Abstract
Background:
We have recently shown that binge or heavy levels of alcohol drinking increases DNA methylation and reduces gene expression of POMC and PER2 in adult human subjects (Gangisetty et al., 2019). One hypothesis would be that methylation of these two genes is consistently associated with alcohol exposure and could be used as biomarkers to predict risk of PAE. Results of the present study provided some support for this hypothesis.
Methods:
We conducted a series of studies to determine DNA methylation changes in stress regulatory genes proopiomelanocortin (POMC) and period 2 (PER2) using biological samples from three separate cohorts of patients i) pregnant women who consumed moderate to high levels of alcohol or low/unexposed controls, ii) children with PAE and non-alcohol exposed controls, and iii) children with PAE treated with or without choline.
Results:
We found pregnant women who consumed moderate to high levels of alcohol and gave birth to PAE children had higher DNA methylation of POMC and PER2. PAE children also had increased methylation of POMC and PER2. The differences in the gene methylation of PER2 and POMC between PAE and controls did not differ by maternal smoking status. PAE children had increased levels of stress hormone cortisol and adrenocorticotropic hormone (ACTH). Choline supplementation reduced DNA hypermethylation and increased expression of POMC and PER2 in children with PAE.
Conclusions:
These data suggest that PAE significantly elevates DNA methylation of POMC and PER2 and increases levels of stress hormones. Furthermore, these results suggest the possibility that measuring DNA methylation levels of PER2 and POMC in biological samples from pregnant women or from children may be useful for identification of a woman or a child with PAE.
Introduction
Despite a number of research reports showing evidence that consuming alcohol during pregnancy is harmful for the developing fetus, more than one in ten women reports alcohol use during pregnancy, many of these women report heavy episodic or binge drinking (Floyd and Sidhu, 2004; Tan et al., 2015). This latter group is at highest risk for having a child with FASD, which can manifest as growth impairment, abnormal facial features, neurobehavioral deficit, or combination of these characteristics. The prevalence of FASD has been estimated to be as high as 24–48 per 1,000 live births in the United States (May et al., 2014). FASD is recurrent within families and incidence of these disorders is about 771 per 1000 in younger siblings of affected children (Abel, 1988). Early detection and rearing in a caring environment has been shown to reduce the severity of impairment (Streissguth et al., 2004). However, in the absence of the completely expressed syndrome, early diagnosis is not easy and would be aided by objective measures that identify risk of prenatal alcohol exposure (PAE) (Bakhireva and Savage, 2011). Given the limitations of self-report, the development of sensitive and reliable measures to detect PAE could be a useful tool in better identification of children with FASD and would increase the opportunities for earlier interventions. Epigenetic-based tools have never been applied thus far to detect ethanol inducible variants in pregnant women and FASD offspring. Epigenetic modifications include DNA methylation, histone modifications and chromatin folding. Epigenetic modifications of a gene have been shown to play a role in maintaining a long-lasting change in gene expression (Govorko et al., 2012; Finegersh et al., 2015).
As stress and circadian physiological systems governing many body functions are often disregulated in alcohol dependent patients and in FASD childern, we sought to test whether epigenetic changes of genes critical for stress and circadian regulation may serve as measures of PAE risk. Research using laboratory animals has shown that chronic alcohol feeding affects expression changes of two genes, Pomc and Per2 in the hypothalamus, important regulators of stress and circadian systems in the adult (Reppert, 2000; Raffin-Sanson et al., 2003; Sarkar, 2012). It has been shwon that the expression levels of Pomc and Per2 genes are reduced and the corticosterone response to a stress challenge is increased in both alcohol dependent animals and in PAE animals (Chen et al., 2006; Agapito et al., 2014). Also shown is that the hyperstress response of PAE animals is reduced following implantation of POMC neurons in the hypothalamus (Sarkar et al., 2008; Logan et al., 2015). Additionally, Per2 genes deletion has been shown to prevent PAE effects on Pomc expression and alter the rhythmic changes in corticosterone levels in the blood (Agapito et al., 2010, 2014). These data suggest that interactions exist between the circadian system, the hypothalamic–pituitary–adrenal axis, and alcohol consumption. We have recently shown that binge or heavy levels of alcohol drinking increases DNA methylation and reduces gene expression of POMC and PER2 in adult human subjects (Gangisetty et al., 2019). One possibility would be that methylation of these two genes is consistently associated with alcohol exposure and could be used as biomarkers to predict risk of PAE. Hence, we determined DNA methylation changes in POMC and PER2 in biological samples of pregnant women who subsequently gave birth to children with or without PAE, and in children with PAE and non-alcohol exposed controls. In order to establish a correlative evidence between POMC and PER2 DNA methylation changes, we measured stress hormone levels in children with PAE and non-alcohol exposed controls. Furthermore, we evaluated the effects of choline supplementation, which is known to alter the epigenetic modification of gene (Kovacheva et al., 2007; Bekdash et al., 2013), on DNA methylation and gene expression of POMC and PER2 in children with PAE.
Methods
Human studies
Study 1:
To test whether DNA methylation of PER2 and POMC are altered by moderate to heavy alcohol use in pregnant women who gave birth to children with prenatal ethanol exposed (PAE; includes both FASD and non-FASD groups), we selected prospectively identified pregnant women from a larger longitudinal cohort study in two regions of Western Ukraine as part of the Collaborative Initiative on Fetal Alcohol Spectrum Disorders (CIFASD). The overall study was approved by the institutional review boards at the University of California San Diego and Lviv Medical University in Ukraine, and all methods were performed in accordance with the relevant guidelines and regulations. Informed consent was obtained from all participants. Pregnant women who reported frequent moderate to heavy drinking in the month around conception and/or in the most recent month of pregnancy and unexposed controls were recruited on average in mid-gestation. Nondrinking woman meeting screening criteria (i.e., no binge episodes, minimal or no alcohol in the month around conception, and no drinking in the most recent month of pregnancy) was recruited as a control. At the time of enrollment and again in the third trimester, mothers provided a blood sample. Infants born to these mothers were followed to one year of age and examined by a trained geneticist for physical features of FASD, growth as well as neurobehavioral impairment as measured using the Bayley Scales of Infant Development, version II. The criteria for classification of FASD have been described elsewhere (Hoyme et al., 2005; Jones et al., 2006). The sample of women selected for this analysis represented moderate to heavily exposed mothers with a FASD-affected or unaffected child; and control mothers (i.e., no binge episodes, minimal or no alcohol in the month around conception, and no drinking in the most recent month of pregnancy). The full protocol and study description are detailed in a previous publication (Chambers et al., 2014).
Study 2:
We measured changes in gene methylation in salivary samples from children with or without PAE. Samples were obtained from participants with PAE and control participants between the ages of 5−-18. History of PAE was obtained at the time of the interview or from review of study data. PAE diagnosis were assigned to the alcohol-exposed group if the mother drank >4 drinks per occasion at least once per week or >3 drinks per week during the pregnancy. Individuals whose mothers did not drink during pregnancy or who drank minimally (<1 drink per week on average and never more than 2 drinks on any one occasion during pregnancy) were designated as controls. The full protocol and study description were described previously (Mattson et al., 2010). Participants were asked to refrain from eating or drinking during the first 15 minutes of the study visit. Saliva was collected via the child expelling into a collection tube. Saliva samples were immediately frozen at −80C, and used for gene methylation or hormone measurement studies.
