Abstract
Background
Some of the metabolic effects of bariatric surgery may be mediated by the gut microbiome.
Objectives
To study the effect of bariatric surgery on changes to gut microbiota composition and bacterial pathways, and their relation to metabolic parameters following bariatric surgery.
Settings
University Hospital, United States and Spain
Methods
Microbial diversity and composition by 16S rRNA sequencing, putative bacterial pathways and targeted circulating metabolites were studied in 26 individuals with severe obesity, with and without type 2 diabetes, before and at 3, 6, and 12 months after either gastric bypass or sleeve gastrectomy.
Results
Bariatric surgery tended to increase alpha diversity, and significantly altered beta diversity, microbiota composition and function up to six months after surgery, but these changes tend to regress to pre-surgery levels by 12 months. Twelve of the 15 bacterial pathways enriched after surgery also regress to pre-surgery levels at 12 months. Network analysis identified groups of bacteria significantly correlated with levels of circulating metabolites over time. There were no differences between study sites, surgery type or diabetes status in terms of microbial diversity and composition at baseline and after surgery.
Conclusions
The association between changes in microbiome with decreased circulating biomarkers of inflammation, increased bile acids and products of choline metabolism and other bacterial pathways suggest that the microbiome partially mediates improvement of metabolism during the first year following bariatric surgery.
Keywords: bariatric surgery, metabolomics, microbiota, 16S rRNA gene, network analysis, predicted metagenome
Introduction
Although most metabolic effects of bariatric surgery (BS) are driven by weight loss, factors such as the gut microbiome can also play a role[1]. For example, microbiota transferred from gastric-bypass (RYGB) operated mice partially confer their phenotypes, suggesting a mediation role of the microbiota[2, 3].
Previous studies have described shifts in the microbiome after BS[1]. While studies have shown changes in circulating biomarkers associated with microbial metabolism such as Trimethylamine N-oxide (TMAO), bile acids, and short-chain fatty acids (SCFA), after BS[4, 5], others have described cross talk between the microbiome and biomarkers of cardiovascular disease (CVD) risk[6–9]. There is however great heterogeneity in changes to the microbiome after BS and few studies go beyond 6 months post-surgery[1]. The primary goal of this study was to identify association between change in microbiota and biomarkers of microbial and whole body metabolism after BS. We hypothesized that: 1) The rapid changes in microbiome diversity and composition after BS are not sustained overtime; 2) There is a temporal association of bacterial clusters with circulating biomarkers following BS.
Methods
Subject Characteristics
26 participants had BS at St. Luke’s Hospital in New York (NYC) or at Parc Taulí Hospital in Sabadell, Barcelona (BCN). A total of 8 participants had type 2 diabetes, 9 took metformin (one for polycystic ovary syndrome), 11 were treated for hypertension, 10 for dyslipidemia; only one participant had a history of cardiovascular disease. Exclusion criteria included: any infection, gastrointestinal disorder, and antibiotics, probiotics or prebiotics two months prior to sampling. All participants signed a written informed consent prior to enrollment and took proton pump inhibitors and vitamins after surgery.
Study Design
Fasting blood samples, and stool samples (Protocult kit, ABC Medical Enterprizes, Inc, Port St. Lucie, FL) were obtained pre-surgery, 3months (3m), 6m and 12m post-surgery, and stored at −80°C. All participants underwent RYGB (n=19) except 7 from BCN who underwent sleeve gastrectomy (SG, n=7). Diet was neither controlled nor recorded.
Biomarkers and Microbial Analyses
Determinations of plasma concentrations of glucose, insulin, serum amyloid A (SAA), high-sensitivity C-reactive protein (hs-CRP), bile acids, TMAO, betaine, choline, SCFA, lipids, liver function tests and white cell count were done in certified Core Laboratories. Methods for DNA extraction and 16S sequencing of stool samples, and for estimation of alpha and beta diversity, pathway and network analyses are detailed in Supplemental.
Statistical Analyses
Differences in diversity were tested using repeated measures ANOVA for alpha diversity and within versus between UniFrac distances for beta diversity. Correlation of microbiome data with biomarkers over time was assessed using pairwise partial nonparametric Spearman correlations in R (v 3.4.2) with the ppcor library (v 1.1), with false discovery rate (FDR) correction and q < 0.2. Co-occurrence network analysis was performed as we have previously shown [10]. Groups of highly connected bacteria (‘clusters’) were identified using the Bron-Kerbosch algorithm.
