Abstract
Antimicrobial peptides (AMPs) have been an area of great interest, due to the high selectivity of these molecules toward bacterial targets over host cells and the limited development of bacterial resistance to these molecules throughout evolution. Previous work showed that when Histidine was incorporated into the peptide C18G it lost antimicrobial activity. The role of pH on activity and biophysical properties of the peptide was investigated to explain this phenomenon. Minimal Inhibitory Concentration (MIC) results demonstrated that decreased media pH increased antimicrobial activity. Tricholorethanol (TCE) quenching and red-edge excitation spectroscopy (REES) showed a clear pH dependence on peptide aggregation in solution. Trp fluorescence was used to monitor binding to lipid vesicles and demonstrated the peptide binds to anionic bilayers at all pH values tested, however, binding to zwitterionic bilayers was enhanced at pH 7 and 8 (above the His pKa). Dual Quencher Analysis (DQA) confirmed the peptide inserted more deeply in PC:PG and PE:PG membranes, but could insert into PC bilayers at pH conditions above the His pKa. Bacterial membrane permeabilization assays which showed enhanced membrane permeabilization at pH 5 and 6 but vesicle leakage assays indicate enhanced permeabilization of PC and PC:PG bilayers at neutral pH. The results indicate the ionization of the His side chain affects the aggregation state of the peptide in solution and the conformation the peptide adopts when bound to bilayers, but there are likely more subtle influences of lipid composition and properties that impact the ability of the peptide to form pores in membranes.
Keywords: Antimicrobial peptides, fluorescence, lipid binding, pH-dependence, membrane permeabilization
Graphical abstarct
1. Introduction
The development of antimicrobial resistance (AMR) in bacteria has been identified as a major threat by a number of international health agencies (1). As bacteria develop resistance to conventional antibiotics, treatment options are restricted for patients who have infections, as well as an increase in the number of deaths worldwide associated with antibiotic resistant bacteria. This threat highlights the need for the development of novel antimicrobials with lower potential for resistance development.
Antimicrobial peptides (AMPs) have been a topic of a significant amount of study since the isolation of magainin from Xenopus laevis in the 1980’s (2). These peptides are naturally occurring in all higher organisms, indicating bacteria have not been able to mount significant evolutionary resistance to AMPs (3,4). The majority of characterized AMPs work via a membrane-active mechanism, utilizing a cationic, facially amphiphilic sequence/structure to selectively bind bacterial membranes and insert hydrophobic residues into the core of the bilayer, disrupting the bacterial cell membrane leading to cell death (5-7). The membrane active mechanism follows the lack of evolutionary resistance as bacteria could not easily evolve away from using lipid bilayers as the cell membrane. As such, AMPs are continually studied as a potential platform for novel antimicrobials with low potential for resistance development(4,6,8,9).
AMPs have been isolated from numerous organisms, been designed de novo, and developed from larger proteins (10-12). C18G is a short, 18 aa peptide derived from the C-terminus of the human platelet factor IV protein (13). This peptide exhibits broad spectrum antibacterial activity, effectively binds anionic lipid bilayers, adopts a helical conformation upon binding bilayers or bilayer mimics, and induces membrane disruption in both Gram positive and Gram negative bacteria (14,15). Notably, the peptide is primarily composed of Leu and Lys residues, 33% and 39% respectively, with a net charge of +8 at neutral pH. Previous experiments on this peptide demonstrated that both the hydrophobic content and the type of residues giving the cationic charge greatly affect the properties and activity of C18G. Notably, replacing the original Lys residues with His resulted in a complete loss of activity against all bacterial strains investigated (14,15).
The loss of activity with His in C18G was interesting as the result was not completely expected. On one hand, the experiments on the C18G-His peptide, in which the Lys residues were replaced with His, were performed at neutral pH, where the peptide should have only a +1 charge, thus likely exhibiting a reduced affinity for anionic bacterial cell surfaces. On the other hand, the C18G-His peptide was shown to interact with anionic bilayers and adopt a helical conformation upon binding, similar to +8 charged analogs with Lys or other cationic residues. Additionally, His is a major amino acid component of the naturally occurring host defense peptides known as histatins, found in mammalian oral cavities and salivary secretions (16,17). Histatins are known to also exhibit strong antifungal activity, and are already in clinical trials for antifungal mouthwashes used in treatment of oral candidiasis in HIV patients (18-20). In general, histatins are thought to act against bacteria and fungi in a similar fashion, disrupting the cell membrane and causing content leakage(18). Additionally, the multitude of His residues in histatins have been shown to chelate heavy metals, leading to the generation of reactive oxygen species in and around target cells (21,22). Beyond histatins, His residues are found in AMPs at approximately the same frequency as in full length proteins: 2.07% in AMPs, 2.27% in mammalian proteins. Indeed, ~36% of sequences in the APD3 antimicrobial peptide database contain at least one histidine residue, and ~16% that contain multiple His residues (23). Still, the role of His in AMPs and the mechanism of action has only been studied for a limited number of sequences compared to Lys and Arg(24,25).
In this work, a more thorough investigation of the C18G-His peptide was carried out to further elucidate the membrane binding properties and potential antibacterial activity using a combination of spectroscopic and microbiological methods. The C18G-His peptide exhibited significantly enhanced activity at pH 5 and 6, indicating that the charge-state of the histidine, and the peptide in general, is an important driver for antibacterial activity. The interactions of the peptide with bilayers was then investigated using the Trp in the C18G-His sequence as a reporter. Binding to lipid bilayers was evaluated under several solution pH conditions and with a “mammalian” and a “bacterial” lipid composition with varied anionic lipid content. Quenching of Trp fluorescence was used to investigate both bilayer orientation and peptide aggregation in solution, indicating that both environmental pH and bilayer composition affect the depth at which the peptides insert into the bilayer. Circular dichroism spectroscopy was used to characterize the secondary structure of the peptides at different pH conditions and when in solution vs. bound to a lipid bilayer. In aggregate, the results show that while C18G-His can bind to lipid bilayers of either composition and at all pH values tested, the depth and orientation of the peptide when bound is significantly different between the electrostatic-enhanced binding (low pH, anionic lipids) and the neutral form (neutral pH, zwitterionic lipids).
2. Experimental Methods
2.1. Materials
Peptides were purchased from Genscript (Piscataway, NJ, USA) or synthesized in-house by solid phase FMOC-chemistry methods. In-house synthesized sequences were synthesized on rink-amide resin with DMF as the main solvent, 20% piperidine in DMF as the deprotecting agent, HATU (1-[Bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium3-oxid hexafluorophosphate) as the activator, with a 5:1 amino acid excess during coupling. Peptide cleavage from the resin was achieved using a standard mixture of 92.5:2.5:2.5:2.5 trifluoroacetic acid (TFA)/water/triisopropylsilane/ethanedithiol. Cleaved peptides were separated from the resin via gravity filtration and subsequently flocculated in ice-cold diethyl ether followed by sedimentation by low speed centrifugation. All peptides were purified by reversed-phase HPLC (RP-HPLC) with a linear gradient of water and acetonitrile each containing 0.1% TFA using a Jupiter 300 C4 column (Phenomenex). Peptide identity was confirmed by ESI-MS analysis. Lipids, (16:0–18:1) 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine (POPC, PC), and (16:0–18:1) 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylglycerol (POPG, PG), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE, PE) were purchased from Avanti Polar Lipids (Alabaster, AL, USA), and stored as stocks in chloroform at −20 °C. Lipid vesicles were prepared were prepared as described previously (26). Small unilamellar vesicles (SUVs) were created through sonication of multilamellar vesicles (MLVs) in a high-power bath sonicator (Avanti Lipids, Alabaster, AL, USA) for 20 min to yield small unilamellar vesicles (SUVs).
The buffers used were ABS (150 mM NaCl, 50 mM NaCH3CO2/CH3CO2H; pH 5.0), CBS (150 mM NaCl, 50 mM C6H8O7/Na3C6H5O7; pH 6.0), PBS (150 mM NaCl, 50 mM Na2HPO4/NaH2PO4; pH 7.0), TBS ((150 mM NaCl, 50 mM NH2C(CH2OH)3; pH 8.0), 10× diluted buffer (for circular dichroism (CD) measurements), or Z-Buffer (0.1 M Na2HPO4/NaH2PO4, 10 mM KC1, 1 mM MgSO4, 0.05 M β-mercaptoethanol, adjusted to appropriate pH).
