The early prediction of deterioration could have an important role in supporting healthcare professionals, as an estimated 11% of deaths in hospital follow a failure to promptly recognize and treat deteriorating patients (1). To achieve this goal requires predictions of patient risk that are continuously updated and accurate, and delivered at an individual level with sufficient context and enough time to act. Here we develop a deep learning approach for the continuous risk prediction of future deterioration in patients, building upon recent work that models adverse events from electronic health records (2–17) and using acute kidney injury—a common and potentially life-threatening condition (18)—as an exemplar. Our model was developed on a large, longitudinal dataset of electronic health records that cover diverse clinical environments, comprising 703,782 adult patients across 172 inpatient and 1,062 outpatient sites. Our model predicts 55.8% of all inpatient episodes of acute kidney injury, and 90.2% of all acute kidney injury that requires subsequent administration of dialysis, with a lead time of up to 48 h and a ratio of 2 false alerts for every true alert. In addition to predicting future acute kidney injury, our model provides confidence assessments and a list of the clinical features that are most salient to each prediction, alongside predicted future trajectories for clinically relevant blood tests (9). Although the recognition and prompt treatment of acute kidney injury is known to be challenging, our approach may offer opportunities for identifying patients at risk within a time window that enables early treatment.
Adverse events and clinical complications are a major cause of mortality and poor patient outcomes, and substantial effort has been made to improve their recognition18,19. Few predictors have found their way into routine clinical practice, either because they lack effective sensitivity and specificity, or because they report already existing damage20. One example relates to AKI, a potentially life threatening condition affecting approximately 1 in 5 US inpatient admissions21. Although a substantial proportion of cases are thought to be preventable with early treatment22, current AKI detection algorithms depend on changes in serum creatinine as a marker of acute decline in renal function. Elevation of serum creatinine lags behind renal injury, resulting in delayed access to treatment. This supports a case for preventative ‘screening’ type alerts, but there is no evidence that current rule based alerts improve outcomes23. For predictive alerts to be effective they must empower clinicians to act before major clinical decline has occurred by: (i) delivering actionable insights on preventable conditions; (ii) being personalised for specific patients; (iii) offering sufficient contextual information to inform clinical decision-making; and (iv) being generally applicable across patient populations24.
Promising recent work on modelling adverse events from EHR2–17 suggests that the incorporation of machine learning may enable early prediction of AKI. Existing examples of sequential AKI risk models have either not demonstrated a clinically-applicable level of predictive performance25 or have focused on predictions across a short time horizon, leaving little time for clinical assessment and intervention26.
Our proposed system is a recurrent neural network that operates sequentially over individual electronic health records, processing the data one step at a time and building an internal memory that keeps track of relevant information seen up to that point. At each time point the model outputs a probability of AKI occurring at any stage of severity within the next 48 hours, although our approach can be extended to other time windows or AKI severities (see Extended Data Table 1). When the predicted probability exceeds a specified operating point threshold, the prediction is considered positive. This model was trained using data curated from a multisite retrospective dataset of 703,782 adult patients from all available sites at the US Department of Veterans Affairs (VA) - the largest integrated health care system in the United States. The dataset consisted of information available from the hospital EHR in digital format. The total number of independent entries in the dataset was approximately 6 billion, including 620,000 features. Patients were randomised across training (80%), validation (5%), calibration (5%) or test (10%) sets. A ground truth label for the presence of AKI at any given point in time was added using the internationally accepted “Kidney Disease: Improving Global Outcomes (KDIGO)” criteria18; the incidence of KDIGO AKI was 13.4% of admissions. (Detailed descriptions of the model and dataset are provided in the Methods, and Extended Data Figures 1, 2 & 3.)
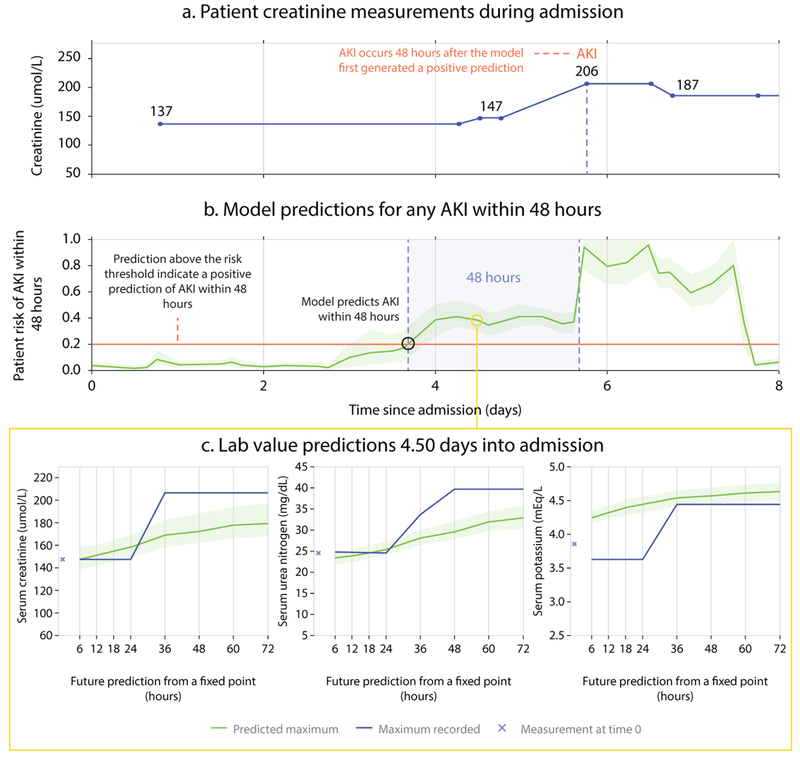
Figure 1 shows the use of our model. At every point throughout an admission the model provides updated estimates of future AKI risk, along with an associated degree of uncertainty. Providing the uncertainty associated with a prediction may help clinicians distinguish ambiguous cases from predictions fully supported by the available data. Identifying an increased risk of future AKI sufficiently in advance is critical, as longer lead times may allow preventative action to be taken. This is possible even when clinicians may not be actively intervening with, or monitoring a patient (Supplementary Information section A for examples)
Figure 1 |. Illustrative example of risk prediction, uncertainty and predicted future laboratory values.
The first 8 days of admission for a male patient aged 65 with a history of chronic obstructive pulmonary disease. (a) Creatinine measurements showing AKI occurring on day 5. (b) Continuous risk predictions; the model predicted increased AKI risk 48 hours before it was observed. A risk above 0.2, corresponding to 33% precision, was the threshold above which AKI was predicted. Lighter green borders on the risk curve indicate uncertainty, taken as the range of 100 ensemble predictions once trimmed for highest and lowest 5 values. (c) Predictions of the maximum future observed values of creatinine, urea, and potassium.
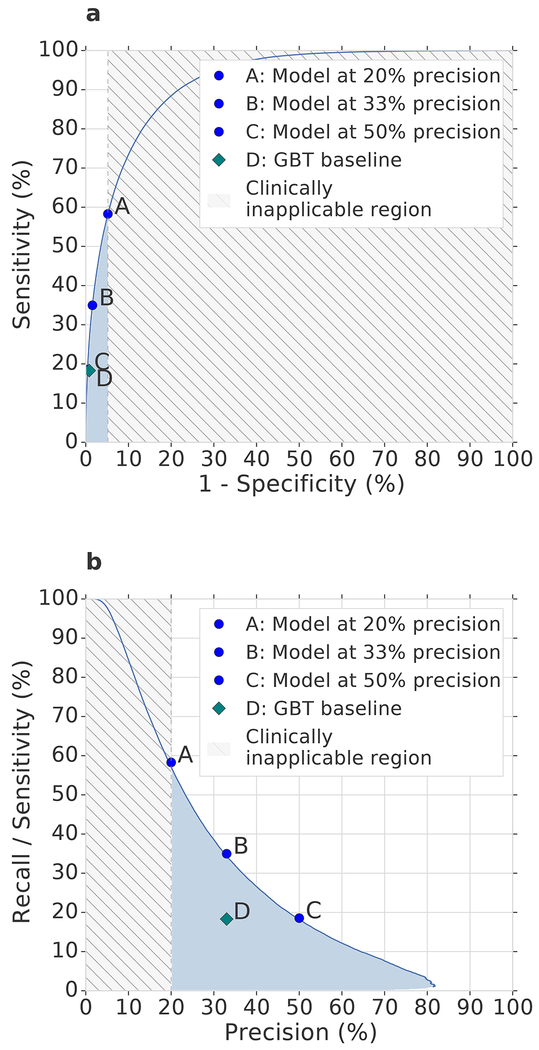
With our approach, 55.8% of inpatient AKI events of any severity were predicted early within a window of up to 48 hours in advance, with a ratio of two false predictions for every true positive. This corresponds to an area under the receiver operating characteristic curve (ROC AUC) of 92.1% and an area under the precision-recall curve (PR AUC) of 29.7%. Set at this threshold our predictive model would, if operationalised, trigger a daily clinical assessment in 2.7% of hospitalised patients in this cohort (Extended Data Table 2). Sensitivity was particularly high in patients who went on to develop lasting complications as a result of AKI. The model provided early predictions correctly in 84.3% of episodes where administration of in-hospital or outpatient dialysis was required within 30 days of the onset of AKI of any stage, and 90.2% of cases where regular outpatient administration of dialysis was scheduled within 90 days of the onset of AKI (Extended Data Table 3). Figure 2 shows the corresponding ROC and PR curves, as well as a spectrum of different operating points of the model. An operating point can be chosen to either further increase the proportion of AKI predicted early, or reduce the percentage of false predictions at each step, according to clinical priority (Figure 3). Applied to stage 3 AKI, 84.1% of inpatient events were predicted up to 48 hours in advance, with a ratio of two false predictions for every true positive (Extended Data Table 4). To respond to these alerts on a daily basis, clinicians would need to attend to approximately 0.8% of in-hospital patients (Extended Data Table 2).
