Abstract
The presence of ribonucleoside monophosphates (rNMPs) in nuclear DNA decreases genome stability. To ensure survival despite rNMP insertions, cells have evolved a complex network of DNA repair mechanisms, in which the ribonucleotide excision repair pathway, initiated by type 2 RNase H (RNase HII/2), plays a major role. We recently demonstrated that eukaryotic RNase H2 cannot repair damage, that is, ribose monophosphate abasic (both apurinic or apyrimidinic) site (rAP) or oxidized rNMP embedded in DNA. Currently, it remains unclear why RNase H2 is unable to repair these modified nucleic acids having either only a sugar moiety or an oxidized base. Here, we compared the endoribonuclease specificity of the RNase HII enzymes from the archaeon Pyrococcus abyssi and the bacterium Escherichia coli, examining their ability to process damaged rNMPs embedded in DNA in vitro. We found that E. coli RNase HII cleaves both rAP and oxidized rNMP sites. In contrast, like the eukaryotic RNase H2, P. abyssi RNase HII did not display any rAP or oxidized rNMP incision activities, even though it recognized them. Notably, the archaeal enzyme was also inactive on a mismatched rNMP, whereas the E. coli enzyme displayed a strong preference for the mispaired rNMP over the paired rNMP in DNA. On the basis of our biochemical findings and also structural modeling analyses of RNase HII/2 proteins from organisms belonging to all three domains of life, we propose that RNases HII/2's dual roles in ribonucleotide excision repair and RNA/DNA hydrolysis result in limited acceptance of modified rNMPs embedded in DNA.
Keywords: ribonuclease, bacteria, Escherichia coli (E. coli), archaea, oxidative stress, abasic-ribose, oxidized-ribonucleotides, Pyrococcus abyssi, Type 2 RNase H
Introduction
A recently recognized type of abundant DNA damage is represented by the incorporation of ribonucleoside monophosphates (rNMPs)3 into genomic DNA (1–7). rNMPs in DNA can impact genome stability in multiple ways (1). Although a helpful effect was hypothesized, in which nicking by RNase H2 at rNMP sites works as a DNA damage signaling event (8, 9), the most established hypothesis considers the presence of rNMPs within the double-helical DNA as harmful for the cells (1, 4, 5, 8). Indeed, it has been demonstrated that the additional hydroxyl group in the 2′ position of the ribose sugar alters and destabilizes the double helix of DNA (10, 11, 4), and its persistent presence can block physiological processes including DNA replication and transcription (1, 12). Several studies have been performed to investigate a putative role of known DNA repair pathways, including nucleotide excision repair (13) and mismatch repair (9, 14), in the removal of rNMPs incorporated in DNA (15). However, to date, only a specific DNA repair pathway, called ribonucleotide excision repair (RER) (16), was shown to have a predominant role in processing single rNMP or short rNMP stretches embedded in DNA. In the RER mechanism, type 2 RNase H proteins cleave the phosphodiester bond at the 5′ side of embedded rNMPs (17, 16). Through metal ion–dependent cleavage, the RNase H2/II processing generates a product bearing a 5′-phosphate and a 3′-OH end that is subsequently repaired by other enzymes, including DNA polymerase, nuclease, and DNA ligase (17, 16). In addition to type 2, RNase H family includes another group called type 1 (18, 14, 7). In mammals, RNase H1 displays incision activity on rNMPs embedded in DNA in a stretch of at least four in series (18). In nonphysiological conditions, in which RER pathway components fail or are genetically inactive, RNase H1 cannot substitute RNase H2 for processing single rNMPs in DNA or tracts shorter than four rNMPs. On the other hand, it was clearly demonstrated how topoisomerase I is able to recognize and cleave single rNMPs embedded in DNA, when RNase H2 is missing (15, 19). Moreover, different studies were carried out to evaluate whether base excision repair (BER) pathway could also work on rNMPs embedded in DNA (9, 12, 15, 20). Our laboratories have recently demonstrated that although BER does not have any role in repairing rNMPs embedded in DNA, apurinic/apyrimidinic endonuclease 1, APE1, the only AP-endonuclease of BER, acts in the removal of modified rNMPs embedded in DNA, such as ribose monophosphate abasic (both apurinic or apyrimidinic) site (rAP) and oxidized rNMP (20). Moreover, contrary to what was expected, rAP and oxidized rNMP sites are not processed by yeast, mouse, and human RNase H2. Considering all these points, using in vitro assays, we evaluated the activity of RNase HII/2 derived from a representative species from all the three domains of life on different rNMP-containing substrates. Although RNase HII/2 is a phylogenetically conserved enzyme, several differences emerge among RNases HII/2 from different organisms during evolution. Structural studies by X-ray crystallography characterized the different structures among bacterial, archaeal, and eukaryotic RNase HII/2 proteins. In both Bacteria and Archaea, RNase HII is a monomeric protein comprising a catalytic core and a small C-terminal helical domain (18). In the hyperthermophilic deep-sea euryarchaeon Pyrococcus abyssi, a single gene, rnh, is present coding for a single enzyme involved in RNA elimination called type 2 RNase H (Pab RNase HII) (21–23). Pab RNase HII works in the resolution of RNA primers at the replication fork and in the repair of single rNMPs embedded in DNA (22, 23). Finally, in Eukarya, RNase H2 is a heterotrimeric protein. Subunit A resembles the bacterial ortholog and comprises the catalytic domain of the whole protein. Moreover, two additional subunits, called B and C, are present (24). Although their functions are still not completely understood, their absence abolishes the catalytic activity of the A subunit (17). In addition, subunit B contains the proliferating cell nuclear antigen–interacting motif (PIP box) that allows recruitment of the enzyme to the replication fork (25). Whether and how the protein structure impacts the protein functions needs further investigation. In this study, we compared the endoribonuclease activities of RNases HII from a bacterial and an archaeon representative species on rAP or oxidized rNMP embedded in DNA. We found that Escherichia coli RNase HII, which is able to cut normal rGMPs embedded in DNA, maintains its endoribonuclease activity on both abasic and oxidized rGMPs present in DNA. On the contrary, P. abyssi RNase HII loses this catalytic specificity. Like the human counterpart, the archaeal RNase HII enzyme exhibits stable substrate binding, although it is unable to process modified rNMPs embedded in DNA. Based on these biochemical data and alignment of amino acid sequences and structures, we speculated that the requirement of the enzymes to recognize rNMPs in DNA and resolve RNA/DNA hybrids limits the optimally recognition process and catalysis of modified rNMPs embedded in DNA.
