Abstract
Pluripotent stem cells represent an attractive cell source for treating muscular dystrophies since they easily allow for the generation of large numbers of highly regenerative myogenic progenitors. Using reprogramming technology, patient-specific pluripotent stem cells have been derived for several types of muscular dystrophies, and genome editing has allowed correction of mutations, opening the opportunity for their therapeutic application in an autologous transplantation setting. However, there has been limited progress on preclinical studies that validate the therapeutic potential of these gene corrected pluripotent stem cell-derived myogenic progenitors. In this review, we highlight the major research advances, challenges and future prospects towards the development of pluripotent stem cell-based therapeutics for muscular dystrophies.
Keywords: pluripotent stem cells, gene correction, myogenic progenitors, cell therapy, muscular dystrophy
Muscle Degeneration and Pluripotent Stem Cells
Muscle degeneration is a condition that affects millions of people worldwide. It is primarily observed in patients suffering from 1 of the more than 30 types of muscular dystrophy (MD) identified so far. Among these, Duchenne Muscular Dystrophy (DMD), caused by mutations in the DMD gene, is the most common and characterized by relentless and catastrophic progression, usually culminating in death by the third decade of life. There is currently no cure for MD disorders, and therefore they represent a serious unmet medical need.
Current research focuses mostly on the development of gene- and cell-based therapeutic approaches to cure MD. Gene therapy involves delivering the missing gene in affected muscles to rescue protein function. Cell therapy (see Glossary) focuses on the replacement of the diseased muscle tissue with muscle stem/progenitor cells, which upon engraftment give rise to new healthy myofibers as well as fuse with regenerating muscle fibers, thus restoring muscle function. Both gene- and cell-based strategies hold great potential, but for the purposes of this review, we will focus on the latter.
Pluripotent stem cells (PSC) are an attractive source for cell-based therapeutics due to their unlimited proliferative potential and their ability to differentiate into all cell types of the body. Considering the development of methodologies that enable the efficient generation of PSC-derived myogenic progenitors endowed with in vivo regenerative potential, combined with the significant recent progress in genome editing technologies, it is plausible to envision the use of the MD patient’s own cells for autologous cell transplantation upon gene correction. This is theoretically feasible since MD progression takes over a decade, allowing more than sufficient time to generate and test iPS cells, correct the mutation, and derive/characterize large numbers of transplantable skeletal muscle derivatives. This review will focus on recent developments in the areas of gene editing and iPS cells and their potential therapeutic applications for MDs.
Muscular Dystrophies and Skeletal Muscle Regeneration
Muscular dystrophy (MD) denotes a large group of heterogeneous genetic diseases characterized by progressive muscle wasting. In addition to the genetic heterogeneity, the age of onset, severity, and types of muscles affected vary significantly among different types of MD (Table 1). In the case of DMD, the most common and severe form of MD, patients are usually wheelchair-bound by their early teens, and rarely survive past their mid-twenties due to severe cardio-respiratory failure. To date, there is no cure for MDs, and current treatments mostly alleviate disease symptoms, which in some cases can slow down disease progression. Recently, exon-skipping with antisense oligonucleotides has been used to treat DMD patients carrying frameshift mutations. However, the clinical data have not shown clear efficacy therefore the conditional approval of the drug Exondys 51, which targets DMD exon 51 skipping [1], has generated controversy.
Table 1:
Types of muscular dystrophies, their origin and phenotype
| Type | Gene associated | Mutation | Inheritance pattern | Age of onset | Muscles affected | Phenotype |
|---|---|---|---|---|---|---|
| Duchenne muscular dystrophy (DMD) | DMD | Loss of function | X-linked (recessive) | 3–5 years | Several muscles of the body and cardiac muscle | Progressive disease, which is fatal due to severe respiratory failure around 20–30 years of age. Patients are wheel chair bound in their early teenage |
| Becker muscular dystrophy (BMD) | DMD | Partial loss of function | X-linked (recessive) | Late childhood | Several muscles of the body and cardiac muscle | Milder than DMD but patients may die after forties if there are respiratory issues |
| Facioscapulo humeral muscular dystrophy (FSHD1 and 2) | DUX4 (type 1 and 2) SMCHD1 (type 2) | Contraction of DNA repeats in chromosome 4 and mutations in SMCHD1 | Autosomal dominant (type 1) Digenic (type 2) | Variable but mostly in 20s | Muscles of the face, shoulder, upper arm and lower legs | Rarely fatal but severely affects the quality of life |
| Myotonic dystrophy (DM1 and DM2) | DMPK (type 1) CNBP (type 2) | Expansion of DNA repeats | Autosomal dominant | Variable for DM1 (birth to 40 years), adult onset for DM2 | Muscles of face, shoulder, lower arms and legs (type 1), Muscles of the neck, shoulders, elbows and hips (type 2) | Difficulty in muscle relaxation and muscle weakness. Congenital onset can be fatal if there are respiratory issues |
| Limb girdle muscular dystrophy (LGMD1 and LGMD2) | MYOT (LGMD1A), (LMNA) LGMD1B, CAV3 (LGMD1C), DNAJB6, (LGMD1D), DES (LGMD1E), CAPN3 (LGMD2A), DYSF (LGMD 2B), SGCG (LGMD2C), SGCA (LGMD2D), SGCB (LGMD2E), SGCD (LGMD2F), TCAP (LGMD2G), TRIM32 (LGMD2H), FKRP (LGMD2I), TTN (LGMD2J), | Usually loss of function mutations | Autosomal dominant (Type 1) or recessive (Type 2) | Variable | Typically muscles around the should and pelvic girdles but some types can affect cardiac muscle | Phenotype is variable. Affects the quality of life. Rarely fatal if there is weakness of cardiac and respiratory muscles |
| Pompe disease | GAA | Usually loss of function mutations | Autosomal recessive | Variable, from birth to adulthood | Respiratory muscles, muscles of hip, upper arms, legs and shoulder | Phenotype is variable. Early onset forms can be fatal |
Many types of MDs are associated with genetic and biochemical defects of the dystrophin-glycoprotein complex (DGC) [2]. These alterations lead to cell membrane damage and apoptosis of muscle cells, resulting in chronic successive cycles of degeneration/regeneration, culminating in compromised regeneration overtime [3]. Nevertheless, the regenerative nature of skeletal muscle provides an opportunity for utilizing cell therapy through delivery of healthy myoblasts that can engraft, fuse to the regenerating muscle fibers and rescue the missing protein function in the dystrophic muscle, as shown by Partridge and colleagues in pioneering studies [4]. Unfortunately, results from early clinical trials did not show clinical benefit [5–8], which was attributed mostly to the poor survival and limited migratory ability of injected myoblasts [9, 10] (see Clinician’s corner). Instead of myoblasts, skeletal muscle stem cells (also known as satellite cells) would be preferable for therapeutic application since these cells have the ability to self-renew and efficiently contribute to muscle regeneration. A major hurdle with muscle tissue is the impossibility of obtaining enough satellite cells without permanently damaging the muscle of the donor. Small muscle biopsies allow for the ex vivo expansion of satellite cell progeny (myoblasts), and therefore cell preparations with reduced engraftment ability [11, 12].
BOX 1, Clinician’s Corner.