Study 3:
To study the effect of choline supplementation on PAE children, we conducted a double-blinded, placebo controlled, randomized trial of choline supplementation in PAE children with FASD. Details of participant characteristics and methodology have been previously described (Wozniak et al., 2013). NCT number for this study was 01149538 and recruitment dates were June, 2010 through May, 2014. Briefly, participants were children ages 2.5 to 5 years at enrollment who had been diagnosed with Fetal Alcohol Syndrome (FAS), Partial FAS, or Alcohol Related Neurodevelopmental Disorder (ARND). Participants were allocated in a 1:1 ratio to either 500 mg choline per day or matching placebo for 9 months. Blood samples were collected by venipuncture at baseline prior to allocation, at 6 months, and at 9 months (only the baseline and 9-month samples are described here). Blood samples were drawn directly into a purple-top tube containing EDTA. The tube was spun in a refrigerated centrifuge. Plasma was drawn off the sample for separate use. The buffy coat was then drawn off with a pipette into a separate plastic tube which was capped and labeled and immediately placed into a −80°C freezer. Buffy coats (PBMC) were sent in batches on dry ice to the lab at Rutgers’ University.
All participants of studies 2 and 3 underwent an Institutional Review Board (IRB)-approved informed consent process involving a parent or guardian as well as a separate assent process with the child for children over the age of 6. All study procedures were approved by the IRBs at each of the four sites (Emory University School of Medicine; University of Minnesota; University of California San Diego/San Diego State University; Children’s Hospital Los Angeles). Participants were compensated for their time.
DNA and RNA extraction, gene expression and DNA methylation measurements
RNA was extracted from buffy coat samples using total RNA purification kit (Norgen Biotek, Ontario, Canada) as per instructions from manufacturer. RNA extracted was treated with DNase to remove any residual DNA using RNase free DNase kit (Qiagen, Valencia, CA). About 1 μg of RNA was converted to first strand complementary DNA (cDNA) using high capacity cDNA reverse transcription kit (Life Technologies) as per instructions from manufacturer.
Real time quantitative PCR (RT-PCR)
POMC and PER2 mRNA levels were measured by quantitative real time PCR SYBR green assay. Primers used for real time PCR was designed using either from primer express 3.0 software (Applied Biosystem) or from literature reference (Wang et al., 2011, 2012). Sequences used for RT primer design were retrieved from NCBI genome browser; POMC (NM_001035256), PER2 (NM_022817), GAPDH (NM_002046), 18S rRNA (NR_145820), RPL19 (NM_000981). Primer sequences are listed in Table 1. The quantity of target genes (PER2, POMC) and three reference genes (GAPDH, 18S, RPL19) were measured using standard curve method. Target gene expression was normalized with the mean of three reference gene expression levels. RT-PCR was performed at 95°C for 5min followed by 40 cycles of 95°C for 15s, 60°C for 30s, 72°C for 40s in Applied Biosystems 7500 Real PCR system.
Table 1.
Showing the primer sequences using for gene methylation and mRNA expression studies.
| Primer Name | Sequence |
|---|---|
| hPER1 MFP | 5’ ATTTAGGTTTACGTGCGTTC 3’ |
| hPER1 MRP | 5’ CGACTCAAAAACGAAAATCG 3’ |
| hPER1 UMFP | 5’ TAGTATTAGTATTTAGGTTTATGTGTGTTT 3’ |
| hPER1 UMRP | 5’ AACAACAATCCAACTCAAAAACAAAAATCA 3’ |
| hPER2 MFP | 5’ GCGGTTTCGTTGCGGTTTAC 3’ |
| hPER2 MRP | 5’ GCCGACGCCGTTTCAAACCG 3’ |
| hPER2 UMFP | 5’ GTGGTGTGGTGTGGTTTTGTTGTGGTTTAT 3’ |
| hPER2 UMRP | 5’ ACACCCCCACACCAACACCATTTCAAACCA 3’ |
| hPOMC MFP | 5’ ACGGGGGTGTTAAGTTTTTC 3’ |
| hPOMC MRP | 5’ ACTATCCTACCGAAAACGCA 3’ |
| hPOMC UMFP | 5’ TATATGGGGGTGTTAAGTTTTTT 3’ |
| hPOMC UMRP | 5’ CACTATCCTACCAAAAACACACC 3’ |
| hPER2 BSP FP | 5’ GTGTGTTTTTGGTTTTGTTTTAGGT 3’ |
| hPER2 BSP RP | 5’-/5Bio/TTAATTTCCTTAACCTTCCCCATAC 3’ |
| hPER2 seq FP | 5’ GAGTGGTCGAGTCGCGCGTAG 3’ |
| hPOMC BSP FP | 5’GTTGGGGTTAAAAATAGGTGGT 3’ |
| hPOMC BSP RP | 5’-/5Bio/TCTATAAAAACCCTCCCTACCC 3’ |
| hPOMC seq FP | 5’GGAGTTGTTTTAGATTTT 3’ |
| hPER2 RT FP | 5’ TCCAGTGGACATGAGACCAA 3’ |
| hPER2 RT RP | 5’ CGCTACTGCAGCCACTTGTA 3’ |
| hPOMC RT FP | 5’ AAGCGCTACGGCGGTTTC 3’ |
| hPOMC RT RP | 5’ TGATGATGGCGTTTTTGAACA 3’ |
| hGAPDH RT FP | 5’ AACCTGCCAAATATGATGAC 3’ |
| hGAPDH RT RP | 5’ ATACCAGGAAATGAGCTTGA 3’ |
| h18S RT FP | 5’ AAACGGCTACCACATCCAAG 3’ |
| h18S RT RP | 5’ CCTCCAATGGATCCTCGTTA 3’ |
| hRPL19 RT FP | 5’ ATCGATCGCCACATGTATCA 3’ |
| hRPL19 RT RP | 5’ GCGTGCTTCCTTGGTCTTAG 3’ |
Forward primer (FP), Reverse primer (RP), Methyl forward primer (MFP), Methyl reverse primer (MRP), Un methyl forward primer (UMFP), Un methyl reverse primer (UMRP).
We used NCBI genome browser to retrieve sequences and used primer express 3.0 software to design primers for Real time PCR. POMC primers we used in our study are from exon 4 which codes for all functional peptides and primers targeted to exon 4 have been used in RT-PCR assay for POMC previously (Shobatake et al., 2018). Exon 1 and 2 are untranslated regions and exon 3 codes for signal peptide (Wang et al., 2012). We always ran the PCR product amplified with a given set of primers on an agarose gel and checked the transcript size before performing the real time PCR to avoid any non-specific amplification. This rules out the possibility of amplifying intronic regions even if primers are from same exonic regions. We also used silicon-based membrane columns in our RNA extraction protocols that efficiently removed most of the DNA. We further employed DNase treatment before converting RNA to cDNA to remove any traces of genomic DNA in the sample. The amplified products in our RT-PCR assay are highly RNA specific.
