Abstract
Differential mobility spectrometry (DMS) has been gaining popularity in small molecule analysis over the last few years due to its selectivity towards a variety of isomeric compounds. While DMS has been utilized in targeted liquid chromatography-mass spectrometry (LC-MS), its use in untargeted discovery workflows has not been systematically explored.
In this contribution, we propose a novel workflow for untargeted metabolomics based solely on DMS separation in a clinically relevant chronic kidney disease (CKD) patient population. We analyzed ten plasma samples from early- and late-stage CKD patients. Peak finding, alignment and filtering steps performed on the DMS-MS data yielded a list of 881 metabolic features (unique mass-to-charge and migration time combinations). Differential analysis by CKD patient group revealed three main features of interest. One of them was putatively identified as bilirubin based on high-accuracy MS data and comparison of its optimum compensation voltage (COV) with that of an authentic standard.
The DMS-MS analysis was 4 times faster than a typical HPLC-MS run, which suggests potential for the utilization of this technique in screening studies. However, its lower separation efficiency and reduced signal intensity make it less suitable for low-abundant features. Fewer features were detected by the DMS-based platform compared to an HPLC-MS-based approach but, importantly, the two approaches resulted in different features. This indicates a high degree of orthogonality between HPLC- and DMS-based approaches and demonstrates the need for larger studies comparing the two techniques.
The workflow described here can be adapted for other areas of metabolomics and has value as a pre-screening method to develop semi-targeted workflows and as a faster alternative to HPLC in large biomedical studies.
Keywords: Differential mobility spectrometry, mass spectrometry, biomarker discovery, untargeted metabolomics, chronic kidney disease
Graphical Abstract

1. Introduction
In this contribution, we propose a workflow for fast metabolomic biomarker screening by differential mobility spectrometry coupled to mass spectrometry (DMS-MS). It is based on the hypothesis that DMS-MS has the potential to significantly reduce analysis time and provide an orthogonal method for the detection of differential features.
Metabolomics has emerged as a valuable tool to identify metabolic perturbations in diabetes [1–4], cardiovascular disease [5–7], cancer [8] and chronic kidney disease (CKD) [9–11]. The latter is a major risk factor for cardiovascular mortality as well as a risk for progression to End Stage Renal Disease (ESRD). Life expectancy becomes progressively shorter with greater severity of CKD in all age groups. The prevalence of CKD in the U.S. adult general population is 15 per cent (United States Renal Data System, Annual Report 2018, accessed via https://www.usrds.org/ on February 2, 2019).
As just a fraction of the total costs of kidney disease care, Medicare expenditures for ESRD exceeded $31.8 billion in 2014, accounting for 7.2% of the overall Medicare paid claims costs. Additionally, Medicare spending for beneficiaries aged 65+ who have CKD exceeded $50 billion in 2014, representing 20% of all Medicare spending in this age group. Although interventions can help to delay the progression of CKD, no single intervention or group of interventions has been found effective in reducing the rate of CKD progression to ESRD. Since 2011, the incidence of CKD has begun rising again and the number of prevalent ESRD cases is rising by >20,000 cases per year (United States Renal Data System, Annual Report 2016, accessed via https://www.usrds.org/ on August 26, 2018).
Effectively addressing the public health concern of CKD necessitates improving the prognostic and diagnostic value of currently used clinical markers for CKD diagnosis and prognosis.
MS-based, untargeted metabolomics is playing a major role in the efforts to identify suitable biomarkers. In CKD, rather than individual biomarkers, panels of metabolites could be used to increase the accuracy and precision of CKD diagnosis and prediction [12]. With this approach, the incremental changes in metabolite levels reflecting CKD onset and progression require large patient cohorts to achieve the necessary statistical power. Fast analytical methods are desirable to achieve the necessary throughput and minimize instrumental drift.
DMS is a newly emerging separation technique that has enriched targeted metabolomics analyses for a wide range of target compounds, among them drug metabolites [13–15] or, more recently, biomarkers of radiation exposure and creatinine [16]. The benefits of DMS include a fast separation mechanism and an easily tunable selectivity that is often complementary to high-performance liquid chromatography (HPLC)-based separations [17]. Furthermore, DMS can be coupled to MS in a straightforward fashion [18]. However, there are no prior reports of an LC-free DMS-MS platform being used in an untargeted metabolomics study.
We recently showed that, using isopropanol (iPrOH) as the modifier, DMS-MS facilitates the detection of a wide range of metabolites with very short analysis times [17]. Based on this finding, we envisioned a DMS-MS- based screening platform capable of identifying metabolic features associated with the progression of CKD in biological samples derived from CKD patients.
For our pilot study, we selected plasma samples of CKD patients from an existing cohort [19]. We utilized the unique selectivity and speed of DMS together with high-accuracy MS detection to extract differentially expressed features in patients with early- compared to late-stage CKD. The simple analytical workflow is based on direct infusion of plasma extracts into the DMS cell, followed by separation of the metabolic features and sequential transfer to the MS. This is achieved by ramping the compensation voltage (COV) over a predetermined range and collecting an MS spectrum at every COV step [17]. Differential analysis of the high-accuracy DMS-MS data by unsupervised and univariate methods revealed features of interest (FOIs) having different intensities in patients with early-stage compared to advanced CKD. Selected FOIs were tentatively identified based on accurate mass and observed optimum COV. This preliminary identification must be further corroborated by targeted LC-MS/MS analysis (outside the scope of this study). Conceptually, our study is a proof-of principle in a small patient population and does not have diagnostic or predictive power. However, it has broader applicability for studies in (clinical) metabolomics across a spectrum of disorders that ranges from metabolic diseases to cancer to cardiovascular disease.
2. Experimental section
Chemicals.
HPLC-MS grade water and acetonitrile (ACN) and HPLC grade isopropanol (iPrOH) were purchased from Fisher Scientific (Hampton, NH). Ammonium acetate (NH4OAc) was purchased from Sigma Aldrich (St. Louis, MO).
Human plasma samples.
Pooled plasma (PP) from healthy volunteers, used in preliminary experiments and for confirming intra-batch consistency in the main experiment, was obtained from the American Red Cross. Ten plasma samples of CKD patients were obtained from the University of Michigan’s Clinical Phenotyping Resource and Biobank Core (CPROBE) [9]. Established under the George O’Brien Kidney Center at the University of Michigan, CPROBE is a multicenter cohort of 1235 adult individuals with CKD. Biologic specimens and clinical data have been collected for translational research. The inclusion criteria were patients from all stages of CKD balanced by sex and race across all stages of CKD. We selected, five patients each from Stages 2 and 5. Baseline clinical and laboratory data as well as stored plasma samples were gathered cross-sectionally at the time of enrollment. We used the CKD Epidemiology Collaboration equation for eGFR calculation [20]. Key patient demographics are summarized in Electronic Supplementary Material (ESM) Table S1. All biological samples were stored at −80°C until processing.
Sample preparation.