2.2. Minimal Inhibitory Concentration / Minimal Bactericidal Concentration Assays
Bacteria were streaked onto LB-Miller agar (BD-Difco, Franklin Lakes, NJ, USA) plates from a frozen permanent stock (E. coli D31(27), S. aureus ATCC: 27660, P. aeruginosa PAO1, and A. baumatmii ATCC: 19606). Overnight cultures were inoculated with a single colony of each bacterial strain into fresh LB or Mueller Hinton broth (MH, BD-Difco) and placed in a 37 °C shaking incubator (250 rpm) for ~18 h. After the overnight incubation, a 1:200 dilution of the culture was made in fresh LB or MH for use in subsequent experiments.
Minimal Inhibitory Concentration (MIC) was determined using a broth microdilution assay as reported previously (28,29). Briefly, bacteria were grown as described above, and upon reaching mid-log phase were subsequently diluted to ~105 cfu/mL in pH-adjusted MH media. Next, 90 μL of this diluted culture was added to wells of a sterile 96-well plate containing serially diluted aliquots of the peptide for a final volume in the well of 100 μL. The plate was covered and incubated at 37 °C for 18 h. The OD600 was measured after incubation with a Spectramax M5 multimode plate reader. The minimal bactericidal concentration (MBC) was determined by taking 1 μL of culture from each well of the MIC plate and spotted on a fresh LB-agar plate which was incubated overnight at 37°C. MBC was determined from the presence or absence of growth on each spot after the overnight incubation.
2.3. Fluorescence Spectroscopy
Fluorescence spectra were recorded on a JY-Horiba Fluoromax4 instrument with emission and excitation slits set to 2.5 nm. Emission spectra for binding assays, samples were excited at 280 nm with emission measured over the range of 300–400 nm. All samples contained 2 μM peptide and were titrated with a lipid vesicle stock ranging in concentration from 1–2 mM. The barycenter of the spectrum is an intensity-weighted average over the wavelengths measured, and Δbarycenter is the difference between the barycenter of the sample lacking lipid, and that of a given sample (14). Spectra were corrected for background and dilution. Red-edge excitation spectra (REES) were excited at 280, 290, 295, 300, 305, or 307 nm and emission spectra were collected from 310-410 nm.
The acrylamide quenching assays were performed as previously described. Briefly samples were prepared containing 2 μM peptide and 250 μM total lipid. Samples were excited at 295 nm (to reduce the inner filter effect of acrylamide) and emission was measured at 340 nm. After measurement, an aliquot of 4M acrylamide was added to the sample, mixed, allowed to equilibrate for ~2 minutes, and the fluorescence emission was re-measured. All data were corrected for dilution, background, and remaining inner filter effects as described previously (14).
The TCE quenching experiments were performed by adding successive aliquots of TCE from a 10M stock to samples containing 2 μM peptide in buffer. Fluorescence was measured after each addition. Samples were excited at 280 nm and emission was measured at 340 nm. All data were corrected for dilution and background fluorescence.
Dual quencher analysis (DQA) was performed as previously described (26,30). Briefly, two sets of samples containing 2μM peptide and 200μM total lipid were created. One set was used as the unquenched sample (F0) and 0.235M acrylamide would subsequently be added to those (Fac). The other set contained 10 mol% 10-doxyl nonadecane (10-DN) in the vesicles (Fdox). The Q-ratio was calculated as described previously (26,30,31).
2.4. Circular Dichroism Spectroscopy
CD spectra were collected using a Jasco J-815 spectropolarimeter. Samples for CD spectroscopy contained 5 μM peptide in 0.1X buffer, 0.1X buffer with 200 μM lipid vesicles, or 0.1X buffer supplemented with 50% (v/v) trifluoroethanol. All spectra shown are the average of 64–128 scans and were corrected for backgrounds by subtracting the spectra of the samples lacking peptide. Lipid vesicles for CD spectra were created using the ethanol dilution method (32). Lipid films were created as described above, but after vacuum the films were first dissolved in 10 μL of ethanol and vortexed vigorously while adding the appropriate volume of buffer.
2.5. Vesicle permeabilization assays
Assays were carried out as previously described (33). Briefly, vesicles were formed in the presence of 60μM of Ru(bpy)32+ and subjected to 15 rounds of extrusion through two stacked 0.2 micron polycarbonate filters using an extruder apparatus (Avanti Polar lipids, Alabaster AL). The vesicle mixture was then loaded onto a Sephadex G-25 column in pH 7 buffer. Vesicle fractions eluted first and were visibly turbid. An aliquot of the vesicles was diluted in pH-adjusted buffer and mixed with 2μM peptide. The Ru(bpy)32+ fluorescence lifetime was measured with a home-built laser-induced fluorescence (LIF) instrument, described previously. Intensity decays were fit with a single exponential decay function where the reciprocal of the decay constant is the fluorescence lifetime. Fractional leakage was determined by comparing sample lifetimes to those of untreated vesicles and free Ru(bpy)32+
Calcein leakage assays were performed similar to Ru(bpy)32+ assays but instead using a solution of 75mM calcein as the fluorophore as described previously (34). Aliquots of vesicles were mixed with pH-adjusted buffer and mixed with serial dilutions of peptide. Fluorescence intensity was measured in a SpectraMax M5 plate reader (Molecular Devices, San Jose CA). Fractional leakage was calculated based on untreated vesicles (0% leakage) and vesicles treated with 20μL Triton X-100 (100% leakage). Notably, intensities varied at each pH as calcein fluorescence is pH sensitive.
2.6. E. coli membrane permeabilization assays
Permeabilization of the E. coli outer membrane was carried out similar to previously described (28,35). Briefly, a single colony of E. coli D31 was inoculated in LB broth supplemented with 100 mg/mL ampicillin and allowed to grow in a shaking 37 °C incubator overnight. The next day, the overnight culture was diluted 1:200 in fresh LB-Amp and grown at 37 °C with shaking until the OD600 was ~0.2. The culture was subsequently centrifuged at 2500 rpm for 15 min. The pellet was re-suspended in an equal volume of pH-adjusted buffer and 80~L of this bacterial suspension was added to a 96-well plate containing serially diluted peptides or detergent (positive control). Immediately before the first measurement, an aliquot of 0.5mg/ml nitrocefin was added to each sample well (final concentration 0.05 mg/mL) and mixed by pipetting to ensure homogeneous distribution and minimize bubble formation. The sample absorbance was measured at 486 nm every 5 min for a total of 90 min. All assays were performed in triplicate.
Permeabilization of the E. coli inner membrane was carried out similar to previously described (28,35). The procedure is similar to that of the outer membrane assay with several modifications. Bacteria were grown overnight in LB media without Amp, and dilutions were made in LB supplemented with 1mM IPTG. Assay plates were prepared the same, except that cells were not centrifuged before the assay, and were added to the plate directly in the culture medium. Each well contained peptide or control molecules to which 57μL of pH-adjusted buffer and then 18μL of culture were added. The ONPG substrate was dissolved in pH-adjusted buffer and added to each well at a final concentration of 0.6 mg/mL. The sample absorbance was measured at 420 nm in 5 min intervals for 90 min. All assays were performed at least in triplicate. Absorbance values were converted to moles of ONP product to account for varying molar absorptivity of the product as a function of pH.
2.7. Hemolysis Assay
Hemolysis of sheep red blood cells (RBCs) was used to quantify membrane destabilization by leakage of hemoglobin. A 5mL aliquot of defibrinated sheep blood (HemoStat Laboratories) was mixed with 5mL sterile PBS and subsequently sedimented via centrifugation. The supernatant was aspirated and the cell pellet was resuspended to a final volume of 10 mL in PBS. This was repeated three times. The final cell pellet was resuspended to 10ml and 135 μL of this cell suspension was added to all wells of a conical-bottom 96-well plate containing 15 μL of serially diluted peptide or the detergent Triton-X 100 as a positive control. The plate was covered and allowed to incubate for 60 min at 37°C while shaking (100rpm). The plate was centrifuged for 15 min at low speed to pellet the in-tact RBCs. Leakage analysis was performed by carefully removing 6 μL of the cell supernatant and mixing with 94 μL of fresh PBS followed by absorbance measurements at 420 nm using a Molecular Devices M5 plate reader. Percent hemolysis was calculated based on the absorbance of each well compared to those wells with no additive and those with Triton-X100.
