Abstract
Light-induced endocytosis of rhodopsin in the retina is critical for preventing photoreceptor hyperactivity and for the survival of photoreceptor cells. In Drosophila, this process is mediated by arrestin1 (Arr1). Because Arr1 lacks a clathrin-binding domain required for receptor internalization and the C-terminal sequence that interacts with the β-subunit of the clathrin adaptor protein AP2, the mechanism of how Arr1 mediates endocytosis of the major rhodopsin Rh1 is unclear. Here, using several approaches, including Arr binding and pulldown assays, immunofluorescence techniques, and EM imaging, we found that Drosophila metallophosphoesterase (dMPPE) is involved in light-induced rhodopsin endocytosis. We observed that the photoreceptor cells of a dmppe mutant exhibit impaired light-induced rhodopsin endocytosis and that this impairment is independent of dMPPE phosphoesterase activity. Furthermore, dMPPE directly interacted with Arr1 and promoted the association of Arr1 with AP2. Of note, genetic dmppe deletion largely prevented retinal degeneration in norpA (encoding phospholipase C) mutants, which were reported previously to contribute to retinal degeneration, by suppressing Rh1 endocytosis. Our findings demonstrate that Arr1 interacts with AP2 and that dMPPE functions as a critical regulator in Rh1 endocytosis and retinal degeneration.
Keywords: G protein-coupled receptor (GPCR), rhodopsin, endocytosis, arrestin, retinal degeneration, clathrin adaptor protein AP2, photoreceptor function, clathrin, metallophosphoesterase (MPPE)
Introduction
G protein–coupled receptors (GPCRs)3 mediate extracellular signals from neurotransmitters, hormones, cytokines, and sensory stimuli to intracellular signaling cascades (1, 2). To control the signaling event, a family of regulatory molecules called arrestins uncouple the activated receptors from the G proteins to deactivate GPCRs (3, 4). When the GPCRs are continuously activated, the bound arrestins will recruit clathrin proteins to internalize the receptor from the cell surface through dynamin-dependent endocytosis. The internalized GPCRs subsequently undergo lysosomal degradation or are recycled back to the plasma membrane (5–8). Both internalization and down-regulation of GPCRs reduce their density on the plasma membrane, thereby causing long-term desensitization of the cell to extracellular stimuli.
In Drosophila photoreceptors, light stimulation activates rhodopsin to form activated metarhodopsin, which in turn activates heterotrimeric G proteins and phospholipase C (PLC) (9). The activation of PLC leads to opening of transient receptor potential and transient receptor potential–like channels and extracellular Ca2+ influx (10, 11). Endocytosis of a major rhodopsin called Rh1 is mediated by two types of arrestins. The major visual arrestin Arr2 interacts with the β-subunit of the clathrin adaptor protein AP2 through its C-terminal tail to induce endocytosis of the photoactivated metarhodopsin in a number of visual system mutants (12–14). The minor visual arrestin Arr1 is essential for light-induced endocytosis of Rh1, even though it lacks the C-terminal AP2-binding sequence (15). Whether Arr1 also mediates Rh1 endocytosis through its interaction with AP2 remains unknown.
Drosophila metallophosphoesterase (dMPPE) is an eye-enriched protein. Our previous study showed that it acts as a phosphoesterase and mediates Rh1 deglycosylation. In this study, we show that dMPPE is involved in light-induced Rh1 endocytosis and that its role in regulating Rh1 endocytosis is independent of its phosphoesterase activity. We further reveal that dMPPE directly interacts with Arr1 and regulates light-induced Rh1 endocytosis by promoting Arr1/AP2 association. We also show that genetic removal of dmppe suppresses retinal degeneration in both norpA and trp mutants. Our study reveals the molecular mechanism of Arr1-mediated endocytosis of Rh1 and identifies a regulator in light-induced Rh1 endocytosis and retinal degeneration.
Results
dMPPE is involved in light-induced Rh1 endocytosis
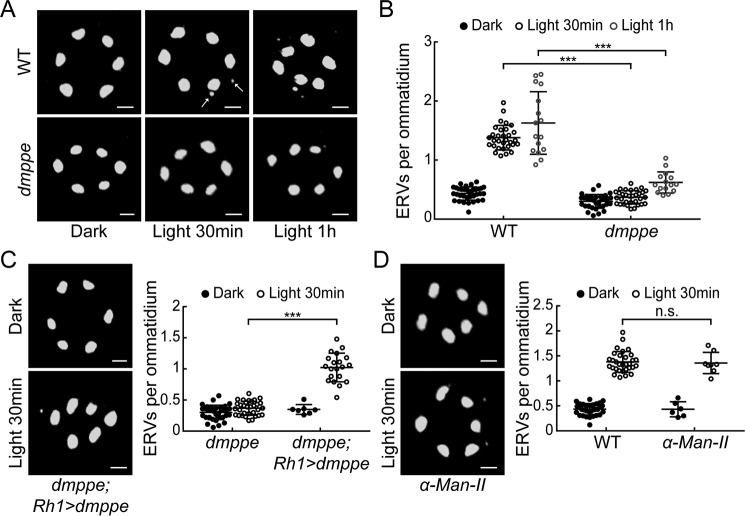
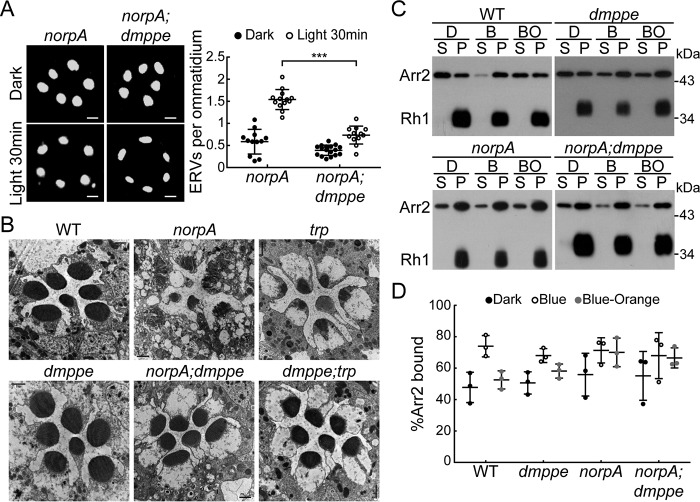
Previous studies have identified some eye-enriched genes through microarray analysis (16, 17). To investigate the potential functions of these genes in light-induced Rh1 endocytosis, we screened flies with eye-enriched gene mutations to characterize novel regulators for light-induced Rh1 endocytosis. These flies were first adapted to a dark environment so that most Rh1 proteins were localized in the rhabdomere (Fig. 1A). Then they were exposed to room light (700 lux) for 30 min to induce Rh1 endocytosis. Our previous study showed that dmppe mutants exhibit defects in Rh1 deglycosylation (18). In this screen, we observed much fewer endocytic Rh1 vesicles (ERVs) in the cell bodies of dmppe mutant photoreceptors compared with WT ones even after prolonged light exposure (Fig. 1, A and B: 30-min exposure, 0.37 ± 0.11 versus 1.38 ± 0.21; 1-h exposure, 0.62 ± 0.18 versus 1.63 ± 0.53; p < 0.001, t test). This phenotype was rescued by expressing dMPPE in the dmppe mutants using p[UAS-dMPPE] transgene (Fig. 1C: 0.37 ± 0.11 versus 1.02 ± 0.23; p < 0.001, t test). These data indicate that dMPPE is involved in light-induced rapid Rh1 endocytosis.