Immunosuppression is considered to be crucial for the success of cell therapies. Lack of proper immunosuppression is proposed to be one of the reasons for the poor engraftment of myoblasts in clinical trials for muscular dystrophy (MD) [84]. Pluripotent stem cells (PSCs) could be used in the future for either allogeneic or autologous cell transplantation for MD. Allogeneic cell transplantation involves the use of cells from a genetically non-identical healthy individual for therapy, which may invoke an immune response in the absence of continual immune suppression. Autologous cell transplantation involves the use of the patient’s own cells for therapy, after the correction of genetic defect in the context of genetic disease. However, restoration of the missing protein’s expression due to gene correction could still elicit an immune response, since it will introduce new epitopes to the patient’s immune system. Therefore, whether by autologous or allogeneic cell transplantation, immune considerations are relevant and the use of immunosuppression to some extent may be necessary even in the autologous setting for some patients. The current proof-of-concept studies test the therapeutic potential of human PSCs in immunodeficient mouse models, as human cells will not engraft in immunocompetent mice, thus they cannot address these important issues. Future research should focus on optimizing the use of immunosuppression in relevant preclinical models for PS- based-cell therapy, for example in non-human primates. The choice of the method, dosage and timing for immunosuppression will need to be determined on a case by case basis. Immunosuppression poses a not insignificant risk of side effects for cell transplant recipients. To avoid the use of immunosuppression, a method to generate hypoimmunogenic iPSCs that cannot be recognized by the immune system has recently been reported [85]. Although these immune evading cells could provide a universal donor cell population for therapeutic applications, they could pose a safety issue in the case of potential tumorigenesis.
Cardio-respiratory failure is the major cause of fatality in the case of DMD, for which there is no cure. Therefore, future research should explore the possibility of iPSC-based cell therapy for the treatment of cardiac failure as well. Differentiation of PSCs into cardiac progenitors that could regenerate the dystrophic hearts will be critical. Recent studies reporting on the use of hESC-derived cardiomyocytes restoring the function in the macaque monkey model of myocardial infarction are encouraging [86]. The possibility of using a similar approach for DMD is yet to be tested.
There have been several studies demonstrating the regenerative potential of mesoangioblasts in mouse and dog models of muscular dystrophy [13, 14]. These encouraging findings prompted a phase I/II clinical trial, consisting of multiple intra-arterial infusions of mesoangioblasts in pediatric DMD patients (Eudract 2011–000176-33)I. Although the therapy was proven to be feasible and relatively safe, there was no demonstration of efficacy or clinical benefit [15]. Therefore, further studies and alternate sources of early skeletal muscle progenitors are still necessary for the development of an effective stem cell therapy for MDs.
Pluripotent Stem Cells
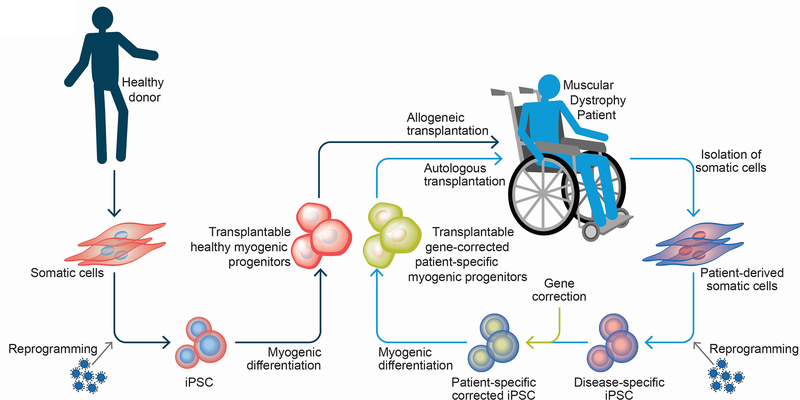
PSCs can differentiate into cells of all three germ layers, ectoderm, mesoderm and endoderm. In addition to this broad differentiation ability, they possess extraordinary self-renewal capacity, allowing for unlimited expansion in vitro. PSCs encompass embryonic and induced pluripotent stem cells (ESCs and iPSCs, respectively). ESCs are established from the inner cell mass of blastocyst stage embryos [16, 17], and were first derived from human embryos in 1998 by James Thomson and colleagues [18]. ESC derivation is inefficient and requires destruction of human embryos, which raises ethical concerns. About a decade later, these issues were overcome with the technology of reprogramming somatic cells to the pluripotent state (iPS cells). This strategy, pioneered by Shinya Yamanaka and colleagues, based on the transient expression of the pluripotency-associated transcription factors, brings PSC-based therapy much closer to reality since it allows for the derivation of patient-specific iPS cells, eliminating the ethical and practical constraints associated with ES cells [19, 20]. For therapeutic application to muscular dystrophies, one could envision the use of healthy human leukocyte antigen (HLA)-matched iPS cell-derived myogenic progenitors (allogeneic transplantation) or the patient’s own iPS cell-derived myogenic progenitors (autologous transplantation). In allogeneic therapy, HLA matching reduces the risk of immune rejection of the transplanted cells and thus universal donor iPS cell banks referred to as the HLA haplobank model are being established [21–24]. The autologous approach requires in vitro genetic correction of dystrophic iPS cells prior to transplantation (Figure 1), allowing for personalized medicine. In any case, some aspects should be taken in to consideration for the development of a PSC-based therapy for muscular dystrophies, including i) the implementation of a controlled differentiation protocol to specifically derive skeletal myogenic progenitors, devoid of residual PSCs and other non-muscle lineages; ii) myogenic progenitors should be endowed with robust in vivo regenerative potential, and iii) for long-term efficacy, it is critical that transplanted cells also seed the satellite cell compartment, therefore allowing for sustained expression of the therapeutic protein in newly formed muscle fibers.
Figure 1: Pluripotent stem cell-based therapeutics for muscular dystrophy.
PSC-derived myogenic progenitors could be used for the potential treatment of MD patients through either autologous or allogeneic cell transplantation. For allogeneic transplantation, iPSCs would be derived from somatic cells of a healthy individual. Upon inducing myogenic differentiation, healthy myogenic progenitors obtained from these iPSCs would be transplanted in the MD patient. In the autologous transplantation setting, iPSCs would be derived from the MD patient’s own cells. These patient-specific iPSCs would be corrected for the genetic defect using genome editing techniques to derive gene corrected iPSCs, which would then be differentiated into transplantable gene corrected myogenic progenitors. These myogenic progenitors would then be used for autologous cell transplantation.
Derivation of PSC-Derived Myogenic Progenitors
Several protocols have been developed for the derivation of myogenic progenitors from PSCs, which may involve the use of transgenes or not. In this section, we will focus mostly on strategies that have documented the in vivo regenerative potential of resulting myogenic progenitors, but we also describe a few in vitro-only publications that were important for progress in the field (Table 2).