Results
Bacterial RNase HII cleaves rAP and oxidized rNMP embedded in dsDNA
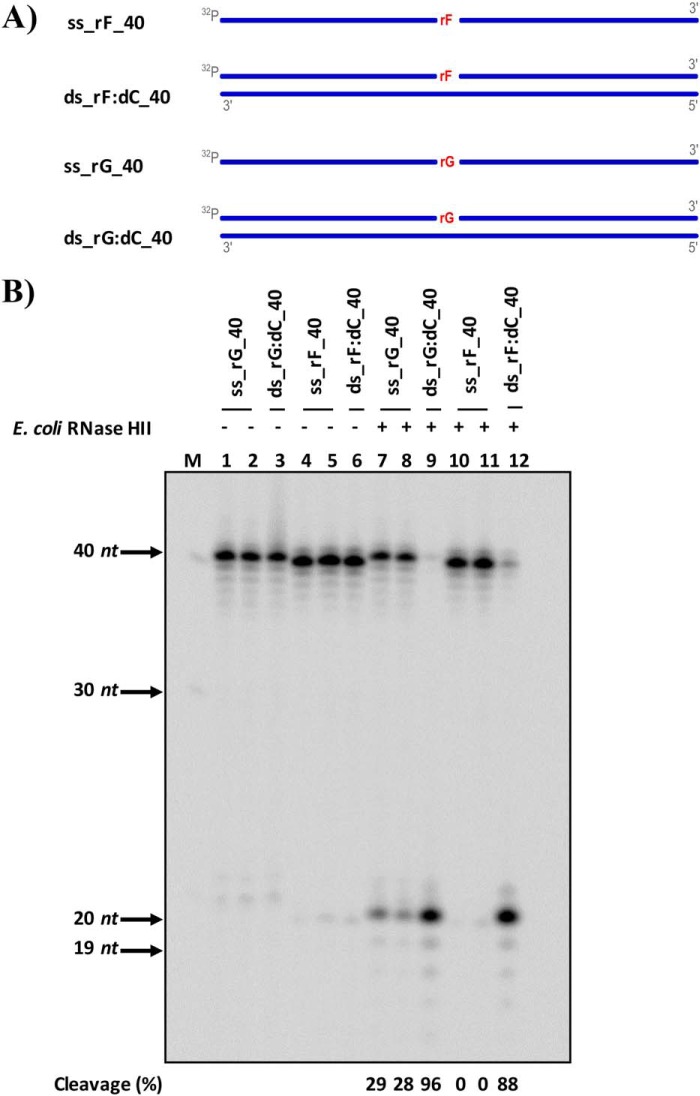
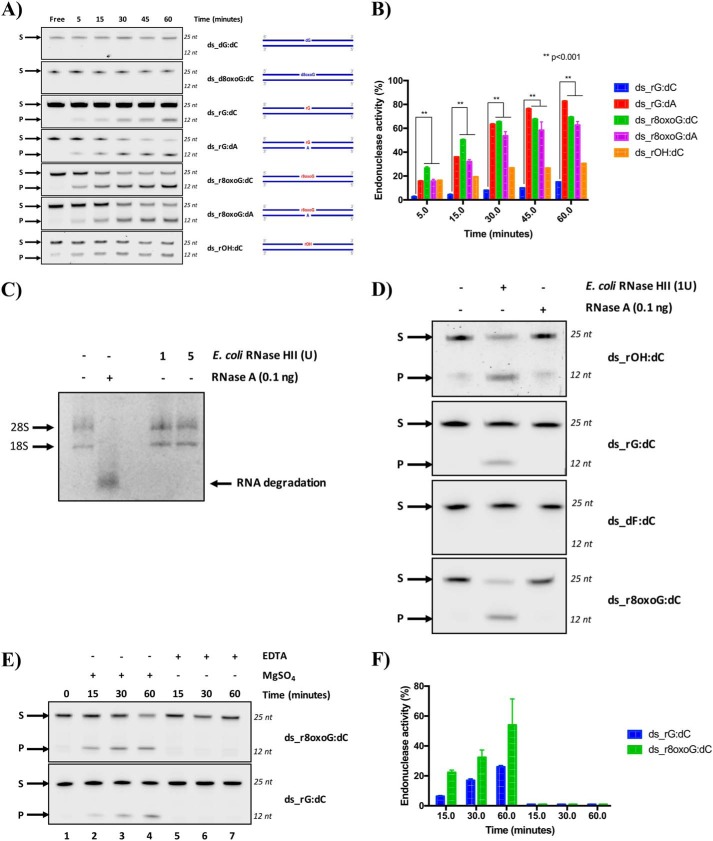
RNase H type 2 is the major enzyme responsible for the removal of unmodified rNMPs embedded in DNA (24); unexpectedly, we recently demonstrated that yeast, mouse, and human RNase H2 enzymes are unable to process rAP or oxidized rNMP embedded in DNA (20). Here, we examined whether RNase HII from E. coli cleaves a rAP embedded in a 40-bp DNA sequence (Fig. 1A). We used single-stranded (ss) or double-stranded (ds) DNA substrates containing either an rGMP (rG) or a THF (rF), analog of the rAP (Fig. 1A and Table 1). Following incubation, denaturing gel electrophoresis was used to visualize the cleavage (Fig. 1B). As expected, E. coli RNase HII cleaves the single rG in the DNA duplex substrate (lane 9 in Fig. 1B). In addition, E. coli RNase HII also cleaves rG when present in the ssDNA substrate, although with less efficiency than the dsDNA substrate (lanes 7 and 8 and lane 9, respectively, in Fig. 1B). Interestingly, E. coli RNase HII cleaves also the rAP in the dsDNA but not in the ssDNA substrate (lanes 12 and lanes 10 and 11, respectively, in Fig. 1B). These results demonstrate that E. coli RNase HII is able to cleave a rAP when this is incorporated in a DNA duplex. Next, we extended our interests on the ability of E. coli RNase HII protein to process a true abasic (ds_rOH:dC), containing an hydroxyl group in C1′ rather than a THF group, and an oxidized rNMP (ds_r8oxoG:dC) within ds oligonucleotides (ODNs) compared with control dsODNs (Fig. 2 and Table 1). Specifically, the control oligonucleotides include the negative controls dGMP- and 8-oxoguanine dNMP–containing ODNs, called ds_dG:dC and ds_d8oxoG:dC, respectively, and the positive control, in which an unmodified rGMP was embedded, called ds_rG:dC (Fig. 2 and Table 1). All modifications are located at the 13th position over a length of 25 ODNs. Moreover, because oxidized G can pair with dC or dA, both rG- and r8oxoG-containing strands, as well as dG control substrate, were annealed with either dC- or dA-containing complementary strands (Fig. 2 and Table 1). To better investigate the cleavage activity of the bacterial RNase HII, each dsODN was first incubated with the recombinant E. coli protein for the indicated times (Fig. 2). Upon reaction completion, denaturing gel electrophoresis was used to quantify the amount of cleaved product (Fig. 2A). Then the percentage of product obtained upon cleavage was calculated and compared among all the ODNs (Fig. 2B). As expected, data obtained showed that bacterial RNase HII efficiently cuts, in a time-dependent manner, its canonical substrate paired with dCMP (ds_rG:dC) and also in a mispaired condition with an dAMP (ds_rG:dA). Under the same conditions, as expected, no effect was observed for RNase HII on the ds_d8oxoG:dC and ds_dG:dC ODNs. Notably, bacterial RNase HII was able to cleave the r8oxoG ODN as efficiently as its positive controls and, compared with the ds_rG:dC, even to a higher extent. In fact, after 60 min of incubation with the enzyme, cleavage of the ds_r8oxoG:dC substrate was 50% higher than that of the ds_rG:dC oligonucleotide. Interestingly, when ds_rG:dC is considered with respect to the ds_rG:dA substrate, the presence of the mismatch with adenine in place of cytosine influenced bacterial RNase HII cleavage capability, which results markedly more active to process the rGMP (Fig. 2). In addition, the efficiency of cleavage of ds_r8oxoG:dA by the bacterial RNase HII was comparable with that of ds_r8oxoG:dC (Fig. 2). Our results also confirmed that E. coli RNase HII is active on rAP embedded in DNA, as observed by the presence of cleavage products accumulating with ds_rOH:dC (see results in Fig. 1). With the purpose of verifying that the observed activity of the E. coli RNase HII was not a consequence of contamination of the commercial enzyme by RNase A, despite having information about the purity of the protein (∼99%), we performed additional control experiments. First, we tested the efficiency of RNase A cleavage on genomic RNA extracted from HeLa cells compared with the activity detectable in the presence of increasing amounts of E. coli RNase HII (Fig. 2C). After 1 h of RNase A incubation with eukaryotic total RNA, in a ratio of [RNase A/genomic RNA] of 2.5*10−4, a complete degradation of RNA was observed. Indeed, the two bands corresponding to the 28S and 18S ribosomal RNAs (rRNAs) were sharply visible when RNase A was not present, whereas a complete rRNA degradation was clearly observed when RNase A was present. On the contrary, E. coli RNase HII treatment did not affect RNA stability at all (Fig. 2C), confirming the absence of RNase A contamination in the E. coli RNase HII samples. Using the same conditions, we compared the cleavage activity, by both RNases, on all the rNMP-containing DNA substrates. As shown in Fig. 2D, whereas E. coli RNase HII cleaved the substrates containing the modified rNMPs, RNase A did not. These data clearly demonstrate the absence of contaminating RNase A having RNase H-like activity in the commercial RNase HII sample, and, more importantly, that the bacterial RNase HII enzyme, but not RNase A, specifically processes lesions including oxidized rNMP and rAP embedded in DNA. It is well-known that the presence of magnesium ion is necessary for promoting the cleavage activity of E. coli RNase HII (20, 26). Here, we investigated the cleavage specificity of the bacterial RNase HII comparing its activity in the presence and absence of magnesium in the reaction buffer. Although in the previous experiments the commercial buffer was used, we prepared a homemade buffer, in which we added or not MgSO4 from the original recipe. As reported in Fig. 2 (E and F), when magnesium is present, the activity of RNase HII is promoted in both unmodified and oxidized rG (lanes 2–4). When the MgSO4 is not added into the reaction buffer and, in addition, any divalent ions are chelated by the presence of EDTA, the RNase HII is inactive on the substrates containing the modified rNMPs (lanes 5–7). Taking all these results together, we demonstrated that E. coli RNase HII, differently from its mammalian and yeast counterparts (20), maintains its ability to process not only single unmodified rNMPs embedded in DNA but also rAP and oxidized rNMP sites in an Mg2+-dependent manner.