200 μL of an ice-cooled, 1:1:1 (v/v/v) mixture of methanol, acetone and acetonitrile were used to precipitate proteins and extract metabolites from 50 μL of plasma. After incubation at +4°C (30 min), and −20°C (60 min) and centrifugation (+4°C, 10 min, 10,000 rpm), the supernatant was transferred to autosampler vials, dried under nitrogen and re-dissolved in ACN/H2O (80:20, v/v). Samples were kept at 4°C in the autosampler until analysis. The samples were analyzed in a block-randomized sequence together with solvent blanks and pooled plasma samples.
Instrumentation.
HPLC and DMS-MS experiments were carried out on a 5600+ Triple TOF quadrupole-time of flight instrument (Sciex, Framingham, MA) coupled to a Shimadzu Nexera X2 UHPLC system with degasser, binary pump and thermostated autosampler via a Turbo V ion source. A SelexION differential mobility cell (Sciex) was installed in the atmospheric pressure region of the MS instrument for the DMS experiments.
Experimental setup.
The reversed-phase column used in the LC runs was a Waters Acquity HSS T3 C-18 (1.7 μm particle size, 2.1 × 100 mm inner diameter) equipped with a 5 mm pre-column. The mobile phase consisted of a mixture of water (A) and methanol (B) with 0.1 % formic acid. Injection volume was 5 μL. The conditions were: flow rate 0.3 mL/min, temperature 55°C, gradient: 0–1.5 min: 5 % B, 23 min: 75 % B, 26 min: 98% B, 34.5 min: 98%B, 34.6 min: 5% B, equilibration time: 10 min, total run time: 45 min. MS/MS data were collected using a data-independent acquisition (IDA) approach.
For the DMS experiments, a zero-dead volume stainless steel union replaced the column. The carrier solvent was 75 % ACN and 25 % H2O (50 mM NH4OAc) and the solvent flow was 20 μL/min. The DMS separation voltage (SV) was 3.8 kV. Each DMS measurement was followed by a two-minute washing step at 200 μL/min.
The “COV cycling” experiments for selecting metabolite features from pooled plasma are described in the ESM (Section 1.1). The “COV ramping” experiments for feature discovery in CPROBE samples were carried out by infusing 40 μL of plasma extract into the DMS cell. The COV range (−40 V - +20 V) was covered in 0.2 V steps. All measurements were carried out in positive mode and the m/z range was 100–1200 (MS cycle time: 1 s). The run time was 5 minutes per sample, not including a 30 s wait time between injection and initiation of the COV ramping.
Data processing and statistical analysis.
For visualization, individual ion traces were extracted with a width of 0.075 Da. First-pass data evaluation, peak-picking and alignment were performed with PeakView 2.2 and MarkerView 1.3.1 (Sciex), respectively. Parameter settings are summarized in the ESM (Section 1.3).
Peak picking and alignment were conducted after transformation of the ionograms from the voltage (COV) into the time (min) domain. 881 features, characterized by a unique combination of m/z and migration time (m/z_tM), were identified from 17 samples (five per group for early- and late-stage CKD and seven technical replicates of PP). Using the online platform MetaboAnalyst [21], 225 features with over 50 % missing values were removed and no missing values remained. Feature filtering was based on the group median. The feature intensity was normalized by the sample median, log transformed and auto-scaled.
Statistical tests were based on assumption of normal distribution and a probability cutoff of 0.05 was employed unless otherwise noted.
METLIN database search.
The METLIN database was searched for the accurate mass of FOIs allowing for proton, sodium and ammonium adducts with 30 ppm mass accuracy. Results shown in Section 3.4 (Table 1) represent a sub-set of the matches.
Table 1.
Primary features. Average compensation voltage (COV) and relative standard deviation (RSD) are based on 10 samples from the CPROBE cohort
| Feature | Stage 5 | m/z | tM/min | COV | Metlin hits (selection) | |
|---|---|---|---|---|---|---|
| Average/V | RSD/% | (number of hits, [Adduct], mass accuracy) | ||||
| A | down | 266.85 | 1.84 | −24.9 | −1.1 | No hits |
| B | down | 586.30 | 3.06 | −10.74 | −0.9 | 9 hits |
| C28H39N7O7 ([M+H], 10 ppm) – peptide, e.g. Phe Arg Tyr Tyr | ||||||
| C28H44N2O8S ([M+NH4], 18 ppm) – leukotriene F4 | ||||||
| C34H36N2O6 ([M+NH4], 23 ppm): y-morphine | ||||||
| C | down | 602.28 | 3.14 | −9.87 | −5.9 | 7 hits |
| C29H39N5O9 ([M+H], 4 ppm): peptide, e.g. Gly Lys Tyr Tyr | ||||||
| C33H36N4O6 [M+NH4], 29 ppm): bilirubin | ||||||
| C26H32N8O8 ([M+NH4], 19 ppm): Asp His Trp Gln | ||||||
| D | up | 232.98 | 2.39 | −18.18 | −2.6 | No hits |
| E | up | 386.70 | 2.1 | −22.1 | −0.6 | No hits |
| F | up | 842.59 | 3.78 | −1.1 | −56.0 | 18 hits [M+H]+, 30 hits [M+NH4]+, 40 hits [M+Na]+ |
| C49H80NO8P ([M+H, 25 ppm): PE_44:9 | ||||||
| C46H84NO10P ([M+H], 0 ppm: PS_40:3 | ||||||
| C46H81O10P ([M+NH4], 0 ppm: PG_40:5 | ||||||
| C47H83NO8P ([M+Na], 27 ppm): PC 39:6 or PC_O-18:0/22:6 | ||||||
Patient protection.
All human studies were approved by the Institutional Review Board (IRB) for the University of Michigan and the CPROBE ancillary studies committee. The samples were de-identified and no personal information was included in the patient data provided by the CPROBE coordinators for this study. As such, the use of human samples in this study did not require separate approval by an IRB or renewal of informed consent. Information on the CPROBE study is available at https://clinicaltrials.gov/ct2/show/NCT01016613.
The pooled plasma samples used as experimental controls were obtained from the American Red Cross and were not subject to IRB approval or informed consent.
3. Results
Sample selection:
This study was conducted using plasma samples of patients with early- and late-stage CKD (N=5 per group) from the CPROBE cohort. ESM Table S1 summarizes the key demographic data of CKD patients in this study.
The patient samples were selected based on CKD stage and the groups were matched for sex, race and incidence of diabetes. In accordance with the clinical phenotype, the first group included early stage CKD (Stage 2; estimated glomerular filtration rate-eGFR- 60–90) and the second group included late-stage CKD (Stage 5; eGFR <15) patients. Stage 5 patients were older (p<0.05) and, as expected by design, had reduced kidney function. Clinically, this is manifested in their lower eGFR and higher serum creatinine compared to Stage 2 CKD patients (p<0.01).
Due to the limited amounts of patient sample available, pooled plasma from healthy controls was employed for the preliminary studies described in the ESM Sections 1.1 and 1.2 and to determine the separation performance in the main experiment (see 3.1). Technical replicates of the pooled plasma extract were processed along with patient samples and analyzed during system equilibration and in between CKD patient samples.
