Abstract
Forkhead box protein O1 (FOXO1) is a transcription factor involved in various cellular processes such as glucose metabolism, development, stress resistance, and tumor suppression. FOXO1's transcriptional activity is controlled by different environmental cues through a myriad of posttranslational modifications. In response to growth factors, the serine/threonine kinase AKT phosphorylates Thr24 and Ser256 in FOXO1 to stimulate binding of 14-3-3 proteins, causing FOXO1 inactivation. In contrast, low nutrient and energy levels induce FOXO1 activity. AMP-activated protein kinase (AMPK), a master regulator of cellular energy homeostasis, partly mediates this effect through phosphorylation of Ser383 and Thr649 in FOXO1. In this study, we identified Ser22 as an additional AMPK phosphorylation site in FOXO1's N terminus, with Ser22 phosphorylation preventing binding of 14-3-3 proteins. The crystal structure of a FOXO1 peptide in complex with 14-3-3 σ at 2.3 Å resolution revealed that this is a consequence of both steric hindrance and electrostatic repulsion. Furthermore, we found that AMPK-mediated Ser22 phosphorylation impairs Thr24 phosphorylation by AKT in a hierarchical manner. Thus, numerous mechanisms maintain FOXO1 activity via AMPK signaling. AMPK-mediated Ser22 phosphorylation directly and indirectly averts binding of 14-3-3 proteins, whereas phosphorylation of Ser383 and Thr649 complementarily stimulates FOXO1 activity. Our results shed light on a mechanism that integrates inputs from both AMPK and AKT signaling pathways in a small motif to fine-tune FOXO1 transcriptional activity.
Keywords: FOXO, 14-3-3 protein, AMP-activated kinase (AMPK), Akt PKB, gene transcription, crystal structure, posttranslational modification (PTM), protein phosphorylation, NMR, 14-3-3 interaction, energy homeostasis, FOXO1 transcription factor, kinase signaling
Introduction
The forkhead box O (FOXO)7 family of transcription factors is well-conserved throughout evolution, and they control the expression of a multitude of genes involved in various cellular processes. FOXO1 (FKHR), one of the family members, has been shown to regulate cellular homeostasis, insulin and glucose metabolism, immune responses, cell cycle progression, apoptosis, and senescence (1). Moreover, FOXO1-null mice are not able to complete embryonic development, confirming its essential role (2).
The transcriptional activity of FOXO1 is tightly regulated through numerous posttranslational modifications, such as phosphorylation, methylation, acetylation, ubiquitination, and glycosylation (reviewed in Refs. 3, 4). Multiple kinases are known to reduce FOXO1 activity via phosphorylation (e.g. AKT (5); CDK5 (6), PKA (7), CDK2 (8), CDK1 (9), and DYRK1, GSK3, and CK1 (10)). Of these, FOXO1 phosphorylation by the PI3K/AKT pathway is extensively studied. In response to a range of extracellular stimuli like growth factors, insulin, and cytokines, AKT phosphorylates Thr24, Ser256, and Ser319 in FOXO1 (11, 12). This allows binding of 14-3-3 scaffolding proteins to FOXO1 and consequent inhibition of transcriptional activity through export from the nucleus to the cytoplasm. 14-3-3 proteins are ubiquitously expressed in mammalian cells, and they mainly regulate the function of binding partners by affecting their subcellular localization (13). Notably, a triple mutant of FOXO1 (T24A/S256A/S319A), which cannot be phosphorylated by AKT, is predominantly nuclear and constitutively active because interactions with 14-3-3 proteins are abolished (14).
In contrast, various kinases activate the transcriptional activity of FOXO1 (AMPK (15), MST1 (16), and extracellular signal-regulated kinase and p38 (17)). AMP-activated protein kinase (AMPK) is a major regulator of cellular energy homeostasis and is activated under conditions such as nutrient deprivation or oxidative stress (18–22). Yun et al. (15) showed that AMPK phosphorylates FOXO1 on residues Ser383 and Thr649; however, the molecular mechanism for increased FOXO1 activity upon phosphorylation of these residues remains elusive. Moreover, the authors suggested that additional AMPK sites might exist that could further explain the regulation of the transcriptional activity of FOXO1 by AMPK.
In this work, we identified Ser22 in FOXO1 as an additional AMPK phosphorylation site. Structural and biophysical data elucidated that phosphorylation of Ser22 reduces binding of 14-3-3 proteins to FOXO1 through steric hindrance and electrostatic repulsion. At the same time, Ser22 phosphorylation indirectly prevented 14-3-3 binding by negatively regulating Thr24 phosphorylation by AKT. Interestingly, phosphorylation of Thr24 did not affect phosphorylation of Ser22 by AMPK to a similar extent, suggesting hierarchical cross-talk between both signaling pathways. Ser22 phosphorylation both directly and indirectly abrogates complex formation with 14-3-3 proteins and plays a central role in controlling the transcriptional activity of FOXO1.
Results
The N-terminal domain of FOXO1 is intrinsically disordered
The FOXO1 protein consists of three major functional domains: the Forkhead box domain (FHD), which is responsible for DNA binding; the C-terminal transcription activation domain (TAD); and the less characterized N-terminal domain (NTD). Moreover, the protein contains signals that mediate nuclear localization and nuclear export (Fig. 1A). Although the FHD and TAD have been extensively studied, little is known about the contribution of the NTD regarding transcriptional activity of FOXO1.
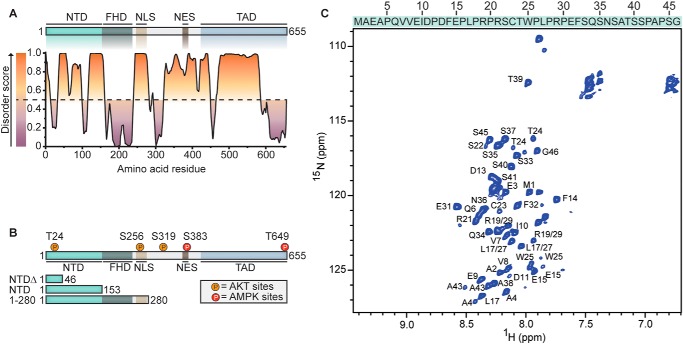
Figure 1.
Analysis of intrinsically disordered regions in FOXO1. A, representation of the functional domains of FOXO1: NTD 1–153, FHD 154–244, TAD 421–655. A disorder score was predicted for each amino acid using PONDR-FIT (23). The dashed line at 0.5 of the y axis separates residues that were predicted to be disordered (score > 0.5, orange) and ordered (score < 0.5, purple). B, the known AKT and AMPK phosphorylation sites in FOXO1 are highlighted in orange and red, respectively. Constructs that were used in this study are indicated: NTDΔ (1–46), NTD (1–153), and a fragment containing the NTD, FHD, and nuclear localization signal (1–280), which was used to analyze interactions with 14-3-3 proteins in cells (Fig. 4A). C, assignment of the 1H-15N HMQC spectrum of NTDΔ. Several amino acids display pairs of signals, as these are preceded by prolines that occur in both cis and trans conformation. Amino acids Leu17/Leu27 and Arg19/Arg29 could not be distinguished because they are flanked by prolines, which do not show signals in a 1H-15N HMQC spectrum. Residues of the purification tag were observed but were not assigned. Amino acid sequence of NTDΔ is indicated on top of the NMR spectrum in one-letter code.
To better understand the structural features of the different domains of FOXO1, we first performed in silico predictions of the disorder tendency (23, 24). This analysis suggested a folded FHD (154–244) that is flanked by predominantly disordered regions (disorder scores ≥ 0.5, Fig. 1A). Of note, crystal structures have been solved for the FOXO1 FHD (25, 26), confirming a structured organization of this region. Next, we focused on the NTD and purified two recombinant proteins for structural examination using 2D NMR. One protein encompassed the full NTD (1–153) and the other a N-terminal region of the NTD (1–46), hereafter referred to as NTD and NTDΔ, respectively (Fig. 1B). The 1H-15N Heteronuclear Single Quantum Coherence (HSQC) NMR spectra of NTDΔ (Fig. 1C) and the NTD (Fig. S1) showed narrow dispersions of 1H-15N chemical shifts in the 1H dimension (7.7–8.6 ppm), a characteristic of proteins lacking secondary structure elements. Although backbone resonances of NTDΔ were fully assigned, individual assignments of amino acids in the entire NTD could not be obtained because of peak overlaps. Nevertheless, both spectra showed a high similarity in chemical shifts and line widths, indicating an intrinsically disordered nature of the NTD. A secondary structure propensity (SSP) analysis (27) further confirmed a predominantly random coil conformation of NTDΔ based on the Cα and Cβ chemical shifts from the 3D 13C-15N HNCαCβ experiment (Fig. S2). However, residues 21–25 and 35–40 had positive average SSP scores, 0.19 and 0.08, respectively, suggesting a weak partial α-helical propensity of those regions.
