Abstract
In this work, we studied the expression kinetics and innate immune response of a self-amplifying mRNA (sa-RNA) after electroporation and lipid-nanoparticle (LNP)-mediated delivery in the skin of mice. Intradermal electroporation of the sa-RNA resulted in a plateau-shaped expression, with the plateau between day 3 and day 10. The overall protein expression of sa-RNA was significantly higher than that obtained after electroporation of plasmid DNA (pDNA) or non-replication mRNAs. Moreover, using IFN-β reporter mice, we elucidated that intradermal electroporation of sa-RNA induced a short-lived moderate innate immune response, which did not affect the expression of the sa-RNA. A completely different expression profile and innate immune response were observed when LNPs were used. The expression peaked 24 h after intradermal injection of sa-RNA-LNPs and subsequently showed a sharp drop. This drop might be explained by a translational blockage caused by the strong innate immune response that we observed in IFN-β reporter mice shortly (4 h) after intradermal injection of sa-RNA-LNPs. A final interesting observation was the capacity of sa-RNA-LNPs to transfect the draining lymph nodes after intradermal injection.
Keywords: self-amplifying mRNA, pDNA, non-replicating mRNA, lipid nanoparticles, innate immune response, electroporation, intradermal, expression kinetics, mice
Graphical Abstract
Introduction
Synthetic mRNAs are currently intensively studied for protein (replacement) therapy, gene editing, stem cell reprogramming, and immunotherapy.1, 2, 3 Each application requires distinct mRNA properties. For instance, protein (replacement) therapy asks for a long-acting and innate immunosilent mRNA, while mRNA vaccination may require opposite characteristics. Multiple synthetic mRNA platforms, such as unmodified mRNA, nucleoside-modified mRNA, and self-amplifying mRNA (sa-RNA) are currently available. To find the right match between the foreseen therapeutic application and the mRNA platform, information on the in vivo expression kinetics and innate immune response of these different mRNA platforms is crucial. Nucleoside-modified mRNAs are considered innate immunosilent,4 and sa-RNAs are long acting as they contain the coding sequences of a viral replicase complex that ensures amplification of the complete sa-RNA strand and, especially, the shorter subgenomic mRNA strand that contains the gene of interest.5, 6, 7 The amplification and abundance of these subgenomic mRNAs engender a high protein production. The viral replicase complex in sa-RNA originates from a single-stranded positive-sense RNA virus like, e.g., Venezuelan equine encephalitis virus (VEEV).7, 8, 9, 10
In vitro-transcribed mRNA (IVT mRNA) can be delivered in vivo using non-viral carriers such as lipid nanoparticles (LNPs)11, 12, 13 and physical methods such as electroporation.14, 15, 16 Non-viral carriers formulate the mRNA into nanoparticles that enter the cells by endocytosis, while it is believed that physical methods like electroporation mainly deliver the mRNA directly in the cytosol via temporal cell-membrane perforations.12, 17 One can expect that these different delivery mechanisms will influence the expression efficacy as well as the extent to which the innate immune system recognizes this synthetic mRNA.18 The innate immune system recognizes IVT mRNA by pathogen recognition receptors (PRRs) that are located in the endosomes (Toll-like receptors [TLRs] 3, 7, and 8) and in the cytoplasm (e.g., retinoic-acid-inducible gene I [RIG-I]-like receptors). Activation of these PRRs results in the induction of nuclear factor κB (NF-κB), caspase 1, and interferon regulatory factors 3 and 7 (IRF3 and IRF7, respectively), which respectively leads to the production of pro-inflammatory cytokines, cell death, and type I interferons (IFNs).19 Type I IFNs are known to activate 2′,5′-oligoadenylate synthetase (OAS) and protein kinase R (PKR).10 The former subsequently activates an endonuclease (RNaseL) that degrades the intracellular mRNA, while the latter inhibits the translation by phosphorylating eIF2.20, 21 These cellular actions will decrease the translation of the introduced synthetic mRNA. Furthermore, extensive activation of caspase-1 by synthetic mRNA can also cause pyroptosis, a form of immunogenic cell death that exhibits features of apoptosis as well as necrosis.19, 22 For certain applications like protein (replacement) therapy, a repeated administration of the mRNA will be required. For these applications, it will be important to control not only the extent of the innate immune response but also the duration of the innate immune response. Indeed, if the innate immune response has not faded away at the moment of the next mRNA injection, a chronical inflammatory condition will be established. The duration of the innate immune response after in vivo delivery of mRNA is not well studied, and most information comes from studies that used RT-PCR or ELISA to quantify key innate immune proteins.23, 24, 25 Although RT-PCR is a very sensitive technique, the correlations between mRNA levels and protein expression are not always linear.26 On the other hand, ELISA may not be sensitive enough to detect moderate innate immune responses in the excised tissues.27, 28 Furthermore, analyzing the in vivo innate immune response in tissues by RT-PCR and ELISA is invasive and, therefore, cannot be used to follow up the innate immune response in one and the same mouse. Therefore, reporter mice like, e.g., the IFN-β luciferase reporter mice may serve as a sensitive and reliable alternative to monitor the extent, duration, and location of the innate immune responses after mRNA delivery in mice.28
Recently, we developed, in collaboration with the laboratory of Ron Weiss (Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA, USA), a new sa-RNA. This sa-RNA has the capacity to overcome innate immune responses as it encodes the non-structural proteins (nsPs) of VEEV, which are known to impair critical signaling events downstream of the type I IFN receptor, leading to disruption of STAT1 signaling.29 In this study, we determined the in vivo efficacy and innate immunity of this novel VEEV-based sa-RNA at different doses and benchmarked it against plasmid DNA (pDNA) as well as unmodified and N1-methylpseudouridine (m1ψ)-modified synthetic mRNA. To avoid carrier-related effects on the efficiency and innate immunogenicity of these vectors, in vivo electroporation was used to compare the in vivo performance of these vectors. Electroporation- and carrier-mediated deliveries of mRNA occur by completely different mechanisms. As it is currently not known to which extend these different delivery mechanisms affect the in vivo performance of synthetic mRNA, we formulated our sa-RNA into LNPs and compared the expression profile and innate immune response with that obtained after in vivo electroporation. Because of our interest in mRNA vaccination, intradermal delivery was used, as the skin is extremely immune competent and easily accessible.30 Our interest in mRNA vaccination also inspired us to study the mRNA expression in the draining lymph nodes as an indicator of mRNA transfection in antigen-presenting cells.