Table 2:
Methods for the differentiation of hPSC into myogenic progenitors and in vivo engraftment
| Method | Purification | In vivo experiment | Engraftment and function | Transgene | Reference |
|---|---|---|---|---|---|
| Adenoviral based delivery of MYOD1; monolayer cultures | N/A | Transplantation of 0.5 × 106 cells per TA muscle of immunodeficient Rag/mdx mice | 150–175 fibers positive for human SPECTRIN per section | MYOD | [27] |
| Lentiviral delivery of MYOD1 in PSC-derived mesoangioblasts | N/A | Transplantation of 106 cells per TA muscle of immunodeficient Sgca-null/scid/beige mice | 50 fibers positive for α-sarcoglycan per section | MYOD | [28] |
| Lentiviral delivery of doxycycline- inducible PAX7; embryoid body cultures | Purification of PAX7+ myogenic progenitors based on GFP transgene | Transplantation of 0.3 × 106 cells per TA muscle of immunodeficient NSG-mdx4Cv mice | 100 fibers positive for human dystrophin per section. Also showed satellite cell engraftment and functional improvement of transplanted muscles in comparison with untreated muscles. | PAX7 | [32] |
| Similar to [32] | Positive selection for surface markers CD54, integrin α9β1, and Syndecan2 for the purification of PAX7+ myogenic progenitors | Transplantation of 0.5 × 106 cells per in TA muscles of immunodeficient NSG and NSGmdx4Cv mice | 50 fibers positive for human dystrophin per section. Also showed satellite cell engraftment | PAX7 | [33] |
| hESC differentiated into mesenchymal precursor and then into myogenic progenitors | Positive selection for CD73 and NCAM | Transplantation of 0.5 × 106 cells per TA muscle of immunodeficient SCID/Beige mice | Few fibers positive for human nuclear antigen and human laminin positive fibers were detected | Transgene- free | [34] |
| hPSC differentiated into embryoid bodies and then into myogenic mesenchymal progenitor cells. | N/A | Transplantation of 0.5 × 106 cells per TA muscle of immunodeficient NOG mice | 10–20% fibers containing human nuclei. Also showed satellite cell engraftment | Transgene- free | [35] |
| Monolayer culture based on the use of GSK3β inhibitor and FGF2 growth factor treatment | Positive selection for surface markers CXCR4 and CMET, and negative selection for ACHR and HNK1 | N/A | N/A | Transgene- free | [36] |
| Similar to [36] | Positive selection for C-MET and negative selection for HNK1 | Transplantation of 0.5 × 106 cells per TA muscle of immunodeficient NSG mice | 50 fibers positive for human spectrin per section | Transgene-free | [38] |
| Similar to [36] | N/A | N/A | N/A | Transgene- free | [37] |
| Monolayer culture based on the use of GSK3β and BMP inhibition, and treatment with FGF2, HGF and IGF1 | N/A | N/A | N/A | Transgene- free | [39, 40] |
| Embryoid body cultures and microRNA cocktail treatment | Positive selection for surface markers CD140a, CD140b, and CD44 | Transplantation of 0.5 × 106 cells in the femoral artery of immunodeficient Rag2/Il2rg null Sgcb null mice | 15–20% fibers positive for human dystrophin. Functional improvement shown in the Extensor Digitorum Longus (EDL) muscle | Transgene- free | [42, 43] |
| Similar to protocols in [37, 39] | Positive selection for surface markers ERBB3 and NGFR | Transplantation of 106 cells per TA muscles of immunodeficient mdx-NSG mice | 150 fibers positive for human dystrophin/spectrin upon co-injection of cells with TGFβ inhibitor | Transgene- free | [44] |
| Monolayer culture based on the use of GSK3β and NOTCH inhibition | Positive selection for NCAM, and negative selection for HNK1 | Transplantation of 1–3 × 106 cells per TA muscles of immunodeficient NOD-Rag1null IL2rgnull and NSG-mdx4cv mice | Approximately 100 fibers positive for human laminin and lamin A/C per section in NSGmdx4cv, and 200300 fibers in NOD-Rag1null IL2rgnull mice. Also showed satellite cell engraftment | Transgene- free | [45] |
| Monolayer culture based on the use of GSK3β, BMP and TGFβ inhibition | Positive selection for CD10, and negative selection for CD24 | Transplantation of 0.25 × 106 cells per TA muscles of in immunodeficient NSG-mdx4cv mice | Approximately 50 fibers positive for human dystrophin/lamin A/C per section. Also showed satellite cell engraftment | Transgene- free | [46–48] |
Transgene-dependent myogenic differentiation
The first description of somatic cell fate transdifferentiation was reported by Weintraub and colleagues [25, 26] when they observed that MYOD could reprogram fibroblasts into myoblasts. Accordingly, several investigators have used this strategy to generate myoblasts from human PS cells. One study utilized adenoviral vector-based delivery of MYOD to generate myoblasts from human ES and DMD iPS cells, which upon transplantation into Rag/mdx mice (an immunodeficient DMD mouse model) fused with existing myofibers [27]. Tedesco and colleagues applied conditional expression of MYOD using lentiviral vectors to promote the muscle differentiation of human iPSC-derived mesoangioblasts. Transplantation of these cells into α-Sgca-null immunodeficient mice led to myofiber engraftment and rescue of SGCA protein [28]. Despite this positive outcome, a caveat with the use of MYOD is the derivation of a more committed myogenic cell (myoblast), which possess limited proliferative capacity and may not contribute to the stem cell pool upon transplantation.
In the transcription factor hierarchy of skeletal myogenesis, the transcription factor PAX7, which is positioned upstream of MYOD, is critical for postnatal muscle regeneration [29, 30], being expressed in satellite cells during their specification, proliferation, and activation [31]. With the premise that PAX7 expression would target a more primitive cell (muscle stem/progenitor cell) within the muscle hierarchy, Darabi and colleagues used conditional expression of PAX7 (iPAX7) to promote the in vitro differentiation of human ES and iPS cells towards the myogenic lineage. Using this strategy, these authors generated a highly expandable population of myogenic progenitors, which upon intramuscular transplantation into NSG/mdx mice resulted in donor-derived myofibers expressing human dystrophin, improvement of muscle contractile parameters, and seeding of the satellite cell compartment, and therefore, contribution to long-term regeneration [32]. Through gene expression profiling of iPAX7 differentiating cells, Magli and colleagues recently identified CD54, integrin α9β1, and SYNDECAN-2 (SDC2) as surface markers to be used for the prospective isolation of human PS cell-derived myogenic progenitors using both fluorescent- and cGMP-compatible magnetic-based sorting technologies [33]. Therefore, the use of PAX7 transgene allows efficient derivation of therapeutically relevant myogenic progenitors from PSCs.
Transgene-free myogenic differentiation
Numerous transgene-free protocols have been published for the in vitro derivation of myogenic cells from PSCs. Two studies described the derivation of myogenic cells through a mesenchymal precursor [34, 35], but engraftment data overall was limited. Most recent protocols make use of small molecules to direct the differentiation of PSCs towards the myogenic lineage. Since most skeletal muscles derive from paraxial mesoderm during development, a key aspect is to recapitulate this process during the in vitro differentiation of PSCs. Initial studies utilized small molecule treatment to induce WNT signaling activation through GSK3β inhibition to differentiate PSCs into paraxial mesoderm, and then fibroblast growth factor 2 (FGF2) to derive myogenic progenitors [36, 37], and transplantation of resulting myogenic progenitors into immunodeficient mice produced myofiber engraftment [38]. This protocol was further improved by Chal and colleagues as they applied inhibition of GSK3β and BMP signaling along with subsequent exposure to pro-myogenic growth factors, including FGF2, hepatocyte growth factor (HGF), and insulin-like growth factor 1 (IGF-1) to induce differentiation of human PSCs towards the myogenic lineage [39, 40]. Even though these improvements enhanced the generation of cells with in vitro myogenic differentiation potential, the heterogeneity within these cultures (presence of non-myogenic cells) and lack of in vivo regenerative potential [41] limit their use for therapeutic application. In 2017, Giacomazzi and colleagues reported that myogenic progenitors derived from mesoangioblast iPSCs engraft better than from fibroblast iPSCs, and that treatment with microRNA cocktail further enhances this engraftment potential [42, 43]. In 2018, Hicks and colleagues documented that myogenic progenitors can be purified from PSC-derived monolayer cultures [36, 37] using the cell surface proteins ERBB3 and NGFR [44], which upon transplantation into NSG/mdx mice, along with TGFβ signaling inhibitor, gave rise to dystrophin-expressing myofibers. Another group reported myofiber and satellite cell engraftment upon the utilization of GSK3β inhibition followed by NOTCH signaling inhibition to generate PSC-derived myoblasts [45]. Most recently, Wu and colleagues reported a transgene-free protocol utilizing a novel PAX7/MYF5 double reporter PSC line [46–48]. By applying inhibition of the GSK3β, TGFβ and BMP signaling pathways during the early stages of PSC, in combination with subsequent purification based on the expression of CD10 and absence of CD24, these authors showed the derivation of myogenic progenitors capable of giving rise to myofibers and satellite cells upon their transplantation into NSG/mdx mice [48]. Despite significant progress on the generation of transgene-free protocols that result in engraftable myogenic progenitors, the heterogeneity within these cultures remain a challenge for therapeutic applications.