The NTD retains unstructured properties upon Thr24 phosphorylation by AKT
The NTD of FOXO1 contains one of the two 14-3-3 binding motifs (amino acids 19–26), and phosphorylation of Thr24 by AKT is required to mediate interactions with 14-3-3 proteins. Interestingly, residues 21–25 showed a weak α-helical propensity (Fig. S2), and, therefore, we hypothesized that phosphorylation of Thr24 might induce structural changes that could influence 14-3-3 binding.
To measure phosphorylation of Thr24 in time-resolved 2D 1H-15N Heteronuclear Multiple Quantum Coherence (HMQC) NMR experiments, either NTDΔ or the full NTD were incubated with purified AKT. Over the course of 3 h, a single chemical shift appeared at 8.95/119.89 (1H/15N) ppm (Fig. 2A, Fig. S3A), a region typical for phosphorylated residues (28). Simultaneously, two chemical shifts assigned to Thr24 gradually disappeared from 7.97/116.45 (1H/15N) and 8.13/116.82 (1H/15N) ppm, highlighting strong phosphorylation of Thr24 by AKT, as expected. The appearance of two distinct individual chemical shifts might be caused by specific cis and trans conformations of the flanking proline residue, as observed with other intrinsically disordered proteins (29). NTDΔ contains 10 prolines, and the presence of 15 Hα/N peaks in a 15N-13C Hα(Cα)N NMR spectra revealed that five prolines occur in both cis and trans conformations (Fig. S4). Apart from the changes in chemical shifts for Thr24, only minor differences were detected for residues in proximity to the phosphorylation site: Leu17/Leu27, Arg19/Arg29, Arg21, Ser22, Cys23, and Tyr25.
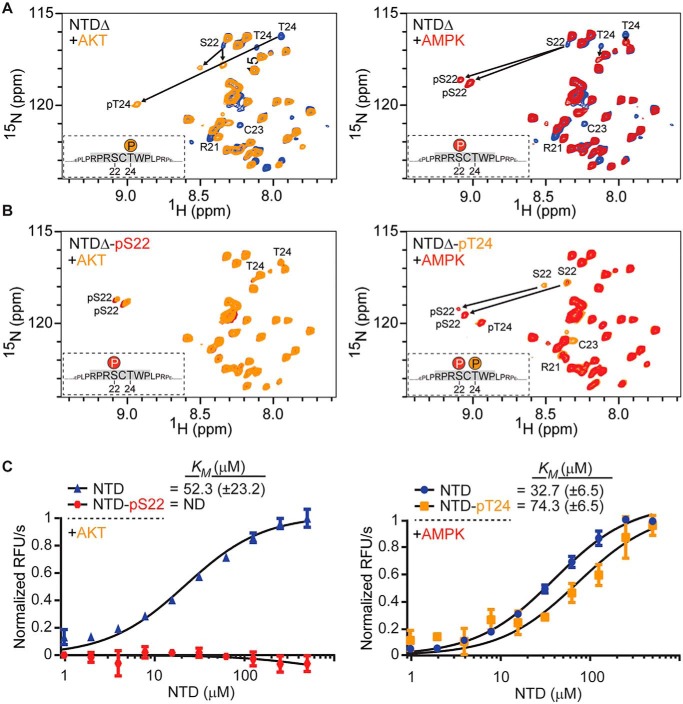
Figure 2.
Phosphorylation of the NTD by AKT and AMPK. A, 2D 1H-15N HMQC NMR spectra of NTDΔ incubated for 3 h with purified AKT (orange) or AMPK (red) to assess Thr24 and Ser22 phosphorylation by AKT and AMPK, respectively. Chemical shift changes of phosphorylated amino acids are indicated by arrows. Doubling of peaks for pSer22 suggests dual conformations. B, 2D 1H-15N HMQC NMR spectra of prephosphorylated NTDΔ incubated with the indicated kinases for 4.5 h; NTDΔ-pSer22 with AKT to monitor Thr24 phosphorylation (orange) and NTDΔ-pT24 with AMPK to analyze Ser22 phosphorylation (red). Chemical shift changes of phosphorylated amino acids are indicated by arrows. C, the activities of both AKT and AMPK kinases on different substrates were analyzed using a fluorescence-based ADP biosensor. The unphosphorylated NTD as substrate for AKT and AMPK is shown in blue. The activity of AKT on NTD-pSer22 is shown in red. The activity of AMPK on NTD-pThr24 is shown in orange. Km values were determined based on the initial reaction rates. Data are mean ± S.D. from three independent experiments. ND, not detectable.
Finally, the SSP scores for both the unphosphorylated and Thr24 phosphorylated NTDΔ were comparable (Fig. S2). This suggested that Thr24 phosphorylation by AKT does not influence the local structural organization of the NTD.
AMPK phosphorylates Ser22 in the NTD
AMPK has been shown to phosphorylate Ser383 and Thr649 in FOXO1, but it was suspected that additional AMPK phosphorylation sites might exist (30). Several AMPK phosphorylation motifs have been described (15, 22, 31, 32), and central elements are two to three basic and/or hydrophobic residues at position 3–7 N-terminally together with one hydrophobic residue at position 3–4 C-terminally of the phosphorylated residue. Interestingly, Ser22 in the NTD of FOXO1 fulfills these criteria and was predicted by the PhosphoNET software (33) as a potential AMPK site.
To analyze whether AMPK can phosphorylate Ser22 in vitro, we first performed an LC-MS measurement and identified one major and one minor phosphorylation site by the occurrence of a +80-Da and +160-Da peak. Subsequent peptide digestion and LC-MS/MS suggested Ser22 as an AMPK phosphorylation site in the NTD of FOXO1 (Fig. S5).
To confirm phosphorylation of Ser22 by AMPK, we incubated NTDΔ as well as the full NTD with purified AMPK and evaluated the chemical shifts in time-resolved 2D 1H-15N HMQC NMR experiments. Within 2 h, gradual disappearance of the Ser22 chemical shift at 8.36/116.64 (1H/15N) ppm was observed in combination with the emergence of two new peaks at 9.04/118.80 (1H/15N) ppm and 9.09/118.65 (1H/15N) ppm, indicating that Ser22 was phosphorylated by AMPK (Fig. 2A and Fig. S3A). Flanking residues next to the phosphorylation site were shifted in the NMR spectra (Leu17/Leu27, Arg19/Arg29, Arg21, Cys23, Thr24, and Tyr25), as observed for the phosphorylation of Thr24 by AKT within the same region. There was no substantial difference between the SSP scores of the unphosphorylated and Ser22-phosphorylated NTDΔ (Fig. S2). Hence, neither Ser22 nor Thr24 phosphorylation changed the secondary structure of this region of the NTD.
Phosphorylation of Ser22 by AMPK decreases phosphorylation of Thr24 by AKT
The proximity of Ser22 and Thr24 led us explore whether the phosphorylation status of one of those residues affects the subsequent phosphorylation of the other site or whether phosphorylation occurs independently. Time-resolved 2D NMR experiments on the prephosphorylated NTDΔ were conducted. NTDΔ-pThr24 was incubated with purified AMPK, and over the course of 4.5 h, two peaks assigned to Ser22 at 8.53/117.87 (1H/15N) ppm and 8.37/117.71 (1H/15N) ppm disappeared (Fig. 2B, red). In parallel, two new peaks appeared at 9.07/119.49 (1H/15N) ppm and 9.11/119.14 (1H/15N) ppm, illustrating successful phosphorylation of Ser22. Full phosphorylation of Ser22 in NTDΔ-pThr24 was achieved after about 4.5 h, approximately twice the time compared with the unphosphorylated protein as a substrate. Next, we incubated NTDΔ-pSer22 with AKT. No peaks were shifted in the NMR spectra after 4.5 h, highlighting that AKT was unable to phosphorylate Thr24 within this time scale (Fig. 2B, orange). After 24 h, a weak signal appeared for pThr24, with a concomitant decrease in intensity of the Thr24 peak, suggesting very weak phosphorylation by AKT (data not shown). Comparable results were observed with the full NTD (Fig. S3B). Taken together, the prior phosphorylation status of Ser22 drastically affected subsequent phosphorylation of Thr24 by AKT, but pThr24 only moderately reduced the phosphorylation rate of Ser22 by AMPK.