3. Results
3.1. Peptide Sequence –
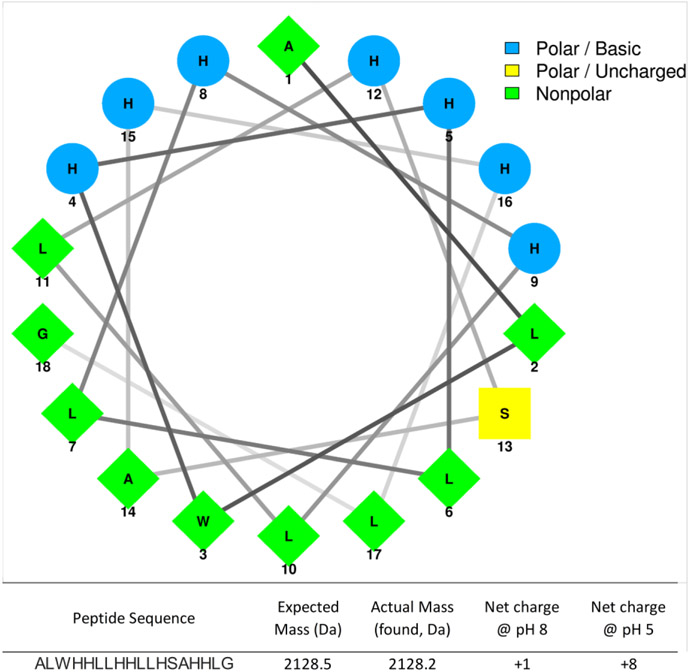
The original sequence of C18G is a modified sequence based on the 13-amino acid C-terminal helix of human platelet factor IV (13). This modified sequence was subsequently shown to activate a number of bacterial signaling pathways and exhibit broad spectrum antimicrobial activity (36,37). The modified C18G peptide sequence in which Lys are replaced with His was first characterized in previous work (14). The peptide is predicted to form a facially amphiphilic α-helix (Figure 1) (38).
Figure 1. Helical wheel diagram of C18G-His.
Hydrophobic residues are shown in green, His in blue, and polar uncharged in yellow. Peptide sequence and physiochemical properties are shown below the wheel.
3.2. Antimicrobial Activity
The effect of pH on antimicrobial activity was investigated using MIC and MBC assays. The MIC is the concentration of peptide which prevents bacterial growth in an overnight culture, while the MBC is the concentration required to kill the bacteria in the overnight culture. This distinction allows for discrimination between bacteriostatic and bactericidal mechanisms, and if these mechanisms are dose dependent. The MICs and MBCs of the C18G-His peptide at pH 5, 6, 7, and 8 are shown in Table 1. In all cases, the C18G-His peptide exhibited strong antibacterial activity at pH 5 which diminished at increasing pH values for all bacteria tested (S. aureus, E. coli, A. baumatmii, and P. aeruginosa). Similarly, the MBCs at all pH values were determined and showed the same pattern of pH dependence.
Table 1:
Antibacterial Activity (μM)
| S. aureus | P. aeruginosa | A. baumannii | E. coli | |||||
|---|---|---|---|---|---|---|---|---|
| pH | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC |
| 5 | 0.47 | 0.94 | 3.75 | 3.75 | 0.23 | 0.94 | 0.47 | 0.94 |
| 6 | 15 | 15 | 7.5 | 7.5 | 1.88 | 3.75 | 1.88 | 1.88 |
| 7 | 15 | >15 | >15 | >15 | 7.5 | 7.5 | 3.75 | 7.5 |
| 8 | 15 | >15 | >15 | >15 | >15 | >15 | 15 | 15 |
3.3. Peptide-peptide interactions in solution
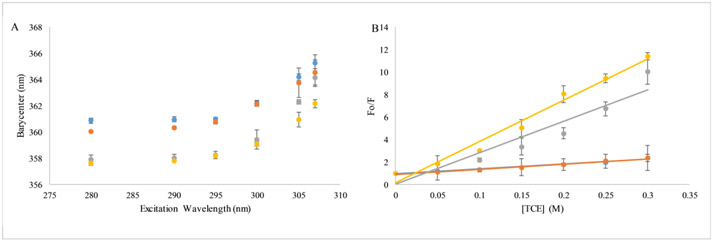
Due to the significant change in peptide charge over the pH range investigated coupled with the relatively high content of hydrophobic residues, aggregation of the peptides in solution was investigated as factor in the decreased antimicrobial activity. First, red-edge excitation spectra (REES) were collected for the peptide at all pH values tested. REES relies on the conformational flexibility of the Trp side chain, with more conformationally restricted Trp residues yielding red-shifted emission spectra as the excitation wavelength increases(39). As shown in Figure 2A, the C18G-His peptide REES exhibits upward curvature in the plot of barycenter vs. excitation wavelength, indicative of restricted motion around the Trp. Notably, although the absolute values of the barycenters vary as a function of pH, but the overall shape of the curve is the same regardless of pH. The REES of C18G which contains Lys instead of His and thus undergoes no change in net charge across the pH range tested as well as NATA (a model of fully conformationally free Trp), are shown in Supplemental Figure 1. These results show that at all pH values the Trp in C18G-His is somewhat restricted in local mobility.
Figure 2. Peptide aggregation assays.
(A) Red Edge Excitation Spectra (REES), (B) Tricholoroethane (TCE) quenching. In both panels colors represent pH 5 (blue), pH 6 (orange), pH 7 (yellow), pH 8 (gray). In all cases samples contained 2μM C18G-His peptide. All data are averages of 3-5 samples and error bars represent the standard deviation.
Peptide aggregation in solution was also interrogated using fluorescence quenching. In this case, TCE is a nonpolar molecule that effectively quenches Trp fluorescence only when the Trp is located in a nonpolar environment. Thus, if the C18G-His aggregates, the Trp would likely be exposed to a less polar local environment compared to the aqueous milieu in the monomeric form, and thus exhibit enhanced quenching by TCE. TCE quenching of C18G-His at all 4 pH values is shown in Figure 2B. Additionally, the TCE quenching of NATA is shown for comparison in Supplemental Figure 2. Overall, there was a significant change in quenching from low pH to neutral, indicating the Trp is in a more nonpolar environment at neutral pH.
3.3. Peptide Interactions with Lipid Membranes
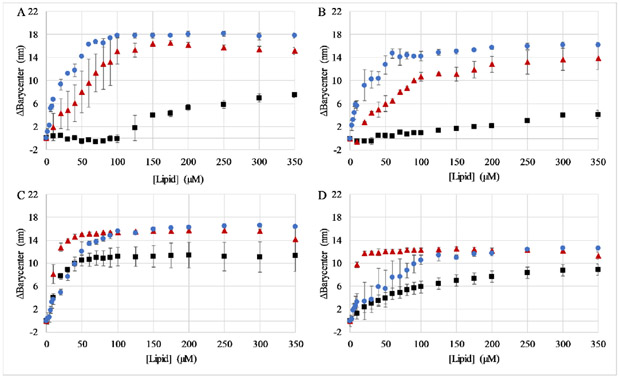
In order to better understand the underlying cause for the differential antimicrobial activity, the peptide interactions with small unilamellar vesicles was investigated. Vesicles were composed of either 100% DOPC lipids resulting in a zwitterionic, net neutral charge or 75:25 DOPC:DOPG and 75:25 DOPE:DOPG, resulting in a net negative surface charge (DOPG has a - 1 charge). The charged groups of PC, PE, and PG lipids have pKa values that are sufficiently far from the pH range being investigated that there should be negligible influence of pH on the charge state of the lipids (40). Binding to the bilayers was determined by changes in the Trp fluorescence emission from the peptide. Briefly, Trp is an environmentally sensitive fluorophore that exhibits a blue-shift in the emission spectrum when the Trp encounters a less polar environment compared to the aqueous milieu. This spectral shift is quantified by calculating the spectral barycenter, or weighted average of intensities over the spectral range, and how this barycenter changes as a function of the concentration of lipids titrated into the sample (Figure 3, Supplemental Figure 3). The results from the titrations show a clear preference for anionic bilayers compared to zwitterionic bilayers. Interestingly, this trend is also apparent at pH 7 and 8 where the peptide should be closer to neutral charge.
Figure 3. Peptide binding to small unilamellar vesicles.
Peptide binding was assayed using Trp fluorescence emission by titrating SUVs into a sample containing 2μM peptide. The spectral barycenter was calculated after each addition of lipid vesicles and the Δbarycenter is the difference between the initial barycenter and that after each addition. Binding was assayed at (A) pH 5, (B) pH 6, (C) pH 7, and (D) pH 8. In all panels red symbols represent vesicles composed of 3:1 PC:PG, blue symbols represent vesicles composed of 3:1 PE:PG, and black symbols represent PC vesicles . All data are averages of 2-3 samples and the error bars represent the standard deviation.