Results
Participant characteristics pre-surgery, and change in weight and metabolic parameters after BS did not differ between NYC (n=14) and BCN (n=12) cohorts, or between RYGB and SG (n=7) in the BCN cohort (Supplemental Table 1, 2 and 3). Neither alpha nor beta diversity differ between the NYC and BCN cohorts, at any time point pre- or post-surgery. A random forest classifier was also unable to distinguish between the two cohorts[11]. Taxa previously reported to be associated with use of metformin (Akkermansia muciniphila, Butyrivibrio, Bacteroides fragilis) were not differentially enriched between metformin users and non-users at pre-surgery (p > 0.05, Wilcoxon Rank Sum test). Study site, surgery type, being on metformin and type 2 diabetes were also not associated with either pre-surgery or overall changes in alpha (p > 0.05, Wilcoxon Rank Sum test) or beta diversity (p > 0.05, PERMANOVA) at any time point. We therefore pooled all patients for the analyses.
Effect of Bariatric Surgery (BS)
With weight loss, all circulating metabolic biomarkers improved, as expected (Table1). The magnitude of the change was largest for hsCRP (−84.69%), SAA (−60.01%), conjugated bile acids (+100.52%) and TMAO (+246.19%) (Supp Figure 1).
Table 1:
Effect of surgical weight loss on clinical and circulating biomarkers for all subjects together (BCN and NYC cohorts).
| Pre-surgery | 3 months | 6 months | 12 months | |
|---|---|---|---|---|
| N | 26 | 22 | 23 | 26 |
| Weight (kg) | 123.1 (17.0) | 97.3 (16.1)*** | 86.8 (13.7)*** ǂ | 81.5 (12.5)*** ǂ # |
| Weight loss (%) | 21.1 (3.9) | 28.3 (5.1)ǂ | 33.5 (6.8)ǂ | |
| Excess Weight Loss (%) | 48.8 (13.5) | 65.8 (16.1)ǂ | 76.1(17.6)ǂ# | |
| BMI (kg/m2) | ||||
| 46.1 (6.3) | 36.5 (5.9)*** | 32.6 (4.8)***ǂ | 30.5 (4.5)*** ǂ # | |
| Systolic Blood Pressure (mmHg) | 125.2 (12.4) | 124.7 (11.1) | 127.1 (15.5) | 122.3 (17) |
| Diastolic Blood Pressure (mmHg) | 71.8 (17.9) | 78.9 (6.1) | 76.6 (5.8) | 72 (9.7) |
| Fasting Glucose (mmol/L) | 6.6 (2.7) | 4.9 (0.9)** | 4.9 (1.0)*** | 4.8 (0.6)*** |
| Fasting Insulin (pmol/L) | 191.7 (91) | 86.1 (36.8)*** | 97.9 (82.0)*** | 75.0 (36.8)*** |
| HOMA-IR | 8.0 (4.8) | 2.9 (1.8)*** | 3.1 (2.4)*** | 2.4 (1.3)*** ǂ |
| A1C (%) | 6.2 (1.0) | 5.5 (0.5)*** | 5.4 (0.4)*** | 5.4 (0.4)*** |
| T-CHOL (mmol/L) | 4.9 (1.1) | 4.5 (1.4) | 4.5 (1.4) | 4.5 (1.2) |
| HDL-CHOL (mmol/L) | 1.2 (0.3) | 1.1 (0.2) | 1.4 (0.3) ǂ | 1.5 (0.4) ǂ # |
| LDL-CHOL (mmol/L) | 2.9 (0.9) | 2.8 (1.2) | 2.6 (1.1) | 2.6 (1.0) |
| Triglycerides (mmol/L) | 1.7 (1.1) | 1.3 (0.7) | 1.1 (0.6)*** | 1.0 (0.4)** ǂ |
| WBC (K/mcL) | 8.1 (2.2) | 6.3 (1.7)* | 6.5 (1.7)** | 6.2 (1.2)** |
