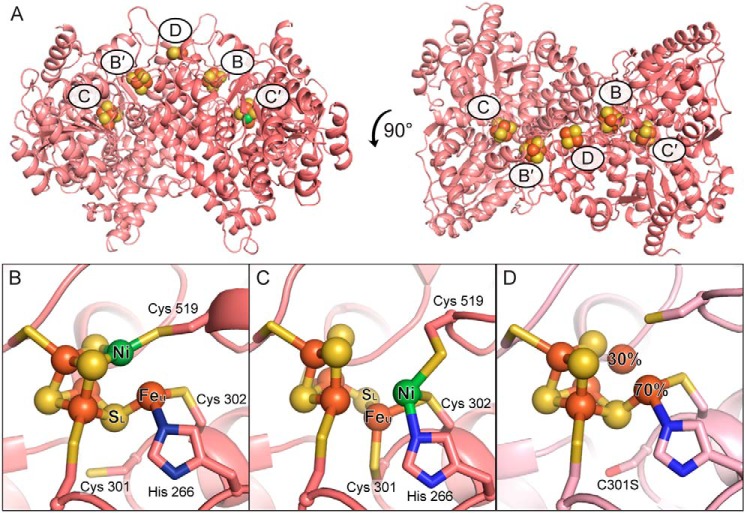
Figure 1.
The metalloclusters of CODH. A, the overall homodimeric structure of DvCODH (PDB code 6B6V). Metalloclusters are shown as spheres and labeled. Note that the B-cluster of one monomer completes the electron transfer pathway of the opposing monomer. B, the C-cluster in its canonical, reduced state (PDB code 6B6V). C, the oxidized C-cluster (PDB code 6B6W). A lysine residue that completes a distorted tetrahedral coordination geometry around the nickel ion has been omitted for simplicity. D, the C-cluster of DvCODH(C301S)+CooC (PDB code 6DC2). Residue numbers correspond to the sequence of DvCODH. Protein is shown in ribbon representation in pink with metalloclusters shown as spheres and sticks with nickel in green, iron in orange, and sulfur in yellow; in B–D, ligating amino acid residue side chains are shown as sticks with sulfur in yellow, nitrogen in blue, and oxygen in red. The structures shown in this figure are described in Ref. 9.
Limited insight into the process of C-cluster assembly has been gleaned from the co-operonic expression of accessory proteins that appear to play roles in cluster maturation, in particular incorporation of nickel. Previous studies have shown that integration of nickel into the C-cluster depends on the accessory protein CooC (15–17). Certain organisms express additional proteins, CooJ and CooT, that have been implicated in C-cluster maturation; however, CooC appears to be the only dedicated and essential maturation factor expressed by all CODH-containing organisms (15, 18–22). CooC is a P-loop ATPase with sequence similarity to UreG and HypB, maturation factors involved in nickel transfer to the active sites of urease and nickel–iron hydrogenase, respectively (15, 23). In analogy to UreG and HypB, CooC has been proposed to use ATP hydrolysis to facilitate nickel insertion into CODH (15, 16, 23, 24). Alternatively, CooC has been proposed to fold or otherwise mediate formation of the proper nickel-binding site in CODH (16, 17, 25).
To gain a further understanding of C-cluster maturation and the role of CooC, we have recently developed a means to heterologously express Desulfovibrio vulgaris CODH (DvCODH) in either the presence (DvCODH+CooC) or the absence (DvCODH−CooC) of the D. vulgaris CooC maturase (DvCooC) using Desulfovibrio fructosovorans as an expression host (17). This differential expression results in substantially different enzymatic phenotypes (see Table 1). As-isolated DvCODH+CooC binds about half of the expected nickel content and exhibits a lag phase in activity followed by a relatively low rate of CO oxidation (160 μmol·min−1·mg−1) as compared with the previously published activities of monofunctional CODHs from other species, which range from ∼4400 to 16,000 μmol·min−1·mg−1 (17, 26, 27). Incubation of the as-isolated DvCODH+CooC with both NiCl2 and the reductant sodium dithionite results in elimination of the lag phase and a 10-fold increase in CO oxidation activity; however, activation does not occur in the presence of NiCl2 or sodium dithionite alone (17). In contrast, as-isolated DvCODH−CooC contains low amounts of nickel (0–0.2 nickel/monomer), has nearly no activity (4% of as-isolated DvCODH+CooC), and undergoes limited activation with NiCl2 and sodium dithionite (17), suggesting that DvCooC is involved in constructing the appropriate nickel-binding site in DvCODH (25).
Table 1.
Metal content and activity of DvCODH variants
ND, not detected.
| DvCODH sample | As-isolated nickel/monomer | As-isolated iron/monomer | As-isolated CO oxidation activity (μmol·min−1·mg−1) | Nickel-reconstituted CO oxidation activity (μmol·min−1·mg−1) | Reference |
|---|---|---|---|---|---|
| WT+CooC | 0.4–0.9 | 8–10.5 | 160 | 1660 | 17 |
| WT−CooC | 0–0.2 | 7.5–8.5 | <5 | 4–60 | 17 |
| 0 | 10 | ND | ND | This work | |
| C301S+CooC | 0 | 13 | ND | ND | 9 |
| 0 | 10 ± 2.5 | ND | ND | This work | |
| ΔD+CooC | 0.02 | 8 ± 1 | ND | <5 | This work |
Interestingly, our previously published crystal structures of DvCODH+CooC revealed that the C-cluster adopts an alternative conformation upon exposure to oxygen in which the Ni, Feu, and SL ions shift by as much as 3 Å, and the Ni and Feu ions adopt new coordination environments (Fig. 1C) (9). Notably, this oxidized conformation of the C-cluster can be converted back to the canonical, reduced conformation by incubation with reducing agent (9). The oxidized conformation involves ligation by a cysteine residue that is strictly conserved in CODHs but that does not serve as a ligand to the active, reduced conformation of the cluster (9). Mutation of this cysteine residue (Cys-301) in DvCODH+CooC to serine (C301S) was shown to result in an inactive enzyme that does not bind nickel (see Table 1) (9), similar to previous results on the CODH from Moorella thermoacetica (28). The crystal structure of DvCODH(C301S)+CooC revealed a partially assembled C-cluster in which Feu adopted a split conformation: Feu was in its canonical binding site at 70% occupancy, and Feu was incorporated into the cubane portion of the cluster at 30% occupancy, taking up the canonical nickel-binding site (Fig. 1D) (9). This split C-cluster conformation combined with the inability of the DvCODH(C301S)+CooC variant to incorporate nickel led us to propose that the oxidized conformation of the cluster could be an intermediate in C-cluster maturation, although how this conformation may participate in the assembly process remained unclear (9).