3.1. Separation performance of DMS
Data acquisition.
In DMS analysis, a given combination of separation voltage (SV) and compensation voltage (COV) facilitates the transfer of compounds of a specific molecular mass, charge and shape through the DMS cell and into the MS. Molecules having different properties collide with the DMS electrodes and are eliminated.
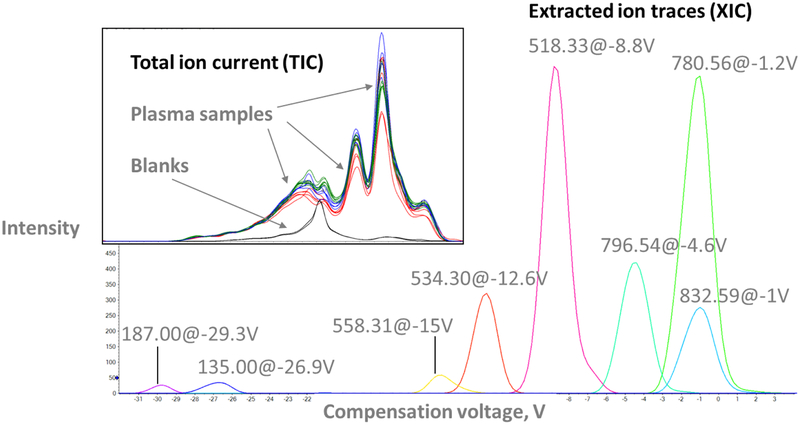
“COV ramping” increases the COV in a stepwise fashion to cover the analytical range of interest for metabolites and transfer them to the MS in a sequential fashion. By collecting an MS spectrum at every COV, ramping experiments provide ionograms, plots of MS signal intensity vs. COV analogous to chromatograms (Fig. 1, insert). Individual ion traces (XICs) can be extracted from ionograms to determine the optimum COV for a given m/z from the apex of the corresponding peak (Fig. 1, main panel). A detailed description of the COV cycling mode, an alternative DMS operation mode, can be found in ESM Sections 1.1 and 1.2).
Fig. 1.
Extracted ion traces for selected features are distributed across the COV range in a representative plasma sample (main panel). Insert: Total ion current (TIC) traces of plasma samples (colored lines) display similar peaks and are clearly distinct from those of blank samples (black traces)
Separation characteristics.
The analytical performance of the DMS was determined based on seven technical replicates of PP extract analyzed along with the patient samples. The XICs of metabolic features known to be present in the PP samples (see ESM Section 1.1) were used to determine that the average peak width in the main experiment was 1.5 V. In combination with the experimental COV range of 60 V, the DMS platform had a peak capacity of 20. This is at least an order of magnitude lower than a typical HPLC separation and raises concerns regarding the accurate quantification and identification of metabolic features in the presence of isobaric, co-migrating species. The total peak capacity of the DMS-MS platform is significantly larger and depends on the scan speed of the mass spectrometer. Here, an m/z range of 1,100 was covered in one second to enhance signal intensity (see Experimental).
The analysis time in the DMS-MS experiments was 5 minutes per sample, plus two minutes of cleaning after each run. This is significantly faster than a typical HPLC separation and has the potential to facilitate higher throughput and reduce issues with instrumental drift.
In addition, the run-to-run reproducibility of the compensation voltage was determined to be excellent, with RSDs for COVs of reference features typically below 1 % (ESM Table S3). This is in line with a highly reproducible LC method. However, the relatively high peak area RSDs for some species are indicative of ion suppression, a common and often prohibitive issue in direct infusion MS analysis.
3.2. Data evaluation and processing
Generating the feature list.
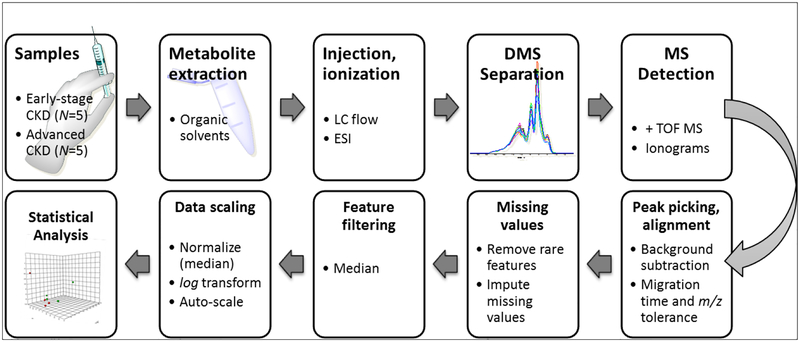
A metabolic feature derived from DMS-MS data is characterized by a unique m/z and corresponding COV or migration time (tM). In this experiment, feature finding with open-access tools failed due to the low MS signal intensity and efficiency of the DMS platform. Therefore, we combined commercial and open-access software solutions to generate peak lists and extract the differential features (Fig. 2).
Fig. 2.
The experimental workflow included extraction of the plasma metabolites with organic solvent, DMSMS analysis, peak finding and alignment, normalization and statistical analysis
The ionograms were transferred from the COV into the migration time domain by importing the .wiff files into MarkerView. Feature finding and peak alignment across samples were performed and881 distinct m/z_tM features with their corresponding intensities were obtained (see also ESM Section 1.2, for details). Each feature is denoted as m/z@COV before the transformation or m/z_tM (after transformation).
The blanks and PP extracts were removed from the original sample list before it was transposed and imported into MetaboAnalyst for differential analysis [21]. 255 features were removed because they either showed constant intensities across samples (non-relevant features for differential analysis) or only appeared in a single sample (low-confidence features likely to be artifacts). The feature intensities were then median-normalized, log-transformed and auto-scaled (ESM Fig. S2). Note that some features still showed non-normal intensity distributions across samples. This is an effect of the small sample size and large intensity differences between CKD stages.
3.3. Selection of features of interest
Unsupervised methods.
“Features of interest” (FOIs) are features present at significantly different levels in samples from early- versus late-stage CKD patients. To select those features, we considered Principal Component Analysis (PCA) and Partial Least Squares Discriminant Analysis (PLS-DA) [23].
As shown in the ESM, 2-dimensional PCA score plots showed a significant overlap between early and late-stage patients (ESM Fig. S3 a). Three components were required to achieve separation of the two groups (ESM Fig. S3 b, cumulative variance explained 34.2+18.2+ 12.0 =64.4%). PCA loading plots were therefore deemed unsuitable for the selection of FOIs.
PLS-DA plots, on the other hand, showed a separation of Stage 2- and Stage 5 CKD patients and, in addition, revealed sub-clusters within each stage (ESM Fig. S3 c). The clustering pattern, which was corroborated by correlation analysis (ESM Fig. S3 d), was not linked to gender, race, incidence of diabetes or statin use (ESM Table S4) but may be related to the run order. The pattern corresponded to a decreasing signal intensity of reference features with run order (not shown), which suggests that instrumental drift may have influenced the experimental results. This was an unexpected finding given the exceptionally short run times.
