Abstract
An early hallmark of type 2 diabetes is a failure of proinsulin-to-insulin processing in pancreatic β-cells, resulting in hyperproinsulinemia. Proinsulin processing is quite sensitive to nutrient flux, and β-cell–specific deletion of the nutrient-sensing protein modifier OGlcNAc transferase (βOGTKO) causes β-cell failure and diabetes, including early development of hyperproinsulinemia. The mechanisms underlying this latter defect are unknown. Here, using several approaches, including site-directed mutagenesis, Click O-GlcNAc labeling, immunoblotting, and immunofluorescence and EM imaging, we provide the first evidence for a relationship between the O-GlcNAcylation of eukaryotic translation initiation factor 4γ1 (eIF4G1) and carboxypeptidase E (CPE)-dependent proinsulin processing in βOGTKO mice. We first established that βOGTKO hyperproinsulinemia is independent of age, sex, glucose levels, and endoplasmic reticulum–CCAAT enhancer-binding protein homologous protein (CHOP)–mediated stress status. Of note, OGT loss was associated with a reduction in β-cell–resident CPE, and genetic reconstitution of CPE in βOGTKO islets rescued the dysfunctional proinsulin-to-insulin ratio. We show that although CPE is not directly OGlcNAc modified in islets, overexpression of the suspected OGT target eIF4G1, previously shown to regulate CPE translation in β-cells, increases islet CPE levels, and fully reverses βOGTKO islet-induced hyperproinsulinemia. Furthermore, our results reveal that OGT O-GlcNAc-modifies eIF4G1 at Ser-61 and that this modification is critical for eIF4G1 protein stability. Together, these results indicate a direct link between nutrient-sensitive OGT and insulin processing, underscoring the importance of post-translational O-GlcNAc modification in general cell physiology.
Keywords: O-GlcNAcylation, O-linked N-acetylglucosamine (O-GlcNAc) transferase (OGT), insulin synthesis, post-translational modification (PTM), diabetes, Chop, eIF4G1, ER-stress, hyperproinsulinemia, islet
Introduction
Hyperproinsulinemia, defined here as an increase in both the absolute and relative concentration of immature proinsulin, is one of the early hallmarks of type 2 diabetes (T2D)2 in humans (1, 2). Hyperproinsulinemia is a marker of pancreatic β-cell dysfunction; elevated serum proinsulin levels are associated with glucose intolerance (3, 4) and future development of T2D in humans (5). The molecular mechanisms involved in the elevated proinsulin-to-insulin ratio are not well-understood and the relationship between nutrient-sensor protein O-GlcNAc transferase (OGT) and insulin processing is unexplored.
Proinsulin processing efficiency is sensitive to the availability and flux of glucose and other nutrients such as lipids and amino acids (6). Loss of the islet-enriched, nutrient-driven enzyme OGT (βOGTKO) in β-cells resulted in hyperproinsulinemia (7), prior to the onset of hyperglycemia and diabetes, suggesting a plausible mechanistic link between dynamic nutrient flux and protein-mediated insulin processing. The nutrient sensor OGT is the sole enzyme that post-translationally modifies its target proteins by adding a single GlcNAc moiety, derived from multiple nutrient sources including glucose and lipids through the hexosamine biosynthesis pathway, onto serine/threonine residues (8–10). The effects of O-GlcNAc modification (O-GlcNAcylation) are target and context-specific but may include multiple protein functions, stability, and localization (9, 11). In the hyperglycemic condition of T2D, excessive O-GlcNAcylation has been associated with β-cell failure and T2D complications such as nephropathy and cardiovascular disease (12, 13). O-GlcNAcase (OGA) exclusively removes O-GlcNAc and is a T2D susceptibility gene (14). In β-cells, crucial transcription factors such as Pdx1 and NeuroD1 are O-GlcNAc–modified to affect their protein function and localization in response to glucose, thus contributing to the maintenance of β-cell mass and function (15, 16).
Subsequent to the transcription and translation of pre-proinsulin, proinsulin processing begins in the endoplasmic reticulum (ER) with post-translational protein folding and ends in the insulin granules with multiple proteolytic cleavage events mediated by prohormone convertases and exopeptidases, such as carboxypeptidase E (CPE) (17). A mutation in CPE resulting in reduced enzyme activity is the driving mechanism behind hyperproinsulinemic fat/fat mice, a model of obesity-precipitated T2D (18, 19), suggesting that loss of CPE is sufficient to trigger physiologically relevant defects in proinsulin processing. CPE protein has shown to be specifically regulated in the β-cell by eukaryotic translation initiation factor 4γ1 (eIF4G1), a scaffolding protein of the translation initiation complex, implicating this protein in the proinsulin to insulin processing pathway (20). Several diabetes mouse models, including β-cell–specific insulin receptor (βIRKO) and mTOR/raptor knockout, show a loss in both CPE and eIF4G1 protein, accompanied by hyperproinsulinemia, highlighting the importance of these proteins in the biosynthesis of properly folded and mature insulin (20, 21).
In the present study, we examined the molecular mechanisms underlying OGT loss-driven hyperproinsulinemia. We discovered that the down-regulation of CPE protein in islets of βOGTKO mice is independent of sex, blood glucose levels, and CHOP-mediated ER stress status. Mechanistically, the loss of CPE protein was not due to direct O-GlcNAcylation or transcriptional regulation on CPE, but, in part, due to translational suppression downstream of a loss of eIF4G1. Subsequently, we show that eIF4G1 is O-GlcNAc–modified at serine 61 in β-cells where its O-GlcNAcylation status is required for protein stability. Overexpression of eIF4G1 in dispersed islet cells improved CPE levels and fully rescued βOGTKO hyperproinsulinemia at a cellular level. Altogether, these data suggest a novel role for β-cell OGT in regulating the translational expression of CPE and eIF4G1, critical proteins in the insulin processing pathway that mediate an early point of failure in the development of diabetes.