Results
Electroporation Increases the Expression of sa-RNA and pDNA after Intradermal Injection
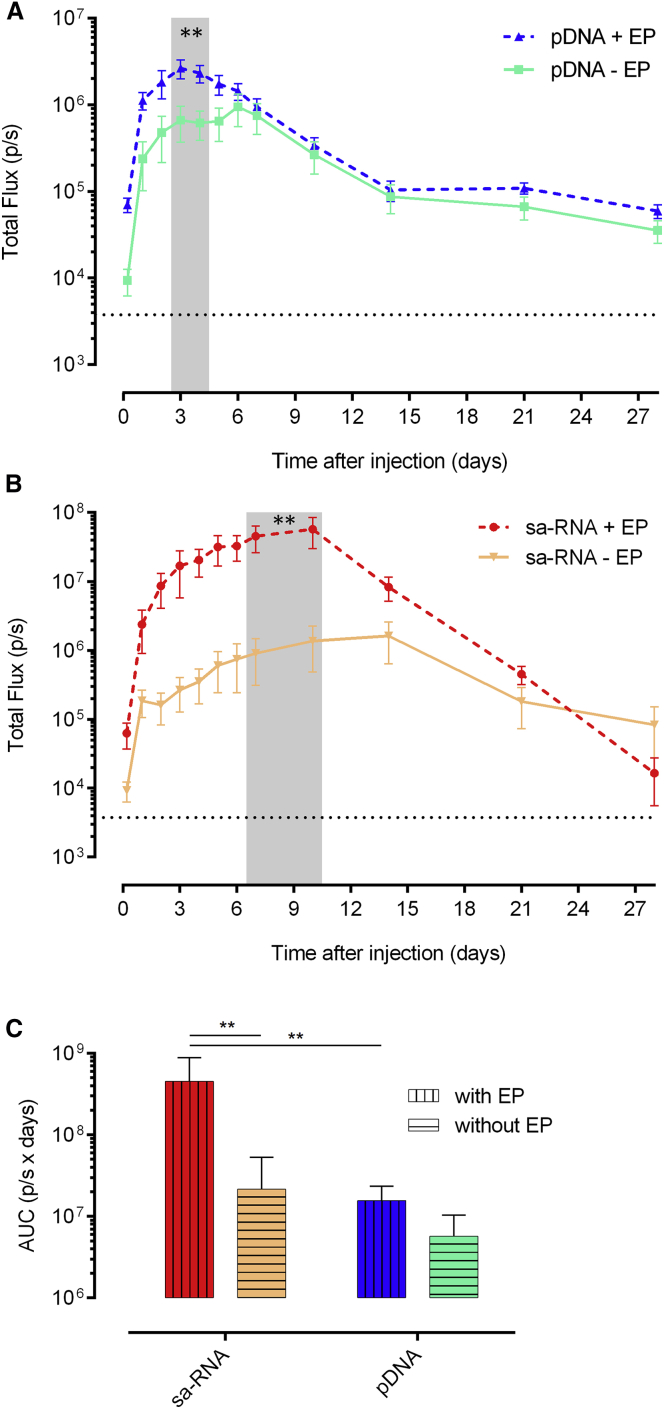
In the first set of experiments, the expression kinetics after intradermal injection of pDNA or sa-RNA with or without subsequent electroporation was studied in BALB/c mice. The intradermal delivery route was selected, as the skin is a large, accessible organ containing many antigen-presenting cells (APCs) for initiating an effective immune response. With both vectors, the luciferase expression could be detected as early as 5 h after injection. Electroporation increased the expression of pDNA as well as sa-RNA. However, the beneficial effect of electroporation on pDNA transfection was only observed during the first 5 days (Figure 1A). The peak in luciferase expression after intradermal electroporation of pDNA was reached after 2 days, and at this time point, electroporation induced a 3.5-fold increase in expression. The beneficial effect of electroporation was much more pronounced for sa-RNA (Figure 1B). The luciferase expression profile of the sa-RNA was also different from that of pDNA. sa-RNA reached its maximal expression 8–10 days after injection, and at this moment, electroporation caused a 40-fold increase in expression compared to naked delivery. After 28 days, the expression of the sa-RNA had declined to background, while the expression of pDNA lasted longer than 28 days. To allow for a better comparison of the expression levels, the areas under the curve (AUCs) were calculated (Figure 1C). After intradermal electroporation of sa-RNA, the total amount of luciferase produced during the follow-up period was significantly higher (30-fold higher) than after intradermal electroporation of pDNA (Figure 1C). Furthermore, the overall expression of intradermally administered pDNA and sa-RNA was increased 3-fold and 21-fold, respectively, by electroporation. These data demonstrate again that, especially, the expression of sa-RNA and, to a lesser extent, pDNA is increased by electroporation and that the sa-RNA clearly outperforms pDNA after intradermal electroporation.
Figure 1.
Effect of Intradermal Electroporation on the Expression Kinetics of sa-RNA and pDNA
(A and B) The in vivo bioluminescence was measured at different time points after intradermal injection of 5 μg of either pDNA (A) or sa-RNA (B), without (− EP) or with (+ EP) electroporation using needle array electrodes. Per group, three mice were injected on each flank (n = 6). Results are indicated as mean ± SEM of the total flux in the regions of interest (ROIs). The dotted black line represents the background signal. The shaded area indicates significant differences between the expression with and without electroporation. (C) AUC of the curves represented in (A) and (B) (mean ± SEM). **p < 0.01.
Comparison of the Dose-Dependent Expression of sa-RNA, Non-replicating mRNAs, and pDNA
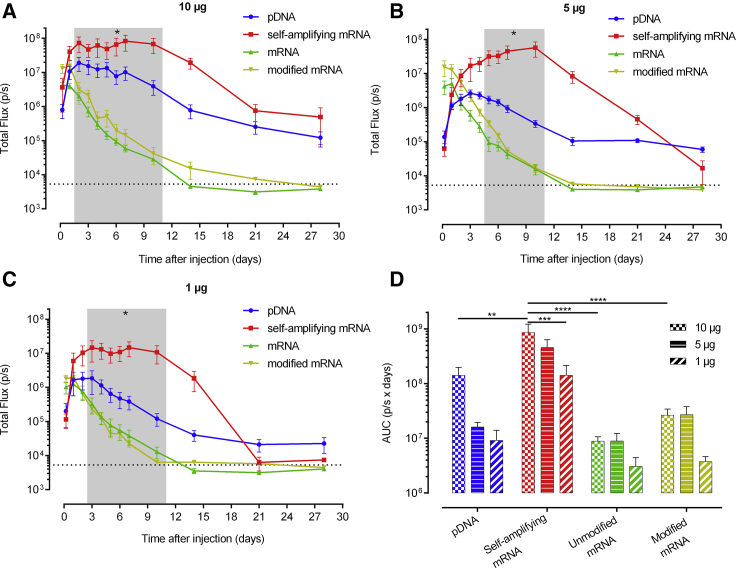
Besides sa-RNA and pDNA, non-replicating mRNAs, regardless of whether they contain modified nucleosides, are currently also intensively studied for vaccination, gene editing, or protein replacement therapy applications. However, a side-by-side comparison of the dose-expression profiles of these different synthetic mRNA platforms and pDNA after in vivo electroporation has not been performed. Therefore, we studied the luciferase expression levels and kinetics after intradermal electroporation of 10, 5, or 1 μg sa-RNA, m1ψ-modified mRNA, and unmodified mRNA or pDNA (Figures 2A–2C). The non-replicating mRNAs and pDNA reached, independent of the dose, their peak expression 5 h and 3 days, respectively, after transfection. After this point, the expression of the non-replicating mRNAs steadily decreased and disappeared at day 14 or day 10, when only 1 μg was used. pDNA showed, after its peak, a less steep drop, and the expression of the two highest doses clearly remained above background until the end of the follow-up period. The expression profile of sa-RNA was different and reached, unrelated to the dose, a maximal plateau expression between day 3 and day 10. After this plateau, the expression showed a gradual drop, and background levels were reached during the follow-up period (i.e., at days 28 and 21) with the two lowest doses (i.e., 5 and 1 μg, respectively). The largest difference was reached 10 days after injection, as sa-RNA was still inducing maximum luciferase expression, while the non-replicating mRNAs had almost reached background level.
Figure 2.
Dose-Dependent Expression Kinetics after Intradermal Electroporation of pDNA, sa-RNA, and Unmodified and m1ψ-Modified mRNA in Mice
(A–C) Three mice were intradermally electroporated in both flanks with 10 μg (A), 5 μg (B), or 1 μg (C) of each vector (n = 6). In vivo bioluminescence was measured over 4 weeks, and mean ± SEM of the total flux in the ROIs is displayed. The dotted line represents the background signal. The shaded area indicates a significant difference between the luciferase expression of sa-RNA compared to pDNA, modified mRNA, and unmodified mRNA, except for 10 μg after 3 days (no significance) and 10 μg and 1 μg after 5 days (only significant difference between sa-RNA and the non-replicating mRNAs). (D) AUC (mean ± SEM) of the expression kinetic curves shown in (A)–(C). *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
To gain more insight into the dose dependence, the total protein expression was evaluated by calculating the AUC of the dose-response curves over a period of 4 weeks after injection (Figure 2D). These AUCs nicely show a significant higher protein expression after sa-RNA transfection compared to pDNA and non-replicating mRNA. The m1ψ modification induced more protein expression compared to the non-modified variant (when 10 or 5 μg was used) but less than pDNA or sa-RNA. A striking observation is that 10 or 5 μg of the non-replicating mRNAs result in a similar expression curve and produce comparable amounts of luciferase during the follow-up period (Figures 2A, 2B, and 2D).
sa-RNA at Nanogram Doses Follows an All-or-Nothing Pattern
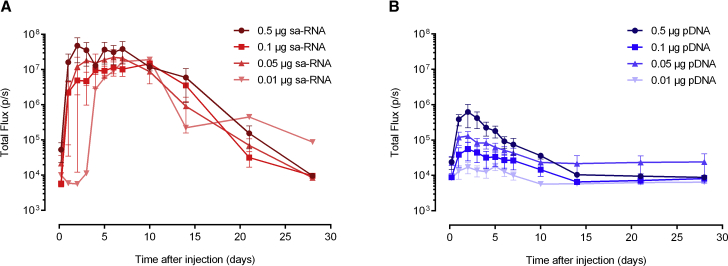
The high expression of 1 μg sa-RNA (Figure 2C) and the fact that sa-RNA can amplify itself after intracellular delivery encouraged us to examine the expression when doses as low as 10 ng were intradermally electroporated (Figure 3). At these submicron doses, the sa-RNA showed an all-or-nothing pattern: injection either resulted in a very high expression of luciferase or no expression at all. An overview of the success rates as a function of the sa-RNA dose is given in Table 1. An intradermal electroporation of sa-RNA was considered successful when the luciferase expression generated a total flux above 106 photons per second (i.e., 200-fold above the background signal) during at least 4 consecutive days. A high dose of 10 μg was successful in 6 out of 6 attempts, while the lowest dose of 10 ng only initiated expression in 1 out of 6 attempts. All successful injections resulted, independent of the dose (except at the lowest dose, i.e., 10 ng), in similar expression levels and curves with a maximal plateau expression between circa days 3 and 10. Submicron doses of pDNA were also assessed and resulted in much lower expression levels than those of sa-RNA, and they showed a clear dose-dependent profile. Ten nanograms of pDNA gave almost no luciferase expression, whereas 10 μg pDNA (Figure 2A) reached peak luciferase levels comparable to those obtained with the successful injection of 10 ng sa-RNA.