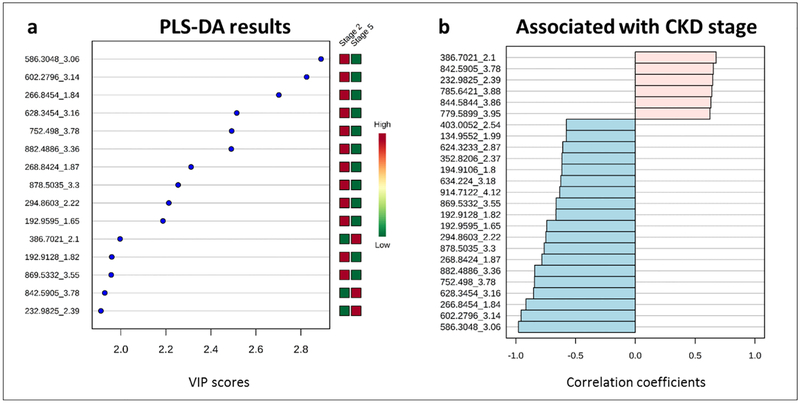
A PLS-DA-based list of FOIs was created based on the top 15 features by VIP score for the first five components. Fig. 3 a shows the top 15 features based on their VIP scores for PLS-DA component 1. Removing redundant features appearing in the top 15 more than once resulted in a shortlist containing 17 features.
Fig. 3.
a Top 15 distinguishing features based on Partial Least Squares-Discriminant Analysis (PLS-DA) Variable Importance In Projection (VIP) scores for component 1. b Top 25 distinguishing features by t-tests
Univariate analysis (t-tests).
To select FOIs for further investigation, we also employed univariate analysis (comparing feature intensity differences within and between the CKD groups) and correlation analysis (changes in feature intensities from stage 2 to stage 5 CKD).
According to fold change analysis, 157 features exceeded a logarithmic intensity difference greater than 2 (ESM Fig. S4). Of the top 25 features correlated to CKD stage, only six showed increased intensities in stage 5 samples compared to stage 2. The other 19 were up-regulated in stage 2 (Fig. 3b).
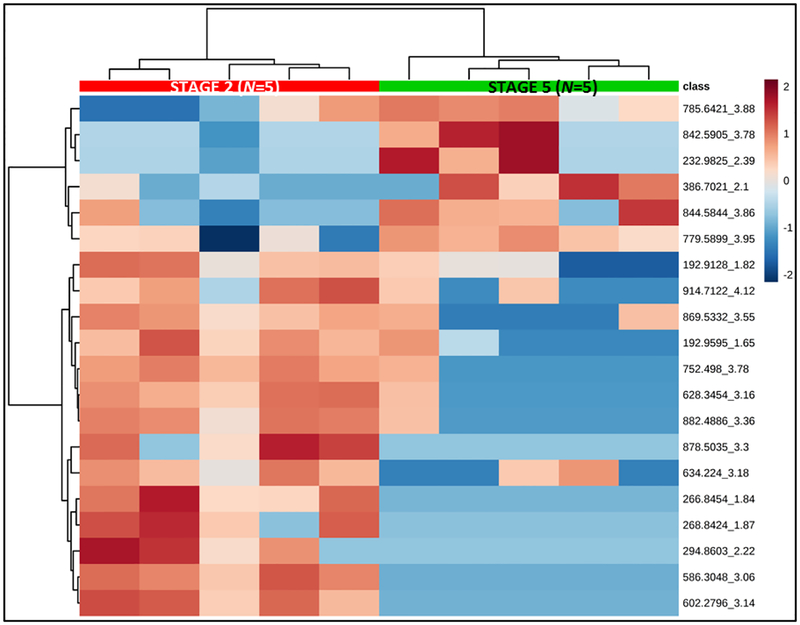
Fig. 4 shows a heat-map based on relative signal intensities of the top 25 features by t-tests (equal variance, parametric tests). In this graph, each column represents a sample and each row represents a feature. Color intensities correspond to relative signal intensity for each differentially regulated feature across samples. The samples are arranged into the two CKD patient groups (marked in red and green) by unsupervised clustering.
Fig. 4.
Heatmap showing samples in columns and top 20 distinguishing features by t-tests in rows. Colors represent relative feature intensities across samples. Samples are arranged by unsupervised clustering
After correcting for multiple testing using the false discovery rate (FDR) method, three primary features of interest emerged from our analysis (see Figure 3): Features A (m/z_tM 266.8454_1.84), B (586.3048_3.06) and C (602.2796_3.14) were all down-regulated in the late-stage CKD samples. They strongly correlated with each other (R2>0.9) and B and C even co-migrated with average COVs of −10.7 V and −9.9 V, respectively (Table 1). Co-migration could be an indication of chemical similarity but the low separation power of the DMS does not facilitate a definitive conclusion.
The three FOIs were also main drivers for the separation of the groups in the PLS-DA analysis (Fig. 3a).
We also selected three additional FOIs for further investigation based on the fact that they were elevated in stage 5 compared to stage 2. Features D (232.9825_2.39), E (386.7021_2.1) and F (842.5905_3.78) had the highest VIP scores in the PLS-DA analysis but did not pass the FDR-corrected fold change analysis due to their relatively high intra-group variability (boxplots not shown). Generally, the raw intensity data showed considerable intra-group variability with RSDs ranging from 26–89 %. Data treatment including normalization, log transformation and auto-scaling, however, successfully corrected these issues.
As observed previously for the reference features in PP (ESM Table S2), the COV reproducibility for FOIs in CPROBE samples was excellent with RSDs below 2% for most features (Table 1). The notable example was FOI F with an exceptionally high RSD of 58%. The extracted ion traces for this feature showed highly heterogeneous peak-shapes ranging from Gaussian in some samples to poorly resolved double peaks in others with no apparent correlation to CKD group or available demographic data. We suspected that Feature F contains multiple, unresolved molecular species (see also Section 3.4, Towards feature identification) and performed a HPLC-MS analysis of the same sample set (see below).
Comparison with HPLC-MS data.
For comparison, the CKD patient samples were also analyzed using a generic RP-HPLC-MS method on same instrument with the DMS cell removed. As expected, approximately 3–4 times more features were detected with the latter method. After removing low-abundance (I<100 cps) and low-confidence features (present in less than 4 samples per group), about 3,000 features remained. In absolute numbers, the HPLC-based platform detects more features than DMS-MS. This is related to a) the higher separation efficiency of the LC compared to DMS, b) the resulting, reduced ion suppression and c) the increased ion path length in DMS. However, relative to the run time (5 minutes for DMS, 20 minutes for HPLC), the DMS performance can be viewed as competitive with 170 features per minute compared to 150 features per minute for HPLC-MS.
Comparison of the differential features extracted from DMS and HPLC data showed that the two analyses were highly complementary.