Figure 1.
dmppe mutants exhibit impaired light-induced Rh1 endocytosis. A, ERVs in WT and dmppe mutant photoreceptor cells were induced by exposure to light for 30 min or 1 h. Cross-sections of the eye were stained with a monoclonal Rh1 antibody. Two ERVs in the light-stimulated WT section are indicated with arrows. Scale bar, 2 μm. B, quantification of the number of ERVs per ommatidium for each genotype and treatment. Data are presented as the mean ± S.D. WT, dark: 34 sections from 14 flies; light 30 min: 31 sections from 14 flies; light 1 h: 16 sections from 6 flies; dmppe, dark: 33 sections from 12 flies; light 30 min: 32 sections from 12 flies; light 1 h: 15 sections from 7 flies. All sections were prepared and stained in parallel. The sections for the same genotype or treatment were grouped together for quantification. Note that the scatter plots for WT flies are also presented in (D) and Figs. 2C and 4C; the scatterplots for dmppe mutants are also presented in (C) and Figs. 2C and 4C. C, ERVs in dmppe; Rh1-GAL4/UAS-dMPPE flies were induced by exposure to light for 30 min. Quantification of the number of ERVs per ommatidium is presented the right panel. Data are presented as the mean ± S.D. dmppe;Rh1-GAL4/UAS-dMPPE, dark: 7 sections from 3 flies; light 30 min: 20 sections from 6 flies. Scale bar, 2 μm. D, ERVs were induced in the α-man-II mutant flies by exposure to light for 30 min. Quantification of the number of ERVs per ommatidium is presented the right panel. Data are presented as the mean ± S.D. α-man-II, dark: 6 sections from 3 flies; light 30 min: 8 sections from 4 flies. Scale bar, 2 μm. n.s., no significance; ***, p < 0.001.
Our previous study showed that dMPPE mediates Rh1 deglycosylation (18). Moreover, the glycosylation status of several membrane receptors has been reported to affect their endocytosis (19). Thus, we hypothesized that defective deglycosylation may affect Rh1 endocytosis. We measured light-induced Rh1 endocytosis in an α-man-II mutant that also displays defective Rh1 deglycosylation (18). Our data demonstrate that the number of ERVs per ommatidium in the α-man-II mutant was comparable to that in WT flies (Fig. 1D: 1.36 ± 0.21 versus 1.38 ± 0.21; p = 0.86, t test), suggesting that the oligosaccharide chain does not significantly interfere with Rh1 endocytosis.
The role of dMPPE in regulating light-induced Rh1 endocytosis is independent of its phosphoesterase activity
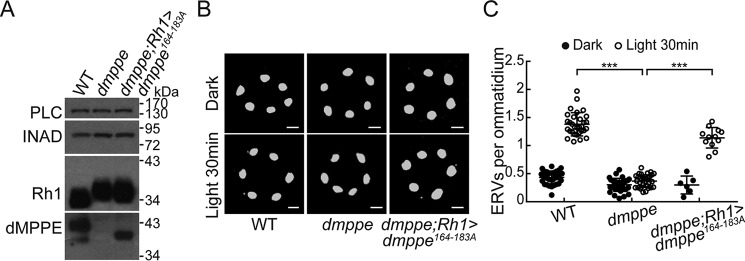
Given that dMPPE functions as a phosphoesterase (18), we next investigated whether its phosphoesterase activity is critical for its role in regulating light-induced Rh1 endocytosis. We replaced the 164–183 residues of dMPPE, which are essential for its phosphoesterase activity (18), with alanine and generated a transgenic fly expressing the mutant dMPPE164–183A protein. We expressed the mutant dMPPE164–183A protein in the dmppe null mutant background (cn,dmppe,bw;rh1-GAL4/p[UAS-dmppe164–183A]). Western blot analysis confirmed the expression of dMPPE164–183A proteins (Fig. 2A). Because phosphoesterase activity is required for Rh1 deglycosylation, dMPPE164–183A proteins failed to rescue the Rh1 deglycosylation defects in the dmppe mutants (cn,dmppe,bw;rh1-GAL4/p[UAS-dmppe164–183A]) (Fig. 2A). Interestingly, we found that defective Rh1 endocytosis was largely rescued by expressing dMPPE164–183A in the dmppe mutant (Fig. 2, B and C: 1.14 ± 0.18 versus 0.37 ± 0.11; p < 0.001, t test). These data demonstrate that the role of dMPPE in regulating light-induced Rh1 endocytosis is independent of its phosphoesterase activity.
Figure 2.
The role of dMPPE in regulating light-induced Rh1 endocytosis is independent of its phosphoesterase activity. A, Western blots show the expression of dMPPE164–183A and the molecular weight of Rh1 in cn,dmppe,bw;rh1-GAL4/p[UAS-dmppe164–183A] flies. B, ERVs in cn,dmppe,bw mutant and cn,dmppe,bw;rh1-GAL4/p[UAS-dmppe164–183A] flies. Cross-sections of the eye were stained with a monoclonal Rh1 antibody. Scale bar, 2 μm. C, quantification of the number of ERVs per ommatidium for each genotype and treatment. Data are presented as the mean ± S.D. WT, dark: 34 sections from 14 flies; light 30 min: 31 sections from 14 flies; dmppe, dark: 33 sections from 12 flies; light 30 min: 32 sections from 12 flies; dmppe;rh1>dmppe164–183A, dark: 6 sections from 3 flies; light 30 min: 12 sections from 6 flies. The scatter plots for WT flies and dmppe mutants are also presented in Figs. 1B and 4C. ***, p < 0.001.
