Abstract
Search for specificity in cancers has been a Holy Grail in cancer immunology. Cancer geneticists have long known that cancers harbor transforming and other mutations. Immunologists have long known that inbred mice can be immunized against syngeneic cancers, indicating the existence of cancer-specific antigens. With the technological advances in high throughput DNA sequencing and bioinformatics, the genetic and immunologic lines of enquiry are now converging to provide definitive evidence that human cancers are vastly different from normal tissues at the genetic level, and that some of these differences are recognized by the immune system. The very vastness of genetic changes in cancers now raises a different question. Which of the many cancer-specific genetic (genomic) changes are actually recognized by the immune system, and why? New observations are now beginning to probe these vital issues with unprecedented resolution and are informing a new generation of studies in human cancer immunotherapy.
Introduction
Understanding of the generation and recognition of specificity has delighted immunologists considerably over the last two centuries. Knowledge of specificity has also brought us arguably the greatest tool to enhance human (and animal) health in the form of vaccines against an array of infections. Immunologists have also therefore been keen to utilize the principle of specificity in treating cancer. For cancers of infectious (viral) etiology, the problem of specificity is straight forward, and has led to spectacular results as in the case of prophylactic vaccines against genital herpes. However, for cancers of apparently non-infectious etiology (i.e. the majority of human cancers) the issue of specificity becomes quite complicated quite quickly. What, if anything, is specific in these cancers?
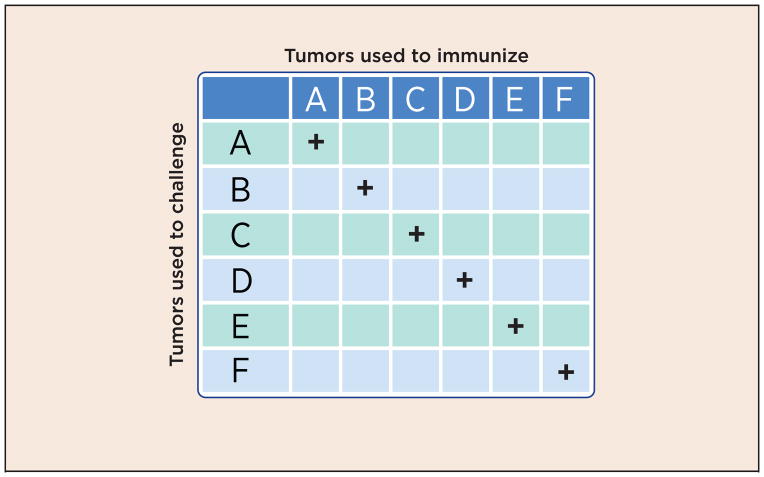
This simple question has been surprisingly difficult to answer, except perhaps now. The first hints that specificity may exist, came from the early experiments of Ludwik Gross, Prehn and Main, and most persuasively, George and Eva Klein (1). Using inbred syngeneic mice, and the tumors that can be induced in them by methylcholanthrene, these early studies noted that mice could be immunized against syngeneic tumors, and rendered resistant to them by such immunization. Remarkably, each individual tumor elicited immunity only against itself, and not against any other tumor of the same type, induced in the same fashion even when large panels of tumors were tested (2) (Fig. 1). These early experiments were quickly replicated and expanded upon. Although the phenomenon was shown most easily in carcinogen-induced and UV-induced tumors; spontaneous tumors also elicited weak but detectable specific immunity (see ref. 3). Such experiments ought not to be, and were not, contemplated in humans. Whether or not human cancers harbor some cancer-specific moieties has therefore remained an open question. Those who have been willing to extrapolate the observations from carcinogen-induced mouse tumors have been inclined to believe that human tumors are similarly immunogenic (i.e. harbor specificity); others have remained atheistic or agnostic. Not surprisingly, this tension between the “believers” and the atheists has echoed through the subsequent history of cancer immunity, and it is this tension that may now begin to be resolved.
Fig. 1. Antigenic individuality of tumors.
A given chemically induced tumor (as an example, A) elicits immunity against itself but not against other tumors (B, C, D, E or F), even though all tumors are induced by the same carcinogen, in genetically identical animals, and are histologically identical.