To further interrogate the process of C-cluster assembly, we have now determined crystal structures of DvCODH produced in the absence of CooC (DvCODH−CooC) and of a DvCODH variant produced in the presence of CooC and engineered to not contain the surface-accessible D-cluster (DvCODH(ΔD)+CooC). Comparison of the DvCODH−CooC structure to that of DvCODH(C301S)+CooC (9) suggests a possible link between CooC-dependent cluster assembly and the ability to adopt the alternative, oxidized cluster arrangement. Furthermore, removal of the D-cluster leads to formation of an incomplete C-cluster, highlighting the importance of this redox-active iron–sulfur cluster for C-cluster maturation. Combined, these results expand our understanding of C-cluster biogenesis, with an emphasis on the importance of accessing different cluster conformations and redox states.
Results
The C-cluster expressed in the absence of CooC is a [3Fe–4S] cluster with a mobile fourth iron
The CODH from D. vulgaris was expressed heterologously in D. fructosovorans in the absence of the C-cluster maturation factor CooC (DvCODH−CooC), as described previously (17). The preparation of protein that was used for crystallization displayed no detectible CO oxidation activity and contained 0 nickel atoms/monomer and 10 iron atoms/monomer (Table 1). The crystal structure of DvCODH−CooC was determined to 1.50 Å resolution (Table 2). The structure aligns well (Cα root-mean-square deviation of 0.18 Å for 1250 Cα atoms within the CODH dimer) with our previously determined structure of DvCODH+CooC (9), and the B- and D-clusters of the enzyme are both present and fully intact. Thus, the overall structure of DvCODH is retained when expressed in the absence of CooC.
Table 2.
Crystallographic data collection and refinement statistics
| WT−CooC as-isolated | WT−CooC as-isolated iron peaka | WT−CooC reduced | WT−CooC reduced iron peaka | (ΔD)+CooC iron peaka | |
|---|---|---|---|---|---|
| Data collection | |||||
| Wavelength (Å) | 0.9792 | 1.7389 | 0.9792 | 1.7389 | 1.7379 |
| Space group | P21 | P21 | P21 | P21 | C2 |
| Cell dimensions | |||||
| a, b, c (Å) | 64.8, 144.1, 123.4 | 64.8, 144.2, 123.4 | 64.7, 143.8, 123.1 | 64.7, 143.7, 123.2 | 110.5, 100.6, 65.3 |
| β (°) | 98.5 | 98.5 | 98.6 | 98.6 | 124.7 |
| Resolution (Å)b | 100–1.50 (1.53–1.50) | 100–1.97 (2.01–1.97) | 100–1.72 (1.75–1.72) | 100–2.32 (2.37–2.32) | 50.0–2.48 (2.57–2.48) |
| Completeness (%)b | 95.0 (93.1) | 90.7 (87.3) | 95.9 (95.6) | 94.2 (93.0) | 93.4 (88.6) |
| Redundancyb | 6.5 (6.0) | 4.4 (4.3) | 5.1 (4.9) | 4.3 (4.2) | 3.0 (2.0) |
| Unique reflectionsb | 338890 (24577) | 283488 (20224) | 225437 (16599) | 178944 (13009) | 38000 (3637) |
| Rsym (%)b | 9.2 (90.4) | 12.2 (60.7) | 9.3 (91.3) | 11.6 (87.5) | 22.0 (78.3) |
| CC½b | 99.8 (71.2) | 99.5 (75.7) | 99.8 (68.5) | 99.6 (65.0) | 95.8 (47.2) |
| <I/σI >b | 11.5 (1.9) | 7.9 (2.0) | 11.3 (2.0) | 9.88 (2.1) | 6.1 (2.1) |
| Refinement | |||||
| Resolution (Å) | 93.3–1.50 | 93.3–1.72 | 45.4–2.48 | ||
| No. reflections | 338812 | 225376 | 37976 | ||
| Rwork/Rfree | 0.154/0.178 | 0.149/0.176 | 0.207/0.248 | ||
| Monomer/asu | 4 | 4 | 1 | ||
| No. atoms | |||||
| Protein | 18977 | 18682 | 4481 | ||
| B-cluster | 32 | 32 | 8 | ||
| C-cluster | 36 | 36 | 8 | ||
| D-cluster | 8 | 8 | |||
| Water | 2673 | 1899 | 25 | ||
| B-factors | |||||
| Protein | 19.6 | 22.6 | 41.5 | ||
| B-cluster | 15.8 | 16.7 | 40.3 | ||
| C-cluster | 21.0 | 24.4 | 45.6 | ||
| D-cluster | 17.9 | 18.9 | |||
| Water | 33.9 | 34.0 | 40.7 | ||
| Root-mean-square bond deviations | |||||
| Lengths (Å) | 0.007 | 0.007 | 0.003 | ||
| Angles (°) | 0.932 | 0.919 | 0.737 | ||
| Rotamer outliers (%) | 0.15 | 0.52 | 0.67 |
a Bijvoet pairs were not merged during data processing.
b The values in parentheses are for the highest-resolution shell.
asu, asymmetric unit.
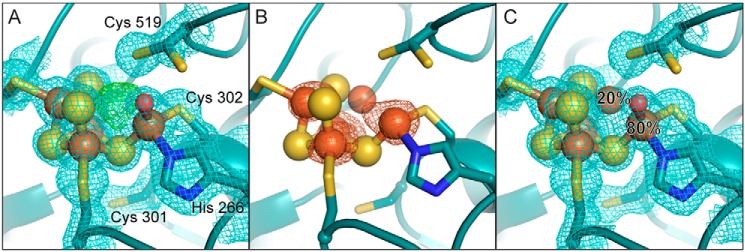
At the C-cluster of DvCODH−CooC, the [3Fe–4S] partial cubane portion of the canonical C-cluster is intact and present at full occupancy (Fig. 2), indicating that CooC is not necessary for formation of this part of the C-cluster. Modeling of the Feu ion, however, was more complicated. When the C-cluster was modeled as a [3Fe–4S]–Feu cluster at full occupancy, residual positive difference electron density was observed in the open cubane position, indicating the presence of an additional atom (Fig. 2A). Further, iron anomalous difference maps (Table 2) reveal a shoulder extending from the canonical Feu-binding site into the cubane position, suggesting the presence of iron at partial occupancy (Fig. 2B). Given the lack of nickel in the sample that was crystallized, the positive difference density in the electron density maps, and the shoulder in the iron anomalous maps, we rationalized that Feu could be present in a split conformation. Refinement of Feu with a split conformation revealed that at 80% occupancy, Feu is in its canonical binding site, ligated by His-266, Cys-302, and a water molecule, whereas at 20% occupancy, Feu is incorporated into the cubane portion of the cluster and ligated by Cys-519, forming a distorted [4Fe–4S] cluster (Fig. 2C). Similar split Feu conformations were observed across multiple crystal structures of DvCODH−CooC samples that lacked nickel. Interestingly, this split Feu conformation is similar to what was observed previously in our structure of DvCODH(C301S)+CooC (9) (Fig. 1D), a CODH variant that is unable to adopt the alternative, oxidized C-cluster conformation because of the absence of the Cys-301 thiol for coordination to Feu. Together, the structural similarity between the C-clusters in DvCODH−CooC and DvCODH(C301S)+CooC (9) suggests a link between Cys-301 and the role of CooC in cluster assembly, perhaps due to a CooC-induced conformational change in which Feu becomes ligated by Cys-301 (see “Discussion”). Additionally, these data suggest that it is not the lack of an open coordination site for nickel that prevents nickel incorporation into the C-cluster. Although there is some Feu in the nickel-binding site, there is not enough to explain the inability to reconstitute the C-cluster with nickel.