dMPPE interacts directly with Arr1
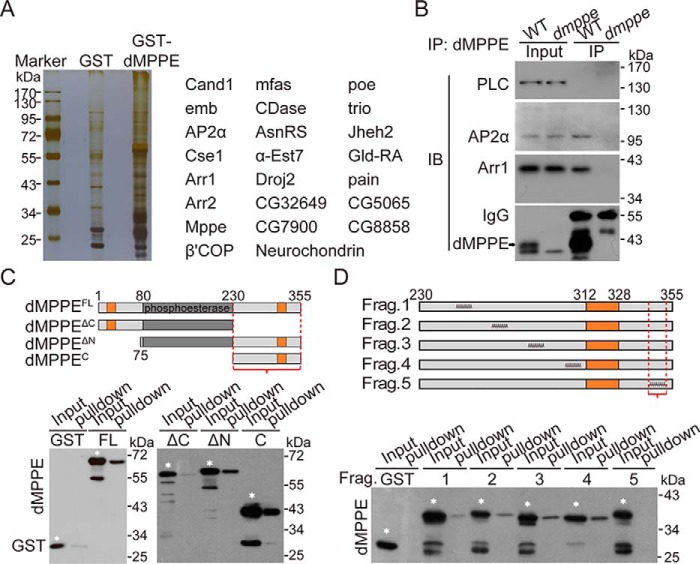
To explore how dMPPE regulates Rh1 endocytosis, we performed pulldown and nano–LC-MS/MS analysis to identify proteins that interact with dMPPE. We found several molecules involved in the processes of Rh1 endocytosis, such as Arr1, Arr2, and the α-subunit of AP2 (Fig. 3A and Table S1). Further co-immunoprecipitation experiments confirmed that dMPPE indeed associated with Arr1 and AP2α in vivo (Fig. 3B).
Figure 3.
dMPPE directly interacts with Arr1. A, pulldown assay followed by mass spectrum analysis revealed proteins associated with dMPPE. The major dMPPE-associated proteins are listed in the right panel. B, co-immunoprecipitation of dMPPE with Arr1 and AP2α in vivo. Fly head extracts from either WT or dmppe mutants were immunoprecipitated with anti-dMPPE antibody. The precipitates, as well as 4% of the head extract, were subjected to Western blotting with antibody against dMPPE, Arr1, AP2α, and PLC (negative control). IP, immunoprecipitated. C, pulldown assays were performed to map the C-terminal region (bracket) as an Arr1-binding site of dMPPE. MBP-Arr1 fusion proteins were coupled to amylase resin for the pulldown assay. The pulldown samples, as well as a portion (10% of the input for pulldown) of the purified GST-fused dMPPE fragments, were loaded for Western blot analysis. Various GST-dMPPE fusion fragments used for the pulldown assay are indicated by white stars. The other low molecular bands in each lane are degraded products. Upper panel, encoded regions of dMPPE fragments; the transmembrane domains are marked in orange. Bottom panel, Western blots of pulldown assays. D, alanine scanning mutagenesis mapped residues 344–352 (bracket) as an Arr1-binding site of dMPPE. MBP-Arr1 fusion proteins were coupled to amylase resin and incubated with various purified GST-dMPPE fusion fragments. The GST-dMPPE fusion fragments used for the pulldown assays are indicated with white stars. The other low molecular bands in each lane are degraded products. Upper panel, schematic depiction of protein sequences substituted with alanines in the mutant dMPPE; the transmembrane domain is marked in tan. Bottom panel, Western blots of pulldown assays.
Next, we used recombinant GST-dMPPE proteins to map the critical Arr1-interacting region. dMPPE has been predicted to contain two transmembrane domains (14–30 aa and 312–328 aa) and both the N and C termini are localized to the intracellular side. Therefore, we generated the N-terminal deletion mutant GST-dMPPEΔN and the C-terminal deletion mutant GST-dMPPEΔC. We found that both full-length GST-dMPPE and GST-dMPPEΔN bound to Arr1, whereas GST-dMPPEΔC did not (Fig. 3C). Furthermore, the C-terminal region of dMPPE (GST-dMPPEC) alone was sufficient for Arr1 binding (Fig. 3C). We further narrowed down the binding epitope to residues 344–352 of dMPPE using alanine scanning mutagenesis (Fig. 3D). Taken together, these data indicate that the C-terminal intracellular tail of dMPPE is essential for Arr1 binding.
dMPPE/Arr1 interaction regulates light-induced Rh1 endocytosis
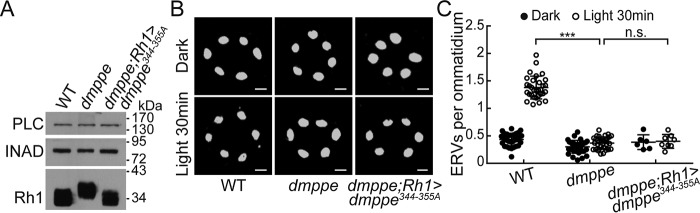
To investigate whether dMPPE/Arr1 interaction is essential for light-induced Rh1 endocytosis, we mutated the 344–355 residues of dMPPE to alanine and generated the p[UAS-dMPPE344–355A] transgenes. The expression of p[UAS-dMPPE344–355A] successfully restored defective Rh1 deglycosylation in dmppe mutants (Fig. 4A), but failed to recover the reduced light-induced Rh1 endocytosis in dmppe mutants (Fig. 4, B and C: 0.40 ± 0.12 versus 0.37 ± 0.11; p = 0.45, t test). These data demonstrate that dMPPE/Arr1 interaction is essential for light-induced Rh1 endocytosis.
Figure 4.
Arr1/dMPPE interaction regulates Rh1 endocytosis. A, Western blots show the molecular weight of Rh1 in cn,dmppe,bw;rh1-GAL4/p[UAS-dmppe344–345A] flies. B, ERVs in dmppe mutants expressing mutant dMPPE344–355A were induced by exposure to light for 30 min. Cross-sections of the eye were stained with a monoclonal Rh1 antibody. Scale bar, 2 μm. C, quantification of the number of ERVs per ommatidium for each genotype and treatment. Data are presented as the mean ± S.D. WT, dark: 34 sections from 14 flies; light 30 min: 31 sections from 14 flies; dmppe, dark: 33 sections from 12 flies; light 30 min: 32 sections from 12 flies; dmppe;rh1>dmppe344–355A, dark: 6 sections from 3 flies; light 30 min: 8 sections from 4 flies. The scatter plots for WT flies and dmppe mutants are also presented in Figs. 1B and 2C. n.s., no significance; ***, p < 0.001.
dMPPE/Arr1 interaction does not affect Rh1/Arr1 association
Given that Arr1 binding to the C-terminal tail of Rh1 is the initial step in Arr1-mediated Rh1 endocytosis (15), we further performed an arrestin-binding and -release assay to examine whether dMPPE affects Rh1/Arr1 association. Rh1 can be photoconverted between the active (metarhodopsin, M) and inactive states (rhodopsin, R) (15). In the dark-adapted WT, Arr1 does not bind to inactive R and mainly exists in the supernatants of head homogenates. After exposure to blue light (480 nm), inactive R is photoconverted to M and most Arr1 associates with rhabdomere membranes. When M is photoconverted back to R by orange light exposure (580 nm), Arr1 is released and detected in the supernatant again (Fig. 5, A and B). In the binding and release assay, dmppe mutants showed a similar Arr1 association pattern as WT files (Fig. 5, A and B: dark, 32.5 ± 6.9% versus 35.7 ± 9.3%, p = 0.66; blue light, 69.1 ± 8.9% versus 70.1 ± 2.0%, p = 0.87; orange light, 40.0 ± 11.3% versus 34.5 ± 3.5%, p = 0.46; t test). Notably, dMPPE itself did not undergo translocation and resided predominately in the membrane fraction in WT flies (Fig. 5, A and C). These results demonstrate that the dMPPE/Arr1 interaction does not affect Rh1/Arr1 association.