In the HPLC-MS dataset, 58 features were determined to be significantly different between early- and late-stage CKD samples based on normalized, log transformed and auto-scaled data (t-tests, unequal variance, FDR-adjusted p<0.05, ESM Table S6). There was no overlap between these 58 features and the top 25 features from t-tests on DMS data. This result suggests that DMS screening might be orthogonal to RP-based analysis. The potential for expansion of the separation space in untargeted metabolomics may be best exploited by coupling the two techniques.
We investigated the m/z traces of the DMS-derived FOIs in the HPLC-based dataset for the presence of isobaric interferences. A B, D and E appeared as single peaks (XICs for m/z ± 0.005 Da) with E showing very low intensity. The extracted ion chromatograms of FOIs C and F showed multiple peaks, which illustrates the limitations of the DMS separation. However, it also demonstrates how DMS experiments can trigger follow-up by HPLC-MS and inform the design of targeted follow-up experiments.
3.4. Towards feature identification by accurate m/z and COV
The accurate mass and optimum COV data collected by the DMS-MS platform can be utilized to assign tentative identities to metabolite features. Accurate mass data facilitates database searches and the observed COV can be compared to that of an authentic standard measured under identical conditions to narrow down candidate identities.
We searched for the accurate m/z of FOIs A-F in the METLIN online database. A 30 ppm window was chosen to account for the potential instrumental drift in between calibration runs. Excluding drugs and toxicants, we found several potential matches for FOIs B, C and F but none for A, D and E. A selection of candidates is included in Table 1.
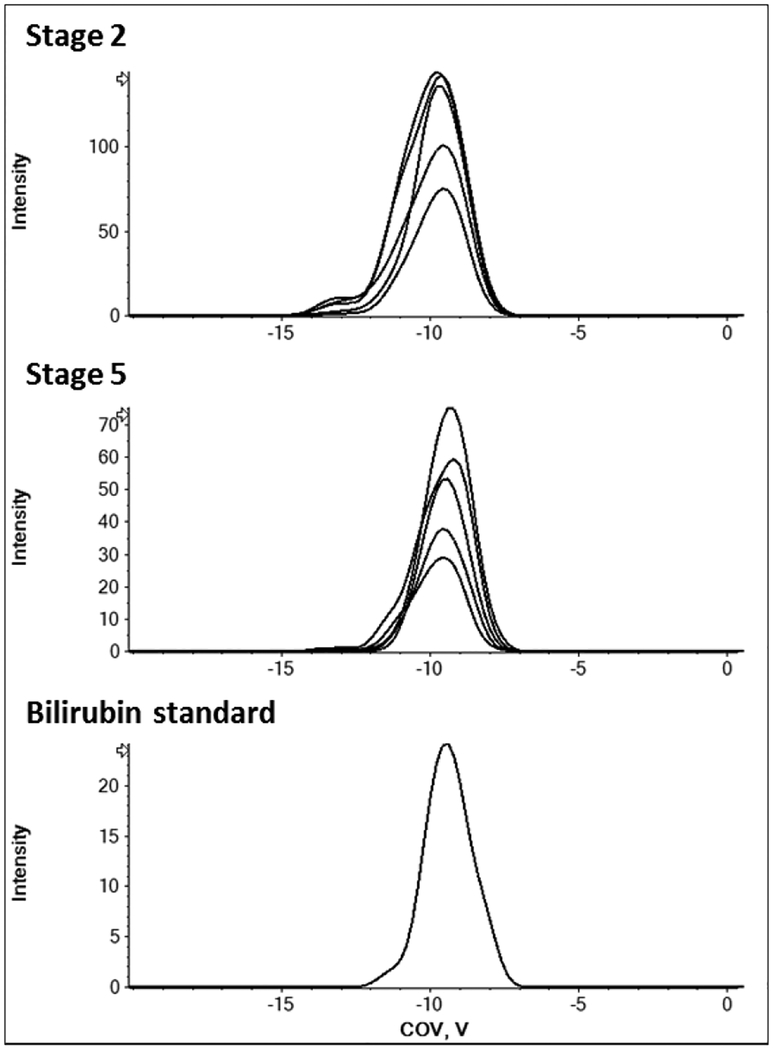
Most potential matches referred to peptides or lipid species for which authentic standards were not readily available. However, FOI C (m/z 602), matched the ammonium adduct of bilirubin. As shown in Fig. 5, the optimum COV of FOI C is identical to that of the authentic bilirubin standard and the ammonium adduct was indeed the main ion detected for bilirubin. It is important to note, while accurate mass and compensation voltage matches can provide useful hints at a feature’s identity, MS/MS and retention time matches with an authentic standard are still the gold standard for metabolite identification. Thus, further experiments would be necessary to unequivocally confirm the identity of FOI C.
Fig. 5.
The COV of the unknown feature 602 matches that of an authentic bilirubin standard measured under identical conditions (separation voltage 3.8 kV, modifier isopropanol). Extracted ion chromatogram at m/z 602.297 +/−0.025 Da
FOI F first captured our attention because of its unusually high COV variability (58 % RSD compared to <2 % for all others). We observed that it appeared as two poorly resolved peaks in several samples, showed multiple peaks in the HPLC analysis (ESM Fig. S5), and yielded a very high number of METLIN hits. The suggestions represented phospholipids of different classes, namely phosphatidylethanolamines (PE), -glycerols (PG), -cholines (PC) and -serines (PS). These findings suggested that FOI F contains multiple molecular species, most likely members of the phospholipid family. The results are consistent with the profile of isomeric phospholipids in plasma samples. As for feature C, MS/MS data would be required to unequivocally establish its identity.
4. Discussion
Significance.
In this contribution, we discuss a workflow for the extraction of dysregulated metabolite features from DMS-MS screening data from biological samples. We used plasma from CKD patients to demonstrate its utility for probing phenotypical differences between disease states and compare the results to HPLC-MS. Importantly, we show how DMS-MS generated data can be processed to extract differential features and putatively identify metabolites. To the best of our knowledge, this is the first report of the application of DMS technology for untargeted metabolomics screening (PubMed search, January 2019). Due to its novelty, some issues remain unresolved in this contribution and further work is needed to make this approach suitable for large-scale studies. An important limitation is the lack of (internal) standards. Potentially, plasma background ions could serve as reference in the absence of suitable standards. Another crucial step would be the incorporation of MS/MS into the DMS-based workflow. Furthermore, it appears that the quality and biological relevance of features of interest discovered with this approach would be greatly enhanced by sample de-salting and pre-concentration steps. Nonetheless, we believe that our work is an important step towards the broader application of DMS as a biomarker discovery tool across a variety of disorders in clinical metabolomics due to its relative simplicity and potential for high throughput.
Experimental design.
We analyzed plasma extracts of CKD patients selected from an existing patient cohort based on previously obtained clinical diagnosis. They corresponded to early (stage 2) or advanced (stage 5) CKD groups. In accordance with our fundamental understanding of CKD as a progressive disease, the DMSMS-based features detected in early- and late stage patients showed significant differences. The distinguishing, dysregulated metabolite features were selected using unsupervised clustering and fold change analysis. Putative identification of individual features was facilitated by a combination of high-accuracy MS data, which gave access to the molecular composition, and the feature-specific COVs, which can be matched with an authentic standard and followed up on by HPLC-MS and –MS/MS analysis.