Methylation-specific real time PCR (MSP) assay for DNA methylation
DNA was extracted from blood and PBMC samples using DNeasy blood and tissue extraction kit (Qiagen, Valencia, CA). DNA samples (2 μg) extracted from saliva, blood or PBMC were bisulfite converted using EZ DNA methylation kit (Zymo Research, Orange, CA). DNA methylation analyses for POMC, PER1 and PER2 were performed by SYBR green real time PCR method of methylation specific PCR (MSPR) using primer sets spanning different CpG sites of POMC, PER1 and PER2 promoters, as described previously (Gangisetty et al., 2014; Govrko et al., 2012; Shih et al., 2006). Promoter sequences were analyzed for CpG island determination and MSP primers were designed using methyl primer sequence v1.0 program or from literature reference (Shih et al., 2006). Primer sequences specific for methylated and unmethylated DNA are listed in Table 1. Human highly methylated and low methylated control DNA (Epigen DX, Worcester MA) were subjected to bisulfite conversion and were used for preparing the standard curve. Real time PCR was performed using SYBR green master mix with specific primers and bisulfite converted DNA as template. RT PCR was performed using a program at 95°C for 5 min, 50 cycles of 95°C for 30sec, 60°C for 1min, and 72°C for 1 min. Relative quantities of methylated and unmethylated DNA were calculated using a standard curve and the ratios of methylated versus unmethylated DNA was determined. Relative DNA methylation level was calculated by normalizing the PAE value with the average value of controls and presented in figures and the text. All samples were run in duplicate.
DNA methylation by Pyrosequencing
The level of DNA methylation at specific CpG site was quantitated by pyrosequencing assay. Briefly 1ug of genomic DNA was subjected to bisulfite conversion using EZ DNA methylation kit (Zymo Research, Orange, CA). Region of interest was amplified using bisulfite specific PCR (BSP) primers. Pyromark PCR kit (Qiagen, Valencia, CA) was used employing forward and biotin labelled reverse primers as per instructions of the manufacturer. Biotinylated PCR product was mixed with streptavidin beads and annealed with sequencing primer. Streptavidin bound biotinylated PCR product was captured using a vacuum filtration sample transfer device (Qiagen, Valencia, CA). Sequencing was performed using Pyromark Gold Q96 CDT reagents (Qiagen, Valencia, CA) on a PSQ HS96A model pyrosequencing machine (Qiagen, Valencia, CA) as per the instructions from the manufacturer. In the pyrosequencing study, we analyzed one control C in non CpG background for efficient C→T conversion. The percent C remaining as C in the target CpG was considered % methylation.
ELISA assays
Human salivary hormone levels were measured using ELISA. For ACTH, we used Human ACTH ELISA Kit (Cloud-Clone Corp, Katy, TX; cat# CEA836Hu) and for cortisol we used Human Cortisol ELISA Kit (Salimetrics, Carlsbad, CA; Cat# 1–3002, Lot# 1502504). These assays were performed following the instruction provided in the kits. All samples were measured in duplicate.
Data Analysis
Data were analyzed using Prism 5.0 (GraphPad Software). The data shown in the figures are mean ± SEM. The data population was first tested for normality using D’Agostino & Pearson omnibus normality test for two groups or Bartlett’s test for equal variance for multiple groups. If the variance differ we used non-parametric test. Differences between two groups with significant variances were compared using Mann-Whitney test (two tailed p values were used). Differences between multiple treatment groups with non-parametric values were assessed using Kruskal-Wallis statistics followed by Dunn’s Multiple Comparison posttest. Differences between multiple treatment groups with parametric values were assessed with one-way analysis of variance (ANOVA) followed by Newman Keuls posttest. P<0.05 was considered significant.
Results
Identification of alcohol epigenetic marks in mother and child
In the first study, we employed blood DNA samples obtained on average in the second trimester from pregnant women enrolled at two sites in Ukraine in a cohort study as part of the CIFASD. Samples were collected from women who reported consuming moderate to heavy amounts of alcohol for at least some part of pregnancy and controls (Chambers et al., 2014; Coles et al., 2015). Demographic information on the overall sample of women has been described previously (Balaraman et al., 2016). DNA methylation levels of PER2 and POMC were measured using MSP assay and represented in Fig 1. Schematic diagram representing hPER2 and hPOMC promoter CpG islands were shown in Fig. 2A and 2D The DNA methylation level of PER2 was significantly elevated in women who reported moderate to heavy alcohol consumption compared to controls (Fig. 1B:Mann-Whitney U test; U=39, n1=11, n2=19, p=0.0052). POMC DNA methylation was also increased in alcohol-exposed women when compared to controls (Fig. 1D; U=12, n1=11, n2=19, p=0.0001).
Figure. 1.
Changes in PER2 and POMC gene methylation in pregnanat women who gave birth to children with PAE. A, C. Gene methylation changes in blood DNA were evaluated for two groups of mothers: a) women who reported low or no alcohol consumption in pregnancy (Control), b) women who reported frequent moderate to heavy drinking and had a child with ethanol exposed (PAE); PER2 DNA methylation (A), POMC DNA methylation (B). Number of samples in each group is shown between brackets under the group heading on the X axis. Differences between groups are shown by lines with p values on the top of bar graphs.
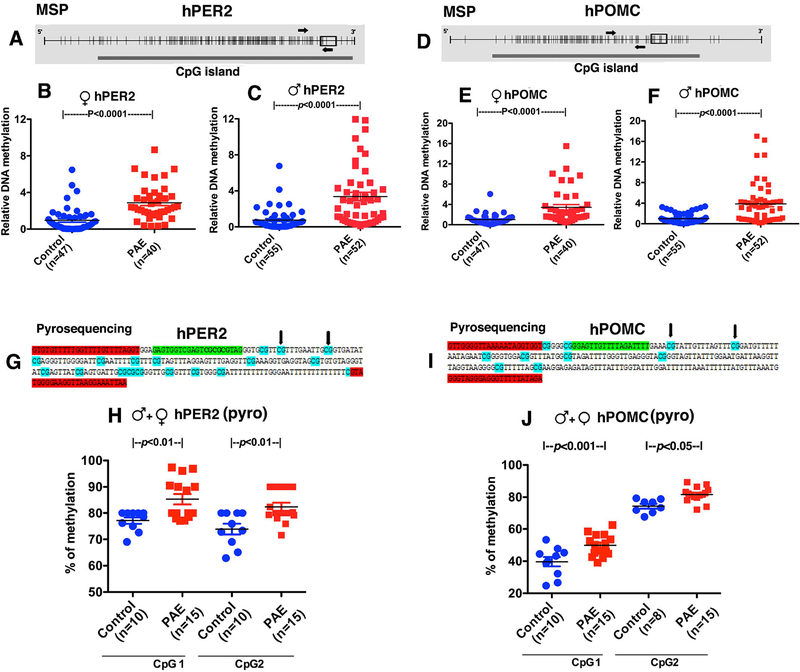
Figure. 2.