The binding to the lipid bilayer is only one component of how the peptides interact with the bilayer. Upon binding, the conformation that the peptide adopts in the bilayer can be impacted by both sequence and the charge state of the ionizable residues. Acrylamide quenching was used to gain deeper insight to how the peptides orient in bilayers. Acrylamide, an aqueous collisional quencher of Trp, is only effective at quenching Trp residues which are exposed to the aqueous environment. Thus, if a peptide is bound such that the Trp is occluded from water, the quenching will be weaker. Quenching experiments were performed and the Stern-Volmer quenching constants were extracted from the data (Table 2, Supplemental Figure 4). At all pH values except pH 8, the results surprisingly show an increase in the Ksv between buffer and bound to PC bilayers, indicating the Trp becomes more exposed upon binding. Uniformly at all pH values the Ksv decreases between peptides bound to PC compared to peptides bound to PC:PG bilayers and a small further decrease when bound to PE:PG bilayers.
Table 2.
Acrylamide Quenching Ksv (M−1)
| pH | Buffer | PC | PC:PG | PE:PG |
|---|---|---|---|---|
| 5 | 3.65 ± 0.36 | 7.24 ± 0.86 | 2.24 ± 1.16 | 1.32 ± 0.57 |
| 6 | 2.68 ± 0.62 | 10.61 ± 0.55 | 5.33 ± 4.11 | 0.88 ± 0.06 |
| 7 | 4.42 ± 0.87 | 6.36 ± 0.59 | 3.52 ± 0.12 | 1.42 ± 0.37 |
| 8 | 22.91 ± 1.96 | 6.12 ± 1.81 | 3.38 ± 0.34 | 2.22 ± 0.38 |
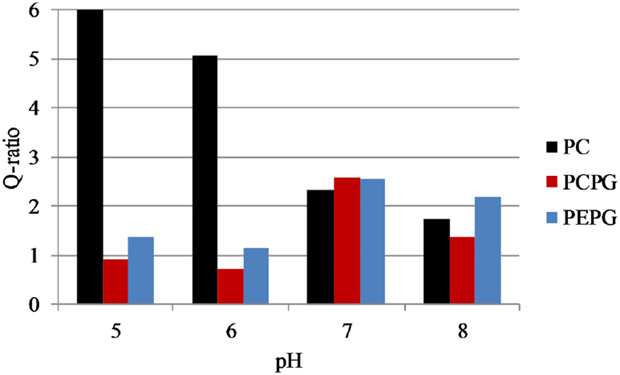
The quenching experiments were expanded using the dual-quencher analysis (DQA) method that incorporates the membrane bound quencher 10-doxyl nonadecane (10-DN). This assay uses both aqueous and membrane bound quenchers, acrylamide and 10-DN respectively, to determine the relative depth of a fluorophore in the bilayer. The output of this assay is referred to as the Q-ratio: a higher Q-ratio indicates a shallow depth in the bilayer while a low Q-ratio indicates a deeper location buried in the nonpolar core of the bilayer. Figure 4 shows the results of DQA in PC and PC:PG bilayers at the 4 different pH values tested. Overall the results show the peptide adopts a more deeply buried conformation in both PC:PG and PE:PG bilayers, consistent with the acrylamide Ksv analysis. Notably, the depth trends for PC is in the opposite directioncompared to PC:PG and PE:PG as a function of pH. According to the Q-ration, the peptide adopts a more deeply buried conformation at high pH in PC but is more shallowly buried in PC:PG and PE:PG at higher pH.
Figure 4. Dual Quencher Analysis (DQA) of C18G-His at different pH values.
Q-ratios were calculated for peptides bound to PC (Black), PC:PG (red), or PE:PG (blue) bilayers. The high Q-ratio for C18G-His with PC vesicles at pH 5 (13.3) is off scale for clarity of presentation purposes. Q-ratio is calculated from the ratio of F0/F values for quenching by 10-Doxyl nonadecane and acrylamide at fixed concentrations. Fluorophore depth in the bilayer is inversely proportional to Q-ratio. Values are calculated from average F0/F values of 3-5 samples.
3.4. Membrane Permeabilization Assays
While there are clear differences in antibacterial activity and peptide-bilayer interactions at different pH values, the mechanism by which the peptide is exerting antibacterial activity is still not clear. As such, a series of bilayer or membrane permeabilization assays were carried out. First, vesicle dye-leakage assays were performed using the Ru(bpy)32+ fluorophore which exhibits differential fluorescence lifetime when it is trapped inside lipid vesicles as opposed to when it is free in solution, such as when released from the vesicle lumen (33). The results of the leakage assay are shown in Figure 5A. Additional dose-dependent leakage assays were performed using the self-quenching fluorophore calcein (Supplemental Figure 9).
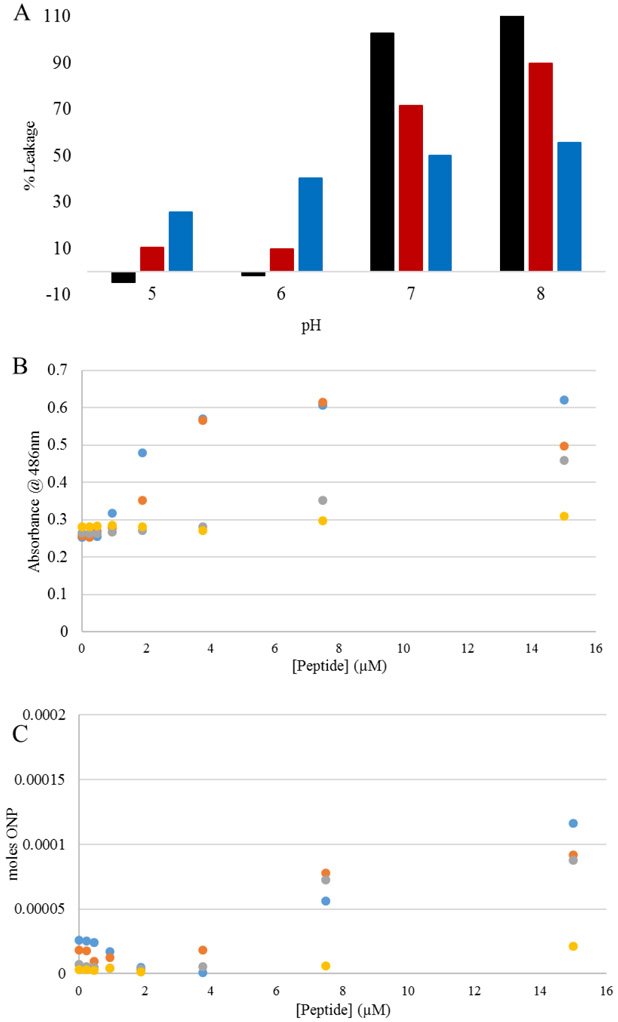
Figure 5. Membrane permeabilization of vesicles or E. coli by C18G-His at different pH conditions.
(A) Ru(bpy)32+ leakage from PC (black), PC:PG (red), or PE:PG (blue) vesicles. (B) Outer membrane permeability assayed by nitrocefin hydrolysis by β-lactamase in the bacterial periplasm measured at 486m. (C) Inner membrane permeability assayed by ONPG hydrolysis by β-galactosidase in the bacterial cytoplasm measured at 420m. In both panels B&C colors represent pH 5 (blue), pH 6 (orange), pH 7 (gray), pH 8 (yellow); data represent hydrolysis measured after 30min of exposure to varying concentrations of peptide. All data in B&C are averages of 3 samples with data in A being the result of fitting of exponential intensity decay functions.
While model membrane studies have revealed differences in binding conformation at different pH values, interactions with the significantly more complex bacterial cell membrane are inherently more representative of the normal functionality of AMPs. As the proposed mechanism of action for many AMPs is membrane destabilization, coupled with previous work from our group demonstrating the membrane activity of C18G, bacterial membrane permeabilization was investigated. Briefly, an enzyme-chromogenic substrate pair were used to monitor the permeability of the outer membrane (β-lactamase and nitrocefin) and the inner membrane (β-galactosidase and ONPG). The substrates are only minimally permeable across the membrane under normal conditions, but if the peptide (or other agent) permeabilizes the membrane, transit across the membrane increases resulting in an increased amount of substrate converted into the chromophoric product. In general, there is little to no permeabilization of PC and PC:PG bilayers at low pH, but increased permeabilization of PE:PG at low pH, consistent with the MIC results. However, leakge is significantly enhanced from PC and PC:PG vesicles at pH 7 and 8.
The results of the membrane permeability assays are shown in Figure 5B-C. The permeabilization of both the outer and inner membranes exhibited the standard dose-dependent permeabilization previously reported for C18G at low pH, but the permeabilzation was significantly reduced at pH 7 and 8. The data in Figure 5 is a snapshot of the 30 minute timepoint, with full kinetics traces shown in Supplemental Figures 5 and 6. Interestingly, in both the outer and inner membrane studies, there was some destabilization of the membranes at pH 7 and 8, but this took significantly longer to develop compared to the lower pH conditions. It should be noted that the chromogenic product of ONPG hydrolysis by β-galactosidase is pH sensitive, so extinction coefficients of the product were determined at the 4 pH values tested and used to convert to moles product in Figures 5B and S6. These calibration curves are shown in Supplemental Figure 7.