Figure 5.
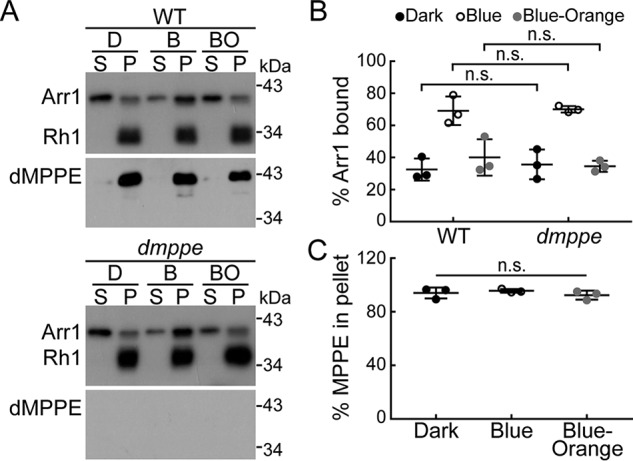
dMPPE/Arr1 interaction does not affect Rh1/Arr1 association. A, Arr1/Rh1 binding in WT and dmppe mutant flies. Arr1/Rh1-binding and -release assays were performed as described in “Experimental Procedures.” Upper panel, Arr1/Rh1 binding in WT flies. Lower panel, Arr1/Rh1 binding in dmppe mutant flies. B, quantification of the percentage of Arr1 bound to rhodopsin-containing membranes in the dark (D), after treatment with blue light (B), or after treatment with blue light followed by orange light (BO). Three sets of independent data were averaged, and the data are presented as the mean ± S.D. C, quantification of relative dMPPE in membrane fraction. Three sets of independent data were averaged, and the data are presented as the mean ± S.D. n.s., no significance.
dMPPE promotes Arr1/AP2 association
The interaction between β-arrestin and the AP2 adaptor complex has been shown to be critical for GPCR endocytosis (20). In Drosophila, point mutations in the C-terminal tail of Arr2 (Arr2K391A and Arr2R393A) completely disrupted its interaction with AP2 (12–14). However, Arr1 lacks the C-terminal sequences that interact with AP2 (Fig. 6A), suggesting that an alternative mechanism must exist for Arr1-mediated Rh1 endocytosis. Interestingly, AP2 was also identified as one of the dMPPE-associated proteins in our MS analysis (Fig. 3A). Our co-immunoprecipitation experiments confirmed that dMPPE can associate with both Arr1 and AP2α in vivo (Fig. 3B). Thus, we hypothesized that dMPPE might function as a linker between Arr1 and AP2α. To test this, we carried out co-immunoprecipitation experiments and found that anti-Arr1 antibodies precipitated much less AP2α from the extracts of dmppe mutant heads than that of WT heads (Fig. 6B: 100% versus 39.4 ± 15.3%; p < 0.001; t test). These data strongly indicate that dMPPE promotes the association between Arr1 and AP2α.
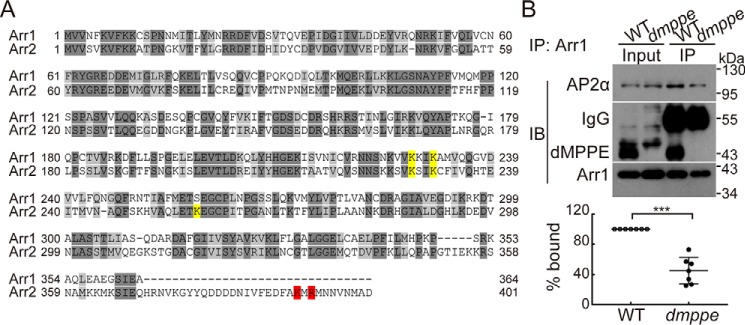
Figure 6.
dMPPE promotes Arr1/AP2 association. A, alignment of Drosophila Arr1 and Arr2. The AP2-binding residues in Arr2 are highlighted in red. The C terminus of Arr1 is short and lacks these AP2-binding residues. The PIs-binding residues are highlighted in yellow. B, Arr1 co-immunoprecipitated with AP2α from WT head extracts, but a much lower quantity than from dmppe mutant head extracts. Fly head extracts from either WT or dmppe mutants were immunoprecipitated with anti-Arr1 antibody. The precipitates, as well as 4% of head extracts, were subjected to Western blot analysis with antibodies against dMPPE, Arr1, and AP2α. IP, immunoprecipitated. Quantification of the relative level of AP2α co-immunoprecipitated by anti-Arr1 antibody is presented under the blots. Seven sets of independent data were averaged, and the data are presented as the mean ± S.D. ***, p < 0.001.
Depletion of dMPPE prevents retinal degeneration in norpA mutants by impairing Arr2/AP2α association
Because Arr1 and Arr2 exhibit high similarity in their amino acid sequence and Arr2 was shown to bind dMPPE in our MS spectrum analysis, we further investigated whether dMPPE also promotes Arr2/AP2α association. Previous reports showed that norpA (encoding phospholipase C) mutants undergo light-dependent retinal degeneration because of abnormal accumulation of Rh1/Arr2 stable complexes (12, 21). Arr2/AP2 association is essential for internalization and loss of the α-subunit of the AP2 adaptor complex can suppress retinal degeneration in the norpA mutant (14). Our data show that genetic removal of dmppe diminishes Arr2/AP2α association and suppresses Rh1 endocytosis. We found that 30-min exposure to room light induced much fewer ERVs in norpA;dmppe photoreceptors compared with norpA photoreceptors (Fig. 7A: 1.54 ± 0.22 versus 0.74 ± 0.20; p < 0.001; t test). EM study further revealed that the norpA;dmppe double mutant largely rescued the retinal degeneration observed in 6-day-old norpA mutants (Fig. 7B). Moreover, arrestin-binding and -release assay showed that stable Rh1/Arr2 complexes accumulated in norpA;dmppe double mutants (Fig. 7, C and D). Taken together, these data indicate that genetic removal of dmppe suppressed retinal degeneration in norpA mutants by inhibiting Rh1 endocytosis but not disrupting stable Rh1/Arr2 complex formation.
Figure 7.