A series of technologies have been brought to bear on the search for specificity in cancers over the past five to six decades. A large body of literature describes the use of electrophoresis of several kinds and dimensions to look for bands or spots that may be unique to cancers. Although these studies have been enormously productive in understanding the biochemical complexity of tissues, highlighting molecular species that may be overexpressed or under expressed in cancers, they failed to turn up something that may be truly cancer-specific. Serology, in its many incarnations from classical polyclonal sera to B cell hybridomas to SEREX (serological analysis of cDNA expression libraries) analyses, has been similarly unsuccessful in identifying a truly cancer-specific molecule (4,5). Regardless, many antibodies whose targets are expressed selectively, or preferentially on cancer cells, but are not truly cancer-specific, are in wide and productive clinical use today.
It is the study of genes that provided the first unambiguous demonstration that cancers harbor truly cancer-specific changes. In a series of path-breaking papers in 1983–84, Stuart Aaronson and others showed that specific point mutations in the ras oncogenes were responsible for their transforming activity (see ref. 6 as an early example). These were the first demonstrations that cancers contained truly cancer-specific moieties. However, such oncogenic genetic changes (or mutations in driver genes in contemporary parlance) have not been shown to be responsible for eliciting cancer immunity, even though T-cell responses to mutant oncogenes have been shown to be immunopreventive (7). The following sections of this Master of Immunology article address the necessarily intertwined (but not identical) questions about what is cancer-specific and what elicits cancer immunity.
Search for a shared cancer antigen
In the 1990s, it became possible to identify the targets of T cells of mouse and human cancers. Such studies, led by Thierry Boon, showed that T cells from tumor-immunized mice and cancer patients often recognized completely un-altered, i.e. normal peptide sequences that were shared among cancers (8). These sequences were either derived from differentiation antigens shared between normal and cancer cells of the particular differentiated lineage or from cancer-testes antigens, which were shared among cancers and germinal tissues but not detectable on normal adult tissues. The realization that T cells of cancer patients easily recognized un-mutated sequences gave rise to high enthusiasm to find cancer vaccines against entire classes of cancers (such as melanomas or lung cancers). Important questions remained: what was the immunologic basis of immunizing against self-antigens without precipitating catastrophic autoimmunity? The un-mutated sequences did not elicit significant tumor rejection in mouse models (9). What was the scientific and preclinical rationale for the human studies?
Regardless, the attention of the field shifted quite dramatically from the search for cancer specificity to trying to break tolerance against self-antigens that were expressed on cancers and some normal cells alike. Taking a page from the vast experience with cancer chemotherapy, a degree of autoimmunity began to be deemed an acceptable price to pay for a certain degree of anti-cancer activity. Although several early studies showed hints of clinical activity, two large randomized, well-controlled, and well-executed Phase 3 clinical trials that tested the efficacy of immunization of melanoma and lung cancer patients with a cancer-testes antigen, MAGE, persuasively failed to show clinical activity (10,11). Nonetheless, reasonable questions remain; for example, these randomized studies looked for efficacy in all patients immunized rather than among patients who generated a CD8 T-cell response to MAGE. In support of the efficacy of T cells against shared antigens, infusion of autologous CD4 T cells against another cancer-testes antigen, NY-ESO1, was shown to mediate a durable clinical remission (12). In another study, Yuan and colleagues (13) showed strong correlation of clinical and immunologic data in metastatic melanoma patients with heterogeneous tumor responses to ipilimumab therapy. Clearly, the final word on the utility of shared cancer antigens in human cancer immunotherapy has not yet been written.
Search for unique cancer antigens
Although the non-mutated or shared antigens became the point of overwhelming focus among the antigens recognized by human and mouse T cells, those studies also revealed that T cells of some patients recognized mutated, cancer-specific peptide sequences as well (see ref. 14 for a comprehensive, well-annotated compilation). This cancer-specificity came with the burden that, in an echo of the original observations of Klein and colleagues, and Prehn and Main (1–3), these mutations were unique to individual cancers. These mutations were not shared across multiple cancers. They could not therefore be realistically used for immunizing patients. Advent of new technologies will later change that perception as discussed later.
But the search for cancer specificity, and cancer vaccination, continued in parallel in other arenas. Building on the intrinsic immunogenicity of autologous cancer cells and on the observations that cancer cells transfected with granulocyte-macrophage colony-stimulating factor (GM-CSF) are far better immunogens than un-transfected cells (15), a series of clinical trials are exploring the efficacy of therapeutic vaccination with GM-CSF-modified autologous colorectal or pancreatic cancer cells (16). This approach aims to harness the individually specific, as well as any shared immunogenicity of cancers.
