Abstract
Nonmelanoma skin cancers (NMSCs) are the most common type of skin cancers. Major risk factors for NMSCs include exposure to ultraviolet (UV) irradiation. Ursolic acid (UA) is a natural triterpenoid enriched in blueberries and herbal medicinal products, and possess anticancer activities. This study focuses on the impact of UA on epigenomic, genomic mechanisms and prevention of UVB-mediated NMSC. CpG methylome and RNA transcriptome alterations of early, promotion and late stages of UA treated on UVB-induced NMSC in SKH-1 hairless mice were conducted using CpG methyl-seq and RNA-seq. Samples were collected at weeks 2, 15 and 25, and integrated bioinformatic analyses were performed to identify key pathways and genes modified by UA against UVB-induced NMSC. Morphologically, UA significantly reduced NMSC tumor volume and tumor number. DNA methylome showed inflammatory pathways IL-8, NF-κB and Nrf2 pathways were highly involved. Anti-oxidative stress master regulator Nrf2, cyclin D1, DNA damage, and anti-inflammatory pathways were induced by UA. Nrf2, cyclin D1, TNFrsf1b and Mybl1 at early (2 weeks) and late (25 weeks) stages were identified and validated by quantitative PCR (qPCR). In summary, integration of CpG methylome and RNA transcriptome studies show UA alters anti-oxidative, anti-inflammatory and anti-cancer pathways in UVB-induced NMSC carcinogenesis. Particularly, UA appears to drive Nrf2 and its upstream/downstream genes, anti-inflammatory (at early stages) and cell cycle regulatory (both early and late stages) genes, of which might contribute to the overall chemopreventive effects of UVB-induced MNSC. This study may provide potential biomarkers/targets for chemoprevention of early-stage of UVB-induced NMSC in human.
Keywords: Nonmelanoma skin carcinoma, ursolic acid, epigenetic modification, skin cancer prevention, next-generation sequencing
1 ǀ. INTRODUCTION
Approximately 1 million new cases of skin cancer are diagnosed annually in the United States, including basal cell carcinomas (BCCs), squamous cell carcinomas (SCCs), and melanomas. BCCs and SCCs are nonmelanoma skin cancers (NMSCs), which comprised 96% of all skin cancers.1 Exposure to ultraviolet B (UVB) is a major causative factor for NMSCs, and inflammation is considered to play the most crucial role during carcinogenesis.2 Short-term UVB exposure causes inflammatory responses with increased expression of cyclooxygenase-2 (COX-2) and production of carcinogenic prostaglandin (PG) metabolites in the early stage of UV-irradiation damage.3 Long-term exposure to UV irradiation induces inflammatory oxidative stress, causes DNA damage and mutation, and disrupts cell cycle and proliferation in nearly all stages of NMSC carcinogenesis.4 Thus, many studies have focused on the roles of both acute and chronic UV-irradiation-induced inflammation in the alteration of skin oncogenes and morphology.5–7 Recently, p53, PETN and other carcinogenesis genes have been reported to respond to UV irradiation in both in vitro and in vivo model.8–10 To comprehensively understand how UV irradiation influences the epidermis and identifies key regulatory genes for diagnosis and therapy development, it is necessary to integratively analyze multi-omic datasets, including the transcriptomes and DNA methylomes of human skin cancer samples treated with either commercial drugs or bioactive ingredients. Epigenetic deregulation has become an emerging hallmark for cancers and provides important insights into novel therapeutic targets and drug development.11–13 The U.S. Food and Drug Administration has approved several histone deacetylase (HDAC) and DNA methyltransferase (DNMT) inhibitors as chemotherapeutics under limited usage.14,15 For example, RAS oncogene inhibitors are currently under clinical evaluation for lung, colon and pancreatic cancers.16 Aberrant epigenetic alterations are observed in the development and progression of skin cancers.17–19 We recently established extensive gene methylation profiles in skin carcinogenesis by conducting global genome-wide epigenomic analysis of UVB-irradiated SKH-1 hairless mice and DMBA/TPA-treated CD-1 mice.20 The inflammatory genes IL-6 and SOCS1 (suppressor of cytokine signaling 1) were found to be significantly hypermethylated in the UVB-irradiation tumor samples. Epigenetic alterations of the Nrf2 and p21 genes are also observed in skin cancer cells.21,22 These findings suggest a promising and effective method to develop skin cancer therapy by modifying the DNA methylation status of specific biomarker genes and pathways.
Dietary compounds isolated from fruits and vegetables show great potential for skin cancer prevention due to relative abundance and lower toxicity. Forte et al. reported that the cruciferous and leafy vegetable-enriched Mediterranean diet protects against cutaneous melanoma.23 In addition, a high intake of cruciferous vegetables and triterpenoids enriched beans showed a strong protective effect in non-melanocytic skin cancer in a case-control study conducted in Melbourne, Australia.24 Topical application of epigallocatechin gallate (EGCG) and sulforaphane extracted from tea and broccoli sprout exhibits a strong protective effect against skin carcinogenesis induced by UVB-irradiation.25,26 Ursolic acid (UA) is a natural pentacyclic triterpenoids enriched in blueberries and cranberries that has shown strong cancer prevention activity.27 Previous studies have demonstrated that UA and its derivatives have strong inhibitory effect on UVB-induced oxidative stress damage and skin carcinogenesis through p53, NF-κB pathway, mitochondrial membrane potential and other inflammatory pathways and cell cycle checking system28–32, as well as via epigenetic modification.33 However, a more comprehensive method of global assessment including integration of epigenome/DNA CpG methylome and transcriptome study of the protective effect of UA against UVB-induced carcinogenesis would be needed.
In this study, for the first time, we explored the protective effects of UA in SKH-1 hairless mice during UVB-induced skin carcinogenesis. Animals were exposed to UVB irradiation (60 mJ/cm2) for 25 weeks. Epidermal and tumor samples were collected at early (2 wk), transition (15 wk) and late (25 wk) stages, and RNA and DNA libraries were prepared according the protocol provided by supplier for next-generation sequencing (NGS). RNA-seq and methyl-seq data were analyzed by multiple Bioconductor R packages including Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), regional methylation patterns, and Ingenuity pathway analysis (IPA) from Qiagen. In summary, our study identified a set of genes significantly modified by UA in both the DNA methylome and transcriptome potentially contributing to the overall chemopreventive effects of NMSCs and revealed unique biomarkers that will benefit future clinical trials of skin cancer prevention and treatment.
2 ǀ. MATERIALS AND METHODS
2.1 ǀ. Materials
UA was purchased from Sigma-Aldrich (Cat. No. U6753, >90% purity; St. Louis, MO, USA). Acetone (HPLC grade) and 10% phosphate-buffered formalin were obtained from Fisher Scientific (Hampton, NH, USA). UV lamps that emit UV0042 (280 – 320 nm; 75–80% of total energy) and UVA (320–375 nm; 20–25% of total energy) were used as described in previous studies.34–37 The dose of UVB was quantified using a UVB Spectra 305 dosimeter (Daavlin Co., Bryan, OH, USA). The radiation was calibrated with an IL-1700 research radiometer/photometer from International Light Inc. (Newburyport, MA, USA).
2.2 ǀ. Animal care
Female SKH-1 hairless mice were purchased from Charles River Laboratories (Wilmington, MA, USA) at 5 weeks of age, as described in previous studies.37,38 These mice were maintained at a controlled temperature (20 – 22°C) and humidity (45 – 55%) under 12-hour light and dark cycles at the Rutgers Animal Facility. Food and water were provided ad libitum. After one week of acclimatization, mice at 6 weeks of age were randomly assigned to three groups and tail-tattooed: Non-UVB with acetone (control) group, UVB with acetone (UVB) group and UVB with UA (UA) group (n = 20). All animal experiments were conducted under the animal protocol (04–003) approved by the Institutional Animal Care and Use Committee (IACUC) of Rutgers University.
2.3 ǀ. Experimental design
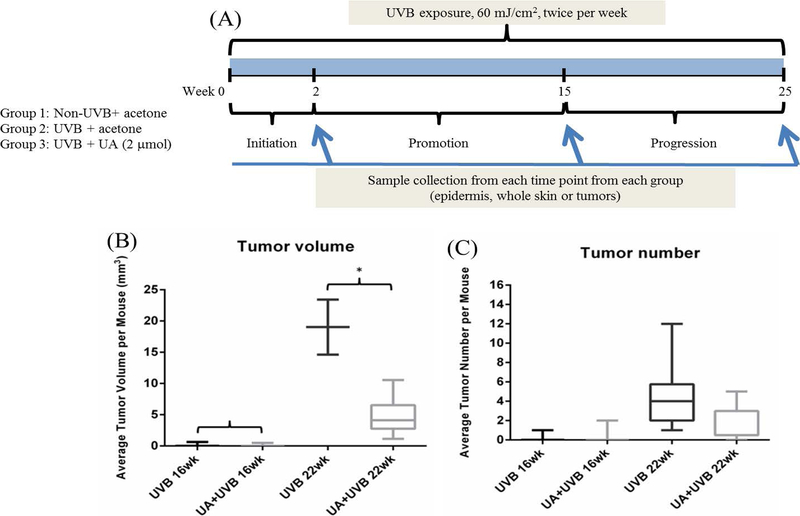
The experimental design is summarized in Figure 1A. In brief, at the age of 8 weeks, all mice began exposure to UVB irradiation at the dose of 60 mJ/cm2, which is considered a medium UVB dosage based on our previous results (data not shown here). After irradiation, mice underwent topical application of: (1) 200 μL of acetone without UVB irradiation as the control group, (2) 200 μL of acetone with UVB irradiation as the UVB group, and (3) 200 μL of 2 μmol UA in acetone with UVB irradiation as the UA group. All UVB-irradiated groups of mice were exposed twice per week and followed by topical applications of the above treatements on the same and the following day for a 25-week period. Body weight, skin condition and tumor growth were monitored daily and recorded biweekly. Tumor volumes were determined and calculated using the formula V = π/6 × Height × Length × Width. Mice were sacrificed by CO2 asphyxiation. The skin and tumor tissues were saved either for histological assays in 10% phosphate-buffered formalin or for molecular assays by being snap frozen and stored at – 80 °C.