Figure 3.
Expression Kinetics of Low Doses of sa-RNA and pDNA after Intradermal Electroporation
(A and B) Doses from 0.5 to 0.01 μg of sa-RNA (A) or pDNA (B) were intradermally electroporated, and luciferase expression was monitored during 4 weeks by in vivo bioluminescence imaging. Only sa-RNA administrations that resulted in a successful expression are used to calculate the mean. Results are indicated as mean ± SEM of the total flux in the ROIs. Six administrations were performed. However, for the sa-RNA, not all intradermal electroporations resulted in a successful expression. The administrations that were successful are shown in (A) (n values were 4, 2, 4, and 1 for 0.5, 0.1, 0.05, and 0.01 μg sa-RNA, respectively).
Table 1.
Rate of Successful sa-RNA Injections
| Dose (μg) | Successful Injections | Total Injections | Rate (%) |
|---|---|---|---|
| 10 | 6 | 6 | 100 |
| 5 | 6 | 7 | 85.7 |
| 1 | 5 | 8 | 62.5 |
| 0.5 | 4 | 6 | 66.7 |
| 0.1 | 2 | 6 | 33.3 |
| 0.05 | 4 | 6 | 66.7 |
| 0.01 | 1 | 6 | 16.7 |
Kinetics of the Type I IFN Response after Intradermal Electroporation of sa-RNA, Non-replicating mRNAs, and pDNA
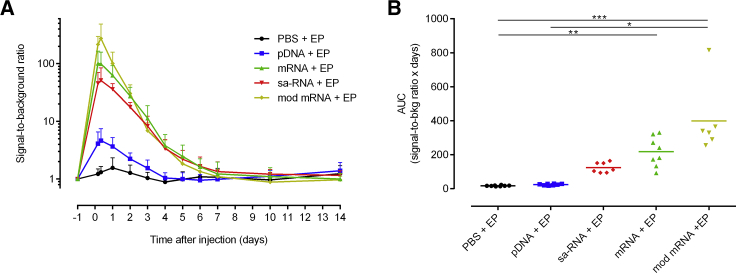
To acquire information about the extent of the induced innate immune response, we evaluated the level and duration of IFN-β induction upon intradermal electroporation of the different synthetic mRNAs, pDNA, and buffer in IFN-β reporter mice (Figure 4). Electroporation of buffer did not provoke a notable increase in IFN-β, while all the vectors caused a clear IFN-β response that reached its maximum 8 h after injection.
Figure 4.
The Extent and Duration of the Innate Immune Responses after Intradermal Electroporation of PBS, pDNA, sa-RNA, and Unmodified and m1ψ-Modified mRNA
Five micrograms of each vector coding for EGFP was injected on the flank of heterozygous IFN-β luciferase reporter mice. (A) In vivo bioluminescence was measured starting 4 h after injection. Signal-to-background ratio was calculated by dividing the bioluminescence signal in the ROIs (total flux, photons per second [p/s]) by its background signal at day −1 for each mouse. Means + SD are displayed (n = 6–8). (B) As a measure of the overall IFN-β expression, the AUCs of the curves in (A) were calculated. The horizontal bars represent the means. *p < 0.05; **p < 0.01; ***p < 0.001.
The highest IFN-β response was observed with the non-replicating mRNAs. Modified mRNA induced the highest average IFN-β induction shortly after electroporation, followed by unmodified mRNA. However, after 3 days, the IFN-β response induced by the mRNAs becomes similar. IFN-β induction returns to baseline level 7 days after electroporation of mRNA. This led to an overall non-significant higher innate immune response induced by modified mRNA (Figure 4B). Self-amplifying mRNA caused, especially shortly after administration, a slightly lower IFN-β response than the non-replicating mRNAs. However, the IFN-β response dropped slower after administration of the sa-RNA, resulting in an IFN-β response similar to that of unmodified mRNA from day 3 onward. No significant induction of IFN-β was noticed after the administration of pDNA. The induction reached its maximum 1 day after administration and subsequently declined to background around day 4.
Electroporation versus Lipid-Nanoparticle-Mediated Delivery of sa-RNA
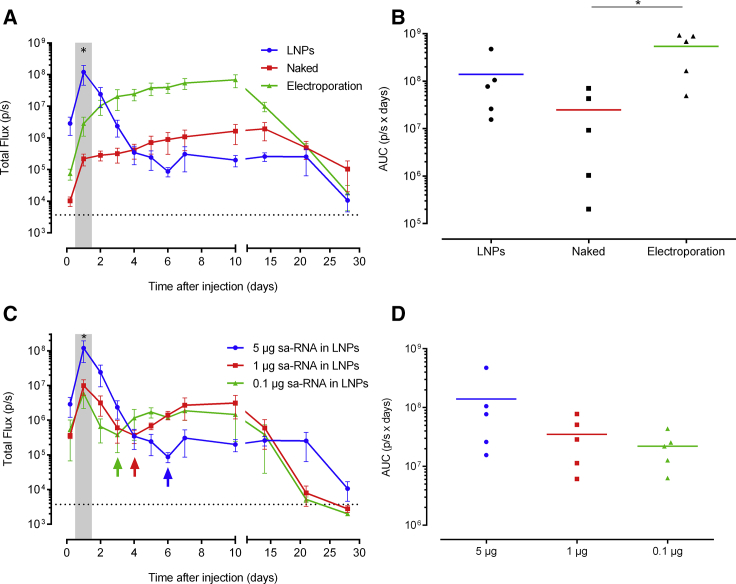
In the next experiment, we compared the expression of 5 μg sa-RNA formulated into state-of-the-art LNPs with that of naked sa-RNA with or without subsequent electroporation. sa-RNAs formulated into LNPs showed completely different expression profiles compared to those of naked or electroporated sa-RNA after intradermal administration (Figure 5A). LNP-mediated transfection of sa-RNA resulted in an early-peak luciferase expression 24 h after injection. After this time point, the expression showed a very steep drop. However, after 7 days, a slight resurgence in luciferase expression occurred. As reported in Figure 1, the expression of the naked sa-RNA was, again, clearly increased by electroporation. The shapes of the expression profiles of non-electroporated and electroporated sa-RNA were similar: a sharp increase during the first 24 h and, subsequently, a steady increase until the maximal expression levels were reached around 8–10 days after injection. After day 10, the luciferase expression gradually dropped, and 28 days after administration, the expression of the naked, electroporated, and LNP-delivered sa-RNA became close to background.
Figure 5.
Lipid Nanoparticle versus Electroporation-Mediated Intradermal Delivery of sa-RNA
(A) Three mice per group were intradermally injected on both flanks with 5 μg of either sa-RNA-LNPs, naked sa-RNA, or naked sa-RNA followed by electroporation. In vivo bioluminescence was measured over 4 weeks, and mean ± SEM of the total flux in the ROIs is displayed. The injection site was chosen as the ROI and did not include the area of the draining lymph nodes. The dotted line represents the background signal, and the shaded area indicates a significant difference between luciferase expression after use of LNPs versus electroporation. (B) AUC (mean ± SEM) of the expression kinetic curves shown in (A) was calculated and displayed to compare total protein expression between the different delivery systems. Individual values are plotted with the mean. (C) In vivo bioluminescence after intradermal injection of 5, 1, or 0.1 μg sa-RNA encapsulated in LNPs. (D) The calculated AUCs of the curves in (C). The shaded area indicates that the luciferase expression after injection of 5 μg sa-RNA-LNPs is significantly higher than 1 μg and 0.1 μg. Mean ± SEM are displayed. The arrows indicate the time points at which the expression resurgences. *p < 0.05.
For certain applications such as protein (replacement) therapy, the total amount of produced protein over time is more relevant than the peak expression level. Therefore, we calculated the AUCs shown in Figures 5A and 5C. The protein production after electroporation of the sa-RNA was, respectively, 22-fold and 4-fold higher than the amount of protein produced after injection of, respectively, naked or LNP-formulated sa-RNA (Figure 5B).