Additionally, the original observations that mice can be immunized against syngeneic chemically induced sarcomas have been pursued biochemically in order to identify the molecular basis of specific cancer immunity. These studies, carried out over about twenty years, pointed to a family of commonly expressed molecules, the heat shock proteins (HSP) (specifically hsp70 and hsp90 members), which could be used to immunize against cancers, provided they were isolated from the same cancer cells (17). HSPs isolated from normal tissues elicited detectable but only minor tumor resistance. Extensive analyses have uncovered the fact that immunogenicity was actually derived from short peptides that were non-covalently associated with the HSP molecules, and not from the HSP molecules per se. These observations were made using a number of viral and model antigen systems (18). The peptides were the immunogens, while the HSP molecules were the adjuvants, deriving their adjuvanticity from the fact that HSPs could target/deliver the peptides to dendritic cells (DC) through interaction with the HSP-receptors on DCs (19). These observations solved an immediate problem from the perspective of autologous cancer vaccination. If the HSP preparations isolated from cancer cells were intrinsically associated with antigenic peptides, one could immunize cancer patients with HSP-peptide complexes isolated from surgically resected cancers of each patient. The isolation of GMP (good manufacturing practices) grade HSP material from individual cancers was logistically and economically feasible. Several early clinical studies showed substantial hints of clinical activity (20,21). However, a randomized Phase 3 trial in patients with non-metastatic renal cell carcinoma failed to show convincing clinical activity, except in a large subset of patients that was identified post hoc (22). (The HSP-peptide vaccine, OncoPhage, was nonetheless approved for treatment in Russia in 2008 for this large subset of patients (n=362), becoming the first cancer vaccine to be approved anywhere in the world.) Based on Phase 2 data demonstrating impressive clinical activity (23), a large NIH/CTEP-supported randomized trial, testing the efficacy of autologous tumor-derived HSP-peptide complexes in patients with glioblastoma multiforme, is currently underway. The HSP-peptide approach aims to harness the individually specific, rather than a shared immunogenicity of cancers. Inherent in these studies is the notion that immune responses against cancer-specific antigens are more likely to have antitumor activity than shared antigens, as originally determined in mouse models.
Molecular definition of cancer specificity
Although approaches of vaccination with autologous cancer cells or autologous cancer-derived HSP-peptide complexes harness the individually specific immunogenicity of cancers, neither approach addresses the original question: what is it that is specific in cancers? It is useful to recall that each individual tumor elicits immunity only against itself, and not against its tumor type, suggesting the incomprehensible possibility that each time a new tumor arises, it carries its own unique antigen! Clearly, there is not sufficient space in the genome for such an unlimited antigenic potential. In order to resolve this paradox, it was suggested over 20 years ago (24), that there are no cancer-specific antigens but only cancer-specific epitopes, which arise because of random passenger mutations in dividing cells. These cancer-specific epitopes arise simply because the processes of DNA replication and repair do not happen with complete fidelity (Fig. 2), and because they are random — each new tumor has a unique set of mutations, and hence epitopes, or neoepitopes (Fig. 2). In this thinking, the immunologic individuality of cancers simply recapitulates the genetic individuality inherent in all cells, normal and cancerous.
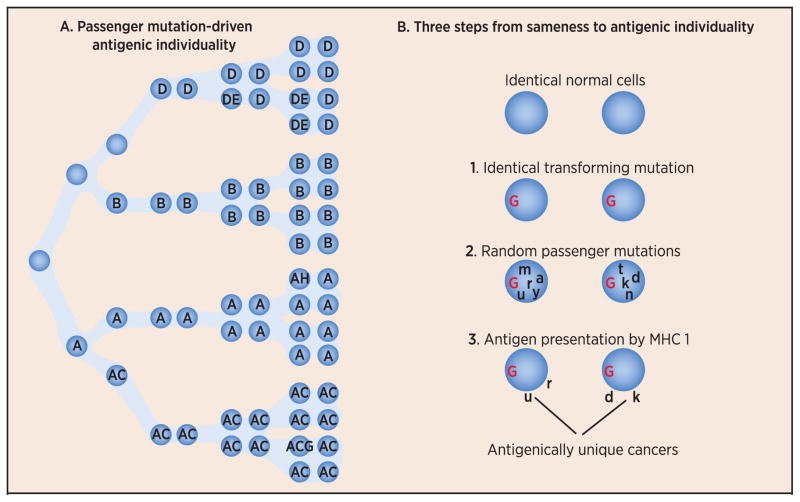
Fig. 2. A mechanism of generation of antigenic individuality in tumors.