| SAA (ng/mL) | 58.6 (51.9) | 37.5 (40.8)* | 29.0 (26.7)** | 19.2 (16.9)*** # |
| hsCRP (mg/L) | 9.5 (7.5) | 9.2 (19.7)* | 3.7 (4.3)*** | 1.2 (1.2)*** # |
| AST (U/L) | 25.9 (10.6) | 20.6 (7.5) | 21 (6.3) | 21 (7) |
| ALT (U/L) | 29.9 (16.2) | 23.3 (18.9) | 22.6 (11.0) | 23.8 (15.4) |
| Acetate (μM) | 78.2 (36.4) | 82.5 (29.7) | 70.3 (25.8) | 94.8 (45.1) |
| Propionate (μM) | 3.5 (2.3) | 3.1 (1.7) | 3.4 (2.0) | 3.7 (2.2) |
| Butyrate (μM) | 1.2 (1.4) | 0.8 (0.9) | 0.7 (0.8) | 0.9 (0.9) |
| TMAO (μM) | 0.5 (0.3) | 2.3 (3.5)* | 1.9 (2.9)** | 1.5 (1.1)*** |
| Betaine (μM) | 2.5 (1.8) | 3.1 (2.5) | 3.1 (2.0) | 3.4 (2.5) |
| Choline (μM) | 26.1 (20.2) | 23.9 (20.9) | 30.4 (29.0) | 36.4 (41.4) ǂ |
| Carnitine (μM) | 7.7 (2.3) | 6.9 (2.1) | 8.0 (2.9) | 7.8 (2.9) ǂ |
| Isobutyrate (μM) | 3.7 (5.8) | 2.9 (5.2) | 4.0 (5.6) | 3.1 (4.4) |
| Isovalerate (μM) | 5.2 (8.4) | 3.7 (7.0) | 5.4 (8.6) | 3.5 (6.0) |
| Valerate (μM) | 0.3(0.2) | 0.3 (0.2) | 0.2 (0.2) | 0.3 (0.1) |
| Hexanoate (μM) | 0.5 (0.7) | 0.4 (0.4) | 0.6 (0.9) | 0.3 (0.6) |
| Heptanoate (μM) | 0.08 (0.1) | 0.09 (0.1) | 0.06 (0.06) | 0.05 (0.07) |
| Octanoate (μM) | 0.5 (0.2) | 0.7 (0.3) | 0.8 (0.9) | 0.5 (0.2) |
| Total Bile Acids (μM) | 2.3 (0.9) | 2.4 (1.4) | 4.6 (3.5) | 3.4 (2.9) |
| Conjugated Bile Acids (μM) | 1.7 (0.8) | 1.4 (0.8) | 3.2 (3.3) | 2.4 (3.0) |
| Unconjugated Bile Acids (μM) | 0.5 (0.2) | 1.0 (1.1) | 1.5 (1.6) | 1.0 (0.9) |
Data are presented as mean (Standard Deviation).
Different from pre-surgery:
P < 0.05
P < 0.01
P < 0.001
Different from 3-months: P < 0.05
Different from 6-months: P < 0.05.
Analysis performed via Friedman test, post-hoc analysis performed via Wilcoxon Signed-Tank tests with Bonferroni correction applied. HOMA-IR: Homeostatic Model Assessment for Insulin Resistance; AIC: glycosylated hemoglobin; T-CHOL: total cholesterol; HDL-CHOL: HDL cholesterol; LDL-CHOL: LDL cholesterol; WBC: white blood count; SAA: serum amyloid A; hsCRP: ultrasensitive C reactive protein; AST: Aspartate Amino Transferase; ALT: Alanine Amino Transferase; TMAO: trimethylamine-N-oxide.
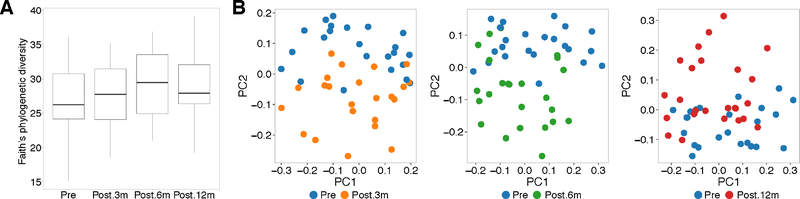
Surgery induced rapid changes in gut microbiome diversity and composition. There was a trend toward a gradual increase of bacterial alpha diversity until 6m, with a decrease at 12m compared to 6m, but still higher than pre-surgery (Figure 1A). Beta diversity was significantly different from pre-surgery at all time-points post-surgery (3m: p=1.83e-06, 6m: p=1.81e-05, 12m: p=0.003, Figure 1B). Again, at 12m the mean distance to pre-surgery microbiome was smaller (0.602 ± 0.05) than at 6m (0.608 ± 0.05) or 3m (0.621 ± 0.05), indicating a partial regression towards pre-surgery microbiome composition.