Figure 1.
25-week UA treatment in UVB-induced skin carcinogenesis on SKH-1 hair-less mice. A, Experimental design of the animal study. Mice were 8 weeks old when the experiment began. B, Tumor volume per mouse measured at week 16 and 22. C, Tumor number per mouse measured at week 16 and 22. Only the tumor diameter greater than 2 mm is considered and recorded
2.4 ǀ. Sequencing library preparation
UVB induces genomic alteration happens mainly at the skin epidermis layer. Therefore, we selected the epidermal tissues from both UVB and UVB+UA groups to perform epigenome/transcriptome analyses. In addition, since whole skin tumor would contain epidermis, dermis and hypodermis layers, we selected tumor adjacent whole skin tissues as negative control for sequencing library preparation. Epidermal tissues from the UVB and UA groups at week 2 (initiation stage), week 15 (promotion stage) and week 25 (progression stage) and tumor/whole skin (adjacent normal) samples at week 25 were selected for RNA-seq and Methyl-seq library preparation and sequencing. In brief, paired RNA and DNA were extracted using the AllPrep DNA/RNA/Protein Mini Kit (Qiagen, Valencia, CA, USA), following the manufacturer’s protocol provided by the supplier. The RNA-seq library was prepared using an Illumina TruSeq RNA preparation kit (Illumina, San Diego, CA, USA) and sequenced on the Illumina NextSeq 500 platform with 75 bp single-end reads to a minimum sequencing depth of 20 – 25 million reads per sample. The methyl-seq library was prepared using the Agilent SureSelect Methyl-Seq kit (Agilent Technologies, Santa Clara, CA, USA) as described in the manufacturer’s protocol. The captured methyl-seq library was bisulfite converted using the EZ DNA Methylation-Gold kit (Zymo Research, Orange, CA, USA) as described in the manufacturer’s protocol before indexing. The methyl-seq library was sequenced on an Illumina NextSeq 500 platform with 75 bp single-end reads to a minimum sequencing depth of 40–50 million reads per sample.
2.5 ǀ. Bioinformatics analyses for RNA-seq
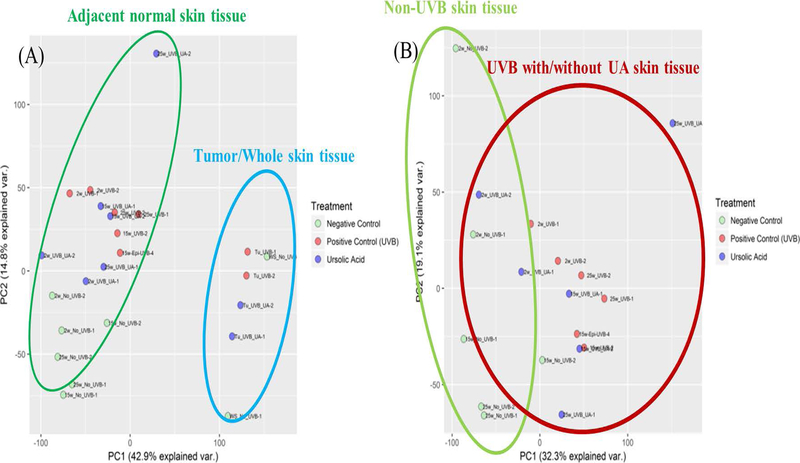
RNA-seq reads were aligned to the reference mouse genome (mm10) provided by the UCSC database and processed by HISAT2 (version 2.1.0)39, SAMtools (Version 1.8)40, and Picard41. Deduplicated reads were then annotated by FeatureCounts with reference genomes from UCSC database42. Statistical analysis were performed by DEseq243. Principle component analysis (PCA) was performed to determine the modes of variance among RNA-seq samples and results are summarized in Figure 2.
Figure 2.
Principle component analysis of RNA-seq of UA and UVB adjacent skin and tumor samples across all time points. A, Adjacent normal skin and tumor/whole skin tissue. B, Adjacent normal skin samples only
2.6 ǀ. Ingenuity pathway analysis (IPA)
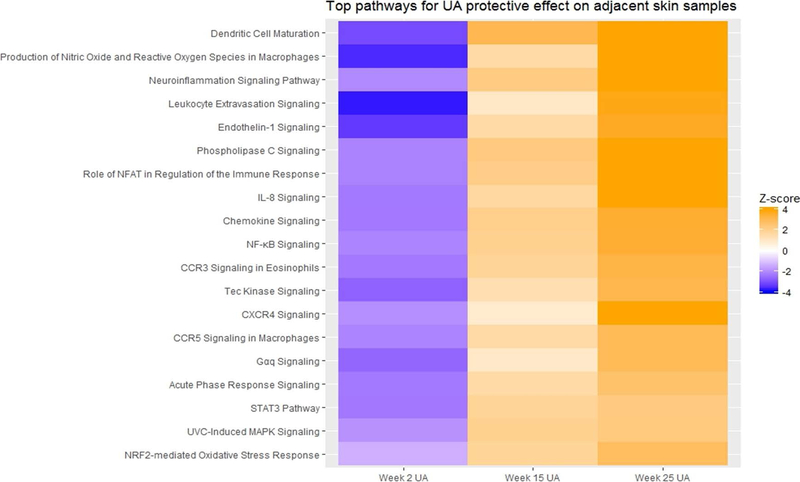
Genes were filtered as the following parameters: absolute value of log2 fold change greater than 2 with a false detection rate (FDR) less than 0.05, and analyzed through the use of IPA core analysis.44 Expression analysis was set up as the core analysis type, and Expr Fold Change was set up as the measurement type. The top canonical pathways associated with UA protective effects against UV-induced carcinogenesis were identified using epidermal samples. The results of all time point comparisons are summarized with Z-scores (activation-scores) on a scale from −4.0 to 4.0 (Figure 3).
Figure 3.
Top pathways induced by UA on adjacent skin samples from IPA. Pathway activation score is used as primary parameters. Multiple pathway comparison is used for RNA-seq results from week 2, 15 and 25 UA group
2.7 ǀ. GO and KEGG enrichment analyses
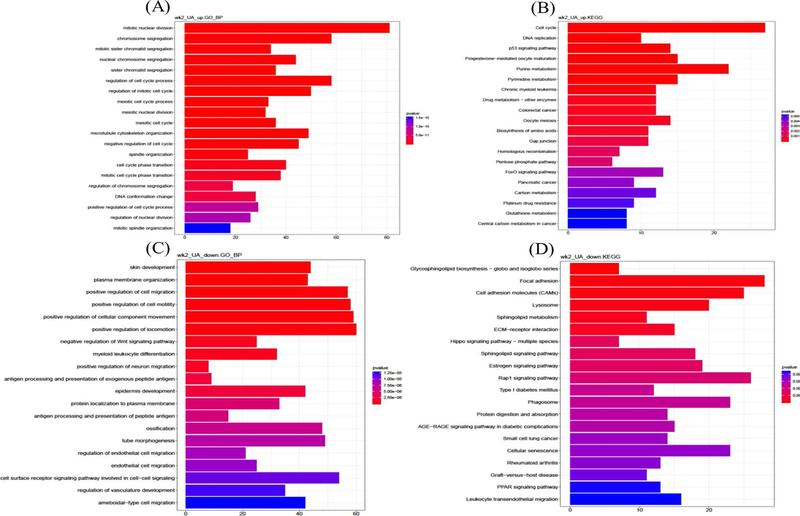
Raw Reads were input into bioconductor R packages clusterProfiler for GO and KEGG gene enrichment analysis and data visualization.45 The differentially expressed genes (DEGs) were filtered from raw reads based on the cutting off log2 fold change greater than 1 or less than −1 with q-value less than 0.05. The most significant GO biological process terms46,47 and KEGG48–50 analysis were performed (Figure 4).
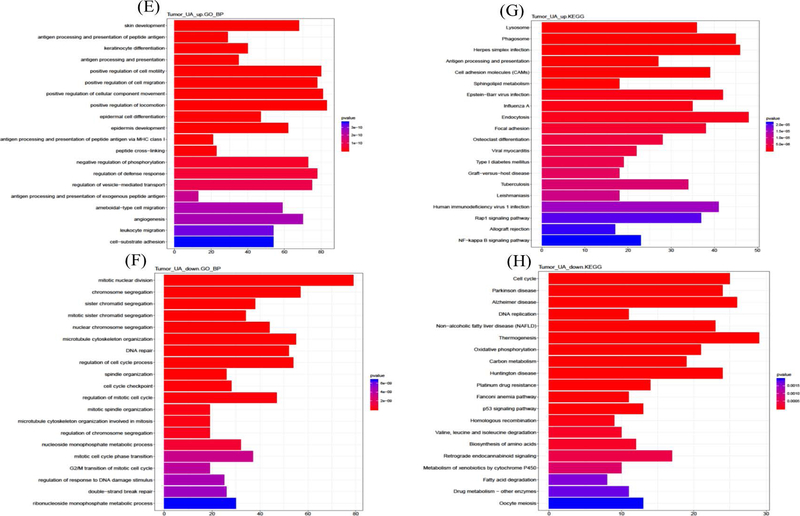
Figure 4.