Changes in PER2 and POMC gene methylation as determined by methylation specific RT-PCR (MSP) (A-F) and pyrosequencing (G, J) in children with PAE. A-F. Schematic diagrams representing hPER2 and hPOMC promoter CpG islands are shown in Fig. 2A and 2D. Each small vertical line represents a single CpG. The thick solid bar below represents a CpG island. Small rectangular open box shows the location of exon1 in the sequence. MSP primers were represented with arrows covering about 15 CpGs for PER2 in Fig. 2A and 9 CpGs for POMC in Fig. 2D. Gene methylation changes in salivary DNA samples were evaluated by first by MSP in male and female children 5–18 years old with prenatal alcohol exposed (PAE) and controls; female patients PER2 (B) and POMC (E), male patients PER2 (C) and POMC (F). Some of the same samples from male and female PAE and control children were assayed by pyrosequencing for determination of methylation changes at two CpG sites of PER2 (H) and POMC (J). Schematic diagrams representing hPER2 (A), hPOMC (D) promoter CpG islands targeted in pyrosequencing assays. The pyrosequencing primers were designed to target two different CpGs for PER2 (G) and two for POMC (I). Bisulfite sequencing primers used were highlighted in red. Sequencing primer is highlighted in light green. Each CpG in the sequence is highlighted in turquoise. The two CpGs whose methylation measured was represented with black arrow. Differences between groups are shown by lines with p values on the top of bar graphs.
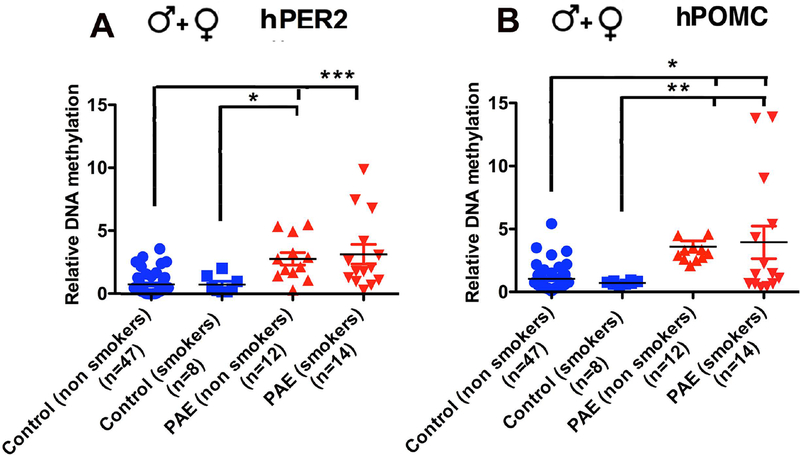
Biological samples of the children of mothers of Ukraine cohort were not available for this study. However, we were able to obtain salivary samples from children ages of 5–18 years with PAE andcontrols from a cross sectional study conducted by CIFASD investigators in the US. DNA methylation levels of PER2 and POMC were measured using MSP assay first. Schematic diagrams representing hPER2 and hPOMC promoter CpG islands were shown in Fig. 2A and 2D. We found that PER2 and POMC DNA methylation levels were higher in both male and female PAE subjects, as compared to control subjects (Fig. 2B, hPER2-female, U=283, n1=47, n2=40, p=0.0001; Fig. 2C, hPER2-male, U=512, n1=55, n2=52, p=0.0001; Fig. 2E, hPOMC-female, U=308, n1=47, n2=40, p=0.0001; Fig. 2F, hPOMC-male, U=570, n1=55, n2=52, p=0.0001). Further validation of gene methylation changes in PAE subjects was accomplished by measuring the methylation level of each of the genes at single CpG resolution using pyrosequencing assays. Schematic diagrams show the two CpGs for PER2 (2G) and two CpGs for POMC (Fig. 2I) whose methylation levels were measured. We found increased methylation in promoters of both PER2 and POMC DNA at two CpG sites (Fig. 2H, J; data were analyzed using one way ANOVA and Newman-Keuls posttest; hPER2 - F (3, 46) = 7.93, p<0.0002, CpG1- control vs PAE p<0.01, CpG2 - control vs PAE p<0.01; hPOMC - F (3, 44) = 113.1, p<0.0001, CpG1- control vs PAE p<0.001, CpG2- control vs PAE p<0.05). The alcohol effect on gene methylation of PER2 appears to have some specificity, since PER2 (Fig. 2) and not PER1 gene (both PER1 and PER2 belongs to the period family of circadian genes) methylation (female- control, 1.00 ± 0.157, PAE, 0.726 ± 0.170, U=654, n1=53, n2=31, two-tailed p=0.1216; male- control, 0.995 ± 0.211, PAE, 0.762 ± 0.171, U=1529, n1=62, n2=51, two-tailed p=0.7664) differs between PAE and control subjects. Additionally, we found that differences in PER2 and POMC DNA methylation between PAE and controls did not differ by maternal smoking status (Fig. 3; data were analyzed by Kruskal-Wallis statistics followed by Dunn’s Multiple Comparison posttest; hPER2- chi-squared = 27.40 with 3 d.f. p<0.0001; hPOMC - chi-squared = 22.15 with 3 d.f. p<0.0001; between group differences are shown on the figure). These data suggest that PAE makes epigenetic changes on PER2 and POMC resulted from maternal exposure of ethanol during pregnancy. Additionally, these data identify that maternal tobacco use did not influence alcohol epigenetic marks on these genes.
Figure 3.
PER2 (A) and POMC (B) gene methylation changes in PAE children (both male and female) were not influenced by their mother smoking habits. Patients population were similar to those in Fig. 2. Data are represented as Mean ± SEM. Number of samples in each group is shown between brackets under the group heading on the X axis. No significant differences between smokers and nonsmokers are observed. Significant differences between control and PAE are found and are shown by lines with p values
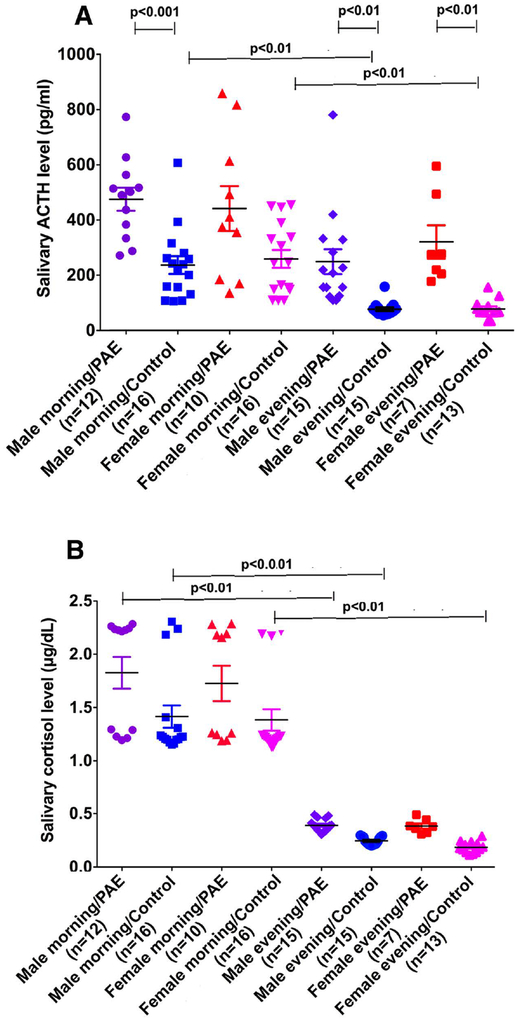
We measured stress hormones adrenocorticotropic hormone (ACTH) and cortisol levels in saliva collected under basal conditions from a cohort of PAE children. Since ACTH and cortisol are released in a circadian pattern, we measured their levels in the morning and in the evening. Figure 4A and B shows that both ACTH and cortisol levels had circadian variations. PAE children had higher levels of ACTH both in the morning and in the evening (chi-squared = 67.64 with 7 d.f. p<0.0001; between group differences are shown on the figure), although morning ACTH levels of PAE female children were not significantly different from the controls due to large sample variations. Mean plasma cortisol levels of PAE were higher in both male and females at all time points (chi-squared = 89.69 with 7 d.f. p<0.0001; between group fifferences are shown on the figure), but were not significantly different from control groups due to large sample variations and small sample numbers. These data identify an association between POMC and PER2 DNA methylation and increased stress hormones levels in saliva and also suggest that moderate to heavy alcohol use during pregnancy induces epigenetic marks to reduce POMC and PER2 gene expression that persist in PAE children.