Figure 1.
RNase HII from E. coli cleaves rAP embedded in dsDNA. A, scheme of the ss and ds 40-mer substrates used to test the activity of E. coli RNase HII including a ribo abasic site mimicked by a tetrafuran residue indicated with rF and an unmodified rGMP (rG) embedded in DNA. DNA strands are depicted in blue, whereas rNMPs are in red. The 5′ radiolabels, indicated by 32P, at the 5′ are in gray. B, representative denaturing polyacrylamide gel of cleavage result obtained on all substrates without (lanes 1–6) and with (lanes 7–12) E. coli RNase HII protein. The far left lane, marked with M, corresponds to the ssDNA ladder. On the left side of the image, molecular weights are indicated, with arrows to the corresponding bands. Lanes 1 and 7 and lanes 4 and 10 are ss substrates containing rG and rF, respectively; lanes 2 and 8 and lanes 5 and 11 are ss substrates containing rG and rF, respectively, which are cooled slowly at room temperature to observe any self-annealing; lanes 3 and 9 are ds substrates containing rG; and lanes 6 and 12 are ds substrates containing rF. The cleavage percentages of reactions are displayed below the image.
Table 1.
List of ODN sequences
The sequences of the 5′ → 3′ ss ODNs and their complements are here reported. All ODNs are composed by deoxyribonucleotides, identified by “d.” In the middle column, a modified base is highlighted in bold type, and when useful, the type of modification is reported as follows: 8oxo indicates oxidation at eighth position of the guanine, OH indicates an abasic site, and F indicates tetrahydrofuran. Moreover, if the modified base is an rNMP, the modification is preceded by “r.” The name of each ssODN is reported in the left column of the table, resuming the type of modification.
| Sequence (5′ → 3′) | Tm | |
|---|---|---|
| °C | ||
| ss_rG_40 | dAdGdTdGdTdGdGdAdTdGdCdCdGdTdGdAdAdGdArGdAdTdCdAdTdCdAdAdGdGdTdTdAdTdCdGdCdCdTdC | 74.0 |
| ss_rF_40 | dAdGdTdGdTdGdGdAdTdGdCdCdGdTdGdAdAdGdArFdAdTdCdAdTdCdAdAdGdGdTdTdAdTdCdGdCdCdTdC | 67.0 |
| ss_comp_40 | dGdAdGdGdCdGdAdTdAdAdCdCdTdTdGdAdTdGdAdTdTdCdTdTdCdAdCdGdGdCdAdTdCdCdAdCdAdCdT | 67.0 |
| ss_dG | dGdGdAdTdCdCdGdGdTdAdGdTdGdTdTdAdGdGdCdCdTdGdAdAdC | 69.0 |
| ss_rG | dGdGdAdTdCdCdGdGdTdAdGdTrGdTdTdAdGdGdCdCdTdGdAdAdC | 74.0 |
| ss_d8oxoG | dGdGdAdTdCdCdGdGdTdAdGdTd8oxoGdTdTdAdGdGdCdCdTdGdAdAdC | 67.0 |
| ss_r8oxoG | dGdGdAdTdCdCdGdGdTdAdGdTr8oxoGdTdTdAdGdGdCdCdTdGdAdAdC | 67.0 |
| ss_dF | dGdGdAdTdCdCdGdGdTdAdGdTdFdTdTdAdGdGdCdCdTdGdAdAdC | 67.0 |
| ss_rOH | dGdGdAdTdCdCdGdGdTdAdGdTrOHdTdTdAdGdGdCdCdTdGdAdAdC | 64.0 |
| ss_comp_dC | dGdTdTdCdAdGdGdCdCdTdAdAdCdAdCdTdAdCdCdGdGdAdTdCdC | 58.0 |
| ss_comp_dA | dGdTdTdCdAdGdGdCdCdTdAdAdAdAdCdTdAdCdCdGdGdAdTdCdC | 59.0 |
Figure 2.
RNase HII from E. coli exclusively cleaves rAP and oxidized rNMP embedded in DNA. A, representative denaturing polyacrylamide gel of incision by E. coli RNase HII on different 25-mer dsODNs. Endoribonuclease activity of E. coli RNase HII protein was investigated in vitro at different time points using the commercial ThermoPol® buffer as specified under “Experimental procedures.” ds_dG:dC and ds_d8oxoG:dC were used as negative control ODNs, whereas ds_rG:dC was used as a positive control. Moreover, rG- and r8oxoG-containing ODNs were tested in two different pairing conditions: paired with dC or with dA. S denotes substrate position, and P denotes product position. Time points, expressed in minutes, are shown on the top of the figure. Alongside each panel, a schematic representation of each substrate is reported. dsODNs are symbolized as blue lines standing for 25-mer in which the 13th base on the 5′ → 3′ strand is a deoxy- or ribo-modified site paired with a base (dC or dA) on the complementary strand (3′ → 5′). If the modified site is a deoxyribonucleotide (d), it was colored in blue, whereas if this is an rNMP (r), it is colored in red. Lastly, a fluorophore IRDye700, IRDye800, or Cy5 dye labels the top 5′ end of each dsODN. B, graph shows the time course kinetics of E. coli RNase HII incision on each substrate. The activity is reported as a percentage of substrate converted to product at the indicated time. The data are expressed as means ± S.D. of three independent replicas. A p value of < 0.001 is marked with two asterisks (**). C, representative RNase-free agarose gel, in which genomic RNA samples extracted from HeLa cells were run after incubation with both ribonucleases, RNase A and E. coli RNase HII, as explained under “Experimental procedures.” On top of the image, the absence or presence of the RNase enzymes is indicated as − or +, respectively. Specifically, the two different amounts of E. coli RNase HII were indicated on top of the figure, expressed in units (U). 28S and 18S rRNAs are pointed at the left of each corresponding band. RNA degradation upon RNase A treatment is indicated by an arrow on the right side of the figure. D, representative denaturing polyacrylamide gel shows the activities of RNase HII (1 unit) and RNase A (0.1 ng) on ds_rOH:dC, ds_rG:dC, ds_dF:dC, and ds_r8oxoG:dC oligonucleotides after a 1-h incubation at 37 °C. At the left of the panel, S and P indicate the substrate and the product, respectively. E, representative denaturing polyacrylamide gel of time-course kinetics cleavage by a fixed amount of E. coli RNase HII on ds_rG:dC and ds_r8oxoG:dC oligonucleotides under different time points, expressed in minutes and indicated on the top of the figure. The reaction was performed in the homemade ThermoPol® buffer as explained under “Experimental procedures.” The MgSO4 (2 mm) effect was investigated, removing its presence from the buffer. Moreover, in the samples in which the MgSO4 was removed, EDTA (5 mm) was added. S and P correspond to substrate and product, respectively. F, relative graph illustrating the time-course kinetics activity. The data are expressed as means ± S.D. of three independent replicas.