Depletion of dMPPE suppresses retinal degeneration in norpA mutant. A, ERVs in norpA mutants and norpA;dmppe double mutants were induced by exposure to the light for 30 min. Cross-sections of the eye were stained with a monoclonal Rh1 antibody. Quantification of the number of ERVs per ommatidium for each genotype and treatment is presented in the bottom panel. Data are presented as the mean ± S.D. norpA, dark: 12 sections from 5 flies; light 30 min: 13 sections from 6 flies; norpA;dmppe, dark: 17 sections from 6 flies; light 30 min: 12 sections from 6 flies. Scale bar, 2 μm. B, EM analyses revealed that genetic removal of dmppe suppresses retinal degeneration in both norpA and trp mutants. Six-day-old flies were raised in 12-h light/12-h dark conditions. Each picture shows a single ommatidium. Scale bar, 1 μm. C, Arr2-Rh1 binding in WT, dmppe, norpA, and norpA;dmppe flies. Arr2-Rh1–binding and –release assays were performed as described in “Experimental Procedures.” D, quantification of the percentage of Arr2 bound to rhodopsin-containing membranes in the dark (D), after treatment with blue light (B), or after treatment with blue light followed by orange light (BO). Three sets of independent data were averaged, and the data are presented as the mean ± S.D. ***, p < 0.001.
trp (encoding transient receptor potential channel) mutants have been shown to undergo light-dependent retinal degeneration. Although depletion of Ptdlns(4,5)P2 but not accumulation of Rh1/Arr2 stable complexes accounts for light-induced retinal degeneration in trp mutants (22), elimination of Arr2 indeed partially suppressed the retinal degeneration in trp mutants (23). Consistently, we found that genetic removal of dmppe also largely suppressed retinal degeneration in trp mutants (Fig. 7B). The molecular mechanism of dMPPE depletion in suppressing retinal degeneration in trp mutants requires further investigation.
Discussion
dMPPE regulates Rh1 endocytosis
Endocytosis is a fundamental cell biological process that is important for various aspects of cellular physiology ranging from nutrient uptake and signal transduction to synaptic transmission and development (24, 25). In Drosophila photoreceptors, light stimulation induces isomerization of chromophores, thereby inducing a conformational change of inactive rhodopsin to active metarhodopsin (9). Under normal physiological conditions, a fraction of metarhodopsin is internalized upon light exposure (15, 26). Endocytosis of Rh1 is thought to scavenge constitutively active or damaged metarhodopsin to induce long-term adaptation and protect the photoreceptors from excessive Ca2+ influx–induced cell toxicity (26, 27). Moreover, endocytosis and exocytosis of Rh1 also maintains Rh1 homeostasis upon long-term light exposure (26–28).
It has been known that C-terminal phosphorylation of Rh1 triggers its endocytosis and that Arr1 binds to phosphorylated Rh1 to initiate endocytosis (15). Structural studies have revealed that the N- and C-terminal domains of arrestin are composed of antiparallel β-sheets that are linked by an unusual polar core (29). A network of intramolecular interactions between charged residues buried within the polar core maintains the basal conformation of arrestin (30). When the N-terminal domain of arrestin binds to activated GPCRs, arrestins undergo a conformational change to expose their C-terminal domain. The C-terminal tail of arrestin interacts with components of the endocytic machinery, including the clathrin heavy-chain and the β-subunit of AP2, thereby facilitating GPCR internalization (31). However, Drosophila Arr2 and Arr1 do not contain a clathrin-binding domain. Although Drosophila Arr2 has been shown to interact with the β-subunit of AP2 (14), Drosophila Arr1 lacks the sequences needed to mediate an interaction with AP2β. In this study, we show that dmppe mutants exhibit reduced light-induced Rh1 endocytosis and reveal that the intracellular tail of dMPPE binds directly to Arr1. Depletion of dMPPE or the Arr1-binding sites in dMPPE largely impairs Arr1/AP2 association and Rh1 endocytosis. Our study identified a novel regulator in Rh1 endocytosis and provides strong evidence that the Arr1/dMPPE interaction regulates Rh1 endocytosis.
Our co-immunoprecipitation assay revealed that Arr1 can still partially associate with AP2 in the absence of dMPPE. These data are consistent with the observation that prolonged light exposure can still induce Rh1 endocytosis in dmppe mutants. These results suggest that other molecules might be able to mediate Arr1/AP2 association. Phosphatidylinositol-bisphosphate (PI(4,5)P2) is an attractive candidate (4, 5). The C-terminal domain of β-arrestin contains a basic region that binds to PI(4,5)P2 and Drosophila Arr2 has been shown to interact with PI(4,5)P2 (32, 33). The N terminus of the AP2 α-subunit can bind to PI(4,5)P2, and this interaction promotes the localization of AP2 to the plasma membrane and regulates GPCR endocytosis (34–37).
Genetic removal of dmppe prevents retinal degeneration in both norpA and trp mutants
Under normal conditions, the interaction between Arr2 and Rh1 is transient, because light-triggered Ca2+ influx activates CaM kinase II, which subsequently phosphorylates Arr2 to cause its release from Rh1 (38, 39). In norpA mutants, photoresponses were abolished, which prevents the normal rise in Ca2+ after light stimulation and results in the accumulation of Rh1/Arr2 stable complexes and retinal degeneration (12, 21). In this study, we show that genetic removal of dmppe prevents retinal degeneration in norpA mutants but does not disrupt the formation of stable Rh1/Arr2 complexes. We further demonstrated that depletion of dMPPE suppresses Rh1 endocytosis in norpA mutants. Consistent with this, inhibition of Rh1 endocytosis using a temperature-sensitive dynamin (Shibirets) or by removing the α-subunit of AP2 has been shown to suppress retinal degeneration in norpA mutants (12, 14). These results support the conclusion that dMPPE regulates Rh1 endocytosis.
In summary, our study provides evidence that the dMPPE/Arr1 interaction is critical for light-induced Rh1 endocytosis and for preventing retinal degeneration. Our findings have identified a novel regulator in Rh1 endocytosis and retinal degeneration. In addition, our results provide a linkage between Arr1 and the endocytic machinery.
Experimental procedures
Fly genetics and light treatment
The mutant alleles of pBacCG8889e02905 were obtained from Harvard Medical School and were crossed into a cn,bw background. WT flies used in this study were cn,bw. The mutant alleles used for each gene in this work were norpAP24, arr25, arr11, and trp9. p[UAS::dMPPE] was generated by our lab (18). The P-element insertion line α-Man-IIαLL01094 was obtained from the Kyoto stock center and genetic mosaics were induced using the FLP-FRT, p[GMR-hid] technique with an ey-FLP driver to generate mitotic clones of a single genotype in the eye (40). All flies were reared at 25 °C on standard medium in a 12-h light/12-h dark cycle incubator, with 60–80% relative humidity. To induce Rh1 endocytosis, white eye flies younger than 3 days old were dark adapted overnight and then exposed to room light (∼700 lux) for 30 min or 1 h.