Figure 1: Change of alpha and beta diversity after bariatric surgery.
A. Alpha diversity (Faith’s phylogenetic diversity) pre- and at 3m (Post.3m), 6m (Post. 6m) and 12m (Post.12m) after surgery (p=0.158, ANOVA). B. Principal coordinate analysis plots on unweighted UniFrac distances (beta diversity) comparing samples pre-surgery versus 3m (p= 1.831e-06, Student’s t-test), 6m (p= 1.816e-05), and 12m (p= 0.003676).
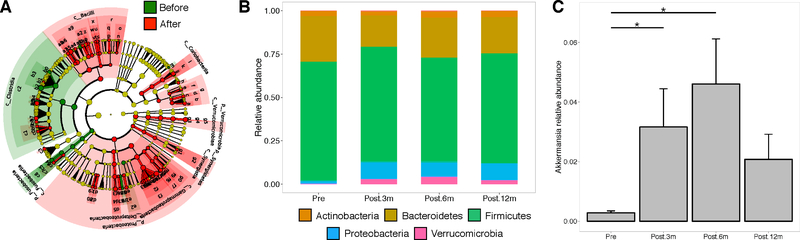
The pre-surgery microbiome exhibited a depletion of Verrucomicrobia and an enrichment of Fusobacteria and Clostridia compared to post-surgery (Figure 2A). The abundance of Verrucomicrobia and Proteobacteria increased after surgery. However, similar to microbial diversity, this trend diminished at 12m compared to 3m and 6m (Figure 2B). The observed changes in Verrucomicrobia were driven primarily by the Akkermansia genus (p<0.05 for 3m, 6m, and 12m compared to pre-surgery; Figure 2C). No change was observed in Firmicutes, Bacteroidetes, Firmicutes/Bacteroidetes, or in any other bacterial taxa tested for differences between pre- and post-surgery. Overall, these results suggest that BS rapidly modifies the diversity and composition of the gut microbiome, although these changes are only partially sustained over time.
Figure 2: Change of bacterial composition after bariatric surgery.
A. LEfSe analysis comparing taxa enriched before and after surgery. B. Relative abundance summarized at the phylum level, at pre-surgery, 3m, 6m, and 12m. C. Relative abundance of Akkermansia muciniphila at each time point (mean ± standard error of the mean).
Effect of BS on bacterial functional pathways
Several bacterial functional pathways related to metabolism differed significantly between pre-surgery and 3m (25 pathways), 6m (29 pathways) and 12m (18 pathways) (Supp Figure 2A–C). In total, 15 pathways were differentially enriched at 3m, 6m and 12m, compared to pre-surgery, including functions related to metabolism of amino acids (tryptophan, tyrosine, histidine, and alanine, aspartate and glutamate), carbohydrates (ascorbate and aldarate, starch and sucrose, propanoate and galactose metabolism), lipids (fatty acid metabolism), cofactors and vitamins (thiamine), terpenoids and polyketides (terpenoid backbone biosynthesis, enzymatic families (protein kinases), and biosynthesis of ansamycins and other secondary metabolites (i.e., organic compound phenylpropanoid). Twelve of these pathways showed partial regression at 12m towards pre-surgery levels (Supp Figure 2D), paralleling our findings in bacterial diversity and composition.
We found eight significant correlations over time between bacterial genera and biomarkers. Bacteroides was significantly correlated with heptanoate. Bifidobacterium was correlated with total cholesterol, LDL-C, and weight loss. Blautia was negatively correlated with butyrate and positively with choline. Finally, Butyricimonas was positively correlated with iso-butyrate and negatively with HDL-C (Supp Figure 3 A–H). Both conjugated and unconjugated bile acids correlated with bacteria (Supplemental Tables 4 and 5).
We performed network analysis and identified groups of bacteria–as opposed to bacteria in isolation– that were associated with circulating biomarkers. Pre-surgery, one cluster composed of Slackia, Anaerostipes, Dorea and Clostridiales was associated with glucose, while Planococcaceae, Haemophilus and Peptoniphilus were associated with TMAO (Supp Figure 4 A). However, at 3m these associations were no longer present, and primary bile acids were significantly correlated with two clusters: one formed by Oscillospira, Lachnospiraceae, and Blautia, and a second cluster formed by Ruminococcaceae, Lachnospiraceae, Blautia, and Coprococcus. Butyrate was also correlated with two clusters (Oscillospira, Eggerthella, and Blautia; Coprococcus, Blautia, Dorea, and Megamonas] (Supp Figure 4B). Bile acids and butyrate were still significantly associated with several bacterial clusters at 6m (Supp Figure 4C). Importantly, some of the correlations between bacteria and biomarkers observed at 12m resembled those found at pre-surgery (Supp Figure 4 D). Overall, these results identify bacterial clusters that might be acting synergistically to modulate circulating biomarkers, and confirm our observations of partial regression towards pre-surgery microbiome and metabolites.