Fig. 4.
Circadian levels of salivary ACTH (A) and cortisol (B). Number of samples in each group is shown between brackets under the group heading on the X axis. Data are represented as Mean ± SEM. Number of samples in each group is shown between brackets under the group heading on the X axis. Differences between groups are shown by lines with p values on the top of bar graphs.
Choline treatment to reverse alcohol epigenetic marks in children with fetal alcohol spectrum disorders
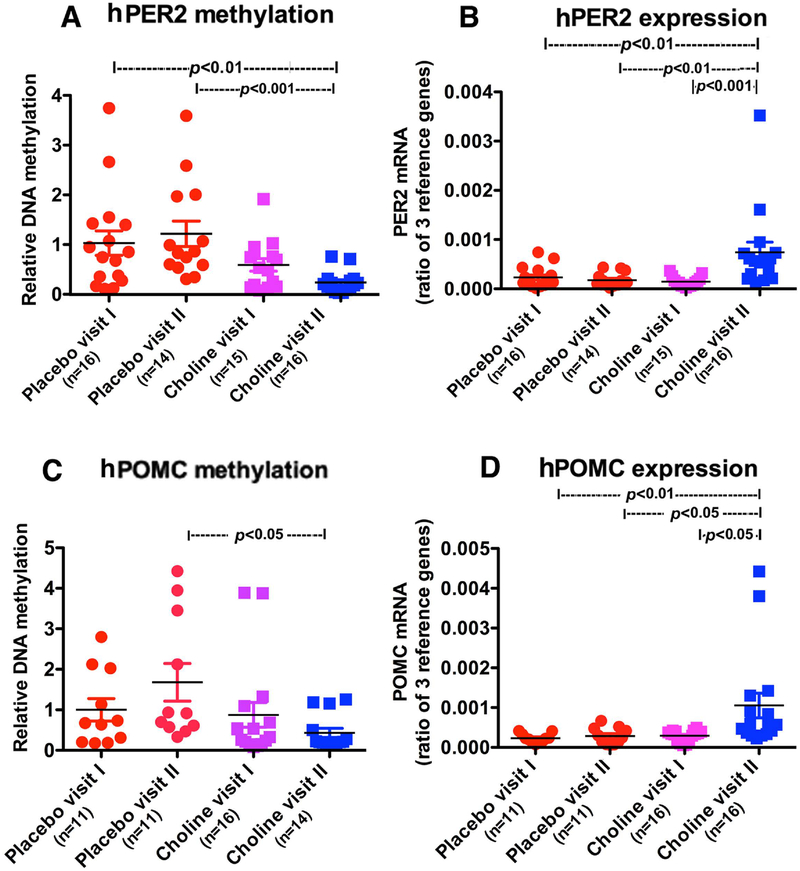
Recently, choline a nutritional supplement known to alter DNA methylation (Niculescu et al., 2006; Kovacheva et al., 2007) has been shown to attenuate some of the PAE effects on learning and memory (Thomas et al., 2010). Choline also alters DNA methylation of POMC and reduces stress axis abnormalities in animal models of PAE (Bekdash et al., 2013). Hence, we measured DNA methylation changes of PER2 and POMC in children (age 2 to 5) with PAE and a diagnosis of FASD who participated in an intervention trial with choline or placebo treatment for 9 months at the University of Minnesota. We obtained blood samples at basal (visit 1), and 9 months (visit II) of choline or placebo treatments. This patient cohort showed reduction in both PER2 (Fig. 5A) and POMC DNA (Fig. 5C) methylation following 9 months of choline treatment (as compared to placebo but not choline Visit 1) when the gene expression values of PER2 (Fig. 5B) and POMC (Fig. 5D) were elevated (hPER2, methylation, chi-squared = 20.02 with 3 d.f. p<0.0002; hPER2 mRNA, chi-squared = 20.02 with 3 d.f. p<0.0002; hPOMC methylation, chi-squared = 10.37 with 3 d.f., p<0.0157; hPOMC mRNA, chi-squared = 16.26 with 3 d.f., p<0.001; between group differences are shown on the figure). These data provide additional support for the concept that PAE makes epigenetic marks on PER2 and POMC genes, and further identify a possible therapeutic value of choline supplementation in reversing alcohol epigenetic marks on stress regulatory genes in children with FASD.
Figure 5.
Effects of a double-blinded, placebo controlled, randomized trial of choline supplementation on PER2 and POMC gene methylation and expression in children with fetal alcohol spectrum disorders (FASD). Participant children with FASD were treated with either 500 mg choline per day or matching placebo for 9 months. A-D. PER 2 DNA methylation (A) and expression (B) and POMC DNA methylation (C) and expression (D). Data are represented as Mean ± SEM. Number of samples in each group is shown between brackets under the group heading on the X axis. Differences between groups are shown by lines with p values on the top of bar graphs.
Discussion
Using different experimental paradigms and subject samples, these data provide evidence that moderate to high amount of alcohol drinking during pregnancy makes epigenetic marks on POMC and PER2 genes that persists in children during the development. Although we did not have samples from children with PAE collected longitudinally from gestation, these data suggest that alcohol exposure in human gestation can be recorded as persistent changes in epigenetic information of key stress regulatory genes in the offspring. Alcohol effects on POMC and PER2 genes in children were not influenced by maternal tobacco use during pregnancy. POMC is a precursor gene known to produce various peptide hormones including β-endorphin, α-melanocyte-stimulating hormone (α-MSH) and ACTH. These hormones regulate many body functions including stress hormone release (Raffin-Sanson et al., 2010). Alcohol also increased DNA methylation of PER2. The PER2 gene is noted for its major role in circadian rhythm. This gene is part of a number of circadian genes that interact with each other to make up an auto-regulatory feedback loop, in which its activation and repression cycle control the body central clock in the hypothalamus and the cellular clock in all cells (Reppert, 2000). We found alcohol makes its epigenetic marks on PER2 gene but not PER1 gene. Whether or not alcohol makes epigenetic marks on other circadian genes needs further investigation. It is not apparent why alcohol alters PER2 but not PER1 DNA methylation. In this regard it is interesting to note that Per2 mutant (Per2Brdm1) mice display enhanced alcohol consumption and preference (Spanagel et al., 2005), whereas Per1Brdm1 mutant mice do not show such an enhancement in alcohol drinking behavior (Zghoul et al., 2007). Also, alcoholics with a specific set of polymorphisms in the Per2 gene consume less alcohol than alcoholics without the polymorphisms (Spanagel et al., 2005a,b). PER2 gene methylation and transcriptional repression may not only affect the circadian clock mechanism but also control specific cellular responses. For example, it has been shown that mutation of mPer2 causes a loss of alcohol’s stimulatory effect on Pomc expression (Agapito et al., 2004). PAE effects on circadian expression of Pomc (Chen et al., 2006) and corticosterone rhythm (Yang et al., 2009) have been reported. Thus, it appears that the PER2 gene is involved in the control of POMC and their cellular responses. We observed an increased level of DNA methylation of POMC in biological fluid samples of PAE children is associated with increased ACTH and cortisol levels. These data are consistent with the data in rodent in which PAE increased Pomc gene methylation in the hypothalamus and elevated ACTH and cortisol levels in plasma (Govorko et al., 2012). Animal studies have connected the stress axis functional abnormalities in PAE with alcohol-induced epigenetic modification of PER2 and POMC genes (Boyadjieva et al., 2009; Logan et al., 2015; Gangisetty et al., 2014).