Generation of transgenic flies
To generate p[UAS-dMPPE164–183A] and p[UAS-dMPPE344–355A] transgenic flies, fragments of CG8889-RA cDNA encoding 164–183 aa and 344–355 aa were replaced with alanines, respectively. The modified cDNAs were subcloned into the pUAST-attB vector and then injected into y1,w67c23;P{CaryP}attP2 flies. The transgenic flies were subsequently crossed into the cn,dmppe,bw background. dMPPE164–183A and dMPPE344–355A proteins were expressed in photoreceptor cells by using Rh1-GAL4.
Western blotting
Western blot analyses were carried out as described previously (41). Fly heads were homogenized in 2× SDS sample buffer. Proteins were fractionated by SDS-PAGE and transferred to PVDF membranes (Millipore) in Tris-glycine buffer. After blocking, the blots were probed with anti-dMPPE antibody (rabbit, 1:1000 dilution), anti-Rh1 antibody (mouse, 1:3000 dilution) (Developmental Studies Hybridoma Bank (DSHB), catalog no. 4C5; RRID: AB_528451), anti-PLC antibody (rabbit, 1:1000 dilution) (from Dr. C. Montell), anti-Arr2 antibody (rabbit, 1:1000 dilution), anti-GST antibody (mouse, 1:2000 dilution) (Amart), anti-Arr1 antibody (rabbit, 1:1000 dilution) (from Dr. H.-S. Li), and anti-AP2α antibody (mouse, 1:1000 dilution) (Abcam, catalog no. ab2730; RRID: AB_303255) at 4 °C overnight. The membranes were washed three times and then incubated with horseradish peroxidase–conjugated anti-mouse or rabbit IgG antibody (1:5000 dilution) (Millipore). The signals were detected using ECL reagents (GE Healthcare).
Co-immunoprecipitation
Two hundred fly heads were collected and homogenized on ice in 1 ml lysis buffer (10 mm imidazole, pH 7.35, 10% sucrose, 5 mm MgCl2, 1 mm DTT, 160 mm KCl, and 1× cOmplete Protease Inhibitor mixture (Roche)) (42). After rotation for 30 min at 4 °C, the lysates were centrifuged at 14,000 rpm at 4 °C for 5 min. The supernatant was incubated with 1 μl anti-dMPPE or 0.5 μl anti-Arr2 or anti-Arr1 antibodies at 4 °C for 2 h. After blocking with 4% BSA in PBS buffer at 4 °C for 30 min, 50 μl protein A beads (Thermo Fisher) were added and incubated at 4 °C overnight. After five washes with PBS, the bound complexes were eluted with 2× SDS sample buffer and subjected to SDS-PAGE and Western blotting.
Pulldown assays
All GST and MBP fusion proteins were expressed in Escherichia coli BL21 cells and purified with GSH-Sepharose (GE Healthcare) or amylose resin (New England Biolabs), respectively. To map the Arr1-binding site in dMPPE, MBP-Arr1 fusion protein–coupled beads were incubated with various purified GST-dMPPE fragments. After washing, the elution was analyzed by Western blotting.
Arrestin-binding assay
Arr1- and Arr2-binding assays were performed as described previously (38, 43). Briefly, eight flies from each group and condition were collected and adapted to the dark overnight. After exposure to pure blue light (480 ± 10 nm) for 30 s, fly heads were homogenized in the dark. After centrifuging at 14,000 rpm for 5 min, pellet and supernatant fractions were separated under dim red light and subjected to SDS-PAGE and Western blot analysis. Arr1- and Arr2-release assays were performed in the same manner, except that the flies were exposed to 30 s of pure blue light followed by 90 s of pure orange light (580 ± 10 nm).
Immunostaining
Immunofluorescence staining was carried out as described previously (44). Briefly, fly heads were hemisected and immersed in 10% formaldehyde fixing solution for 2 h on ice. The eyes were dehydrated with acetone and embedded with LR White resin. One-micrometer sections were cut across the top one third of the eye and stained with anti-Rh1 antibody (1:300) (DSHB, catalog no. 4C5). The stained sections were examined under an LSM 700 confocal microscope (Zeiss, Germany). The activity of Rh1 endocytosis was quantitated as the number of ERVs per ommatidium. At least six sections from three flies were examined and averaged for each genotype and light treatment.
EM
EM was performed as described previously (45). Briefly, fly heads were hemisected and fixed in 0.1 m sodium cacodylate buffer (pH 7.2) with 2.5% glutaraldehyde and 4% paraformaldehyde at 4 °C for 2 h. After rinsing with 0.1 m sodium cacodylate for three times, the eyes were stained with 1% osmium tetroxide for 1 h at room temperature. After ethanol dehydration and propylene oxide washing, the samples were embedded using standard procedures. The ultrathin sections (90 nm) were cut, collected, and stained with uranyl acetate followed by lead citrate. Micrographs were taken at 80 kV on a Hitachi-7650.
Identification of dMPPE-associated proteins by MS
The GST fusion proteins were expressed in E. coli BL21 cells and purified with GSH-Sepharose (GE Healthcare). Approximately 500 μg of each protein was coupled to 5 ml Sepharose 4B agarose and incubated with head extracts of 1000 flies (in PBS containing 1% Triton X-100 and protease inhibitors). After three washes with PBS, the proteins were eluted and dissolved in ammonium bicarbonate buffer solution (25 mm, pH 8.0) and digested with trypsin at 37 °C for 16 h. The nano–LC-MS/MS experiments were performed with LTQ Orbitrap MS (Thermo Fisher) equipped with a nanoelectrospray ion source as described previously (46). The LTQ Orbitrap instrument was operated in positive ion mode. An Agilent 1100 series LC system equipped with a reverse-phase microcapillary column (0.075 × 150 mm, C18 column, Dionex) was used, with the mobile phase consisting of buffer A (0.1% formic acid in H2O) and buffer B (0.1% formic acid in acetonitrile). The analytical condition was set at a linear gradient from 0 to 60% buffer B in 60 min, and the flow rate was adjusted to 200 nl/min. The column was re-equilibrated at initial condition for 10 min. The MS/MS spectra acquired from precursor ions were submitted to MaxQuant.
MaxQuant was used for identification and peptide quantification. A spectrum selector was used to import spectrums from raw files. The search engine was used to cross-reference identified peptides against Drosophila UniProt fasta database. The mass tolerance was set to be 10 ppm for precursors and 0.01 Da for fragment ions. Oxidation on methionine and deamination on asparagine and glutamine were chosen as variable modifications. Carbamidomethyl on cysteine was set as static modification. Trypsin was set as the proteolytic enzyme with up to two missed cleavages. The precursor ion area detector in MaxQuant was used for quantification. Each protein was considered identified when at least three unique peptides were identified. The list of identified proteins is included in the supporting information.
Experimental design and statistical analysis
Immunostaining and EM studies on fly eye sections were conducted to explore the function of dMPPE in Rh1 endocytosis and retinal degeneration. Biochemical studies on fly eyes were conducted to explore the detailed mechanism.