Discussion
The microbiome plays an important role in CVD[12], and changes in composition after BS are associated with decreased CVD risk[13]. However, the exact mechanisms through which the microbiota contributes to these outcomes are poorly understood.
Our main findings are: 1) changes in microbiome composition and diversity occur rapidly (3m) after surgery, are generally sustained over time, but tend toward a regression to pre-surgery values by 12m; 2) these changes associate with alterations in gene expression related to bacterial metabolic pathways and with circulating biomarkers of CVD; 3) the associations of clusters of bacteria with circulating biomarkers show temporal change. These results support a strong interaction of the gut microbiome with the improvement of whole body metabolism in the first year after BS.
The lack of differences before and after surgery in the microbiome of the NYC and BCN cohorts suggests that the severe obesity phenotype and surgical weight loss may over-ride genetic, environmental or dietary influences on the microbiome[14]. Geographical differences in gut microbiome[15] may not apply to the extreme phenotype of morbid obesity.
As expected, BS resulted in rapid weight loss sustained at 12m, associated with improvement in circulating biomarkers related with insulin resistance and CVD risk, and with changes in metabolites derived from microbial metabolism (secondary bile acids, SCFA, TMAO, betaine and choline), as shown previously[5, 16], some of them (TMAO) associated with increased CVD[17]. The paradoxical increase of TMAO with decreased overall CVD risk post-RYGB[4] is difficult to explain, suggesting a complex association of TMAO and cardiovascular risk after BS.
The greater diversity of the microbiome after BS, previously shown after SG or after RYGB, could result from changes in diet, accelerated nutrient transit time, luminal pH[18], exposure of undigested nutrients to the lower intestine, altered bile acids metabolism[5], or increased presence of oxygen in the distal intestine[19]. Microbial composition was also significantly distinct post-surgery. Fusobacteria and Clostridiales were enriched in the gut microbiome pre-surgery, consistent with prior associations with obesity[20]. The genus Fusobacterium is composed mostly of oral bacteria that may play pathogenic roles in esophageal cancer[21] and inflammatory bowel disease[22]. We observed an increase in abundance of the Proteobacteria phylum following surgery, as shown before, after RYGB or SG. The abundance of Akkermansia, low in obesity[23], increased after surgery, similar to previous reports in RYGB and SG[7], but transiently with a decrease at 12m (.(Figure 2C) Akkermansia, via Toll-like receptors[8], may be a mediator of the ameliorated glucose metabolism and decreased inflammation after surgery. Despite previous reports of obesity being associated with the ratio of Bacteroidetes to Firmicutes and of changes in this ratio after surgery, we found no such association in our cohort, consistent with a more recent meta-analysis[24]. Similarly, we also did not find major changes associated with surgery type, type 2 diabetes, or metformin use. The effect of surgery type on the microbiome in humans is contradictory: while a study of 14 obese patients found RYGB to be associated with a decrease in Bacteroidetes compared to SG[25], a different study on 19 patients observed the opposite effect[26]. A third study on 26 patients even found that both RYGB and SG were associated with an increase in Bacteroidetes[27]. A literature review on the effect of surgery type in the microbiome proposed that SG is associated with less alterations than RYGB[28], although none of the seven articles referenced included both RYGB and SG subjects, and so conclusions can only be hypothesized. Overall, differences in the populations under study and experimental methods could explain these inconsistencies.
Two large studies have reported an effect of type 2 diabetes status in the gut microbiome[29, 30]; however, the effect seemed cohort specific, and, distinct to our cohort, the studied individuals were non-obese.
A randomized study of metformin administration in individual with type 2 diabetes showed not only differences in stool microbiome of metformin users, but also that metformin-transformed microbiome conferred a metabolic advantage when transferred to germ free mice[31]. We did not find microbial differences between metformin users and non-users at pre-surgery; this could partially be due to lack of control in our study for pre-surgery diet changes, and adherence to metformin use.