Results
Hyperproinsulinemia in βOGTKO mice is associated with reduced CPE protein in β-cells
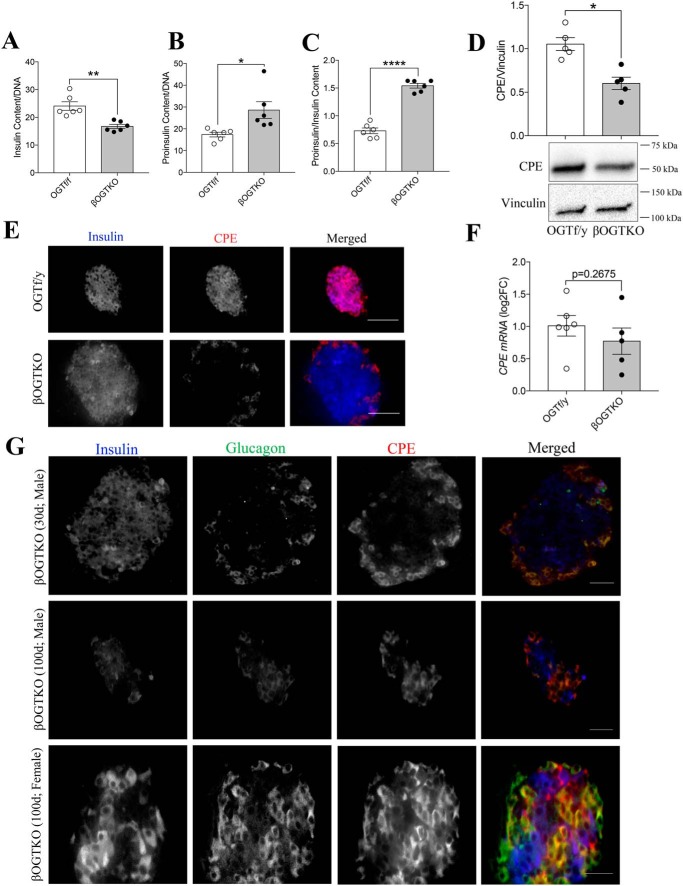
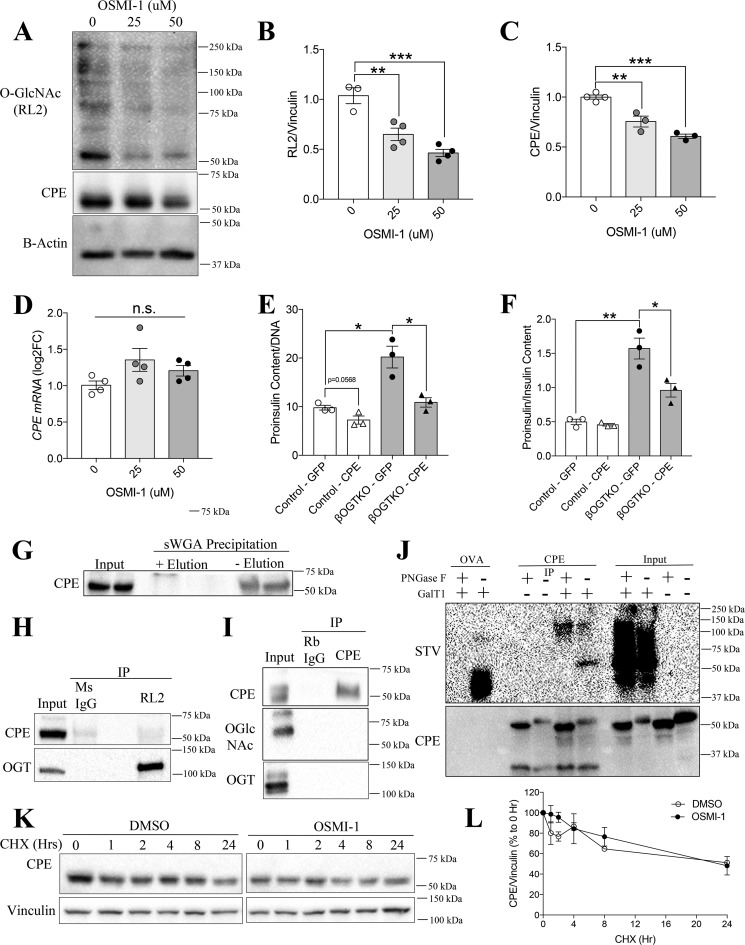
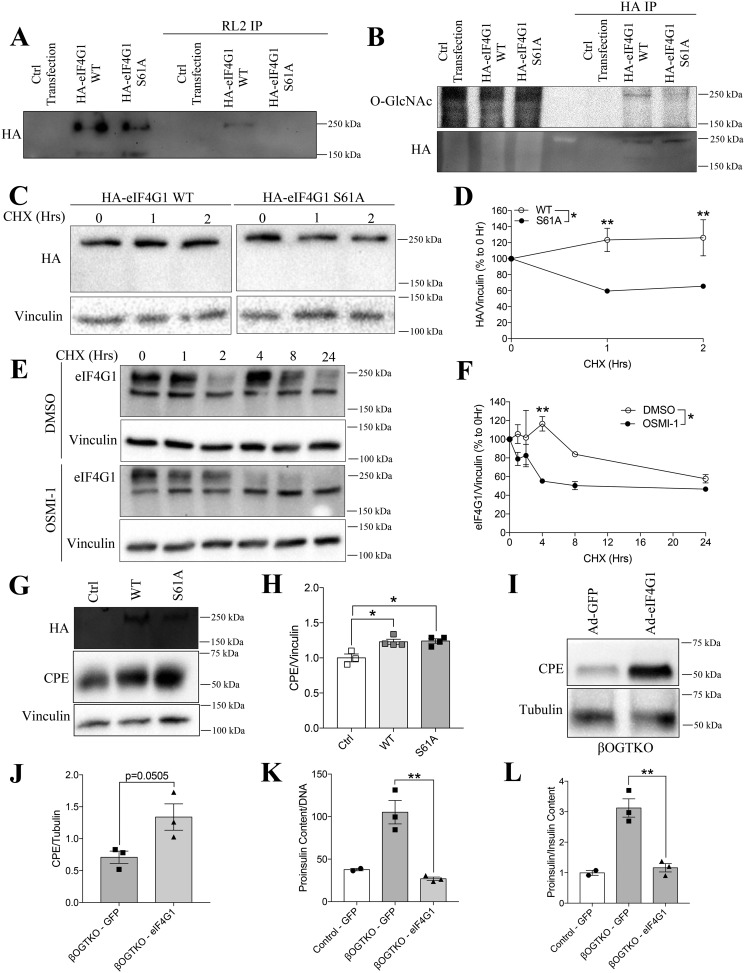
Islets from normoglycemic βOGTKO female mice display reduced insulin content, increased proinsulin content, and proinsulin/insulin ratio (Fig. 1, A–C). To study the underlying mechanisms, we measured the protein level of CPE, a critical proinsulin cleavage enzyme, in βOGTKO islets. We observed significant reduction in total CPE protein in βOGTKO islets compared with littermate control-derived islets (Fig. 1D). Because the islet is composed of multiple cells (22), we assessed CPE protein presence via immunofluorescence staining. By immunofluorescence staining we showed that CPE loss is preferentially in β-cells (Fig. 1E). By Western blotting, we observed about ∼50% of CPE protein reduction in whole islets, this is explained in part by the normal level of CPE in glucagon-expressing α-cells (Fig. 1, D and E). In another model with partial OGT loss (βOGTHET), islets also demonstrated less CPE staining intensity (Fig. S1A) as did βOGTKO islets from mice of varying ages and sex (Fig. 1G). Furthermore, an OGT inhibitor, OSMI-1, decreased levels of both global O-GlcNAcylation, a measure of OGT activity, and CPE protein level in INS-1 cells (Fig. 2, A–C). Islets with complete or partial OGT loss showed no significant changes in CPE mRNA (Fig. 1F and Fig. S1B) nor did INS-1 cells treated with OGT inhibitor in vitro (Fig. 2D), suggesting the defect was occurring after the gene transcription. To test the direct role of CPE in the OGT-loss phenotype, we genetically reconstituted CPE levels by transiently infecting dispersed WT and βOGTKO primary islet cells with either adenoviral GFP or CPE, achieving ∼70–80% infection rate (Fig. S2A). Although CPE overexpression had a minimal effect on WT proinsulin levels (Fig. 2, E and F), it fully rescued βOGTKO proinsulin cell content (Fig. 2E) and dramatically lowered the proinsulin/insulin content ratio (Fig. 2F), suggesting that down-regulation of CPE is a major contributor to hyperproinsulinemic dysfunction in βOGTKO mice.