To further substantiate this observation, Michaelis–Menten kinetics (Km) were determined for AMPK and AKT with the unphosphorylated and prephosphorylated NTD. For AMPK, the Km of the unphosphorylated NTD and NTD-pThr24 as a substrate was 32 μm and 74 μm, respectively (Fig. 2C, right panel). This statistically significant (p = 0.0016) 2.3-fold increase of Km was in a similar range as observed in the NMR measurements. For AKT, the Km was 52 μm for the unphosphorylated NTD, whereas the Km could not be determined for the NTD-pSer22, implying that it is a poor AKT substrate (Fig. 2C, left panel). Overall, time-resolved 2D NMR measurements combined with the biochemical assay demonstrate a hierarchical order where the AMPK-mediated phosphorylation of Ser22 in the NTD has a strong regulatory effect on Thr24 phosphorylation by AKT but not vice versa.
Phosphorylation of Ser22 prevents interactions of FOXO1 with 14-3-3 proteins
A major mechanism to control FOXO1 transcriptional activity is the binding of 14-3-3 proteins. Different 14-3-3 interaction motifs have been described in the literature, and FOXO1 contains two mode I motifs, RXX(pS/pT)XP (34). A prerequisite for complex formation is phosphorylation of one residue in each of the 14-3-3 interaction motifs by AKT: Thr24 and Ser256 (11). Interestingly, Ser22 is located in the N-terminal 14-3-3 binding interface (Fig. S8B). To examine the impact of Ser22 phosphorylation on 14-3-3 binding, we first studied the interaction of the NTD and 14-3-3 proteins in vitro using surface plasmon resonance (SPR) direct binding assays. 14-3-3 ζ was immobilized to the SPR sensor surface, and binding affinities of the unphosphorylated NTD or the NTD phosphorylated on Ser22, Thr24, or both were measured. As expected, an AKT-phosphorylated NTD (pThr24) caused a strong interaction with 14-3-3 ζ (Kd, 5.1 μm), confirming the central role of pThr24 as binding platform for 14-3-3 proteins (Fig. 3A). An AMPK-phosphorylated NTD (pSer22) showed a very weak affinity to 14-3-3 ζ. Interestingly, the double-phosphorylated NTD (pSer22/pThr24) also possessed a reduced affinity for 14-3-3 ζ of more than 50-fold (Kd, 260 μm). Similar results were observed with the θ isoform of 14-3-3 (Fig. S6A), and it appeared that phosphorylation of Ser22 strongly blocked 14-3-3 binding even in the context of phosphorylated Thr24.
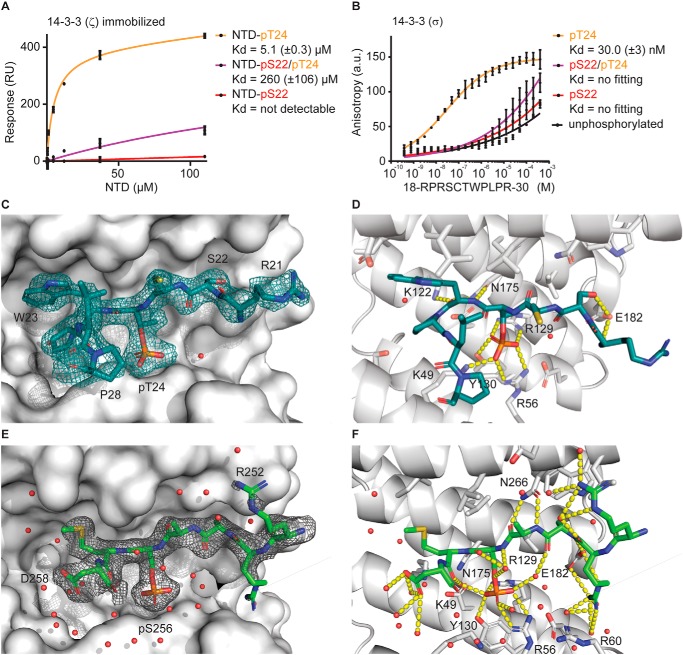
Figure 3.
Phosphorylation of Ser22 in the NTD of FOXO1 hinders 14-3-3 interactions in vitro. A, surface plasmon resonance binding analysis of 14-3-3 ζ and the NTD of FOXO1 phosphorylated at the indicated residues. NTD-pThr24 served as positive control, and the unphosphorylated NTD was used as a linear component for data correction. Data are expressed as mean ± S.D. of technical triplicates from one representative experiment. B, fluorescence anisotropy binding analysis of interactions between 14-3-3 σ and the FOXO1 peptide (18RPRSCTWPLPR30) in different phosphorylation states. Data are expressed as means ± S.D. from three independent experiments and fitted with Prism (specific binding with the Hill slope). C, representative FOXO1/14-3-3 complex with displayed electron density (final 2Fo-Fc, contoured at 1σ) with 14-3-3 as a gray surface, FOXO1-pThr24 as sticks, and water as a red sphere. Amino acids of the FOXO1-pThr24 peptide are indicated in one-letter code. D, details of the interactions between FOXO1-pThr24 and 14-3-3 σ. Amino acids of 14-3-3 σ are indicated in one-letter code, and hydrogen bonds are illustrated by yellow dashed lines. E, FOXO1-pSer256/14-3-3 complex with displayed electron density (Final 2Fo-Fc, contoured at 1σ) with 14-3-3 as a gray surface, FOXO1-pSer256 as sticks, and water as a red sphere. Amino acids of the FOXO1-pSer256 peptide are indicated in one-letter code. F, details of the interactions between FOXO1-pSer256 and 14-3-3 σ. Amino acids of 14-3-3 σ are indicated in one-letter code, and hydrogen bonds are illustrated by yellow dashed lines.
Crystal structures of pThr24 and pSer256 peptides in complex with 14-3-3 σ
To fully understand the consequence of Ser22 phosphorylation on 14-3-3 binding, we aimed to crystallize the interface of the NTD and 14-3-3. However, no crystals could be obtained with the full NTD, and, therefore, peptides were used to represent the different phosphorylated 14-3-3 recognition sites of FOXO1. The peptide representing the pThr24 site consists of 11 amino acids with the sequence 18RPRSCTWPLPR30.
First, Fluorescein amidite (FAM)-labeled versions of the peptides in different phosphorylation states were used in fluorescence anisotropy measurements to test whether they reflect the binding behavior between the NTD and 14-3-3 proteins observed in SPR experiments. The highest affinity was measured between 14-3-3 σ and the pThr24 peptide (30 nm), whereas the pSer22 and double-phosphorylated peptides had weaker binding affinities (Fig. 3B). For 14-3-3 ζ, the pThr24 peptide had an affinity of 18.3 nm, whereas the double-phosphorylated peptide had a binding affinity of 12.9 μm (Fig. S6B). Moreover, the binding of the pSer22 and unphosphorylated peptide could not be quantified. Together, these results are in line with the findings of the SPR experiments employing the entire NTD.
Next, unlabeled peptides were used for structure analysis. The pThr24 peptide in complex with 14-3-3 σ produced crystals of the space group P21 that diffracted to 2.3 Å and harbored eight 14-3-3/peptide molecules in the asymmetric unit (Fig. S7A). Up to nine residues of the 11 amino acids comprising the pThr24 peptide could be built into the electron density (Fig. 3C). The peptide binds into the amphipathic groove of the 14-3-3 protein, where the side chain of Trp25 establishes an extensive hydrophobic contact with the “hydrophobic roof” (35) of the 14-3-3 monomer binding channel. The two prolines (Pro26 and P28) induce a strong turn in the peptide, guiding the chain away from the Trp25 position toward the 14-3-3 dimer interface and out of the central binding channel. The pThr24 is coordinated by Lys49, Arg57, Arg129, and Tyr130 of the protein. Additional polar contacts were mainly established via the peptide backbone and polar residues of 14-3-3 (carbonyl of Trp25 with Lys122, nitrogen of Trp25 with Asn175, and nitrogen of Arg21 with Glu182) (Fig. 3D). Interestingly, the hydroxyl group of Ser22 forms a polar contact with Glu182, locking this residue in all eight monomers in a similar conformation (Fig. S7A). A close-up of that conformation explains how a phosphorylation of this residues disrupts the binding of the double-phosphorylated peptide and the NTD. The serine fills a flat pocket of the 14-3-3 surface; any extension of that residue will cause a steric clash with the protein and impair binding.