Recent Progress in Genome Editing Technologies
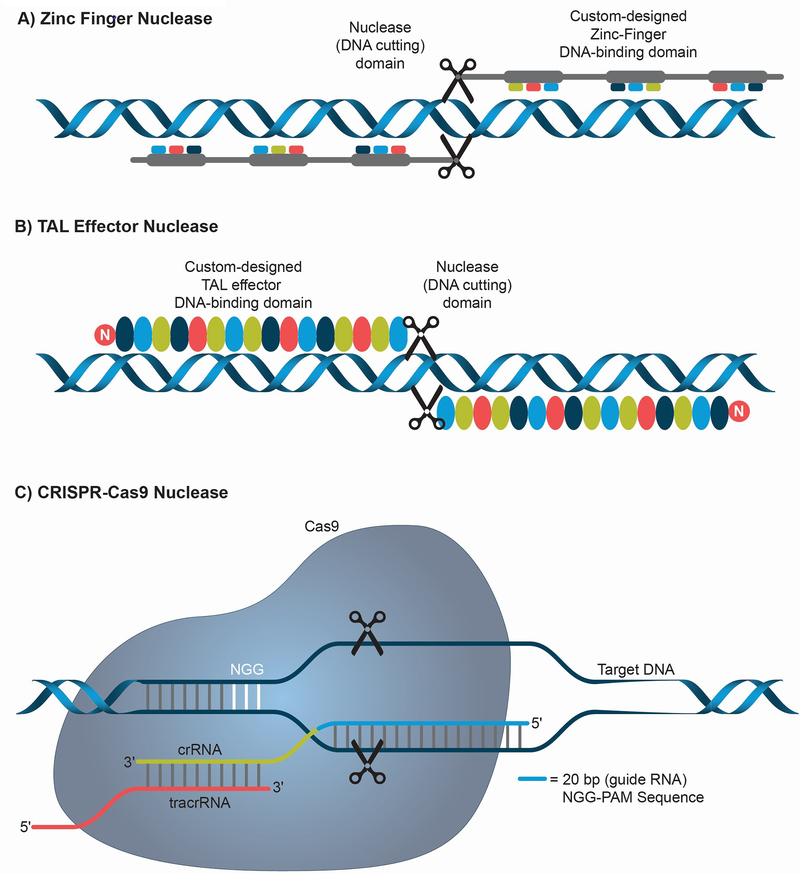
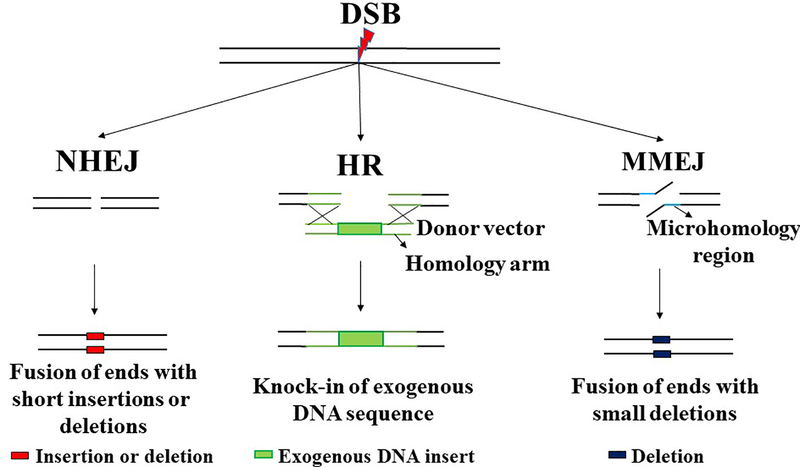
The recent and rapid progress in the development of genome-editing technologies has allowed investigators to easily introduce sequence-specific modifications into the human genome. Genome editing exploits endogenous DNA repair mechanisms to induce these modifications. Zinc finger nucleases (ZFN), transcription activator-like effector nucleases (TALENs) and clustered regularly interspaced short palindromic repeats-CRISPR associated protein 9 (CRISPR-Cas9) are the three sequence-specific nucleases commonly utilized for genome editing (Figure 2). ZFNs are artificial restriction enzymes, which consist of DNA binding zinc finger domains fused to a nuclease to achieve sequence specific DNA cleavage [49]. The rules underlying sequence specificity of ZFNs are imprecise, meaning design involves a large empirical component, therefore effective ZFNs generally require a long phase of trial and error optimization to obtain sufficient specificity. TALENs are fusions of a nuclease to TAL effector DNA domains, whose sequence specificity is predetermined and therefore TALENs can be easily designed and quickly synthesized [50–52]. Their principal disadvantage is their large size. CRISPR-Cas9 was developed from a bacterial immune defense mechanism targeting DNA of viruses. Sequence specificity is defined by a short RNA that is homologous to and derived from viral sequences, and the RNA-protein complex targets a nuclease to destroy viral DNA. CRISPR-Cas9 nuclease has been adapted for genome editing wherein short RNA complementary to the DNA sequence of interest can direct the Cas9 to cleave genomic DNA [53–56]. CRISPR-Cas9 is much easier to design than TALENs and ZFN as it requires a short RNA as opposed to designing a protein, however its principal weakness is the complexity introduced by the need to employ both a protein and an RNA for sequence specificity. To induce genome editing using these sequence specific nucleases, endogenous DNA repair mechanisms, such as non-homologous end joining (NHEJ), homologous recombination (HR) and microhomology-mediated end joining (MMEJ) are exploited (Figure 3). NHEJ involves repair of the double-strand break (DSB) by ligation of the ends without the need for a homologous template [57]. This method of DNA repair can induce short insertions or deletions, which can change the reading frame – useful in cases where disease is due to a nonsense mutation that shifted reading frame. HR makes use of a homologous template to repair the DSB through exchange of sequence information, and thus it can introduce an exactly defined sequence at the site of interest. This strategy can also be used to insert large DNA sequences if a template containing left and right homology arms is provided [58, 59]. Alternatively, short single-stranded oligonucleotides can be used as a template. MMEJ repairs the DSB by fusing the ends of broken DNA based on micro-homology of about 2–25 base pair (bp), leading to deletion of sequence flanking the DSB [60].
Figure 2: Sequence-specific nucleases used for genome editing.