Figure 1.
Carboxypeptidase E is reduced in OGT-deficient β-cells. Total islet content of insulin (A), proinsulin (B), and proinsulin/insulin ratio (C) taken from 10 islet/animal, normalized to DNA, were isolated from 4–5–week–old female OGT f/f (OGT flox/flox) and βOGTKO (Rip-cre: OGT flox/flox) mice (n = 5). Islet CPE levels in 4–5–week–old male OGT f/y and βOGTKO (n = 5) were as assessed by Western blotting (relative to vinculin, normalized to OGT f/y avg.) D, immunofluorescence staining for insulin (blue) and CPE (red) of fixed pancreatic sections at ×20 magnification (E) and qPCR gene expression (relative to β-Actin) of CPE transcript in isolated islets (n = 5) are shown as log2-fold change relative to control average (F). Scale bar = 50 μm. Images are representative of five different mice. Immunofluorescence staining of CPE (red), insulin (blue), and glucagon (green) of pancreata of normoglycemic 30-day-old male βOGTKO, hyperglycemic 100-day-old male βOGTKO, 100-day-old female βOGTKO (G) are shown. Scale bar = 30 μm. Images are representative of three different mice. Statistical analyses were conducted using unpaired, two-tailed Student's t test with significance; *, p < 0.05.
Figure 2.
Carboxypeptidase E is not regulated at the post-translational level through O-GlcNAc modification in β-cells. Protein O-GlcNAcylation (RL2), CPE protein (relative to vinculin, normalized to control or 0 μm OSMI) (A–C), and CPE gene expression (relative to β-Actin, log2-fold change to 0 μm) (D) in INS-1 cells were treated for 8 h with the OGT inhibitor OSMI-1 (0, 25, 50 μm, n = 4 per group). Islet proinsulin content (E) and proinsulin-to-insulin ratio (F) from WT and βOGTKO dispersed islet cells were infected with Ad-GFP or Ad-CPE (n = 3). MIN6 CPE protein with sWGA precipitation (±glycoprotein elution buffer (N-acetylglucosamine)) (G), and mouse RL2 (pan-OGlcNAc) (H) or CPE (I) immunoprecipitation followed by immunoblot against CPE, RL2, or OGT are shown. The blots are representative of three independent experiments. INS1 lysates, followed by CPE immunoprecipitation or purified ovalbumin with enzymatically labeled GlcNAc in the presence or absence of PNGase F, were immunoblotted against streptavidin (STV) against Biotin or CPE (J). Representative blot (K) and densitometry quantification relative to vinculin (as % of 0 h baseline) (L) of INS-1 CPE protein after 0–24 h of treatment with protein synthesis blocker CHX (50 μg/ml) was following a 2-h pre-treatment with OSMI-1 (30 μm) or DMSO control (n = 3). Statistical analyses were conducted using unpaired, two-tailed Student's t test, one-way or two-way ANOVA, with significance; *, p < 0.05.
CPE is not O-GlcNAc-modified nor a binding partner of OGT in β-cells
The localization of CPE remains controversial. Earlier studies report that CPE is mainly localized in secretory granules, near but not inside of Golgi apparatus, and pro-CPE in the ER (19, 23, 24). Another proposed model of CPE localization places the protein inside the insulin vesicles/granules with the C-terminal tail, a putative O-GlcNAcylation region based on YinOYang prediction software (25), projecting into the cytoplasm where OGT could theoretically modify it (26) to influence proinsulin processing. To assess for the presence of an O-GlcNAc moiety on CPE, we precipitated all O-GlcNAcylated proteins from MIN6 β-cell lysates using agarose beads conjugated to either succinylated wheat germ agglutinin (sWGA) (Fig. 2G) or the O-GlcNAc–specific RL2 antibody (Fig. 2H). Through sWGA precipitation, we found CPE to be post-translationally modified with glycosylation containing N-acetylglucosamine (Fig. 2G). Due to nonspecificity of the sWGA precipitation technique to O-GlcNAcylation by OGT, we performed immunoprecipitation using O-GlcNAc–specific antibody and found CPE to lack O-GlcNAcylation (Fig. 2H). Here, we used OGT, as positive control for this experiment, as OGT has previously shown to O-GlcNAcylate itself (27). Next, we validated the absence of CPE O-GlcNAcylation using the inverse technique, a CPE antibody IP followed by immunoblot against O-GlcNAc (Fig. 2I). Using co-IP, we were also unable to detect any protein–protein interaction between OGT and CPE (Fig. 2I). Additionally, to verify the lack of O-GlcNAcylation on CPE, we utilized the Click-IT O-GlcNAc Enzymatic system to label GlcNAc moiety using the Gal-T1 (Y289L), in the presence or absence of PNGase F in INS1 cell lysates. Purified ovalbumin was used as a positive control for PNGase F treatment. We observed that CPE is indeed GlcNAc modified, but with PNGase F treatment, we failed to detect any band at the expected size of 50 kDa, verifying that CPE is not O-GlcNAcylated by OGT (Fig. 2J). Furthermore, we found that in vitro OGT inhibition of INS1 β-cells with OSMI-1 (30 μm) had no effect on CPE degradation (Fig. 2, K and L). We verified the translational blockade by cycloheximide (CHX) through immunoblotting against c-myc, a protein with a short half-life (Fig. S1F). Together these data suggest that CPE is not post-translationally regulated by OGT.