Animal studies identified that alcohol induced epigenetic marks involve methylation of Pomc gene via suppression of H3 lysine 9 acetylation, activation of HDACs (e.g., HDAC1), and various methylation promoting genes (G9a, Setdb1). Dnmt1 and Mbd1/MeCP2 assist in spreading the silencing signal (Govorko et al., 2012; Beakdash et al., 2013). Although, the mechanism by which alcohol epigenetically modifies Per2 gene has yet to be established, similar mechanisms may also be involved in alcohol-induced alteration of this gene methylation.
We showed here that choline supplementation in children with an FASD reduced the DNA methylation and increased the expression levels of PER2 and POMC. Our data in children with an FASD are in agreement with the rodent study where choline supplementation during the developmental period normalized PAE-increased global DNA methylation in the hippocampus and prefrontal cortex (Balaraman et al., 2017) and Pomc gene methylation in the hypothalamus (Beakdash et al., 2013). However, it should be mentioned the manner in which choline impacts methylation has varied across studies (Niculescu et al., 2006; Kovacheva et al., 2007; Balaraman et al., 2016). The variability of DNA methylation changes due to choline availability may have been the result of the initial state of the animals as was observed in a rodent study and some extent in the present study where choline supplementation led to opposite changes in DNA methylation in PAE and control subjects. This raises the question is whether choline is reversing gene-specific effects of alcohol or whether it impacted genes differ. For example, it has been shown that choline supplementation alters the levels of histone deacetylases, DNA methyl transferases and DNA binding protein MeCP2 to decrease Pomc gene methylation in the hypothalamus in PAE rats (Beakdash et al., 2013). Choline supplementation also has been shown to normalize disturbances in microRNAs (miRNAs) expression following developmental alcohol exposure and can protect specific miRNAs from induction by ethanol (Balaraman et al., 2016). Further research is required to define the mechanism by which choline affect PAE-induced POMC and PER2 gene methylation changes. Choline may also contribute to the etiology of stress-related disorders and age-related decline in memory later in life (Kovacheva et al., 2007; Bekdash, 2016). Rodent studies suggest that choline supplementation enhances performance in memory-related tasks during adulthood in prenatal alcohol exposed rats (Schneider and Thomas, 2016; Idrus et al., 2017). Studies are currently ongoing to determine if children with an FASD who participated in the choline supplementation treatment improve their performance in memory-related tasks and stress coping behaviors.
We have used methylation specific PCR (MSP) assay for measurement of promoter gene methylation. MSP determines methylation patterns of CpG island regions of the gene and is widely used to analyze promoter specific methylation of genes from tissues and biological fluids (Yoshioka et al., 2018; Husseiny et al., 2012; Mehta et al., 2014; Parrella et al., 2009). We have previously used this method to detect Pomc promoter methylation in various tissues of fetal alcohol exposed rats (Govorko et al., 2012; Gangisetty et al., 2014; Bekdash et al., 2013). This method detects the relative quantity of methyl DNA and unmethyl DNA in two different PCR reactions using highly methylated DNA as standard for methyl DNA and low methylated DNA as standard for unmethyl DNA. Although this method does not provide information at single CpG resolution, it gives relative quantification of methylation of a number of CpGs within the PCR product regions. This is because the primers used are specific to methyl and unmethyl DNAs. Furthermore, to validate the data obtained from MSP assays, we have conducted pyrosequencing measurements of POMC and PER2 genes promoter methylations in some of the same samples used in MSP assays. Pyrosequencing assay determines methylation at single CpG resolution. Using pyrosequencing assays, we analyzed two CpGs within a CpG island on the gene promoter and found similar increase in DNA methylation of POMC and PER2 genes in PAE compared to control as we found using MSP assays (Fig. 2).
We anticipate that the alcohol effect on POMC gene methylation is not cell specific. This is because studies conducted in laboratory animals revealed that alcohol increases POMC gene methylation in the hypothalamus, pituitary gland and in sperm samples (Govorko et al., 2012; Bekdash et al., 2013; Gangisetty et al., 2014). In human, alcohol also increases POMC gene methylation in peripheral blood (Muschler et al., 2010; Zhang et al., 2013; Gangisetty et al., 2019; Figs. 1 and 5) and in saliva samples (Fig. 2).
Recently, a number of studies have been conducted to determine the DNA methylation signature of human PAE by determining the genome-wide DNA methylation patterns of buccal epithelial cells in PAE and control children. However, these studies were conducted at a single timepoint and the identified changes in genes of PAE children were not verified in mothers who gave birth to PAE children. The current study presents the first example of an association between binge and high drinking levels and a change in DNA methylation of a specific group of genes that persist both in mother and children. Diseases that have been associated with gestational exposure to ethanol and related POMC and PER2 gene hypermethylation, such as sleep apnea (Chen et al., 2012), depression (Turner et al., 2015), anxiety (Wengel et al., 2011), stress hyperresponse (Keiver et al., 2014), and alcohol abuse (Kendler et al., 2013) are of particular interest in this respect for children and adults with FASD. In summary, this study identified stable epigenetic marks on two important genes regulating various biological functions that may serve as markers of ethanol consumption in pregnant women. Epigenetic epidemiologic studies may need to be conducted to establish a connection between the epigenetic modifications of these groups of genes and FASD related diseases. Also, understanding how epigenetic control is modified by early ethanol exposure may shed light on the link between development and health over the lifespan and ultimately suggest new ways to prevent FASD and other alcohol related diseases.
Acknowledgements
The authors thank Shaima Jabbar for technical assistance in the ELISA assays for stress hormones. This work was supported by National Institutes of Health Grants U24 AA014811, U01 AA014835, U01AA014809, R21AA019580, R33AA019580. We are grateful to the OMNI-Net Birth Defects Prevention Program and to the women and their children who participated in this study. We also acknowledge the Collaborative Initiative on Fetal Alcohol Spectrum Disorders (CIFASD.org) for intellectual and administrative support. All authors reported no financial interests or potential conflicts of interest.