Up- and Down-regulated gene GO terms and KEGG enriched pathway analysis on week 2 adjacent normal skin samples and week 25 tumor tissues. A, Week 2 epidermis biological process GO term by up-regulated gene of UA treatment. B, Week 2 epidermis KEGG pathways by up-regulated genes of UA treatment. C, Week 2 epidermis biological process GO term by down-regulated gene of UA treatment. D, Week 2 epidermis KEGG pathways by down-regulated genes of UA treatment. E, Week 25 tumor biological process GO term by up-regulated gene of UA treatment. F, Week 25 tumor KEGG pathways by up-regulated genes of UA treatment. G, Week 25 tumor biological process GO term by down-regulated gene of UA treatment. H, Week 25 tumor KEGG pathways by down-regulated genes of UA treatment
2.8 ǀ. Bioinformatics analyses for Methyl-seq
Methyl-seq reads were mapped to reference mouse genome (mm10) with the Bismark (version 0.19.1).51 DMRfinder (version 0.1) was used to extract methylation counts and cluster CpG sites into DMRs.52 Genomic annotation was performed with ChIPseeker (version 1.16.0) in R (version 3.5.0).53
2.9 ǀ. Validation by qPCR
The mRNAs significantly expressed by the induction of UA were determined using qPCR analysis. RNA (previously used for RNA-seq library preparation) was reverse transcribed by TaqMan and analyzed by the QuantStudio 5 Real-time PCR system using SYBR Green PCR Master Mix (ThermoFisher, Waltham, MA, USA). The primers for gene validation were selected from PrimerBank54–56 and ordered from Integrated DNA Technologies (IDT, Coralville, IA, USA). The relative fold change was normalized to the internal standard gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH).
2.10 ǀ. Statistical analysis
The data are presented as the mean ± standard deviation (SD), except as otherwise stated. One-way analysis of variance (ANOVA) was performed with Tukey’s multiple comparison test for multiple group comparisons. Student’s t-test was performed for simple comparison. Methylation differences were analyzed by the Mann–Whitney U-test. p-value less than 0.05, and False Discovery Rate (FDR) less than 10% were considered significant, except as otherwise stated.
3 ǀ. RESULTS
3.1 ǀ. Morphological Observations
The experimental design of animal study was shown in Figure 1A. 8-week old female SKH-1 hairless mice were used and body weight, tumor number and volume were measured biweekly for 25 weeks. At week 2, 15 and 25, mice were sacrificed and both epidermis and whole skin/tumor samples were collected. During the experimental period, UA significantly reduced the tumor volume and tumor number. As shown in Figure 1B, C, we observed the minor changes between UA and UVB group on tumor volume and number starting at week 16, with significant reduction starting at week 22.
3.2 ǀ. Observations of RNA-seq data
We performed PCA on epidermis alone, epidermis and tumor/whole skin tissues combined to identify the clustering profiles of the transcriptomic alterations induced by UA. All the epidermal samples from weeks 2, 15 and 25 clustered together but were significantly separated from tumor/whole skin tissues (Figure 2A). However, the epidermal samples without UVB or UA treatment (control groups) clustered together and were slightly separated from the UA and UVB groups, which were not significantly separated (Figure 2B). These results reveal some fundamental facts about carcinogenesis induced by UVB irradiation: during the process, UVB induced larger-scale gene expression alterations, and UA exhibited protective effects via a small proportion of genes that may reverse the damage. Notably, these results indicate that the discovery of driver genes regulated by UA in UVB-induced carcinogenesis may be very feasible.
To further explore the UA induced signal transduction pathways, IPA was performed to obtain signaling pathway information. Log2 fold change was set up as greater than 2 or less than −2 with FDR less than 0.05, and 1585, 1383, 1013 and 615 unique genes were obtained from week 2, 15 and 25 adjacent epidermis and week 25 tumor, respectively. These genes were analyzed by IPA and top canonical pathways and upstream regulators are summarized in Table 1. Then, a three-way combination analysis was performed to identify signaling pathways consistently and significantly activated or inhibited by UA across all time points, and they were visualized as a heatmap with activation Z-scores (Figure 3). Interestingly, UA plays multiple roles in the initiation, promotion and progression phases during UVB-induced carcinogenesis. Nrf2 is a major regulatory gene that responds to oxidative stress and is well known for its double-edged sword effects in the different phases of carcinogenesis.57–60 UA activates the Nrf2-mediated oxidative stress response in the initiation stage (week 2) but inhibits it during the promotion (week 15) and progression (week 25) stages. Nuclear transcription factor-kappa B (NF-κB)61,62 and acute phase responses signaling pathways63,64 are associated with external stimuli, such as UV irradiation, oxidative and DNA damaging agents. UA also showed regulatory effects on these signaling pathways. Interleukin 8 (IL-8) plays key roles in inflammation, tumor proliferation and angiogenesis65,66, activates CXCR1/267 and vascular endothelial growth factor receptor (VEGFR)68. IL-8 also regulates NF-κB signaling pathway in some benign prostatic hyperplasia stromal cells.69 Similar to the Nrf2 and NF-κB signaling pathways, UA activates the IL-8 signaling pathway in the initiation phase but is inhibited during the promotion and progression phases. The IPA analysis also indicated other signaling pathways, but to fully interpret the RNA-seq data of UA’s preventive effects during UVB-induced carcinogenesis, future additional bioinformatics tools related to gene functions will be needed to further explore more potential clinical biomarkers.
Table 1.
Summary of Top 5 canonical pathways and upstream regulators of week 2, 15 and 25 and tumor/whole skin tissues
| Top canonical pathways | p-value | Overlap | Top upstream regulators | p-value | |
|---|---|---|---|---|---|
| Week2 | |||||
| Antioxidant Action of Vitamin C | 8.40E-04 | 19.6% | 20/102 | PLA2G10 | 1.79E-27 |
| ATM signaling | 1.10E-04 | 19.8% | 19/96 | CSF2 | 3.61E-25 |
| Cell cycle: G2/M DNA damage checkpoint regulation | 3.15E-04 | 24.0% | 12/50 | PTGER2 | 1.38E-17 |
| GP6 Signaling Pathway | 3.99E-04 | 16.8% | 22/131 | GSTP1 | 3.28E-17 |
| p53 Signaling | 4.75E-04 | 17.8% | 19/107 | TCF4 | 7.77E-15 |
| Week15 | |||||
| Antigen Presentation Pathway | 5.72E-13 | 53.6% | 15/28 | TRIM24 | 4.37E-22 |
| Cdc42 Signaling | 9.49E-07 | 16.3% | 22/135 | STAT1 | 5.89E-20 |
| OX40 Signaling Pathway | 3.85E-06 | 21.2% | 14/66 | IRF7 | 1.19E-17 |
| Dendritic Cell Maturation | 1.38E-05 | 13.5% | 23/170 | ACKR2 | 2.05E-17 |
| Interferon Signaling | 1.42E-05 | 29.0% | 9/31 | IRF3 | 7.63E-17 |
| Week25 | |||||
| GP6 Signaling Pathway | 7.34E-12 | 26.0% | 34/131 | IFNG | 5.79E-29 |
| Hepatic Fibrosis/Hepatic Stellate Cell Activation | 2.19E-11 | 22.2% | 40/180 | Alpha catenin | 1.43E-21 |
| Dendritic Cell Maturation | 4.26E-08 | 19.4% | 33/170 | IRF3 | 5.02E-20 |
| Antigen Presentation Pathway | 1.08E-07 | 42.9% | 12/28 | STAT1 | 8.17E-20 |
| Complement System | 9.16E-07 | 36.4% | 12/33 | IRF7 | 2.15E-18 |
| Tumor/Whole skin | |||||
| Antigen Presentation Pathway | 1.33E-06 | 28.6% | 8/28 | IKBKG | 8.32E-08 |
| Graft-versus-Host Disease Signaling | 1.60E-05 | 21.1% | 8/38 | CHUK | 9.51E-08 |
| Altered T Cell and B Cell Signaling in Rheumatoid Arthritis | 3.95E-05 | 13.4% | 11/82 | IKBKB | 1.25E-06 |
| Type I Diabetes Mellitus Signaling | 6.68E-05 | 11.8% | 12/102 | TP73 | 3.19E-06 |
| Dendritic Cell Maturation | 7.03E-05 | 9.4% | 16/170 | STAT6 | 5.41E-06 |
GO is widely used to evaluate the biological functions systematically and usually applied on high-throughput sequencing dataset. Gene ontology enrichment analysis helps to identify both the gene and function by annotating sequencing raw reads to reference genome database. Multiple Bioconductor R packages such as clusterProfiler45, Goseq70, GOexpress71, are designed to interpret RNA-seq data and obtain information related to three types of gene ontology: biological process, molecular function and cellular component. KEGG is the database used for understanding and mapping large-scale datasets and exploring significant pathway information. The combined analyses would shed light on determining both the unique target genes and the pathways associated with RNA-seq reads. Hence, to identify representative genes and develop early-stage potential targets, the protective effects of UA were explored with a focus on the initiation phase and tumor/whole skin samples, using both GO and KEGG analysis. 2413 and 2798 unique genes were selected by filtration from the RNA-seq UA group at week 2 and tumor samples at week 25, respectively, based on log2 fold changes and p-values (Figure 4). Then, these genes were divided into UA up- and down-regulated genes, and GO and KEGG analysis were performed separately for each group. The top 20 significant GO terms (biological process) activated (or enhanced) by UA are listed in Figure 4A. Several UA-regulated biological processes that positively control cell division and the cell cycle; mitotic nuclear division, cell cycle phase transition, mitotic cell cycle process, etc. are among the pathways present. Cell division and the cell cycle play crucial roles in maintaining homeostasis in normal tissues but are disrupted in the carcinogenic process and cancer.72–74 Studies have determined that dysfunctions in certain genes, such as BRAC1 and BRAC2, as well as other cell cycle checkpoint genes, such as cyclin-dependent kinase 1, increase the risk of carcinogenesis.75,76 KEGG pathway enrichment analysis revealed that UA positively regulates the cell cycle and the p53 and FoxO signaling pathways (Figure 4B), as reported previously.77–79 These pathways are highly associated with reactive oxygen species (ROS) and chronic inflammation driven carcinogenesis.80–82 The top 20 significant GO terms (biological process) deactivated or inhibited by UA are listed in Figure 4C. Wnt family of proteins are involved in a series of regulatory roles related to cell development, homeostasis and cell apoptosis.83 The Wnt/Ca+ signaling pathway is one of three major Wnt signaling pathways that respond to ROS both in vitro and in vivo.84–86 Other significant biological processes are associated with cell migration and cell motility, as well as skin and epidermis development. KEGG analysis of UA down-regulated genes revealed several signaling pathways including cellular senescence and focal adhesion (Figure 4D). Cellular senescence is defined as the termination step of cell differentiation triggered by multiple factors, including oncogenic activation87, DNA damage88, ROS89 and UVB irradiation90–92. Focal adhesion kinases have been shown serving important roles in cell cycle, and cell motility in cancers.93,94 p5395, PTEN96, and pp125 FAK97 are linked to cellular senescence and focal adhesion signaling pathways. In tumor samples at week 25, UA up-regulated genes show opposing effects, activating or enhancing skin development, epidermal development and differentiation, and cell motility and migration compared to week 2 epidermis samples (Figure 4E). KEGG analysis revealed that UA also activated the focal adhesion, Rap1 signaling pathway and NF-κB signaling pathways, which are related to ROS-induced inflammation and carcinogenesis (Figure 4F).98–100 The KEGG analysis of UA down-regulated genes shows that one possible mechanism underlying the protective effects of UA is DNA repair, including regulation of response to DNA damage stimulus and double-strand break repair signaling pathways.