In addition to bacterial membrane permeabilization, the pH dependence of red blood cell (RBC) membrane disruption by C18G-His was also examined. Notably, the peptide was shown to induce minimal RBC lysis (< 2%) under all pH conditions at all concentrations tested except pH 5, 15μM peptide, which resulted in ~ 9% RBC lysis. The results of these experiments are shown in Supplemental Figure 8.
3.5. Peptide Secondary Structure
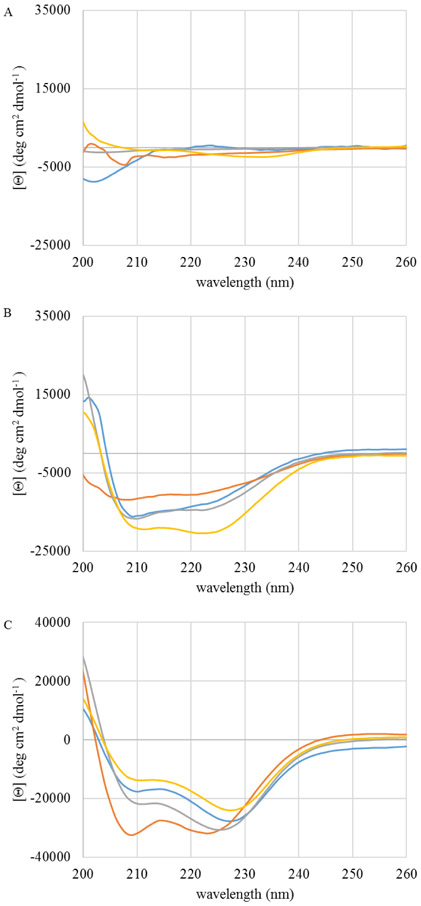
As many AMPs, including C18G, are known to adopt an α-helical conformation when bound to lipid bilayers, CD spectroscopy was used to investigate the structure of C18G-His in solution and when bound to PC:PG bilayers as a function of pH (Figure 6). Generally, the peptide did not display significant α-helical spectral signatures in buffer, nor did the spectra reflect β-strand or PPII. Upon binding to bilayers, the spectra display a more traditional α-helical pattern at all pH values.
Figure 6. Circular Dichroism (CD) spectroscopy of C18G-His.
In all panels colors represent pH 5 (blue), pH 6 (orange), pH 7 (gray), pH 8 (yellow). Spectra of samples containing 5 μM peptide were recorded in (A) buffer, (B) buffer containing PC:PG vesicles (250 μM total lipid), or (C) in buffer supplemented with 50% TFE (v:v). All spectra are the average of 64 scans and have been background corrected by subtraction of spectra lacking peptide.
4. Discussion
4.1. Solution properties of C18G-His
Although the near neutral pKa of His is well known and well studied in the context of enzyme catalytic mechanisms and biological sensors, the role in AMPs and membrane active peptides is less understood. Many studies on pH-triggered membrane active peptides such as pHLIP (41-43) rely on carboxylic acid containing side chains as the ionizable group driving changes in bilayer binding or orientation. However, the net cationic charge coupled with the decreased frequency of Asp and Glu in AMPs make this mode of pH trigger less viable. Considering the mechanism of many AMPs involves an electrostatic interaction between the cationic AMP and the anionic bacterial cell surface, the His pKa inherently yields a neutral side chain at physiological pH and cationic at low pH. However, the hydrophobic moieties of the AMPs are unaffected by pH and could thus still contribute to activity/binding to bilayers.
The REES and TCE quenching results indicate that the C18G-His peptide is aggregating to some extent in solution. While specific aggregation numbers cannot be determined, there appears to be some differential aggregation based on the comparison of these experiments. On the one hand, the REES data indicate that the peptide aggregates despite the pH and thus the net charge of the peptide. The upward curvature of barycenter vs. excitation wavelength is the hallmark of restricted Trp motion, similar to that seen in melittin(39,44-46). However, TCE quenching clearly shows a pH dependence, indicating there is some difference in local environment at low pH and neutral pH. This change in exposure of the Trp to TCE is similar to that seen for different aggregated states of melittin (39). Another AMP, dermaseptin PD-3-7, has also been shown to undergo pH-dependent oligomerization at pH 5-6 (47) while the pHlip peptide has also exhibited pH-dependent aggregation state changes (48,49). The discrepancy between REES and TCE may be the result of different aggregation number, changes in local conformation within the aggregate, or variable exposure of the Trp to solution. The GALA peptide, a designed amphipathic peptide, showed a similar pH-dependent change in aggregation state in solution by Trp emission changes (50). The comparison to the original C18G sequence containing Lys as the cationic amino acids (Supplemental Figure 1) indicates that the aggregation is directly linked to the presence of the His amino acids. Regarding the REES data, an alternative explanation may include more restricted local motion of the Trp side chain as the bulky His residue is in a neighboring position.
In addition, evidence of C18G-His aggregation is also seen in the acrylamide quenching experiments. This is evident when comparing the Ksv values obtained for acrylamide quenching in solution vs. those when the peptide was interacting with 100% PC (zwitterionic) bilayers. At pH 5-7, the Ksv values increase upon binding to lipid bilayers, indicating the Trp becomes more exposed to the aqueous acrylamide upon binding to the bilayer. This is contrary to what was observed for C18G and other AMPs containing Lys and other cationic amino acids (14,26) and what would be expected for a peptide transitioning from a monomeric, soluble state to a membrane bound state. However, this change in exposure is consistent with an aggregated conformation in which the Trp is buried at the interior of the aqueous aggregate but then becomes more exposed as the peptide interacts with the bilayer in a shallow position. Alternatively, if there is an equilibrium between aggregate, monomer, and membrane bound conformations, the quenching in the presence of PC bilayers may reflect incomplete binding and some subpopulation of peptides existing in a monomeric state which would be highly susceptible to quenching by acrylamide.
The likelihood of C18G-His aggregation could add an additional step in the binding & insertion pathway and could impact both the equilibrium and kinetics of the process. If the peptide must undergo a de-oligomerization before interaction with the bacterial membrane, this would impact the apparent affinity for bilayers and potentially the time required for the antibacterial activity.
4.2. Membrane interactions of C18G-His
The interactions of the C18G-His with lipid bilayers is also a likely contributing factor to the loss of antimicrobial activity at neutral pH. The first and most obvious area of interest is the binding to anionic bilayers which mimic bacterial membranes. The neutralization of the peptide above the His pKa should reduce or remove the electrostatic driving force that contributes to binding and is often cited as a major factor in AMP selectivity for bacterial cells over host cells(51). However, the hydrophobic amino acids are unaffected by this pH change, and can still be a driving force for membrane binding. Using the MPEx software suite, at neutral pH the total hydrophobic moment of the peptide is calculated at 6.17 and the ΔG of partitioning from water to bilayer is slightly favorable (−1.91 kcal·mol−1), however when the His residues are charged at lower pH, the hydrophobic moment increases to 16.44 and most importantly the ΔG of partitioning increases to an unfavorable 13.63 kcal·mol−1(52). These calculations are based on an experimentally determined hydrophobicity scale based on partitioning from water to a zwitterionic PC bilayer, which is consistent with the binding titrations (48). At neutral pH, the C18GHis bound to PC bilayers, presumably through favorable hydrophobic interactions between the peptide and the nonpolar core of the bilayer. As such, the mode of binding likely changes between lower pH and neutral pH as well as between zwitterionic and anionic bilayers. It should be noted that quantification of the binding affinity, traditionally reported as Kd, is inappropriate to calculate in the reported experiments. There is some slight variability in the final emission barycenter in the bound state, likely due to local environmental differences around the Trp arising from either Trp depth in the bilayer, local polarity from the various lipid headgroups used, vesicle curvature differences as a result of bilayer composition, or some combination of all of these. As such, global fitting of the data to compare binding affinities at different pH and lipid conditions would be less than appropriate.
Further analyzing the lipid dependence on binding shows some enhancement of binding to PE:PG vesicles compared to PC:PG vesicles. The enhancement could result from the inherent packing differences between PC and PE, resulting in easier access to the hydrophobic core, or from curvature differences in the vesicles. MD simulations support differential binding and conformational arrangements of some AMPs in PC vs. PE bilayers (53,54) while others indicate a more complex balance of headgroup and acyl-chain composition driving selectivity (55). Similarly, experimental studies have demonstrated a clear preference of some AMPs for PE-containing bilayers (56-58), however other peptides do not show this PE-dependent preference (59,60).