Reference
- Agapito M, Mian N, Boyadjieva NI, Sarkar DK (2010) Period 2 gene deletion abolishes beta-endorphin neuronal response to ethanol. Alcohol Clin Exp Res 34:1613–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agapito MA, Zhang C, Murugan S, Sarkar DK (2014) Fetal alcohol exposure disrupts metabolic signaling in hypothalamic proopiomelanocortin neurons via a circadian mechanism in male mice. Endocrinology 155:2578–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhireva LN, Savage DD (2011) Focus on: biomarkers of fetal alcohol exposure and fetal alcohol effects. Alcohol Res Health 34:56–63. [PMC free article] [PubMed] [Google Scholar]
- Balaraman S, Schafer JJ, Tseng AM, Wertelecki W, Yevtushok L, Zymak-Zakutnya N, Chambers CD, Miranda RC (2016) Plasma miRNA profiles in pregnant women predict infant outcomes following prenatal alcohol exposure. PLoS One 11:e0165081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaraman S, Idrus NM, Miranda RC, Thomas JD (2017) Postnatal choline supplementation selectively attenuates hippocampal microRNA alterations associated with developmental alcohol exposure. Alcohol 60:159–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekdash RA, Zhang C, Sarkar DK (2013) Gestational choline supplementation normalized fetal alcohol-induced alterations in histone modifications, DNA methylation, and proopiomelanocortin (POMC) gene expression in β-endorphin-producing POMC neurons of the hypothalamus. Alcohol Clin Exp Res 37:1133–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekdash RA (2016) Choline and the brain: an epigenetic perspective. Adv Neurobiol 12:381–399. [DOI] [PubMed] [Google Scholar]
- Boyadjieva NI, Ortigüela M, Arjona A, Cheng X, Sarkar DK (2009) Beta-endorphin neuronal cell transplant reduces corticotropin releasing hormone hyperresponse to lipopolysaccharide and eliminates natural killer cell functional deficiencies in fetal alcohol exposed rats. Alcohol Clin Exp Res 33:931–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers CD, Yevtushok L, Zymak-Zakutnya N, Korzhynskyy Y, Ostapchuk L, Akhmedzhanova D, Chan PH, Xu R, Wertelecki W (2014) Prevalence and predictors of maternal alcohol consumption in 2 regions of Ukraine. Alcohol Clin Exp Res 38:1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CP, Kuhn P, Advis JP, Sarkar DK (2006) Prenatal ethanol exposure alters the expression of period genes governing the circadian function of beta-endorphin neurons in the hypothalamus. J Neurochem 97:1026–1033. [DOI] [PubMed] [Google Scholar]
- Chen ML, Olson HC, Picciano JF, Starr JR, Owens J (2012) Sleep problems in children with fetal alcohol spectrum disorders. J Clin Sleep Med 8:421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles CD, Kable JA, Keen CL, Jones KL, Wertelecki W, Granovska IV, Pashtepa AO, Chambers CD (2015) Dose and timing of prenatal alcohol exposure and maternal nutritional supplements: developmental effects on 6-month-old infants. Matern Child Health J 19:2605–2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finegersh A, Rompala GR, Martin DI, Homanics GE (2015) Drinking beyond a lifetime: New and emerging insights into paternal alcohol exposure on subsequent generations. Alcohol. 49:461–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd RL, Sidhu JS (2004) Monitoring prenatal alcohol exposure. Am J Med Genet C Semin Med Genet 127C:3–9. [DOI] [PubMed] [Google Scholar]
- Gangisetty O, Bekdash R, Maglakelidze G, Sarkar DK (2014) Fetal alcohol exposure alters proopiomelanocortin gene expression and hypothalamic-pituitary-adrenal axis function via increasing MeCP2 expression in the hypothalamus. PLoS One 9:e113228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangisetty O, Sinha R, Sarkar DK (2019) Hypermethylation of proopiomelanocortin and period 2 genes are associated with greater subjective and behavioral motivation for alcohol in humans. Alcohol Clin Exp Res. 43 (2) : 212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianoulakis C, de Waele JP, Thavundayil J (1996) Implication of the endogenous opioid system in excessive ethanol consumption. Alcohol 13:19–23. [DOI] [PubMed] [Google Scholar]
- Govorko D, Bekdash RA, Zhang C, Sarkar DK (2012) Male germline transmits fetal alcohol adverse effect on hypothalamic proopiomelanocortin gene across generations. Biol Psychiatry 72:378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyme HE, May PA, Kalberg WO, Kodituwakku P, Gossage JP, Trujillo PM, Buckley DG, Miller JH, Aragon AS, Khaole N, Viljoen DL, Jones KL, Robinson LK (2005) A practical clinical approach to diagnosis of fetal alcohol spectrum disorders: clarification of the 1996 institute of medicine criteria. Pediatrics. 115:39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husseiny MI, Kuroda A, Kaye AN, Nair I, Kandeel F, Ferreri K (2012) Development of a quantitative methylation-specific polymerase chain reaction method for monitoring beta cell death in type 1 diabetes. PLoS One 7:e47942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idrus NM, Breit KR, Thomas JD (2017) Dietary choline levels modify the effects of prenatal alcohol exposure in rats. Neurotoxicol Teratol. 59:43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KL, Robinson LK, Bakhireva LN, Marintcheva G, Storojev V, Strahova A, Sergeevskaya S, Budantseva S, Mattson SN, Riley EP, Chambers CD (2006) Accuracy of the diagnosis of physical features of fetal alcohol syndrome by pediatricians after specialized training. Pediatrics 118:e1734–1738. [DOI] [PubMed] [Google Scholar]
- Keiver K, Bertram CP, Orr AP, Clarren S (2015) Salivary cortisol levels are elevated in the afternoon and at bedtime in children with prenatal alcohol exposure. Alcohol 49:79–87. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Gardner CO, Edwards A, Hickman M, Heron J, Macleod J, Lewis G, Dick DM (2013) Dimensions of parental alcohol use/problems and offspring temperament, externalizing behaviors, and alcohol use/problems. Alcohol Clin Exp Res 37:2118–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacheva VP, Mellott TJ, Davison JM, Wagner N, Lopez-Coviella I, Schnitzler AC, Blusztajn JK (2007) Gestational choline deficiency causes global and Igf2 gene DNA hypermethylation by up-regulation of Dnmt1 expression. J Biol Chem 282:31777–31788. [DOI] [PubMed] [Google Scholar]