To complete the picture of FOXO1 binding to 14-3-3, the structure was also solved of the complex between pSer256 and the C-terminal 14-3-3 binding site to 1.9 Å and in the space group P212121, with two molecules found in the asymmetric unit (Fig. S7C). Seven residues (Arg252-Asp258) of the 12 amino acids comprising the FOXO1-pSer256 peptide (251RRRAASMDNNSK262) could be built into the electron density (Fig. 3E). The phosphorylated serine (pSer256) is found in the same pocket as pThr24, coordinated by Arg57, Arg129, and Tyr130 of 14-3-3 (Fig. 3F). The hydrophobic roof of the 14-3-3 monomer binding channel is occupied by the side chain of Met257, whereas residues Asp258 and Asn259 extend out of the binding groove. The small side chain of the x-2 residue, Ala254, is in a similar position as the corresponding residue Ser22; however, it lacks the opportunity to engage in a polar contact with Glu182 in 14-3-3. These results emphasize a distinct feature of the N-terminal 14-3-3 binding site, where interactions can be directly regulated through phosphorylation of both Thr24 and Ser22.
Ser22 phosphorylation hinders 14-3-3 interactions in cells
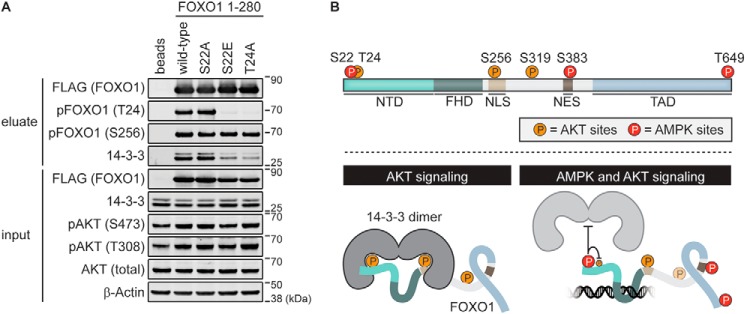
To further investigate the significance of Ser22 and Thr24 phosphorylation regarding 14-3-3 interactions in a cellular context, we used FOXO1 (1–280) constructs (Fig. 1B) bearing either phosphodeficient (S22A and T24A) or phosphomimetic (S22E) mutations for immunoprecipitation in HEK293 cells. Amino acids 1–280 of FOXO1 contained both 14-3-3 interaction motifs, but it was observed that a single point mutation (T24A) in the N-terminal binding site is sufficient to strongly reduce the overall 14-3-3 interactions because of loss of avidity (13). The WT construct was phosphorylated on Thr24 and precipitated, as expected, various isoforms of 14-3-3 proteins (Fig. 4A). Phosphorylation of Thr24 was a consequence of continuous AKT activity under optimal growth conditions, confirmed with pAKT antibodies (Thr308 and Ser473). For our experiments, we selected a HEK293 cell line with low endogenous FOXO1 levels; therefore, we could ensure that the signal of the pFOXO1 (Thr24) antibody represented the phosphorylation status of the ectopically expressed proteins. The T24A mutation that prevented phosphorylation by AKT strongly reduced 14-3-3 interactions, as described previously (13, 36). The S22E mutation reflected an intracellular state of continuous AMPK activity, where Ser22 is phosphorylated, and strikingly showed attenuated 14-3-3 binding comparable with the T24A mutation. The structural data propose that mimicking the Ser22 phosphorylation directly prevents 14-3-3 binding through steric hindrance and electrostatic repulsion in the binding pocket. Moreover, despite active AKT, Thr24 phosphorylation was not detectable, and it seemed that Ser22 phosphorylation additionally controls 14-3-3 binding indirectly by preventing AKT from accessing its phosphorylation site. This is consistent with our in vitro data showing that NTD-pSer22 is a poor AKT substrate. Overall, our results substantiate the central role of the phosphorylation state of Ser22 to control binding of 14-3-3 proteins, thus regulating the transcriptional activity of FOXO1.
Figure 4.
The phosphorylation state of Ser22 regulates binding of 14-3-3 proteins to FOXO1 in HEK293 cells. A, FLAG-FOXO1(1–280)–EGFP fusion proteins were expressed in HEK293 cells for 20 h. After cell lysis, complexes were immunoprecipitated using anti-FLAG-agarose beads and analyzed by Western blotting with the indicated antibodies. Eluates were adjusted according to amounts of bait proteins (anti-FLAG). The T24A construct served as a positive control for a reduced 14-3-3 interaction. The activity of AKT kinase was monitored with pAKT (Ser473/Thr308) antibodies. B, schematic overview of the regulation of transcriptional activity of FOXO1 by the AKT and AMPK signaling pathways. Optimal growth conditions result in active AKT, and phosphorylation of Thr24 as well as Ser256 in FOXO1 stimulates binding of 14-3-3 proteins to inhibit its transcriptional activity. On the other hand, phosphorylation of Ser22 by AMPK is likely to increase the activity of FOXO1 by preventing 14-3-3 binding both directly through steric hindrance and electrostatic repulsion and indirectly by attenuating Thr24 phosphorylation by AKT.
Discussion
FOXO transcription factors orchestrate the expression of genes involved in central cellular processes such as apoptosis, cell cycle progression, and stress resistance. Among the plethora of posttranslational modifications that regulate FOXO1 transcriptional activity, phosphorylation by AKT and the energy-sensing AMPK act in an opposing fashion. AKT-mediated phosphorylation of FOXO1 induces binding of 14-3-3 proteins, its nuclear export, and consequent transcriptional inactivation (13), whereas phosphorylation by AMPK has been shown to increase FOXO1 activity (37). In this work, we expand the interaction network of AKT and AMPK on FOXO1 by demonstrating that AMPK can phosphorylate the NTD of FOXO1 on Ser22 to affect 14-3-3 interactions.
Attenuated binding of 14-3-3 proteins to FOXO1, caused by phosphorylation of Ser22, could be explained by conformational changes within the NTD of FOXO1, steric and electrostatic effects decreasing the binding affinity between FOXO1 and 14-3-3, or a reduction in AKT-mediated phosphorylation of Thr24 in FOXO1.
Conformational switching driven by posttranslational modification of intrinsically disordered proteins has been shown, for example, for Nucleophosmin (NPM1), where successive multiple phosphorylations induce a change from an ordered pentamer to a disordered monomeric state to control its function (30). Another example includes 4EBP2, where phosphorylation by ERK2 induces a conformational change from a disordered to a folded structure to form a regulatory switch (38). Our NMR results showed that no conformational changes occur in the NTD of FOXO1 by Ser22 or Thr24 phosphorylation and exclude this as a probable cause to modulate 14-3-3 binding.
Another possibility could be that phosphorylation of Ser22 indirectly prevents 14-3-3 interactions by diminishing Thr24 phosphorylation by AKT. In a cellular context, we observed that AKT was unable to phosphorylate Thr24 in the phosphomimetic S22E FOXO1, whereas the phosphodeficient S22A as well as WT FOXO1 were unaffected. Moreover, time-resolved 2D NMR and measuring the Michaelis–Menten kinetics also showed that pSer22 impedes Thr24 phosphorylation by AKT. Interestingly, the crystal structure of AKT with bound substrate (39) suggests that a phosphoserine in the −2 position of the AKT phosphorylation site would be unfavorable electrostatically, leading to steric hindrance and obstructing binding of AKT. A similar sensitivity to mutations in the AKT binding motif was observed in patients with diffuse large B cell lymphoma (36). It was shown that the presence of point mutations at Arg19 and Arg21 in the NTD of FOXO1 diminished phosphorylation of Thr24 by AKT, as we observed for pSer22. Together, this emphasizes that phosphorylation of Ser22 renders the NTD a poor substrate for AKT and diminishes Thr24 phosphorylation. This, in turn, will reduce 14-3-3 interactions and activate FOXO1.
Besides the effect on phosphorylation of Thr24 by AKT, we observed that phosphorylation of Ser22 directly reduces the affinity for the ζ and θ isoforms of 14-3-3 in SPR measurements. Our structure of 14-3-3 σ and a pT24 FOXO1 peptide further showed that phosphorylation of Ser22 at the −2 position of the canonically phosphorylated serine or threonine in the 14-3-3 motif would lead to both electrostatic repulsion and steric hindrance. Comparable alleviation of 14-3-3 binding through phosphorylation at the same position within the binding surface has been reported previously for RGS18 (40) and kinesin-14 (41). The effect of steric hindrance at the −2 position is additionally illustrated by a mutation in C-Raf, where a mutation of serine to a leucine (S257L) reduces 14-3-3 binding (42). Together, the phosphorylation status of Ser22 will translate into regulatory effects on 14-3-3 binding through steric and electrostatic forces in the binding pocket.