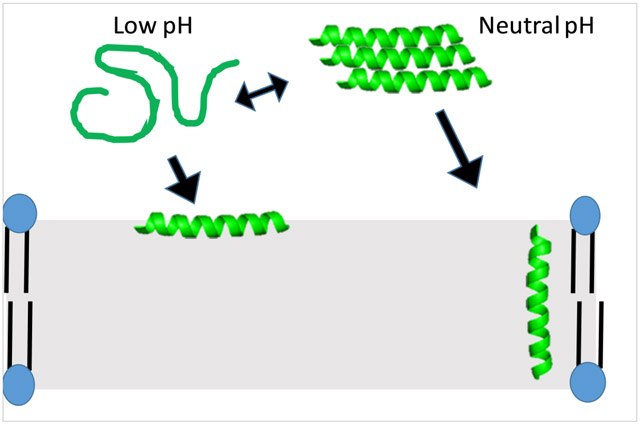
The DQA measurements (Figure 4) provide evidence that although the peptide binds to both neutral and anionic bilayers, the conformation is likely different. At low pH, the peptide exhibited a very high DQA with PC vesicles, consistent with the barycenter results. This indicates extremely weak binding to zwitterionic vesicles, which is consistent with a model that has the peptide favoring the aqueous soluble and potentially aggregated form under these conditions. However, once the pH favors neutralization of the His side chains, the peptide is able to bind to bilayers, although likely in a shallow orientation. Conversely, with anionic vesicles the peptide adopts a more deeply inserted topology at low pH, consistent with the electrostatic driving forces promoting interactions between the peptide and the bilayer. As the side chains are neutralized, the peptide appears to adopt a more shallowly-inserted orientation quenching, mimicking that observed in zwitterionic bilayers. This indicates that once the electrostatic binding force is eliminated, the peptide adopts a similar conformation in zwitterionic and anionic vesicles, likely driven through hydrophobic interactions. From this analysis there seems to be little effect of the PE vs. PC headgroup on peptide conformation in the bilayer with respect to Trp penetration depth.
The impact of pH and amino acid ionization state on topography of membrane associated peptides and proteins has been documented in a number of systems. London and coworkers showed that ionization of Asp or Lys in polyLeu peptides could alter both transmembrane insertion, peptide orientation, and oligomerization state(32,61,62) as well as in the Erb b2 TM helix(63). Similarly, Asp and Glu protonation events are the primary drivers of TM insertion and behavior in the well characterized pHLIP peptide system(41-43). Additionally, simulations have shown that protonation events can also affect the secondary structure of bilayer embedded pHLIP (64). Recent findings from the Hristova and Wimley labs have demonstrated highly potent, pH-dependent membrane permeabilizing peptides which are designed to become active at low pH (65).
Histidine mediated changes in membrane orientation have been reported in both model peptide systems and natural proteins. Koeppe and coworkers demonstrated that histidine protonation/deprotonation can impact the helical tilt as well as overall transmembrane conformation of model GWALP23 peptides, with charged histidine side chains driving orientations to reduce charge exposure to the nonpolar core of the bilayer (66). In natural proteins, Gillet and coworkers showed that the protonation of several histidine residues in the T-domain of diphtheria toxin serve as the trigger for T-domain interaction with bilayers, serving as an initiator for membrane insertion and translocation of the catalytic domain(67).
In antibacterial peptides, the closest sequence to C18G-His reported in the literature is the LAH4 peptide, characterized by Bechinger and coworkers. The LAH4 peptide exhibits similar pH-dependent antibacterial action to C18G-His, however retains more antibacterial activity at neutral pH (68). This difference can likely be attributed to the inclusion of 4 Lys residues in LAH4, yielding a more strongly cationic sequence at neutral pH which should enhance binding to bacterial membranes. Similarly, LAH4 disrupts lipid bilayers and exhibits higher degrees of aggregation at neutral pH compared to acidic pH (69,70). Notably, the LAH4 peptide undergoes a pH-dependent topographical transition from a shallow, surface topography to a TM orientation(71,72). A topographical shift was observed in PC:PG vesicles for C18G-His between low pH and neutral pH, which could be a similar phenomenon. The quenching data cannot unambiguously distinguish between a surface orientation with the Trp buried in the bilayer and a TM orientation in which the Trp is in the bilayer but near the headgroup region (61). Due to the position of the Trp in the C18G-His sequence, a surface conformation with a buried Trp and TM conformation with a slightly more aqueous exposed Trp is possible.
Finally, the membrane permeabilizing activity of this peptide demonstrates clear differences between model membrane systems and in-tact bacteria. The results of the vesicle leakage assays demonstrate both a clear enhancement of bilayer disruption at low pH for those vesicles containing PE vs. PC, but also a dose-dependent activity for both PE:PG and PC:PG vesicles. These results are consistent with findings by Leber et. al. who demonstrated a clear lipid dependence on permeabilization (57) as well as numerous reports of dose-dependent vesicle permeabilizing activity of AMPs (15,73). The differences in activity with PE containing vesicles may again be linked to binding differences (56-58) or due to the inherent differences in curvature strain associated with bilayers composed of PC vs. PE lipids (56,57). Nonetheless, the activity of C18G-His in vesicles is not a perfect mimic of the activity shown with in-tact bacteria which exhibited extensive peptide-induced permeabilization of the E. coli outer and inner membranes at low pH, and reduced permeabilization at neutral pH. This difference may be attributed to the inherently more complex bacterial membrane composition, size and curvature differences of the LUVs used in this study (spherical with d = 0.2 μm) vs. the average E. coli cell size (rod-like 0.5 × 2 μm), or increased susceptibility due to the bacterial membrane or LPS remodeling as part of the pH stress response (74-77). Alternatively, the different orientations of the peptide at neutral pH may enhance the ability to permeabilize PC or PC:PG bilayers compared to the binding/orientation at low pH. It is worth noting that the lack of permeabilization of ovine RBCs (Supplemental Figure 8) which are enriched in PE lipids indicates that PE alone is not sufficient to allow C18G-His to permeabilize bilayers. There may be combinatorial effects that include peptide orientation (driven by binding and electrostatics) and lipid curvature preferences that play into the final, membrane active conformation.
Taken together, the C18G-His peptide clearly demonstrates pH-dependent antibacterial activity which is likely related to the binding and orientation of the peptide when bound to bilayers under different pH conditions. Lipid dependent effects such as anionic charge and curvature propensity appear to modulate this activity. Moving forward, His-containing AMPs may be of significant interest in targeting acidic tissues or acidic infection sites. The low membrane activity at neutral pH will minimize deleterious interactions with host cells and commensal bacterial flora. These His-containing peptides can be models for the design and development of potential peptidomimetic therapeutics.
Supplementary Material
HIGHLIGHTS.