Figure 2.
The DvCODH−CooC C-cluster is a [3Fe–4S] cluster with a mobile Feu. A, refinement of a [3Fe–4S]–Feu C-cluster results in positive Fo − Fc electron density (green mesh, contoured to +3σ) at the nickel-binding site. 2Fo − Fc electron density (blue mesh) contoured to 1σ. A water molecule (red sphere) is bound to Feu. Cys-519 adopts alternative conformations. B, iron anomalous difference map (orange mesh, contoured to 6σ) indicates the presence of iron at partial occupancy in the canonical nickel-binding site. The Feu-ligating water molecule has been omitted for simplicity. C, the C-cluster refined with an alternative conformation of Feu. At 80% occupancy, Feu is ligated by His-266 and Cys-302 in its canonical binding site. At 20% occupancy, Feu is incorporated into the cubane portion of the cluster and ligated by Cys-519. 2Fo − Fc electron density (blue mesh) contoured to 1σ. Protein is shown in ribbon representation in teal with ligating amino acid residue side chains in sticks; cluster ions shown as spheres and sticks; iron in orange, sulfur in yellow, nitrogen in blue, and oxygen in red.
Reduction of DvCODH−CooC induces movement of Feu into the cubane position
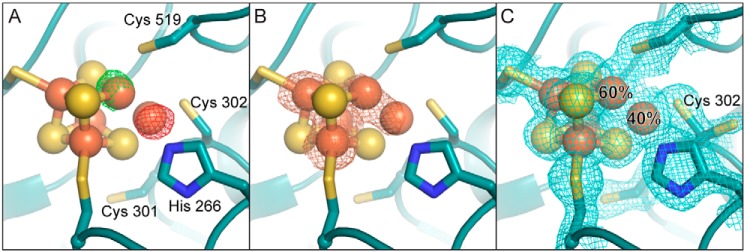
The presence of Feu at partial occupancy in the nickel-binding site of the C-cluster in both our new structure, DvCODH−CooC, and the previous structure, DvCODH(C301S)+CooC (9), is intriguing. Notably, the [3Fe–4S] clusters of aconitase and ferredoxins, as well as synthetic model compounds, are well-known to incorporate exogenous metal into their open cubane site upon reduction due to the increased nucleophilicity of the open sulfide ions in the reduced state (29–33). Therefore, we hypothesized that reduction of the immature pre–C-cluster, which in part resembles a [3Fe–4S] cluster, could have led to movement of Feu from its canonical binding site into the cubane position in some CODH molecules. To test this hypothesis, crystals of as-isolated DvCODH−CooC were soaked in the reductant sodium dithionite prior to cryo-cooling and X-ray data collection. The structure of reduced DvCODH−CooC was determined to 1.72 Å resolution (Table 2), revealing greater incorporation of Feu into the cubane position relative to the structure of the as-isolated enzyme (Fig. 3, A and B). Here, the Feu ion resides in its canonical position at 40% occupancy and in the cubane portion at 60% occupancy (Fig. 3C). Together, these data suggest that reduction of the pre–C-cluster before nickel is inserted can lead to mismetallation of the nickel site, and therefore careful control of cluster redox state is likely essential during the C-cluster maturation process in vivo.
Figure 3.
Reduction of DvCODH−CooC induces movement of Feu into the iron–sulfur cubane portion of the C-cluster. A, isomorphous difference map (Fo(reduced) − Fo(as-isolated)) reveals increased electron density at the canonical nickel-binding site of the cubane (green mesh, contoured to +5σ) and decreased electron density at the canonical Feu binding site (red mesh, contoured to −5σ) in the structure of reduced DvCODH−CooC relative to as-isolated DvCODH−CooC. B, iron anomalous difference map (orange mesh, contoured to 6σ) reveals a strong peak of iron anomalous signal in the canonical nickel-binding site of the cubane (compare with Fig. 2B). C, the C-cluster of DvCODH−CooC refined with an alternative conformation of Feu. At 60% occupancy, Feu is incorporated into the cubane portion of the cluster and ligated by Cys-519. At 40% occupancy, Feu is ligated by His-266 and Cys-302 in its canonical binding site. Cys-302 adopts alternative conformations. 2Fo − Fc electron density (blue mesh) contoured to 1σ. Protein is shown in ribbon representation in teal with ligating amino acid residue side chains in sticks; cluster ions shown as spheres and sticks; iron in orange, sulfur in yellow, and nitrogen in blue.
The D-cluster is necessary for proper C-cluster assembly in the presence of CooC
To test the hypothesis that control of redox state is essential to C-cluster maturation, we sought to disrupt electron transfer between the C-cluster and external redox partners by removal of the solvent-exposed D-cluster, which serves as an electron conduit during CO/CO2 interconversion. Toward this goal, we designed a DvCODH double-mutant variant in which the D-cluster–ligating cysteine residues (Cys-42 and Cys-45) were replaced with alanine resides to abolish binding of the D-cluster (DvCODH(ΔD)). This variant was expressed in the presence of CooC (DvCODH(ΔD)+CooC) and purified to homogeneity. Similar to DvCODH−CooC and DvCODH(C301S)+CooC, DvCODH(ΔD)+CooC is inactive as-isolated and does not contain appreciable amounts of nickel (Table 1). No increase in activity is observed upon incubation with nickel (Table 1). These observations indicate that the D-cluster is essential for C-cluster maturation. To characterize the impact of a D-cluster deletion on C-cluster architecture, the crystal structure of DvCODH(ΔD)+CooC was determined to 2.48 Å resolution (Table 2). The overall structure aligns well (Cα root-mean-square deviation of 0.29 Å for 1242 Cα atoms within the CODH dimer) with that of DvCODH+CooC. The structure contains both the B- and C-clusters and confirms that the D-cluster is not present in this protein variant (Fig. 4A). The absence of the D-cluster leads to local disorder, and residues 41–44 could not be modeled (Fig. 4A, inset). At the C-cluster of DvCODH(ΔD)+CooC, we observe an intact [3Fe–4S]–Feu scaffold with Feu present at 100% occupancy in its canonical binding site (Fig. 4, B and C). This result is consistent with the above-mentioned idea that movement of Feu into the cubane is induced by reduction and that the D-cluster mediates that reduction. Additionally, it is notable that the C-cluster of DvCODH(ΔD)+CooC, in which 100% of Feu is in the canonical location, cannot be activated by incubation with nickel. Further, the fact that the structure of DvCODH(ΔD)+CooC is largely unchanged by D-cluster loss suggests that it is the D-cluster's redox role, rather than a structural role, that is required for nickel insertion.
Figure 4.