Figure 2 |. Model performance illustrated by Receiver Operating Characteristic (ROC) and Precision/Recall (PR) curves.
(a) ROC and (b) PR curves for the risk that AKI of any severity will occur within 48 hours. Blue dots: different model operating points (A, 20% precision; C, 33% precision; E, 50% precision; see Extended Data Table 4). Grey shading: area corresponding to operating points with greater than four false positives for each true positive. Blue shading: performance in the more clinically applicable part of the operating space. The model significantly (p-value of <1e-6 outperformed the gradient-boosted tree baseline, shown in (b) for operating point C using two-sided Mann–Whitney U test on 200 samples per model (see Methods).
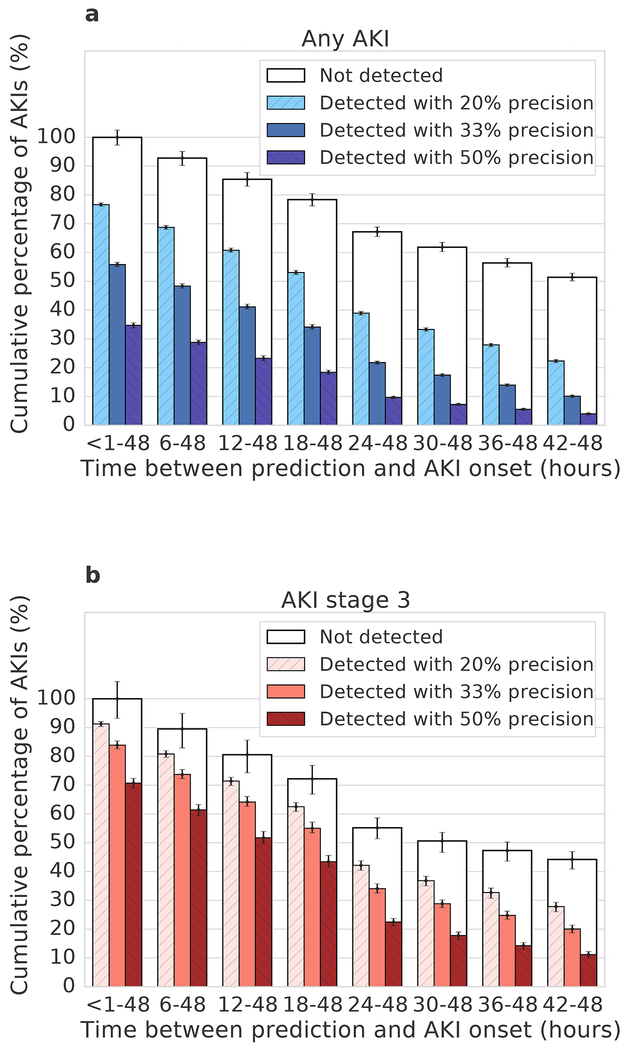
Figure 3 |. The time between model prediction and actual AKI event.
The models predict AKI risk within a particular time window. Within this the time in hours between prediction and AKI can vary (error bars: bootstrap pivotal 95% confidence intervals; n=200). a, b, Prediction performance for any AKI (a) and AKI stage 3 (b) 48 h ahead of time, shown for different precisions. A greater proportion were correctly predicted closer to the time step immediately prior to the AKI. The available time window for prediction is shortened in AKI events which occur <48 hours after admission; for each column the boxed area shows the upper limit on possible predictions.
The model correctly identifies substantial future increases in seven auxiliary biochemical tests in 88.5% of cases (Supplement B), and provides information about the factors that are most salient to the computation of each risk prediction. The greatest saliency was identified for laboratory tests known to be relevant to renal function (see Supplement C) The predictive performance of our model was maintained across time and hospital sites, demonstrated by additional experiments that show generalisability to data acquired at time points after the model was trained (Extended Data Table 5).
Our approach significantly outperformed (p < 0.001) established state-of-the-art baseline models (Supplement D). For example, we implemented a baseline model with gradient-boosted trees using manually curated features that are known to be relevant for modelling kidney function and in the delivery of routine care (Supplementary Information, sections E and F), combined with aggregate statistical information on trends observed in the recent history of the patient. This yielded 3599 clinically relevant features provided to the baselines at each step (see Methods). For the same level of precision, this baseline model was able to detect 36.0% of all inpatient AKI episodes up to 48 hours ahead of time, compared to 55.8% for our model.
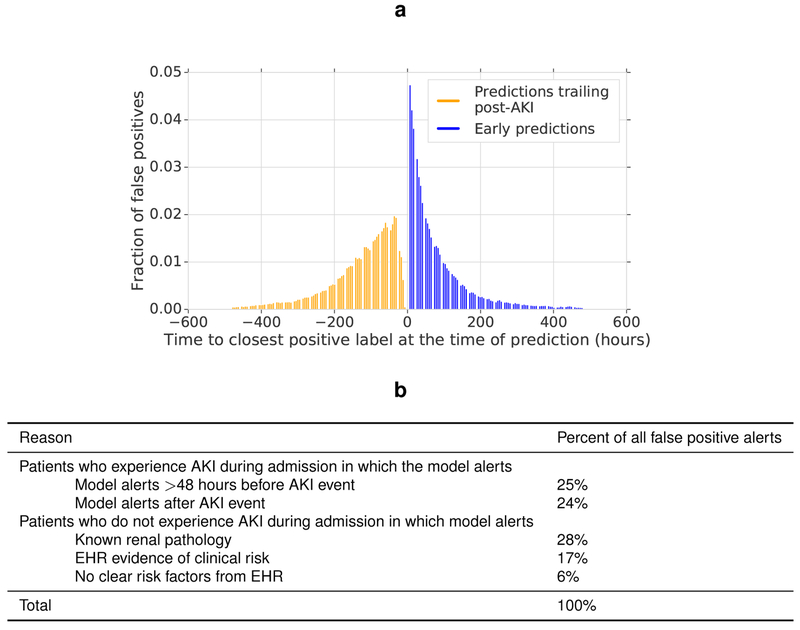
Of the false positive alerts made by our model, 24.9% were positive predictions made even earlier than the 48 hour window in patients who subsequently developed AKI (Extended Data Figure 4). 57.1% of these occurred in patients with pre-existing chronic kidney disease (CKD), who are at a higher risk of developing AKI. Of the remaining false positive alerts, 24.1% were trailing predictions that occurred after an AKI episode had already begun; such alerts can be filtered out in clinical practice. For positive risk predictions where no AKI was subsequently observed in this retrospective dataset, it is probable that many occurred in patients at risk of AKI where appropriate preventative treatment was administered which averted subsequent AKI. In addition to these early and trailing predictions, 88% of the remaining false positive alerts occurred in patients with severe renal impairment, known renal pathology, or evidence in the EHR that the patient required clinical review (Extended Data Figure 4).
Our aim is to provide risk predictions that enable personalized preventative action to be delivered at a large scale. The way these predictions are used may vary by clinical setting: a trainee doctor could be alerted in real time to each patient under their care, while a specialist nephrologist or rapid response teams27 can identify high risk patients to prioritise their response. This is possible because performance was consistent across multiple clinically important groups, notably those at an elevated risk of AKI (Supplement G). Our model is designed to complement existing routine care, as it is trained specifically to predict AKI that happened in this retrospective dataset despite existing best practices.
Although we demonstrate a model trained and evaluated on a clinically representative set of patients from the entire VA health care system, the demographic is not representative of the global population. Female patients comprised 6.38% of patients in the dataset, and model performance was lower for this demographic (Extended Data Table 6). Validating the predictive performance of the proposed system on a general population would require training and evaluating the model on additional representative datasets. Future work will need to address the under-representation of sub-populations in the training data28 and overcome the impact of potential confounding factors related to hospital processes29. KDIGO is an indicator of AKI that lags long after the initial renal impairment, and model performance could be enhanced by improvements in the ground-truth definition of AKI and data quality30.
Despite the state-of-the-art retrospective performance of our model compared to existing literature, future work should now prospectively evaluate and independently validate the proposed model to establish its clinical utility and effect on patient outcomes, as well as explore the role of the model in researching strategies for delivering preventative care for AKI.
In summary, we demonstrate a deep learning approach for the continuous prediction of AKI within a clinically-actionable window of up to 48 hours in advance. We report performance on a clinically diverse population and across a large number of sites to show that our approach may allow for the delivery of potentially preventative treatment, prior to the physiological insult itself in a large number of the cases. Our results open up the possibility for deep learning to guide the prevention of clinically important adverse events. With the possibility of risk predictions delivered in clinically-actionable windows alongside the increasing size and scope of EHR datasets, we now shift to a regime where the role for machine learning in clinical care can grow rapidly, supplying new tools to enhance the patient and clinician experience, and potentially becoming a ubiquitous and integral part of routine clinical pathways.
Methods
Data Description
The clinical data used in this study was collected by the US Department of Veterans Affairs and transferred to DeepMind in de-identified format. No personal information was included in the dataset, which met HIPAA “Safe Harbor” criteria for de-identification.
The US Department of Veterans Affairs (VA) serves a population of over nine million veterans and their families across the entire United States of America. The VA is composed of 1,243 health care facilities (sites), including 172 VA Medical Centers and 1,062 outpatient facilities31. Data from these sites is aggregated into 130 data centres, of which 114 had data of inpatient admissions that we used in this study. Four sites were excluded since they had fewer than 250 admissions during the five year time period. No other patients were excluded based on location.
The data comprised all patients aged between 18 and 90 admitted for secondary care to medical or surgical services from the beginning of October 2011 to the end of September 2015, including laboratory data, and where there was at least one year of EHR data prior to admission. The data included medical records with entries up to 10 years prior to each admission date and up to two years afterwards, where available. Where available in the VA database, data included outpatient visits, admissions, diagnoses as International Statistical Classification of Diseases and Related Health Problems (ICD9) codes, procedures as Current Procedural Terminology (CPT) codes, laboratory results (including but not limited to biochemistry, haematology, cytology, toxicology, microbiology and histopathology), medications and prescriptions, orders, vital signs, health factors and note titles. Free text, and diagnoses that were rare (fewer than 12 distinct patients with at least one occurrence in the VA database), were excluded to ensure all potential privacy concerns were addressed. In addition, conditions that were considered sensitive were excluded prior to transfer, such as patients with HIV/AIDS, sexually transmitted diseases, substance abuse, and those admitted to mental health services.