We found several changes in bacterial functions over time; 15 pathways were identified with sustained change after BS (Supplemental Figure 2D). Circulating branched chain (BCAA) and aromatic amino acids (AAA) are associated with insulin resistance and decrease after RYGB, in parallel with improved insulin sensitivity[32]. The uptake of BCAA from the circulation by the gut bacteria could possibly modulate their circulating concentrations[33].
Circulating essential polyunsaturated fatty acids and specific phosphotidylcholine species, ceramides and sphingomyelins[34], decreased after RYGB. The parallel increase in bacterial pathways related to the biosynthesis of fatty acid metabolism may be in response to, and to compensate for, these decreased levels. Pathways involved in carbohydrate metabolism, enriched in obesity[7], decrease after BS. In line with our results, Liu et al. showed that bacterial pathways involved in carbohydrate fermentation, citrate cycle, glycosaminoglycan degradation and LPS synthesis, and production of AAA and BCAA, changed 3m after BS, trending towards those of lean individuals[7]. Our data also show a decreased microbial capacity for the biosynthesis of ansamycins, a family of bacterial metabolites with antimicrobial activity, and for the biosynthesis of some B vitamins. This could predispose certain patients to gastrointestinal infections or vitamin deficiencies after BS. Overall, our results indicate that specific bacterial pathways change significantly, some in a sustained manner after surgery, but their clinical significance remains to be further investigated.
Microbes can act synergistically in the production and degradation of metabolites[35], reflecting interactions between species that are often physiologically important[36]. We identified correlations over time between specific bacteria (both in isolation and as clusters) and circulating biomarkers derived in part from bacterial metabolism (secondary bile acids, TMAO, SCFA), and biomarkers of CVD. These correlations change early after surgery with a trend to the return at pre-surgery levels after 12m, while whole body metabolism remains significantly improved and/or normalized. Others have shown persistent low microbial richness one year after BS when metabolic improvement is achieved[6]. The lack of sustainability of some of the changes observed in the microbiome and/or the lack of full rescue of obesity-associated microbial characteristics suggest that mechanisms others than the microbiome are implicated in metabolic improvement after bariatric surgery, and that the richness of the microbiome alone does may not predict the response to the surgery in the long term[37]. It also suggests that additional therapeutic strategies may be needed to maintain beneficial changes of the microbiome after BS. Our data question the clinical impact of microbial changes on improved whole body metabolism 12 months after weight loss surgery; alternatively, the reversal of microbiome towards pre-surgery characteristics could be a predictor of future deterioration of whole body metabolism. The clinical relevance of the gut microbiome may lay in its other effects on bile acids[5] and vitamins metabolism, metformin effect[31], and, importantly, the susceptibility to infections.
Our study has several strengths and innovative aspects. It is one of few studies[7] that combines microbiome characterization, bacterial function/pathway analysis and circulating biomarkers, and one of few studies[6] that assess changes up to 12m after surgery. Furthermore, our analysis is the first to identify clusters of synergistic bacteria associated with biomarkers that were significantly and temporally altered after BS. Our study also has some limitations. We use predicted rather than sequenced bacterial functions, although we have previously shown that such predictions are highly accurate. Also, we did not control for diet, a limitation shared by most, if not all, human bariatric studies.
In conclusion, we show a strong effect of surgical weight loss on microbiome composition, diversity, and function trending to regress to pre-surgery levels at 12m post-surgery. Future studies will assess the predictive value of pre-surgery microbiome quality on metabolic outcomes after BS, sustainability of changes after 12m, and possible rescue strategies.
Supplementary Material
Highlights.
Surgical weight loss results in increased diversity and change in pathways of the microbiome.
Most changes tend to revert to pre-surgery levels at 12 months after surgery.
There is a temporal variability of bacterial clusters association with cardiovascular biomarkers.
The clinical relevance of microbial changes after bariatric surgery is unknown.
Acknowledgements
The study was supported in part by grants from the NIH (R01DK067561, P30DK26687-30, P30DK063608, R01MH110418, R01DK114038, R01DK090989, T32 DK007559-25), and by the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant Number UL1 TR000040. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. AC was supported by Intensifica’t al Taulí 2016–2017 from Fundació Parc Taulí-Banc de Sabadell. We thank our participants and Daniel Baron-Brenner, Rocío Pareja and Marta Hurtado for helping in sample and data collection.