3.3 ǀ. Observations of methyl-seq data
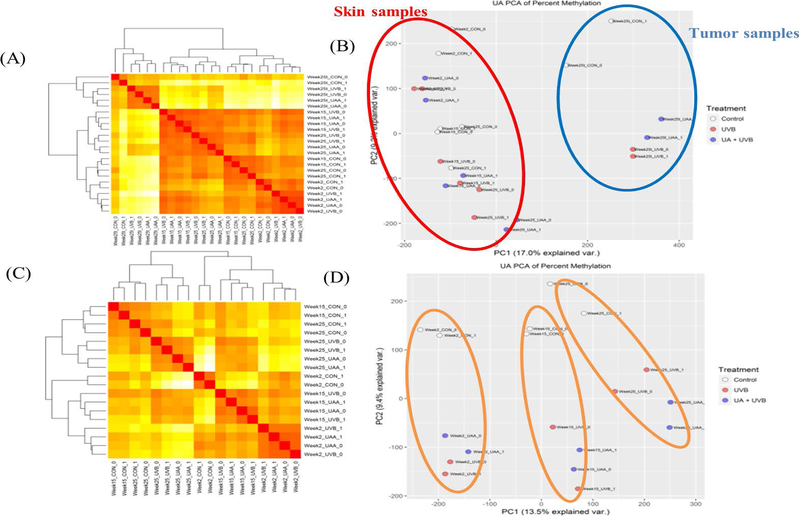
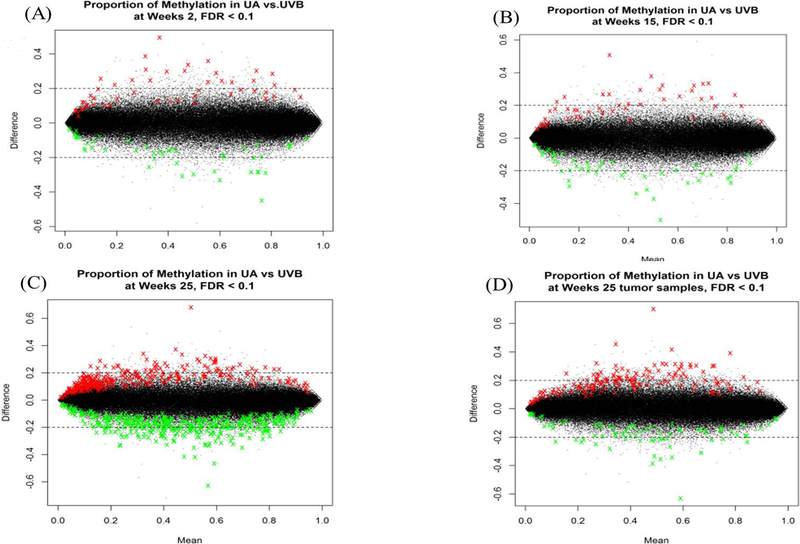
One of the main purposes of interrogating the methyl-seq data is to determine the CpG methylation alteration induced by UA treatment following UVB exposure. The methyl-seq results were expressed as heatmap plots with Euclidean distance clustering and PCA analysis with and without tumor/whole skin tissue (Figure 5). Epidermal tissues at the initiation, promotion and progression phases are clustered together, while all tumor/whole skin tissues are separated from them in both the heatmap and PCA analysis (Figure 5A, B). The methyl-seq data show not only a clear division by UVB irradiation but also a significant separation by duration (Figure 5C, D). The results of RNA-seq and methyl-seq showed a consistent pattern in both analyses. In addition, the MA plot of DNA methylation alteration by UA treatment indicated that, over time, UA induced dramatic global changes in epidermal tissues. A significant methylation difference of 20% or more was observed between the UA group and the UVB group at each time point in the epidermal tissue, with FDR < 0.1 (Figure 6). Each colored dot represents one significant DMR, and green and red dots represent DNA clusters that are significantly hypo or hypermethylated with UA treatment. The amount of significant DMRs were increased with time in adjacent epidermis samples, and also increased with tumor samples (Figure 6A–C). These observations suggest that the protective effects of UA occur gradually by altering the methylation status of genes during UVB-irradiated carcinogenesis. Potent biomarker genes can thus be discovered by integrating RNA-seq and methyl-seq data to reveal the potential mechanism of skin carcinogenesis caused by UVB exposure.
Figure 5.
Global DNA methylation status heatmap with Euclidean distance clustering. A, DNA methylation of adjacent normal skin and tumor heatmap with Euclidean distance clustering. B, Principle component analysis of adjacent normal skin and tumors. C, DNA methylation of adjacent normal skin heatmap with Euclidean distance clustering. D, Principle component analysis of adjacent normal skins
Figure 6.
MA plot of DNA methylation status and alteration by UA treatment cross all time points. A-D, Week 2, 15, 25 epidermis and week 25 tumor tissues, respectively. The thresholds are set up as methylation different over 20% and false positive ratio (FDR) < 0.1. Dot represents raw sequence reads and annotated to specific gene. Green dots represent that at these gene promoter regions, CpG sites are significantly hypomethylated by UA. Red dots represent that at these gene promoter regions, CpG sites are significantly hypermethylated by UA. Black dots represent at these genes promoter region, CpG sites are not significantly hyper or hypomethylated
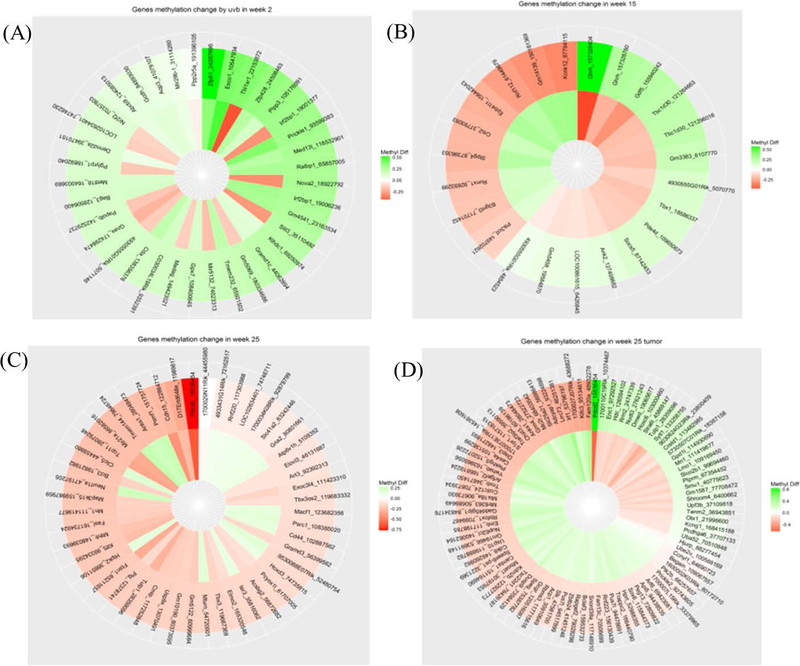
Examining the methyl-seq data of CpG methylation alteration induced by UA treatment, shows the opposite pattern to that caused by UVB-induced carcinogenesis. All the significant CpG methylation changes from weeks 2, 15, and 25 and tumor tissues induced by UVB and reversed by UA are displayed in the circular heatmap plots (Figure 7A–D). The outer layer represents the genes that underwent UVB-induced methylation based on reference genome annotation, and the inner layer represents UA-induced methylation reversal. The full list of genes is provided in Supplemental Table S1.
Figure 7.