Following this set of inclusion criteria, the final dataset comprised 703,782 patients, providing 6,352,945,637 clinical event entries. Each clinical entry denoted a single procedure, laboratory test result, prescription, diagnosis etc, with 3,958,637,494 coming from outpatient events and the remaining 2,394,308,143 events from admissions. Extended Data Table 6 contains an overview of patient demographics in the data as well as prevalence of conditions associated with AKI across the data splits. The final dataset was randomly divided into training (80% of observations), validation (5%), calibration (5%) and testing (10%) sets. All data for a single patient was assigned to exactly one of these splits.
Data Preprocessing
Feature Representation
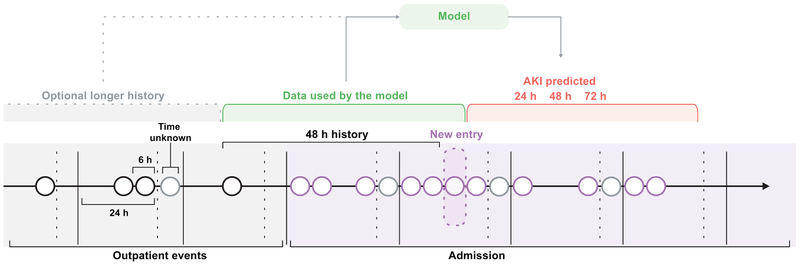
Every patient in the dataset was represented by a sequence of events, with each event providing the patient information that was recorded within a 6 hour period, i.e. each day was broken into four 6 hour periods and all records occurring within the same 6 hour period were grouped together. The available data within these six-hour windows, along with additional summary statistics and augmentations, formed a feature set that was used as input to our predictive models. Extended Data Figure 1 provides a diagrammatic view of a patient sequence and its temporal structure.
We did not perform any imputation of missing numerical values, because explicit imputation of missing values does not always provide consistent improvements to predictive models based on electronic health records32. Instead, we associated each numerical feature with one or more discrete presence features to enable our models to distinguish between the absence of a numerical value and an actual value of zero. Additionally, these presence features encoded whether a particular numerical value is considered to be normal, low, high, very low or very high. For some data points, the explicit numerical values were not recorded (usually when the values were considered normal), and the provision of this encoding of the numerical data allowed our models to process these measurements even in their absence. Discrete features like diagnostics or procedural codes were also encoded as binary presence features.
All numerical features were normalised to the [0, 1] range after capping the extreme values at the 1st and 99th percentile. This prevents the normalisation from being dominated by potentially large data entry errors while preserving most of the signal.
Each clinical feature was mapped onto a corresponding high-level concept, such as procedure, diagnosis, prescription, lab test, vital sign, admission, transfer etc. A total of 29 such high-level concepts were present in the data. At each step, a histogram of frequencies of these concepts among the clinical entries that take place at that step was provided to the models along with the numerical and binary presence features.
The approximate age of each patient in days, as well as which 6 hour period in the day the data is associated with, were provided as explicit features to the models. In addition, we provided some simple features that make it easier for the models to predict the risk of developing AKI. In particular, we provided the median yearly creatinine baseline and the minimum 48 hours creatinine baseline as additional numerical features. These are the baseline values that are used in the KDIGO criteria and help give important context to the models on how to interpret new serum creatinine measurements as they become available.
We additionally computed three historical aggregate feature representations at each step: one for the past 48 hours, one for the past 6 months, and one for the past 5 years. All histories were optionally provided to the models and the decision on which combination of historical data to include was based on the model performance on the validation set. We did this historical aggregation for discrete features by including whether they were observed in the historical interval or not. For numerical features we included the count, mean, median, standard deviation, minimum and maximum value observed in the interval, as well as simple trend features like the difference between the last observed value and the minimum or maximum and the average difference between subsequent steps (which measures the temporal short-term variability of the measurement). Supplementary Information section H provides the effect of volume and recency of available data on model performance.
Because patient measurements are made irregularly, not all 6-hour time periods in a day will have new data associated with them. Our models operate at regular time intervals regardless, and all time periods without new measurements include only the available metadata, and optionally the historical aggregate features. This approach makes continuous risk predictions possible, and allows our models to utilise the patterns of missingness in the data during the training process.
For about 35% of all entries, the day on which they occurred was known, but not the specific time during the day. For each day in the sequence of events, we aggregated these unknown-time entries into a specific bucket that was appended to the end of the day. This ensured that our models could iterate over this information without potentially leaking information from the future. Our models were not allowed to make predictions from these surrogate points and they were not factored into the evaluation. The models can utilise the information contained within the surrogate points on the next time step, corresponding to the first interval of the following day.
Diagnoses in the data are sometimes known to be recorded in the EHR prior to the time when an actual diagnosis was made clinically. To avoid leaking future information to the models, we shifted all of the diagnoses within each admission to the very end of that admission and only provided them to the models at that point, where they can be factored in for future admissions. This discards potentially useful information, so the performance obtained in this way is conservative by design and it is possible that in reality the models would be able to perform better with this information provided in a consistent way.
Ground Truth Labels using KDIGO
The patient AKI states were computed at each time step based on the KDIGO18 criteria, the recommendations of which are based on systematic reviews of relevant trials. KDIGO accepts three definitions of AKI: an increase in serum creatinine of 0.3mg/dl (26.5 μmol/l) within 48 hours; an increase in serum creatinine of 1.5 times a patient’s baseline creatinine level, known or presumed to have occurred within the prior 7 days; or a urine output of <0.5 ml/kg/h over 6 hours18. The first two definitions were used to provide ground truth labels for the onset of an AKI; the third definition could not be used as urine output was not recorded digitally in the majority of sites that formed part of this work. A baseline of median annualised creatinine was used where previous measurements where available; where these were not present the Modification of Diet in Renal Disease (MDRD) formula was applied to estimate a baseline creatinine. Using the KDIGO criteria based on serum creatinine and its corresponding definitions for AKI severity, three AKI categories were obtained: ‘all AKI’ (KDIGO stages 1, 2 & 3), ‘moderate and severe AKI’ (KDIGO stages 2 & 3), and ‘severe AKI’ (KDIGO stage 3).
The AKI stages were computed at times when there was a serum creatinine measurement present in the sequence and then copied forward in time until the next creatinine measurement, at which time the ground truth AKI state was updated accordingly. To avoid basing the current estimate of the KDIGO AKI stage on a previous measurement that may no longer be reliable, the AKI states were propagated for at most 4 days forward in case no new creatinine measurements were observed. From that point onwards, AKI states were marked as unknown. Patients experiencing acute kidney injury tend to be closely monitored and their levels of serum creatinine are measured regularly, so an absence of a measurement for multiple days in such cases is uncommon. A gap of 4 days between subsequent creatinine measurements represents the 95th percentile in the distribution of time between two consecutive creatinine measurements.
The prediction target at each point in time is a binary variable that is positive if the AKI category of interest (e.g., all AKI) occurs within a chosen future time horizon. If no AKI state was recorded within the chosen horizon, this was interpreted as a negative. We use eight future time horizons, 6h,12h, 18h, 24h, 36h, 48h, 60h, and 72h ahead, which are all available at each time point.
Event sequences of patients undergoing renal replacement therapy (RRT) were excluded from the target labels heuristically based on the data entries of RRT procedures being performed in the EHR, for the duration of dialysis administration. We have excluded entire subsequences of events between RRT procedures that occur within a week of each other. The edges of the subsequence were also appropriately excluded from label computations.
Models for predicting AKI
Our predictive system operates sequentially over the electronic health record. At each time point, input features, which we described above, were provided to a statistical model whose output is a probability of any-severity stage of AKI occurring in the next 48 hours. If this probability exceeds a chosen operating threshold, we make a positive prediction that can then trigger an alert. This is a general framework within which existing approaches also fit, and we describe the baseline methods in the next section. The novelty of this work is in the design of the particular model that is used and its training procedure, and the demonstration of its effectiveness - on a large-scale EHR dataset and across many different regimes - in making useful predictions of future AKI.
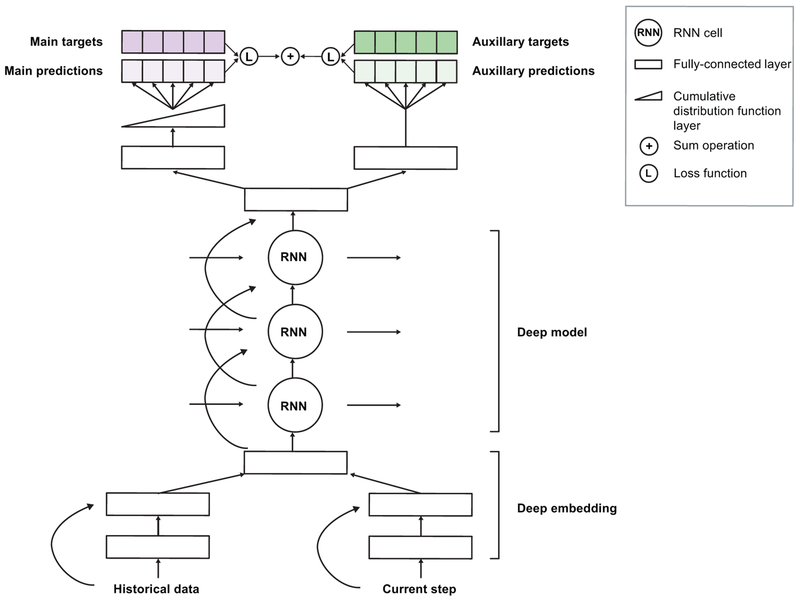
Extended Data Figure 2 gives a schematic view of our model, which makes predictions by first transforming the input features using an embedding module. This embedding is fed into a multi-layer recurrent neural network, the output of which at every time point is fed into a prediction module that provides the probability of future AKI at the time horizon for which the model will be trained. The entire model can be trained end-to-end, i.e. the parameters can be learned jointly without pretraining any parts of the model. To provide useful predictions, we train an ensemble of predictors to estimate the model’s confidence, and the resulting ensemble predictions are then calibrated using isotonic regression to reflect the frequency of observed outcomes33.