We also studied the dose-dependent expression after intradermal injection of sa-RNA-LNPs. Doses lower than 5 μg, i.e., 1 and 0.1 μg, of the LNP-formulated sa-RNA resulted in similar but lower expression profiles (Figures 5C and 5D). With the lower doses (i.e., 0.1 and 1 μg), a similar resurgence of the luciferase is noticed as with the 5-μg dose. However, this resurgence was more pronounced, and the lower the dose, the earlier this resurgence was noticed. All injections of sa-RNA-LNPs resulted in a successful expression.
sa-RNA Formulated in LNPs Cause Expression in the Draining Lymph Node after Intradermal Injection
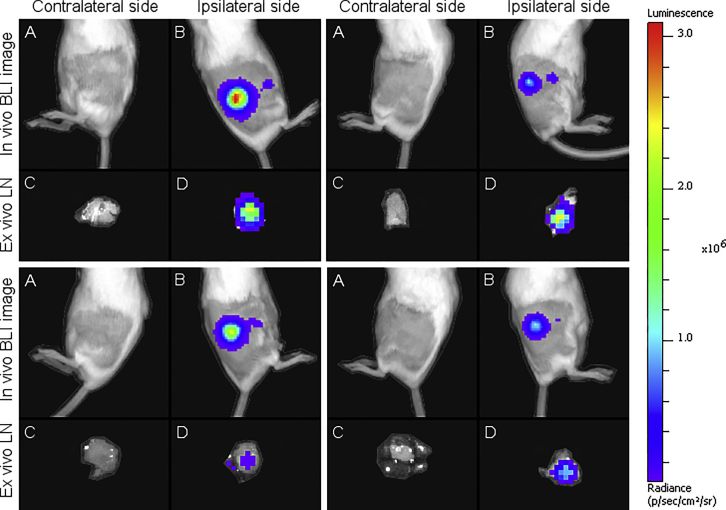
During evaluation of the LNP-formulated sa-RNA, we observed a second, clearly delineated, smaller bioluminescent spot next to the injection spot (Figure 6). This spot was located at the position where the subiliac lymph node can be found and, hence, suggested expression of the sa-RNA in the draining lymph node.31 This additional expression spot could be seen until 48 h after administration (data not shown).
Figure 6.
Bioluminescence Images of Four Mice after Intradermal Injection of 5 μg sa-RNA Encapsulated in LNPs
(A and B) In vivo bioluminescence images of (A) the untreated contralateral (negative) flanks and (B) the treated ipsilateral (positive) flanks, respectively, 5 h after injection of sa-RNA-LNPs. Next to the bright bioluminescent spot, which is the injection spot, a small bioluminescent spot is visible at the location of the draining subiliac lymph node (B). Twenty-four hours after injection of the sa-RNA-LNPs, the subiliac lymph nodes were excised, and ex vivo bioluminescent images were taken. (C) and (D) show, respectively, the draining subiliac lymph nodes of the untreated contralateral (negative) flanks and treated ipsilateral (positive) flanks that received sa-RNA-LNPs.
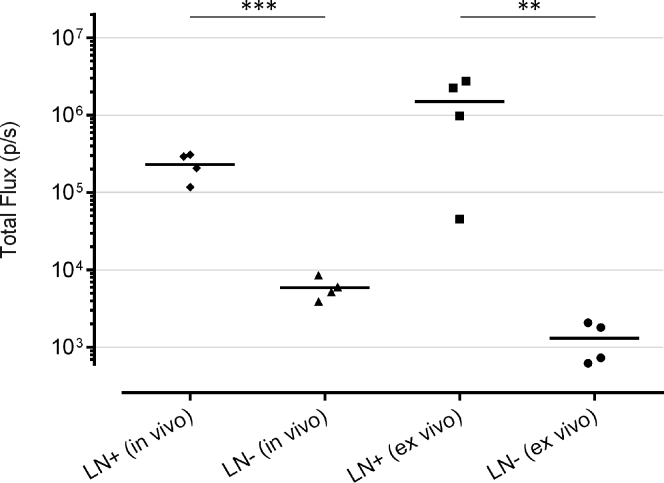
To confirm this observation, mice were euthanized 24 h after intradermal injection of sa-RNA-LNPs, and the ipsilateral and contralateral (negative) subiliac lymph nodes were excised to be imaged ex vivo (Figures 6 and 7). The ipsilateral (positive) subiliac lymph nodes showed significantly higher luciferase expression compared to the contralateral (negative) lymph nodes. This indicates either that transfected APCs traveled from the injection spot to the draining ipsilateral subiliac lymph node or that the sa-RNA-LNPs traveled to the ipsilateral subiliac lymph node and transfected APCs and/or lymph node stromal cells at this site. Such transfection in the draining lymph nodes was not observed after injection of the same dose of naked sa-RNA with or without electroporation.
Figure 7.
Comparison of the In Vivo and Ex Vivo Bioluminescence Signals in the Draining Lymph Nodes after Intradermal Injection of 5 μg sa-RNA-LNPs
The in vivo bioluminescence signals were measured 5 h after intradermal injection. Twenty-four hours after injection of the sa-RNA-LNPs, mice were euthanized, and the subiliac lymph nodes were excised and dripped with luciferin immediately before measurement. LN+, ipsilateral lymph node draining the injection spot. LN−, the contralateral lymph node. Horizontal bars represent the mean (n = 4). **p < 0.01; ***p < 0.001.
Kinetics of the Type I IFN Immune Response after Lipid-Nanoparticle- and Electroporation-Mediated Delivery of sa-RNA
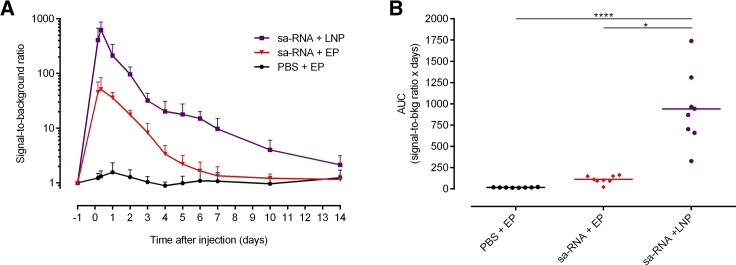
Self-amplifying mRNA formulated in LNPs will enter the cells through endosomes, which will likely lead to stimulation of the endosomal TLRs. It is expected that these TLRs get shunned when electroporation is used. To study whether this difference in cellular uptake affects the IFN-β response, we compared the kinetics of the IFN-β response after LNP- and electroporation-mediated delivery of the sa-RNA (Figure 8). Intradermal injection of 5 μg sa-RNA-LNPs caused, compared to intradermal electroporation, a higher IFN-β response that also lasted longer: after LNP-mediated delivery of sa-RNA, IFN-β induction persisted for 14 days, whereas electroporation-mediated delivery only induced an IFN-β response during 7 days after injection. During the 14-day follow-up period, the IFN-β response was about 8-fold higher after LNP-mediated delivery.
Figure 8.
Kinetics of the Type I Interferon Immune Response after Lipid-Nanoparticle-Mediated Delivery of sa-RNA
(A) The extent and kinetics of the IFN-β induction in reporter mice after LNP- and electroporation-mediated intradermal delivery of 5 μg sa-RNA encoding EGFP was measured by in vivo bioluminescence imaging. The in vivo bioluminescence was measured starting 4 h after injection and was further monitored for 2 weeks. Signal-to-background ratios were calculated by dividing the bioluminescence signals in the ROIs (total flux, p/s), with its background signal at day −1 for each mouse. Means ± SD are displayed (n = 8). (B) As a measure of the overall IFN-β expression, the AUCs of the curves in (A) were calculated. Individual values are plotted with the mean. *p < 0.05; ****p < 0.0001.