Archaeal RNase HII, though being able to recognize both rAP and oxidized rNMP embedded in dsDNA, is unable to process them
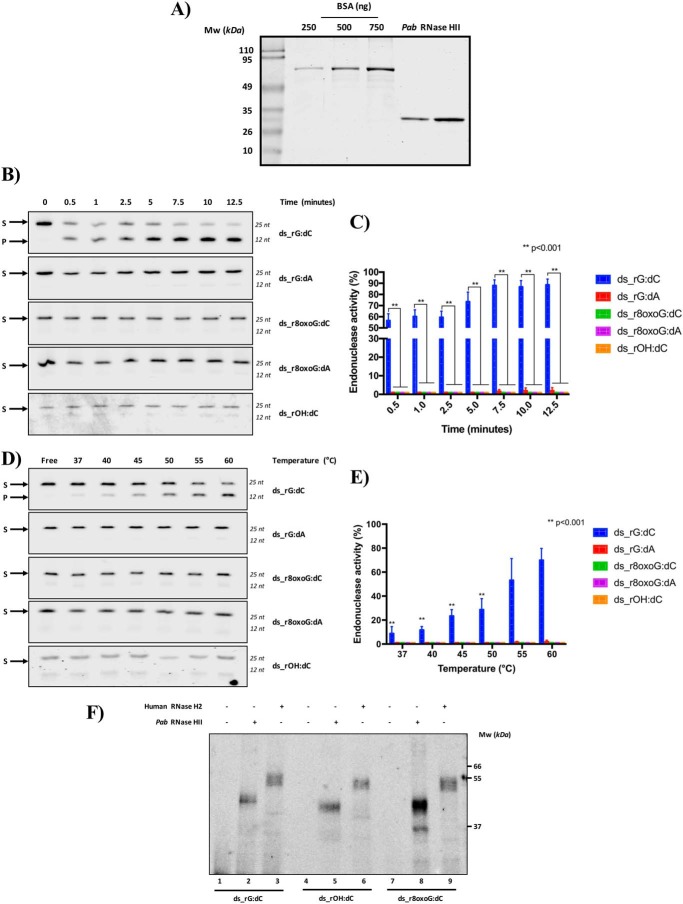
In our recent study (20), we demonstrated that yeast, mouse, and human RNase H2, although able to cleave at a single rGMP embedded in DNA, are not capable of cleaving modified rGMPs including oxidized and abasic dsODNs. On the contrary, the type 2 RNase H E. coli counterpart is active on these same substrates. We thought that a loss of this processing activity in eukaryotes could have been fixed during evolution. For this reason, we were intrigued to examine the activity of another type 2 RNase H enzyme present in Archaea, specifically from the P. abyssi (Pab) organism. As described under “Experimental procedures,” the recombinant Pab RNase HII monomeric protein was expressed and purified to homogeneity and carefully quantified (Fig. 3A). Considering that this protein exerts its activity at 60 °C in a pH condition at ∼ 8.0 (22), we incubated the Pab protein with each rNMP-containing DNA substrate and stopped the reaction at different time points. As shown in Fig. 3 (B and C), the Pab RNase HII protein was highly active on the ds_rG:dC, whereas it was completely inactive on oxidized and abasic dsODNs, ds_r8oxoG:dC, and ds_rOH:dC. Markedly, and unlike the E. coli RNase HII (Fig. 2), the Pab RNase HII protein was also unable to cleave the rG-containing dsODN when this was mismatched with adenine in place of cytosine (Fig. 3, B and C). Because the optimal working temperature of the archaeal RNase HII is ∼60 °C, close to the temperature of melting (Tm) of the ODNs used in this study (Table 1), we hypothesized that the absence of cleavage activity was due to a possible denaturation of the dsODNs that could hamper the interpretation of the results obtained. To exclude this possibility, we performed a temperature-dependent experiment to monitor the processing activity of the Pab RNase HII enzyme when the temperature of reaction was lower than that corresponding to the Tm of the dsODNs (Fig. 3, D and E). Under optimal conditions for the protein (60 °C), Pab RNase HII efficiently cleaved the control substrate ds_rG:dC. The product formation decreased when the temperature was lower than 60 °C in a linear manner. These data show that the Pab RNase HII enzyme works more slowly at temperatures lower than its optimal temperature. The activity of Pab RNase HII on the dsODNs containing modified rNMPs was undetectable at all the temperatures tested. These results show that the optimal high temperature of Pab RNase HII is not solely responsible for its inability to process oxidized rNMP or rAP sites, as a consequence of a putative nonoptimal dsDNA structural conformation. Our results demonstrate that archaeal RNase HII from P. abyssi is not able to process rAP or oxidized rNMP embedded in DNA, like the eukaryotic RNase H2 (20). We then investigated whether the lack of processing activity of Pab RNase HII on rAP and on oxidized rNMP embedded in DNA was due to an inability of binding these substrates. In a recent study, we showed that human RNase H2 is unable to process an rAP and also an oxidized rNMP in DNA (20). Here, we measured the ability of archaeal RNase HII and human RNase H2 to bind the abasic, ds_rOH:dC, and the oxidized, ds_r8oxoG:dC dsODNs, as well as the positive control ds_rG:dC through cross-linking experiments (Fig. 3F). As shown in the representative gel, the cross-linking analysis allowed us to detect the formation of a stable protein-DNA complex migrating at ∼40 and 55 kDa for Pab RNase HII (lanes 2, 5, and 8) and human RNase H2 (lanes 3, 6, and 9), respectively, with all the substrates used in this study. In light of these results, we conclude that although the eukaryotic and archaeal RNase HII/2 proteins have no capability to process rAP and oxidized rNMP embedded in DNA, these enzymes maintain the ability to bind them, generating a stable DNA-protein -complex.
Figure 3.
RNase HII from P. abyssi does not cleave rAP and oxidized rNMP embedded in DNA. A, different volumes (1 and 2 μl) of the recombinant RNase HII protein from P. abyssi were quantified as described under “Experimental procedures.” Both bands corresponding to different amounts of the protein (apparent molecular mass, 27,600 Da) were quantified and normalized on a standardization curve (250, 500, or 750 ng) of BSA protein. The molecular mass (MW) expressed in kDa is shown on the left side of the image. B, representative denaturing polyacrylamide gel of time course kinetics cleavage by a fixed amount of Pab RNase HII protein on each substrate. Different time points, expressed in minutes, are indicated on the top of the figure. ds_rG:dC oligonucleotide was used as positive control. S and P correspond to substrate and product, respectively. C, relative graph illustrating the time-course kinetics activity of the protein on different ODNs. The data are expressed as means ± S.D. of three independent replicas. A p value of < 0.001 is marked with two asterisks (**). D, representative denaturing polyacrylamide gel of temperature-response cleavage by Pab RNase HII on each ODN. The reaction was performed for 15 min at different temperatures indicated on the top of the figure and expressed in °C. S and P correspond to substrate and product, respectively. E, relative graph illustrating the Pab RNase HII cleavage activity upon different temperatures on all the used dsODNs. The data are expressed as means ± S.D. of three independent replicas. A p value of <0.001 is marked with two asterisks (**). F, RNases HII/2 from human and Pab recognize oxidized rNMP and rAP embedded in DNA but are unable to process them. After the incubation between dsODNs and human or Pab type 2 RNase H proteins, binding complexes were cross-linked upon UV exposure as explained under “Experimental procedures” and run on a denaturing 8% (w/v) SDS-PAGE gel. The bands correspond to the complex DNA protein, migrating in a different way following the molecular mass of both proteins and dsODNs. Molecular mass, expressed in kDa, is shown on the right side of the image.