(A) Zinc Finger Nucleases (ZFN) are artificial restriction enzymes that possess a DNA binding protein domain fused with a nuclease domain to achieve sequence-specific cleavage. ZFNs are designed in pairs to produce double stranded breaks (DSB). (B) TAL Effector Nucleases (TALEN) are restriction enzymes that possess TAL effector DNA binding domains fused to a nuclease domain to achieve sequence-specific cleavage. TALENs are also designed in pairs to enable a highly specific DSB. (C) CRISPR-Cas9 (Clustered Regularly Interspaced Palindromic Repeats-CRISPR associated protein 9) are sequence specific nucleases discovered as part of a bacterial antiviral immune mechanism. A short RNA (guide RNA, usually 20 bp) complementary to the target DNA sequence is utilized to direct the Cas9 protein to a specific genomic region to create DSB. The system requires the presence of a short sequence in the genomic DNA adjacent to the guide RNA target sequence called the protospacer adjacent motif (PAM) which in the case of Streptococcus pyogenes Cas9 is NGG (N can be any nucleotide). crRNA and tracrRNA associate with the Cas9 to help in recognizing and cleaving target DNA.
Figure 3: DNA repair mechanisms utilized for genome editing.
(A) NHEJ (Non-Homologous End Joining) is a double stranded break repair (DSB) pathway which causes fusion of the two ends directly without requiring a template for repair. NHEJ could be error prone as it usually creates short insertions or deletions (in red). For genome editing, NHEJ is used to create large deletions (2 DSBs) or short deletions (1 DSB) usually for correcting frameshift mutations and thereby restoring the reading frame in a subset of targeted cells. (B) HR (Homologous recombination) is a DSB repair pathway that utilizes a homologous template to exchange sequence information and is thus usually error free. For genome editing, HR is used for knocking in an exogenous DNA sequence by providing exogenous donor vector with the insert (green box) flanked by homology arms (green line). Single stranded oligonucleotides can also be used as a template for HR. (C) MMEJ (Microhomology Mediated End Joining) is an error prone DSB repair pathway that utilizes the microhomology region (2–25 bp) to repair and thereby cause deletion of the flanking sequence. For genome editing, MMEJ can be utilized to create small deletions (blue box) for correcting frame shift mutations caused by microduplication. Alternatively, MMEJ can also be used for knock-in of exogenous DNA sequence.
Gene Correction of MD Patient-Specific iPS Cells
In the past decade, several reports have documented the gene correction of MD patient-specific iPS cells (Table 3). Kazuki et al delivered wild-type DMD gene to DMD patient-specific iPS cells using a human artificial chromosome (HAC) as a gene delivery vector. Because HAC delivery in the cell type of interest requires fusion with microcells carrying the HAC, there is potential for the transfer of other genetic information [61]. Studies that use sequence specific nucleases focus mostly on exon skipping approaches aimed at reverting mutations caused by frameshift, leading to rescue by expression of functional dystrophin protein bearing a small deletion. Among these, Li and colleagues applied TALENs and CRISPR-Cas9 to correct a deletion mutation of DMD exon 44. To correct this mutation, they applied three different approaches, i) NHEJ-based deletion of exon 45 splice acceptor, ii) the entire exon 45, and iii) HR-based knock-in of exon 44. Although no transplantation studies were performed, all three of these approaches showed rescue of protein expression upon the in vitro differentiation of gene corrected iPS cells into myotubes [62]. Young et al. reported a genome editing approach to correct DMD mutations between exons 45 to 55 (known as a “hot spot”), and thus applicable to around 60% of all DMD mutations in humans. This was achieved by designing a pair of CRISPR-Cas9 constructs able to delete everything from exon 45 to 55, which then resulted in restoration of the DMD reading frame. Utilizing this approach, the authors showed correction of DMD mutations in three patient-specific iPSCs, and rescue of dystrophin protein expression both in vitro and in vivo [63]. Long and colleagues designed and validated guide RNAs for CRISPR-Cas9 to induce deletion of splice acceptor or donor sites that can lead to skipping of 12 different exons of DMD by NHEJ, thereby correcting frameshift mutations. This approach was shown to rescue DMD protein in gene edited DMD iPSC-derived cardiomyocytes [64].
Table 3:
Gene correction studies in MD patient-specific iPS cells
| Gene correction tool | DNA repair mechanism | Type of MD and mutation | Validation of gene correction | Reference |
|---|---|---|---|---|
| HAC | N/A | DMD | Dystrophin protein rescue was detected in teratoma formed from the gene corrected iPS cells in immunodeficient mice. | [61] |
| TALEN, CRISPR-Cas9 | NHEJ for deletion of either splice acceptor or the entire sequence of exon 45 or HR for rescue of fulllength exon 44. | DMD (exon 44 deletion) | Dystrophin protein rescue was detected in the myotubes derived from the gene corrected iPS cells. | [62] |
| CRISPR-Cas9 | NHEJ for deletion of exons 45 to 55 to restore reading frame | DMD (any frameshift mutation between exon 45–55) | Dystrophin protein rescue was detected in vitro in myotubes derived from gene corrected iPS cells and in vivo upon transplantation of the gene corrected iPSC derived myogenic progenitors in immunodeficient mdx mice. | [63] |
| CRISPR-Cas9 | NHEJ for deletion of splice donor or acceptor sites to restore the reading frame | DMD (mutations in 12 different exons causing frameshift) | Dystrophin protein rescue was shown in vitro in gene corrected iPSC-derived cardiomyocytes. | [64] |
| Lentiviral vector for SGCA delivery | N/A | LGMD2D (SGCA mutations) | α-sarcoglycan protein expression was detected in vitro in myotubes derived from corrected iPSC and in vivo upon transplantation into SGCA null immunodeficient mice. | [28] |
| TALEN, CRISPR-Cas9 | HR with ssODN donor for correction or HR for knock-in of DYSF cDNA in H11 safe harbor locus | LGMD2B (DYSF, nonsense mutation), LGMD2D (SGCA, missense mutation) | Dysferlin and α-sarcoglycan protein rescue was shown in gene corrected iPSC- derived myotubes | [65] |
| CRISPR-Cas9 | MMEJ on the microduplication site to restore the reading frame | LGMD2G (TCAP microduplication causing frameshift) | TCAP protein rescue was shown in gene corrected iPSC derived myotubes | [66] |
| TALEN | HR for knock-in of polyA signal sequence in intron 9 of DMPK to prevent transcription of the repeats | DM1 (expansion of CTG repeats in the 3’ UTR of DMPK gene) | Elimination of nuclear foci and rescue of splicing defects was shown in gene corrected iPSC derived cardiomyocytes and neural stem cells. | [67] |
| CRISPR-Cas9 | HR for knock-in of polyA signal sequence in the 3’UTR upstream of the repeats to prevent their transcription | DM1 (expansion of CTG repeats in the 3’ UTR of DMPK gene) | Elimination of nuclear foci in gene corrected iPSC derived cardiomyocytes, neural stem cells and myotubes. | [68] |
| CRISPR-Cas9 | NHEJ for deletion of CTG repeats | DM1 (expansion of CTG repeats in the 3’UTR of DMPK gene) | Elimination of nuclear foci and splicing defects in gene corrected iPSC derived myotubes. | [69] |
| CRISPR-Cas9 | HR from exogenous donor vector for knockin of GAA cDNA in the AAVS1 safe harbor locus | Pompe disease (GAA mutation) | GAA protein and activity rescue in gene corrected iPSC derived myotubes. | [38] |
Several types of limb-girdle MD (LGMD) have also been targeted for gene correction. In the case of LGMD type 2D (LGMD2D), investigators have engineered lentiviral vectors to deliver SGCA gene to LGMD2D iPS cells, and transplantation of muscle derivatives in a mouse model of LGMD2D led to rescue of α-sarcoglycan protein expression [28]. Turan and colleagues reported correction of LGMD2B and LGMD2D iPS cells using CRISPR-Cas9 and TALENs. They used two HR based approaches, one used short oligonucleotide donor to correct a point mutation and the second applied knock-in of the corrected cDNA in a safe harbor locus. Both approaches resulted in rescue of respective missing dysferlin and α-sarcoglycan proteins in corrected LGMD2B and LGMD2D iPSC-derived myotubes respectively in vitro [65]. Iyer et al. reported the gene correction of LGMD2G patient-specific iPS cells. The mutation was an 8-bp microduplication in the TCAP gene, which leads to frameshift. They utilized CRISPR-Cas9 induced MMEJ-based approach to delete the microduplication and rescue wild-type TCAP protein expression in iPSC-derived myotubes in vitro [66].