DNA methylation alteration induced by UA treatment across all time points and tumor samples. A-D, Week 2, 15, 25 epidermis and week 25 tumor tissues, respectively. All significant CpG methylation changes induced by both UVB alone and UA treatment, and shows reserving pattern have been displayed in the circular heatmap plots. The outer layer represents the UVB induced methylation genes after annotated on genome reference, and the inner layer represents the UA induced methylation reversing
3.4 ǀ. Cross-referencing the RNA-seq and methyl-seq data
Genes from the RNA-seq dataset with transcriptional differences of over 4-fold and p-values <0.05 were selected. Genes with CpG methylation changes of over 30% in promoter and distal intergenic regions with p-value < 0.05 were selected and cross-referenced with genes filtered from RNA-seq. In the week 2 epidermal tissue, a total of 305 genes were found in which CpG sites at the promoter and/or intergenic regions were highly hypermethylated, while transcriptional expression was significantly down-regulated. Fifty-three genes were identified as hypomethylated CpG sites at the promoter and/or intergenic regions, while transcriptional levels were significantly up-regulated. Nrf2 is shown in the hypo-/up-regulated methyl-seq and RNA-seq crosstalk list, with significant transcriptional alteration (log2 fold change = 2.02) and 4 CpG sites significantly hypomethylated (between 30.6% to 50.8% hypomethylated). These results indicate that, during the early phase of UVB-induced carcinogenesis, Nrf2 expression was significantly up-regulated potentially by the protective effects of UA in response to this small UVB dosage acute exposure. In the week 25 tumor/whole skin tissue treated with UA, 88 genes were hypermethylated at the promoter and/or intergenic region, with transcriptional expression significantly down-regulated. A total of 159 genes were found to be highly hypomethylated at the promoter and/or intergenic region, while transcript levels were significantly up-regulated. The gene expression of Nrf2 was significantly inhibited by 2.8-fold. Other genes following by similar patterns were also identified and top 10 genes from the week 2 and week 25 tumor samples are summarized in Table 2.
Table 2.
Methyl-seq and RNA-seq data cross-talking summary of week 2 epidermis and week 25 tumor tissues. Top 10 hyper-down and hypo-up regulated genes are listed
| Week2 epidermis | hypo-up | hyper-down | ||||
|---|---|---|---|---|---|---|
| Gene ID | Hypomethylation (%) | RNA expression (log2fold-change) | Gene ID | Hypermethylation (%) | RNA expression (log2fold-change) | |
| Npbwr1 | 35.3 | 2.52 | Slco5a1 | 35.3 | −9.05 | |
| Plekhb2 | 57.8 | 2.17 | Ogfrl1 | 43.8 | −2.72 | |
| Klf7 | 44.8 | 3.19 | Bend6 | 53.3 | −2.84 | |
| Mgat5 | 57.6 | 2.05 | Mgat4a | 46.4 | −2.06 | |
| Ube2t | 48.5 | 4.33 | Creg2 | 36.3 | −3.56 | |
| Phlda3 | 40.1 | 2.05 | Gm973 | 31.2 | −2.77 | |
| Kcnj9 | 38.8 | 9.41 | Slc4a3 | 32.7 | −4.20 | |
| Fbxo5 | 40.9 | 2.46 | Arl4c | 50.5 | −3.66 | |
| Plagl1 | 67.1 | 2.20 | Mlph | 67.0 | −3.03 | |
| Myb | 34.1 | 2.65 | Twist2 | 54.7 | −5.32 | |
| Gene ID | Hypomethylation (%) | RNA expression (log2fold-change) | Gene ID | Hypermethylation (%) | RNA expression (log2fold-change) | |
| Rgs20 | 52.6 | 2.45 | Gdap1 | 41.6 | −1.88 | |
| Kcnq5 | 54.2 | 2.46 | 1500015O10R ik |
47.0 | −5.49 | |
| Cnnm4 | 50.5 | 2.73 | Mettl21e | 34.5 | −7.72 | |
| Col3a1 | 45.2 | 4.57 | Dnah7b | 52.0 | −2.19 | |
| Ankrd44 | 35.4 | 3.83 | Hspe1 | 63.1 | −3.45 | |
| Cd28 | 58.5 | 4.49 | Bard1 | 32.3 | −3.93 | |
| Ctla4 | 63.6 | 9.35 | Atic | 33.8 | −2.50 | |
| Cxcr2 | 45.1 | 2.65 | Prkag3 | 37.2 | −2.29 | |
| Slc4a3 | 30.2 | 3.37 | Fam124b | 39.6 | −2.75 | |
| Pax3 | 24.2 | 2.37 | Pid1 | 44.6 | −2.12 |
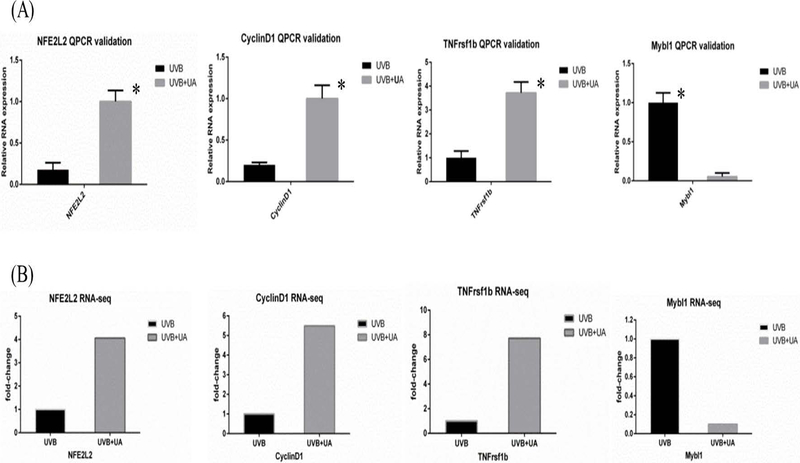
3.5 ǀ. qPCR validation
Nrf2 (or NFE2L2), cyclinD1, TNF-receptor superfamily 1B (tnfrsf1b), and Mybl1 were selected for qPCR validation. The oligo primers are summarized in Supplemental Table S2. The relative mRNA expression levels of the genes are presented in Figure 8A. The RNA-seq data based on FPKM (fragments per kilobase of exon per million fragments mapped) of these genes are presented in Figure 8B. qPCR showed similar pattern consistent with the RNA-seq results, suggesting that the NGS approach used in this study can appropriately represent transcriptional expression.
Figure 8.
UA protective representing gene validated by qPCR and cross referenced with RNA-seq. A, qPCR validation of relative RNA expression results in UVB and UA groups. B, RNA-seq fold-change results in UVB and UA groups
4. ǀ. Discussion
4.1 ǀ. UA protective effect in UVB induced carcinogenesis
Over the last decade, the number of new cases of nonmelanoma skin cancer diagnosed in the United States has increased dramatically potentially due to chronic overexposure to UVB.101 Conventional protective methods, including sunscreen and protective clothing, have been employed inadequately.102 In the present study, we performed topical application of UA to the skin of UVB-irradiated SKH-1 hairless mice, and the epidermis and tumor/whole skin tissues at weeks 2, 15 and 25 were collected for RNA-seq and methyl-seq multi-omic analyses. Computational Bioconductor R packages were utilized for comprehensive analyses of the transcriptome and methylome datasets, including PCA, GO, IPA and KEGG pathway analyses, and methylation patterns. Morphologically, UA reduced both the tumor volume and tumor number, suggesting the potent chemopreventive effect of UA. We next performed the significant grouping of epidermis and tumor/whole skin samples by PCA and clustering analyses. Results show that not only are the tumor/whole skin samples epigenetically distinct from the epidermis, but UVB also induced dramatic changes and caused significant separation between the epidermal tissue groups in both the RNA-seq and methyl-seq data. UA appears to target a relatively small number set of genes to accomplish its protective effects when compared with the number of genes altered by UVB irradiation. For example, the Nrf2 and NQO-1 key antioxidant genes were significantly up-regulated by UA in the early phase of UVB carcinogenesis. Meanwhile, methylation analysis also revealed that the CpG sites of these genes were significantly hypomethylated. Furthermore, other potential driver genes of cell proliferation, differentiation and cell cycle regulatory pathways were also found to be impacted. Previously, we and others have conducted numerous studies on elucidating UA anti-inflammatory mechanisms in vitro32,103–107 and in vivo108–110, mainly through oral administration. Topical application of UA is also applied on mouse ear model and back side of skin for anti-inflammation activity measurement.111–113 However, our current study raises another level with the multi-omic high-throughput approaches on the UVB-irradiated skin carcinogenesis process, and explored the potential epigenetics/epigenomic target genes impacted by chemopreventive agent UA from early-phase to late stage tumors using integrative analyses of transcriptome and CpG methylome.
4.2 ǀ. Nrf2 is a key regulatory gene associated with ROS in UVB-induced carcinogenesis
Previously, we have reported several papers showing the important role of Nrf2-mediated signaling pathways in respond to ROS-induced inflammation in cancers.114–118 Epigenetic study with mouse skin epidermal JB6 P+ cell transformation found epigenetic modification of Nrf2 by cancer chemopreventive/natural phytochemicals including isothiocyanate119, focoxanthin120, taxifolin121 and reserpine122. Cross-examination of RNA-seq and DNA methyl-seq data shows many genes have cause-and-effect relationship between DNA CpG methylation versus RNA expression, including Nrf2 gene. Based on the results, Nrf2 is hypomethylated at 4 CpG sites coupled with concomitant increase in Nrf2 RNA expression during the early phase (2 weeks) of UVB-induced carcinogenesis development. It is a clear evidence to support that UA could modulate antioxidative stress/inflammation pathways such as Nrf2 with UVB-induced CpG modification in the skin at early phase. Further study of Nrf2 related genes such as Keap1, NQO-1 and HO-1 would be necessary to comprehensively understand the functional role of Nrf2 gene in the initial stage of NMSCs development including using Nrf2 knockout mice and Keap1 knockout or overexpression mouse models.