Embedding modules.
The embedding layers transform the high-dimensional and sparse input features into a lower-dimensional continuous representation that makes subsequent prediction easier. We use a deep multilayer perceptron with residual connections and rectified-linear (ReLU) activations. We use L1 regularisation on the embedding parameters to prevent overfitting and to ensure that our model focuses on the most salient features. We compared simpler linear transformations, which did not perform as well as the multi-layer version we used. We also compared unsupervised approaches such as factor analysis, standard auto-encoders and variational auto-encoders, but did not find any significant advantages in using these methods.
Recurrent neural network core.
Recurrent neural networks (RNNs) run sequentially over the EHR entries and are able to implicitly model the historical context of a patient by modifying an internal representation (or state) through time. We use a stacked multiple-layer recurrent network with highway connections between each layer34, which at each time step takes the embedding vector as an input. We use the Simple Recurrent Unit (SRU) network as the RNN architecture, with tanh activations. We chose this from a broad range of alternative RNN architectures, specifically the long short-term memory (LSTM)35, update gate RNN (UGRNN) and Intersection RNN36, simple recurrent units (SRU)37,38, gated recurrent units (GRU)39, the Neural Turing Machine (NTM)40, memory-augmented neural network (MANN)41, the Differentiable Neural Computer (DNC)42, and the Relational Memory Core (RMC)43. These alternatives did not provide significant performance improvements over the SRU architecture (see Supplement D).
Prediction targets and training objectives.
The output of the RNN is fed to a final linear prediction layer that makes predictions over all 8 future prediction windows (6 hour windows from 6 hours ahead to 72 hours ahead). We use a cumulative distribution function layer (CDF) across different time windows to encourage monotonicity, since the presence of AKI within a shorter time window implies a presence of AKI within a longer time window. Each of the resulting eight outputs provides a binary prediction for AKI severity at a specific time window and is compared to the ground truth label using the cross-entropy loss function (Bernoulli log-likelihood).
We also make a set of auxiliary numerical predictions, where at each step we also predict the maximum future observed value of a set of laboratory tests over the same set of time intervals as used to make the future AKI predictions. The laboratory tests predicted are ones known to be relevant to kidney function, specifically: creatinine, urea nitrogen, sodium, potassium, chloride, calcium and phosphate. This multitask approach results in better generalisation and more robust representations, especially under class imbalance44–46. The overall improvement we observed from including the auxiliary task was around 3% PR AUC in most cases (see Supplement A for more details).
Our overall loss function is the weighted sum of the cross-entropy loss from the AKI-predictions and the squared loss for each of the seven laboratory test predictions. We investigated the use of oversampling and overweighting of the positive labels to account for class imbalance. For oversampling, each mini-batch contains a larger percentage of positive samples than average in the entire dataset. For overweighting, prediction for positive labels contributes proportionally more to the total loss.
Training and hyperparameters.
We selected our proposed model architecture among several alternatives based on the validation set performance (see Supplement D) and have subsequently performed an ablation analysis of the design choices (see Supplement I). All variables are initialised via normalised (Xavier) initialisation47 and trained using the Adam optimisation scheme48. We employ exponential learning rate decay during training. The best validation results were achieved using an initial learning rate of 0.001 decayed every 12,000 training steps by a factor of 0.85, with a batch size of 128 and a backpropagation through time window of 128. The embedding layer is of size 400 for each of the numerical and presence input features (800 in total when concatenated) and uses 2 layers. The best performing RNN architecture used a cell size of 200 units per layer and 3 layers. A detailed overview of different hyperparameter combinations evaluated in the experiments is available in Supplement J. We conducted extensive hyperparameter explorations of dropout rates for different kinds of dropout to determine the best model regularisation. We have considered input dropout, output dropout, embedding dropout, cell state dropout and variational dropout. None of these had led to improvements, so dropout is not included in our model.
Competitive Baseline Methods
Established models for future AKI prediction make use of L1-regularised logistic regression or gradient boosted trees (GBTs), trained on a clinically relevant set of features known to be important either for routine clinical practice or the modelling of kidney function. A curated set of clinically-relevant features was chosen using existing AKI literature (see Supplement F) and the consensus opinion of six clinicians: three senior attending physicians with over twenty years expertise, one nephrologist and two intensive care specialists; and three clinical residents with expertise in nephrology, internal medicine and surgery. This set was further extended to include 36 of the most salient features discovered by our deep learning model that were not in the original list, to give further predictive signal to the baseline. The final curated dataset contained 315 base features of demographics, admission information, vital sign measurements, select laboratory tests and medications, and diagnoses of chronic conditions directly associated with an increased risk of AKI. The full feature set is listed in Supplement E We additionally computed a set of manually engineered features (yearly and 48-hourly baseline creatinine levels (consistent with KDIGO guidelines), the ratio of blood urea nitrogen to serum creatinine, grouped severely reduced glomerular filtration rate (corresponding to stages 3a to 5), and flagging diabetic patients by combining ICD9 codes and values of measured haemoglobin A1c) and a representation of the short-term and long-term history of a patient (see ‘Feature representation’). These features were provided explicitly, since the interaction terms and historical trends might not have been recovered by simpler models. This resulted in a total of 3599 possible features for the baseline model. We provide a table with a full set of baseline comparison in Supplement D.
Evaluation
The data was split into training, validation, calibration and test sets in such a way that information from a given patient is present only in one split. The training split was used to train the proposed models. The validation set was used to iteratively improve the models by selecting the best model architectures and hyperparameters.
The models selected on the validation set were recalibrated on the calibration set in order to further improve the quality of the risk predictions. Deep learning models with softmax or sigmoid output trained with cross-entropy loss are prone to miscalibration, and recalibration ensures that consistent probabilistic interpretations of the model predictions can be made49. For calibration we considered Platt scaling50 and Isotonic Regression33. To compare uncalibrated predictions to recalibrated ones we used the Brier score51 and reliability plots52. The best models were finally evaluated on the independent test set that was held out during model development.
The main metrics used in model selection and the final report are: the AKI episode sensitivity, the area under the precision-recall curve (PR AUC), the area under the receiver-operating curve (ROC AUC), and the per-step precision, per-step sensitivity and per-step specificity. The AKI episode sensitivity corresponds to the percentage of all AKI episodes that were correctly predicted ahead of time within the corresponding time windows of up to 48 hours. In contrast, the precision is computed per-step since the predictions are made at each step, to account for the rate of false alerts over time.
Due to the sequential nature of making predictions, the total number of positive steps does not directly correspond to the total number of distinct AKI episodes. Multiple positive alerting opportunities may be associated with a single AKI episode and different AKI episodes may offer a different number of such early alerting steps depending on how late they occur within the admission. AKIs occurring later during in-hospital stay can be predicted earlier than those that occur immediately upon admission. To better assess the clinical applicability of the proposed model we explicitly compute the AKI episode sensitivity for different levels of step-wise precision.
Given that the models were designed for continuous monitoring and risk prediction, they were evaluated at each 6-hour time step within all of the admissions for each patient except for the steps within AKI episodes which were ignored. The models were not evaluated on outpatient events. All steps where there was no record of AKI occurring in the relevant future time window were considered as negative examples.
Approximately 2% of individual time steps presented to the models sequentially were associated with a positive AKI label, so the AKI prediction task is class-imbalanced. For per-step performance metrics, we report both the area under the receiver operating characteristic curve (ROC AUC) as well as the area under the precision-recall curve (PR AUC). PR AUC is known to be more informative for class-imbalanced predictive tasks53, as it is more sensitive to changes in the number of false positive predictions.
To gauge uncertainty on a trained model’s performance we calculated 95% confidence intervals with the pivot bootstrap estimator54. This was done by sampling the entire validation and test dataset with replacement 200 times. Because bootstrapping assumes the resampling of independent events, we resample entire patients instead of resampling individual admissions or time steps. Where appropriate we also compute a Mann–Whitney U test (two-sided)55 on the samples for the respective models.
To quantify the uncertainty on model predictions (versus overall performance) we trained an ensemble of 100 models with a fixed set of hyperparameters but different initial seeds. This follows similar uncertainty approaches in supervised learning56 and medical imaging predictions57. The prediction confidence was assessed by inspecting the variance over the 100 model predictions from the ensemble. This confidence reflected the accuracy of a prediction: the mean standard deviation of false positive predictions was higher than the mean standard deviation of true positive predictions and similarly for false negative versus true negative predictions (p-value < 0.01, see Supplement K).
Reporting Summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Ethics and Information Governance
This work, and the collection of data on implied consent, received Tennessee Valley Healthcare System Institutional Review Board (IRB) approval from the US Department of Veterans Affairs. De-identification was performed in line with the Health Insurance Portability and Accountability Act (HIPAA), and validated by the US Department of Veterans Affairs Central Database and Information Governance departments. Only de-identified retrospective data was used for research, without the active involvement of patients.
Code Availability
We make use of several open-source libraries to conduct our experiments, namely the machine learning framework TensorFlow (https://github.com/tensorflow/tensorflow) along with the TensorFlow library Sonnet (https://github.com/deepmind/sonnet) which provides implementations of individual model components58. Our experimental framework makes use of proprietary libraries and we are unable to publicly release this code. We detail the experiments and implementation details in the methods section and in the supplementary figures to allow for independent replication.
Data Availability
The clinical data used for the training, validation and test sets was collected at the US Department of Veterans Affairs and transferred to a secure data centre with strict access controls in de-identified format. Data was used with both local and national permissions. It is not publicly available and restrictions apply to its use. The de-identified dataset, or a test subset, may be available from the US Department of Veterans Affairs subject to local and national ethical approvals.