Discussion
The use of synthetic mRNA for the in vivo production of therapeutic proteins like, e.g., antibodies, erythropoietin, or blood-clotting factors, is recently gaining more and more attention.32, 33, 34 For these applications, a long-acting mRNA that can produce therapeutic proteins during several weeks would be ideal. Additionally, activation of the innate immune system by the delivered synthetic mRNA should be as low as possible, or at least it should not affect the translation of the synthetic mRNA. Our data demonstrate that in vivo electroporation of a VEEV-based sa-RNA meets the aforementioned criteria. Electroporation of this sa-RNA caused a high and stable expression over 2 weeks. In contrast, non-replicating unmodified and modified mRNAs resulted in a much lower and shorter protein production. sa-RNAs based on, e.g., VEEV have been evaluated in the past as a possible new vaccination platform.6, 35, 36, 37, 38 However, the in vivo expression kinetics and, especially, the inherent innate immunity of VEEV-based sa-RNAs have not been studied in detail. We found that the sa-RNA caused, despite its intracellular amplification, a lower innate immune response than the non-replicating unmodified and modified mRNAs. Nevertheless, the kinetics of the IFN-β response was similar between sa-RNA and non-replicating mRNAs, with a maximal IFN-β induction 5 h after administration followed by a steady decline. This may indicate that mainly the introduced sa-RNA, and not the replication of the sa-RNA, is causing the innate immune response. The absence of a strong and prolonged innate immune response after in vivo delivery of our VEEV-based sa-RNA can be explained by the capacity of the nsPs of VEEV to impair the signaling downstream of the type I IFN receptor.29 Additionally, it has been reported that nsP2 can shut off the translation of host mRNA by blocking its nuclear export.39, 40, 41 These actions of the nsPs of VEEV may temper the innate immune response and prevent it from going into full swing during the replication of our VEEV-based sa-RNA.
sa-RNAs generate a replicase complex that is composed of the nsP1–4 of a single-stranded RNA virus. This raises the concern that these viral proteins may elicit an adaptive immune response, which would impede repeated administration of sa-RNAs. However, we have strong indications that such neutralizing adaptive immune response is not established. Indeed, we observed that re-administration of our VEEV-based sa-RNA after 4 weeks did not result in a lower protein expression (unpublished data).
We showed that sa-RNAs are very potent vectors, as they can cause gene expression at doses as low as 10 ng after local administration in mice. However, it is important to mention that electroporation of 10 ng sa-RNA was only successful in 1 out of 6 injections. By evaluating the success rate as a function of the dose, we found that the percentage of successful injections gradually dropped when the dose of sa-RNA was lowered. We recently showed that this observation may be attributed to the presence of RNases and that the repeatability and efficacy of intradermal electroporated sa-RNA can be increased by adding a protein-based RNase inhibitor to the sa-RNA just before injection.42 RNases are abundantly present on the skin,43 and these RNases can contaminate the tip of the needle during injection. In this way, minuscule amounts of RNases may be introduced and especially low doses of sa-RNAs will be very sensitive to such contaminating RNases.
To overcome activation of the RNA sensors, the mRNA can be rendered less immunogenic through incorporation of modified nucleotides like pseudouridine, 5-methylcytidine, and N1-methylpseudouridine.44, 45, 46, 47 However, incorporation of N1-methylpseudouridine-modified nucleosides in our non-replicating mRNA did not reduce the innate immune response; hence, no significant increases in protein production were observed after electroporation of nucleoside-modified mRNAs. Similar results have been reported by Kauffman et al.,48 who found that incorporation of pseudouridine-modified nucleosides had no significant effect on either immunogenicity or protein expression of mRNA-LNPs after systemic injection. In our work and in that of Kauffman et al., the mRNA was not purified by high-pressure liquid chromatography (HPLC); hence, double-stranded mRNA or short aborted mRNA species that are known to be highly immunogenic were probably not completely removed, causing the innate immune response.
Although the expression of pDNA is lower than that of sa-RNA during the first weeks, pDNA is still an interesting vector for protein (replacement) therapy, as its expression lasts longer than that of sa-RNA (up to 5 months after intradermal injection).49 CpG motifs in the pDNA as well as LPS contamination during pDNA preparation can result in, respectively, TLR9 and TLR4 activation, leading to IFN-β induction. However, in our hands, the innate immune response after intradermal pDNA electroporation was very low and short-lived compared to mRNA vectors (Figure 4A), which further supports the use of pDNA for protein therapy. Since we use an endotoxin-removing purification kit for the preparation of the pDNA, most of the IFN-β response observed in Figure 4A is probably due to the CpG motifs in the pDNA. Nevertheless, the use of pDNA has some drawbacks like, e.g., the theoretical risk of genomic integration and oncogenic mutagenesis; the presence of antibiotic-resistance genes; and the fact that, for certain therapeutic proteins, an uncontrolled expression during several months is not warranted.
To study the effect of different delivery methods on the in vivo performance of our VEEV-based sa-RNA, we encapsulated the sa-RNA into LNPs. Intradermal injection of these sa-RNA-LNPs resulted in a completely different expression profile than obtained after intradermal electroporation. Indeed, the expression of LNP formulated sa-RNA peaked shortly (i.e., 24 h) after their administration. After this peak, the expression dropped sharply. Formulation of sa-RNAs into LNPs resulted, thus, in a faster and initial higher expression than obtained after electroporation mediated delivery. This indicates that LNPs cause a much more efficient intracellular delivery of the sa-RNA than electroporation (Figure 5A). LNPs escort all mRNA into the cells without (excessive) degradation by extracellular RNases, while injection of naked mRNA in combination with electroporation presumably suffers from mRNA loss due to RNase degradation. Furthermore, electroporation does not direct all mRNA into the cells, resulting in a lower delivery efficiency. However, a strong innate immune response is induced as a consequence of the massive intracellular delivery of LNP-formulated sa-RNAs (Figure 8). This strong IFN-β induction is most likely responsible for the sharp drop in expression 24 h after administration of the sa-RNA-LNPs (Figure 5C). However, at days 3–6, the drop in expression stopped, and a resurgence of the expression was noticed. This resurgence of the expression was more pronounced at lower doses of the sa-RNA-LNPs. Lower doses of the sa-RNA-LNPs are expected to induce a lower innate immune response. Hence, it takes less time for the innate immune response to drop below the “mRNA translation blockage threshold,” and also, more RNAs are expected to survive this shorter and less intense innate immune response. Due to this more pronounced and faster resurgence of the expression, low doses of sa-RNA-LNPs resulted, from day 5 onward until day 15, in a higher expression than high doses of sa-RNA-LNPs. Nevertheless, the highest dose (5 μg) of the sa-RNA-LNPs still resulted in a much higher expression shortly after administration (i.e., day 1 until day 3). The remarkable expression profiles in Figure 5C is, thus, due to a dose-depended innate immune response and the capacity of the replicase complex to generate many subgenomic mRNA copies from a few intact sa-RNA strands. A similar resurgence, however, was not observed after electroporation of the sa-RNA. Therefore, the IFN-β response induced by electroporation of sa-RNA is, most likely, not strong enough to cause a decrease in luciferase expression.
A very interesting observation was the presence of a short-lived expression (up to 48 h after administration; data not shown) in the draining subiliac lymph node after intradermal delivery of sa-RNA-LNPs (Figures 6 and 7). Expression of luciferase in the lymph nodes can be the result of the transport of the sa-RNA-LNPs toward the draining lymph nodes or due to migration of transfected immune cells at the injection site toward the draining lymph nodes. The luciferase protein expressed at the site of injection can also be transported to the lymph nodes. Further research is necessary to determine how the lymph nodes get luciferase positive and which types of cells are transfected. Nevertheless, this expression in the lymph nodes most likely indicates transfection of APCs. Therefore, formulation of sa-RNAs into LNPs is expected to genuinely boost the efficacy of sa-RNA vaccines as was also shown by Geall et al.50, 51
In conclusion, we demonstrated that in vivo intradermal electroporation of a VEEV-based sa-RNA outperformed the expression obtained after electroporation of pDNA or non-replication mRNAs. Furthermore, in vivo electroporation of our sa-RNA resulted in a short-lived and moderate innate immune response that did not affect the expression of the sa-RNA. When the VEEV-based sa-RNA was encapsulated in LNPs, a completely different expression and innate immune response profile was obtained. The expression rapidly peaked 24 h after intradermal injection of sa-RNA-LNPs and subsequently showed a sharp drop that can be attributed to a massive induction of the innate immune system. Interestingly, intradermal injection of sa-RNA-LNPs also resulted in a protein expression in the lymph nodes, which supports the potential use of sa-RNA-LNPs as vaccines. However, it needs to be examined whether the induced innate immune response after administration of the sa-RNA-LNPs is balanced enough to potentiate adaptive immune responses. Indeed, it has been shown that a too-high innate immune response can also be detrimental for the adaptive immune response.52, 53
Materials and Methods
Mice
Female wild-type BALB/cJRj mice were purchased from Janvier Labs (Le Genest-Saint-Isle, France) and housed in individual ventilated cages in a climate-controlled facility under a 14-h/10-h light/dark cycle. Heterozygous IFN-β reporter mice with a BALB/c background were a kind gift of Johan Grooten,54 and the breed was further maintained in house. Shortly, the mice were genetically modified by replacing the IFNΒ gene with the luciferase reporter gene, placing the reporter under the control of an IFNB promoter. The IFN-β reporter mice used in our study were heterozygotes, which implies that they can produce IFN-β and have a functional IFN-β pathway. We measured the induction of IFN-β by in vivo bioluminescence imaging after systemic administration of luciferin. All mice were aged between 7 and 10 weeks at the start of the experiments and kept in individually ventilated cages with ad libitum access to food and water. The in vivo mouse experiments were conducted with the approval of the ethical committee of the Faculty of Veterinary Medicine, Ghent University (EC2016/17).