The Western blotting and fluorescence images were analyzed using the Image J software (National Institutes of Health). Data from more than three independent experiments were averaged. Statistical analysis was performed using Prism 5.0 (GraphPad Software). All data are presented as the mean ± S.D. Student's t test was used to compare the genotypes or conditions. Statistical significance was set at p < 0.05 (*); p < 0.01 (**); p < 0.001 (***); and no significance (n.s.).
Author contributions
Y. M. data curation; Y. M. formal analysis; Y. M. investigation; Y. T. and Z. C. Z. writing-review and editing; Z. C. Z. and J. H. project administration; J. H. conceptualization; J. H. supervision; J. H. writing-original draft.
Supplementary Material
Acknowledgments
We thank Craig Montell for anti-PLC antibody and anti-INAD antibody; Hong-sheng Li for anti-TRP antibody and anti-Arr1 antibody; Bloomington Stock Center and Kyoto stock center for the flies; people in the Han laboratory for critical comments on the manuscript.
This work was supported by National Natural Science Foundation of China Grants 31471031 and 31771171 (to J. H.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1 and supporting information.
- GPCR
- G protein–coupled receptor
- PLC
- phospholipase C
- dMPPE
- Drosophila metallophosphoesterase
- ERV
- endocytic Rh1 vesicles
- aa
- amino acids
- M
- metarhodopsin
- R
- rhodopsin
- PI(4,5)P2
- phosphatidylinositol-bisphosphate
- RRID
- Research Resource Identifiers
- DSHB
- Developmental Studies Hybridoma Bank.
References
- 1. Pierce K. L., Premont R. T., and Lefkowitz R. J. (2002) Seven-transmembrane receptors. Nat. Rev. Mol. Cell Biol. 3, 639–650 10.1038/nrm908 [DOI] [PubMed] [Google Scholar]
- 2. Barry D. M., Munanairi A., and Chen Z. F. (2018) Spinal mechanisms of itch transmission. Neurosci. Bull. 34, 156–164 10.1007/s12264-017-0125-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shenoy S. K., and Lefkowitz R. J. (2011) β-Arrestin-mediated receptor trafficking and signal transduction. Trends Pharmacol. Sci. 32, 521–533 10.1016/j.tips.2011.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lefkowitz R. J., and Shenoy S. K. (2005) Transduction of receptor signals by β-arrestins. Science 308, 512–517 10.1126/science.1109237 [DOI] [PubMed] [Google Scholar]
- 5. Rajagopal S., and Shenoy S. K. (2018) GPCR desensitization: Acute and prolonged phases. Cell. Signal. 41, 9–16 10.1016/j.cellsig.2017.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Irannejad R., and von Zastrow M. (2014) GPCR signaling along the endocytic pathway. Curr. Opin. Cell Biol. 27, 109–116 10.1016/j.ceb.2013.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kang D. S., Tian X., and Benovic J. L. (2014) Role of β-arrestins and arrestin domain-containing proteins in G protein-coupled receptor trafficking. Curr. Opin. Cell Biol. 27, 63–71 10.1016/j.ceb.2013.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Smith J. S., and Rajagopal S. (2016) The β-arrestins: Multifunctional regulators of G protein-coupled receptors. J. Biol. Chem. 291, 8969–8977 10.1074/jbc.R115.713313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tian Y., Hu W., Tong H., and Han J. (2012) Phototransduction in Drosophila. Sci. China Life Sci. 55, 27–34 10.1007/s11427-012-4272-4 [DOI] [PubMed] [Google Scholar]
- 10. Hardie R. C., and Minke B. (1992) The trp gene is essential for a light-activated Ca2+ channel in Drosophila photoreceptors. Neuron 8, 643–651 10.1016/0896-6273(92)90086-S [DOI] [PubMed] [Google Scholar]
- 11. Peretz A., Suss-Toby E., Rom-Glas A., Arnon A., Payne R., and Minke B. (1994) The light response of Drosophila photoreceptors is accompanied by an increase in cellular calcium: Effects of specific mutations. Neuron 12, 1257–1267 10.1016/0896-6273(94)90442-1 [DOI] [PubMed] [Google Scholar]
- 12. Alloway P. G., Howard L., and Dolph P. J. (2000) The formation of stable rhodopsin-arrestin complexes induces apoptosis and photoreceptor cell degeneration. Neuron 28, 129–138 10.1016/S0896-6273(00)00091-X [DOI] [PubMed] [Google Scholar]
- 13. Kiselev A., Socolich M., Vinós J., Hardy R. W., Zuker C. S., and Ranganathan R. (2000) A molecular pathway for light-dependent photoreceptor apoptosis in Drosophila. Neuron 28, 139–152 10.1016/S0896-6273(00)00092-1 [DOI] [PubMed] [Google Scholar]
- 14. Orem N. R., Xia L., and Dolph P. J. (2006) An essential role for endocytosis of rhodopsin through interaction of visual arrestin with the AP-2 adaptor. J. Cell Sci. 119, 3141–3148 10.1242/jcs.03052 [DOI] [PubMed] [Google Scholar]
- 15. Satoh A. K., and Ready D. F. (2005) Arrestin1 mediates light-dependent rhodopsin endocytosis and cell survival. Curr. Biol. 15, 1722–1733 10.1016/j.cub.2005.08.064 [DOI] [PubMed] [Google Scholar]
- 16. Xu H., Lee S. J., Suzuki E., Dugan K. D., Stoddard A., Li H. S., Chodosh L. A., and Montell C. (2004) A lysosomal tetraspanin associated with retinal degeneration identified via a genome-wide screen. EMBO J. 23, 811–822 10.1038/sj.emboj.7600112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xu Y., and Wang T. (2016) CULD is required for rhodopsin and TRPL channel endocytic trafficking and survival of photoreceptor cells. J. Cell Sci. 129, 394–405 10.1242/jcs.178764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cao J., Li Y., Xia W., Reddig K., Hu W., Xie W., Li H. S., and Han J. (2011) A Drosophila metallophosphoesterase mediates deglycosylation of rhodopsin. EMBO J. 30, 3701–3713 10.1038/emboj.2011.254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Partridge E. A., Le Roy C., Di Guglielmo G. M., Pawling J., Cheung P., Granovsky M., Nabi I. R., Wrana J. L., and Dennis J. W. (2004) Regulation of cytokine receptors by Golgi N-glycan processing and endocytosis. Science 306, 120–124 10.1126/science.1102109 [DOI] [PubMed] [Google Scholar]
- 20. Laporte S. A., Oakley R. H., Holt J. A., Barak L. S., and Caron M. G. (2000) The interaction of β-arrestin with the AP-2 adaptor is required for the clustering of β2-adrenergic receptor into clathrin-coated pits. J. Biol. Chem. 275, 23120–23126 10.1074/jbc.M002581200 [DOI] [PubMed] [Google Scholar]