This work is intended as a proof-of-concept study illustrating the data acquisition and analysis. Due to the small sample size, the differential features do not have diagnostic or prognostic power in the context of CKD. Follow-up in a larger, appropriately powered patient cohort representing all CKD stages is required to achieve clinically relevant results.
DMS vs. HPLC.
Compared to HPLC, DMS has advantages in terms of easy method development, analysis speed and throughput and its often complementary selectivity [17]. The former is the result of a faster separation mechanism on the microsecond scale [18] and shorter regeneration and cleaning time between runs. Our setup, in which five minutes of analysis time were followed by a two-minute wash step, compared favorably to typical HPLC analysis times of 20–30 minutes per sample. The fast analysis time was the major driver behind this work, as (pre-)clinical studies in CKD and other clinical areas would greatly benefit from faster screening procedures.
On the other hand, important performance characteristics of the DMS, such as efficiency, peak capacity and absolute signal intensity, are inferior to those of a typical HPLC separation. The direct infusion approach is susceptible to ion suppression, and further signal loss occurs in the DMS cell. Therefore, low-abundant species and metabolites suffering from competitive ionization are likely to be underrepresented in DMS-based experiments. Co-migration of critical metabolites, a result of the lower separation efficiency of the DMS, can potentially cause erroneous quantitation in DMS-based analysis. Studies utilizing well-characterized sets of metabolite standards are required to improve our understanding of these phenomena and to better quantify the differences between HPLC and DMS performance.
Ion suppression could be greatly reduced by implementing a clean-up or desalting step such as solid phase extraction or a very short (5 mm) LC column. Removing salts would enhance metabolite identification by reducing the number of different adducts for a single precursor. In addition, it would reduce competitive ionization in the ESI source and neutralization effects such as proton transfers in the DMS cell. We had not included these approaches in our experimental design to keep the workflow as simple as possible as it was a proof-of-concept study. Based on our results, however, we recommend that follow-up studies include measures to reduce the matrix and ion load.
Some limitations of the DMS, such as the ion path length, are not readily addressable by the end user. Instrumental developments are expected to improve ion transmission and facilitate higher signal intensity and improve the detection of low-abundance species as well as reproducibility.
Data evaluation.
Data evaluation and processing constituted major bottlenecks in this study. No commercial tools or common guidelines exist for performing automated peak picking and alignment operations on DMS-generated data and the low DMS efficiency and signal intensity were prohibitive to using open-access tools throughout the whole process. To arrive at the peak picking and data processing workflow reported here, we used a combination of commercial and open-access software went through several iterations to optimize the parameters and confirm consistent analysis results.
We also faced data analysis challenges related to the MS detection. Specifically, a reduction in signal intensity over time was noted for some features. Initial evaluation of the data for 8 reference features suggested that instrumental drift might influence the peak area results. Decreasing mass accuracy was suspected as a potential source of the issue. An MS calibration had been performed immediately prior to the sample analysis where the average mass accuracy for m/z range 50–1200 was found to be < 1 ppm and continuous internal calibration was performed during runs. Thus, MS precision was ruled out as the cause for declining peak intensity.
Since no column was used in the DMS-MS runs, low system backpressure could have led to higher-than-expected run-to-run variability compared to HPLC. UHPLC pumps deliver more constant flow at higher backpressures. Even though we were operating within instrument specifications, small differences in the flow rate could have affected the detected signal. However, this cause was also ruled out after confirming that the TIC intensities were constant across all samples. It is worth noting that increasing the length of PEEK tubing connecting the injector to the ESI source or placing a short column in the flow path would prevent pump stability concerns altogether. We concluded that the most likely causes for the observed variability were ion suppression and co-migration of isobaric compounds. Studies involving multi-analyte sets of known composition and concentration will have to be investigated together with different biological matrices in order to quantify these effects. As indicated in the “DMS vs. HPLC” section, a sample clean-up step would greatly reduce matrix effects and could improve signal intensity. Suitable isotope-labeled standards or reference background ions should be employed in the future to facilitate normalization across samples and batches. Before attempting to draw biological conclusions from a study, the method should be validated in larger cohorts. Further work will need to focus on overcoming the pitfalls demonstrated in our pilot study before DMSMS can be used as a more routine application for clinical biomarker discovery.
Differential analysis.
We performed partially redundant data reduction and analysis steps to select the main FOIs from the initial list of 881 unique m/z and COV (migration time) pairs. In the absence of suitable reference signals and internal standards, mean and median-based approaches were used to filter and normalize feature intensities across samples. Rigorous removal of low-confidence features drastically reduced the initial list, and only the six most influential features emerging from PLS-DA analysis were carried forward.
The results of univariate analysis (t-tests, corrected for multiple testing) confirmed 3 of the 6 FOIs. However, the validity of p-value cutoffs in this study is limited due to the small sample size. The large discrepancy between the high number of features and the small number of patients is typical for a low-powered pilot study. It is important to keep in mind that this study was designed as a proof of concept. In order to obtain biologically relevant information and high-confidence biomarker candidates, it must be repeated in a larger cohort and, ideally, also include patients with intermediate stage CKD.
The goal of untargeted analysis is the identification of differentiating features. With DMS-MS data, identification relies on accurate mass and feature-specific COV. We performed database searches based on the average m/z value determined from 10 MS scans with a relatively wide window of 30 ppm to increase the chance of hits. Thus, multiple potential chemical formulas and structure matches were obtained for 4 out of 8 FOIs. MS/MS data were not collected in this study, meaning that identification was limited to the level of the chemical composition.
As a notable exception, FOI C was identified as bilirubin based on its m/z and COV as well as an adduct match with the authentic standard. The tentative identification of this feature demonstrates that, similar to the HPLC retention factor, the COV reflects each feature’s molecular properties in a composite fashion. In the case of feature F, which showed exceptionally high intra-group variability, we were able to gain useful hints from a follow-up HPLC run. The unknown feature produced multiple, strongly retained peaks, suggesting a hydrophobic, structurally heterogeneous mixture. In combination with the accurate mass, we classified FOI F as a phospholipid isomer mixture. The latter, narrower classification shows that DMS-MS can be a time- and cost-effective tool to guide biomarker discovery by HPLC.
Limitations of this study and future directions.
The small sample size of N=5 per group is an important limitation of our study. While appropriate for a proof-of-concept, it prevents any biological conclusions to be drawn or biomarker candidates be selected from the acquired data. In addition, age was recognized as a potential confounder in our patient population.
For clinically meaningful CKD research, a method validation must be undertaken and the study replicated in a larger cohort that includes intermediate CKD stages. Nonetheless, we expect our pilot study to pave the way for future, appropriately powered large-scale studies to follow up on this work.