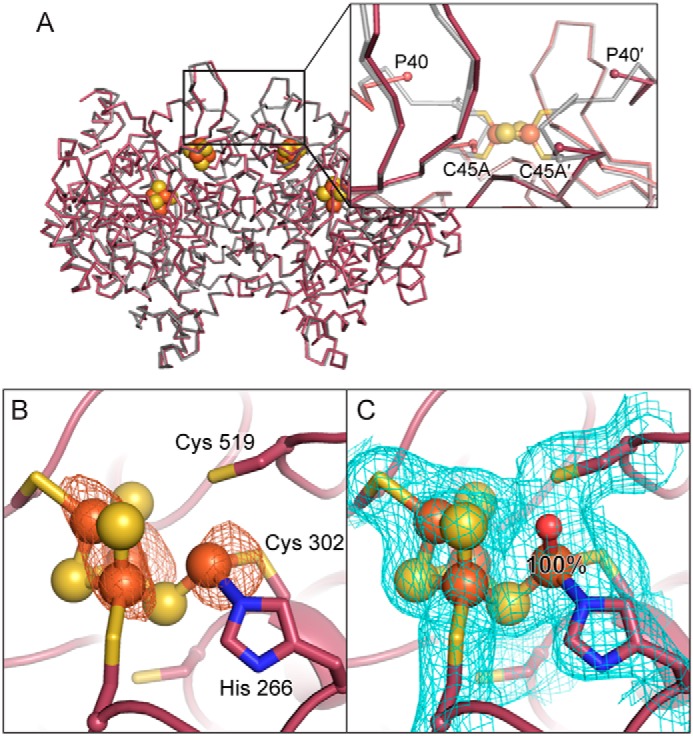
Removal of the D-cluster does not alter the overall structure but leads to incomplete C-cluster assembly. A, structural alignment of DvCODH(ΔD)+CooC (maroon) with DvCODH+CooC (gray; PDB code 6B6V) (9). The inset shows disorder in the vicinity of the D-cluster in DvCODH(ΔD)+CooC. Proteins are shown as the Cα trace of each structure. B- and C-clusters of DvCODH(ΔD)+CooC are shown as spheres. B, iron anomalous difference map (orange mesh, contoured to 5σ) suggests the presence of Feu at full occupancy in its canonical binding site. C, refinement of DvCODH(ΔD)+CooC confirms the location and occupancy of Feu. 2Fo − Fc electron density (blue mesh) contoured to 1σ. In B and C, protein is shown in ribbon representation in maroon with ligating amino acid residue side chains in sticks; cluster ions shown as spheres and sticks; iron in orange, sulfur in yellow, nitrogen in blue, and oxygen in red.
Discussion
Here we present a series of crystal structures of DvCODH to provide insight into the process of C-cluster assembly and maturation, the mechanisms of which remained largely elusive. Our structures suggest that the C-cluster maturase CooC is primarily involved in nickel insertion rather than in formation of the [3Fe–4S]–Feu scaffold and reveal that nickel insertion is additionally dependent on the D-cluster, likely because of its role in mediating electron transfer. Together, these findings allow us to propose a model for C-cluster assembly and maturation involving multiple cluster conformations and redox states.
In our structure of as-isolated DvCODH−CooC, we observe a largely (80%) intact iron–sulfur scaffold that contains four iron ions and four sulfur ions arranged as a [3Fe–4S]–Feu pre–C-cluster that lacks nickel. The presence of this prearranged iron–sulfur scaffold in the absence of dedicated C-cluster assembly machinery suggests that the [3Fe–4S]–Feu cluster arrangement can be formed using general iron–sulfur cluster biogenesis pathways, such as the sulfur utilization factor (SUF) nitrogen fixation (NIF) systems, both of which are present in the D. fructosovorans expression host, as well as D. vulgaris itself. Two possibilities for the formation of the iron–sulfur scaffold can be envisioned (Fig. 5A). First, the pre–C-cluster could be inserted in two pieces: a single iron ion inserted into the unique His-266/Cys-302 site and a [3Fe–4S] cluster inserted into the cubane site. Linkage of Feu and the [3Fe–4S] cluster via the cubane sulfide (SL) could occur subsequently (Fig. 5A, upper pathway). Alternatively, the C-cluster–binding site of CODH could become loaded with a [4Fe–4S] cluster that is distorted by CODH concomitant with the insertion step or is acted upon by an unknown maturation factor to remove an iron ion from the cubane, forming Feu (Fig. 5A, lower pathway). Regardless of the exact assembly mechanism, our data indicate that CooC is not necessary for formation of a four-iron–containing iron–sulfur scaffold and that its primary role is likely in facilitating nickel insertion.
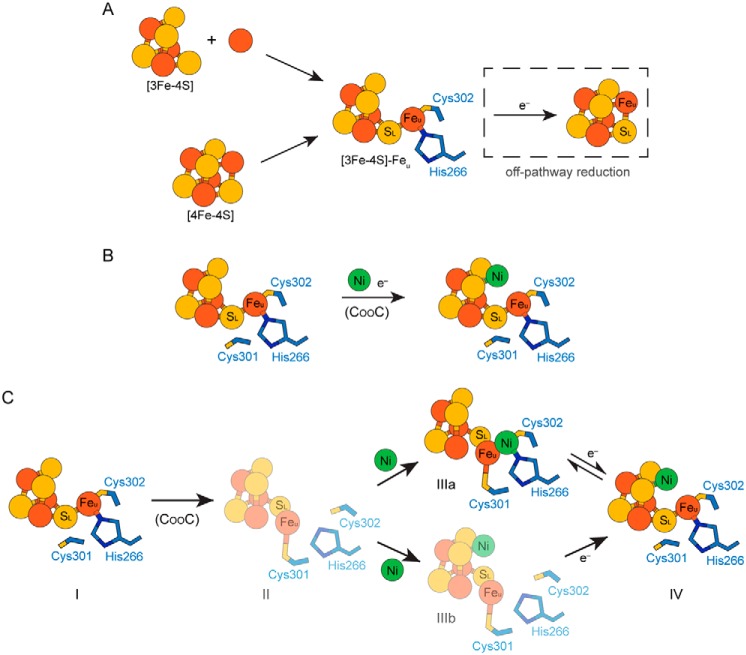
Figure 5.
Models of C-cluster assembly. A, formation of the C-cluster iron–sulfur scaffold. The iron–sulfur scaffold could be assembled through two different pathways. First, the components of the C-cluster could be inserted as a [3Fe–4S] cluster that combines with a mononuclear iron ion (upper pathway). Alternatively, the C-cluster site could become loaded with a [4Fe–4S] cluster followed by removal of an iron ion from the cubane to form Feu (lower pathway). In either case, an off-pathway reduction event could (re)convert the [3Fe–4S]–Feu scaffold into a [4Fe–4S] cluster. B and C, two independent models for nickel insertion into the C-cluster. B, nickel could be inserted directly into a reduced [3Fe–4S]–Feu pre–C-cluster. C, alternative model for nickel insertion involving multiple C-cluster conformations. Starting from the [3Fe–4S]–Feu pre–C-cluster (state I), CooC may be involved in inducing a conformational change in the C-cluster in which Feu becomes ligated by Cys-301 (state II). Nickel could then bind in either the canonical Feu-binding site (as observed in structures of the oxidized C-cluster (9); state IIIa) or in the cubane position (state IIIb). Cluster reduction could then result in formation of the fully mature C-cluster (state IV). Electrons (e−) indicate reduction events. In C, conformations of the C-cluster that have not been characterized crystallographically are shown in faded colors.