Extended Data
Extended Data Figure 1 |. The sequential representation of EHR data.
All EHR data available for each patient was structured into a sequential history for both inpatient and outpatient events in six hourly blocks, shown here as circles. In each 24 hour period events without a recorded time were included in a fifth block. Apart from the data present at the current time step, the models optionally receive an embedding of the previous 48 hours and the longer history of 6 months or 5 years.
Extended Data Figure 2 |. The proposed model architecture.
The best performance was achieved by a multitask deep recurrent highway network architecture on top of an L1-regularised deep residual embedding component that learns the best data representation end-to-end without pre-training.
Extended Data Figure 3 |. Calibration.
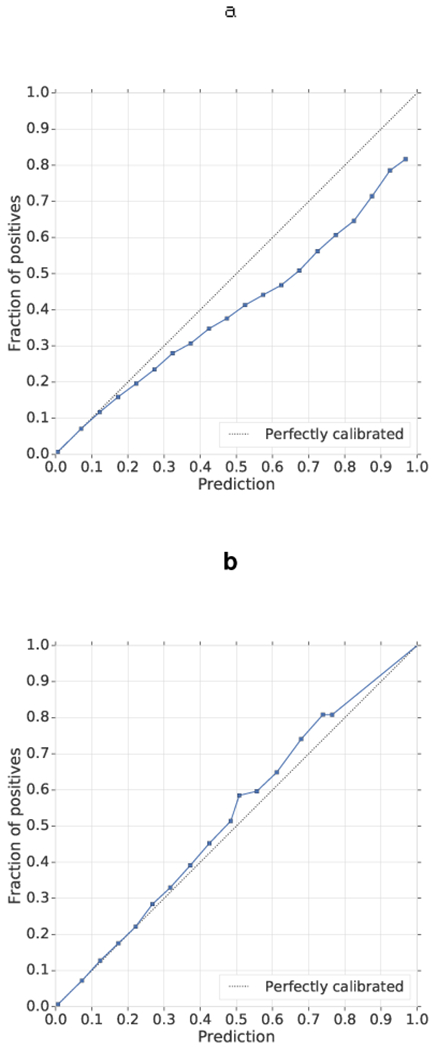
a, b, The predictions were recalibrated using isotonic regression before (a) and after (b) calibration. Model predictions were grouped into 20 buckets, with a mean model risk prediction plotted against the percentage of positive labels in that bucket. The diagonal line demonstrates the ideal calibration.
Extended Data Figure 4 |. Analysis of false positive predictions.
a, For prediction of any AKI within 48 h at 33% precision, nearly half of all predictions are trailing, after the AKI has already occurred (orange bars) or early, more than 48 h prior (blue bars). The histogram shows the distribution of these trailing and early false positives for prediction. Incorrect predictions are mapped to their closest preceding or following episode of AKI (whichever is closer) if that episode occurs in an admission. For ±1 day, 15.2% of false positives correspond to observed AKI events within 1 day after the prediction (model reacted too early) and 2.9% correspond to observed AKI events within 1 day before the prediction (model reacted too late). b, Subgroup analysis for all false-positive alerts. In addition to the 49% of false-positive alerts that were made in admissions during which there was at least one episode of AKI, many of the remaining false-positive alerts were made in patients who had evidence of clinical risk factors present in their available electronic health record data. These risk factors are shown here for the proposed model that predicts any stage of AKI occurring within the next 48 h.
Extended Data Table 1 |. Model performance for predicting AKI within the full range of possible prediction windows from 6-72 hours.
On shorter time windows, closer to the actual onset of AKI, the model achieves a higher ROC AUC (a), but lower PR AUC (b). This stems from different numbers of positive steps within windows of different length. These differences affect both the model precision and the false positive rate. When making predictions across shorter time windows there is more uncertainty in the exact time of the AKI onset due to minor physiological fluctuations and this results in a lower precision being needed in order to achieve high sensitivity. 95% bootstrap pivot confidence intervals are calculated using n=200 bootstrap samples.
| a | |||
|---|---|---|---|
| ROC AUC [95% CI] | |||
| Time windows | Any AKI | AKI stages 2 and 3 | AKI stage 3 |
| 24h | 93.4% [93.3, 93.6] | 97.1% [96.9, 97.3] | 98.8% [98.7, 98.9] |
| 48h | 92.1% [91.9, 92.3] | 95.7% [95.5, 96.0] | 98.0% [97.8, 98.2] |
| 72h | 91.4% [91.1, 91.6] | 94.7% [94.4, 95.0] | 97.3% [97.2, 97.6] |
| b | |||
| PR AUC [95% CI] | |||
| Time windows | Any AKI | AKI stages 2 and 3 | AKI stage 3 |
|
| |||
| 24h | 25.9% [24.6, 27.0] | 36.8% [35.1, 38.7] | 47.6% [45.1,49.7] |
| 48h | 29.7% [28.5, 30.8] | 37.8% [36.1, 39.6] | 48.7% [46.4, 51.1] |
| 72h | 31.7% [30.6, 32.8] | 37.4% [35.6, 39.1] | 48.0% [46.1,49.9] |
Extended Data Table 2 |. Daily frequency of true and false positive alerts when predicting different stages of AKI.
The frequency of alerts and its standard deviation are shown for a time window of 48 hours an operating point corresponding to a 1:2 TP:FP ratio (N=5101 days). On an average day, clinicians would receive true positive alerts of AKI predicted to occur within a window of 48 hours ahead in 0.85% of all in-hospital patients, and a false positive prediction of a future AKI in 1.89% of patients, when predicting the future AKI of any severity. Assuming none of the false positives can be filtered out and immediately discarded, clinicians would need to attend to approximately 2.7% of all in-hospital patients. For the most severe stages of AKI, the model alerts on an average day in 0.8% of all patients. Of those, 0.27% are true positives and 0.56% are false positives. Note that there are multiple time steps at which the predictions are made within each day, so the TP:FP ratio of the daily alerts differs slightly from the step-wise ratio. (a) Daily frequency of true and false positive alerts when predicting any stage of AKI. (b) Daily frequency of true and false positive alerts when predicting KDIGO AKI stages two and above. (c) Daily frequency of true and false positive alerts when predicting the most severe stage of AKI - KDIGO AKI stage 3.
| a | |
|---|---|
| Alert type | Frequency predicting any stage of AKI |
| True positive alerts | 0.85% ± 0.71 |
| False positive alerts | 1.89% ± 1.20 |
| No alerts | 97.26% ± 1.63 |
| b | |
| Alert type | Frequency predicting KDIGO AKI stages 2 and above |
| True positive alerts | 0.30% ± 0.35 |
| False positive alerts | 0.64% ± 0.55 |
| No alerts | 99.06% ± 0.75 |
| c | |
| Alert type | Frequency predicting KDIGO AKI stage 3 |
| True positive alerts | 0.27% ± 0.33 |
| False positive alerts | 0.56% ± 0.85 |
| No alerts | 99.17% ± 0.96 |
Extended Data Table 3 |. Model performance on patients requiring subsequent dialysis.
Model performance only in AKI cases where either in-hospital or outpatient administration of dialysis is required within 30 days of the onset of AKI, or where regular outpatient administration of dialysis is scheduled within 90 days. The model successfully predicts a large proportion of these AKI cases early, 84.3% of AKI cases where there is any dialysis administration occurring within 30 days and 90.2% of cases where regular outpatient administration of dialysis occurs within 90 days.
| Subgroup name | Sensitivity (AKI episode) | PRAUC | ROC AUC | Sensitivity (step) | Specificity (step) |
|---|---|---|---|---|---|
| In-hospital/outpatient dialysis within 30 days | 84.3% | 70.5% | 83.5% | 67.7% | 83.3% |
| Outpatient dialysis within 90 days | 90.2% | 71.9% | 83.8% | 76.5% | 76.3% |
Extended Data Table 4 |. Operating points for predicting AKI up to 48 hours ahead of time.
(a) For prediction of any AKI, the model correctly identifies 55.8% of all AKI episodes early if allowing for two false positives for every true positive, and 34.7% if allowing for one false positive for every true positive. For more severe AKI stages it is possible to achieve a higher sensitivity for any fixed level of precision. Performance increases for prediction of (b) AKI stages 2 & 3, and (c) AKI stage 3 alone. 95% bootstrap pivot confidence intervals are calculated using n=200 bootstrap samples for all tables.