CPE loss is independent of ER stress-mediated Chop in OGT-deficient β-cells
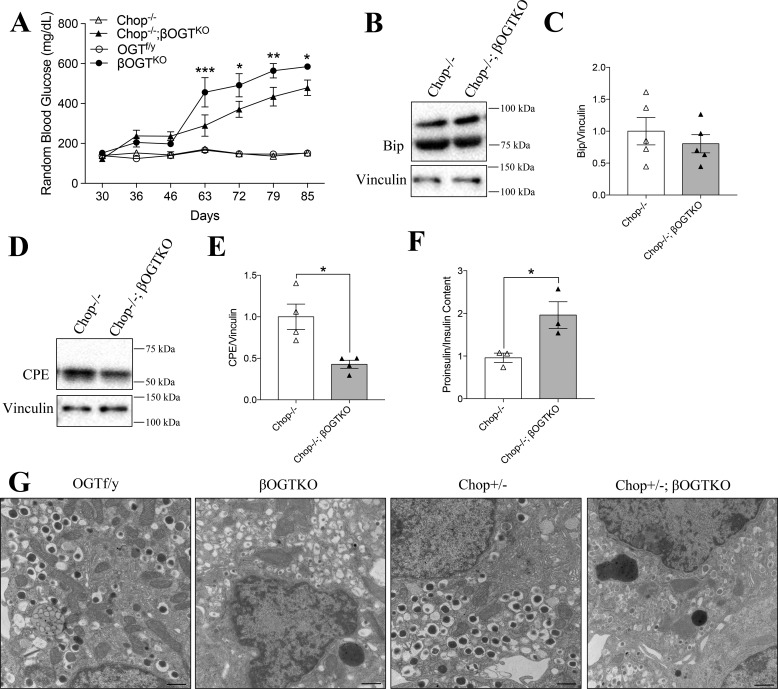
Because CPE was not regulated via transcription or post-translational modification by OGT, we considered the possibility that ER stress, a well-known translational contributor to protein biosynthesis dysfunction (28), drives CPE degradation. We previously observed elevated levels of the ER stress-mediating Chop protein in βOGTKO islets and its partial deletion improved the diabetic phenotype (i.e. delayed the onset of hyperglycemia and improved β-cell mass) in these mice (7). To examine whether a reduction in ER stress could also rescue CPE protein expression, we generated βOGTKO mice in a full Chop null background. Full deletion of Chop significantly but incompletely improved hyperglycemia in the Chop−/−;βOGTKO was compared with littermate βOGTKO mice (Fig. 3A). We previously showed increased Bip, an ER stress marker and regulator of unfolded protein response (UPR) activation, in βOGTKO islets (7). In islets from 1-month-old, normoglycemic mice, Chop−/−;βOGTKO islets, we showed a comparable level of Bip to the control Chop−/− islets, suggestive of normalization of the ER stress level in Chop−/−;βOGTKO model (Fig. 3, B and C). Although the Chop−/−;βOGTKO mice had reduced glucose levels compared with that of βOGTKO, the islet CPE protein level remained significantly reduced and proinsulin/insulin ratio high (Fig. 3, D–F), revealing the persistence of an insulin processing defect despite the reduction in ER stress mediated by Chop. We previously showed that Chop−/+;βOGTKO was sufficient to reduced glucose levels and rescue β-cell mass in βOGTKO mice. Interestingly, EM images revealed fewer insulin granules in Chop−/+;βOGTKO β-cells were compared with littermate controls, regardless of Chop status, and granules with a less dense core (Fig. 3G), a phenomena that has been linked to reduced mature insulin content (29). This is consistent with our previous finding that total insulin content is reduced in βOGTKO (7) despite restoration of blood glucose levels. These data led us to conclude that Chop-mediated ER stress is not a major contributor to CPE loss or hyperproinsulinemia in β-cells deficient of OGT.
Figure 3.
βOGTKO CPE loss persists in model without ER stress-mediated Chop gene. Random blood glucose from postnatal days 30–85 in βOGTKO, Chop−/−;βOGTKO, and control mice (Chop−/−, OGTf/y) (n = 4) (A). Immunoblot of isolated islets from Chop−/− and Chop−/−;βOGTKO mice, measuring ER-stress marker Bip (B and C; n = 5) and CPE (D and E; n = 4), relative to vinculin; y axis shows fold-change relative to Chop−/−. Proinsulin/insulin content ratio of islets from 4- to 5-week-old female Chop−/− and Chop−/−;βOGTKO mice (n = 3) (F). EM images at ×25,000 direct magnification of a β-cell from each genotype group (G). Scale bar, 600 nm. The EM images are representative of 6–8 distinct β-cell images from 2 individual animals per group. Statistical analyses were conducted using unpaired, two-tailed student t test or two-way ANOVA (A; between βOGTKO and Chop−/−;βOGTKO), with significance, *, p < 0.05.
eIF4G1 is O-GlcNAc-modified in β-cells and is reduced in OGT-deficient β-cells
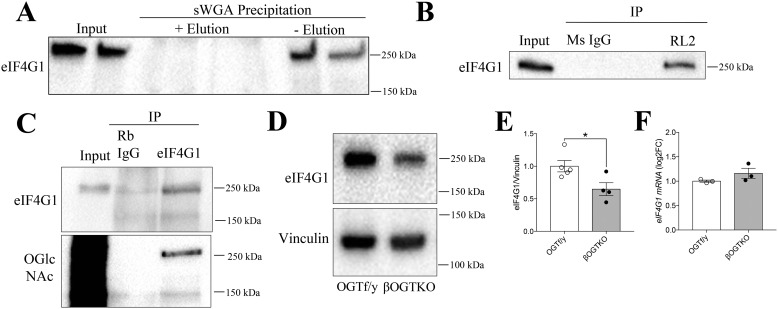
In the absence of a direct OGT-CPE relationship, we hypothesized that CPE may be translationally modulated indirectly through OGT regulation of an upstream factor. One possibility is through eIF4G1, a requisite scaffolding protein in the translation initiation complex that has been recently implicated to regulate CPE specifically in β-cells (20). We first used an sWGA-pulldown in MIN6 cells to show that eIF4G1 is glycosylated (Fig. 4A) and confirmed its O-GlcNAcylation status both by RL2 IP with eIF4G1 immunoblotting (Fig. 4B) and through eIF4G1 IP with RL2 immunoblot, observing a clear band at the expected size of 250 kDa (Fig. 4C). We also found that eIF4G1 protein levels were significantly reduced in βOGTKO islets compared with control, without any changes in eIF4G1 transcript level (Fig. 4, D–F). Combined with the novel finding of eIF4G1 as a direct OGT target in β-cells, we hypothesized that the O-GlcNAcylation status of this protein may impact its translational function or stability.
Figure 4.
An OGT target, eIF4G1, is reduced in OGT-deficient β-cells. Glycosylation of MIN6 eIF4G1 by sWGA precipitation + eIF4G1 immunoblot (A) and RL2 IP (versus mouse IgG control) + eIF4G1 immunoblot (B) or eIF4G1 IP (versus rabbit IgG) + eIF4G1 and RL2 immunoblot (C). The blots are representative of two independent experiments. eIF4G1 levels in 4–5–week–old OGT f/y (n = 5) and βOGTKO (n = 4) mice, shown for immunoblot protein (relative to vinculin, normalized to OGT f/y avg.) (D and E) and qPCR gene expression (relative to β-actin, log2-fold change to OGT f/y avg.) (n = 3) (F). Statistical analyses were conducted using unpaired, two-tailed Student's t test; *, p < 0.05.
O-GlcNAc modification of eIF4G1 at serine 61 supports its protein stability
Based on YinOYang software prediction and proteomics studies (25, 30, 31), we projected serine 61 in human eIF4G1 as the residue with the highest potential for O-GlcNAcylation. Through site-directed mutagenesis on an HA-tagged human eIF4G1 plasmid construct, substituting serine 61 for a non-O-GlcNAcylatable alanine residue (S61A), we showed a complete loss of O-GlcNAcylated eIF4G1 in transfected 293T cells through both RL2-enrichment/HA immunoblot and HA-enrichment/RL2 immunoblot, strongly suggesting that Ser-61 is the sole OGT target site on eIF4G1 (Fig. 5, A and B). On a functional level, loss of O-GlcNAcylation capacity led to a significant destabilization of the eIF4G1 S61A mutant (∼50% decrease) under conditions of total protein synthesis blockade (cycloheximide, 40 μg/ml) (Fig. 5, B and C). We observed a similar loss of eIF4G1 stability in INS-1 β-cells following pharmacological OGT inhibition (OSMI-1, 30 μm; cycloheximide, 50 μg/ml) (Fig. 5, D and E). Interestingly, overexpression of either WT or S61A eIF4G1 in INS1 cells increased CPE protein levels, suggesting that an adequate eIF4G1 level is sufficient to drive CPE translation, independent of its O-GlcNAc status (Fig. 5, F and G). Overall, these data support the hypothesis that O-GlcNAc–dependent eIF4G1 protein stability is a contributing mechanism behind the loss of eIF4G1 in βOGTKO islets.