Wang et al. reported knock-in of polyA signal sequence in the 3’UTR, directly upstream of the DNA repeats as a gene correction approach in DM1 patient-specific iPS cells. They used CRISPR-Cas9 and HR for this knock-in to prevent the transcription of the repeats but retain the transcription of full-length DMPK gene. This gene correction led to the elimination of nuclear foci in the corrected DM1 patient-specific iPSC-derived cardiomyocytes, neural stem cells and myotubes [67] [68]. Another group reported an approach for complete deletion of the DNA repeats in the DM1 patient-specific iPS cells. They used a pair of CRISPR-Cas9 to induce NHEJ based deletion of the repeats. This correction led to elimination of nuclear foci and rescue of splicing defects in corrected iPSC derived myotubes [69]. van der Waal and colleagues have shown the gene correction of Pompe disease patient-specific iPS cells. They used CRISPR-Cas9 and HR to knock-in GAA cDNA to the AAVS1 safe harbor locus [38]. This led to the rescue of the GAA protein expression in myotubes derived from gene corrected iPSC. Therefore, as discussed here and outlined in Table 3, gene correction of several and different types of MD patient-specific iPS cells have been reported in recent years, providing the scope for future autologous cell therapy applications.
Preclinical Studies and the Current Challenges
Although several studies have shown engraftment of PSC-derived myogenic progenitors, very few studies show significant myofiber engraftment, and importantly, functional recovery of transplanted dystrophic muscles. Validation of functional improvement is critical for confirming the therapeutic efficacy of the PSC-derived myogenic progenitors. Furthermore, those studies that have tested cells in vivo have utilized intramuscular transplantation in mice. However, it will be quite challenging to perform intramuscular injections in humans, as this would require an enormous number of injections due to the size and number of muscles affected. Ideally, systemic transplantation will be the method of choice to deliver cells to all the affected muscles. However, so far there is no proof of concept for efficient systemic delivery of human PSC derived myogenic progenitors. The other critical aspect to validate is survivability of the transplanted muscle progenitors in a dystrophic environment, which is quite hostile to the incoming cells. This was a major issue in clinical trials with myoblasts [70, 71].
Safety of PSC-based cell therapeutics is another aspect to consider. Delivery of transgenes by lentiviral vector poses a risk of mutagenesis due to random genomic integrations, but the third-generation lentiviral vector is significantly safer than the previous versions [72, 73], and are currently being used in gene therapy clinical trials (NCT01745120II, NCT01896102III, NCT01515462IV, NCT01560182V) [74–77]. Another critical issue is the potential for formation of teratoma from contaminating undifferentiated pluripotent stem cells [78].
To translate the PSC-based cell therapeutics from small animal studies to human, scaling up and preparation of clinically compatible cell preparation are important aspects. Although PSCs possess unlimited proliferative potential, their differentiation from pluripotent cells into myogenic progenitors is complex and somewhat variable. It would therefore be ideal if the derived myogenic progeny were expandable without sacrificing engraftment potential as this would allow scale up post-differentiation batches of cells. The procedure for derivation of myogenic progenitors from PSCs must be GMP optimized for clinical compatibility [79]. Thus, many preclinical studies are required to address these important aspects before PSC-based cell therapeutics for MD are realized. Importantly, planned/ongoing iPSC-based clinical trials for macular degeneration (UMIN000011929)VI [80], Parkinson’s disease (UMIN000033564)VII [81], spinal cord injury [82] and ischemic heart disease [83] will provide key lessons for the future PSC-based cell therapeutics.
Concluding Remarks
Proof-of-concept transplantation studies in mouse models of MD have shown that human PSC-derived myogenic progenitors possess a certain capacity for in vivo regenerative potential. The significant progress in the fields of iPSC differentiation and genome editing technologies in the past decade has brought the concept of autologous stem cell therapy closer to reality for MD patients. Nevertheless, many challenges remain (see Outstanding Questions). In addition to the scientific challenges summarized in this review, it is important to consider the high cost of manufacturing a cell preparation that is suitable for only one patient. In addition, despite significant progress, the amount of time it takes to generate, genetically correct, screen, as well as characterize clones can be lengthy, which further significantly increases the cost. The development of universal gene correction strategies to correct all mutations of a gene of interest for each type of MD will make it more amenable to develop autologous cell therapy. Thus, we believe that translation of allogeneic cell therapy may be more feasible in the near future, as it would allow a single cell line to be used in many patients with different types of MD. Although many challenges exist, the past decade has provided grounds for great optimism that PSC based cell therapeutics for MD will eventually be translated to the clinic.
Outstanding Questions.
Do PSC-derived myogenic progenitors mature in vivo upon engraftment?
Will there be limitations in the survival and migratory ability of PSC-derived myogenic progenitors upon transplantation in the dystrophic muscle environment?
Will the transplanted PSC-derived myogenic progenitors contribute sufficiently to the satellite cell pool to maintain the long-term efficacy of potential cell therapy applications?
Will it be possible to develop universal gene correction methods applicable for all mutations in a given gene to develop efficient autologous cell therapy with iPSC?
Is it possible to perform systemic transplantation of human PSC-derived myogenic progenitors and have them engraft in the affected skeletal muscles?
Highlights.
Several methodologies have been reported for the derivation of myogenic progenitors from human pluripotent stem cells
Proof-of-concept studies have shown the therapeutic potential of pluripotent derived myogenic progenitors in mouse models of muscular dystrophy
Induced pluripotent stem cell technology allows for the generation of muscular dystrophy patient-specific pluripotent stem cells
Progress in genome editing techniques has enabled gene correction of mutations in muscular dystrophy patient-specific induced pluripotent stem cells allowing for their potential therapeutic application in autologous cell transplantation settings.
Acknowledgements
RCRP is supported by NIH grants R01 AR055299 and R01 AR071439, ADVault, Inc and MyDirectives.com. MK is supported by NHI R01 AR055685. We are thankful to Cynthia Faraday for graphical design.
Glossary
- Allogeneic cell transplantation
The use of cells from a healthy donor for cell therapy usually from an individual genetically non-identical to the patient.
- Autologous cell transplantation
The use of the patient’s own cells for transplantation.
- Cell therapy
Transfer of live cells into a patient for the purpose of mitigating or curing a disease.
- Engraftment
The ability of transplanted cells to reside and function as part of the host environment. If the transplanted cells become part of the muscle fiber and seed the satellite cell pool, it is denoted by the terms myofiber and satellite cell engraftment, respectively.
- Gene correction
The technique of altering a genetic mutation, either by exactly reversing it or by introducing a sequence that will result in equivalent-to-normal function, in order to cure the disease in the patient derived cells.
- Genome editing
The technique used to modify the genome of the cells in order to fix or create a genetic defect.
- Mesoangioblasts
also known as pericytes, reside in association with blood vessels and are multipotent cells can give rise to different mesodermal cell types.
- Myoblasts
Proliferating muscle progenitors that derive from activated muscle stem cells and give rise to multinucleated myotubes in vitro and muscle fibers in vivo.