| a | ||||
|---|---|---|---|---|
| Operating points for predicting any AKI up to 48 hours ahead of time | ||||
| Precision | True positive / False positive | Sensitivity [95% Cl] (AKI episode) | Sensitivity [95% Cl] (step) | Specificity [95% Cl] (step) |
| 20.0% | 1:4 | 76.7% [75.6, 77.8] | 58.3% [56.9, 59.8] | 94.8% [94.6, 95.1] |
| 25.0% | 1:3 | 68.2% [66.9, 69.7] | 47.7% [46.1,49.4] | 96.8% [96.6, 97.0] |
| 33.0% | 1:2 | 55.8% [53.9, 57.7] | 35.0% [33.3, 36.7] | 98.4% [98.3, 98.5] |
| 40.0% | 2:3 | 46.6% [44.5, 49.0] | 27.1% [25.2, 28.9] | 99.1% [99.0, 99.2] |
| 50.0% | 1:1 | 34.7% [32.0, 37.2] | 18.5% [16.7, 20.3] | 99.6% [99.5, 99.6] |
| 60.0% | 3:2 | 24.7% [21.8, 27.3] | 12.4% [10.5, 13.9] | 99.8% [99.8, 99.8] |
| 75.0% | 3:1 | 12.0% [9.3, 14.6] | 5.5% [3.9, 7.0] | 100.0% [99.9, 100.0] |
| b | ||||
| Operating points for predicting AKI stages 2 and 3 up to 48 hours ahead of time | ||||
| Precision | True positive / False positive | Sensitivity [95% Cl] (AKI episode) | Sensitivity [95% Cl] (step) | Specificity [95% Cl] (step) |
| 20.0% | 1:4 | 82.0% [80.6, 83.5] | 65.8% [64.0, 67.9] | 98.5% [98.4, 98.6] |
| 25.0% | 1:3 | 77.8% [76.3, 79.7] | 60.4% [58.3, 62.8] | 99.0% [98.9, 99.1] |
| 33.0% | 1:2 | 71.4% [69.6, 73.7] | 51.8% [49.6, 54.8] | 99.4% [99.4, 99.5] |
| 40.0% | 2:3 | 65.2% [63.0, 67.7] | 44.6% [42.1,47.3] | 99.6% [99.6, 99.7] |
| 50.0% | 1:1 | 56.2% [54.0, 59.2] | 35.8% [33.5, 38.9] | 99.8% [99.8, 99.8] |
| 60.0% | 3:2 | 45.1% [42.2, 48.6] | 26.3% [23.8, 29.4] | 99.9% [99.9, 99.9] |
| 75.0% | 3:1 | 27.5% [24.2, 31.5] | 13.8% [11.7, 16.3] | 100.0% [100.0, 100.0] |
| c | ||||
| Operating points for predicting AKI stage 3 up to 48 hours ahead of time | ||||
| Precision | True positive / False positive | Sensitivity [95% Cl] (AKI episode) | Sensitivity [95% Cl] (step) | Specificity [95% Cl] (step) |
| 20.0% | 1:4 | 91.2% [90.4, 92.3] | 80.3% [78.4, 82.4] | 98.8% [98.7, 98.9] |
| 25.0% | 1:3 | 88.8% [87.7, 90.1] | 75.8% [73.7, 78.3] | 99.1% [99.0, 99.2] |
| 33.0% | 1:2 | 84.1% [82.4, 85.9] | 68.3% [65.7, 71.0] | 99.5% [99.4, 99.5] |
| 40.0% | 2:3 | 79.5% [77.4, 81.8] | 61.1% [57.9, 64.5] | 99.7% [99.6, 99.7] |
| 50.0% | 1:1 | 71.3% [68.3, 74.4] | 50.2% [46.4, 53.8] | 99.8% [99.8, 99.8] |
| 60.0% | 3:2 | 61.2% [57.6, 64.9] | 39.9% [35.7, 43.8] | 99.9% [99.9, 99.9] |
| 75.0% | 3:1 | 40.5% [36.5, 46.1] | 23.2% [19.6, 27.2] | 100.0% [100.0, 100.0] |
Extended Data Table 5 |. Future and cross-site generalisability experiments.
(a) Model performance when trained before the time point tP and tested after tP, both on the entirety of the future patient population as well as subgroups of patients for which the model has or hasn’t seen historical information during training. The model maintains a comparable level of performance on unseen future data, with a higher level of sensitivity of 59% for a time window of 48 hours ahead of time and a precision of two false positives per step for each true positive. The ranges correspond to bootstrap pivotal 95% confidence intervals with n=200. Note that this experiment is not a replacement for a prospective evaluation of the model. (b) Cohort statistics for (a), shown for both before and after the temporal split tP that was used to simulate model performance on future data. (c) Comparison of model performance when applied to data from previously unseen hospital sites. Data was split across sites so that 80% of the data was in group A and 20% in group B. No site from group B was present in group A and vice versa. The data was split into training, validation, calibration and test in the same way as in the other experiments. The table reports model performance when trained on site group A when evaluating on the test set within site group A versus the test set within site group B for predicting all AKI severities up to 48 hours ahead of time. Comparable performance is seen across key all key metrics. 95% bootstrap pivot confidence intervals are calculated using n=200 bootstrap samples. Note that the model would still need to be retrained to generalise outside of the VA population to a different demographic and a different set of clinical pathways and hospital processes elsewhere.
| a | ||||
|---|---|---|---|---|
| Patient cohorts |
||||
| Metric [95% CI] | Before tp (test) | New admissions after tp (test) | Subsequent admissions after tp | All patients after tp |
| Sensitivity (AKI episode) | 55.09 [54.01, 56.06] | 59 [57.11, 60.71] | 59.04 [58.38, 59.63] | 58.97 [58.33, 59.52] |
| ROC AUC | 92.25 [92.01, 92.42] | 90.19 [89.76, 90.77] | 89.98 [89.83, 90.17] | 89.98 [89.81, 90.14] |
| PRAUC | 29.97 [28.61, 31.15] | 30.75 [28.65, 32.81] | 31.54 [30.87, 32.30] | 31.28 [30.44, 32.02] |
| Sensitivity (step) | 34.26 [33.17, 35.28] | 36.87 [35.2, 38.85] | 37.23 [36.67, 37.88] | 37.08 [36.40, 37.65] |
| Specificity (step) | 98.55 [98.50, 98.60] | 97.66 [97.54, 97.76] | 97.63 [97.58, 97.68] | 97.64 [97.59, 97.68] |
| Precision | 32.51 [31.44, 33.21] | 32.66 [31.2, 34.03] | 32.97 [32.52, 33.47] | 32.84 [32.28, 33.33] |
| b | ||||
| Before tp | After tp | |||
| Patients | ||||
| Number of patients | 599,871 | 246,406 | ||
| Average age* | 61.3 | 64.2 | ||
| Admissions within a given period | ||||
| Unique admissions | 2,134,544 | 364,778 | ||
| ICU admissions | 226,585(10.62%) | 40,102 (10.99%) | ||
| Medical admissions | 1,040,923 (48.77%) | 170,383 (46.71%) | ||
| Surgical admissions | 373,823(17.51%) | 67,617 (18.54%) | ||
| No creatinine measured | 458,486 (21.48%) | 52,115 (14.29%) | ||
| Any Chronic Kidney Disease | 774,883 (36.30%) | 156,181 (42.82%) | ||
| Any AKI present | 282,398(13.23%) | 41,950 (14.59%) | ||
| c | ||||
| Metric [95% Cl] | Site group A | Site group B | ||
| Sensitivity (AKI episode) | 55.6% [54.5, 56.6] | 54.6% [52.8, 56.3] | ||
| ROC AUC | 91.8% [91.6, 92.1] | 91.3% [90.8, 91.7] | ||
| PRAUC | 30.0% [28.6, 31.2] | 30.6% [28.3, 32.7] | ||
| Sensitivity (step) | 34.3% [33.1, 35.2] | 34.7% [32.6, 36.2] | ||
| Specificity (step) | 98.5% [98.4, 98.5] | 98.3% [98.2, 98.4] | ||
Extended Data Table 6 |. Summary statistics for the data.
A breakdown of training (80%), validation (5%), calibration (5%) and test (10%) datasets by both unique patients and individual admissions. Where appropriate, percent of total dataset size is reported in parentheses. The dataset was representative of the overall VA population for clinically relevant demographics and diagnostic groups associated with renal pathology. *Average age after taking into account exclusion criteria and statistical noise added to meet HIPAA Safe Harbor criteria. **CKD stage 1 is evidence of renal parenchymal damage with a normal glomerular filtration rate (GFR). This is rarely recorded in our dataset; instead the numbers for stage 1 CKD have been estimated from admissions that carried an ICD-9 code for CKD, but where GFR was normal. For this reason these numbers may under-represent the true prevalence in the population. ***172 VA inpatient sites and 1,062 outpatient sites were eligible for inclusion. 130 data centres aggregate data from one or more of these facilities, of which 114 such data centres had data for inpatient admissions used in this study. While the exact number of sites included was not provided in the dataset for this work, no patients were excluded based on location.
| Training | Validation | Calibration | Test | ||
|---|---|---|---|---|---|
| Patients | |||||
| Unique patients | 562,507 | 35,277 | 35,317 | 70,681 | |
| Average age* | 62.4 | 62.5 | 62.4 | 62.3 | |
| Ethnicity | Black | 106,299(18.9%) | 6,544(18.6%) | 6,675(18.6%) | 13,183 (18.7%) |
| Other | 456,208 (81.1%) | 28,733 (81.4%) | 28,642 (81.4%) | 57,498 (81.3%) | |
| Gender | Female | 35,855 (6.4%) | 2,300 (6.5%) | 2,252 (6.4%) | 4,519 (6.4%) |
| Male | 526,652 (93.6%) | 32,977 (93.5%) | 33,065 (93.6%) | 66,162 (93.6%) | |
| Diabetes | 56,958(10.1%) | 3,599(10.2%) | 3,702(10.5%) | 7,093 (10.0%) | |
| Admissions within a five year period | |||||
| Data center sites | 130*** | 130*** | 130*** | 130*** | |
| Unique admissions per patient | 2,004,217 | 124,255 | 125,928 | 252,492 | |
| Average | 3.6 | 3.5 | 3.6 | 3.6 | |
| Median | 2 | 2 | 2 | 2 | |
| Duration (days) | Average | 9.6 | 9.6 | 9.6 | 9.6 |
| Median | 3.2 | 3.2 | 3.2 | 3.2 | |
| ICU admissions | 214,644(10.7%) | 13,161 (10.6%) | 13,411 (10.6%) | 26,739 (10.6%) | |
| Medical admissions | 971,527 (48.5%) | 60,762 (48.9%) | 61,281 (48.7%) | 121,675 (48.2%) | |
| Surgical admissions | 354,008(17.7%) | 21,857(17.6%) | 22,093(17.5%) | 44,766 (17.7%) | |
| Renal replacement therapy | 22,284(1.1%) | 1,367(1.1%) | 1,384(1.1%) | 2,784 (1.1%) | |
| No creatinine measured | 408,927 (20.4%) | 25,162 (20.3%) | 25,503 (20.3%) | 51,484 (20.4%) | |
| Chronic Kidney Disease | Any | 746,692 (37.3%) | 46,677 (37.5%) | 46,622 (37.0%) | 94,105 (37.3%) |
| Stage 1** | 8,409 (0.4%) | 515 (0.4%) | 576 (0.5%) | 1,103 (0.4%) | |
| Stage 2 | 429,990 (21.5%) | 27,162 (21.9%) | 26,927 (21.4%) | 54,476 (21.6%) | |
| Stage 3A | 156,720 (7.8%) | 9,837 (7.9%) | 9,803 (7.8%) | 19,548 (7.7%) | |
| Stage 3B | 77,801 (3.9%) | 4,675 (3.8%) | 4,823 (3.7%) | 9,760 (3.9%) | |
| Stage 4 | 50,535 (2.5%) | 3,004 (2.5%) | 3,066 (2.5%) | 6,223 (2.5%) | |
| Stage 5 | 31,646(1.6%) | 1,999(1.6%) | 2,003(1.6%) | 4,098 (1.6%) | |
| AKI present | Any AKI | 267,396(13.3%) | 16,671 (13.4%) | 16,760(13.3%) | 33,759 (13.4%) |
| Stage 1 | 207,441 (10.4%) | 12,794(10.3%) | 12,951 (10.3%) | 26,215(10.4%) | |
| Stage 2 | 43,446 (2.2%) | 2,780 (2.2%) | 2,783 (2.2%) | 5,575 (2.2%) | |
| Stage 3 | 66,734 (3.3%) | 4,267 (3.4%) | 4,162 (3.3%) | 8,453 (3.3%) | |
Supplementary Material
Acknowledgements
We thank the Veterans and their families under the care of the US Department of Veterans Affairs. We would also like to thank A. Graves, O. Vinyals, K. Kavukcuoglu, S. Chiappa, T. Lillicrap, R. Raine, P. Keane, A. Schlosberg, O. Ronneberger, J. De Fauw, K. Ruark, M. Jones, J. Quinn, D. Chou, C. Meaden, G. Screen, W. West, R. West, P. Sundberg and the Google AI team, J. Besley, M. Bawn, K. Ayoub and R. Ahmed. Finally, we thank the many VA physicians, administrators and researchers who worked on the data collection, and the rest of the DeepMind team for their support, ideas and encouragement.