Once the iron–sulfur framework of the C-cluster has been assembled in CODH, nickel insertion can occur to form the fully mature and active cluster. Here we consider two possibilities for nickel insertion. In the first, C-cluster maturation in vivo involves the CooC-dependent insertion of nickel into a preformed [3Fe–4S]–Feu scaffold that resembles our DvCODH−CooC structures with Feu in its canonical site coordinated by His-266 and Cys-302 (Fig. 5B). In the second, nickel is inserted into a [3Fe–4S]–Feu scaffold in which Cys-301 coordinates Feu (Fig. 5C), a state that is reminiscent of the metal positions observed in our previous structure of fully oxidized DvCODH+CooC (Fig. 1C) (9).
For scenario I (Fig. 5B), the key role of CooC, in addition to nickel insertion, may be to control the redox state of the pre–C-cluster, allowing for nickel insertion without mismetallation of the nickel site. In analogy to metal capture by [3Fe–4S] clusters in other systems, nickel insertion into the scaffold as shown in Fig. 5B would likely require that the [3Fe–4S] framework be in a reduced state to increase the nucleophilicity of the open cubane site, allowing for binding of exogenous metal (29–33). In the case of the C-cluster, however, addition of exogenous nickel is likely complicated by the presence of Feu, which we have shown can migrate into the open coordination site of the reduced cubane (Figs. 2 and 3). In the context of C-cluster assembly, this observation indicates that the redox state of the pre–C-cluster must be tightly regulated to avoid mismetallation. One strategy for ensuring nickel incorporation in vivo could be to couple binding of CooC with cluster reduction, such that cluster reduction occurs just prior to nickel insertion. For example, binding of CooC could in some way facilitate interaction of CODH with a low-potential electron transfer protein, such as a reduced ferredoxin.
Although control of cluster reduction provides one route to prevent mismetallation, the previously reported structure of a fully oxidized C-cluster (Fig. 1C) (9) suggests another possible strategy for avoiding incorporation of Feu into the cubane in vivo (Fig. 5C). In particular, the position of Feu in the oxidized cluster, ligated by Cys-301, could represent an alternative binding site in which Feu is positioned prior to nickel insertion, such that Feu is not ligated in immediate proximity to the remainder of the [3Fe–4S] scaffold (Fig. 5C, state II). In this model, CooC could be involved in inducing a conformational change in the C-cluster prior to nickel insertion such that Feu becomes ligated by Cys-301 (Fig. 5C, state I to state II). Given the inability of the C-cluster to incorporate nickel in the absence of the D-cluster, this conformational change could additionally be redox-dependent. In any case, nickel could then be inserted into the His-266/Cys-302 binding site that is normally occupied by Feu, resulting in formation of the oxidized C-cluster conformation (Fig. 5C, state IIIa). Subsequent reduction, possibly facilitated by a change in reduction potential as a result of nickel binding, would then trigger formation of the active C-cluster via the three-atom migration of Feu, SL, and nickel that we have described previously and that occurs upon reduction of the oxidized cluster conformation (Fig. 5C, state IIIa to state IV) (9).
With these two proposals in mind (Fig. 5, B and C), we revisited the CODH literature. In addition to our previous characterization of DvCODH(C301S)+CooC (9), several additional mutagenesis studies on the CODHs from M. thermoacetica (MtCODH) (28), R. rubrum (RrCODH) (34–36), and C. hydrogenoformans (ChCODH-II) (37) are better explained by the mechanism shown in Fig. 5C than that in Fig. 5B. First, the proposal in Fig. 5B does not explain why substitution of the noncanonical C-cluster ligand Cys-301 in DvCODH and MtCODH results in inactive CODH variants that lack nickel (9, 28), whereas the mechanism in Fig. 5C provides a role for Cys-301 in nickel insertion. Second, substitution of the C-cluster ligating histidine residue with valine (in RrCODH) or alanine (in ChCODH-II) resulted in CODHs with iron contents that were indistinguishable from WT but that were impaired in their ability to incorporate nickel in vivo (34, 37). Additional mutagenesis experiments in which each of the canonical C-cluster ligating cysteine residues were mutated to alanine or serine revealed that His-266 and Cys-302 (D. vulgaris numbering) are in fact the only protein-based ligands to the canonical C-cluster that are necessary for nickel incorporation (34–37). Together, these data support the hypothesis that the His-266/Cys-302 site serves as the binding site for nickel during nickel incorporation (Fig. 5C). Second, the kinetics of nickel activation in nickel-deficient RrCODH (produced in the presence of RrCooC) suggest a two-step mechanism in which nickel first binds to the enzyme reversibly and then is seated into its active and stable site (26, 36).
Collectively, these findings support a model in which Cys-301 binds Feu while nickel is first inserted into the His-266/Cys-302 site, followed by rearrangement to form the active C-cluster (Fig. 5C, upper pathway), consistent with structures of the C-cluster that we have observed experimentally (9). We note that an alternative assembly pathway could also involve coordination of Feu by Cys-301 while nickel is inserted into its canonical site in the iron–sulfur cubane, although such a state has not been observed crystallographically (Fig. 5C, state IIIb) and does not explain the RrCODH mutational data mentioned above. One caveat of the model presented in Fig. 5C is that we have only observed Feu coordinated to Cys-301 in the oxidized state of the C-cluster (9), whereas the presence of reducing agent is known to be essential for nickel-dependent activation in vitro (17, 26). That being said, it has not yet been possible to visualize a nickel-deficient form of DvCODH+CooC in either an oxidized or reduced state to know whether Feu movement occurs and/or is redox-dependent in the absence of nickel.
Overall, our data begin to reveal the requirements for assembly of a fully intact and activatable C-cluster: 1) the C-cluster maturase CooC (this work and Refs. 15–17), 2) the D-cluster (this work), and 3) the noncanonical Feu ligand Cys-301 (9, 28). Together, these observations begin to expand our understanding of the complex and tightly regulated process of C-cluster biogenesis. In particular, given the varied metal-binding sites that we have observed within the C-cluster scaffold, the insertion of nickel is not a straightforward process and appears to be more complicated than originally thought.