Structural modeling and alignment analyses highlight subtle structural differences in RNase H type 2 enzymes that may affect the spatial positioning of essential amino acids for cleavage
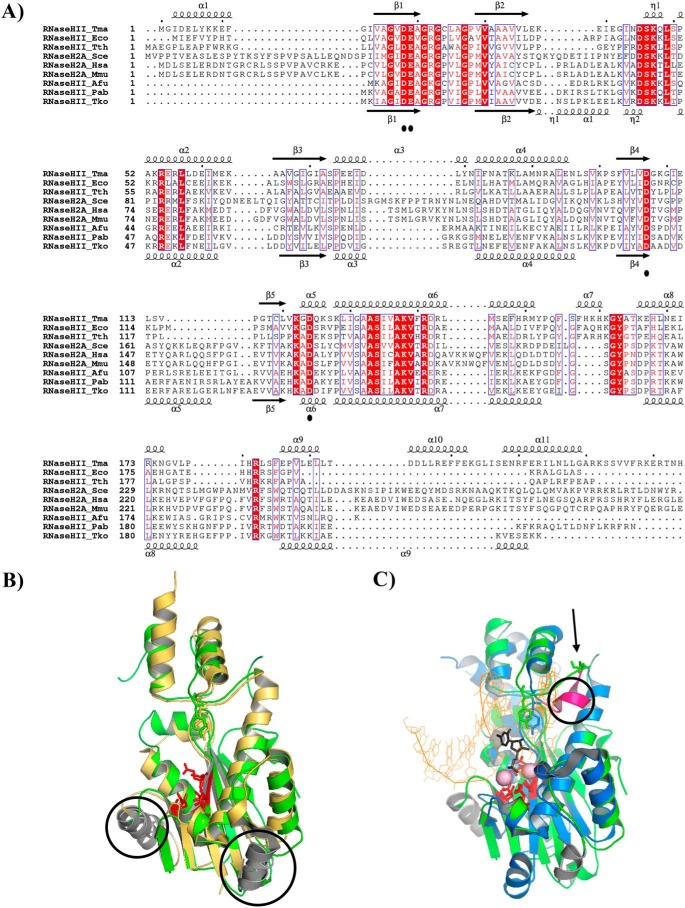
To provide a molecular explanation for the functional differences observed for bacterial, archaeal, and eukaryotic RNase HII/2 proteins, we performed an in silico structural analysis of the bacterial Thermotoga maritima (Tma), Thermus thermophilus (Tth), E. coli (Eco), Saccharomyces cerevisiae (Sce), Homo sapiens (Hsa), Mus Musculus (Mmu), Archaeoglobus fulgidus (Afu), P. abyssi (Pab), and Thermococcus kodakarensis (Tko). The alignment was generated using three representative proteins of RNases HII/2 from each kingdom of life, whose sequence diversity was chosen to best illustrate the sequence variability. Moreover, structural modeling is based on the availability of the three-dimensional structures of one representative RNase HII/2 (TmaRnaseHII, HsaRNaseH2, and TkoRNaseHII) from each domain of life. Comparison of secondary structure elements of RNases HII/2 proteins points out conserved residues involved in catalysis, consisting of four highly conserved carboxylates (DEDD motif), the GRG 2′-OH sensing motif, and residues involved in substrate binding (Fig. 4A) (27–30). Interestingly, archaeal and eukaryotic RNases HII/2 contain 2 α-helical structures (namely α1 and α2, according to the secondary structure of Tko RNase HII), which are absent in the bacterial ortholog. Additionally, a short one-turn helix flanked by two other helices is unique to bacterial RNase HII (namely α7 according to the secondary structure of Tma RNase HII). Likewise, the three-dimensional structure of RNases HII/2 in Archaea (Tko RNase HII), Bacteria (Tma RNase HII) and eukaryotes (Hsa RNase H2) depicts a conserved catalytic core, termed RNase H fold, consisting, of a five-stranded β sheet with three antiparallel and two parallel strands (54123, ↑↑↑↓↑) surrounded by α-helices (Fig. 4, B and C). When Tko RNase HII was superimposed on the structure of Hsa RNase HII using the positions of 140 pairs of C-α atoms, the resulting root mean square deviation was 1.278 Å (Fig. 4B). In this model, the secondary elements superimpose very well. The active site geometry and the conserved tyrosine residue (Tyr-210 in H. sapiens and Tyr-170 in T. kodakarensis) strikingly overlap. This suggests that the cleavage mechanism is conserved in eukaryotes and Archaea. When Hsa RNase HII was superimposed to the structure of the Tma RNase HII substrate complex, using the positions of 128 pairs of C-α atoms, the resulting root mean square deviation was 2.414 Å (Fig. 4C). In such a superimposition, the RNase H fold overlaps quite well, and the active-site residues adopt a similar geometry as also observed with the archaeal ortholog, suggesting that a two-metal ion catalytic mechanism operates in RNases HII/2. However, the spatial clustering of residues surrounding the conserved tyrosine (Tyr-163 in T. maritima) is clearly different between the bacterial and eukaryotic/archaeal RNases HII/2. Indeed, a bulky secondary structural element, consisting of a short one-turn helix (α7 in T. maritima at the top of the secondary structure alignment; Fig. 4A and α7 in magenta; Fig. 4C), locates in close proximity of the active site in the bacterial structure. This region, in the archaeal/eukaryotic structures, corresponds to a linear straight segment in which an aspartic residue points toward the cleaved DNA_1RNA_DNA strand (Fig. 4C). No counterpart acidic residue is found in the bacterial structure. Therefore, these subtle structural differences may affect the spatial positioning of essential amino acids for cleavage.
Figure 4.
Type 2 RNases H from Archaea and eukaryotes display the highest structural similarities. A, structure-based sequence alignment of T. maritima (Tma), T. thermophilus (Tth), E. coli (Eco), S. cerevisiae (Sce), H. sapiens (Hsa), M. musculus (Mmu), A. fulgidus (Afu), P. abyssi (Pab), and T. kodakarensis (Tko). Sequence alignment is based on the three-dimensional structure of RNase HII_Tma (corresponding secondary structure elements are shown at the top) (PDB code 3O3F) and RNase HII_Tko (corresponding secondary structure elements are shown at the bottom) (PDB code 1IO2). Conserved active site residues are marked with black dots. Strictly conserved residues are highlighted in red. Structure-based sequence alignment was generated using ESPript 3. B, superposition of structures of RNase HII/2 from T. kodakarensis (PDB code 1IO2) in yellow and H. sapiens (PDB code 3PUF) in green on the C-α atoms from the central β-sheet of the RNase H fold. Active site residues are in red, and the conserved tyrosine residue are shown as sticks. The two conserved α-helices (α1 and α5 secondary elements from RNase HII_Tko depicted at the bottom of the alignment) in T. kodakarensis and H. sapiens but absent in T. maritima are circled. C, superposition of structures of RNase HII/2 from T. maritima (PDB code 3O3F) in blue and H. sapiens (PDB code 3PUF) in green on the C-α atoms from the central β-sheet of the RNase H fold. Active site residues in red and the conserved tyrosine residue are shown as sticks. ds_DNA_1RNA_DNA is shown as orange lines excepted for the 1RNA, which is shown as black sticks. Magnesium ions are shown as pale pink spheres. The unique magenta α-helix (α7 secondary elements from RNase HII_Tma depicted at the top of the alignment) is circled. The arrow indicates the missing aspartic residue in T. maritima. Superpositions and structural figures were prepared in PyMOL (Schrödinger).
Discussion
A frequent phenomenon, having profound, detrimental effects on genome stability of both prokaryotes and eukaryotes, is the failure to remove rNMPs from DNA (8). In Bacteria, including Archaea, as well as in mammalians and in yeast cells, the main processing mechanism responsible for repairing these lesions in DNA is the RER pathway, which involves the RNase H type 2 enzyme (15, 18, 24, 31, 32). Similarly to deoxyribonucleotides, rNMPs are susceptible to modifications and oxidative insults (33, 34). Moreover, a significant generation of abasic site formation has been demonstrated upon RNA oxidation and alkylation (35). Therefore, among the many rNMPs that are introduced in the nuclear genome per cell cycle (36), a significant amount of incorporated rNMPs could be oxidized or become abasic sites, thus affecting genome stability. In a recent work, we found that eukaryotic RNases H2 derived from human, mouse, and budding yeast were unable to process rAP or oxidized rNMP embedded in DNA, which, on the contrary, were readily leaved by the APE1 protein of the BER pathway (20). Here, we evaluated whether rAP or oxidized rNMPs embedded in DNA could be processed by the prokaryotic orthologous of RNase H type 2 proteins. We demonstrated that although bacterial RNase HII from E. coli was proficient in processing both oxidized rNMP and rAP embedded in DNA, in a Mg2+-dependent manner, the archaeal ortholog from P. abyssi was unable to incise at either of these sites (Fig. 5). Interestingly, Cilli et al. (9) showed that the commercial prokaryotic RNase HII has the ability to remove a single rNMP when paired to an oxidized base or to incise an oxidized rNMP in a DNA duplex. Moreover, Sassa et al. (37) showed that the commercially available prokaryotic E. coli RNase HII preserves the ability to remove an oxidized rNMP in a DNA duplex. In line with these studies, our present findings confirm that commercially available E. coli RNase HII retains the ability to process both rAP and oxidized rNMPs embedded in DNA, whereas eukaryotic RNase H2 is completely inactive on these substrates (20). Our data suggest that the ability to process abasic residues or r8oxoG embedded in DNA might have been lost during evolution. We also noticed that the ability of E. coli RNase HII to process the rNMPs embedded in DNA does not absolutely depend on base pairing, as demonstrated by the fact that the rG is processed significantly more efficiently when it is paired with dA than with dC. Interestingly, the human RNase H2 protein retains some preference for cleavage of rG:dA versus rG:dC (20), although not as prominent as observed for E. coli RNase HII (Fig. 2, A and B). Archaeal RNase HII from P. abyssi has undetectable ability to cleave rG opposite to dA, whereas it cleaves the canonical rG:dC pair (Fig. 3, B and C). It is possible that the rG:dA pair causes a local helix distortion, which may facilitate protein recognition of the lesion in the bacterial and marginally in the eukaryotic form of RNase H type 2. It will be interesting to investigate whether the archaeal mismatch repair system can