G.R. & H.M. were supported by University College London and the National Institute for Health Research (NIHR) University College London Hospitals Biomedical Research Centre. The views expressed are those of these author(s) and not necessarily those of the NHS, the NIHR or the Department of Health.
Abbreviations
- AE
Autoencoder
- AKI
Acute Kidney Injury
- AKIN
Acute Kidney Injury Network
- AUC
Area Under Curve
- BIDMC
Beth Israel Deaconess Medical Center
- CDF
Cumulative Distribution Function
- CKD
Chronic Kidney Disease
- CNN
Convolutional Neural Network
- COPD
Chronic Obstructive Pulmonary Disease
- CPT
Current Procedural Terminology
- DNC
Differentiable Neural Computer
- ED
Emergency Department
- EHR
Electronic Health Record
- ER
Emergency Room
- GAM
Generalised Additive Model
- GBT
Gradient Boosted Trees
- GFR
Glomerular Filtration Rate
- GRU
Gated Recurrent Unit
- GP
Gaussian Processes
- HIPAA
Health Insurance Portability and Accountability Act
- ICD-9
International Statistical Classification of Diseases and Related Health Problems
- ICU
Intensive Care Unit
- IRB
Institutional Review Board
- ITU
Intensive Treatment Unit
- IV
Intravenous Therapy
- KDIGO
Kidney Disease: Improving Global Outcomes guidelines
- LOINC
Logical Observation Identifiers Names and Codes
- LR
Logistic Regression
- LSTM
Long Short-Term Memory Network
- MANN
Memory-Augmented Neural Network
- MDP
Markov Decision Process
- MLP
Multilayer Perceptron
- NHSE
National Health Service England
- NPV
Negative Predictive Value
- NTM
Neural Turing Machine
- PPV
Positive Predictive Value
- PR
Precision/Recall
- ReLU
Rectified Linear Unit
- RF
Random Forest
- RIFLE
Risk, Injury, Failure, Loss of kidney function, and End-stage kidney disease
- RNN
Recurrent Neural Network
- RMC
Relational Memory Core
- ROC
Receiver Operating Characteristic
- RRT
Renal Replacement Therapy
- SMC
Stanford Medical Centre
- SRU
Simple Recurrent Unit
- TRIPOD
Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis
- UGRNN
Update Gate Recurrent Neural Network
- VA
US Department of Veterans Affairs
- VAE
Variational Autoencoder
- WCC
White Cell Count
Footnotes
Supplementary Information
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
G.R., H.M. and C.L. are paid contractors of DeepMind. The authors have no other competing interests to disclose.
References
- 1.Thomson R, Luettel D, Healey F, and Scobie S, “Safer care for the acutely ill patient: Learning from serious incidents”, National Patient Safety Agency, 2007. [Google Scholar]
- 2.Henry KE, Hager DN, Pronovost PJ, and Saria S, “A targeted real-time early warning score (trewscore) for septic shock”, Science Translational Medicine, vol. 7, no. 299, pp. 299ra122–299ra122, 2015. [DOI] [PubMed] [Google Scholar]
- 3.Rajkomar A, Oren E, Chen K, Dai AM, Hajaj N, Hardt M, Liu PJ, Liu X, Marcus J, Sun M, Sundberg P, Yee H, Zhang K, Zhang Y, Flores G, Duggan GE, Irvine J, Le Q, Litsch K, Mossin A, Tansuwan J, Wang D, Wexler J, Wilson J, Ludwig D, Volchenboum SL, Chou K, Pearson M, Madabushi S, Shah NH, Butte AJ, Howell M, Cui C, Corrado G, and Dean J, “Scalable and accurate deep learning with electronic health records”, NPJ Digital Medicine, vol. 1, no. 1, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koyner JL, Adhikari R, Edelson DP, and Churpek MM, “Development of a multicenter ward based AKI prediction model”, Clinical Journal of the American Society of Nephrology, pp. 1935–1943, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng P, Waitman LR, Hu Y, and Liu M, “Predicting inpatient acute kidney injury over different time horizons: How early and accurate?”, in AMIA Annual Symposium Proceedings, vol. 2017, p. 565, American Medical Informatics Association, 2017. [PMC free article] [PubMed] [Google Scholar]
- 6.Koyner JL, Carey KA, Edelson DP, and Churpek MM, “The development of a machine learning inpatient acute kidney injury prediction model”, Critical Care Medicine, vol. 46, no. 7, pp. 1070–1077, 2018. [DOI] [PubMed] [Google Scholar]
- 7.Komorowski M, Celi LA, Badawi O, Gordon A, and Faisal A, “The artificial intelligence clinician learns optimal treatment strategies for sepsis in intensive care”, Nature Medicine, vol. 24, pp. 1716–1720, 2018. [DOI] [PubMed] [Google Scholar]
- 8.Avati A, Jung K, Harman S, Downing L, Ng AY, and Shah NH, “Improving palliative care with deep learning,” 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), pp. 311–316, 2017. [Google Scholar]
- 9.Lim B and van der Schaar M, “Disease-Atlas: Navigating disease trajectories with deep learning,” Proceedings of Machine Learning Research, vol. 85, 2018. [Google Scholar]
- 10.Futoma J, Hariharan S, and Heller KA, “Learning to detect sepsis with a multitask gaussian process RNN classifier,” in Proceedings of the International Conference on Machine Learning, (Precup D and Teh YW, eds.), pp. 1174–1182, 2017. [Google Scholar]
- 11.Miotto R, Li L, Kidd B, and Dudley JT, “Deep Patient: An unsupervised representation to predict the future of patients from the electronic health records,” Scientific Reports, vol. 6, no. 26094, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lipton ZC, Kale DC, Elkan C, and Wetzel R, “Learning to diagnose with LSTM recurrent neural networks,”International Conference on Learning Representations, 2016. [Google Scholar]
- 13.Yu Cheng PZJH, Wang Fei, “Risk prediction with electronic health records a deep learning approach,” in Proceedings of the SIAM International Conference on Data Mining, pp. 432–440, 2016. [Google Scholar]
- 14.Soleimani H, Subbaswamy A, and Saria S, “Treatment-response models for counterfactual reasoning with continuous-time, continuous-valued interventions,” arXiv Preprint, arXiv:1704.02038, 2017.
- 15.Alaa AM, Yoon J, Hu S, and van der Schaar M, “Personalized risk scoring for critical care patients using mixtures of gaussian process experts,” arXiv Preprint, arXiv:1605.00959, 2016.