Random mutations (point mutations, translocations, insertions or deletions) occur in normal cells as a result of un-repaired errors in DNA replication (see panel A). The mutations will include one or more oncogenic (or driver) mutation (not shown), as well as many passenger mutations (colored circles within cells). These mutations occur at every division cycle and mark the lineage of that mutant cell as long as the mutation does not disadvantage the cell’s survival, or mutations in other cells do not give their bearers a survival advantage. Note how the tumor mass resulting from these divisions (and ongoing mutations) becomes highly heterogeneous with respect to mutations. The immunological consequences of these events were hypothesized in 1993 (Srivastava 1993) and are shown in panel B, which is adapted from a figure in that publication. Starting from two identical normal cells, which get transformed (Step 1) by the same driver oncogenic mutation (denoted in red as G), the progeny of each of the two transformed cells begin to differentiate from each other as a result of passenger mutations (denoted by random letters of the alphabet in black), which are random and hence unique to each tumor (Step 2). A subset of these mutations (mis-sense mutations) alters the sequences of expressed transcripts which are translated into mutated proteins and peptides derived from such mutated proteins. Some tumor cells may have more mutations than others (Vogelstein). Because of antigen processing (proteasomal activity, chaperoning of peptides by other molecules) and presentation, a subset of the mutated peptides are presented by the MHC I molecules of the tumor (Step 3). A subset of the MHC I-mutant peptide complexes will be recognized by the T cell receptors of T cells of the host. Peptide corresponding to an individual driver or passenger mutation may or may not be recognized by the immune system, but as a class, peptides encoded by passenger mutations are far more likely to be recognized by the immune system simply because passenger mutations are far more numerous. It is also conceivable teleologically that peptides derived from driver mutations are highly unlikely to be effective targets of immune response; should that be the case, the tumors would be eliminated very early during their progression. The heterogeneity of tumors with respect to passenger mutations also results in immunogenic heterogeneity of tumors.
At the time this suggestion was made (1993), identification of random passenger mutations in individual cancers was not technologically testable. That changed with the advent of high throughput DNA sequencing and bioinformatics capabilities that now make it possible to identify mutations and predict neoepitopes on an unprecedented scale. When it rains, it pours. From a long era in which we could not begin to define what is immunologically specific in cancers, we have now come into the era in which the literature is replete with theoretical possibilities of vast numbers of neoepitopes in a large array of cancers. Thousands of human cancer genomes have now been compared with matching normal genomes, and thousands of potential neoepitopes have now been predicted (25–28). These findings are entirely consistent with earlier anecdotal observations: (a) that neoepitopes are present in human cancers, and that (b) the neoepitopes are unique to individual cancers.
Search for the “true” neoepitopes: starting with certain assumptions
How many of the potential neoepitopes, predicted by DNA sequencing and bioinformatic analyses, are real, and are all neoepitopes equal? Earlier anecdotal studies with mutant neoepitopes of human cancers had already drawn the correlation between immune response to neoepitopes and clinical outcomes (29). More recently, Brown and colleagues (30) identified hundreds of predicted neoepitopes in a wide array of human cancers, and correlative data suggested that the mutational burden of cancers was indicative of higher infiltration of tumors by T cells and better clinical outcomes. Fritsch and colleagues (31) have similarly compiled detailed data that suggest that the presence of neoepitopes with a higher binding affinity to their cognate HLA alleles, is associated with better clinical outcomes. Such correlations are useful in generating testable hypotheses, but do not prove causality. Specifically, they cannot help us identify prospectively, which of the many predicted neoepitopes shall elicit immunity that will protect the host from tumor growth. That burden must, for now, be borne by the murine models in which predictions may be tested.
A recent flurry of studies attempts to approach this problem. All of the studies use the same initial steps; with some important variations, they all compare the expressed sequences of tumors to the normal genomes and identify the single nucleotide variants (SNV). All make predictions of neoepitopes based on algorithms that predict MHC I-peptide binding, and all test the peptides for their ability to elicit CD8 T cells and tumor rejection. All studies begin with the following key assumptions, which have a solid experimental and theoretical basis (32–34), based primarily on our experience with viral epitopes of mouse and human MHC alleles:
MHC I-peptide affinity is a key predictor of a peptide’s ability to elicit CD8+ T-cell response, and to confer protective immunity.
CD8 T-cell responses, as measured in vitro, are reliable surrogates for antitumor activity in vivo.