4.3 ǀ. The cell cycle plays crucial roles in UVB-induced carcinogenesis
The cell cycle signaling pathways in general plays critical biological events in maintaining the homeostasis of cell division and cell fate.123,124 Our current integrative epigenome/transcriptome study of UA treated UVB-induced mouse model shows many of these pathways and genes are modified epigenetically with CpG methylation and the corresponding alterations in RNA expression, including cyclinD1, CDK1/2 and CDK6. The functional significance of these pathways and genes would require further in-depth study down the road.
In summary, in this study, we integrated the DNA methylome and RNA transcriptome in UA treated UVB-induced nonmelanoma carcinogenesis using NGS technology. RNA-seq and methyl-seq data were analyzed by multiple computational pipelines and R packages in UVB exposed SKH-1 hairless mice epidermis and tumor tissues. To identify potential epigenome modification, biomarker genes and the associated pathways impacted by UA, bioinformatic tools including PCA, GO and KEGG enrichment pathway analyses, IPA and DNA methylation patterns were applied to comprehensively understand the potential mechanisms associated with UA. Our results show several inflammatory pathways IL-6, NF-κB, Nrf2 and cell cycle regulatory pathways are highly regulated by UA during UVB irradiation. Potential biomarker genes such as Nrf2, cyclin D1 are identified by RNA-seq and methyl-seq integrative analyses and validated. Overall, our study reveals that UA protects UVB-induced skin carcinogenesis potentially via epigenome modifications including DNA CpG methylome coupled with RNA transcriptome alterations. These pathways and genes could potentially be utilized for future clinical studies of UA or other cancer chemopreventive agents in prevention of early phase of UVB-induced NMSC in human.
Supplementary Material
ACHKNOLWEDGEs
We thank all members of Dr. Ah-Ng Kong’s lab for helpful discussions and preparation of the manuscript.
FUNDING
This study was supported in part by R01 CA200129, R01-CA118947, and R01-CA152826 from the National Cancer Institute (NCI), R01 AT009152 from the National Center for Complementary and Integrative Health (NCCIH), and institutional funds awarded to Dr. Ah-Ng Kong.
ABBREVIATIONS
- UVB
ultraviolet B
- UA
ursolic acid
- ROS
reactive oxidative stress
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- IPA
ingenuity pathway analysis
- TNF-a
tumor necrosis factor-alpha
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflict of interest
DATA AVAILABILITY STATEMENT
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
REFERENCE
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. [DOI] [PubMed] [Google Scholar]
- 2.Kim Y, He YY. Ultraviolet radiation-induced non-melanoma skin cancer: Regulation of DNA damage repair and inflammation. Genes Dis. 2014;1(2):188–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mukhtar H, Elmets CA. Photocarcinogenesis: mechanisms, models and human health implications. Photochemistry and photobiology. 1996;63(4):356–357. [DOI] [PubMed] [Google Scholar]
- 4.Katiyar SK, Matsui MS, Mukhtar H. Kinetics of UV light-induced cyclobutane pyrimidine dimers in human skin in vivo: an immunohistochemical analysis of both epidermis and dermis. Photochemistry and photobiology. 2000;72(6):788–793. [DOI] [PubMed] [Google Scholar]
- 5.Martin J, Duncan FJ, Keiser T, et al. Macrophage migration inhibitory factor (MIF) plays a critical role in pathogenesis of ultraviolet-B (UVB) -induced nonmelanoma skin cancer (NMSC). FASEB J. 2009;23(3):720–730. [DOI] [PubMed] [Google Scholar]
- 6.Buckman SY, Gresham A, Hale P, et al. COX-2 expression is induced by UVB exposure in human skin: Implications for the development of skin cancer. Carcinogenesis. 1998;19(5):723–729. [DOI] [PubMed] [Google Scholar]
- 7.Thomas-Ahner JM, Wulff BC, Tober KL, Kusewitt DF, Riggenbach JA, Oberyszyn TM. Gender differences in UVB-induced skin carcinogenesis, inflammation, and DNA damage. Cancer Res. 2007;67(7):3468–3474. [DOI] [PubMed] [Google Scholar]
- 8.Kress S, Sutter C, Strickland PT, Mukhtar H, Schweizer J, Schwarz M. Carcinogen-specific mutational pattern in the p53 gene in ultraviolet B radiation-induced squamous cell carcinomas of mouse skin. Cancer Res. 1992;52(22):6400–6403. [PubMed] [Google Scholar]
- 9.Ming M, Han W, Maddox J, et al. UVB-induced ERK/AKT-dependent PTEN suppression promotes survival of epidermal keratinocytes. Oncogene. 2010;29(4):492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao C, Lu S, Kivlin R, et al. SIRT1 confers protection against UVB- and H2O2-induced cell death via modulation of p53 and JNK in cultured skin keratinocytes. J Cell Mol Med. 2009;13(9B):3632–3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esteller M Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8(4):286–298. [DOI] [PubMed] [Google Scholar]
- 12.Jeronimo C, Henrique R. Epigenetic biomarkers in urological tumors: A systematic review. Cancer letters. 2014;342(2):264–274. [DOI] [PubMed] [Google Scholar]
- 13.Wang F, Zhang N, Wang J, Wu H, Zheng X. Tumor purity and differential methylation in cancer epigenomics. Brief Funct Genomics. 2016;15(6):408–419. [DOI] [PubMed] [Google Scholar]
- 14.Fenaux P, Ades L. Review of azacitidine trials in Intermediate-2-and High-risk myelodysplastic syndromes. Leuk Res. 2009;33 Suppl 2:S7–11. [DOI] [PubMed] [Google Scholar]
- 15.Golabek K, Strzelczyk JK, Wiczkowski A, Michalski M. Potential use of histone deacetylase inhibitors in cancer therapy. Contemp Oncol (Pozn). 2015;19(6):436–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baines AT, Xu D, Der CJ. Inhibition of Ras for cancer treatment: the search continues. Future Med Chem. 2011;3(14):1787–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Doorn R, Gruis NA, Willemze R, van der Velden PA, Tensen CP. Aberrant DNA methylation in cutaneous malignancies. Semin Oncol. 2005;32(5):479–487. [DOI] [PubMed] [Google Scholar]
- 18.Bachman AN, Curtin GM, Doolittle DJ, Goodman JI. Altered methylation in gene-specific and GC-rich regions of DNA is progressive and nonrandom during promotion of skin tumorigenesis. Toxicol Sci. 2006;91(2):406–418. [DOI] [PubMed] [Google Scholar]
- 19.Schinke C, Mo Y, Yu Y, et al. Aberrant DNA methylation in malignant melanoma. Melanoma research. 2010;20(4):253–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang AY, Lee JH, Shu L, et al. Genome-wide analysis of DNA methylation in UVB- and DMBA/TPA-induced mouse skin cancer models. Life Sci. 2014;113(1–2):45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paredes-Gonzalez X, Fuentes F, Su ZY, Kong ANT. Apigenin Reactivates Nrf2 Anti-oxidative Stress Signaling in Mouse Skin Epidermal JB6 P + Cells Through Epigenetics Modifications. Aaps Journal. 2014;16(4):727–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin JR, Qin HH, Wu WY, He SJ, Xu JH. Vitamin C Protects Against UV Irradiation-Induced Apoptosis Through Reactivating Silenced Tumor Suppressor Genes p21 and p16 in a Tet-Dependent DNA Demethylation Manner in Human Skin Cancer Cells. Cancer Biotherapy and Radiopharmaceuticals. 2014;29(6):257–264. [DOI] [PubMed] [Google Scholar]
- 23.Fortes C, Mastroeni S, Melchi F, et al. A protective effect of the Mediterranean diet for cutaneous melanoma. Int J Epidemiol. 2008;37(5):1018–1029. [DOI] [PubMed] [Google Scholar]
- 24.Kune GA, Bannerman S, Field B, et al. Diet, alcohol, smoking, serum beta-carotene, and vitamin A in male nonmelanocytic skin cancer patients and controls. Nutrition and cancer. 1992;18(3):237–244. [DOI] [PubMed] [Google Scholar]
- 25.Lu YP, Lou YR, Xie JG, et al. Topical applications of caffeine or (−)-epigallocatechin gallate (EGCG) inhibit carcinogenesis and selectively increase apoptosis in UVB-induced skin tumors in mice. P Natl Acad Sci USA. 2002;99(19):12455–12460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dinkova-Kostova AT, Jenkins SN, Fahey JW, et al. Protection against UV-light-induced skin carcinogenesis in SKH-1 high-risk mice by sulforaphane-containing broccoli sprout extracts. Cancer Lett. 2006;240(2):243–252. [DOI] [PubMed] [Google Scholar]
- 27.Yin R, Li T, Tian JX, Xi P, Liu RH. Ursolic acid, a potential anticancer compound for breast cancer therapy. Crit Rev Food Sci Nutr. 2018;58(4):568–574. [DOI] [PubMed] [Google Scholar]
- 28.Lee YH, Wang E, Kumar N, Glickman RD. Ursolic acid differentially modulates apoptosis in skin melanoma and retinal pigment epithelial cells exposed to UV-VIS broadband radiation. Apoptosis: an international journal on programmed cell death. 2014;19(5):816–828. [DOI] [PubMed] [Google Scholar]
- 29.Tokuda H, Ohigashi H, Koshimizu K, Ito Y. Inhibitory effects of ursolic and oleanolic acid on skin tumor promotion by 12-O-tetradecanoylphorbol-13-acetate. Cancer letters. 1986;33(3):279–285. [DOI] [PubMed] [Google Scholar]