- 16.Perotte A, Elhadad N, Hirsch JS, Ranganath R, and Blei D, “Risk prediction for chronic kidney disease progression using heterogeneous electronic health record data and time series analysis,” Journal of the American Medical Informatics Association, vol. 22, no. 4, pp. 872–880, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bihorac A, Ozrazgat-Baslanti T, Ebadi A, Motaei A, Madkour M, Pardalos PM, Li-230pori G, Hogan WR, Efron PA, Moore F, et al. , “MySurgeryRisk: Development and validation of a machine-learning risk algorithm for major complications and death after surgery,”Annals of Surgery, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khwaja A, “KDIGO clinical practice guidelines for acute kidney injury,” Nephron Clinical Practice, vol. 120, no. 4, pp. c179–c184, 2012. [DOI] [PubMed] [Google Scholar]
- 19.Stenhouse C, Coates S, Tivey M, Allsop P, and Parker T, “Prospective evaluation of a modified early warning score to aid earlier detection of patients developing critical illness on a general surgical ward,” The British Journal of Anaesthesia, vol. 84, no. 5, p. 663P, 2000. [Google Scholar]
- 20.Alge JL and Arthur JM, “Biomarkers of AKI: A review of mechanistic relevance and potential therapeutic implications,” Clinical Journal of the American Society of Nephrology, vol. 10, no. 1, pp. 147–155, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang HE, Muntner P, Chertow GM, and Warnock DG, “Acute kidney injury and mortality in hospitalized patients,” American Journal of Nephrology, vol. 35, pp. 349–355, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MacLeod A, “NCEPOD report on acute kidney injury—must do better,” The Lancet, vol. 374, no. 9699, pp. 1405–1406, 2009. [DOI] [PubMed] [Google Scholar]
- 23.Lachance P, Villeneuve PM, Rewa OG, Wilson FP, Selby NM, Featherstone RM, and Bagshaw SM, “Association between e-alert implementation for detection of acute kidney injury and outcomes: a systematic review,” Nephrology Dialysis Transplantation, vol. 32, no 2, pp. 265–272, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson AEW, Ghassemi MM, Nemati S, Niehaus KE, Clifton DA, and Clifford GD, “Machine learning and decision support in critical care,” Proceedings of the IEEE, vol. 104, no. 2, pp. 444–466, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohamadlou H, Lynn-Palevsky A, Barton C, Chettipally U, Shieh L, Calvert J, Saber NR, and Das R, “Prediction of acute kidney injury with a machine learning algorithm using electronic health record data,”Canadian Journal of Kidney Health And Disease, vol. 5, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan Z, Du H, Yuan Ngiam K, Wang F, Shum P, and Feng M, “A self-correcting deep learning approach to predict acute conditions in critical care,” arXiv Preprint, arXiv:1901.04364, 2019. [Google Scholar]
- 27.Park S, Baek SH, Ahn S, Lee K-H, Hwang H, Ryu J, Ahn SY, Chin HJ, Na KY, Chae D-W, and Kim S, “Impact of electronic acute kidney injury (AKI) alerts with automated nephrologist consultation on detection and severity of AKI: A quality improvement study,” American Journal of Kidney Diseases, vol. 71, no. 1, pp. 9–19, 2018. [DOI] [PubMed] [Google Scholar]
- 28.Chen I, Johansson FD, and Sontag D, “Why is my classifier discriminatory?,” arXiv Preprint, arXiv:1805.12002, 2018.
- 29.Schulam P and Saria S, “Reliable decision support using counterfactual models,” in Advances in Neural Information Processing Systems, (Guyon I, Luxburg U, Bengio S, Wallach H, Fergus R, Vishwanathan S, and Garnett R, eds.), vol. 30, pp. 1697–1708,2762017. [Google Scholar]
- 30.Telenti A, Steinhubl SR, and Topol EJ, “Rethinking the medical record,” The Lancet, vol. 391, no. 10125, p. 1013, 2018. [DOI] [PubMed] [Google Scholar]
Methods-only References
- 31.Department of Veterans Affairs, “Veterans Health Administration: Providing health care for Veterans.” https://www.va.gov/health/, 2018. (Accessed November 9, 2018).
- 32.Razavian N and Sontag D, “Temporal convolutional neural networks for diagnosis from lab tests,” arXiv Preprint, arXiv:1511.07938, 2015.
- 33.Zadrozny B and Elkan C, “Transforming classifier scores into accurate multiclass prob-ability estimates,” in Proceedings of the 8th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, pp. 694–699, ACM, 2002. [Google Scholar]
- 34.Zilly JG, Srivastava RK, Koutník J, and Schmidhuber J, “Recurrent highway net-works,” in Proceedings of the International Conference on Machine Learning (Precupand D Teh YW, eds.), vol. 70, pp. 4189–4198, 2017. [Google Scholar]
- 35.Hochreiter S and Schmidhuber J, “Long short-term memory,” Neural Computation, vol. 9, no. 8, pp. 1735–1780, 1997. [DOI] [PubMed] [Google Scholar]
- 36.Collins J, Sohl-Dickstein J, and Sussillo D, “Capacity and learnability in recurrent neural networks,” International Conference on Learning Representations, 2017. [Google Scholar]
- 37.Bradbury J, Merity S, Xiong C, and Socher R, “Quasi-recurrent neural networks,” International Conference on Learning Representations, 2017. [Google Scholar]
- 38.Lei T and Zhang Y, “Training RNNs as fast as CNNs,” arXiv Preprint, arXiv:1709.02755,2017.
- 39.Chung J, Gulcehre C, Cho K, and Bengio Y, “Empirical evaluation of gated recurrent neural networks on sequence modeling,” arXiv Preprint, arXiv:1412.3555, 2014.
- 40.Graves A, Wayne G, and Danihelka I, “Neural turing machines,” arXiv Preprint, arXiv:1410.5401, 2014.
- 41.Santoro A, Bartunov S, Botvinick M, Wierstra D, and Lillicrap T, “Meta-learning with memory-augmented neural networks,” in Proceedings of the International Conference on Machine Learning (Balcan MF and Weinberger KQ, eds.), pp. 1842–1850, 2016. [Google Scholar]
- 42.Graves A, Wayne G, Reynolds M, Harley T, Danihelka I, Grabska-Barwi ´nska A, Colmenarejo SG, Grefenstette E, Ramalho T, Agapiou J, et al. , “Hybrid computing using a neural network with dynamic external memory,” Nature, vol. 538, no. 7626, pp. 471–476, 2016. [DOI] [PubMed] [Google Scholar]
- 43.Santoro A, Faulkner R, Raposo D, Rae J, Chrzanowski M, Weber T, Wierstra D,Vinyals O, Pascanu R, and Lillicrap T, “Relational recurrent neural networks,” arXiv Preprint, arXiv:1806.01822, 2018.
- 44.Caruana R, Baluja S, and Mitchell T, “Using the future to “sort out” the present: Rankprop and multitask learning for medical risk evaluation,” in Advances in Neural Infor-mation Processing Systems (Mozer M, Jordan M, and Petsche T, eds.), vol. 9, pp. 959–965, 1996. [Google Scholar]
- 45.Wiens J, Guttag J, and Horvitz E, “Patient risk stratification with time-varying param-eters: A multitask learning approach,” Journal of Machine Learning Research, vol. 17,no. 1, pp. 2797–2819, 2016. [Google Scholar]
- 46.Ding DY, Simpson C, Pfohl S, Kale DC, Jung K, and Shah NH, “The effectiveness of multitask learning for phenotyping with electronic health records data,” arXiv Preprint, arXiv:1808.03331, 2018. [PMC free article] [PubMed] [Google Scholar]
- 47.Glorot X and Bengio Y, “Understanding the difficulty of training deep feed forward neural networks,” in International Conference on Artificial Intelligence and Statistics (Tehand YW Titterington M, eds.), vol. 9, pp. 249–256, 2010. [Google Scholar]
- 48.Kingma DP and Ba J, “Adam: A method for stochastic optimization,” International Conference on Learning Representations, 2015. [Google Scholar]
- 49.Guo C, Pleiss G, Sun Y, and Weinberger KQ, “On calibration of modern neural net-works,” in Proceedings of the International Conference on Machine Learning (Precupand D Teh YW, eds.), pp. 1321–1330, 2017. [Google Scholar]
- 50.Platt JC, “Probabilistic outputs for support vector machines and comparisons to regular-ized likelihood methods,” in Advances in Large-Margin Classifiers, pp. 61–74, MIT Press,1999. [Google Scholar]
- 51.Brier GW, “Verification of forecasts expressed in terms of probability,” Monthly Weather Review, vol. 78, no. 1, pp. 1–3, 1950. [Google Scholar]
- 52.Niculescu-Mizil A and Caruana R, “Predicting good probabilities with supervised learning,” in Proceedings of the International Conference on Machine Learning (Raedtand LD Wrobel S, eds.), pp. 625–632, ACM, 2005. [Google Scholar]
- 53.Saito T and Rehmsmeier M, “The precision recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets,” PLOS One, vol. 10, no. 3, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Efron B and Tibshirani RJ,An introduction to the bootstrap. CRC press, 1994. [Google Scholar]
- 55.Mann HB and Whitney DR, “On a test of whether one of two random variables is stochastically larger than the other,” The Annals of Mathematical Statistics, vol. 18, no. 1,pp. 50–60, 1947. [Google Scholar]
- 56.Lakshminarayanan B, Pritzel A, and Blundell C, “Simple and scalable predictive uncer-tainty estimation using deep ensembles,” in Advances in Neural Information Processing Systems (Guyon I, Luxburg U, Bengio S, Wallach H, Fergus R, Vishwanathan S, and Garnett R, eds.), vol. 30, pp. 6402–6413, 2017. [Google Scholar]
- 57.Fauw JD, Ledsam JR, Romera-Paredes B, Nikolov S, Tomasev N, Blackwell S,Askham H, Glorot X, O’Donoghue B, Visentin D, van den Driessche G, Laksh-minarayanan B, Meyer C, Mackinder F, Bouton S, Ayoub KW, Chopra R, King D,Karthikesalingam A, Hughes CO, Raine RA, Hughes JC, Sim DA, Egan CA,Tufail A, Montgomery H, Hassabis D, Rees G, Back T, Khaw PT, Suleyman M, Cornebise J, Keane PA, and Ronneberger O, “Clinically applicable deep learning for diagnosis and referral in retinal disease,” Nature Medicine, vol. 24, pp. 1342–1350, 2018. [DOI] [PubMed] [Google Scholar]
- 58.Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, Corrado GS, Davis A,Dean J, Devin M, Ghemawat S, Goodfellow IJ, Harp A, Irving G, Isard M, Jia Y,Józefowicz R, Kaiser L, Kudlur M, Levenberg J, Mané D, Monga R, Moore S,Murray DG, Olah C, Schuster M, Shlens J, Steiner B, Sutskever I, Talwar K, Tucker PA, Vanhoucke V, Vasudevan V, Viégas FB, Vinyals O, Warden P, Wattenberg M,Wicke M, Yu Y, and Zheng X, “Tensorflow: Large-scale machine learning on heteroge-neous distributed systems,” 2015.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The clinical data used for the training, validation and test sets was collected at the US Department of Veterans Affairs and transferred to a secure data centre with strict access controls in de-identified format. Data was used with both local and national permissions. It is not publicly available and restrictions apply to its use. The de-identified dataset, or a test subset, may be available from the US Department of Veterans Affairs subject to local and national ethical approvals.