recognize and process mispaired rNMPs in DNA as the E. coli and S. cerevisiae mismatch repair factors do (14). Our findings clearly reveal the inability of archaeal RNase HII to incise at oxidized rNMPs or at abasic residues and are similar to results obtained using eukaryotic RNases H2 from budding yeast, mouse, and human (20), but contrary to those obtained using E. coli RNase HII. The differences between bacterial RNase HII and archaeal/eukaryotic RNases HII/2 in their capacity to cleave rAP and oxidized rNMP embedded in DNA might be linked to different positioning of residues surrounding the active site. In our study, archaeal/eukaryotic RNases HII/2 were able to bind both oligonucleotides containing embedded abasic and oxidized residues in dsDNA like the bacterial counterpart (Fig. 3F). Despite the overall structural conservation between archaeal, bacterial, and eukaryotic RNases HII/2, we demonstrate that specific ribonucleotide enzyme catalysis has not been similarly preserved among Bacteria, Archaea, and Eukarya. Our results are in agreement with previous studies showing differential substrate preferences between bacterial and archaeal/eukaryotic RNases HII/2 (28, 29). Although the precise evolutionary relationship between eukaryotes and Archaea continues to be a subject of debate (38), these findings clearly highlight a common evolutionary scenario between Archaea and eukaryotes in keeping inactive RNases HII/2 on modified rNMPs embedded in DNA. The inability to process modified rNMPs by the RER mechanism might have led to alternative removal pathways. In eukaryotes, embedded abasic residues and oxidized rNMPs in dsDNA can be processed by APE1 (20). Moreover, damage tolerance pathways involving translesion DNA polymerases can be employed to prevent the stalling of DNA replication at difficult lesions in eukaryotic cells (37). It is commonly accepted that genetic traits are gained by duplication, which permits separate evolution into two functions, as a consequence of a selective pressure in favor of a new trait that gives an adaptive advantage to the organism and/or because of the lack of pressure to retain that trait. In Archaea, the observed absence of ability of RNase HII to cleave rAP and oxidized rNMP sites might be explained on the basis of adaptive molecular mechanisms to survive at extremely high temperatures. Moreover, we cannot exclude, at present, that an interacting partner might render P. abyssi RNase HII or similarly eukaryotic RNase H2 active or, on the other hand, that another protein might be involved in this process. In light of our previous work (20) showing that APE1 is, in eukaryotic cells, able to actively cleave rAP-containing substrates and, to a lesser extent, r8oxoG-containing substrates, some important questions arise, including whether the APE1 pathway could be less error-prone than the RER pathway and what could be the biological advantage. More investigations are now required to elucidate these issues in both archaeal and mammalian cells to further support the hypothesis that the mechanisms observed could be correlated with evolutionary processes.
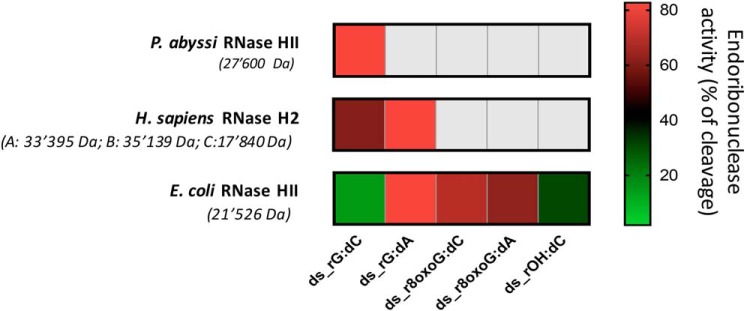
Figure 5.
Difference in cleavage activity of modified ribonucleotides embedded in DNA among RNase HII/2 from the indicated species. The endoribonuclease activities of the indicated enzymes, expressed as percentages of cleavage, on each of the substrates used in our study, are represented as a heat map, in which the gray color represents lack of any enzymatic activity, whereas the different colors from green to red represent increasing processing activity. Specifically, in the case of E. coli activity, the percentage of cleavage obtained from data shown in Fig. 2 was considered; in the case of P. abyssi activity, the percentage of cleavage obtained from data shown in Fig. 3 was considered; in the case of human RNase H2 activity, the percentage of cleavage obtained from data published in Malfatti et al. (20) was considered. Experimental conditions for each enzyme were the same when testing the different substrates.
Experimental procedures
Double-stranded synthetic oligonucleotides
All unmodified and modified double-stranded ODNs sequences are described in Table 1. The rNMP-containing 40-mers ss_rG_40 and ss_rF_40 were purchased from Dharmacon (Lafayette, CO) and annealed with complementary sequence, ss_comp_40, purchased from Invitrogen. The oligonucleotides ss_rG_40 and ss_rF_40 were 5′ end-labeled with [γ-32P]ATP (PerkinElmer Life Sciences) by T4 polynucleotide kinase (New England Biolabs, Ipswich, MA) in a reaction mixture containing 10 μm ATP using 10× polynucleotide kinase buffer (New England Biolabs). This labeling reaction was carried on at 37 °C for 1 h, followed by inactivation at 65 °C for 20 min. The reactions were purified by using Illustra MicroSpin G-25 column (GE Healthcare, Buckinghamshire, UK). ss_dG, ss_rG, and ss_d8oxoG ODNs labeled with IRDye700 fluorophore at 5′ end, and their complementary strand, ss_comp, were synthesized from Metabion International AG (Steinkirchen, Germany). In parallel, ss_dF probe was synthesized from IDT Technologies (Coralville, IA) and labeled with IRDye800 fluorophore at 5′ end. Finally, 1′ rOH ODN, called ss_rOH, was synthesized from Dharmacon and labeled with Cy5 fluorophore at 5′ end (Table 1). All ODNs were purified by reverse phase–high-performance liquid chromatography. Finally, synthesis of the oligonucleotide containing an internal r8oxoG and an IRDye700 fluorophore at 5′ end was carried out in-house as already described (20, 39). For performing the annealing reaction, all ssODNs were resuspended in RNase- and DNase-free water at 100 μm. Then 100 pmol of each probe was annealed with 150 pmol of its complementary DNA strand (as indicated in Table 1) in 10 mm Tris-HCl, pH 7.4, and 10 mm MgCl2, heated at 95 °C, and cooled down overnight in the dark.
Expression and quantification of recombinant proteins
Recombinant E. coli RNase HII was purchased from New England Biolabs. Recombinant P. abyssi (Pab) RNase HII was produced as described by Le Laz et al. (22). Different amounts of Pab RNase HII were loaded onto a 10% (w/v) SDS-PAGE electrophoresis gel, which was subsequently stained using Coomassie Brilliant Blue stain (ThermoFisher) and visualized with an Odyssey CLx IR Imaging system (LI-COR, Bad Homburg vor der Höhe, Germany). The signal of each band was quantified and normalized with a BSA standardization curve using Image Studio software (LI-COR) (Fig. 3A). Recombinant human RNase H2 was produced as described by Chon et al. (24, 40) and tested for its activity and purity in our previous publication (20).
RNase HII activity assays
All 32P-labeled substrates (10 nm) were treated with 1 unit of E. coli RNase HII enzyme (New England Biolabs) for 1 h at 37 °C in 10× Thermopol® buffer (New England Biolabs). This was followed by stopping the reaction by adding 2× denaturing PAGE gel buffer (0.01% bromphenol blue, 95% formamide, and 20 mm EDTA, pH 8.0) and heating to 95 °C for 5 min. After dilutions, the products of this reaction were analyzed by 15% (w/v) polyacrylamide, 8 m urea gel electrophoresis (urea-PAGE). 20–100 oligonucleotide length standard (Integrated Device Technology) was used as a ladder. After electrophoresis, the gels were exposed to a phosphor screen overnight. The images were taken with Typhoon Trio+ (GE Healthcare) and obtained with ImageQuant (GE Healthcare). The band intensities were quantified by Multi Gauge V3.0 (Fujifilm). Endoribonuclease activity of E. coli RNase HII on 25-mer dsODNs was investigated by reacting 1 unit (0.1 ng/μl) of the protein with 250 fmol of DNA or RNA/DNA oligonucleotides (25 nm) in the commercial 10× ThermoPol® reaction buffer (New England Biolabs) or in the equivalent homemade buffer containing 10 mm KCl, 20 mm Tris-HCl, pH 8.8, 10 mm (NH4)2SO4, 2 mm MgSO4, 0.1% Triton X-100, in a final volume of 10 μl for the indicated times at 37 °C. Endoribonuclease activity of P. abyssi RNase HII on 25-mer dsODNs was tested by reacting 200 fmol (20 nm) of the protein with 250 fmol of DNA or RNA/DNA oligonucleotides (25 nm) in a buffer containing 50 mm Tris-HCl, pH 8.0, and 5 mm MgCl2 in a final volume of 10 μl for the indicated times at the optimal temperature of 60 °C. At the end of all reactions, the samples were blocked with a stop solution, containing 99.5% (v/v) formamide (Sigma–Aldrich) and 10× Orange Loading Dye (LI-COR) and heated at 95 °C for 5 min. Then all samples were loaded onto a 7 M denaturing 20% polyacrylamide gel and were visualized with an Odyssey CLx IR Imaging system (LI-COR). The signals of the nonincised substrate and the incision product bands were quantified using Image Studio software (LI-COR).