- Logan RW, Wynne O, Maglakelidze G, Zhang C, O’Connell S, Boyadjieva NI, Sarkar DK (2015) β-Endorphin neuronal transplantation into the hypothalamus alters anxiety-like behaviors in prenatal alcohol-exposed rats and alcohol-non-preferring and alcohol-preferring rats. Alcohol Clin Exp Res 39:146–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson SN, Foroud T, Sowell ER, Jones KL, Coles CD, Fagerlund A, Autti-RämöI, May PA, Adnams CM, Konovalova V, Wetherill L, Arenson AD, Barnett WK, Riley EP (2010) Collaborative initiative on fetal alcohol spectrum disorders: methodology of clinical projects. Alcohol 44:635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May PA, May PA, Baete A, Russo J, Elliott AJ, Blankenship J, Kalberg WO, Buckley D, Brooks M, Hasken J, Abdul-Rahman O, Adam MP, Robinson LK, Manning M, Hoyme HE (2014) Prevalence and characteristics of fetal alcohol spectrum disorders. Pediatrics 134:855–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A, Mielnik A, Schlegel PN, Paduch DA (2014) Novel methylation specific real-time PCR test for the diagnosis of Klinefelter syndrome. Asian J Androl. 16:684–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschler MA, Hillemacher T, Kraus C, Kornhuber J, Bleich S, Frieling H (2010) DNA methylation of the POMC gene promoter is associated with craving in alcohol dependence. J Neural Transm (Vienna). 117:513–519; [DOI] [PubMed] [Google Scholar]
- Niculescu MD, Craciunescu CN, Zeisel SH (2006) Dietary choline deficiency alters global and gene-specific DNA methylation in the developing hippocampus of mouse fetal brains. FASEB J 20:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrella P, la Torre A, Copetti M, Valori VM, Barbano R, Notarangelo A, Bisceglia M, Gallo AP, Balsamo T, Poeta ML, Carella M, Catapano D, Parisi S, Dallapiccola B, Maiello E, D’Angelo V, Fazio VM (2009) High specificity of quantitative methylation-specific PCR analysis for MGMT promoter hypermethylation detection in gliomas. J Biomed Biotechnol. 2009:531692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotsky PM (1986) Opioid inhibition of immunoreactive corticotropin-releasing factor secretion into the hypophysial-portal circulation of rats. Regul Pept 16:235–242. [DOI] [PubMed] [Google Scholar]
- Raffin-Sanson ML, de Keyzer Y, Bertagna X (2003) Proopiomelanocortin, a polypeptide precursor with multiple functions: from physiology to pathological conditions. Eur J Endocrinol 149:79–90. [DOI] [PubMed] [Google Scholar]
- Reppert SM (2000) Cellular and molecular basis of circadian timing in mammals. Semin Perinatol 24:243–246. [DOI] [PubMed] [Google Scholar]
- Roth-Deri I, Green-Sadan T, Yadid G (2008) Beta-endorphin and drug-induced reward and reinforcement. Prog Neurobiol 86:1–21. [DOI] [PubMed] [Google Scholar]
- Sarkar DK, Boyadjieva NI, Chen CP, Ortigüela M, Reuhl K, Clement EM, Kuhn P, Marano J (2008) Cyclic adenosine monophosphate differentiated beta-endorphin neurons promote immune function and prevent prostate cancer growth. Proc Natl Acad Sci U S A. 105:9105–9110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar DK (2012) Circadian genes, the stress axis, and alcoholism. Alcohol Res 34:362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider RD, Thomas JD (2016) Adolescent choline supplementation attenuates working memory deficits in rats exposed to alcohol during the third trimester equivalent. Alcohol Clin Exp Res 40:897–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih MC, Yeh KT, Tang KP, Chen JC, Chang JG (2006) Promoter methylation in circadian genes of endometrial cancers detected by methylation-specific PCR. Mol Carcinog 45:732–740. [DOI] [PubMed] [Google Scholar]
- Shobatake R, Takasawa K, Ota H, Itaya-Hironaka A, Yamauchi A, Sakuramoto-Tsuchida S, Uchiyama T, Makino M, Sugie K, Takasawa S, Ueno S (2018) Up-regulation of POMC and CART mRNAs by intermittent hypoxia via GATA transcription factors in human neuronal cells. Int J Biochem Cell Biol 95:100–107. [DOI] [PubMed] [Google Scholar]
- Spanagel R, Pendyala G, Abarca C, Zghoul T, Sanchis-Segura C, Magnone MC, Lascorz J, Depner M, Holzberg D, Soyka M, Schreiber S, Matsuda F, Lathrop M, Schumann G, Albrecht U (2005a) The clock gene Per2 influences the glutamatergic system and modulates alcohol consumption. Nat Med 11:35–42. [DOI] [PubMed] [Google Scholar]
- Spanagel R, Rosenwasser AM, Schumann G, Sarkar DK (2005b) Alcohol consumption and the body’s biological clock. Alcohol Clin Exp Res 29:1550–1557. [DOI] [PubMed] [Google Scholar]
- Streissguth AP, Bookstein FL, Barr HM, Sampson PD, O’Malley K, Young JK (2004) Risk factors for adverse life outcomes in fetal alcohol syndrome and fetal alcohol effects J Dev Behav Pediatr. 25:228–238. [DOI] [PubMed] [Google Scholar]
- Tan CH, Denny CH, Cheal NE, Sniezek JE, Kanny D (2015) Alcohol use and binge drinking among women of childbearing age - United States, 2011–2013. MMWR Morb Mortal Wkly Rep 64:1042–1046. [DOI] [PubMed] [Google Scholar]
- Thomas JD, Idrus NM, Monk BR, Dominguez HD (2010) Prenatal choline supplementation mitigates behavioral alterations associated with prenatal alcohol exposure in rats. Birth Defects Res A Clin Mol Teratol 88:827–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner K, Reynolds JN, McGrath P, Lingley-Pottie P, Huguet A, Hewitt A, Green C, Wozney L, Mushquash C, Muhajarine N, Sourander A, Caughey H, Roane J (2015) Guided internet-based parent training for challenging behavior in children with fetal alcohol spectrum disorder (strongest families FASD): study protocol for a randomized controlled trial. JMIR Res Protoc 4:e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Gelernter J, Kranzler HR, Zhang H (2012) Identification of POMC exonic variants associated with substance dependence and body mass index. PLoS One 7:e45300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Hua L, Lu C, Chen Z (2011) Expression of circadian clock gene human Period2 (hPer2) in human colorectal carcinoma. World J Surg Oncol. 9:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengel T, Hanlon-Dearman AC, Fjeldsted B (2011) Sleep and sensory characteristics in young children with fetal alcohol spectrum disorder. J Dev Behav Pediatr 32:384–92. [DOI] [PubMed] [Google Scholar]
- Wozniak JR, Fuglestad AJ, Eckerle JK, Kroupina MG, Miller NC, Boys CJ, Brearley AM, Fink BA, Hoecker HL, Zeisel SH, Georgieff MK (2013) Choline supplementation in children with fetal alcohol spectrum disorders has high feasibility and tolerability. Nutr Res 33:897–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Liu A, Weidenhammer A, Cooksey RC, McClain D, Kim MK, Aguilera G, Abel ED, Chung JH (2009) The role of mPer2 clock gene in glucocorticoid and feeding rhythms. Endocrinology 150(5):2153–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka M, Matsutani T, Hara A, Hirono S, Hiwasa T, Takiguchi M, Iwadate Y (2018) Real-time methylation-specific PCR for the evaluation of methylation status of MGMT gene in glioblastoma. Oncotarget. 9:27728–27735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zghoul T, Abarca C, Sanchis-Segura C, Albrecht U, Schumann G, Spanagel R (2007) Ethanol self-administration and reinstatement of ethanol-seeking behavior in Per1(Brdm1) mutant mice. Psychopharmacology (Berl) 190:13–19. [DOI] [PubMed] [Google Scholar]
- Zhang H, Herman AI, Kranzler HR, Anton RF, Zhao H, Zheng W, Gelernter J. Array-based profiling of DNA methylation changes associated with alcohol dependence. Alcohol Clin Exp Res. 2013. January;37 Suppl 1:E108–15. [DOI] [PMC free article] [PubMed] [Google Scholar]