Plasmid Constructs
Bacteria containing the pGL4.13 plasmid (GenBank: AY738225) were a kind gift of Katrien Remaut (Ghent University). This plasmid from Promega (Madison, WI, USA) encodes the reporter gene luciferase (luc2) from P. pyralis, controlled by the SV40 promoter, and was used to examine expression kinetics. Similarly, a pDNA (pEGFP-N1, GenBank: U55762.1) encoding EGFP and containing 321 CpG motifs55 was used to study the IFN-β response after intradermal electroporation of pDNA in reporter mice. E. coli containing the pEGFP-N1 were also a kind gift of Katrien Remaut. This plasmid contains the EGFP gene controlled by a cytomegalovirus (CMV) promoter.
Further, we used 4 different plasmids for the production of RNA by IVT. pTK160 (11,519 bp) and pMC15 (10,586 bp) are derived from VEEV strain TC-83 containing a substitution in the 5′ UTR (r.3a > g) and in nsP2 (p.Q739L). The sequence coding for the structural proteins was replaced by the reporter gene luc2 or EGFP, respectively, corresponding to the sequences used in pGL4.13 and pEGFP-N1. Additionally, the vectors contained restriction sites for the I-SceI endonuclease prior to a T7 polymerase promoter and downstream of a short poly(A) sequence. pTK305 (4,112 bp)45 and pMC13 (3,179 bp) plasmids were used to produce (modified) mRNA containing, respectively, the luc2 or the EGFP reporter gene. These plasmids contain, besides the reporter genes, VEEV-derived 5′ and 3′ UTRs and were constructed using standard Gateway cloning procedures. In short, the EGFP sequence was derived from the peGFP-N1 plasmid (a kind gift of Dr. Katrien Remaut) using attB-containing primers. The EGFP cassette of the entry clone (pENTR221-EGFP) was inserted into destination vector pTK155 or pTK318 for the sa-RNA or (un)modified mRNA, respectively, using LR clonase II (Thermo Fisher Scientific, Waltham, MA, USA). These destination vectors (kind gifts from Tasuku Kitada and Ron Weiss) contain an attR1-ccdB-ChloramphenicolR-attR2 sequence that allowed site-specific recombination with the attL1-eGFP-attL2 from pENTR221-EGFP to generate pMC15 and pMC13, respectively. The pTK160 and pTK305 plasmids, containing the Fluc2 sequence from pGL4 (Promega), were produced in a similar fashion and were also a kind gift of Tasuku Kitada and Ron Weiss.
E. coli bacteria containing the plasmids were cultivated in lysogeny broth (LB; Invitrogen, Waltham, MA, USA), and the plasmids were subsequently isolated using the EndoFree Plasmid Maxi Kit (QIAGEN) when pDNA was used for injection or the Plasmid Plus Midi Kit (QIAGEN) when further processed to mRNA.
mRNA Synthesis
Template DNA was generated by linearizing the aforementioned plasmids using I-SceI endonuclease (New England Biolabs, Ipswich, MA, USA) before IVT with the MEGAscript T7 Transcription Kit (Life Technologies, Waltham, MA, USA). Post-transcriptional modifications were applied using the ScriptCap m7G Capping System, 2′-O-Methyltransferase Kit, and the A-Plus Poly(A) Polymerase Tailing Kit (CELLSCRIPT, Madison, WI, USA). All mRNAs were purified with the RNeasy Mini Kit (QIAGEN) after IVT and after each modification. For production of modified mRNA, the uridine nucleotide was completely replaced by N1-methylpseudouridine (tebu-bio, Boechout, Belgium) during IVT. Correct translation of EGFP-encoding IVT mRNAs and pDNA was verified after transfection of baby hamster kidney (BHK) cells using Lipofectamine MessengerMAX (Thermo Fisher Scientific) and subsequent evaluation using the Nikon Eclipse Ti-S fluorescent microscope (Nikon, Leuven, Belgium).
Injection, Electroporation, and LNPs
Different doses of pDNA and mRNA (ranging from 0.01 to 10 μg) were dissolved in 50 μL PBS, and 1 U RNasin Plus RNase Inhibitor (Promega) per microliter of solution was added to the mRNA before storing the solution at −80°C. Mice were sedated using the inhalation anesthetic isoflurane, at 5% for induction and 2% for maintenance, and were shaved to decrease photon scattering and absorption by the fur during the luciferase-mediated conversion of luciferin. Injections were performed intradermally on one or both flanks of the mice using a 29G insulin needle (VWR, Amsterdam, the Netherlands). Electroporation, when used, was executed immediately after each injection with a 2-needle array electrode containing 4 needles per row of 4 mm (AgilePulse, BTX Harvard Apparatus, Holliston, MA, USA) as previously described.56 In brief, 2 short high-voltage pulses were given (450 V, 0.05 ms), followed by 8 long low-voltage pulses (100 V, 10 ms) and an interval of 300 ms between each pulse. sa-RNAs coding for EGFP or luciferase were also encapsulated in LNPs, as previously described.57 Briefly, an ethanolic lipid solution was rapidly mixed with an aqueous solution containing sa-RNA (pH 4). The lipid solution consists of ionizable lipids, phosphatidylcholine, cholesterol, and PEG in a 50/10/38.5/1.5 ratio, respectively. The RNA-loaded particles were characterized and subsequently stored at −80°C at a concentration of 1 μg/μL. The mean hydrodynamic diameter of these sa-RNA-LNPs was 72 ± 3 nm, with a polydispersity index of 0.032 ± 0.009, and the zeta potential equaled −6 ± 1 mV (Zetasizer Nano ZS90, Malvern Panalytical, Worcestershire, UK). The encapsulation efficiency was determined by RiboGreen RNA Assay Kit (as described previously)58 and equaled 95.8% ± 1.0%.
Bioluminescence Imaging
Mice were intraperitoneally injected with 200 μL D-luciferin (15 mg/mL, Gold Biotechnology, Olivette, MO, USA), and in vivo bioluminescent imaging was performed 15 min later using an IVIS Lumina II (PerkinElmer, Zaventem, Belgium). The total flux in the region of interest was determined using the Living Image Software v4.3.1. In vivo bioluminescence imaging was repeated at different time points to study the expression kinetics of pDNA and the different mRNA vectors.
Statistical Analysis
Statistical analysis and development of the graphs were performed with the software GraphPad Prism 6 (GraphPad Software, San Diego, CA, USA). Longitudinal experiments were analyzed with repeated-measures two-way ANOVA, followed by Sidak’s or Tukey’s multiple comparisons test. Fixed-time point analysis was performed using a ratio paired t test, and outliers were determined using Grubbs’ test. Differences are found to be significant when the p value was <0.05 (*p < 0.05; **p < 0.01; ***p < 0.001; and ****p < 0.0001).
Author Contributions
H.H. and N.N.S. designed the experiments and drafted the manuscript. H.H., J.D.T., Z.Z., S.M.C., and A.G. conducted the experiments. Y.K.T. and B.L.M. encapsulated the sa-RNA in LNPs. N.N.S. supervised all experimental work. D.V. helped with the supervision of the work. All authors read, improved, and approved the manuscript.
Acknowledgments
We would like to thank Tasuku Kitada and Ron Weiss of the Massachusetts Institute of Technology for kindly providing us with the pTK160 and pTK305 plasmids. This research has also benefitted from a statistical consult with Ghent University FIRE (Fostering Innovative Research based on Evidence). This work was supported by the concerted research action (GOA) fund of Ghent University (Project Code BOF15/GOA/013). We are also grateful for the financial support by the China Scholarship Council (grant number 201607650018).