- Myogenic progenitors
Precursor cells which can differentiate to form muscle in vitro and in vivo.
- Pluripotent stem cells
Undifferentiated cells that can be differentiated into any cell type of the body.
- Satellite cells
Adult muscle stem cells whose nomenclature is based on their location beneath the basal lamina of the muscle fiber. Satellite cells regenerate muscle in response to injury or disease.
- Transgene
An exogenous DNA sequence introduced into the genome generally for the purpose of expressing a protein of interest.
Resources (clinical trials)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Charleston JS et al. (2018) Eteplirsen treatment for Duchenne muscular dystrophy: Exon skipping and dystrophin production. Neurology 90 (24), e2146–e2154. [DOI] [PubMed] [Google Scholar]
- 2.Lapidos KA et al. (2004) The dystrophin glycoprotein complex: signaling strength and integrity for the sarcolemma. Circ Res 94 (8), 1023–31. [DOI] [PubMed] [Google Scholar]
- 3.Wallace GQ and McNally EM (2009) Mechanisms of muscle degeneration, regeneration, and repair in the muscular dystrophies. Annu Rev Physiol 71, 37–57. [DOI] [PubMed] [Google Scholar]
- 4.Partridge TA et al. (1989) Conversion of mdx myofibres from dystrophin-negative to -positive by injection of normal myoblasts. Nature 337 (6203), 176–9. [DOI] [PubMed] [Google Scholar]
- 5.Mendell JR et al. (1995) Myoblast transfer in the treatment of Duchenne’s muscular dystrophy. N Engl J Med. 333, 832–838. [DOI] [PubMed] [Google Scholar]
- 6.Partridge T et al. (1998) Is myoblast transplantation effective? Nat Med. 4, 1208–1209. [DOI] [PubMed] [Google Scholar]
- 7.Tremblay JP et al. (1993) Results of a triple blind clinical study of myoblast transplantations without immunosuppressive treatment in young boys with Duchenne muscular dystrophy. Cell Transplant. 2, 99–112. [DOI] [PubMed] [Google Scholar]
- 8.Vilquin JT (2005) Myoblast transplantation: clinical trials and perspectives. Acta Myol. 24, 119–127. [PubMed] [Google Scholar]
- 9.Gussoni E et al. (1997) The fate of individual myoblasts after transplantation into muscles of DMD patients. Nat Med. 3, 970–977. [DOI] [PubMed] [Google Scholar]
- 10.Qu Z et al. (1998) Development of approaches to improve cell survival in myoblast transfer therapy. J Cell Biol. 142, 1257–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montarras D et al. (2005) Direct isolation of satellite cells for skeletal muscle regeneration. Science 309 (5743), 2064–7. [DOI] [PubMed] [Google Scholar]
- 12.Ikemoto M et al. (2007) Autologous transplantation of SM/C-2.6(+) satellite cells transduced with micro-dystrophin CS1 cDNA by lentiviral vector into mdx mice. Mol Ther 15 (12), 2178–85. [DOI] [PubMed] [Google Scholar]
- 13.Sampaolesi M et al. (2006) Mesoangioblast stem cells ameliorate muscle function in dystrophic dogs. Nature 444, 574–579. [DOI] [PubMed] [Google Scholar]
- 14.Sampaolesi M et al. (2003) Cell therapy of alpha-sarcoglycan null dystrophic mice through intra-arterial delivery of mesoangioblasts. Science 301, 487–492. [DOI] [PubMed] [Google Scholar]
- 15.Cossu G et al. (2015) Intra-arterial transplantation of HLA-matched donor mesoangioblasts in Duchenne muscular dystrophy. EMBO Mol Med 7 (12), 1513–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans MJ and Kaufman MH (1981) Establishment in culture of pluripotential cells from mouse embryos. Nature 292 (5819), 154–6. [DOI] [PubMed] [Google Scholar]
- 17.Martin GR (1981) Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A 78 (12), 7634–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomson JA et al. (1998) Embryonic stem cell lines derived from human blastocysts. Science 282 (5391), 1145–7. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi K et al. (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131 (5), 861–72. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi K and Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126 (4), 663–76. [DOI] [PubMed] [Google Scholar]
- 21.Taylor CJ et al. (2012) Generating an iPSC bank for HLA-matched tissue transplantation based on known donor and recipient HLA types. Cell Stem Cell 11 (2), 147–52. [DOI] [PubMed] [Google Scholar]
- 22.Turner M et al. (2013) Toward the development of a global induced pluripotent stem cell library. Cell Stem Cell 13 (4), 382–4. [DOI] [PubMed] [Google Scholar]
- 23.de Rham C and Villard J (2014) Potential and limitation of HLA-based banking of human pluripotent stem cells for cell therapy. J Immunol Res 2014, 518135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Solomon S et al. (2015) Banking on iPSC--is it doable and is it worthwhile. Stem Cell Rev 11 (1), 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis RL et al. (1987) Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51 (6), 987–1000. [DOI] [PubMed] [Google Scholar]
- 26.Lassar AB et al. (1986) Transfection of a DNA locus that mediates the conversion of 10T1/2 fibroblasts to myoblasts. Cell 47 (5), 649–56. [DOI] [PubMed] [Google Scholar]
- 27.Goudenege S et al. (2012) Myoblasts derived from normal hESCs and dystrophic hiPSCs efficiently fuse with existing muscle fibers following transplantation. Mol Ther 20 (11), 2153–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tedesco FS et al. (2012) Transplantation of genetically corrected human iPSC-derived progenitors in mice with limb-girdle muscular dystrophy. Sci Transl Med 4 (140), 140ra89. [DOI] [PubMed] [Google Scholar]
- 29.Oustanina S et al. (2004) Pax7 directs postnatal renewal and propagation of myogenic satellite cells but not their specification. EMBO J 23 (16), 3430–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chal J and Pourquie O (2017) Making muscle: skeletal myogenesis in vivo and in vitro. Development 144 (12), 2104–2122. [DOI] [PubMed] [Google Scholar]
- 31.Comai G and Tajbakhsh S (2014) Molecular and cellular regulation of skeletal myogenesis. Curr Top Dev Biol 110, 1–73. [DOI] [PubMed] [Google Scholar]
- 32.Darabi R et al. (2012) Human ES- and iPS-derived myogenic progenitors restore DYSTROPHIN and improve contractility upon transplantation in dystrophic mice. Cell Stem Cell 10 (5), 610–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Magli A et al. (2017) PAX7 Targets, CD54, Integrin α9β1, and SDC2, Allow Isolation of Human ESC/iPSC-Derived Myogenic Progenitors. Cell Rep 19 (13), 2867–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barberi T et al. (2007) Derivation of engraftable skeletal myoblasts from human embryonic stem cells. Nat Med 13 (5), 642–8. [DOI] [PubMed] [Google Scholar]
- 35.Awaya T et al. (2012) Selective development of myogenic mesenchymal cells from human embryonic and induced pluripotent stem cells. PLoS One 7 (12), e51638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borchin B et al. (2013) Derivation and FACS-mediated purification of PAX3+/PAX7+ skeletal muscle precursors from human pluripotent stem cells. Stem Cell Reports 1 (6), 620–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shelton M et al. (2014) Derivation and expansion of PAX7-positive muscle progenitors from human and mouse embryonic stem cells. Stem Cell Reports 3 (3), 516–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van der Wal E et al. (2018) Large-Scale Expansion of Human iPSC-Derived Skeletal Muscle Cells for Disease Modeling and Cell-Based Therapeutic Strategies. Stem Cell Reports 10 (6), 1975–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chal J et al. (2015) Differentiation of pluripotent stem cells to muscle fiber to model Duchenne muscular dystrophy. Nat Biotechnol 33 (9), 962–9. [DOI] [PubMed] [Google Scholar]
- 40.Chal J et al. (2016) Generation of human muscle fibers and satellite-like cells from human pluripotent stem cells in vitro. Nat Protoc 11 (10), 1833–50. [DOI] [PubMed] [Google Scholar]