Second, we provided direct comparison with traditional, untargeted RP LC-MS in this small subset of patient samples but did not investigate the effect the biological matrix has on the detection of key analytes in direct infusion experiments. In order to better understand the potential of DMS-MS for non-targeted metabolites, we must conduct experiments that clearly establish which metabolites can be reliably targeted with this technique. In particular, we have to make sure that ion suppression, co-migration of isobaric compounds or the formation of multiple adducts do not interfere with quantitative analysis.
The lack of MS/MS data impeded the full identification of unknown features. Efforts must be made to integrate tandem MS measurements (data-dependent or data-independent), into DMS-MS workflows. This is essential if DMS-MS is to be used for larger-cohort studies as the elucidation of the chemical identity of differentiating metabolites and metabolite panels is the declared goal of untargeted metabolomics. The lower signal intensity, rather than the fast time scale of the separation, is the major challenge for such endeavors, especially in data-independent MS/MS workflows. Hopefully, instrumental developments will help tackle this issue.
Finally, new concepts are needed to address the particularities of DMS separation on the data analysis side. For example, the state-of-the-art in data reduction for LC-MS-based, untargeted metabolomics includes grouping of features and adducts into compounds based on their identical elution times. This approach may not be feasible for LC-free DMS-MS experiments, as different adducts may have different migration times in DMS separations [17, 24]. Our suggestion is to develop workflows that favor the formation of single adducts by careful choice of ionic additives or, if feasible, use LC retention time to align DMS-separated features. Both of these approaches may limit the application range of DMS for untargeted metabolomics or at least counterbalance some of the benefits we discussed here.
As outlined in the Results, the DMS-MS data did not align as well as expected with RP-LC-MS results of the same samples. Since this is the first report of a DMS-based untargeted metabolomics platform and large-scale studies using DMS are scarce, we are unable to provide a definitive explanation for our observations at this point. However, we were able to extract single ion traces for the DMS-generated FOIs from the HPLC-MS data. This shows that the two platforms are partially “orthogonal” and target overlapping, but not necessarily identical, subsets of the metabolome. The extent of this overlap has important implications for the choice of applying DMS as an alternative to HPLC, in exploratory studies intended to yield targeted HPLC-MS/MS approaches, or as an additional dimension in multi-dimensional metabolomics platforms. More in-depth studies of DMS vs. HPLC separations are required to expand our understanding in this area. We believe that this work provides a foundation for broader application of this exciting and accessible separation technique.
5. Conclusions and Outlook
In this contribution, we propose a simple workflow for the application of DMS as a separation technique for biomarker screening in metabolomics. A total of 881 features, characterized by unique m/z and COV pairs, were detected in plasma extracts of 10 CKD patients.
The DMS-MS platform facilitated the selection of features of interest and putative identification based on compensation voltage and accurate mass. In our small test set, differential analysis of early- and late-stage CKD patients led to three main features of interest. A METLIN database search based on accurate mass produced several matches. For one feature, comparison of the observed COV with an authentic standard pointed at bilirubin. For another feature, follow-up HPLC-MS/MS analysis confirmed the presence of multiple isobaric species, possibly phospholipids.
Compared to HPLC-MS, the DMS-MS platform operated much faster but limitations arose from its lower separation efficiency and sensitivity. Peak capacity and signal intensity were an order of magnitude below HPLC, which resulted in relatively high sample consumption and challenges regarding data analysis (peak picking). The number of differential features distinguishing early- and late-stage CKD patients was significantly higher in HPLC. In addition, DMS-derived features of interest did not correspond well with HPLC-MS results. This demonstrates the need for more, larger and more systematic comparative studies investigating the selectivity overlap between the two techniques and the effects of different biological matrices. Importantly, the sample matrix is expected to have a large, possibly detrimental influence on the quality of data obtained by direct-infusion DMS-MS. For better comparability of DMS and HPLC-based workflows, we recommend de-salting and pre-concentration, for example using a trapping column. Future work will also focus on optimizing sensitivity, the selection of suitable internal standards to improve quantitation, and the incorporation of tandem MS experiments to increase confidence in metabolite identification. Technological advances will bring better sensitivity, which in turn will result in decreased sample consumption and reduced instrument contamination.
Supplementary Material
Acknowledgements:
The authors thank Dr. Farsad Afshinnia (University of Michigan) for help with statistical data analysis and Dr. J. Larry Campbell (Sciex) for providing access to MarkerView software. We also appreciate the assistance of the Michigan Kidney Translational Core Clinical Phenotyping Resource and Biobank Core Investigator Group. It includes Matthias Kretzler and Debbie Gipson (University of Michigan, Ann Arbor), Keith Bellovich (St. Clair Nephrology Research, Detroit), Zeenat Bhat (Wayne State University, Detroit), Crystal Gadegbeku (Temple University Health System, Philadelphia), Susan Massengill (Levin Children’s Hospital, Charlotte), and Kalyani Perumal (JH Stroger Hospital, Chicago).
This work was supported by the Postdoctoral Translational Science Program from the Michigan Institute for Clinical and Health Research UL1TR000433 (to S.W.) and National Institutes of Health grants P30DK089503, DK082841, P30DK081943, U2C ES026553 and DK097153 (to S.P.).
Abbreviations
- ACN
Acetonitrile
- BMI
Body mass index
- CKD
Chronic kidney disease
- COV
Compensation voltage
- CPROBE
Clinical Phenotyping Resource and Biobank Core at the University of Michigan
- cps
Counts per second
- DMS
Differential mobility spectrometry
- eGFR
Estimated glomerular filtration rate
- FC
Fold change
- FDR
False discovery rate
- FOI
Feature of interest
- HPLC
High-performance liquid chromatography
- iPrOH
Isopropanol
- MS
Mass spectrometry
- PCA
Principal component analysis
- PLS-DA
Partial least squares-discriminant analysis
- PP
Pooled plasma
- RP
Reversed-phase
- (R)SD
(Relative) standard deviation
- SV
Separation voltage
- TIC
Total ion current
- UPCR
Urinary protein to creatinine ratio
- VIP
Variable importance in projection (PLS-DA)
- XIC
Extracted ion chromatogram
Footnotes
Conflict of interest statement: The authors state that no conflicts of interest exist.