A sequence alignment of the human FOXO proteins shows full conservation of the serine corresponding to Ser22 in FOXO1 within the N-terminal 14-3-3 recognition motif (Fig. S8B). This offers the possibility that other FOXO family members might be subjected to a similar modulation of 14-3-3 binding through phosphorylation of the conserved serine. However, the kinases involved could vary between different FOXO family members because of sequence differences in the proximity of the phosphorylation site. AMPK is capable to recognize promiscuous substrates, but a hydrophobic residue N-terminal of its phosphorylation site is a core feature in the recognition motif. Strikingly, this hydrophobic residue in position −7 (Leu17 in FOXO1) is only present in FOXO1, whereas FOXO3, FOXO4, and FOXO6 have a polar glutamine residue in the corresponding position that is likely to prevent AMPK binding (Fig. S8B). In fact, upon inhibition of AMPK, decreased transcriptional activity of FOXO1 was observed, whereas the activity of FOXO3 was unaffected (19). Whether AKT-mediated binding of 14-3-3 proteins can be modified through cross-talk between diverse kinases in different FOXO proteins awaits further analysis. The key components of both PI3K/AKT and AMPK signaling pathways are well-conserved throughout evolution. Interestingly, sequence alignments of different FOXO1 homologs between various species suggest that the additional regulation of 14-3-3 binding by AMPK might have evolved in vertebrates, as indicated by a conserved AMPK recognition motif. In Drosophila melanogaster, two residues of the AMPK recognition motif are substituted with amino acids that contain different chemical properties. This makes it unlikely that dFoxO is subjected to a comparable regulation by AMPK. Finally, the direct impairment of 14-3-3 binding through phosphorylation at the −2 position by specific kinases may be a general regulation mechanism among 14-3-3 substrates because a bioinformatic analysis revealed a serine as the most prevalent residue at this position (34).
AMPK and AKT pathways are heavily intertwined by negative feedback loops. This makes it challenging to quantify the specific contribution of Ser22 phosphorylation regarding FOXO1 regulation in a cellular context. Furthermore, interpretation of data and literature is complicated, as tool compounds used to influence AMPK activity (i.e. resveratrol and AICAR) can lead to secondary effects. Cell- and tissue-related differences in signaling pathways, and particularly AMPK and AKT activity, further hamper interpretation of the data. Even though AMPK and AKT act antagonistically on each other and FOXO1, cellular conditions have been described where both kinases are simultaneously active (43–45). Zou et al. (45) suggested that metformin activates FOXO1 via AMPK signaling through impeding phosphorylation by AKT and concomitantly increasing its nuclear localization. Although phosphorylation of FOXO1 was only analyzed on Ser256 and not Thr24 in that study, the mechanism where phosphorylation of Ser22 by AMPK leads to loss of 14-3-3 interactions could in part explain the described nuclear retention of FOXO1. Another report described enhanced FOXO1 transcriptional activity after H2O2 treatment, which is known to activate AMPK. Paradoxically, the increased transcriptional activity was accompanied by a higher level of AKT-phosphorylated pFOXO1 (46). This could be a consequence of increased AMPK activity under these conditions that might overrule the AKT-mediated export of FOXO1 from the nucleus to the cytoplasm. Although further studies are required to fully understand the observed cross-talk between AMPK and AKT signaling to regulate FOXO1 activity as well as the relevant cellular context, we demonstrate a hierarchy in its kinase regulation. Thereby, AKT and AMPK pathways directly integrate on FOXO1 with the potential to fine-tune its transcriptional activity through interactions with 14-3-3 proteins.
Experimental procedures
Vector construction
FOXO1 constructs were synthesized by GeneArt and codon-optimized for expression in respective host cells. FOXO1 constructs (NTDΔ and NTD) contained an N-terminal 6× His-Asn tag as well as a tobacco etch virus protease cleavage site for purification of recombinant proteins and were cloned into a pET-24 vector. FOXO1 (1–280) constructs (WT, S22A, S22E, and T24A) that were used in cell experiments carried an N-terminal 3×FLAG-tag as well as C-terminal enhanced GFP and were cloned into a pcDNA3.1 vector.
Expression and purification of FOXO1 proteins
NTDΔ and NTD were expressed in Escherichia coli BL21 Star (DE3). Terrific broth (TB) medium was supplemented with kanamycin (100 mg/l), 3 mm MgCl2, 0.8% glycerol, 0.02% glucose, and 0.2% lactose for autoinduction (47). The cells were grown at 37 °C for 4 h, followed by 20-h growth at 20 °C before lysis in HEPES buffer (pH 7.4), 150 mm NaCl, 1 mm TCEP, and 1× protease inhibitors (Roche, 26733200) using a cell disruptor at 25 kilopound per square inch (ksi) (Constant Systems). The protein was captured on a HisTrap FF column (GE Healthcare) and subsequently eluted in increasing imidazole concentrations (10–400 mm). The protein was further purified on a size exclusion S75 column (GE Healthcare) that was pre-equilibrated in 20 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1 mm TCEP, and 1× protease inhibitors.
The double-isotopically labeled protein (13C/15N) was expressed in E. coli BL21 Star (DE3) and grown in TB medium to an optical density of 2. Then cells were harvested by centrifugation at 4500 × g for 5 min and resuspended in half of the volume of optimized high cell density minimal medium (48) supplemented with 15NH4Cl (Cambridge Isotope Lab, NLM-467-PK) and 13C glucose (Cambridge Isotope Lab, CLM-1396-PK). After 1 h of adaptation at 37 °C, the temperature was lowered to 20 °C, and protein expression was induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside for 16–48 h. Labeled proteins were purified as described above, with exception of the final buffer that was for the NTD (50 mm NaPi (pH 6.8), 120 mm NaCl, 1 mm TCEP, and 10% D2O) and for NTDΔ (20 mm HEPES (pH 6.7), 100 mm NaCl, 1 mm TCEP, and 10% D2O).
Peptides
EPLPRPRSCTWPLPRPE, EPLPRPR(pS)CTWPLPRPE (pSer22), and EPLPRPRSC(pT)WPLPRPE (pThr24) were purchased from Cambridge Research Biochemicals and diluted in H2O. AKT1-PKB-α S473D (amino acids 118–480, referred to as AKT) and 14-3-3 proteins (isoforms θ and ζ) were produced by the Reagents and Services Division of the MRC Protein Phosphorylation and Ubiquitylation Unit of the University of Dundee and stored at −80 °C. AMPK (His6-α2, S108A-β2, γ1) was expressed and purified as described previously (18) and used at a stock concentration of 6 mg/ml in 120 mm Tris-HCl (pH 8.0), 200 mm NaCl, and 10% glycerol. The ADP biosensor 5-ATR-ParM (tetramethylrhodamine-labeled ParM protein) was expressed, purified, and labeled as described previously (49).
Phosphorylation of FOXO1
The NTD of FOXO1 in 20 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1 mm TCEP, 1× protease inhibitor (Roche, 26733200), and 0.1 mm Na3VO4 was supplemented with AKT or AMPK to a final concentration of 0.19 μm and 0.33 μm, respectively. For sequential phosphorylation of FOXO1, AKT was added overnight before the reaction could proceed with AMPK for one additional night at 20 °C. Prior to surface plasmon resonance experiments, the FOXO1 preparations were buffer-exchanged to 10 mm HEPES (pH 7.4), 350 mm NaCl, 1 mm TCEP, and 0.05% (v/v) Tween 20 using a NAP-10 column. Samples were separated with Novex 4–12% BisTris gels (Invitrogen) in 1× MES running buffer, and phosphorylation was assessed by staining with Pro-Q Diamond Phosphoprotein Gel Stain (Invitrogen) followed by SYPRO Ruby Gel Stain (Invitrogen) and Phos-tag gels (Wako Pure Chemical Industries Ltd.) stained with Instant Blue (Expedeon). Further confirmation of phosphorylation was performed by MS, where the molecular mass of each FOXO1 sample was measured by Electrospray Ionisation-Mass Spectrometry (ESI-MS) micromass ZQ analysis (Waters). Samples were diluted in an equal volume of buffer A (0.1% formic acid and 3% acetonitrile) and separated over a C4 column (Dionex) in a chromatography from 100% buffer A to 100% buffer B (0.1% formic acid and 90% acetonitrile). Mass spectroscopic data were analyzed with the MassLynx software.
Surface plasmon resonance
Experiments were performed on a Biacore 3000 (GE Healthcare) and either 14-3-3 θ or ζ were covalently immobilized on a low-density CM5 chip (Xantec) using standard amine-coupling procedures (1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC)/N-hydroxysulfosuccinimide (NHS) activation) with proteins in acetate buffer (pH 5.5). The various FOXO1 preparations were injected at a concentration of 330 μm to 17 nm with 10 3× dilution steps. The resonance units (RU) at steady state were recorded, and data were adjusted for nonspecific binding observed with unphosphorylated FOXO1. Steady-state data were fitted to a model with Rmax fixed at 600 RU or 400 RU for 14-3-3 θ or ζ, respectively, using nonlinear regression in Prism (GraphPad). The kinetic parameters Kon and Koff of the SPR raw data were extracted using curve fitting in the BiaEvaluation 4.1 software.