Figure 5.
O-GlcNAcylation at serine 61 is critical for eIF4G1 stability. Confirmation of O-GlcNAc-null mutant in 293T cell transfection with control plasmid, HA-eIF4G1 WT or HA-eIF4G1 S61A by RL2 IP + HA-tag immunoblot (A) and HA IP + RL2 immunoblot (B). Protein stability of WT and S61A eIF4G1 (n = 3) in transfected 293T cells treated with CHX (50 μg/ml), shown by HA quantification and a representative blot (relative to vinculin, % 0 h) (C and D). E and F, similar experiments showing native eIF4G1 levels in INS-1 cells pre-treated with DMSO or OSMI-1 (30 μm) (n = 3). G and H, protein levels of HA and CPE (relative to vinculin, normalized to control) in similarly transfected INS-1 cells (n = 4). Western blotting of CPE and tubulin with densitometry (I and J), proinsulin content (K), and proinsulin-to-insulin ratio (L) from GFP or eIF4G1 adenovirus were infected in isolated dispersed βOGTKO islets (n = 3 βOGTKO for Western blotting and islet content; n = 2 WT for islet content data). Statistical analyses were conducted using unpaired, two-tailed Student's t test, one-way or two-way ANOVA, with significance; *, p < 0.05.
Re-expression of eIF4G1 in islets of βOGTKO elevates CPE and rescues hyperproinsulinemia
To determine whether loss of the eIF4G1 protein level led to the decrease in CPE levels in βOGTKO islets, we infected dispersed βOGTKO islet cells with adenoviral eIF4G1, achieving an optimal 80% infection rate post-60 h (Fig. S2B). With the overexpression of eIF4G1 in primary islets, we observed a strong trend (p = 0.05) toward increased CPE protein levels in βOGTKO islet cells compared with GFP-transfected control cells (Fig. 5, H and I). Most importantly, however, both the proinsulin content and proinsulin-to-insulin ratio defects in βOGTKO islets were fully restored to control levels (Fig. 5, J and K). Taken together, our findings reveal a novel role for β-cell OGT in supporting the protein level of eIF4G1, which is critical to the maintenance of CPE and normal insulin processing.
Discussion
In the present study, we reveal a previously unidentified function of the nutrient-sensor enzyme OGT in regulating β-cell insulin processing through the stabilization of eIF4G1, and thereby indirectly regulating CPE protein level. CPE, an enzyme needed for efficient proinsulin-to-insulin conversion was persistently reduced in the islets of young and normoglycemic mice or aged and diabetic βOGTKO mice of both sexes. The idea that the nutrient-sensor OGT affects CPE protein is consistent with other reports that CPE loss is nutrient sensitive, including in response to glucose starvation (32) and high-concentration palmitate (33), reflecting the nutrient-responsive nature of OGT-mediated protein O-GlcNAcylation. Although CPE itself is neither O-GlcNAcylated nor an OGT-binding partner, we show that O-GlcNAc–modified eIF4G1 status contribute to the translational regulation of CPE protein levels, thus proinsulin processing in β-cells.
ER stress is one of the early underlying factors causing β-cell failure in βOGTKO (7), and thus, may contribute to CPE loss in OGT-deficient β-cells. We found that a complete deletion of the ER stress-responsive protein Chop failed to rescue β-cell CPE protein levels, ruling out Chop as an indirect mediator between OGT loss and CPE. However, we cannot completely rule out the contribution of the other arms of the UPR, or non-UPR–mediated ER stress in regulating CPE. Therefore, it remains possible that non-Chop–mediated arms of the UPR may play a role here. Alternatively, the loss of CPE may precede or even contribute to βOGTKO ER stress, as reduction of CPE has been previously shown to induce ER stress and apoptosis in β-cells (33). We also cannot rule out the contribution of non-CPE–mediated hyperproinsulinemia in βOGTKO mice. Preliminary analysis on the transcriptome suggest that another proinsulin cleavage protein PC1/3 message is reduced,3 however, mass spectrometry-based proteome analysis of islets from βOGTKO mice reveal no changes in either PC1/3 or PC2.4
The insulin-signaling pathway was recently suggested to regulate CPE protein translation via translation initiation factor eIF4G1 (20). Mice lacking β-cell insulin receptors show hyperproinsulinemia associated with loss of CPE and a specific decrease in eIF4G1 protein (and not any other isoform of the eIF4G family) (20). Similar to the findings in the βIRKO model (20), transient overexpression of eIF4G1 in βOGTKO cells increases the CPE protein level and reverses the hyperproinsulinemia at the islet level, together suggesting that enhanced eIF4G1 stimulates CPE translation, thus ameliorating insulin processing. Islets from T2D patients as well as mice fed a high-fat diet showed reduction of CPE and eIF4G1, hinting at the roles of these proteins in diabetes progression (20). Liew et al. (20) showed that insulin signaling impacts eIF4G1 via transcription factor Pdx1. Both βIRKO and βOGTKO models show a decrease in Pdx1 (7), which has the potential to regulate eIF4G1 transcription (7, 20), although we did not detect any changes in eIF4G1 transcripts in βOGTKO islets.
A possible mechanism supported by the current data suggests that post-translational modification on eIF4G1 by OGT indirectly drives CPE-mediated insulin processing. In the current paper, we show that human eIF4G1 is O-GlcNAc-modified at serine 61 in β-cells. These data are supported by a recent report from Zhang et al. (34) showing mouse eIF4G1 is O-GlcNAcylated at serine 68 (homolog of human serine 61) in mouse embryonic fibroblast cells. One impact of O-GlcNAcylation on target proteins is prevention of protein degradation. For example, p53, PGC1α, and EZH2's O-GlcNAcylation is important for their protein stability (35–37). Here we showed that the functional importance of this modification seems to be in stabilizing the eIF4G1 protein, thereby enhancing its availability to interact with other translation initiation factors under nutrient conditions that support OGT activation. With glucose and insulin acting upstream of OGT, our proposed mechanism of eIF4G1 stabilization with O-GlcNAcylation may contribute to the molecular mechanisms for efficient protein translation and insulin processing for β-cell physiology (20, 38).
In summary, we provide evidence that perturbation of β-cell O-GlcNAcylation compromises cell physiology and alters cellular signaling and molecular processes contributing to the dysfunction of insulin processing consistent with known defects in islets from patients with T2D. Our data point to a mechanism of OGT regulation of CPE and the post-translational stability of critical translation factor eIFG41. A greater understanding of the complex functions of the nutrient-sensor OGT on factors of the translation initiation machinery such as eIF4G1 could provide a useful therapeutic path to improve β-cell function in pathological conditions contributing to human disease.
Experimental procedures
Animal models and in vivo mouse procedures
The following mice were used for the experiments: OGT flox/flox and Chop−/− (purchased from Jackson Laboratories), and mice harboring one allele of Cre-recombinase under the rat insulin 2 promoter (Rip-cre, generously provided by Dr. Pedro Herrera, University of Geneva, Switzerland). We generated βOGTKO (Rip-cre; OGT flox/y or flox/flox), by breeding OGT flox/flox and Rip-cre mice as previously described (7). Glucose and insulin tolerance tests were performed as previously described (7, 39). Random-fed serum was collected via tail-vein. All studies were approved by the Institutional Animal Care and Use Committee (protocol number 1806-36072A) at the University of Minnesota.