Experimental procedures
Cloning and purification of DvCODH−CooC and DvCODH(ΔD)+CooC
Protein was expressed and purified as described previously (17). Briefly, the D. vulgaris gene encoding CODH (cooS) was cloned into a modified pBGF4 shuttle vector under the control of the promoter of the D. fructosovorans nickel–iron hydrogenase operon and included an N-terminal Strep-tag. For DvCODH(ΔD)+CooC, the expression vector also contained the gene for the CooC maturase (cooC), and mutations encoding C42A and C45A were introduced into the cooS gene by site-directed mutagenesis. To perform mutagenesis by PCR, the HindIII–SacI fragment of the modified pBGF4 plasmid, containing the 5′ end of cooS, was subcloned into pUC19 to serve as a DNA template. The primers GAACAGACGCCGGCCGCCAAATTCGCCGAATTGGGCACCACC (forward, mutations underlined) and GGTGGTGCCCAATTCGGCGAATTTGGCGGCCGGCGTCTGTTC (reverse, mutations underlined) were used to generate the C42A/C45A variant. The mutated HindIII–SacI fragment was then reintroduced into the HindIII–SacI–digested expression vector. The final mutated plasmid was verified by DNA sequencing. Protein was expressed in D. fructosovorans and purified under strictly anaerobic conditions in a Jacomex anaerobic chamber (100% N2 atmosphere) by affinity chromatography on Strep-Tactin Superflow resin. Protein concentrations were determined by amino acid analysis at the Centre for Integrated Structural Biology (Grenoble, France). Metal content was analyzed by inductively coupled plasma optical emission spectroscopy. The as-isolated samples contained nickel and iron as follows: DvCODH−CooC: 0 nickel atoms/monomer, 10 iron atoms/monomer; DvCODH(ΔD)+CooC: 0.02 nickel atoms/monomer, 8.5 iron atoms/monomer. CO oxidation activity was assayed at 37 °C by monitoring the reduction of methyl viologen at 604 nm (ϵ = 13.6 mm−1·cm−1), as described previously (17). Neither DvCODH−CooC nor DvCODH(ΔD)+CooC exhibited detectable CO oxidation activity. Reconstitution of either sample with NiCl2 under reducing conditions did not lead to an increase in activity.
Crystallization of DvCODH variants
DvCODH−CooC and DvCODH(ΔD)+CooC were crystallized in an N2 atmosphere at 21 °C by hanging-drop vapor diffusion in an MBraun anaerobic chamber. A 1 μl aliquot of protein (10 mg/ml in 100 mm Tris-HCl, pH 8) was combined with 1 μl of a precipitant solution (200–275 mm MgCl2, 14–20% PEG 3350) on a glass coverslip and sealed over a reservoir containing 500–700 μl of precipitant solution. Diffraction quality crystals grew in 4–7 days. The crystals were soaked in a cryo-protectant solution containing 200 mm MgCl2, 20–30% PEG 3350, and 10–16% glycerol and cryo-cooled in liquid nitrogen. For structures of reduced DvCODH−CooC, the crystals were soaked in 250 mm MgCl2, 18% (w/v) PEG 3350, 5 mm sodium dithionite for 30 min prior to cryo-protecting and cryo-cooling in liquid nitrogen.
Data collection, model building, and refinement
Diffraction data were collected at the Advanced Photon Source (Argonne, IL) on Beamline 24-ID-C using a Pilatus 6M pixel detector and at a temperature of 100 K. Native and iron peak data were collected on the same crystal for each sample, where applicable. The DvCODH(ΔD)+CooC structure was determined and refined using data collected at the iron peak wavelength. The data for DvCODH−CooC (as-isolated and reduced) were integrated in XDS and scaled in XSCALE (38). The data for DvCODH(ΔD)+CooC were integrated and scaled in HKL2000 (39). All data collection statistics are summarized in Table 2.
Structures were determined by molecular replacement in the program Phaser (40) using our previously published structure of DvCODH (PDB code 6B6V) as a search model. Following molecular replacement, 10 cycles of simulated annealing refinement were performed in Phenix (41) to eliminate existing model bias. Refinement of atomic coordinates and atomic displacement parameters (B-factors) was performed in Phenix, and models were completed by iterative rounds of model building in Coot (42) and refinement in Phenix. Metal cluster geometries were restrained during refinement using custom parameter files. In advanced stages of refinement, water molecules were added automatically in Phenix (41) and modified in Coot (42) with placement of additional water molecules until their number was stable. For the DvCODH−CooC structures, final stages of refinement included translation-libration-screw (TLS) parameterization with one TLS group per monomer (43). Side chains without visible electron density were truncated to the last atom with electron density, and amino acids without visible electron density were not included in the model. Final models contain the following residues (of 629 total): as-isolated DvCODH−CooC: 4–628 (chain A), 4–629 (chain B), 4–629 (chain C), 3–628 (chain D); reduced DvCODH−CooC: 4–629 (chain A), 4–629 (chain B), 4–629 (chain C), 3–629 (chain D); and DvCODH(ΔD)+CooC: 4–40, 45–628 (chain A).
Final refinement yielded models with low free R-factors, excellent stereochemistry, and small root-mean-square deviations from ideal values for bond lengths and angles. The models were validated using simulated annealing composite omit maps calculated in Phenix (41). Model geometry was analyzed using MolProbity (44). Analysis of Ramachandran statistics indicated that each structure contained the following percentages of residues in the favored, allowed, and disallowed regions, respectively: as-isolated DvCODH−CooC: 96.7%, 3.0%, 0.3%; reduced DvCODH−CooC: 96.9%, 2.8%, 0.3%; and DvCODH(ΔD)+CooC: 95.8%, 4.0%, 0.2%. Refinement and geometry statistics are summarized in Table 2. The figures were generated in PyMOL (45). Crystallography packages were compiled by SBGrid (46).
Author contributions
E. C. W., S. E. C., M. M., V. F., S. D., and C. L. D. formal analysis; E. C. W., S. E. C., M. M., V. F., S. D., and C. L. D. validation; E. C. W., S. E. C., M. M., V. F., S. D., and C. L. D. investigation; E. C. W. visualization; E. C. W., C. L., V. F., S. D., and C. L. D. methodology; E. C. W., S. D., and C. L. D. writing-original draft; E. C. W., C. L., V. F., S. D., and C. L. D. writing-review and editing; C. L., V. F., S. D., and C. L. D. conceptualization; C. L., V. F., S. D., and C. L. D. resources; C. L., V. F., S. D., and C. L. D. supervision; C. L., V. F., S. D., and C. L. D. funding acquisition; C. L., V. F., S. D., and C. L. D. project administration; E. C. W. performed crystallographic experiments, analyzed the crystallographic data, and wrote the manuscript.; S. E. C. performed crystallographic experiments, analyzed the crystallographic data.; M. M. purified protein and performed activity assays.; C. L. and V. F. assisted with editing the manuscript.; S. D. purified protein, performed activity assays, and assisted with editing the manuscript.; C. L. D. helped analyze the crystallographic data and wrote the manuscript.
Acknowledgments
We thank Marco Jost and David Born for helpful conversations and critical reading of the manuscript. This work is based on research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which are funded by the NIGMS, National Institutes of Health Grant P41 GM103403. The Pilatus 6M detector on Beamline 24-ID-C is funded by National Institutes of Health Office of Research Infrastructure Programs High End Instrumentation Grant S10 RR029205. This research used resources of the Advanced Photon Source, a U.S. Department of Energy Office of Science User Facility operated for the Department of Energy Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357.