Footnotes
Competing interests
The authors declare no competing interests.
Availability of data and materials
Sequence data for this study will be available upon publication in EBI/ENA under ID PRJEB28869. Study metadata will be provided as a separate file upon request.
Supplementary information is available on the SOARD’s website and include methods for DNA extraction and 16S sequencing, methods for estimation of alpha and beta diversity, pathway and network analyses, and for assays, 3 Tables and 4 Figures.
Clinical Trial: NCT01516320
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Davies Naomi K, O’Sullivan JM, Plank LD, Murphy R. Altered gut microbiome after bariatric surgery and its association with metabolic benefits: A systematic review. Surgery for Obesity and Related Diseases. 2019; S1550-7289:30042–5. [DOI] [PubMed] [Google Scholar]
- [2].Tremaroli V, Karlsson F, Werling M, Ståhlman M, Kovatcheva-Datchary P, Olbers T, et al. Roux-en-Y Gastric Bypass and Vertical Banded Gastroplasty Induce Long-Term Changes on the Human Gut Microbiome Contributing to Fat Mass Regulation. Cell Metabolism. 2015;22:228–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Liou AP, Paziuk M, Luevano JM Jr., Machineni S, Turnbaugh PJ, Kaplan LM. Conserved shifts in the gut microbiota due to gastric bypass reduce host weight and adiposity. Sci Transl Med. 2013;5:178ra41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Trøseid M, Hov JR, Nestvold TK, Thoresen H, Berge RK, Svardal A, et al. Major Increase in Microbiota-Dependent Proatherogenic Metabolite TMAO One Year After Bariatric Surgery. Metabolic Syndrome and Related Disorders. 2016;14:197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Dutia R, Embrey M, O’Brien S, Haeusler RA, Agenor KK, Homel P, et al. Temporal changes in bile acid levels and 12alpha-hydroxylation after Roux-en-Y gastric bypass surgery in type 2 diabetes. Int J Obes (Lond). 2016;40:554. [DOI] [PubMed] [Google Scholar]
- [6].Aron-Wisnewsky J, Prifti E, Belda E, Ichou F, Kayser BD, Dao MC, et al. Major microbiota dysbiosis in severe obesity: fate after bariatric surgery. Gut. 2019;68:70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Liu R, Hong J, Xu X, Feng Q, Zhang D, Gu Y, et al. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nature Medicine. 2017;23:859–68. [DOI] [PubMed] [Google Scholar]
- [8].Caesar R, Tremaroli V, Kovatcheva-Datchary P, Cani Patrice D, Bäckhed F. Crosstalk between Gut Microbiota and Dietary Lipids Aggravates WAT Inflammation through TLR Signaling. Cell Metabolism. 2015;22:658–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ilhan ZE, DiBaise JK, Isern NG, Hoyt DW, Marcus AK, Kang D-W, et al. Distinctive microbiomes and metabolites linked with weight loss after gastric bypass, but not gastric banding. The Isme Journal. 2017;11:2047–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Segal LN, Clemente JC, Tsay J-CJ, Koralov SB, Keller BC, Wu BG, et al. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nature Microbiology. 2016;1:16031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Knights D, Parfrey LW, Zaneveld J, Lozupone C, Knight R. Human-associated microbial signatures: examining their predictive value. Cell host & microbe. 2011;10:292–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Jie Z, Xia H, Zhong S-L, Feng Q, Li S, Liang S, et al. The gut microbiome in atherosclerotic cardiovascular disease. Nature Communications. 2017;8:845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Li JV, Ashrafian H, Bueter M, Kinross J, Sands C, le Roux CW, et al. Metabolic surgery profoundly influences gut microbial–host metabolic cross-talk. Gut. 2011;60:1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210–5. [DOI] [PubMed] [Google Scholar]
- [15].Clemente JC, Pehrsson EC, Blaser MJ, Sandhu K, Gao Z, Wang B, et al. The microbiome of uncontacted Amerindians. Science Advances. 2015;1:e1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Clemente-Postigo M, Roca-Rodriguez MdM, Camargo A, Ocaña-Wilhelmi L, Cardona F, Tinahones FJ. Lipopolysaccharide and lipopolysaccharide-binding protein levels and their relationship to early metabolic improvement after bariatric surgery. Surgery for Obesity and Related Diseases. 2015;11:933–9. [DOI] [PubMed] [Google Scholar]