In early studies, Castle and colleagues (35) predicted several MHC I neoepitopes in a spontaneous mouse melanoma, observed that a good proportion of these neoepitopes elicited CD8 T cells upon immunization, and demonstrated that some predicted neoepitopes also altered the course of tumor growth. Using a broadly similar approach and assumptions, Matsushita and colleagues (36) showed that a single neoepitope is a target of immunological editing in a chemically induced mouse sarcoma. The issue of predicting the true tumor rejection neoepitopes from among a large number of potential candidates became the sharp focus of three subsequent reports (37–39) that examined this question in syngeneic tumors in inbred mice, and coincidentally, in tumors induced by the same carcinogen, 2-methylchlanthrene. All three papers use roughly the same initial steps, and made roughly the same assumptions. Some of their conclusions were identical, and others widely divergent. It is instructive to discuss these three studies in some detail.
Duan and colleagues (37) use the NetMHC algorithm to predict the neoepitopes restricted by the Kd allele in two BALB/c tumors. They ranked the predicted neoepitopes in a descending order of NetMHC matrix scores (i.e. affinity to Kd), and tested tens of the highest scorers for their ability to elicit tumor rejection. Surprisingly, none of them did. They noticed that the NetMHC scores of the non-mutated counterparts of these high-NetMHC-scoring neoepitopes were also high, leading to the suggestion that the CD8 response to these wildtype sequences must be deleted or tolerized, and that the CD8 response to the mutant neoepitopes may too have been tolerized because of the similarity to germline. In this light, a genuine neoepitope would need to be sufficiently different from self, in addition to having a reasonable affinity for MHC I. Duan and colleagues gave a numerical definition of difference-from-self by subtracting the NetMHC matrix scores of the wildtype counterparts from those of potential neoepitopes, and named it the Differential Agretopic Index (DAI). Not surprisingly in hindsight, the highest DAI-ranking neoepitopes were the ones that had mutations in the anchor residues, since these residues contribute disproportionately to the affinity to MHC I. Ranking the predicted neoepitopes on the basis of DAI rather than NetMHC scores, Duan and colleagues identified 15–30% of the predicted neoepitopes to actually elicit tumor rejection. Most surprisingly, they observed that these rejection-eliciting neoepitopes had a poor affinity (IC50 far worse than 500 nM) for Kd or indeed Dd or Ld, even though their activity was CD8-dependent. They concluded that mutations in anchor residues, rather than the T-cell receptor (TCR)-facing residues, create the most dissimilar-from-self neoepitopes and that the neoepitopes with low affinity to MHC I may still elicit protective anttumor immunity. Both conclusions challenge some aspects of our current beliefs about MHC I-peptide-TCR interactions.
In defining the immunome of a fibrosarcoma of the b haplotype, Gubin and colleagues (38) prioritized mutation-derived neoepitopes based on their predicted Class I binding affinity, their likelihood of being proteasomally processed, and their equivalent or preferential binding to MHC I compared to the respective wildtype sequences. These analyses revealed that the top two (and only two) neoepitopes, each of which had a mutation in a non-MHC anchor residue, represented the major epitopes seen by CD8 T cells that developed in tumor-bearing mice either spontaneously or following treatment with checkpoint-blocking antibodies to CTLA-4 and/or PD-1. When used together, the two neoepitopes induced tumor rejection as efficiently as checkpoint blockade therapy. This study additionally demonstrated that blockade of different immune checkpoints leads to largely distinct alterations in tumor-specific CD8 T cells. Broadly speaking, the conclusions of this study are robustly consistent with our understanding of MHC I-peptide-TCR interactions, and enhance our understanding of the molecular mechanisms underlying checkpoint blockade.
Yadav and colleagues (39) add another dimension to the analysis of neoepitopes primarily by adding mass spectrometry to their toolkit. They identify a large number of potential neoepitopes of a fibrosarcoma of the b haplotype by application of the NetMHC algorithm. They also identify the neoepitopes actually presented by the tumor using mass spectroscopy. Upon cross-checking the two databases, they find seven common neoepitopes. Based on affinity to Kb, and on the assumption that the relevant altered residues face the TCR, they predict 2/7 neoepitopes to elicit tumor rejection. This is experimentally confirmed; in addition, one neoepitope of the remaining five, which was not predicted to elicit immunity, and which results from an anchor-residue mutation, also elicits tumor rejection.
The observation that individual neoepitopes arising out of passenger mutations can mediate T cell-mediated tumor immunity is a common thread among the three studies; these observations were indeed hypothesized over 20 years ago (24). All three papers agree that only a tiny minority of predicted neoepitopes actually elicits protective tumor immunity. However, the discordance among the three is enlightening with respect to the two key assumptions made by all three studies, and is discussed in the context of other related findings, in the next section.
Affinity for MHC I: key predictor of immune response against mutated self?