NMR
Measurements were recorded on an 800-MHz Bruker Avanze III spectrometer equipped with a cryogenic probe (Bruker) at 20 °C with 330-μl sample in medium-walled tubes. Spectral assignment was obtained by using 2D 1H-15N Selective Optimized Flip-Angle Short-Transient heteronuclear multiple quantum coherence (SOFAST-HMQC) HMQC and a standard Bruker pulse sequence for triple resonance spectra 3D 1H-15N-13C HNCαCβ, HNCα, and HNCoCαCβ. All spectra were processed in TopSpin (Bruker), and Collaborative Computational Project for NMR (CCPNMR) (50) was used for sequential assignment of chemical shifts.
To follow the phosphorylation of the NTD by AKT or AMPK, a series of 2D 1H-15N SOFAST HMQC experiments were used for either 100 μm uniformly labeled 13C-15N NTD or 400 μm uniformly labeled 13C-15N NTDΔ. After complete phosphorylation, a 3D HNCαCβ spectrum was recorded for assignment, followed by addition of the second kinase to obtain the other series of 2D 1H-15N SOFAST HMQC measurements. At completion of the phosphorylation, or after 24 h, a 3D HNCαCβ spectrum was recorded for assignment of chemical shifts. 15N-13C Hα(Cα)N NMR spectra were recorded to assess conformation of prolines in the NTD, and Hα/N peaks were identified. The integrity of all proteins was monitored by SDS-PAGE after NMR analysis to assure that no proteolytic cleavage occurred during the measurements.
Secondary structure propensity analysis
The secondary chemical shifts (Δδ) were calculated as Δδ = δ − δrc, where δ is the observed chemical shift, and δrc is the random coil chemical shift (27, 51). The presented SSP score is a weighted average of the Δδ shifts over five residues. Negative SSP values indicate a preference for the β-strand or extended structure, whereas positive values indicate an α-helical propensity. A weighted SSP score of 1 represents a full secondary structure. Prolines as well as residues preceding prolines were excluded from analysis because prolines do not have the backbone amide proton. This makes them invisible in 2D 1H-15N HMQC, and they strongly affect the chemical shifts of flanking residues. The phosphorylated residues are not part of the reference database; hence, a correct comparison could not be made. These residues were excluded from the SSP analysis.
Biochemical enzymatic assay
The ADP-sensing 5-ATR-ParM (49) was used to measure the kinase activity of AKT and AMPK. The assay was carried out in 30 mm Tris-HCl (pH 7.5), 25 mm KCl, and 10 mm MgCl2 buffer supplemented with fresh 1 mm TCEP, 5 μm BSA, and 1× protease inhibitors (Roche, 26733200) on the day of the experiment. FOXO1 peptides were dissolved in H2O to a concentration of 20 mm. The assay was performed with a total assay volume of 12 μl in a microtiter plate (Greiner, 784900). Under the assay conditions, the biosensor showed a linear response to ADP in the range of 1–400 μm ADP (Ultra-Pure, Promega). First, 0.05 μm 5-ATR-ParM, 1 mm ATP, and varying concentrations of FOXO1 peptide (0.5 mm to 0.25 μm) were diluted in running buffer, centrifuged at 200 × g for 1 min, and incubated on ice. After 1 h, enzyme was added as well as AKT (5 nm to 200 nm) for the NTD or NTD-pSer22 or AMPK (5 nm to 25 nm) for the NTD or NTD-pThr24. Substrates were pre-equilibrated. The plates were centrifuged at 200 × g for 1 min, followed by kinetics measurements. The accumulation of ADP was assessed as an increased fluorescent signal with excitation at 540 nm and an emission filter at 590 nm using a PheraStar microplate reader (BMG Labtech). Data were recorded with an interval of 120 s for 1.5 h and analyzed with the MARS software (BMG Labtech). The initial linear velocity was extracted and plotted against substrate concentration in Prism (GraphPad). The initial velocity was normalized to the maximum initial velocity, except for AKT, with NTD-pSer22. No activity could be measured; therefore, data were scaled using normalization parameters from the measurement of AKT with the NTD. All experiments were performed in triplicate at least three separate times. Data were analyzed using a nonlinear regression Michaelis–Menten model. A Km for one run in a single experiment (AKT with the NTD, third of four runs) was flagged as an outlier by a Grubb's outlier test and excluded from further analysis. Differences in Km values were tested for significance with one-way analysis of variance. The Km shows a significant difference between AMPK with NTD and NTD-pThr24 as substrates (p = 0.0016); the Km differences are nonsignificant between AKT NTD and AMPK NTD or NTD-pThr24 (p = 0.0795 and p = 0,1592, respectively). The Km value could not be determined for NTD-pSer22 with AKT.
Crystallography
The 14-3-3σΔC protein (C-terminally truncated 14-3-3 σ, amino acids 1–231) was mixed in a 1:1 ratio with either the FOXO1-pThr24 or the FOXO1-pSer256 peptide to a final concentration of 12 mg/ml of protein in buffer (20 mm HEPES (pH 7.5), 2 mm MgCl2, and 2 mm β-mercaptoethanol). The 14-3-3σΔC/FOXO1-pThr24 mixture was crystallized using the sitting drop method in a 1:1 ratio of 14-3-3σΔC/FOXO1-pThr24 complex to crystallization buffer (0.1 m BisTris propane (pH 7), 0.2 m sodium nitrate, 28% (v/v) PEG 400, and 5% (w/v) glycerol) and incubated at 4 °C. The 14-3-3σΔC/FOXO1-pSer256 mixture was crystallized using the sitting drop method in a 1:1 ratio of 14-3-3σΔC/FOXO1-pSer256 complex to crystallization buffer (0.1 m BisTris propane (pH 7), 0.2 m sodium citrate tribasic dehydrate, 20% (w/v) PEG 3350, and 10% (w/v) glycerol) and incubated at 4 °C.
Diffraction data for the crystals were collected at the synchrotron (PETRA III, Hamburg, Germany) and processed with Diffraction Integration for Advanced Light Sources (DIALS) using xia2. The structures were solved by molecular replacement with a previously solved 14-3-3/peptide complex (PDB code 3MHR), using molrep for the 14-3-3σΔC/FOXO1-pThr24 structure and Phaser-MR for the 14-3-3σΔC/FOXO1-pSer256 structure. Model building and refinement of both structures were performed using Coot, REFMAC5, and phenix.refine.
Cell culture and plasmid DNA transfection
HEK293 cells were free of mycoplasma and authenticated by STR DNA profiling analysis. Cells were grown in DMEM (Gibco, 31966) supplemented with 10% FBS (Gibco, 10270-106) without antibiotics at 37 °C in 5% CO2. Plasmid DNA was transfected with FuGENE HD transfection reagent (Promega) according to the manufacturer's protocol.
FLAG immunoprecipitation
Anti-FLAG M2-agarose affinity gel (A2220) was purchased from Sigma-Aldrich, and immunoprecipitations were performed according to the manufacturer's instructions. In brief, HEK293 cells were transfected in 10-cm dishes with 10 μg of FLAG-tagged fusion proteins. 20 h after transfection, cells were washed with ice-cold PBS and resuspended in 1 ml of lysis buffer (20 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1% Triton X-100 (v/v), 2 mm EDTA, 25 mm NaF, 1 mm Na3VO4, 1× cOmplete protease inhibitor mixture (Roche, 26733200)). After incubation on ice for 20 min, lysates were sonicated and cleared by centrifugation (12,000 × g for 10 min) before cell extracts were incubated with anti-FLAG M2 beads for 90 min while rotating. Beads were washed with 20 mm Tris-HCl (pH 7.4), 150 mm NaCl and protein complexes were eluted with SDS sample buffer (without DTT). DTT was added to 50 mm final concentration before SDS-PAGE. All steps were performed at 4 °C.
Western blotting and antibodies
Equal amounts of proteins were separated on 4%–12% BisTris gels (Invitrogen) in MES buffer and transferred to nitrocellulose membranes with a Trans-Blot Turbo system (Bio-Rad).
pFOXO1-Thr24 (9464), pFOXO1-Ser256 (9461), 14-3-3 (8312), AKT (2938), pAKT-Ser473 (4060), and pAKT-Thr308 (13038) antibodies were purchased from Cell Signaling Technology. β-Actin (A1978) and FLAG (F3165) antibodies were from Sigma-Aldrich. IRDye secondary antibodies for Western blotting were from LI-COR, and membranes were scanned using an Odyssey CLx imaging system (LI-COR).