This work was supported by National Institutes of Health Grants T32 GM008334 (to E. C. W. and S. E. C.) and R01 GM069857 and R35 GM126982 (to C. L. D.) and the work in Marseille was funded by the French Agence Nationale de la Recherche (ANR-15-CE05-0020 and ANR-17-CE11-0027). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The atomic coordinates and structure factors (codes 6ONC, 6OND, and 6ONS) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- CODH
- carbon monoxide dehydrogenase
- SL
- linking sulfide
- Feu
- unique iron
- PDB
- Protein Data Bank.
References
- 1. Uffen R. L. (1976) Anaerobic growth of a Rhodopseudomonas species in the dark with carbon monoxide as sole carbon and energy substrate. Proc. Natl. Acad. Sci. U.S.A. 73, 3298–3302 10.1073/pnas.73.9.3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Svetlichny V. A., Sokolova T. G., Gerhardt M., Ringpfeil M., Kostrikina N. A., and Zavarzin G. A. (1991) Carboxydothermus hydrogenoformans gen. nov., sp. nov., a CO-utilizing thermophilic anaerobic bacterium from hydrothermal environments of Kunashir Island. Syst. Appl. Microbiol. 14, 254–260 10.1016/S0723-2020(11)80377-2 [DOI] [Google Scholar]
- 3. Bartholomew G. W., and Alexander M. (1979) Microbial metabolism of carbon monoxide in culture and in soil. Appl. Environ. Microbiol. 37, 932–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Can M., Armstrong F. A., and Ragsdale S. W. (2014) Structure, function, and mechanism of the nickel metalloenzymes, CO dehydrogenase, and acetyl-CoA synthase. Chem. Rev. 114, 4149–4174 10.1021/cr400461p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Drennan C. L., Heo J., Sintchak M. D., Schreiter E., and Ludden P. W. (2001) Life on carbon monoxide: X-ray structure of Rhodospirillum rubrum Ni–Fe–S carbon monoxide dehydrogenase. Proc. Natl. Acad. Sci. U.S.A. 98, 11973–11978 10.1073/pnas.211429998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dobbek H., Svetlitchnyi V., Gremer L., Huber R., and Meyer O. (2001) Crystal structure of a carbon monoxide dehydrogenase reveals a [Ni–4Fe–5S] cluster. Science 293, 1281–1285 10.1126/science.1061500 [DOI] [PubMed] [Google Scholar]
- 7. Kumar M., Lu W. P., Liu L., and Ragsdale S. W. (1993) Kinetic evidence that carbon monoxide dehydrogenase catalyzes the oxidation of carbon monoxide and the synthesis of acetyl-CoA at separate metal centers. J. Am. Chem. Soc. 115, 11646–11647 10.1021/ja00077a093 [DOI] [Google Scholar]
- 8. Anderson M. E., and Lindahl P. A. (1994) Organization of clusters and internal electron pathways in CO dehydrogenase from Clostridium thermoaceticum: relevance to the mechanism of catalysis and cyanide inhibition. Biochemistry 33, 8702–8711 10.1021/bi00195a011 [DOI] [PubMed] [Google Scholar]
- 9. Wittenborn E. C., Merrouch M., Ueda C., Fradale L., Léger C., Fourmond V., Pandelia M. E., Dementin S., and Drennan C. L. (2018) Redox-dependent rearrangements of the NiFeS cluster of carbon monoxide dehydrogenase. Elife 7, e39451 10.7554/eLife.39451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hu Z. G., Spangler N. J., Anderson M. E., Xia J., Ludden P. W., Lindahl P. A., and Münck E. (1996) Nature of the C-cluster in Ni-containing carbon monoxide dehydrogenases. J. Am. Chem. Soc. 118, 830–845 10.1021/ja9528386 [DOI] [Google Scholar]
- 11. DeRose V. J., Telser J., Anderson M. E., Lindahl P. A., and Hoffman B. M. (1998) A multinuclear ENDOR study of the C-cluster in CO dehydrogenase from Clostridium thermoaceticum: evidence for HxO and histidine coordination to the [Fe4S4] center. J. Am. Chem. Soc. 120, 8767–8776 10.1021/ja9731480 [DOI] [Google Scholar]
- 12. Jeoung J. H., and Dobbek H. (2007) Carbon dioxide activation at the Ni,Fe-cluster of anaerobic carbon monoxide dehydrogenase. Science 318, 1461–1464 10.1126/science.1148481 [DOI] [PubMed] [Google Scholar]
- 13. Kung Y., Doukov T. I., Seravalli J., Ragsdale S. W., and Drennan C. L. (2009) Crystallographic snapshots of cyanide- and water-bound C-clusters from bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase. Biochemistry 48, 7432–7440 10.1021/bi900574h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shepard E. M., Boyd E. S., Broderick J. B., and Peters J. W. (2011) Biosynthesis of complex iron–sulfur enzymes. Curr. Opin. Chem. Biol. 15, 319–327 10.1016/j.cbpa.2011.02.012 [DOI] [PubMed] [Google Scholar]
- 15. Kerby R. L., Ludden P. W., and Roberts G. P. (1997) In vivo nickel insertion into the carbon monoxide dehydrogenase of Rhodospirillum rubrum: molecular and physiological characterization of cooCTJ. J. Bacteriol. 179, 2259–2266 10.1128/jb.179.7.2259-2266.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jeon W. B., Cheng J., and Ludden P. W. (2001) Purification and characterization of membrane-associated CooC protein and its functional role in the insertion of nickel into carbon monoxide dehydrogenase from Rhodospirillum rubrum. J. Biol. Chem. 276, 38602–38609 10.1074/jbc.M104945200 [DOI] [PubMed] [Google Scholar]
- 17. Hadj-Saïd J., Pandelia M. E., Léger C., Fourmond V., and Dementin S. (2015) The carbon monoxide dehydrogenase from Desulfovibrio vulgaris. Biochim. Biophys. Acta 1847, 1574–1583 10.1016/j.bbabio.2015.08.002 [DOI] [PubMed] [Google Scholar]
- 18. Watt R. K., and Ludden P. W. (1998) The identification, purification, and characterization of CooJ: a nickel-binding protein that is co-regulated with the Ni-containing CO dehydrogenase from Rhodospirillum rubrum. J. Biol. Chem. 273, 10019–10025 10.1074/jbc.273.16.10019 [DOI] [PubMed] [Google Scholar]
- 19. Jeoung J. H., Goetzl S., Hennig S. E., Fesseler J., Wörmann C., Dendra J., and Dobbek H. (2014) The extended reductive acetyl-CoA pathway: ATPases in metal cluster maturation and reductive activation. Biol. Chem. 395, 545–558 [DOI] [PubMed] [Google Scholar]