- 30.Cho J, Rho O, Junco J, et al. Effect of Combined Treatment with Ursolic Acid and Resveratrol on Skin Tumor Promotion by 12-O-Tetradecanoylphorbol-13-Acetate. Cancer prevention research. 2015;8(9):817–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dinkova-Kostova AT, Liby KT, Stephenson KK, et al. Extremely potent triterpenoid inducers of the phase 2 response: correlations of protection against oxidant and inflammatory stress. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(12):4584–4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim H, Ramirez CN, Su ZY, Kong AN. Epigenetic modifications of triterpenoid ursolic acid in activating Nrf2 and blocking cellular transformation of mouse epidermal cells. J Nutr Biochem. 2016;33:54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang AY, Kim H, Li W, Kong AN. Natural compound-derived epigenetic regulators targeting epigenetic readers, writers and erasers. Curr Top Med Chem. 2016;16(7):697–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lou YR, Peng QY, Li T, et al. Effects of high-fat diets rich in either omega-3 or omega-6 fatty acids on UVB-induced skin carcinogenesis in SKH-1 mice. Carcinogenesis. 2011;32(7):1078–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu YP, Lou YR, Yen P, Mitchell D, Huang MT, Conney AH. Time course for early adaptive responses to ultraviolet B light in the epidermis of SKH-1 mice. Cancer Res. 1999;59(18):4591–4602. [PubMed] [Google Scholar]
- 36.Saw CL, Huang MT, Liu Y, Khor TO, Conney AH, Kong AN. Impact of Nrf2 on UVB-induced skin inflammation/photoprotection and photoprotective effect of sulforaphane. Molecular carcinogenesis. 2011;50(6):479–486. [DOI] [PubMed] [Google Scholar]
- 37.Yang Y, Wu R, Sargsyan D, et al. UVB drives different stages of epigenome alterations during progression of skin cancer. Cancer Lett. 2019;449:20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu YP, Lou YR, Peng QY, Nghiem P, Conney AH. Caffeine decreases phospho-Chk1 (Ser317) and increases mitotic cells with cyclin B1 and caspase 3 in tumors from UVB-treated mice. Cancer prevention research. 2011;4(7):1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Picard. http://broadinstitute.github.io/picard/
- 42.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. [DOI] [PubMed] [Google Scholar]
- 43.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kramer A, Green J, Pollard J, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30(4):523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu GC, Wang LG, Han YY, He QY. clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. Omics. 2012;16(5):284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ashburner M, Ball CA, Blake JA, et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carbon S, Dietze H, Lewis SE, et al. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Research. 2017;45(D1):D331–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Research. 2016;44(D1):D457–D462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research. 2000;28(1):27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27(11):1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gaspar JM, Hart RP. DMRfinder: efficiently identifying differentially methylated regions from MethylC-seq data. BMC Bioinformatics. 2017;18(1):528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu G, Wang LG, He QY. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31(14):2382–2383. [DOI] [PubMed] [Google Scholar]
- 54.Spandidos A, Wang X, Wang H, Seed B. PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 2010;38(Database issue):D792–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spandidos A, Wang X, Wang H, Dragnev S, Thurber T, Seed B. A comprehensive collection of experimentally validated primers for Polymerase Chain Reaction quantitation of murine transcript abundance. BMC Genomics. 2008;9:633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang X, Seed B. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res. 2003;31(24):e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jeong WS, Jun M, Kong AN. Nrf2: a potential molecular target for cancer chemoprevention by natural compounds. Antioxid Redox Signal. 2006;8(1–2):99–106. [DOI] [PubMed] [Google Scholar]
- 58.Wang H, Khor TO, Shu LM, et al. Plants vs. Cancer: A Review on Natural Phytochemicals in Preventing and Treating Cancers and Their Druggability. Anti-Cancer Agent Me. 2012;12(10):1281–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mandlekar S, Hong JL, Kong ANT. Modulation of metabolic enzymes by dietary phytochemicals: A review of mechanisms underlying beneficial versus unfavorable effects. Curr Drug Metab. 2006;7(6):661–675. [DOI] [PubMed] [Google Scholar]
- 60.Nair S, Li W, Kong AN. Natural dietary anti-cancer chemopreventive compounds: redox-mediated differential signaling mechanisms in cytoprotection of normal cells versus cytotoxicity in tumor cells. Acta Pharmacol Sin. 2007;28(4):459–472. [DOI] [PubMed] [Google Scholar]
- 61.Negi G, Kumar A, Sharma SS. Melatonin modulates neuroinflammation and oxidative stress in experimental diabetic neuropathy: effects on NF-kappaB and Nrf2 cascades. Journal of pineal research. 2011;50(2):124–131. [DOI] [PubMed] [Google Scholar]
- 62.Liu X, Sun J. Endothelial cells dysfunction induced by silica nanoparticles through oxidative stress via JNK/P53 and NF-kappaB pathways. Biomaterials. 2010;31(32):8198–8209. [DOI] [PubMed] [Google Scholar]
- 63.Park JS, Chyun JH, Kim YK, Line LL, Chew BP. Astaxanthin decreased oxidative stress and inflammation and enhanced immune response in humans. Nutrition & metabolism. 2010;7:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pini M, Rhodes DH, Fantuzzi G. Hematological and acute-phase responses to diet-induced obesity in IL-6 KO mice. Cytokine. 2011;56(3):708–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sparmann A, Bar-Sagi D. Ras-induced interleukin-8 expression plays a critical role in tumor growth and angiogenesis. Cancer Cell. 2004;6(5):447–458. [DOI] [PubMed] [Google Scholar]
- 66.Li A, Dubey S, Varney ML, Dave BJ, Singh RK. IL-8 directly enhanced endothelial cell survival, proliferation, and matrix metalloproteinases production and regulated angiogenesis. J Immunol. 2003;170(6):3369–3376. [DOI] [PubMed] [Google Scholar]
- 67.Schraufstatter IU, Chung J, Burger M. IL-8 activates endothelial cell CXCR1 and CXCR2 through Rho and Rac signaling pathways. American journal of physiology Lung cellular and molecular physiology. 2001;280(6):L1094–1103. [DOI] [PubMed] [Google Scholar]
- 68.Bancroft CC, Chen Z, Dong G, et al. Coexpression of proangiogenic factors IL-8 and VEGF by human head and neck squamous cell carcinoma involves coactivation by MEK-MAPK and IKK-NF-kappaB signal pathways. Clin Cancer Res. 2001;7(2):435–442. [PubMed] [Google Scholar]
- 69.Penna G, Fibbi B, Amuchastegui S, et al. The vitamin D receptor agonist elocalcitol inhibits IL-8-dependent benign prostatic hyperplasia stromal cell proliferation and inflammatory response by targeting the RhoA/Rho kinase and NF-kappaB pathways. The Prostate. 2009;69(5):480–493. [DOI] [PubMed] [Google Scholar]
- 70.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome biology. 2010;11(2):R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rue-Albrecht K, McGettigan PA, Hernandez B, et al. GOexpress: an R/Bioconductor package for the identification and visualisation of robust gene ontology signatures through supervised learning of gene expression data. Bmc Bioinformatics. 2016;17:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Bio. 2010;11(3):196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461(7267):1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pihan GA, Doxsey SJ. The mitotic machinery as a source of genetic instability in cancer. Semin Cancer Biol. 1999;9(4):289–302. [DOI] [PubMed] [Google Scholar]
- 75.Chen JJ, Silver DP, Walpita D, et al. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Molecular Cell. 1998;2(3):317–328. [DOI] [PubMed] [Google Scholar]
- 76.Schvartzman JM, Sotillo R, Benezra R. Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat Rev Cancer. 2010;10(2):102–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li L, Hou Y, Yu J, et al. Synergism of ursolic acid and cisplatin promotes apoptosis and enhances growth inhibition of cervical cancer cells via suppressing NE-kappa B p65. Oncotarget. 2017;8(57):97416–97427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang CY, Lin CS, Hua CH, et al. Cis-3-O-p-hydroxycinnamoyl Ursolic Acid Induced ROS-Dependent p53-Mediated Mitochondrial Apoptosis in Oral Cancer Cells. Biomol Ther (Seoul). 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Arden KC. Multiple roles of FOXO transcription factors in mammalian cells point to multiple roles in cancer. Exp Gerontol. 2006;41(8):709–717. [DOI] [PubMed] [Google Scholar]
- 80.Bragado P, Armesilla A, Silva A, Porras A. Apoptosis by cisplatin requires p53 mediated p38alpha MAPK activation through ROS generation. Apoptosis. 2007;12(9):1733–1742. [DOI] [PubMed] [Google Scholar]
- 81.Tothova Z, Kollipara R, Huntly BJ, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128(2):325–339. [DOI] [PubMed] [Google Scholar]
- 82.Liu JW, Chandra D, Rudd MD, et al. Induction of prosurvival molecules by apoptotic stimuli: involvement of FOXO3a and ROS. Oncogene. 2005;24(12):2020–2031. [DOI] [PubMed] [Google Scholar]