- [17].Tang WHW, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, et al. Intestinal Microbial Metabolism of Phosphatidylcholine and Cardiovascular Risk. New England Journal of Medicine. 2013;368:1575–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chung WSF, Walker AW, Louis P, Parkhill J, Vermeiren J, Bosscher D, et al. Modulation of the human gut microbiota by dietary fibres occurs at the species level. BMC Biology. 2016;14:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hartman AL, Lough DM, Barupal DK, Fiehn O, Fishbein T, Zasloff M, et al. Human gut microbiome adopts an alternative state following small bowel transplantation. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:17187–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Andoh A, Nishida A, Takahashi K, Inatomi O, Imaeda H, Bamba S, et al. Comparison of the gut microbial community between obese and lean peoples using 16S gene sequencing in a Japanese population. Journal of Clinical Biochemistry and Nutrition. 2016;59:65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yamamura K, Baba Y, Nakagawa S, Mima K, Miyake K, Nakamura K, et al. Human Microbiome Fusobacterium Nucleatum in Esophageal Cancer Tissue Is Associated with Prognosis. Clinical Cancer Research. 2016;22:5574. [DOI] [PubMed] [Google Scholar]
- [22].Paramsothy S, Kamm MA, Kaakoush NO, Walsh AJ, van den Bogaerde J, Samuel D, et al. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. The Lancet. 2017;389:1218–28. [DOI] [PubMed] [Google Scholar]
- [23].Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Human gut microbes associated with obesity. Nature. 2006;444:1022–3. [DOI] [PubMed] [Google Scholar]
- [24].Walters WA, Xu Z, Knight R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS letters. 2014;588:4223–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Murphy R, Tsai P, Jullig M, Liu A, Plank L, Booth M. Differential Changes in Gut Microbiota After Gastric Bypass and Sleeve Gastrectomy Bariatric Surgery Vary According to Diabetes Remission. Obes Surg. 2017;27:917–25. [DOI] [PubMed] [Google Scholar]
- [26].Medina DA, Pedreros JP, Turiel D, Quezada N, Pimentel F, Escalona A, et al. Distinct patterns in the gut microbiota after surgical or medical therapy in obese patients. PeerJ. 2017;5:e3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wang FG, Bai RX, Yan WM, Yan M, Dong LY, Song MM. Differential composition of gut microbiota among healthy volunteers, morbidly obese patients and post-bariatric surgery patients. Exp Ther Med. 2019;17:2268–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ejtahed H-S, Angoorani P, Hasani-Ranjbar S, Siadat S-D, Ghasemi N, Larijani B, et al. Adaptation of human gut microbiota to bariatric surgeries in morbidly obese patients: A systematic review. Microbial Pathogenesis. 2018;116:13–21. [DOI] [PubMed] [Google Scholar]
- [29].Karlsson FH, Tremaroli V, Nookaew I, Bergstrom G, Behre CJ, Fagerberg B, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103. [DOI] [PubMed] [Google Scholar]
- [30].Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. [DOI] [PubMed] [Google Scholar]
- [31].Wu H, Esteve E, Tremaroli V, Khan MT, Caesar R, Manneras-Holm L, et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med. 2017;23:850–8. [DOI] [PubMed] [Google Scholar]
- [32].Laferrère B, Reilly D, Arias S, Swerdlow N, Gorroochurn P, Bawa B, et al. Differential metabolic impact of gastric bypass surgery versus dietary intervention in obese diabetic subjects despite identical weight loss. Sci Transl Med. 2011;3:80re2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Pedersen HK, Gudmundsdottir V, Nielsen HB, Hyotylainen T, Nielsen T, Jensen BAH, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535:376–81. [DOI] [PubMed] [Google Scholar]
- [34].Kayser BD, Lhomme M, Dao MC, Ichou F, Bouillot JL, Prifti E, et al. Serum lipidomics reveals early differential effects of gastric bypass compared with banding on phospholipids and sphingolipids independent of differences in weight loss. International Journal Of Obesity. 2017;41:917–25. [DOI] [PubMed] [Google Scholar]
- [35].Hansen SK, Rainey PB, Haagensen JAJ, Molin S. Evolution of species interactions in a biofilm community. Nature. 2007;445:533–6. [DOI] [PubMed] [Google Scholar]
- [36].Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–6. [DOI] [PubMed] [Google Scholar]
- [37].Cani PD. Severe obesity and gut microbiota: does bariatric surgery really reset the system? Gut. 2019;68:5–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.