References
- 1.Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E Lewis GD, Fox CS, Jacques PF, Fernandez C, O’Donnell CJ, Carr SA, Mootha VK, Florez JC, Souza A, Melander O, Clish CB, Gerszten RE. Metabolite profiles and the risk of developing diabetes. Nat Med 2011;17:448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang TJ, Ngo D, Psychogios N, Dejam A, Larson MG, Vasan RS, Ghorbani A, O’Sullivan J, Cheng S, Rhee EP, Sinha S, McCabe E, Fox CS, O’Donnell CJ, Ho JE, Florez JC, Magnusson M, Pierce KA, Souza AL, Yu Y, Carter C, Light PE, Melander O, Clish CB, Gerszten RE. 2-Aminoadipic acid is a biomarker for diabetes risk. J Clin Invest 2013;123(10):4309–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang-Sattler R, Yu Z, Herder C, Messias AC, Floegel A, He Y, Heim K, Campillos M, Holzapfel C, Thorand B, Grallert H, Xu T, Bader E, Huth C, Mittelstrass K, Döring A, Meisinger C, Gieger C, Prehn C, Roemisch-Margl W, Carstensen M, Xie L, Yamanaka-Okumura H, Xing G, Ceglarek U, Thiery J, Giani G, Lickert H, Lin X, Li Y, Boeing H, Joost HG, de Angelis MH, Rathmann W, Suhre K, Prokisch H, Peters A, Meitinger T, Roden M, Wichmann HE, Pischon T, Adamski J, Illig T. Novel biomarkers for pre‐diabetes identified by metabolomics. Mol Syst Biol 2012;8(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sas KM, Kayampilly P, Byun J, Nair V, Hinder LM, Hur J, Zhang H, Lin C, Qi NR, Michailidis G, Groop PH, Nelson RG, Darshi M, Sharma K, Schelling JR, Sedor JR, Pop-Busui R, Weinberg JM, Soleimanpour SA, Abcouwer SF, Gardner TW, Burant CF, Feldman EL, Kretzler M, Brosius FC 3rd, Pennathur S.Tissue-specific metabolic reprogramming drives nutrient flux in diabetic complications. JCI Insight 2016;1(15). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, Smith JD, DiDonato JA, Chen J, Li H, Wu GD, Lewis JD, Warrier M, Brown JM, Krauss R, Wilson Tang WH, Bushman FD, Lusis AJ, Hazen SL. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med 2013;19(5):576–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y, Hazen SL. Intestinal Microbial Metabolism of Phosphatidylcholine and Cardiovascular Risk. New Engl J Med 2013;368(17):1575–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, Wu Y, Schauer P, Smith JD, Allayee H, Tang WH, DiDonato JA, Lusis AJ, Hazen SL. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 2011;472:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Putluri N, Shojaie A, Vasu VT, Vareed SK, Nalluri S, Putluri V, Thangjam GS, Panzitt K, Tallman CT, Butler C, Sana TR, Fischer SM, Sica G, Brat DJ, Shi H, Palapattu GS, Lotan Y, Weizer AZ, Terris MK, Shariat SF, Michailidis G, Sreekumar A. Metabolomic Profiling Reveals Potential Markers and Bioprocesses Altered in Bladder Cancer Progression. Cancer Res 2011;71(24):7376–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Afshinnia F, Rajendiran TM, Soni T, Byun J, Wernisch S, Sas KM, Hawkins J, Bellovich K, Gipson D, Michailidis G, Pennathur S; Michigan Kidney Translational Core CPROBE Investigator Group. Impaired β-Oxidation and Altered Complex Lipid Fatty Acid Partitioning with Advancing CKD. J Am Soc Nephrol 2018;29(1):295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niewczas MA, Sirich TL, Mathew AV, Skupien J, Mohney RP, Warram JH, Smiles A, Huang X, Walker W, Byun J, Karoly ED, Kensicki EM, Berry GT, Bonventre JV, Pennathur S, Meyer TW, Krolewski AS. Uremic solutes and risk of end-stage renal disease in type 2 diabetes: metabolomic study. Kidney Int 2014;85(5):1214–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Afshinnia F, Rajendiran TM, Karnovsky A, Soni T, Wang X, Xie D, Yang W, Shafi T, Weir MR, He J, Brecklin CS, Rhee EP, Schelling JR, Ojo A, Feldman H, Michailidis G, Pennathur S. Lipidomic Signature of Progression of Chronic Kidney Disease in the Chronic Renal Insufficiency Cohort. Kidney Int Reports 2016;1(4):256–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hocher B, Adamski J. Metabolomics for clinical use and research in chronic kidney disease. Nat Rev Nephrol 2017;13:269. [DOI] [PubMed] [Google Scholar]
- 13.Campbell JL, Blanc JYL, Kibbey RG. Differential mobility spectrometry: a valuable technology for analyzing challenging biological samples. Bioanalysis 2015;7(7):853–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu C, Gómez-Ríos GA, Schneider BB, Le Blanc JCY, Reyes-Garcés N, Arnold DW, Covey TR, Pawliszyn J. Fast quantitation of opioid isomers in human plasma by differential mobility spectrometry/mass spectrometry via SPME/open-port probe sampling interface. Anal Chim Acta 2017;991:89–94. [DOI] [PubMed] [Google Scholar]
- 15.Ayodeji I, Vazquez T, Bailey R, Evans-Nguyen T. Rapid pre-filtering of amphetamine and derivatives by direct analysis in real time (DART)-differential mobility spectrometry (DMS). Anal Methods 2017;9(34):5044–51. [Google Scholar]
- 16.Chen Z, Coy SL, Pannkuk EL, Laiakis EC, Fornace AJ, Vouros P. Differential Mobility Spectrometry-Mass Spectrometry (DMS-MS) in Radiation Biodosimetry: Rapid and High-Throughput Quantitation of Multiple Radiation Biomarkers in Nonhuman Primate Urine. J Am Soc Mass Spectr 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wernisch S, Afshinnia F, Rajendiran T, Pennathur S. Probing the application range and selectivity of a differential mobility spectrometry–mass spectrometry platform for metabolomics. Anal Bioanal Chem 2018;410(12):2865–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanu AB, Dwivedi P, Tam M, Matz L, Hill HH. Ion mobility–mass spectrometry. J Mass Spectrom 2008;43(1):1–22. [DOI] [PubMed] [Google Scholar]
- 19.Wernisch S, Afshinnia F, Rajendiran TM, Pennathur S. Differential mobility – mass spectrometry metabolomics platform for biomarker discovery in chronic kidney disease Annual Meeting of the American Society for Mass Spectrometry; 2017; Indianapolis, IN. [Google Scholar]
- 20.Stevens LA, Schmid CH, Greene T, Zhang YL, Beck GJ, Froissart M, Hamm LL, Lewis JB, Mauer M, Navis GJ, Steffes MW, Eggers PW, Coresh J, Levey AS. Comparative Performance of the CKD Epidemiology Collaboration (CKD-EPI) and the Modification of Diet in Renal Disease (MDRD) Study Equations for Estimating GFR Levels Above 60 mL/min/1.73 m(2). Am J Kidney Dis 2010;56(3):486–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0—making metabolomics more meaningful. Nucleic Acids Res 2015;43(W1):W251–W7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wernisch S, Afshinnia F, Rajendiran T, Pennathur S. Probing the application range and selectivity of a differential mobility spectrometry–mass spectrometry platform for metabolomics. Anal Bioanal Chem 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brereton RG, Lloyd GR. Partial least squares discriminant analysis: taking the magic away. J Chemometr 2014;28(4):213–25. [Google Scholar]
- 24.Anwar A, Psutka J, Walker SWC, Dieckmann T, Janizewski JS, Larry Campbell J, Hopkins WS. Separating and probing tautomers of protonated nucleobases using differential mobility spectrometry. Int J Mass Spectrom 2017. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.