Author contributions
M. S., L. B., T. J., T. N., C. O., and A. S. conceptualization; M. S., L. B., M. W., R. L., A. G., T. J., and A. S. investigation; M. S., L. B., M. W., R. L., and A. S. writing-original draft; A. G., T. J., T. N., and C. O. writing-review and editing; C. O. and A. S. supervision; A. S. funding acquisition.
Supplementary Material
Acknowledgments
We thank Rachel Rowlinson, Anna Hoyle, and Melanie Snow for help with LC-MS/MS (Discovery Sciences, R&D, AstraZeneca, UK); Puneet Ahuja for NMR setup (Discovery Sciences, R&D, AstraZeneca, Sweden); Mats Ormö for support with the ADP biosensor assay (Discovery Sciences, R&D, AstraZeneca, Sweden); and Ryan Guillbert for providing the ParM protein (Discovery Sciences, R&D, AstraZeneca, UK).
L. B., A. G., T. N., and A. S. are employees and shareholders of AstraZeneca. T. J. is an employee and shareholder of Nuevolution.
This article contains Figs. S1–S9.
The atomic coordinates and structure factors (codes 6QZR and 6QZS) have been deposited in the Protein Data Bank (http://wwpdb.org/).
The NMR data reported in this paper have been submitted to the BMRB under accession numbers 27336 (NTDΔ), 27337 (NTDΔ-pSer22), and 27320 (NTDΔ-pThr24).
- FOXO
- forkhead box O
- FKHR
- forkhead in rhabdomyosarcoma
- AMPK
- AMP-activated protein kinase
- FHD
- forkhead box domain
- TAD
- transcription activation domain
- NTD
- N-terminal domain
- SSP
- secondary structure propensity
- SPR
- surface plasmon resonance
- TCEP
- tris(2-carboxyethyl)phosphine
- HMQC
- Heteronuclear Multiple Quantum Coherence
- RU
- resonance units
- SOFAST-HMQC
- Selective Optimized Flip-Angle Short-Transient heteronuclear multiple quantum coherence.
References
- 1. Martins R., Lithgow G. J., and Link W. (2016) Long live FOXO: unraveling the role of FOXO proteins in aging and longevity. Aging Cell 15, 196–207 10.1111/acel.12427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hosaka T., Biggs W. H. 3rd, Tieu D., Boyer A. D., Varki N. M., Cavenee W. K., and Arden K. C. (2004) Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc. Natl. Acad. Sci. U.S.A. 101, 2975–2980 10.1073/pnas.0400093101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Klotz L. O., Sanchez-Ramos C., Prieto-Arroyo I., Urbanek P., Steinbrenner H., and Monsalve M. (2015) Redox regulation of FoxO transcription factors. Redox Biol. 6, 51–72 10.1016/j.redox.2015.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhao Y., Wang Y., and Zhu W. G. (2011) Applications of post-translational modifications of FoxO family proteins in biological functions. J. Mol. Cell Biol. 3, 276–282 10.1093/jmcb/mjr013 [DOI] [PubMed] [Google Scholar]
- 5. Rena G., Guo S., Cichy S. C., Unterman T. G., and Cohen P. (1999) Phosphorylation of the transcription factor forkhead family member FKHR by protein kinase B. J. Biol. Chem. 274, 17179–17183 10.1074/jbc.274.24.17179 [DOI] [PubMed] [Google Scholar]
- 6. Zhou J., Li H., Li X., Zhang G., Niu Y., Yuan Z., Herrup K., Zhang Y. W., Bu G., Xu H., and Zhang J. (2015) The roles of Cdk5-mediated subcellular localization of FOXO1 in neuronal death. J. Neurosci. 35, 2624–2635 10.1523/JNEUROSCI.3051-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee J. W., Chen H., Pullikotil P., and Quon M. J. (2011) Protein kinase A-α directly phosphorylates FoxO1 in vascular endothelial cells to regulate expression of vascular cellular adhesion molecule-1 mRNA. J. Biol. Chem. 286, 6423–6432 10.1074/jbc.M110.180661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huang H., Regan K. M., Lou Z., Chen J., and Tindall D. J. (2006) CDK2-dependent phosphorylation of FOXO1 as an apoptotic response to DNA damage. Science 314, 294–297 10.1126/science.1130512 [DOI] [PubMed] [Google Scholar]
- 9. Yuan Z., Becker E. B., Merlo P., Yamada T., DiBacco S., Konishi Y., Schaefer E. M., and Bonni A. (2008) Activation of FOXO1 by Cdk1 in cycling cells and postmitotic neurons. Science 319, 1665–1668 10.1126/science.1152337 [DOI] [PubMed] [Google Scholar]
- 10. Rena G., Woods Y. L., Prescott A. R., Peggie M., Unterman T. G., Williams M. R., and Cohen P. (2002) Two novel phosphorylation sites on FKHR that are critical for its nuclear exclusion. EMBO J. 21, 2263–2271 10.1093/emboj/21.9.2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brunet A., Bonni A., Zigmond M. J., Lin M. Z., Juo P., Hu L. S., Anderson M. J., Arden K. C., Blenis J., and Greenberg M. E. (1999) Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96, 857–868 10.1016/S0092-8674(00)80595-4 [DOI] [PubMed] [Google Scholar]
- 12. Tang E. D., Nuñez G., Barr F. G., and Guan K. L. (1999) Negative regulation of the forkhead transcription factor FKHR by Akt. J. Biol. Chem. 274, 16741–16746 10.1074/jbc.274.24.16741 [DOI] [PubMed] [Google Scholar]
- 13. Rena G., Prescott A. R., Guo S., Cohen P., and Unterman T. G. (2001) Roles of the forkhead in rhabdomyosarcoma (FKHR) phosphorylation sites in regulating 14-3-3 binding, transactivation and nuclear targeting. Biochem. J. 354, 605–612 10.1042/0264-6021:3540605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Matsuzaki H., Daitoku H., Hatta M., Tanaka K., and Fukamizu A. (2003) Insulin-induced phosphorylation of FKHR (Foxo1) targets to proteasomal degradation. Proc. Natl. Acad. Sci. U.S.A. 100, 11285–11290 10.1073/pnas.1934283100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yun H., Park S., Kim M. J., Yang W. K., Im D. U., Yang K. R., Hong J., Choe W., Kang I., Kim S. S., and Ha J. (2014) AMP-activated protein kinase mediates the antioxidant effects of resveratrol through regulation of the transcription factor FoxO1. FEBS J. 281, 4421–4438 10.1111/febs.12949 [DOI] [PubMed] [Google Scholar]
- 16. Yuan Z., Lehtinen M. K., Merlo P., Villén J., Gygi S., and Bonni A. (2009) Regulation of Neuronal Cell Death by MST1-FOXO1 Signaling. J. Biol. Chem. 284, 11285–11292 10.1074/jbc.M900461200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Asada S., Daitoku H., Matsuzaki H., Saito T., Sudo T., Mukai H., Iwashita S., Kako K., Kishi T., Kasuya Y., and Fukamizu A. (2007) Mitogen-activated protein kinases, Erk and p38, phosphorylate and regulate Foxo1. Cell. Signal. 19, 519–527 [DOI] [PubMed] [Google Scholar]
- 18. Xiao B., Sanders M. J., Carmena D., Bright N. J., Haire L. F., Underwood E., Patel B. R., Heath R. B., Walker P. A., Hallen S., Giordanetto F., Martin S. R., Carling D., and Gamblin S. J. (2013) Structural basis of AMPK regulation by small molecule activators. Nat. Commun. 4, 3017 10.1038/ncomms4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Awad H., Nolette N., Hinton M., and Dakshinamurti S. (2014) AMPK and FoxO1 regulate catalase expression in hypoxic pulmonary arterial smooth muscle. Pediatr. Pulmonol. 49, 885–897 10.1002/ppul.22919 [DOI] [PubMed] [Google Scholar]
- 20. Chen B. L., Ma Y. D., Meng R. S., Xiong Z. J., Wang H. N., Zeng J. Y., Liu C., and Dong Y. G. (2010) Activation of AMPK inhibits cardiomyocyte hypertrophy by modulating of the FOXO1/MuRF1 signaling pathway in vitro. Acta Pharmacol. Sin. 31, 798–804 10.1038/aps.2010.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee K., Ochi E., Song H., and Nakazato K. (2015) Activation of AMP-activated protein kinase induce expression of FoxO1, FoxO3a, and myostatin after exercise-induced muscle damage. Biochem. Biophys. Res. Commun. 466, 289–294 [DOI] [PubMed] [Google Scholar]
- 22. Greer E. L., Dowlatshahi D., Banko M. R., Villen J., Hoang K., Blanchard D., Gygi S. P., and Brunet A. (2007) An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr. Biol. 17, 1646–1656 10.1016/j.cub.2007.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xue B., Dunbrack R. L., Williams R. W., Dunker A. K., and Uversky V. N. (2010) PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta 1804, 996–1010 10.1016/j.bbapap.2010.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Obradovic Z., Peng K., Vucetic S., Radivojac P., Brown C. J., and Dunker A. K. (2003) Predicting intrinsic disorder from amino acid sequence. Proteins 53, 566–572 10.1002/prot.10532 [DOI] [PubMed] [Google Scholar]