- 83.Willert K, Nusse R. Wnt proteins. Cold Spring Harb Perspect Biol. 2012;4(9):a007864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang Q, Symes AJ, Kane CA, et al. A novel role for Wnt/Ca2+ signaling in actin cytoskeleton remodeling and cell motility in prostate cancer. PLoS One. 2010;5(5):e10456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miller JR, Hocking AM, Brown JD, Moon RT. Mechanism and function of signal transduction by the Wnt/beta-catenin and Wnt/Ca2+ pathways. Oncogene. 1999;18(55):7860–7872. [DOI] [PubMed] [Google Scholar]
- 86.Wang X, Mandal AK, Saito H, et al. Arsenic and chromium in drinking water promote tumorigenesis in a mouse colitis-associated colorectal cancer model and the potential mechanism is ROS-mediated Wnt/beta-catenin signaling pathway. Toxicol Appl Pharmacol. 2012;262(1):11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Di Micco R, Fumagalli M, Cicalese A, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444(7119):638–642. [DOI] [PubMed] [Google Scholar]
- 88.d’Adda di Fagagna F, Reaper PM, Clay-Farrace L, et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426(6963):194–198. [DOI] [PubMed] [Google Scholar]
- 89.Ito K, Hirao A, Arai F, et al. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nature medicine. 2006;12(4):446–451. [DOI] [PubMed] [Google Scholar]
- 90.Debacq-Chainiaux F, Borlon C, Pascal T, et al. Repeated exposure of human skin fibroblasts to UVB at subcytotoxic level triggers premature senescence through the TGF-beta1 signaling pathway. Journal of cell science. 2005;118(Pt 4):743–758. [DOI] [PubMed] [Google Scholar]
- 91.Helenius M, Makelainen L, Salminen A. Attenuation of NF-kappaB signaling response to UVB light during cellular senescence. Exp Cell Res. 1999;248(1):194–202. [DOI] [PubMed] [Google Scholar]
- 92.Chainiaux F, Magalhaes JP, Eliaers F, Remacle J, Toussaint O. UVB-induced premature senescence of human diploid skin fibroblasts. The international journal of biochemistry & cell biology. 2002;34(11):1331–1339. [DOI] [PubMed] [Google Scholar]
- 93.McLean GW, Carragher NO, Avizienyte E, Evans J, Brunton VG, Frame MC. The role of focal-adhesion kinase in cancer - a new therapeutic opportunity. Nature reviews Cancer. 2005;5(7):505–515. [DOI] [PubMed] [Google Scholar]
- 94.Weiner TM, Liu ET, Craven RJ, Cance WG. Expression of focal adhesion kinase gene and invasive cancer. Lancet. 1993;342(8878):1024–1025. [DOI] [PubMed] [Google Scholar]
- 95.Chen Z, Trotman LC, Shaffer D, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436(7051):725–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li J, Yen C, Liaw D, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275(5308):1943–1947. [DOI] [PubMed] [Google Scholar]
- 97.Agochiya M, Brunton VG, Owens DW, et al. Increased dosage and amplification of the focal adhesion kinase gene in human cancer cells. Oncogene. 1999;18(41):5646–5653. [DOI] [PubMed] [Google Scholar]
- 98.Remans PH, Gringhuis SI, van Laar JM, et al. Rap1 signaling is required for suppression of Ras-generated reactive oxygen species and protection against oxidative stress in T lymphocytes. Journal of immunology. 2004;173(2):920–931. [DOI] [PubMed] [Google Scholar]
- 99.Remans PH, Wijbrandts CA, Sanders ME, et al. CTLA-4IG suppresses reactive oxygen species by preventing synovial adherent cell-induced inactivation of Rap1, a Ras family GTPASE mediator of oxidative stress in rheumatoid arthritis T cells. Arthritis and rheumatism. 2006;54(10):3135–3143. [DOI] [PubMed] [Google Scholar]
- 100.Chen AC, Arany PR, Huang YY, et al. Low-level laser therapy activates NF-kB via generation of reactive oxygen species in mouse embryonic fibroblasts. PloS one. 2011;6(7):e22453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kwa RE, Campana K, Moy RL. Biology of cutaneous squamous cell carcinoma. J Am Acad Dermatol. 1992;26(1):1–26. [DOI] [PubMed] [Google Scholar]
- 102.Baade PD, Balanda KP, Lowe JB. Changes in skin protection behaviors, attitudes, and sunburn: in a population with the highest incidence of skin cancer in the world. Cancer Detect Prev. 1996;20(6):566–575. [PubMed] [Google Scholar]
- 103.Tsai SJ, Yin MC. Antioxidative and anti-inflammatory protection of oleanolic acid and ursolic acid in PC12 cells. J Food Sci. 2008;73(7):H174–H178. [DOI] [PubMed] [Google Scholar]
- 104.Checker R, Sandur SK, Sharma D, et al. Potent anti-inflammatory activity of ursolic acid, a triterpenoid antioxidant, is mediated through suppression of NF-kappaB, AP-1 and NF-AT. PLoS One. 2012;7(2):e31318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Baricevic D, Sosa S, Della Loggia R, et al. Topical anti-inflammatory activity of Salvia officinalis L. leaves: the relevance of ursolic acid. J Ethnopharmacol. 2001;75(2–3):125–132. [DOI] [PubMed] [Google Scholar]
- 106.Ikeda Y, Murakami A, Ohigashi H. Ursolic acid: an anti- and pro-inflammatory triterpenoid. Mol Nutr Food Res. 2008;52(1):26–42. [DOI] [PubMed] [Google Scholar]
- 107.Wang C, Shu L, Zhang C, et al. Histone Methyltransferase Setd7 Regulates Nrf2 Signaling Pathway by Phenethyl Isothiocyanate and Ursolic Acid in Human Prostate Cancer Cells. Mol Nutr Food Res. 2018;62(18):e1700840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tapondjou LA, Lontsi D, Sondengam BL, et al. In vivo anti-nociceptive and anti-inflammatory effect of the two triterpenes, ursolic acid and 23-hydroxyursolic acid, from Cussonia bancoensis. Arch Pharm Res. 2003;26(2):143–146. [DOI] [PubMed] [Google Scholar]
- 109.Zhang C, Wang C, Li W, et al. Pharmacokinetics and Pharmacodynamics of the Triterpenoid Ursolic Acid in Regulating the Antioxidant, Anti-inflammatory, and Epigenetic Gene Responses in Rat Leukocytes. Mol Pharm. 2017;14(11):3709–3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kim SH, Hong JH, Lee YC. Ursolic acid, a potential PPAR gamma agonist, suppresses ovalbumin-induced airway inflammation and Penh by down-regulating IL-5, IL-13, and IL-17 in a mouse model of allergic asthma. European Journal of Pharmacology. 2013;701(1–3):131–143. [DOI] [PubMed] [Google Scholar]
- 111.Aguirre MC, Delporte C, Backhouse N, et al. Topical anti-inflammatory activity of 2alpha-hydroxy pentacyclic triterpene acids from the leaves of Ugni molinae. Bioorg Med Chem. 2006;14(16):5673–5677. [DOI] [PubMed] [Google Scholar]
- 112.Banno N, Akihisa T, Tokuda H, et al. Anti-inflammatory and antitumor-promoting effects of the triterpene acids from the leaves of Eriobotrya japonica. Biological & Pharmaceutical Bulletin. 2005;28(10):1995–1999. [DOI] [PubMed] [Google Scholar]
- 113.Cho J, Rho O, Junco J, et al. Effect of Combined Treatment with Ursolic Acid and Resveratrol on Skin Tumor Promotion by 12-O-Tetradecanoylphorbol-13-Acetate. Cancer Prev Res. 2015;8(9):817–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li W, Kong AN. Molecular mechanisms of Nrf2-mediated antioxidant response. Mol Carcinog. 2009;48(2):91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Khor TO, Huang MT, Kwon KH, Chan JY, Reddy BS, Kong AN. Nrf2-deficient mice have an increased susceptibility to dextran sulfate sodium-induced colitis. Cancer Res. 2006;66(24):11580–11584. [DOI] [PubMed] [Google Scholar]
- 116.Fang RM, Wu RY, Zuo Q, et al. Sophora flavescens Containing-QYJD Formula Activates Nrf2 Anti-Oxidant Response, Blocks Cellular Transformation and Protects Against DSS-Induced Colitis in Mouse Model. Am J Chinese Med. 2018;46(7):1609–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zuo Q, Wu RY, Xiao X, et al. The dietary flavone luteolin epigenetically activates the Nrf2 pathway and blocks cell transformation in human colorectal cancer HCT116 cells. Journal of Cellular Biochemistry. 2018;119(11):9573–9582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Guo Y, Wu RY, Gaspar JM, et al. DNA methylome and transcriptome alterations and cancer prevention by curcumin in colitis-accelerated colon cancer in mice. Carcinogenesis. 2018;39(5):669–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang C, Wu R, Sargsyan D, et al. CpG methyl-seq and RNA-seq epigenomic and transcriptomic studies on the preventive effects of Moringa isothiocyanate in mouse epidermal JB6 cells induced by the tumor promoter TPA. J Nutr Biochem. 2019;68:69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yang Y, Yang I, Cao M, et al. Fucoxanthin Elicits Epigenetic Modifications, Nrf2 Activation and Blocking Transformation in Mouse Skin JB6 P+ Cells. AAPS J. 2018;20(2):32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kuang H, Tang Z, Zhang C, et al. Taxifolin Activates the Nrf2 Anti-Oxidative Stress Pathway in Mouse Skin Epidermal JB6 P+ Cells through Epigenetic Modifications. Int J Mol Sci. 2017;18(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hong B, Su ZY, Zhang CY, et al. Reserpine Inhibit the JB6 P+ Cell Transformation Through Epigenetic Reactivation of Nrf2-Mediated Anti-oxidative Stress Pathway. Aaps Journal. 2016;18(3):659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schwartz GK, Shah MA. Targeting the cell cycle: a new approach to cancer therapy. J Clin Oncol. 2005;23(36):9408–9421. [DOI] [PubMed] [Google Scholar]
- 124.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9(3):153–166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.