Histidine containing antimicrobial peptide C18G-His loses activity at neutral pH
C18G-His exhibits pH-dependent changes in aggregation state in solution
Membrane binding and conformation is affected both by pH and lipid composition
Peptide induced bacterial membrane permeabilization is enhanced at low pH
5. Acknowledgements
The authors would like to thank Alexandria Kaminski for helpful discussions and training. Funding for this work was provided in part by NIH R15 GM094330 to G.A.C. and L.E.S-O. was supported by NIH R25GM119973. M.A.H. and T.J.P. were supported in part by the honors research program at Rowan College of Gloucester County.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6. References
- 1.Organization, W. H. (2018) Monitoring Global Progress On Addressing Antimicrobial Resistance (AMR). [Google Scholar]
- 2.Zasloff M (1987) Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proceedings of the National Academy of Sciences of the United States of America 84, 5449–5453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang SS, Prodhan ZH, Biswas SK, Le CF, and Sekaran SD (2018) Antimicrobial peptides from different plant sources: Isolation, characterisation, and purification. Phytochemistry 154, 94–105 [DOI] [PubMed] [Google Scholar]
- 4.Yeaman MR, and Yount NY (2003) Mechanisms of antimicrobial peptide action and resistance. Pharmacological reviews 55, 27–55 [DOI] [PubMed] [Google Scholar]
- 5.Avci FG, Akbulut BS, and Ozkirimli E (2018) Membrane Active Peptides and Their Biophysical Characterization. Biomolecules 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marquette A, and Bechinger B (2018) Biophysical Investigations Elucidating the Mechanisms of Action of Antimicrobial Peptides and Their Synergism. Biomolecules 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bechinger B, and Gorr SU (2017) Antimicrobial Peptides: Mechanisms of Action and Resistance. Journal of dental research 96, 254–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuroda K, and Caputo GA (2013) Antimicrobial polymers as synthetic mimics of host-defense peptides. Wiley interdisciplinary reviews. Nanomedicine and nanobiotechnology 5, 49–66 [DOI] [PubMed] [Google Scholar]
- 9.Nuti R, Goud NS, Saraswati AP, Alvala R, and Alvala M (2017) Antimicrobial Peptides: A Promising Therapeutic Strategy in Tackling Antimicrobial Resistance. Current medicinal chemistry 24, 4303–4314 [DOI] [PubMed] [Google Scholar]
- 10.Primon-Barros M, and Jose Macedo A (2017) Animal Venom Peptides: Potential for New Antimicrobial Agents. Current topics in medicinal chemistry 17, 1119–1156 [DOI] [PubMed] [Google Scholar]
- 11.Mishra B, Reiling S, Zarena D, and Wang G (2017) Host defense antimicrobial peptides as antibiotics: design and application strategies. Current opinion in chemical biology 38, 87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Torres MDT, Sothiselvam S, Lu TK, and de la Fuente-Nunez C (2019) Peptide Design Principles for Antimicrobial Applications. Journal of molecular biology [DOI] [PubMed] [Google Scholar]
- 13.Darveau RP, Blake J, Seachord CL, Cosand WL, Cunningham MD, Cassiano-Clough L, and Maloney G (1992) Peptides related to the carboxyl terminus of human platelet factor IV with antibacterial activity. The Journal of clinical investigation 90, 447–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kohn EM, Shirley DJ, Arotsky L, Picciano AM, Ridgway Z, Urban MW, Carone BR, and Caputo GA (2018) Role of Cationic Side Chains in the Antimicrobial Activity of C18G. Molecules 23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saint Jean KD, Henderson KD, Chrom CL, Abiuso LE, Renn LM, and Caputo GA (2018) Effects of Hydrophobic Amino Acid Substitutions on Antimicrobial Peptide Behavior. Probiotics and antimicrobial proteins 10, 408–419 [DOI] [PubMed] [Google Scholar]
- 16.Puri S, and Edgerton M (2014) How does it kill?: understanding the candidacidal mechanism of salivary histatin 5. Eukaryotic cell 13, 958–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Den Hertog AL, Wong Fong Sang HW, Kraayenhof R, Bolscher JG, Van't Hof W, Veerman EC, and Nieuw Amerongen AV (2004) Interactions of histatin 5 and histatin 5-derived peptides with liposome membranes: surface effects, translocation and permeabilization. The Biochemical journal 379, 665–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fitzgerald DH, Coleman DC, and O'Connell BC (2003) Binding, internalisation and degradation of histatin 3 in histatin-resistant derivatives of Candida albicans. FEMS microbiology letters 220, 247–253 [DOI] [PubMed] [Google Scholar]
- 19.Gorr SU (2009) Antimicrobial peptides of the oral cavity. Periodontology 2000 51, 152–180 [DOI] [PubMed] [Google Scholar]
- 20.Khurshid Z, Naseem M, Sheikh Z, Najeeb S, Shahab S, and Zafar MS (2016) Oral antimicrobial peptides: Types and role in the oral cavity. Saudi pharmaceutical journal: SPJ : the official publication of the Saudi Pharmaceutical Society 24, 515–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melino S, Rufini S, Sette M, Morero R, Grottesi A, Paci M, and Petruzzelli R (1999) Zn(2+) ions selectively induce antimicrobial salivary peptide histatin-5 to fuse negatively charged vesicles. Identification and characterization of a zinc-binding motif present in the functional domain. Biochemistry 38, 9626–9633 [DOI] [PubMed] [Google Scholar]
- 22.Grogan J, McKnight CJ, Troxler RF, and Oppenheim FG (2001) Zinc and copper bind to unique sites of histatin 5. FEBS letters 491, 76–80 [DOI] [PubMed] [Google Scholar]
- 23.Wang G, Li X, and Wang Z (2016) APD3: the antimicrobial peptide database as a tool for research and education. Nucleic acids research 44, D1087–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDonald M, Mannion M, Pike D, Lewis K, Flynn A, Brannan AM, Browne MJ, Jackman D, Madera L, Power Coombs MR, Hoskin DW, Rise ML, and Booth V (2015) Structure-function relationships in histidine-rich antimicrobial peptides from Atlantic cod. Biochim Biophys Acta 1848, 1451–1461 [DOI] [PubMed] [Google Scholar]
- 25.Kharidia R, Tu Z, Chen L, and Liang JF (2012) Activity and selectivity of histidine-containing lytic peptides to antibiotic-resistant bacteria. Arch Microbiol 194, 769–778 [DOI] [PubMed] [Google Scholar]
- 26.Shirley DJ, Chrom CL, Richards EA, Carone BR, and Caputo GA (2018) Antimicrobial activity of a porphyrin binding peptide. Peptide science 110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burman LG, Nordstrom K, and Boman HG (1968) Resistance of Escherichia coli to penicillins. V. Physiological comparison of two isogenic strains, one with chromosomally and one with episomally mediated ampicillin resistance. Journal of bacteriology 96, 438–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi H, Palermo EF, Yasuhara K, Caputo GA, and Kuroda K (2013) Molecular design, structures, and activity of antimicrobial peptide-mimetic polymers. Macromolecular bioscience 13, 1285–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiegand I, Hilpert K, and Hancock RE (2008) Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nature protocols 3, 163–175 [DOI] [PubMed] [Google Scholar]
- 30.Caputo GA, and London E (2003) Using a novel dual fluorescence quenching assay for measurement of tryptophan depth within lipid bilayers to determine hydrophobic alpha-helix locations within membranes. Biochemistry 42, 3265–3274 [DOI] [PubMed] [Google Scholar]
- 31.Caputo GA, and London E (2013) Analyzing transmembrane protein and hydrophobic helix topography by dual fluorescence quenching. Methods in molecular biology 974, 279–295 [DOI] [PubMed] [Google Scholar]
- 32.Caputo GA, and London E (2003) Cumulative effects of amino acid substitutions and hydrophobic mismatch upon the transmembrane stability and conformation of hydrophobic alpha-helices. Biochemistry 42, 3275–3285 [DOI] [PubMed] [Google Scholar]
- 33.Hanna SL, Huang JL, Swinton AJ, Caputo GA, and Vaden TD (2017) Synergistic effects of polymyxin and ionic liquids on lipid vesicle membrane stability and aggregation. Biophys Chem 227, 1–7 [DOI] [PubMed] [Google Scholar]
- 34.Chrom CL, Renn LM, and Caputo GA (2019) Characterization and Antimicrobial Activity of Amphiphilic Peptide AP3 and Derivative Sequences. Antibiotics (Basel) 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Z, Ergene C, Lee JY, Shirley DJ, Carone BR, Caputo GA, and Palermo EF (2018) Sequence and Dispersity are Determinants of Photodynamic Antibacterial Activity Exerted by Peptidomimetic Oligo(thiophene)s. ACS applied materials & interfaces [DOI] [PubMed] [Google Scholar]
- 36.Moskowitz SM, Ernst RK, and Miller SI (2004) PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. Journal of bacteriology 186, 575–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guina T, Yi EC, Wang H, Hackett M, and Miller SI (2000) A PhoP-regulated outer membrane protease of Salmonella enterica serovar typhimurium promotes resistance to alpha-helical antimicrobial peptides. Journal of bacteriology 182, 4077–4086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mol ARC, M.S.; Fontes W. (2018) NetWheels: A web application to create high quality peptide helical wheel and net projections. BioRxiv [Google Scholar]
- 39.Raghuraman H, and Chattopadhyay A (2006) Effect of ionic strength on folding and aggregation of the hemolytic peptide melittin in solution. Biopolymers 83, 111–121 [DOI] [PubMed] [Google Scholar]