- 25. Brent M. M., Anand R., and Marmorstein R. (2008) Structural basis for DNA recognition by FoxO1 and its regulation by posttranslational modification. Structure 16, 1407–1416 10.1016/j.str.2008.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Singh P., Han E. H., Endrizzi J. A., O'Brien R. M., and Chi Y.-I. (2017) Crystal structures reveal a new and novel FoxO1 binding site within the human glucose-6-phosphatase catalytic subunit 1 gene promoter. J. Struct. Biol. 198, 54–64 10.1016/j.jsb.2017.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Marsh J. A., Singh V. K., Jia Z., and Forman-Kay J. D. (2006) Sensitivity of secondary structure propensities to sequence differences between α- and γ-synuclein: implications for fibrillation. Protein Sci. 15, 2795–2804 10.1110/ps.062465306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Selenko P., Frueh D. P., Elsaesser S. J., Haas W., Gygi S. P., and Wagner G. (2008) In situ observation of protein phosphorylation by high-resolution NMR spectroscopy. Nat. Struct. Mol. Biol. 15, 321–329 10.1038/nsmb.1395 [DOI] [PubMed] [Google Scholar]
- 29. Reimer U., and Fischer G. (2002) Local structural changes caused by peptidyl-prolyl cis/trans isomerization in the native state of proteins. Biophys. Chem. 96, 203–212 10.1016/S0301-4622(02)00013-3 [DOI] [PubMed] [Google Scholar]
- 30. Mitrea D. M., Grace C. R., Buljan M., Yun M.-K., Pytel N. J., Satumba J., Nourse A., Park C.-G., Madan Babu M., White S. W., and Kriwacki R. W. (2014) Structural polymorphism in the N-terminal oligomerization domain of NPM1. Proc. Natl. Acad. Sci. U.S.A. 111, 4466–4471 10.1073/pnas.1321007111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hardie D. G. (2011) AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes Dev. 25, 1895–1908 10.1101/gad.17420111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marin T. L., Gongol B., Martin M., King S. J., Smith L., Johnson D. A., Subramaniam S., Chien S., and Shyy J. Y. (2015) Identification of AMP-activated protein kinase targets by a consensus sequence search of the proteome. BMC Systems Biol. 9, 13 10.1186/s12918-015-0156-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Safaei J., Maňuch J., Gupta A., Stacho L., and Pelech S. (2011) Prediction of 492 human protein kinase substrate specificities. Proteome Sci. 9, S6 10.1186/1477-5956-9-S1-S6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Johnson C., Crowther S., Stafford M. J., Campbell D. G., Toth R., and MacKintosh C. (2010) Bioinformatic and experimental survey of 14-3-3-binding sites. Biochem. J. 427, 69–78 10.1042/BJ20091834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Würtele M., Jelich-Ottmann C., Wittinghofer A., and Oecking C. (2003) Structural view of a fungal toxin acting on a 14-3-3 regulatory complex. EMBO J. 22, 987–994 10.1093/emboj/cdg104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Trinh D. L., Scott D. W., Morin R. D., Mendez-Lago M., An J., Jones S. J., Mungall A. J., Zhao Y., Schein J., Steidl C., Connors J. M., Gascoyne R. D., and Marra M. A. (2013) Analysis of FOXO1 mutations in diffuse large B-cell lymphoma. Blood 121, 3666–3674 10.1182/blood-2013-01-479865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Greer E. L., Banko M. R., and Brunet A. (2009) AMP-activated protein kinase and FoxO transcription factors in dietary restriction-induced longevity. Ann. N.Y. Acad. Sci. 1170, 688–692 10.1111/j.1749-6632.2009.04019.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bah A., Vernon R. M., Siddiqui Z., Krzeminski M., Muhandiram R., Zhao C., Sonenberg N., Kay L. E., and Forman-Kay J. D. (2015) Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature 519, 106–109 10.1038/nature13999 [DOI] [PubMed] [Google Scholar]
- 39. Yang J., Cron P., Good V. M., Thompson V., Hemmings B. A., and Barford D. (2002) Crystal structure of an activated Akt/protein kinase B ternary complex with GSK3-peptide and AMP-PNP. Nat. Struct. Biol. 9, 940–944 10.1038/nsb870 [DOI] [PubMed] [Google Scholar]
- 40. Gegenbauer K., Elia G., Blanco-Fernandez A., and Smolenski A. (2012) Regulator of G-protein signaling 18 integrates activating and inhibitory signaling in platelets. Blood 119, 3799–3807 10.1182/blood-2011-11-390369 [DOI] [PubMed] [Google Scholar]
- 41. Beaven R., Bastos R. N., Spanos C., Romé P., Cullen C. F., Rappsilber J., Giet R., Goshima G., and Ohkura H. (2017) 14-3-3 regulation of Ncd reveals a new mechanism for targeting proteins to the spindle in oocytes. J. Cell Biol. 216, 3029–3039 10.1083/jcb.201704120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Molzan M., Schumacher B., Ottmann C., Baljuls A., Polzien L., Weyand M., Thiel P., Rose R., Rose M., Kuhenne P., Kaiser M., Rapp U. R., Kuhlmann J., and Ottmann C. (2010) Impaired binding of 14-3-3 to C-RAF in Noonan syndrome suggests new approaches in diseases with increased Ras signaling. Mol. Cell. Biol. 30, 4698–4711 10.1128/MCB.01636-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dalle Pezze P., Ruf S., Sonntag A. G., Langelaar-Makkinje M., Hall P., Heberle A. M., Razquin Navas P., van Eunen K., Tölle R. C., Schwarz J. J., Wiese H., Warscheid B., Deitersen J., Stork B., Fäßler E., et al. (2016) A systems study reveals concurrent activation of AMPK and mTOR by amino acids. Nat. Commun. 7, 13254 10.1038/ncomms13254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Leclerc G. M., Leclerc G. J., Fu G., and Barredo J. C. (2010) AMPK-induced activation of Akt by AICAR is mediated by IGF-1R dependent and independent mechanisms in acute lymphoblastic leukemia. J. Mol. Signal. 5, 15–15 10.1186/1750-2187-5-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zou J., Hong L., Luo C., Li Z., Zhu Y., Huang T., Zhang Y., Yuan H., Hu Y., Wen T., Zhuang W., Cai B., Zhang X., Huang J., and Cheng J. (2016) Metformin inhibits estrogen-dependent endometrial cancer cell growth by activating the AMPK–FOXO1 signal pathway. Cancer Sci. 107, 1806–1817 10.1111/cas.13083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ning Y., Li Z., and Qiu Z. (2015) FOXO1 silence aggravates oxidative stress-promoted apoptosis in cardiomyocytes by reducing autophagy. J. Toxicol. Sci. 40, 637–645 10.2131/jts.40.637 [DOI] [PubMed] [Google Scholar]
- 47. Studier F. W. (2005) Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 10.1016/j.pep.2005.01.016 [DOI] [PubMed] [Google Scholar]
- 48. Sivashanmugam A., Murray V., Cui C., Zhang Y., Wang J., and Li Q. (2009) Practical protocols for production of very high yields of recombinant proteins using Escherichia coli. Protein Sci. 18, 936–948 10.1002/pro.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kunzelmann S., and Webb M. R. (2010) A fluorescent, reagentless biosensor for ADP based on tetramethylrhodamine-labeled ParM. ACS Chem. Biol. 5, 415–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vranken W. F., Boucher W., Stevens T. J., Fogh R. H., Pajon A., Llinas M., Ulrich E. L., Markley J. L., Ionides J., and Laue E. D. (2005) The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59, 687–696 10.1002/prot.20449 [DOI] [PubMed] [Google Scholar]
- 51. Rosenlöw J., Isaksson L., Mayzel M., Lengqvist J., and Orekhov V. Y. (2014) Tyrosine phosphorylation within the intrinsically disordered cytosolic domains of the B-cell receptor: an NMR-based structural analysis. PLoS ONE 9, e96199 10.1371/journal.pone.0096199 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.