Cell culture, genomic RNA extraction, and RNase activity assay
HeLa cells (from ATCC) were cultured in DMEM (EuroClone, Milan, Italy), supplemented with 10% (v/v) fetal bovine serum (EuroClone), 100 units/ml penicillin, 100 mg/ml streptomycin sulfate, and 2 mm l-glutamine (EuroClone) at 37 °C and 5% CO2. Total RNA from HeLa cells was extracted with the SV Total RNAisolation system kit (Promega, Madison, WI). RNase A enzymatic assay was performed by incubating 400 ng of total RNA with 0.1 ng of RNase A from bovine pancreas (Sigma–Aldrich). Prokaryotic RNase HII enzymatic assay was performed incubating 400 ng of total RNA with increasing amounts of E. coli RNase HII. The reactions were carried on 1 h at 37 °C in a final volume of 10 μl. Upon adding 2× RNA loading dye (ThermoFisher), the samples were heated at 65 °C for 5 min and then moved on ice. The samples were run in a 1% (w/v) agarose gel, prepared by thawing 1 g of RNase-free agarose powder (Sigma–Aldrich) in 85 ml of 1× MOPS (20 mm MOPS, 10 mm sodium acetate, 1 mm EDTA) and 15 ml of formaldehyde solution (Sigma–Aldrich). The run was carried on in 1× MOPS buffer, at 3–4 V/cm. The gels were visualized with Gel Doc 2000 (Bio-Rad).
Cross-linking analyses
20 pmol of human RNase H2 (2 μm) was co-incubated with 250 fmol of the probe (25 nm) for 30 min at 37 °C in a buffer containing 10 mm Tris-HCl, pH 7.4, 25 mm KCl, 1 mm MgCl2, and 10 mm EDTA in a final volume of 10 μl. Similarly, 20 pmol of P. abyssi RNase HII (2 μm) was co-incubated with 250 fmol of the probe (25 nm) for 30 min at 60 °C in a buffer containing 50 mm Tris-HCl, pH 8.0, 5 mm MgCl2, and 10 mm EDTA in a final volume of 10 μl. After binding reaction, the samples were exposed to 0.2 J/cm2 UV rays and then stopped in 4× Laemmli, heated at 95 °C for 5 min, and loaded onto an 8% (w/v) SDS-PAGE gel. After the run, the gel was visualized with an Odyssey CLx IR Imaging system (LI-COR).
Statistical analysis
Statistical analyses were performed by using the Student's t test in GraphPad Prism software. When p < 0.05, the data were considered as statistically significant.
Author contributions
M. C. M. data curation; M. C. M., G. H., R. U., F. S., and G. T. formal analysis; M. C. M. and G. T. validation; M. C. M., G. H., S. B., K. D. K., G. N., F. S., and G. T. methodology; M. C. M., G. H., F. S., and G. T. writing-original draft; G. H., R. J. C., F. S., and G. T. conceptualization; G. H., R. J. C., F. S., and G. T. writing-review and editing; R. J. C., F. S., and G. T. supervision; F. S. and G. T. funding acquisition; F. S. and G. T. investigation.
Acknowledgments
We thank the Tell and Storici laboratories for constructive feedbacks during the development of this work.
This work was supported by Associazione Italiana per la Ricerca sul Cancro Grant IG19862 (to G. T.), by NIEHS, National Institutes of Health Grant 1R01ES026243-01 (to F. S.), and Howard Hughes Medical Institute Faculty Scholar Grant 55108574 (to F. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- rNMP
- ribonucleoside monophosphate
- rAP
- ribose monophosphate abasic (both apurinic or apyrimidinic) site
- r8oxoG
- oxidized ribonucleoside monophosphate
- RER
- ribonucleotide excision repair
- RNase HII/2
- type 2 ribonuclease H
- ODN
- oligonucleotide
- BER
- base excision repair
- ss
- single-stranded
- ds
- double-stranded
- rRNA
- ribosomal RNA
- PDB
- Protein Data Bank.
References
- 1. Dalgaard J. Z. (2012) Causes and consequences of ribonucleotide incorporation into nuclear DNA. Trends Genet. 28, 592–597 10.1016/j.tig.2012.07.008 [DOI] [PubMed] [Google Scholar]
- 2. Clausen A. R., Zhang S., Burgers P. M., Lee M. Y., and Kunkel T. A. (2013) Ribonucleotide incorporation, proofreading and bypass by human DNA polymerase δ. DNA Repair 12, 121–127 10.1016/j.dnarep.2012.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Caldecott K. W. (2014) Molecular biology. Ribose: an internal threat to DNA. Science 343, 260–261 10.1126/science.1248234 [DOI] [PubMed] [Google Scholar]
- 4. Evich M., Spring-Connell A. M., Storici F., and Germann M. W. (2016) Structural impact of single ribonucleotide residues in DNA. ChemBioChem. 17, 1968–1977 10.1002/cbic.201600385 [DOI] [PubMed] [Google Scholar]
- 5. Cerritelli S. M., and Crouch R. J. (2016) The balancing act of ribonucleotides in DNA. Trends Biochem. Sci. 41, 434–445 10.1016/j.tibs.2016.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Koh K. D., Balachander S., Hesselberth J. R., and Storici F. (2015) Ribose-seq: global mapping of ribonucleotides embedded in genomic DNA. Nat. Methods. 12, 251–257 10.1038/nmeth.3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. El Hage A., Webb S., Kerr A., and Tollervey D. (2014) Genome-wide distribution of RNA–DNA hybrids identifies RNase H targets in tRNA genes, retrotransposons and mitochondria. PLoS Genet. 10, e1004716 10.1371/journal.pgen.1004716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Potenski C. J., and Klein H. L. (2014) How the misincorporation of ribonucleotides into genomic DNA can be both harmful and helpful to cells. Nucleic Acids Res. 42, 10226–10234 10.1093/nar/gku773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cilli P., Minoprio A., Bossa C., Bignami M., and Mazzei F. (2015) Formation and repair of mismatches containing ribonucleotides and oxidized bases at repeated DNA sequences. J. Biol. Chem. 290, 26259–26269 10.1074/jbc.M115.679209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Egli M., Usman N., and Rich A. (1993) Conformational influence of the ribose 2′-hydroxyl group: crystal structures of DNA-RNA chimeric duplexes. Biochemistry 32, 3221–3237 10.1021/bi00064a004 [DOI] [PubMed] [Google Scholar]
- 11. Chiu H.-C., Koh K. D., Evich M., Lesiak A. L., Germann M. W., Bongiorno A., Riedo E., and Storici F. (2014) RNA intrusions change DNA elastic properties and structure. Nanoscale 6, 10009–10017 10.1039/C4NR01794C [DOI] [PubMed] [Google Scholar]
- 12. Crespan E., Furrer A., Rösinger M., Bertoletti F., Mentegari E., Chiapparini G., Imhof R., Ziegler N., Sturla S. J., Hübscher U., van Loon B., and Maga G. (2016) Impact of ribonucleotide incorporation by DNA polymerases β and λ on oxidative base excision repair. Nat. Commun. 7, 10805 10.1038/ncomms10805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lindsey-Boltz L. A., Kemp M. G., Hu J., and Sancar A. (2015) Analysis of ribonucleotide removal from DNA by human nucleotide excision repair. J. Biol. Chem. 290, 29801–29807 10.1074/jbc.M115.695254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shen Y., Koh K. D., Weiss B., and Storici F. (2011) Mispaired rNMPs in DNA are mutagenic and are targets of mismatch repair and RNases H. Nat. Struct. Mol. Biol. 19, 98–104 [DOI] [PubMed] [Google Scholar]