References
- 1.Petsch B., Schnee M., Vogel A.B., Lange E., Hoffmann B., Voss D., Schlake T., Thess A., Kallen K.J., Stitz L., Kramps T. Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza A virus infection. Nat. Biotechnol. 2012;30:1210–1216. doi: 10.1038/nbt.2436. [DOI] [PubMed] [Google Scholar]
- 2.Ramaswamy S., Tonnu N., Tachikawa K., Limphong P., Vega J.B., Karmali P.P., Chivukula P., Verma I.M. Systemic delivery of factor IX messenger RNA for protein replacement therapy. Proc. Natl. Acad. Sci. USA. 2017;114:E1941–E1950. doi: 10.1073/pnas.1619653114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilboa E., Vieweg J. Cancer immunotherapy with mRNA-transfected dendritic cells. Immunol. Rev. 2004;199:251–263. doi: 10.1111/j.0105-2896.2004.00139.x. [DOI] [PubMed] [Google Scholar]
- 4.Karikó K., Weissman D. Naturally occurring nucleoside modifications suppress the immunostimulatory activity of RNA: implication for therapeutic RNA development. Curr. Opin. Drug Discov. Devel. 2007;10:523–532. [PubMed] [Google Scholar]
- 5.Strauss J.H., Strauss E.G. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petrakova O., Volkova E., Gorchakov R., Paessler S., Kinney R.M., Frolov I. Noncytopathic replication of Venezuelan equine encephalitis virus and eastern equine encephalitis virus replicons in mammalian cells. J. Virol. 2005;79:7597–7608. doi: 10.1128/JVI.79.12.7597-7608.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim D.Y., Atasheva S., McAuley A.J., Plante J.A., Frolova E.I., Beasley D.W., Frolov I. Enhancement of protein expression by alphavirus replicons by designing self-replicating subgenomic RNAs. Proc. Natl. Acad. Sci. USA. 2014;111:10708–10713. doi: 10.1073/pnas.1408677111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ulmer J.B., Mason P.W., Geall A., Mandl C.W. RNA-based vaccines. Vaccine. 2012;30:4414–4418. doi: 10.1016/j.vaccine.2012.04.060. [DOI] [PubMed] [Google Scholar]
- 9.Ljungberg K., Liljeström P. Self-replicating alphavirus RNA vaccines. Expert Rev. Vaccines. 2015;14:177–194. doi: 10.1586/14760584.2015.965690. [DOI] [PubMed] [Google Scholar]
- 10.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines – a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hajj K.A., Whitehead K.A. Tools for translation: non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater. 2015;2:17056. [Google Scholar]
- 12.Li B., Zhang X., Dong Y. Nanoscale platforms for messenger RNA delivery. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2018;11:e1530. doi: 10.1002/wnan.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kulkarni J.A., Cullis P.R., van der Meel R. Lipid nanoparticles enabling gene therapies: from concepts to clinical utility. Nucleic Acid Ther. 2018;28:146–157. doi: 10.1089/nat.2018.0721. [DOI] [PubMed] [Google Scholar]
- 14.Knudsen M.L., Ljungberg K., Liljeström P., Johansson D.X. Intradermal electroporation of RNA. Methods Mol. Biol. 2014;1121:147–154. doi: 10.1007/978-1-4614-9632-8_13. [DOI] [PubMed] [Google Scholar]
- 15.Johansen P., Storni T., Rettig L., Qiu Z., Der-Sarkissian A., Smith K.A., Manolova V., Lang K.S., Senti G., Müllhaupt B. Antigen kinetics determines immune reactivity. Proc. Natl. Acad. Sci. USA. 2008;105:5189–5194. doi: 10.1073/pnas.0706296105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cu Y., Broderick K.E., Banerjee K., Hickman J., Otten G., Barnett S., Kichaev G., Sardesai N.Y., Ulmer J.B., Geall A. Enhanced delivery and potency of self-amplifying mRNA vaccines by electroporation in situ. Vaccines (Basel) 2013;1:367–383. doi: 10.3390/vaccines1030367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Broderick K.E., Humeau L.M. Enhanced delivery of DNA or RNA vaccines by electroporation. Methods Mol. Biol. 2017;1499:193–200. doi: 10.1007/978-1-4939-6481-9_12. [DOI] [PubMed] [Google Scholar]
- 18.Zhong Z., Mc Cafferty S., Combes F., Huysmans H., De Temmerman J., Gitsels A., Vanrompay D., Portela Catani J., Sanders N. mRNA therapeutics deliver a hopeful message. Nano Today. 2018;23:16–39. [Google Scholar]
- 19.Andries O., De Filette M., De Smedt S.C., Demeester J., Van Poucke M., Peelman L., Sanders N.N. Innate immune response and programmed cell death following carrier-mediated delivery of unmodified mRNA to respiratory cells. J. Control. Release. 2013;167:157–166. doi: 10.1016/j.jconrel.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 20.Chakrabarti A., Jha B.K., Silverman R.H. New insights into the role of RNase L in innate immunity. J. Interferon Cytokine Res. 2011;31:49–57. doi: 10.1089/jir.2010.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vattem K.M., Staschke K.A., Wek R.C. Mechanism of activation of the double-stranded-RNA-dependent protein kinase, PKR: role of dimerization and cellular localization in the stimulation of PKR phosphorylation of eukaryotic initiation factor-2 (eIF2) Eur. J. Biochem. 2001;268:3674–3684. doi: 10.1046/j.1432-1327.2001.02273.x. [DOI] [PubMed] [Google Scholar]
- 22.Kepp O., Galluzzi L., Zitvogel L., Kroemer G. Pyroptosis – a cell death modality of its kind? Eur. J. Immunol. 2010;40:627–630. doi: 10.1002/eji.200940160. [DOI] [PubMed] [Google Scholar]
- 23.Leyman B., Huysmans H., Mc Cafferty S., Combes F., Cox E., Devriendt B., Sanders N.N. Comparison of the expression kinetics and immunostimulatory activity of replicating mRNA, non-replicating mRNA and pDNA after intradermal electroporation in pigs. Mol. Pharm. 2018;15:377–384. doi: 10.1021/acs.molpharmaceut.7b00722. [DOI] [PubMed] [Google Scholar]
- 24.Edwards D.K., Jasny E., Yoon H., Horscroft N., Schanen B., Geter T., Fotin-Mleczek M., Petsch B., Wittman V. Adjuvant effects of a sequence-engineered mRNA vaccine: translational profiling demonstrates similar human and murine innate response. J. Transl. Med. 2017;15:1. doi: 10.1186/s12967-016-1111-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alberer M., Gnad-Vogt U., Hong H.S., Mehr K.T., Backert L., Finak G., Gottardo R., Bica M.A., Garofano A., Koch S.D. Safety and immunogenicity of a mRNA rabies vaccine in healthy adults: an open-label, non-randomised, prospective, first-in-human phase 1 clinical trial. Lancet. 2017;390:1511–1520. doi: 10.1016/S0140-6736(17)31665-3. [DOI] [PubMed] [Google Scholar]
- 26.Shebl F.M., Pinto L.A., García-Piñeres A., Lempicki R., Williams M., Harro C., Hildesheim A. Comparison of mRNA and protein measures of cytokines following vaccination with human papillomavirus-16 L1 virus-like particles. Cancer Epidemiol. Biomarkers Prev. 2010;19:978–981. doi: 10.1158/1055-9965.EPI-10-0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosengren S., Firestein G.S., Boyle D.L. Measurement of inflammatory biomarkers in synovial tissue extracts by enzyme-linked immunosorbent assay. Clin. Diagn. Lab. Immunol. 2003;10:1002–1010. doi: 10.1128/CDLI.10.6.1002-1010.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lienenklaus S., Cornitescu M., Zietara N., Łyszkiewicz M., Gekara N., Jabłónska J., Edenhofer F., Rajewsky K., Bruder D., Hafner M. Novel reporter mouse reveals constitutive and inflammatory expression of IFN-beta in vivo. J. Immunol. 2009;183:3229–3236. doi: 10.4049/jimmunol.0804277. [DOI] [PubMed] [Google Scholar]
- 29.Simmons J.D., White L.J., Morrison T.E., Montgomery S.A., Whitmore A.C., Johnston R.E., Heise M.T. Venezuelan equine encephalitis virus disrupts STAT1 signaling by distinct mechanisms independent of host shutoff. J. Virol. 2009;83:10571–10581. doi: 10.1128/JVI.01041-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zehrung D., Jarrahian C., Wales A. Intradermal delivery for vaccine dose sparing: overview of current issues. Vaccine. 2013;31:3392–3395. doi: 10.1016/j.vaccine.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 31.Van den Broeck W., Derore A., Simoens P. Anatomy and nomenclature of murine lymph nodes: descriptive study and nomenclatory standardization in BALB/cAnNCrl mice. J. Immunol. Methods. 2006;312:12–19. doi: 10.1016/j.jim.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 32.Kauffman K.J., Dorkin J.R., Yang J.H., Heartlein M.W., DeRosa F., Mir F.F., Fenton O.S., Anderson D.G. Optimization of lipid nanoparticle formulations for mRNA delivery in vivo with fractional factorial and definitive screening designs. Nano Lett. 2015;15:7300–7306. doi: 10.1021/acs.nanolett.5b02497. [DOI] [PubMed] [Google Scholar]