Islet isolation
After sacrifice through CO2 overdose and cervical dislocation, the pancreas was inflated through the common bile duct with 0.75 mg/ml of ice-cold collagenase (Millipore). The inflated pancreas was microdissected from the cavity and trimmed of fat before digesting for ∼9 min at 37 °C with manual agitation. The digested tissue pellet was washed with Hanks' balanced salt solution (Gibco) containing 2% FBS (GenClone), strained to remove undigested debris, and filtered through a 70-μm cell strainer (Fisher Scientific). After washing the retained digest off the filter screen, islets were handpicked clean into warm islet media (RPMI supplemented with l-glutamine (Corning), 10% FBS, 100 IU/ml of penicillin, and 100 g/ml of streptomycin, and 5 mm glucose). Islets were cultured in complete media overnight in a humidified 37 °C, 5% CO2 incubator before testing or collection.
Insulin and proinsulin ELISA
Insulin and proinsulin from random-fed serum and lysed isolated islets or INS-1 cells were measured using Mouse Ultrasensitive Insulin and Mouse Proinsulin ELISA (ALPCO), per the manufacturer's instructions. Cell and islet content data were normalized to DNA as previously described (7).
Cell culture
MIN6 (a gift from Dr. Peter Arvan, University of Michigan) and HEK 293T (purchased from ATCC) cells were maintained in Dulbecco's modified Eagle's medium-Glutamax (ThermoFisher), with 10% FBS and penicillin/l-streptomycin. INS-1 (a gift from Dr. Peter Arvan) was maintained in RPMI 1640 (Corning), with 10% FBS, penicillin/l-streptomycin, 10 mm HEPES, and 50 μm 2-mercaptoethanol. Cycloheximide (Santa Cruz (SCBT)) and OSMI-1 (Sigma) were dissolved in DMSO, prior to use. For in vitro protein stability experiments, INS-1 cells were pre-treated with OSMI-1 for 2 h prior to treatment with cycloheximide. Cells were maintained in a humidified 37 °C, 5% CO2 incubator.
Plasmid construct/site-directed mutagenesis/transfection
pcDNA3 HA eIF4G1(1–1599) was purchased from Addgene (donated by Dr. Nahum Sonenberg, number 45640). Site-directed mutagenesis was performed on the eIF4G1 plasmid to generate a serine to alanine mutation at residue 61, using QuikChange II XL (Agilent), as per the manufacturer's protocol, with the following primers: forward 5′-cagcccccgagcgctgcagcctcccg-3′ and reverse 5′-cgggaggctgcagcgctcgggggctg-3′. Accuracy of the mutagenesis was confirmed via sequencing (service by UMN Genomics Center). Plasmid transfections were performed using Lipofectamine 2000 (for 293T) or Lonza Nucleofection Kit V (for INS-1 cells), following the manufacturer's instructions. Collection or further testing was carried out 48 h post-transfection. Adenoviral vector for HA-eIF4G1 (with provided plasmid) and mouse CPE (V456014) were generated from Welgen, Inc. For viral infection, dispersed islets were infected with a viral titer of multiplicity of infection 200, and collected 60-h post-infection.
Western blotting
25–80 μg of protein lysates in RIPA buffer + 1% SDS (Bio-Rad) + protease and phosphatase inhibitors (CST), followed by Pierce BCA protein quantitation (ThermoScientific), were resolved by SDS-PAGE, transferred to polyvinylidene difluoride membrane, blocked with 5% nonfat dry milk, and probed with primary antibodies, followed by horseradish peroxidase-conjugated secondary antibodies (Table S1). The blot was visualized with SuperSignal West Pico PLUS (ThermoScientific), per the manufacturer's protocol. Densitometry analysis was performed with NIH ImageJ software.
Immunoprecipitation and sWGA precipitation
Cells were lysed in cold RIPA buffer (+ protease and phosphatase inhibitors) with periodic agitation. After normalizing protein content, lysates were incubated overnight with 1–5 μg/ml of protein-specific antibodies or with an equal amount of control antibodies (Table S1). The protein-antibody complex was captured using protein A/G-agarose beads (Invitrogen). For sWGA precipitation, the lysates were incubated overnight with 25 μl of sWGA-agarose (Vector). After incubation, the sWGA-agarose was washed extensively. Half the precipitate was incubated in RIPA buffer and the other half in glycoprotein eluting solution (N-acetylglucosamine, Vector) to elute GlcNAc protein. The precipitates and input lysates were resolved as described.
Click-IT OGlcNAc labeling experiment
INS1 cell lysates (200 μg) or purified ovalbumin (50 μg; Neta Scientific) were treated with or without PNGase F (New England Biolabs) under denaturing conditions, according to the manufacturer's protocol. Then, the proteins were labeled on GlcNAc moiety using Click-IT O-GlcNAc Enzymatic Labeling System (Invitrogen), and subsequently with a Click-IT Biotin Protein Analysis Detection Kit (Invitrogen), following the manufacturer's instructions. Following immunoprecipitation against CPE and Western blotting as described, horseradish peroxidase-conjugated streptavidin was used to detect Biotin/GlcNAc-labeled proteins.
Immunofluorescence and EM imaging and analysis
Formalin-fixed and paraffin-embedded pancreata were sectioned into 5-μm thick slices and classically prepared for staining with deparaffinization, antigen retrieval and blocking, as described previously (6). Sections were incubated in primary antibodies, followed by secondary antibodies conjugated to fluorophores (Table S1) and DAPI solution (ThermoScientific). Stained slides were imaged on a motorized microscope (Nikon ECLIPSE NI-E). EM images were acquired as previously described (7).
Statistical analysis
Data are presented as mean ± S.E. and were analyzed using unpaired, two-tailed Student's t tests and one-way ANOVA. Multiple outcome data were assessed using repeated measures two-way ANOVA. Statistical analyses were performed in GraphPad Prism version 7 with a significance threshold of p < 0.05.
Author contributions
S. J., A. L., and E. U. A. data curation; S. J. and E. U. A. formal analysis; S. J. and E. U. A. validation; S. J., A. L., and E. U. A. investigation; S. J. and E. U. A. visualization; S. J., A. L., and E. U. A. methodology; S. J. and E. U. A. writing-original draft; S. J., A. L., and E. U. A. writing-review and editing; E. U. A. conceptualization; E. U. A. resources; E. U. A. supervision; E. U. A. funding acquisition.
Supplementary Material
Acknowledgments
We acknowledge Regina Schlichting, Daniel Baumann, Dr. Ramkumar Mohan, and Ingrid Bender for technical support. We thank Drs. Peter Arvan, James Johnson, Ernesto Bernal-Mizrachi, David Bernlohr and Lauren Ball for discussion. We thank Dr. Thomas Pengo for his assistance in Fiji and the University of Minnesota Imaging Center for technical support. We thank Dr. Michael Kyba for access Electroporation and Nucleofector Device. The tissue processing and embedding was performed at the laboratory of Dr. Jop van Berlo.