- 15. Sassa A., Yasui M., and Honma M. (2019) Current perspectives on mechanisms of ribonucleotide incorporation and processing in mammalian DNA. Genes Environ. 41, 3 10.1186/s41021-019-0118-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sparks J. L., Chon H., Cerritelli S. M., Kunkel T. A., Johansson E., Crouch R. J., and Burgers P. M. (2012) RNase H2-initiated ribonucleotide excision repair. Mol. Cell. 47, 980–986 10.1016/j.molcel.2012.06.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cerritelli S. M., and Crouch R. J. (2009) Ribonuclease H: the enzymes in eukaryotes: ribonucleases H of eukaryotes. FEBS J. 276, 1494–1505 10.1111/j.1742-4658.2009.06908.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tadokoro T., and Kanaya S. (2009) Ribonuclease H: molecular diversities, substrate binding domains, and catalytic mechanism of the prokaryotic enzymes: prokaryotic RNases H. FEBS J. 276, 1482–1493 10.1111/j.1742-4658.2009.06907.x [DOI] [PubMed] [Google Scholar]
- 19. Huang S. Y., Ghosh S., and Pommier Y. (2015) Topoisomerase I alone is sufficient to produce short DNA deletions and can also reverse nicks at ribonucleotide sites. J. Biol. Chem. 290, 14068–14076 10.1074/jbc.M115.653345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Malfatti M. C., Balachander S., Antoniali G., Koh K. D., Saint-Pierre C., Gasparutto D., Chon H., Crouch R. J., Storici F., and Tell G. (2017) Abasic and oxidized ribonucleotides embedded in DNA are processed by human APE1 and not by RNase H2. Nucleic Acids Res. 45, 11193–11212 10.1093/nar/gkx723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cohen G. N., Barbe V., Flament D., Galperin M., Heilig R., Lecompte O., Poch O., Prieur D., Quérellou J., Ripp R., Thierry J.-C., Van der Oost J., Weissenbach J., Zivanovic Y., and Forterre P. (2003) An integrated analysis of the genome of the hyperthermophilic archaeon Pyrococcus abyssi. Mol. Microbiol. 47, 1495–1512 10.1046/j.1365-2958.2003.03381.x [DOI] [PubMed] [Google Scholar]
- 22. Le Laz S., Le Goaziou A., and Henneke G. (2010) Structure-specific nuclease activities of Pyrococcus abyssi RNase HII. J. Bacteriol. 192, 3689–3698 10.1128/JB.00268-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Henneke G. (2012) In vitro reconstitution of RNA primer removal in Archaea reveals the existence of two pathways. Biochem. J. 447, 271–280 10.1042/BJ20120959 [DOI] [PubMed] [Google Scholar]
- 24. Chon H., Vassilev A., DePamphilis M. L., Zhao Y., Zhang J., Burgers P. M., Crouch R. J., and Cerritelli S. M. (2009) Contributions of the two accessory subunits, RNASEH2B and RNASEH2C, to the activity and properties of the human RNase H2 complex. Nucleic Acids Res. 37, 96–110 10.1093/nar/gkn913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bubeck D., Reijns M. A., Graham S. C., Astell K. R., Jones E. Y., and Jackson A. P. (2011) PCNA directs type 2 RNase H activity on DNA replication and repair substrates. Nucleic Acids Res. 39, 3652–3666 10.1093/nar/gkq980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ohtani N., Haruki M., Morikawa M., Crouch R. J., Itaya M., and Kanaya S. (1999) Identification of the genes encoding Mn2+-dependent RNase HII and Mg2+-dependent RNase HIII from Bacillus subtilis: classification of RNases H into three families. Biochemistry 38, 605–618 10.1021/bi982207z [DOI] [PubMed] [Google Scholar]
- 27. Muroya A., Tsuchiya D., Ishikawa M., Haruki M., Morikawa M., Kanaya S., and Morikawa K. (2001) Catalytic center of an archaeal type 2 ribonuclease H as revealed by X-ray crystallographic and mutational analyses. Protein Sci. 10, 707–714 10.1110/ps.48001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rychlik M. P., Chon H., Cerritelli S. M., Klimek P., Crouch R. J., and Nowotny M. (2010) Crystal structures of RNase H2 in complex with nucleic acid reveal the mechanism of RNA–DNA junction recognition and cleavage. Mol. Cell. 40, 658–670 10.1016/j.molcel.2010.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Figiel M., Chon H., Cerritelli S. M., Cybulska M., Crouch R. J., and Nowotny M. (2011) The structural and biochemical characterization of human RNase H2 complex reveals the molecular basis for substrate recognition and Aicardi–Goutieres syndrome defects. J. Biol. Chem. 286, 10540–10550 10.1074/jbc.M110.181974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reijns M. A., Bubeck D., Gibson L. C., Graham S. C., Baillie G. S., Jones E. Y., and Jackson A. P. (2011) The structure of the human RNase H2 complex defines key interaction interfaces relevant to enzyme function and human disease. J. Biol. Chem. 286, 10530–10539 10.1074/jbc.M110.177394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Heider M. R., Burkhart B. W., Santangelo T. J., and Gardner A. F. (2017) Defining the RNaseH2 enzyme-initiated ribonucleotide excision repair pathway in Archaea. J. Biol. Chem. 292, 8835–8845 10.1074/jbc.M117.783472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lemor M., Kong Z., Henry E., Brizard R., Laurent S., Bossé A., and Henneke G. (2018) Differential activities of DNA polymerases in processing ribonucleotides during DNA synthesis in Archaea. J. Mol. Biol. 430, 4908–4924 10.1016/j.jmb.2018.10.004 [DOI] [PubMed] [Google Scholar]
- 33. Randerath K., Reddy R., Danna T. F., Watson W. P., Crane A. E., and Randerath E. (1992) Formation of ribonucleotides in DNA modified by oxidative damage in vitro and in vivo: characterization by 32P-postlabeling. Mutat. Res. 275, 355–366 10.1016/0921-8734(92)90038-Q [DOI] [PubMed] [Google Scholar]
- 34. Moreira P. I., Nunomura A., Nakamura M., Takeda A., Shenk J. C., Aliev G., Smith M. A., and Perry G. (2008) Nucleic acid oxidation in Alzheimer disease. Free Radic. Biol. Med. 44, 1493–1505 10.1016/j.freeradbiomed.2008.01.002 [DOI] [PubMed] [Google Scholar]
- 35. Loeb L. A., and Preston B. D. (1986) Mutagenesis by apurinic/apyrimidinic sites. Annu. Rev. Genet. 20, 201–230 10.1146/annurev.ge.20.120186.001221 [DOI] [PubMed] [Google Scholar]
- 36. Williams J. S., Lujan S. A., and Kunkel T. A. (2016) Processing ribonucleotides incorporated during eukaryotic DNA replication. Nat. Rev. Mol. Cell Biol. 17, 350–363 10.1038/nrm.2016.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sassa A., Cağlayan M., Rodriguez Y., Beard W. A., Wilson S. H., Nohmi T., Honma M., and Yasui M. (2016) Impact of ribonucleotide backbone on translesion synthesis and repair of 7,8-dihydro-8-oxoguanine. J. Biol. Chem. 291, 24314–24323 10.1074/jbc.M116.738732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Eme L., Spang A., Lombard J., Stairs C. W., and Ettema T. J. G. (2017) Archaea and the origin of eukaryotes. Nat. Rev. Microbiol. 15, 711–723 10.1038/nrmicro.2017.133 [DOI] [PubMed] [Google Scholar]
- 39. Gasparutto D., Livache T., Bazin H., Duplaa A. M., Guy A., Khorlin A., Molko D., Roget A., and Téoule R. (1992) Chemical synthesis of a biologically active natural tRNA with its minor bases. Nucleic Acids Res. 20, 5159–5166 10.1093/nar/20.19.5159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chon H., Sparks J. L., Rychlik M., Nowotny M., Burgers P. M., Crouch R. J., and Cerritelli S. M. (2013) RNase H2 roles in genome integrity revealed by unlinking its activities. Nucleic Acids Res. 41, 3130–3143 10.1093/nar/gkt027 [DOI] [PMC free article] [PubMed] [Google Scholar]