The assumptions that a neoepitope with a high to intermediate affinity (IC50 of <500 nM) for its cognate MHC I, is very likely to be a good neoepitope, and the converse, that a neoepitope with a low affinity (IC50 of >500 nM) is a poor epitope, have achieved the status of a consensus based on solid experimental data mainly from viral systems (in which epitopes do not have a self-counterpart) (32,33). Such quantitative measurements are immensely helpful in making predictions of immunogenicity. The study of Assarsson and colleagues (34) in the vaccinia virus system is a useful case in point: starting from all possible 9–10 amino acid peptides, 2.5% are high affinity binders to a given HLA allele. Of the actual high affinity binders, half elicit a CD8 response. Of these, only 15% are naturally processed and presented. Thus, if one did not know the status of actual antigen presentation (and for the cancer sequencing data, there is no bioinformatic or experimental high throughput method yet, to make that determination), about 7.5% of the predicted high affinity binding epitopes shall be found to be immunogenic. There is some similarity between this study and that of Van Rooij and colleagues (40), who used the combined criteria of sufficient expression, proteasomal processing, and MHC binding to predict good neoepitopes in a human melanoma; the analysis yielded 448 potential neoepitopes, which were systematically tested. Only two were found to be recognized by the patient’s T cells. The proportion of actual neoepitopes from among the total tested is about a log scale lower than that in the Assarsson study. Regardless, the enormity of this difference between the two studies is not very significant considering the many differences among them. Both studies show that only a very small proportion of good binders are true epitopes. In contrast, Gubin and colleagues (38) and Yadav and colleagues (39) observe that the predictions fit perfectly, and they are able to correctly predict the neoepitope that mediates tumor rejection with complete or near complete accuracy. Duan and colleagues (37), on the other hand, find that the assumptions about the predictive power of affinity of neoepitope with MHC I are insufficient, and they go on to develop an additional algorithm of difference-from-self. In terms of the frequency of true neoepitopes among the many predicted neoepitopes, the observations of Duan and colleagues are consistent with those of Assarsson and colleagues (34) and of Van Rooij and colleagues (40).
One aspect of the observations of Duan colleagues (37) is inconsistent with much of our understanding of immunogenicity of epitopes. They report that the most effective neoepitopes have a low affinity for their cognate (and other) MHC I allele. As mentioned earlier, there are strong data in support of the consensus that good neoepitopes have an intermediate to high affinity for their MHC I alleles (32,33). It is worthwhile examining if that consensus should apply equally to the mutated-self epitopes (which do have a self-counterpart). In fact, Sette and colleagues discussed this possibility precociously in their landmark paper (32): “It is also possible that self-derived antigenic peptides may be characterized by relatively low MHC binding affinity because of selective elimination by thymic education and / or T cell tolerance of T cells reactive against high affinity MHC binding peptides.” Since testable cancer neoepitopes have begun to be available only during the last 2–3 years, data testing this proposition shall soon emerge. For now, except for the study of Duan and colleagues (37), such low affinity binders have been excluded from testing for biologic activity, and the proportion of good epitopes among them is therefore unknown. Should a systematic examination of immunogenicity of poor MHC I-binders confirm the antitumor immunogenicity of a significant proportion of these low binders, a vast unexplored repertoire of neoepitopes as cancer antigens shall be available for investigation.
While discussing MHC I-peptide affinity, it is useful to recognize how such affinity is measured and predicted. Since predictions are based on measurements, the latter is the key. They are generally measured by the ability of a given peptide to compete with another peptide for binding to a given MHC I allele. Such measurements have been amply validated through a very large number of studies and are immensely useful (32–34). It is important to bear in mind though that the methods of MHC I-peptide binding in vitro bear little resemblance to the highly choreographed process of such assembly in vivo (41.) Indeed, peptides that are unable to bind cell surface MHC I molecules, but can be presented by endogenously expressed protein, have been occasionally reported (42). It is conceivable that a binding assay in vitro, which recapitulates the biologic assembly of MHC I-peptide complexes in vivo more closely, will provide somewhat different estimates of MHC I-peptide binding, at least for the peptides that seem to bind poorly in the current assays.
Checkpoint blockade and neoepitopes
Checkpoint blockade has emerged as a game changer in human cancer immunotherapy, and it is certain that immunization of cancer patients with neoepitopes shall be tested in combination with checkpoint blockade (43). Three distinct strains of ideas, and supporting data, have emerged that connect these two modalities.