This work was supported by National Institutes of Health, NIDDK Grants K01 DK103823, R21 DK112144, R03 DK114465, and R01 DK115720 (to E. U. A.), and F31 DK113694, 5T32DK083250 (to A. L). E. U.A. is the guarantor of this work. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Table S1 and Figs. S1–S2.
A. Lockridge et al., unpublished data.
R. Mohan et al., unpublished data.
- T2D
- type 2 diabetes
- OGT
- O-GlcNAc transferase
- OGA
- O-GlcNAcase
- ER
- endoplasmic reticulum
- CPE
- carboxypeptidase E
- eIF4G1
- eukaryotic translation initiation factor 4γ1
- sWGA
- succinylated wheat germ agglutinin
- CHX
- cycloheximide
- IP
- immunoprecipitation
- PNGase F
- peptide N-glycosidase F
- CHOP
- CCAAT enhancer-binding protein homologous protein
- UPR
- unfolded protein response
- HA
- hemagglutinin
- FBS
- fetal bovine serum
- ANOVA
- analysis of variance.
References
- 1. Røder M. E., Dinesen B., Hartling S. G., Houssa P., Vestergaard H., Sodoyez-Goffaux F., and Binder C. (1999) Intact proinsulin and beta-cell function in lean and obese subjects with and without type 2 diabetes. Diabetes Care 22, 609–614 10.2337/diacare.22.4.609 [DOI] [PubMed] [Google Scholar]
- 2. Gelding S. V., Andres C., Niththyananthan R., Gray I. P., Mather H., and Johnston D. G. (1995) Increased secretion of 32,33 split proinsulin after intravenous glucose in glucose-tolerant first-degree relatives of patients with non-insulin dependent diabetes of European, but not Asian, origin. Clin. Endocrinol. 42, 255–264 10.1111/j.1365-2265.1995.tb01873.x [DOI] [PubMed] [Google Scholar]
- 3. Pfützner A., Kunt T., Hohberg C., Mondok A., Pahler S., Konrad T., Lubben G., and Forst T. (2004) Fasting intact proinsulin is a highly specific predictor of insulin resistance in type 2 diabetes. Diabetes Care 27, 682–687 10.2337/diacare.27.3.682 [DOI] [PubMed] [Google Scholar]
- 4. Mykkänen L., Zaccaro D. J., Hales C. N., Festa A., and Haffner S. M. (1999) The relation of proinsulin and insulin to insulin sensitivity and acute insulin response in subjects with newly diagnosed type II diabetes: the insulin resistance atherosclerosis study. Diabetologia 42, 1060–1066 10.1007/s001250051271 [DOI] [PubMed] [Google Scholar]
- 5. Zethelius B., Byberg L., Hales C. N., Lithell H., and Berne C. (2003) Proinsulin and acute insulin response independently predict Type 2 diabetes mellitus in men: report from 27 years of follow-up study. Diabetologia 46, 20–26 10.1007/s00125-002-0995-2 [DOI] [PubMed] [Google Scholar]
- 6. Nagamatsu S., Bolaffi J. L., and Grodsky G. M. (1987) Direct effects of glucose on proinsulin synthesis and processing during desensitization. Endocrinology 120, 1225–1231 10.1210/endo-120-4-1225 [DOI] [PubMed] [Google Scholar]
- 7. Alejandro E. U., Bozadjieva N., Kumusoglu D., Abdulhamid S., Levine H., Haataja L., Vadrevu S., Satin L. S., Arvan P., and Bernal-Mizrachi E. (2015) Disruption of O-linked N-acetylglucosamine signaling induces ER stress and beta cell failure. Cell Rep. 13, 2527–2538 10.1016/j.celrep.2015.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang X., and Qian K. (2017) Protein O-GlcNAcylation: emerging mechanisms and functions. Nat. Rev. Mol. Cell Biol. 18, 452–465 10.1038/nrm.2017.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hart G. W. (2014) Three decades of research on O-GlcNAcylation: a major nutrient sensor that regulates signaling, transcription and cellular metabolism. Front. Endocrinol. (Lausanne) 5, 183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hanover J. A., Krause M. W., and Love D. C. (2010) The hexosamine signaling pathway: O-GlcNAc cycling in feast or famine. Biochim. Biophys. Acta 1800, 80–95 10.1016/j.bbagen.2009.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bond M. R., and Hanover J. A. (2013) O-GlcNAc cycling: a link between metabolism and chronic disease. Annu. Rev. Nutr. 33, 205–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ma J., and Hart G. W. (2013) Protein O-GlcNAcylation in diabetes and diabetic complications. Expert Rev. Proteomics 10, 365–380 10.1586/14789450.2013.820536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Issad T., Masson E., and Pagesy P. (2010) O-GlcNAc modification, insulin signaling and diabetic complications. Diabetes Metab. 36, 423–435 10.1016/j.diabet.2010.09.001 [DOI] [PubMed] [Google Scholar]
- 14. Lehman D. M., Fu D. J., Freeman A. B., Hunt K. J., Leach R. J., Johnson-Pais T., Hamlington J., Dyer T. D., Arya R., Abboud H., Göring H. H., Duggirala R., Blangero J., Konrad R. J., and Stern M. P. (2005) A single nucleotide polymorphism in MGEA5 encoding O-GlcNAc-selective N-acetyl-β-d-glucosaminidase is associated with type 2 diabetes in Mexican Americans. Diabetes 54, 1214–1221 10.2337/diabetes.54.4.1214 [DOI] [PubMed] [Google Scholar]
- 15. Andrali S. S., Qian Q., and Ozcan S. (2007) Glucose mediates the translocation of NeuroD1 by O-linked glycosylation. J. Biol. Chem. 282, 15589–15596 10.1074/jbc.M701762200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gao Y., Miyazaki J., and Hart G. W. (2003) The transcription factor PDX-1 is post-translationally modified by O-linked N-acetylglucosamine and this modification is correlated with its DNA binding activity and insulin secretion in min6 beta-cells. Arch. Biochem. Biophys. 415, 155–163 10.1016/S0003-9861(03)00234-0 [DOI] [PubMed] [Google Scholar]
- 17. Liu M., Wright J., Guo H., Xiong Y., and Arvan P. (2014) Proinsulin entry and transit through the endoplasmic reticulum in pancreatic beta cells. Vitam. Horm. 95, 35–62 10.1016/B978-0-12-800174-5.00002-8 [DOI] [PubMed] [Google Scholar]
- 18. Naggert J. K., Fricker L. D., Varlamov O., Nishina P. M., Rouille Y., Steiner D. F., Carroll R. J., Paigen B. J., and Leiter E. H. (1995) Hyperproinsulinaemia in obese fat/fat mice associated with a carboxypeptidase E mutation which reduces enzyme activity. Nat. Genet. 10, 135–142 10.1038/ng0695-135 [DOI] [PubMed] [Google Scholar]
- 19. Varlamov O., Fricker L. D., Furukawa H., Steiner D. F., Langley S. H., and Leiter E. H. (1997) Beta-cell lines derived from transgenic Cpe(fat)/Cpe(fat) mice are defective in carboxypeptidase E and proinsulin processing. Endocrinology 138, 4883–4892 10.1210/endo.138.11.5506 [DOI] [PubMed] [Google Scholar]