- 41.Kim J et al. (2017) Expansion and Purification Are Critical for the Therapeutic Application of Pluripotent Stem Cell-Derived Myogenic Progenitors. Stem Cell Reports 9 (1), 12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giacomazzi G et al. (2017) MicroRNAs promote skeletal muscle differentiation of mesodermal iPSC-derived progenitors. Nat Commun 8 (1), 1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quattrocelli M et al. (2015) Mesodermal iPSC-derived progenitor cells functionally regenerate cardiac and skeletal muscle. J Clin Invest 125 (12), 4463–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hicks MR et al. (2018) ERBB3 and NGFR mark a distinct skeletal muscle progenitor cell in human development and hPSCs. Nat Cell Biol 20 (1), 46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi IY et al. (2016) Concordant but Varied Phenotypes among Duchenne Muscular Dystrophy Patient-Specific Myoblasts Derived using a Human iPSC-Based Model. Cell Rep 15 (10), 2301–2312. [DOI] [PubMed] [Google Scholar]
- 46.Wu J et al. (2016) Generation and validation of PAX7 reporter lines from human iPS cells using CRISPR/Cas9 technology. Stem Cell Res 16 (2), 220–8. [DOI] [PubMed] [Google Scholar]
- 47.Wu J et al. (2016) Generation and Characterization of a MYF5 Reporter Human iPS Cell Line Using CRISPR/Cas9 Mediated Homologous Recombination. Sci Rep 6, 18759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu J et al. (2018) A Myogenic Double-Reporter Human Pluripotent Stem Cell Line Allows Prospective Isolation of Skeletal Muscle Progenitors. Cell Rep 25 (7), 1966–1981.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim YG et al. (1996) Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci U S A 93 (3), 1156–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller JC et al. (2011) A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 29 (2), 143–8. [DOI] [PubMed] [Google Scholar]
- 51.Christian M et al. (2010) Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186 (2), 757–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mussolino C et al. (2011) A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res 39 (21), 9283–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jinek M et al. (2013) RNA-programmed genome editing in human cells. Elife 2, e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cho SW et al. (2013) Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 31 (3), 230–2. [DOI] [PubMed] [Google Scholar]
- 55.Cong L et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339 (6121), 819–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mali P et al. (2013) RNA-guided human genome engineering via Cas9. Science 339 (6121), 823–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moore JK and Haber JE (1996) Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol 16 (5), 2164–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Capecchi MR (1989) Altering the genome by homologous recombination. Science 244 (4910), 1288–92. [DOI] [PubMed] [Google Scholar]
- 59.Smithies O et al. (1985) Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature 317 (6034), 230–4. [DOI] [PubMed] [Google Scholar]
- 60.McVey M and Lee SE (2008) MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet 24 (11), 529–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kazuki Y et al. (2010) Complete genetic correction of ips cells from Duchenne muscular dystrophy. Mol Ther 18 (2), 386–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li HL et al. (2015) Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Reports 4 (1), 143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Young CS et al. (2016) A Single CRISPR-Cas9 Deletion Strategy that Targets the Majority of DMD Patients Restores Dystrophin Function in hiPSC-Derived Muscle Cells. Cell Stem Cell 18 (4), 533–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Long C et al. (2018) Correction of diverse muscular dystrophy mutations in human engineered heart muscle by single-site genome editing. Sci Adv 4 (1), eaap9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Turan S et al. (2016) Precise Correction of Disease Mutations in Induced Pluripotent Stem Cells Derived From Patients With Limb Girdle Muscular Dystrophy. Mol Ther 24 (4), 685–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Iyer S et al. (2019) Precise therapeutic gene correction by a simple nuclease-induced double-stranded break. Nature 568 (7753), 561–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gao Y et al. (2016) Genome Therapy of Myotonic Dystrophy Type 1 iPS Cells for Development of Autologous Stem Cell Therapy. Mol Ther 24 (8), 1378–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Y et al. (2018) Therapeutic Genome Editing for Myotonic Dystrophy Type 1 Using CRISPR/Cas9. Mol Ther 26 (11), 2617–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dastidar S et al. (2018) Efficient CRISPR/Cas9-mediated editing of trinucleotide repeat expansion in myotonic dystrophy patient-derived iPS and myogenic cells. Nucleic Acids Res 46 (16), 8275–8298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Skuk D et al. (2003) Resetting the problem of cell death following muscle-derived cell transplantation: detection, dynamics and mechanisms. J Neuropathol Exp Neurol 62 (9), 951–67. [DOI] [PubMed] [Google Scholar]
- 71.Beauchamp JR et al. (1999) Dynamics of myoblast transplantation reveal a discrete minority of precursors with stem cell-like properties as the myogenic source. J Cell Biol 144 (6), 1113–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dull T et al. (1998) A third-generation lentivirus vector with a conditional packaging system. J Virol 72 (11), 8463–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zufferey R et al. (1998) Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol 72 (12), 9873–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cavazzana-Calvo M et al. (2010) Transfusion independence and HMGA2 activation after gene therapy of human beta-thalassaemia. Nature 467 (7313), 318–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cartier N et al. (2009) Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science 326 (5954), 818–23. [DOI] [PubMed] [Google Scholar]
- 76.Aiuti A et al. (2013) Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science 341 (6148), 1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Biffi A et al. (2013) Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science 341 (6148), 1233158. [DOI] [PubMed] [Google Scholar]
- 78.Knoepfler PS (2009) Deconstructing stem cell tumorigenicity: a roadmap to safe regenerative medicine. Stem Cells 27 (5), 1050–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Carpenter MK and Rao MS (2015) Concise review: making and using clinically compliant pluripotent stem cell lines. Stem Cells Transl Med 4 (4), 381–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mandai M et al. (2017) Autologous Induced Stem-Cell-Derived Retinal Cells for Macular Degeneration. N Engl J Med 376 (11), 1038–1046. [DOI] [PubMed] [Google Scholar]
- 81.Kikuchi T et al. (2017) Human iPS cell-derived dopaminergic neurons function in a primate Parkinson’s disease model. Nature 548 (7669), 592–596. [DOI] [PubMed] [Google Scholar]
- 82.Tsuji O et al. (2019) Concise Review: Laying the Groundwork for a First-In-Human Study of an Induced Pluripotent Stem Cell-Based Intervention for Spinal Cord Injury. Stem Cells 37 (1), 6–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cyranoski D (2018) ‘Reprogrammed’ stem cells approved to mend human hearts for the first time. Nature 557 (7707), 619–620. [DOI] [PubMed] [Google Scholar]
- 84.Tremblay JP et al. (2009) A case for immunosuppression for myoblast transplantation in duchenne muscular dystrophy. Mol Ther 17 (7), 1122–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Deuse T et al. (2019) Hypoimmunogenic derivatives of induced pluripotent stem cells evade immune rejection in fully immunocompetent allogeneic recipients. Nat Biotechnol 37 (3), 252–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu YW et al. (2018) Human embryonic stem cell-derived cardiomyocytes restore function in infarcted hearts of non-human primates. Nat Biotechnol 36 (7), 597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]