Schumacher and colleagues (40) analyzed the tumor exome of a melanoma patient who had responded to CTLA4-blockade, and reported that T-cell response to a neoepitope had increased significantly after CTLA-4-blockade. They suggested that this observation might reflect “a more general bias toward recognition of neoantigens from highly expressed genes with a high predicted HLA binding affinity”. Interestingly in this regard, Gubin and colleagues (38), who identified two biologically active neoepitopes in a mouse sarcoma, observed, “when used together, the two neoepitopes induced tumor rejection as efficiently as checkpoint blockade therapy.” The prospect that checkpoint blockade may operate even partially through amplification of the T-cell responses to neoepitopes, is enormously appealing for aficionados of cancer specificity.
Duan and colleagues (37) who identified several biologically active neoepitopes of mouse sarcomas, tested the activity of a neoepitope alone, CTLA-4 blockade alone, or both regimens together, and noted that the combination was significantly more effective than either agent alone. This observation suggests that exploration of synergy between immunotherapy with neoepitopes and checkpoint blockade might be productive.
Snyder and colleagues (44) attempted to identify a genomic biomarker in the cancers of melanoma patients who responded to CTLA-4 blockade. They observed that patients whose tumor-neoepitopes had a homology to viral and bacterial antigens (in form of a conserved tetrapeptide motif) were far more likely to respond clinically to CTLA-4 blockade than patients whose tumor-neoepitopes did not have such homology. At first glance, this conclusion is difficult to comprehend. What is a possible mechanistic basis for homology between random passenger mutations in the mammalian genome and pathogens? It is conceivable that in lack of a substantial population of naïve T cells in the adult human (as a consequence of thymic involution), most of the anti-neoepitope T-cell responses are indeed cross-reactive memory responses to pathogens past. If this interpretation of the results of Snyder and colleagues (44) is true, incorporations of lessons from viral immunology to cancer immunology shall be critical. However, another aspect of the hypothesis presented by Snyder and colleagues requires consideration. More recently, combined checkpoint blockade of PD-1 and CTLA-4 has been used in patients with metastatic melanoma; the dual checkpoint blockade results in a dramatic increment (more than two-fold) in the proportion of patients who show clinical responses (45). This observation suggests that it is almost certain that a significant proportion of patients who do not have the tetrapeptide motif (44), and do not respond to CTLA-4 blockade alone, are responding to dual blockade.
Neoepitopes recognized by CD4+ T cells
The entire preceding discussion has focused exclusively on the MHC I-restricted neoepitopes. The neoepitopes presented by MHC II and recognized by CD4+ T cells too have a long past, and now are a focus of renewed attention. The MHC II-restricted neoepitopes have been reported previously in human (46) and mouse (47,48) tumors. Their ability to mediate tumor rejection has also been shown clearly in mouse models. Using genomic and bioinformatic approaches, recent studies now demonstrate that a large proportion of mutations in human and mouse cancers can generate MHC II-restricted neoepitopes that can elicit CD4+ T-cell responses (49). In mouse models tested, such responses contribute to tumor regression, by themselves, or in collaboration with CD8+ T cells. Kreiter and colleagues (49) suggest that “the less stringent length and sequence requirements for peptide binding to MHC II” may be responsible for the high proportion of mutations that generate MHC II-restricted neoepitopes. The mechanism(s) through which neoepitope-elicited CD4+ responses contribute to tumor rejection is currently only vaguely understood, and shall no doubt be further scrutinized.
The path ahead
This Masters of Immunology article began with the question of defining that which is specific in cancers. Recent discoveries, as elucidated here, have answered that question: we now know in surprising abundance what is specific in cancers. It is perhaps reasonable to entertain the possibility that we shall be able to translate the knowledge of specificity into successful immunotherapy of human cancers. Learning from the past, it is useful to be humble in moments of high enthusiasm, and judging from the flood of opinions and reviews on the subject (perhaps more in number than the primary papers), this indeed is a time of high enthusiasm for the neoepitopes. It has been nearly 25 years since the idea of random-passenger-mutations-as-neoepitopes was first proposed (24); the time for its critical testing has arrived. How do we translate the random-passenger-mutations-as-neoepitopes idea into clinical studies? Certain steps in the process are straight forward, while others pose significant hurdles (Fig. 3). The rest of this article is devoted to identifying the hurdles. This list does not include the very obvious steps of combination of immunization with neoepitopes with one or more means of checkpoint blockade, inhibition of tumor-induced immune suppression, and other modalities of cancer therapy that are or will be on the horizon. This list focuses instead on the steps that are needed for us to obtain a more perfect prediction of the true tumor-rejecting neoepitopes from among the many that can now be easily identified.