- 40.Marsh D (2013) Handbook of lipid bilayers, 2nd ed., CRC Press, Taylor & Francis Group, Boca Raton, FL. [Google Scholar]
- 41.Karabadzhak AG, Weerakkody D, Wijesinghe D, Thakur MS, Engelman DM, Andreev OA, Markin VS, and Reshetnyak YK (2012) Modulation of the pHLIP transmembrane helix insertion pathway. Biophysical journal 102, 1846–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weerakkody D, Moshnikova A, Thakur MS, Moshnikova V, Daniels J, Engelman DM, Andreev OA, and Reshetnyak YK (2013) Family of pH (low) insertion peptides for tumor targeting. Proceedings of the National Academy of Sciences of the United States of America 110, 5834–5839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wyatt LC, Moshnikova A, Crawford T, Engelman DM, Andreev OA, and Reshetnyak YK (2018) Peptides of pHLIP family for targeted intracellular and extracellular delivery of cargo molecules to tumors. Proceedings of the National Academy of Sciences of the United States of America 115, E2811–E2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guha S, Rawat SS, Chattopadhyay A, and Bhattacharyya B (1996) Tubulin conformation and dynamics: a red edge excitation shift study. Biochemistry 35, 13426–13433 [DOI] [PubMed] [Google Scholar]
- 45.Schuh MD, and Baldwin MC (2006) Alpha-helix formation in melittin and beta-lactoglobulin A induced by fluorinated dialcohols. The journal of physical chemistry. B 110, 10903–10909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rawat SS, Kelkar DA, and Chattopadhyay A (2004) Monitoring gramicidin conformations in membranes: a fluorescence approach. Biophysical journal 87, 831–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gossler-Schofberger R, Hesser G, Muik M, Wechselberger C, and Jilek A (2009) An orphan dermaseptin from frog skin reversibly assembles to amyloid-like aggregates in a pH-dependent fashion. The FEBS journal 276, 5849–5859 [DOI] [PubMed] [Google Scholar]
- 48.Fendos J, Barrera FN, and Engelman DM (2013) Aspartate embedding depth affects pHLIP's insertion pKa. Biochemistry 52, 4595–4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rao BD, Chakraborty H, Keller S, and Chattopadhyay A (2018) Aggregation Behavior of pHLIP in Aqueous Solution at Low Concentrations: A Fluorescence Study. Journal of fluorescence 28, 967–973 [DOI] [PubMed] [Google Scholar]
- 50.Subbarao NK, Parente RA, Szoka FC Jr., Nadasdi L, and Pongracz K (1987) pH-dependent bilayer destabilization by an amphipathic peptide. Biochemistry 26, 2964–2972 [DOI] [PubMed] [Google Scholar]
- 51.Brown KL, and Hancock RE (2006) Cationic host defense (antimicrobial) peptides. Current opinion in immunology 18, 24–30 [DOI] [PubMed] [Google Scholar]
- 52.Snider C, Jayasinghe. S, Hristova K, and White SH (2009) MPEx: a tool for exploring membrane proteins. Protein science: a publication of the Protein Society 18, 2624–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.von Deuster CI, and Knecht V (2012) Antimicrobial selectivity based on zwitterionic lipids and underlying balance of interactions. Biochim Biophys Acta 1818, 2192–2201 [DOI] [PubMed] [Google Scholar]
- 54.Bennett WF, Hong CK, Wang Y, and Tieleman DP (2016) Antimicrobial Peptide Simulations and the Influence of Force Field on the Free Energy for Pore Formation in Lipid Bilayers. J Chem Theory Comput 12, 4524–4533 [DOI] [PubMed] [Google Scholar]
- 55.Jafari M, Mehrnejad F, and Doustdar F (2017) Insight into the interactions, residue snorkeling, and membrane disordering potency of a single antimicrobial peptide into different lipid bilayers. PLoS One 12, e0187216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koller D, and Lohner K (2014) The role of spontaneous lipid curvature in the interaction of interfacially active peptides with membranes. Biochim Biophys Acta 1838, 2250–2259 [DOI] [PubMed] [Google Scholar]
- 57.Leber R, Pachler M, Kabelka I, Svoboda I, Enkoller D, Vacha R, Lohner K, and Pabst G (2018) Synergism of Antimicrobial Frog Peptides Couples to Membrane Intrinsic Curvature Strain. Biophysical journal 114, 1945–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jin Y, Hammer J, Pate M, Zhang Y, Zhu F, Zmuda E, and Blazyk J (2005) Antimicrobial activities and structures of two linear cationic peptide families with various amphipathic beta-sheet and alpha-helical potentials. Antimicrobial agents and chemotherapy 49, 4957–4964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ishitsuka Y, Pham DS, Waring AJ, Lehrer RI, and Lee KY (2006) Insertion selectivity of antimicrobial peptide protegrin-1 into lipid monolayers: effect of head group electrostatics and tail group packing. Biochim Biophys Acta 1758, 1450–1460 [DOI] [PubMed] [Google Scholar]
- 60.Bozelli JC Jr., Sasahara ET, Pinto MR, Nakaie CR, and Schreier S (2012) Effect of head group and curvature on binding of the antimicrobial peptide tritrpticin to lipid membranes. Chem Phys Lipids 165, 365–373 [DOI] [PubMed] [Google Scholar]
- 61.Caputo GA, and London E (2004) Position and ionization state of Asp in the core of membrane-inserted alpha helices control both the equilibrium between transmembrane and nontransmembrane helix topography and transmembrane helix positioning. Biochemistry 43, 8794–8806 [DOI] [PubMed] [Google Scholar]
- 62.Lew S, Caputo GA, and London E (2003) The effect of interactions involving ionizable residues flanking membrane-inserted hydrophobic helices upon helix-helix interaction. Biochemistry 42, 10833–10842 [DOI] [PubMed] [Google Scholar]
- 63.Shahidullah K, Krishnakumar SS, and London E (2010) The effect of hydrophilic substitutions and anionic lipids upon the transverse positioning of the transmembrane helix of the ErbB2 (neu) protein incorporated into model membrane vesicles. Journal of molecular biology 396, 209–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gupta C, and Mertz B (2017) Protonation Enhances the Inherent Helix-Forming Propensity of pHLIP. ACS omega 2, 8536–8542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim SY, Pittman AE, Zapata-Mercado E, King GM, Wimley WC, and Hristova K (2019) Mechanism of Action of Peptides That Cause the pH-Triggered Macromolecular Poration of Lipid Bilayers. J Am Chem Soc [DOI] [PubMed] [Google Scholar]
- 66.Martfeld AN, Greathouse DV, and Koeppe RE 2nd. (2016) Ionization Properties of Histidine Residues in the Lipid Bilayer Membrane Environment. The Journal of biological chemistry 291, 19146–19156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Perier A, Chassaing A, Raffestin S, Pichard S, Masella M, Menez A, Forge V, Chenal A, and Gillet D (2007) Concerted protonation of key histidines triggers membrane interaction of the diphtheria toxin T domain. The Journal of biological chemistry 282, 24239–24245 [DOI] [PubMed] [Google Scholar]
- 68.Mason AJ, Gasnier C, Kichler A, Prevost G, Aunis D, Metz-Boutigue MH, and Bechinger B (2006) Enhanced membrane disruption and antibiotic action against pathogenic bacteria by designed histidine-rich peptides at acidic pH. Antimicrobial agents and chemotherapy 50, 3305–3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marquette A, Mason AJ, and Bechinger B (2008) Aggregation and membrane permeabilizing properties of designed histidine-containing cationic linear peptide antibiotics. Journal of peptide science : an official publication of the European Peptide Society 14, 488–495 [DOI] [PubMed] [Google Scholar]
- 70.Vogt TC, and Bechinger B (1999) The interactions of histidine-containing amphipathic helical peptide antibiotics with lipid bilayers. The effects of charges and pH. The Journal of biological chemistry 274, 29115–29121 [DOI] [PubMed] [Google Scholar]
- 71.Wolf J, Aisenbrey C, Harmouche N, Raya J, Bertani P, Voievoda N, Suss R, and Bechinger B (2017) pH-Dependent Membrane Interactions of the Histidine-Rich Cell-Penetrating Peptide LAH4-L1. Biophysical journal 113, 1290–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Perrone B, Miles AJ, Salnikov ES, Wallace BA, and Bechinger B (2014) Lipid interactions of LAH4, a peptide with antimicrobial and nucleic acid transfection activities. European biophysics journal: EBJ 43, 499–507 [DOI] [PubMed] [Google Scholar]
- 73.Chen CH, Starr CG, Troendle E, Wiedman G, Wimley WC, Ulmschneider JP, and Ulmschneider MB (2019) Simulation-Guided Rational de Novo Design of a Small Pore-Forming Antimicrobial Peptide. J Am Chem Soc 141, 4839–4848 [DOI] [PubMed] [Google Scholar]
- 74.Vivijs B, Aertsen A, and Michiels CW (2016) Identification of Genes Required for Growth of Escherichia coli MG1655 at Moderately Low pH. Front Microbiol 7, 1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Krulwich TA, Sachs G, and Padan E (2011) Molecular aspects of bacterial pH sensing and homeostasis. Nat Rev Microbiol 9, 330–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rowlett VW, Mallampalli V, Karlstaedt A, Dowhan W, Taegtmeyer H, Margolin W, and Vitrac H (2017) Impact of Membrane Phospholipid Alterations in Escherichia coli on Cellular Function and Bacterial Stress Adaptation. Journal of bacteriology 199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Eguchi Y, and Utsumi R (2014) Alkali metals in addition to acidic pH activate the EvgS histidine kinase sensor in Escherichia coli. Journal of bacteriology 196, 3140–3149 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.