- 21. Wang T., and Montell C. (2007) Phototransduction and retinal degeneration in Drosophila. Pflugers Arch. 454, 821–847 10.1007/s00424-007-0251-1 [DOI] [PubMed] [Google Scholar]
- 22. Sengupta S., Barber T. R., Xia H., Ready D. F., and Hardie R. C. (2013) Depletion of PtdIns(4,5)P2 underlies retinal degeneration in Drosophila trp mutants. J. Cell Sci. 126, 1247–1259 10.1242/jcs.120592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang T., Jiao Y., and Montell C. (2005) Dissecting independent channel and scaffolding roles of the Drosophila transient receptor potential channel. J. Cell Biol. 171, 685–694 10.1083/jcb.200508030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Doherty G. J., and McMahon H. T. (2009) Mechanisms of endocytosis. Annu. Rev. Biochem. 78, 857–902 10.1146/annurev.biochem.78.081307.110540 [DOI] [PubMed] [Google Scholar]
- 25. Schink K. O., Tan K. W., and Stenmark H. (2016) Phosphoinositides in control of membrane dynamics. Annu. Rev. Cell Dev. Biol. 32, 143–171 10.1146/annurev-cellbio-111315-125349 [DOI] [PubMed] [Google Scholar]
- 26. Han J., Reddig K., and Li H. S. (2007) Prolonged Gq activity triggers fly rhodopsin endocytosis and degradation, and reduces photoreceptor sensitivity. EMBO J. 26, 4966–4973 10.1038/sj.emboj.7601929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xiong B., and Bellen H. J. (2013) Rhodopsin homeostasis and retinal degeneration: Lessons from the fly. Trends Neurosci. 36, 652–660 10.1016/j.tins.2013.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang X., Mu Y., Sun M., and Han J. (2017) Bidirectional regulation of fragile X mental retardation protein phosphorylation controls rhodopsin homoeostasis. J. Mol. Cell Biol. 9, 104–116 10.1093/jmcb/mjw041 [DOI] [PubMed] [Google Scholar]
- 29. Hirsch J. A., Schubert C., Gurevich V. V., and Sigler P. B. (1999) The 2.8 Å crystal structure of visual arrestin: A model for arrestin's regulation. Cell 97, 257–269 10.1016/S0092-8674(00)80735-7 [DOI] [PubMed] [Google Scholar]
- 30. Gurevich V. V., and Gurevich E. V. (2006) The structural basis of arrestin-mediated regulation of G-protein-coupled receptors. Pharmacol. Ther. 110, 465–502 10.1016/j.pharmthera.2005.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kaksonen M., and Roux A. (2018) Mechanisms of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 19, 313–326 10.1038/nrm.2017.132 [DOI] [PubMed] [Google Scholar]
- 32. Gaidarov I., Krupnick J. G., Falck J. R., Benovic J. L., and Keen J. H. (1999) Arrestin function in G protein-coupled receptor endocytosis requires phosphoinositide binding. EMBO J. 18, 871–881 10.1093/emboj/18.4.871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee S. J., Xu H., Kang L. W., Amzel L. M., and Montell C. (2003) Light adaptation through phosphoinositide-regulated translocation of Drosophila visual arrestin. Neuron 39, 121–132 10.1016/S0896-6273(03)00390-8 [DOI] [PubMed] [Google Scholar]
- 34. Gaidarov I., and Keen J. H. (1999) Phosphoinositide-AP-2 interactions required for targeting to plasma membrane clathrin-coated pits. J. Cell Biol. 146, 755–764 10.1083/jcb.146.4.755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Padrón D., Wang Y. J., Yamamoto M., Yin H., and Roth M. G. (2003) Phosphatidylinositol phosphate 5-kinase Iβ recruits AP-2 to the plasma membrane and regulates rates of constitutive endocytosis. J. Cell Biol. 162, 693–701 10.1083/jcb.200302051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Page L. J., and Robinson M. S. (1995) Targeting signals and subunit interactions in coated vesicle adaptor complexes. J. Cell Biol. 131, 619–630 10.1083/jcb.131.3.619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Höning S., Ricotta D., Krauss M., Späte K., Spolaore B., Motley A., Robinson M., Robinson C., Haucke V., and Owen D. J. (2005) Phosphatidylinositol-(4,5)-bisphosphate regulates sorting signal recognition by the clathrin-associated adaptor complex AP2. Mol. Cell 18, 519–531 10.1016/j.molcel.2005.04.019 [DOI] [PubMed] [Google Scholar]
- 38. Alloway P. G., and Dolph P. J. (1999) A role for the light-dependent phosphorylation of visual arrestin. Proc. Natl. Acad. Sci. U.S.A. 96, 6072–6077 10.1073/pnas.96.11.6072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Matsumoto H., Kurien B. T., Takagi Y., Kahn E. S., Kinumi T., Komori N., Yamada T., Hayashi F., Isono K., and Pak W. L. (1994) Phosrestin I undergoes the earliest light-induced phosphorylation by a calcium/calmodulin-dependent protein kinase in Drosophila photoreceptors. Neuron 12, 997–1010 10.1016/0896-6273(94)90309-3 [DOI] [PubMed] [Google Scholar]
- 40. Stowers R. S., and Schwarz T. L. (1999) A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics 152, 1631–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu A. H., Chu M., and Wang Y. P. (2019) Up-regulation of Trem2 inhibits hippocampal neuronal apoptosis and alleviates oxidative stress in epilepsy via the PI3K/Akt pathway in mice. Neurosci. Bull. 35, 471–485 10.1007/s12264-018-0324-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lee S. J., and Montell C. (2004) Light-dependent translocation of visual arrestin regulated by the NINAC myosin III. Neuron 43, 95–103 10.1016/j.neuron.2004.06.014 [DOI] [PubMed] [Google Scholar]
- 43. Hu W., Wan D., Yu X., Cao J., Guo P., Li H. S., and Han J. (2012) Protein Gq modulates termination of phototransduction and prevents retinal degeneration. J. Biol. Chem. 287, 13911–13918 10.1074/jbc.M112.339895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu L., Tian Y., Zhang X. Y., Zhang X., Li T., Xie W., and Han J. (2017) Neurexin restricts axonal branching in columns by promoting ephrin clustering. Dev. Cell 41, 94–106.e4 10.1016/j.devcel.2017.03.004 [DOI] [PubMed] [Google Scholar]
- 45. Hu W., Wang T., Wang X., and Han J. (2015) Ih channels control feedback regulation from amacrine cells to photoreceptors. PLoS Biol. 13, e1002115 10.1371/journal.pbio.1002115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wan D., Zhang Z. C., Zhang X., Li Q., and Han J. (2015) X chromosome-linked intellectual disability protein PQBP1 associates with and regulates the translation of specific mRNAs. Hum. Mol. Genet. 24, 4599–4614 10.1093/hmg/ddv191 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.