- 20. Timm J., Brochier-Armanet C., Perard J., Zambelli B., Ollagnier-de-Choudens S., Ciurli S., and Cavazza C. (2017) The CO dehydrogenase accessory protein CooT is a novel nickel-binding protein. Metallomics 9, 575–583 10.1039/C7MT00063D [DOI] [PubMed] [Google Scholar]
- 21. Alfano M., Pérard J., Miras R., Catty P., and Cavazza C. (2018) Biophysical and structural characterization of the putative nickel chaperone CooT from Carboxydothermus hydrogenoformans. J. Biol. Inorg. Chem. 23, 809–817 10.1007/s00775-018-1576-2 [DOI] [PubMed] [Google Scholar]
- 22. Alfano M., Perard J., Carpentier P., Basset C., Zambelli B., Timm J., Crouzy S., Ciurli S., and Cavazza C. (2019) The carbon monoxide dehydrogenase accessory protein CooJ is a histidine-rich multidomain dimer containing an unexpected Ni(II)-binding site. J. Biol. Chem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jeoung J. H., Giese T., Grünwald M., and Dobbek H. (2009) CooC1 from Carboxydothermus hydrogenoformans is a nickel-binding ATPase. Biochemistry 48, 11505–11513 10.1021/bi901443z [DOI] [PubMed] [Google Scholar]
- 24. Jeoung J. H., Giese T., Grünwald M., and Dobbek H. (2010) Crystal structure of the ATP-dependent maturation factor of Ni,Fe-containing carbon monoxide dehydrogenases. J. Mol. Biol. 396, 1165–1179 10.1016/j.jmb.2009.12.062 [DOI] [PubMed] [Google Scholar]
- 25. Merrouch M., Benvenuti M., Lorenzi M., Léger C., Fourmond V., and Dementin S. (2018) Maturation of the [Ni-4Fe–4S] active site of carbon monoxide dehydrogenases. J. Biol. Inorg. Chem. 23, 613–620 10.1007/s00775-018-1541-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ensign S. A., Campbell M. J., and Ludden P. W. (1990) Activation of the nickel-deficient carbon monoxide dehydrogenase from Rhodospirillum rubrum: kinetic characterization and reductant requirement. Biochemistry 29, 2162–2168 10.1021/bi00460a029 [DOI] [PubMed] [Google Scholar]
- 27. Svetlitchnyi V., Peschel C., Acker G., and Meyer O. (2001) Two membrane-associated NiFeS-carbon monoxide dehydrogenases from the anaerobic carbon-monoxide-utilizing eubacterium Carboxydothermus hydrogenoformans. J. Bacteriol. 183, 5134–5144 10.1128/JB.183.17.5134-5144.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim E. J., Feng J., Bramlett M. R., and Lindahl P. A. (2004) Evidence for a proton transfer network and a required persulfide-bond-forming cysteine residue in Ni-containing carbon monoxide dehydrogenases. Biochemistry 43, 5728–5734 10.1021/bi036062u [DOI] [PubMed] [Google Scholar]
- 29. Zhou J., Raebiger J. W., Crawford C. A., and Holm R. H. (1997) Metal ion incorporation reactions of the cluster [Fe3S4(LS3)]3−, containing the cuboidal [Fe3S4]0 core. J. Am. Chem. Soc. 119, 6242–6250 10.1021/ja9704186 [DOI] [Google Scholar]
- 30. Moura J. J., Moura I., Kent T. A., Lipscomb J. D., Huynh B. H., LeGall J., Xavier A. V., and Münck E. (1982) Interconversions of [3Fe–3S] and [4Fe–4S] clusters: Mössbauer and electron paramagnetic resonance studies of Desulfovibrio gigas ferredoxin-II. J. Biol. Chem. 257, 6259–6267 [PubMed] [Google Scholar]
- 31. Kent T. A., Dreyer J. L., Kennedy M. C., Huynh B. H., Emptage M. H., Beinert H., and Münck E. (1982) Mossbauer studies of beef heart aconitase: evidence for facile interconversions of iron–sulfur clusters. Proc. Natl. Acad. Sci. U.S.A. 79, 1096–1100 10.1073/pnas.79.4.1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Robbins A. H., and Stout C. D. (1989) Structure of activated aconitase: formation of the [4Fe–4S] cluster in the crystal. Proc. Natl. Acad. Sci. U.S.A. 86, 3639–3643 10.1073/pnas.86.10.3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Conover R. C., Park J. B., Adams M. W. W., and Johnson M. K. (1990) Formation and properties of an iron-nickel sulfide (NiFe3S4) cluster in Pyrococcus furiosus ferredoxin. J. Am. Chem. Soc. 112, 4562–4564 10.1021/ja00167a074 [DOI] [Google Scholar]
- 34. Spangler N. J., Meyers M. R., Gierke K. L., Kerby R. L., Roberts G. P., and Ludden P. W. (1998) Substitution of valine for histidine 265 in carbon monoxide dehydrogenase from Rhodospirillum rubrum affects activity and spectroscopic states. J. Biol. Chem. 273, 4059–4064 10.1074/jbc.273.7.4059 [DOI] [PubMed] [Google Scholar]
- 35. Staples C. R., Heo J., Spangler N. J., Kerby R. L., Roberts G. P., and Ludden P. W. (1999) Rhodospirillum rubrum CO-dehydrogenase. Part 1. Spectroscopic studies of CODH variant C531A indicate the presence of a binuclear [FeNi] cluster. J. Am. Chem. Soc. 121, 11034–11044 10.1021/ja990396i [DOI] [Google Scholar]
- 36. Jeon W. B., Singer S. W., Ludden P. W., and Rubio L. M. (2005) New insights into the mechanism of nickel insertion into carbon monoxide dehydrogenase: analysis of Rhodospirillum rubrum carbon monoxide dehydrogenase variants with substituted ligands to the [Fe3S4] portion of the active-site C-cluster. J. Biol. Inorg. Chem. 10, 903–912 10.1007/s00775-005-0043-z [DOI] [PubMed] [Google Scholar]
- 37. Inoue T., Takao K., Yoshida T., Wada K., Daifuku T., Yoneda Y., Fukuyama K., and Sako Y. (2013) Cysteine 295 indirectly affects Ni coordination of carbon monoxide dehydrogenase-II C-cluster. Biochem. Biophys. Res. Commun. 441, 13–17 10.1016/j.bbrc.2013.09.143 [DOI] [PubMed] [Google Scholar]
- 38. Kabsch W. (2010) XDS. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- 40. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Painter J., and Merritt E. A. (2006) Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 62, 439–450 10.1107/S0907444906005270 [DOI] [PubMed] [Google Scholar]
- 44. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. DeLano W. L. (2015) The PyMOL Molecular Graphics System, version 1.8, Schroedinger, LLC, New York [Google Scholar]
- 46. Morin A., Eisenbraun B., Key J., Sanschagrin P. C., Timony M. A., Ottaviano M., and Sliz P. (2013) Collaboration gets the most out of software. Elife 2, e01456 10.7554/eLife.01456 [DOI] [PMC free article] [PubMed] [Google Scholar]