- 33.DeRosa F., Guild B., Karve S., Smith L., Love K., Dorkin J.R., Kauffman K.J., Zhang J., Yahalom B., Anderson D.G., Heartlein M.W. Therapeutic efficacy in a hemophilia B model using a biosynthetic mRNA liver depot system. Gene Ther. 2016;23:699–707. doi: 10.1038/gt.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pardi N., Secreto A.J., Shan X., Debonera F., Glover J., Yi Y., Muramatsu H., Ni H., Mui B.L., Tam Y.K. Administration of nucleoside-modified mRNA encoding broadly neutralizing antibody protects humanized mice from HIV-1 challenge. Nat. Commun. 2017;8:14630. doi: 10.1038/ncomms14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pushko P., Parker M., Ludwig G.V., Davis N.L., Johnston R.E., Smith J.F. Replicon-helper systems from attenuated Venezuelan equine encephalitis virus: expression of heterologous genes in vitro and immunization against heterologous pathogens in vivo. Virology. 1997;239:389–401. doi: 10.1006/viro.1997.8878. [DOI] [PubMed] [Google Scholar]
- 36.Balasuriya U.B., Heidner H.W., Hedges J.F., Williams J.C., Davis N.L., Johnston R.E., MacLachlan N.J. Expression of the two major envelope proteins of equine arteritis virus as a heterodimer is necessary for induction of neutralizing antibodies in mice immunized with recombinant Venezuelan equine encephalitis virus replicon particles. J. Virol. 2000;74:10623–10630. doi: 10.1128/jvi.74.22.10623-10630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perri S., Greer C.E., Thudium K., Doe B., Legg H., Liu H., Romero R.E., Tang Z., Bin Q., Dubensky T.W., Jr. An alphavirus replicon particle chimera derived from Venezuelan equine encephalitis and Sindbis viruses is a potent gene-based vaccine delivery vector. J. Virol. 2003;77:10394–10403. doi: 10.1128/JVI.77.19.10394-10403.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lundstrom K. Replicon RNA viral vectors as vaccines. Vaccines (Basel) 2016;4:E39. doi: 10.3390/vaccines4040039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carrasco L., Sanz M.A., González-Almela E. The regulation of translation in alphavirus-infected cells. Viruses. 2018;10:E70. doi: 10.3390/v10020070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin J., Gardner C.L., Burke C.W., Ryman K.D., Klimstra W.B. Similarities and differences in antagonism of neuron alpha/beta interferon responses by Venezuelan equine encephalitis and Sindbis alphaviruses. J. Virol. 2009;83:10036–10047. doi: 10.1128/JVI.01209-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhalla N., Sun C., Metthew Lam L.K., Gardner C.L., Ryman K.D., Klimstra W.B. Host translation shutoff mediated by non-structural protein 2 is a critical factor in the antiviral state resistance of Venezuelan equine encephalitis virus. Virology. 2016;496:147–165. doi: 10.1016/j.virol.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huysmans H., De Temmerman J., Zhong Z., Mc Cafferty S., Combes F., Haesebrouck F., Sanders N.N. Improving the repeatability and efficacy of intradermal electroporated self-replicating mRNA. Mol. Ther. Nucleic Acids. 2019;17:388–395. doi: 10.1016/j.omtn.2019.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Probst J., Brechtel S., Scheel B., Hoerr I., Jung G., Rammensee H.G., Pascolo S. Characterization of the ribonuclease activity on the skin surface. Genet. Vaccines Ther. 2006;4:4. doi: 10.1186/1479-0556-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li B., Luo X., Dong Y. Effects of chemically modified messenger RNA on protein expression. Bioconjug. Chem. 2016;27:849–853. doi: 10.1021/acs.bioconjchem.6b00090. [DOI] [PubMed] [Google Scholar]
- 45.Andries O., Mc Cafferty S., De Smedt S.C., Weiss R., Sanders N.N., Kitada T. N(1)-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control. Release. 2015;217:337–344. doi: 10.1016/j.jconrel.2015.08.051. [DOI] [PubMed] [Google Scholar]
- 46.Karikó K., Muramatsu H., Ludwig J., Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011;39:e142. doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kormann M.S., Hasenpusch G., Aneja M.K., Nica G., Flemmer A.W., Herber-Jonat S., Huppmann M., Mays L.E., Illenyi M., Schams A. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 2011;29:154–157. doi: 10.1038/nbt.1733. [DOI] [PubMed] [Google Scholar]
- 48.Kauffman K.J., Mir F.F., Jhunjhunwala S., Kaczmarek J.C., Hurtado J.E., Yang J.H., Webber M.J., Kowalski P.S., Heartlein M.W., DeRosa F., Anderson D.G. Efficacy and immunogenicity of unmodified and pseudouridine-modified mRNA delivered systemically with lipid nanoparticles in vivo. Biomaterials. 2016;109:78–87. doi: 10.1016/j.biomaterials.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roos A.K., Eriksson F., Timmons J.A., Gerhardt J., Nyman U., Gudmundsdotter L., Bråve A., Wahren B., Pisa P. Skin electroporation: effects on transgene expression, DNA persistence and local tissue environment. PLoS ONE. 2009;4:e7226. doi: 10.1371/journal.pone.0007226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Geall A.J., Verma A., Otten G.R., Shaw C.A., Hekele A., Banerjee K., Cu Y., Beard C.W., Brito L.A., Krucker T. Nonviral delivery of self-amplifying RNA vaccines. Proc. Natl. Acad. Sci. USA. 2012;109:14604–14609. doi: 10.1073/pnas.1209367109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Magini D., Giovani C., Mangiavacchi S., Maccari S., Cecchi R., Ulmer J.B., De Gregorio E., Geall A.J., Brazzoli M., Bertholet S. Self-amplifying mRNA vaccines expressing multiple conserved influenza antigens confer protection against homologous and heterosubtypic viral challenge. PLoS ONE. 2016;11:e0161193. doi: 10.1371/journal.pone.0161193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Beuckelaer A., Grooten J., De Koker S. Type I interferons modulate CD8+ T cell immunity to mRNA vaccines. Trends Mol. Med. 2017;23:216–226. doi: 10.1016/j.molmed.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 53.Pepini T., Pulichino A.M., Carsillo T., Carlson A.L., Sari-Sarraf F., Ramsauer K., Debasitis J.C., Maruggi G., Otten G.R., Geall A.J. Induction of an IFN-mediated antiviral response by a self-amplifying RNA vaccine: implications for vaccine design. J. Immunol. 2017;198:4012–4024. doi: 10.4049/jimmunol.1601877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lambricht L., Vanvarenberg K., De Beuckelaer A., Van Hoecke L., Grooten J., Ucakar B., Lipnik P., Sanders N.N., Lienenklaus S., Préat V., Vandermeulen G. Coadministration of a plasmid encoding HIV-1 gag enhances the efficacy of cancer DNA vaccines. Mol. Ther. 2016;24:1686–1696. doi: 10.1038/mt.2016.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hashimoto Y., Uehara Y., Abu Lila A.S., Ishida T., Kiwada H. Activation of TLR9 by incorporated pDNA within PEG-coated lipoplex enhances anti-PEG IgM production. Gene Ther. 2014;21:593–598. doi: 10.1038/gt.2014.32. [DOI] [PubMed] [Google Scholar]
- 56.Denies S., Cicchelero L., Van Audenhove I., Sanders N.N. Combination of interleukin-12 gene therapy, metronomic cyclophosphamide and DNA cancer vaccination directs all arms of the immune system towards tumor eradication. J. Control. Release. 2014;187:175–182. doi: 10.1016/j.jconrel.2014.05.045. [DOI] [PubMed] [Google Scholar]
- 57.Pardi N., Tuyishime S., Muramatsu H., Kariko K., Mui B.L., Tam Y.K., Madden T.D., Hope M.J., Weissman D. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Control. Release. 2015;217:345–351. doi: 10.1016/j.jconrel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leung A.K., Hafez I.M., Baoukina S., Belliveau N.M., Zhigaltsev I.V., Afshinmanesh E., Tieleman D.P., Hansen C.L., Hope M.J., Cullis P.R. Lipid nanoparticles containing siRNA synthesized by microfluidic mixing exhibit an electron-dense nanostructured core. J. Phys. Chem. C Nanomater. Interfaces. 2012;116:18440–18450. doi: 10.1021/jp303267y. [DOI] [PMC free article] [PubMed] [Google Scholar]