- 20. Liew C. W., Assmann A., Templin A. T., Raum J. C., Lipson K. L., Rajan S., Qiang G., Hu J., Kawamori D., Lindberg I., Philipson L. H., Sonenberg N., Goldfine A. B., Stoffers D. A., Mirmira R. G., Urano F., and Kulkarni R. N. (2014) Insulin regulates carboxypeptidase E by modulating translation initiation scaffolding protein eIF4G1 in pancreatic beta cells. Proc. Natl. Acad. Sci. U.S.A. 111, E2319–E2328 10.1073/pnas.1323066111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Blandino-Rosano M., Barbaresso R., Jimenez-Palomares M., Bozadjieva N., Werneck-de-Castro J. P., Hatanaka M., Mirmira R. G., Sonenberg N., Liu M., Rüegg M. A., Hall M. N., and Bernal-Mizrachi E. (2017) Loss of mTORC1 signalling impairs beta-cell homeostasis and insulin processing. Nat. Commun. 8, 16014 10.1038/ncomms16014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gu G., Dubauskaite J., and Melton D. A. (2002) Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development 129, 2447–2457 [DOI] [PubMed] [Google Scholar]
- 23. Fricker L. D., Das B., and Angeletti R. H. (1990) Identification of the pH-dependent membrane anchor of carboxypeptidase E (EC 3.4.17.10). J. Biol. Chem. 265, 2476–2482 [PubMed] [Google Scholar]
- 24. Chu K. Y., Briggs M. J., Albrecht T., Drain P. F., and Johnson J. D. (2011) Differential regulation and localization of carboxypeptidase D and carboxypeptidase E in human and mouse beta-cells. Islets 3, 155–165 10.4161/isl.3.4.15767 [DOI] [PubMed] [Google Scholar]
- 25. Gupta R., and Brunak S. (2002) Prediction of glycosylation across the human proteome and the correlation to protein function. Pac. Symp. Biocomput. 2002, 310–322 [PubMed] [Google Scholar]
- 26. Dhanvantari S., Arnaoutova I., Snell C. R., Steinbach P. J., Hammond K., Caputo G. A., London E., and Loh Y. P. (2002) Carboxypeptidase E, a prohormone sorting receptor, is anchored to secretory granules via a C-terminal transmembrane insertion. Biochemistry 41, 52–60 10.1021/bi015698n [DOI] [PubMed] [Google Scholar]
- 27. Kreppel L. K., Blomberg M. A., and Hart G. W. (1997) Dynamic glycosylation of nuclear and cytosolic proteins: cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J. Biol. Chem. 272, 9308–9315 10.1074/jbc.272.14.9308 [DOI] [PubMed] [Google Scholar]
- 28. Bravo R., Parra V., Gatica D., Rodriguez A. E., Torrealba N., Paredes F., Wang Z. V., Zorzano A., Hill J. A., Jaimovich E., Quest A. F., and Lavandero S. (2013) Endoplasmic reticulum and the unfolded protein response: dynamics and metabolic integration. Int. Rev. Cell Mol. Biol. 301, 215–290 10.1016/B978-0-12-407704-1.00005-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Furuta M., Carroll R., Martin S., Swift H. H., Ravazzola M., Orci L., and Steiner D. F. (1998) Incomplete processing of proinsulin to insulin accompanied by elevation of Des-31,32 proinsulin intermediates in islets of mice lacking active PC2. J. Biol. Chem. 273, 3431–3437 10.1074/jbc.273.6.3431 [DOI] [PubMed] [Google Scholar]
- 30. Khidekel N., Ficarro S. B., Clark P. M., Bryan M. C., Swaney D. L., Rexach J. E., Sun Y. E., Coon J. J., Peters E. C., and Hsieh-Wilson L. C. (2007) Probing the dynamics of O-GlcNAc glycosylation in the brain using quantitative proteomics. Nat. Chem. Biol. 3, 339–348 10.1038/nchembio881 [DOI] [PubMed] [Google Scholar]
- 31. Xu S. L., Chalkley R. J., Maynard J. C., Wang W., Ni W., Jiang X., Shin K., Cheng L., Savage D., Hühmer A. F., Burlingame A. L., and Wang Z. Y. (2017) Proteomic analysis reveals O-GlcNAc modification on proteins with key regulatory functions in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 114, E1536–E1543 10.1073/pnas.1610452114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Höring E., Harter P. N., Seznec J., Schittenhelm J., Bühring H. J., Bhattacharyya S., von Hattingen E., Zachskorn C., Mittelbronn M., and Naumann U. (2012) The “go or grow” potential of gliomas is linked to the neuropeptide processing enzyme carboxypeptidase E and mediated by metabolic stress. Acta Neuropathol. 124, 83–97 10.1007/s00401-011-0940-x [DOI] [PubMed] [Google Scholar]
- 33. Jeffrey K. D., Alejandro E. U., Luciani D. S., Kalynyak T. B., Hu X., Li H., Lin Y., Townsend R. R., Polonsky K. S., and Johnson J. D. (2008) Carboxypeptidase E mediates palmitate-induced beta-cell ER stress and apoptosis. Proc. Natl. Acad. Sci. U.S.A. 105, 8452–8457 10.1073/pnas.0711232105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang X., Shu X. E., and Qian S. B. (2018) O-GlcNAc modification of eIF4GI acts as a translational switch in heat shock response. Nat. Chem. Biol. 14, 909–916 [DOI] [PubMed] [Google Scholar]
- 35. de Queiroz R. M., Madan R., Chien J., Dias W. B., and Slawson C. (2016) Changes in O-linked N-acetylglucosamine (O-GlcNAc) homeostasis activate the p53 pathway in ovarian cancer cells. J. Biol. Chem. 291, 18897–18914 10.1074/jbc.M116.734533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chu C. S., Lo P. W., Yeh Y. H., Hsu P. H., Peng S. H., Teng Y. C., Kang M. L., Wong C. H., and Juan L. J. (2014) O-GlcNAcylation regulates EZH2 protein stability and function. Proc. Natl. Acad. Sci. U.S.A. 111, 1355–1360 10.1073/pnas.1323226111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ruan H. B., Han X., Li M. D., Singh J. P., Qian K., Azarhoush S., Zhao L., Bennett A. M., Samuel V. T., Wu J., Yates J. R. 3rd, and Yang X. (2012) O-GlcNAc transferase/host cell factor C1 complex regulates gluconeogenesis by modulating PGC-1α stability. Cell Metab. 16, 226–237 10.1016/j.cmet.2012.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gomez E., Powell M. L., Greenman I. C., and Herbert T. P. (2004) Glucose-stimulated protein synthesis in pancreatic beta-cells parallels an increase in the availability of the translational ternary complex (eIF2-GTP. Met-tRNAi) and the dephosphorylation of eIF2α. J. Biol. Chem. 279, 53937–53946 10.1074/jbc.M408682200 [DOI] [PubMed] [Google Scholar]
- 39. Lockridge A. D., Baumann D. C., Akhaphong B., Abrenica A., Miller R. F., and Alejandro E. U. (2016) Serine racemase is expressed in islets and contributes to the regulation of glucose homeostasis. Islets 8, 195–206 10.1080/19382014.2016.1260797 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.