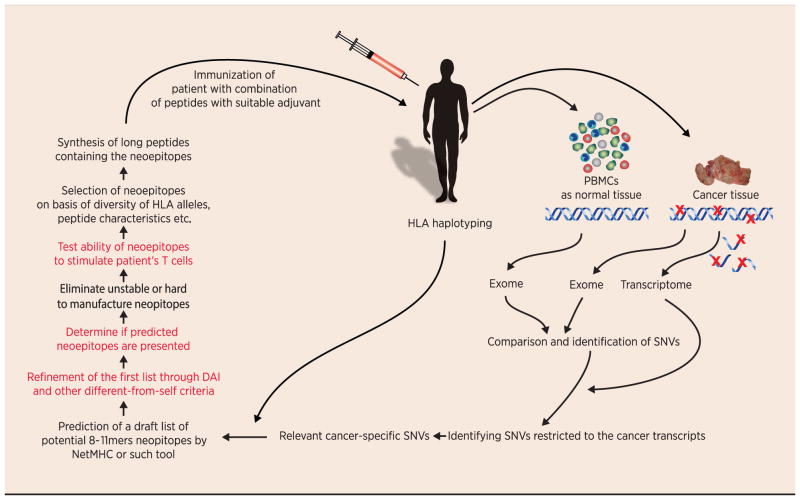
Fig. 3. Pipeline for creation of a personalized set of cancer neoepitopes for each patient.
Steps in red indicate those which are conceptually unclear, or difficult to accomplish in the high throughput manner required for clinical translation today. See text for detailed explanation.
Calling tumor-specific variants and characterizing tumor heterogeneity: It would seem that comparison of the tumor and normal sequences and identification of mutations would be a straightforward process. It is actually anything but, for a number of reasons. Unlike chemically induced cancer cell lines, spontaneous tumors exhibit a high degree of intra-tumor heterogeneity characterized by complex patterns of clonal and regional variation. Accurate identification of somatic variants within such heterogeneous samples remains a challenge. Although many somatic variant calling algorithms have been published, the low overlap between variant calls made from the same sequencing data (50,51) underscores the lack of robustness and inherent biases of current methods. Recognizing this challenge, The Cancer Genome Atlas (TCGA) and the International Cancer Genomics Consortium (ICGC) have teamed up to launch the ICGC-TCGA DREAM Somatic Mutation Calling Challenge, a crowd-sourcing effort aiming to identify best analysis pipelines and accelerate adoption of best practices in the field (52). Accurate calling of mutations is a necessary foundation for accurate characterization of the clonal diversity of highly heterogeneous tumors. Comprehensive characterization of somatic variants by single nucleus whole genome and whole exome sequencing has already been demonstrated, with the nuc-seq protocol in (53) achieving a mean coverage breadth of over 90%. The relevant clonal structure can also be obtained using more cost-effective single-cell sequencing protocols targeted at predicted variants. These bioinformatics-intensive approaches will be essential for creating the necessary edifice for any and all approaches on identification of neoepitopes.
Predicting antigen presentation: Mass spectroscopy of MHC-eluted peptides is the only method available today to identify the neoepitopes that are actually presented (39). At the current level of sophistication, this is not a high throughput approach. Taking vaccinia as an example, as few as 7.5% of all potential epitopes may be actually presented (34). Prediction of presentability by MHC molecules requires a bioinformatic strategy; it is conceivable that we may now have accumulated enough of a database of mouse and human epitopes that it may be possible to algorithmically predict the presented epitopes better than before.
Defining dissimilarity from self: Since most of our learning of T-cell immunity has occurred on basis of viral or model antigens (where no corresponding “self” exists), the need to define a neoepitope’s dissimilarity from “self” is unique to cancer immunity. Duan and colleagues (37) have made an attempt to quantify this dissimilarity in form of the DAI algorithm. However, that algorithm is obviously biased towards anchor residue mutations, and is incomplete. Broader quantitative approaches to definition of dissimilarity from self are sorely needed.
Measuring T-cell response: This basic immunologic tool has made great strides in sensitivity and ease from the chromium release assays to flow cytometric and ELISPOT assays. Although it is unambiguously clear that immune responses to cancer are largely T-cell mediated/driven, there has historically been and there still remains a gap between the antitumor activity as seen in vivo and the T-cell activity measured in vitro. Specifically in the current context, neoepitopes that elicit tumor rejection in a CD8-dependent manner may still not be able to elicit CD8 responses that can be measured in vitro. Improved high throughput assays that measure T-cell activation to large numbers of antigens in a highly sensitive and multi-factorial manner shall go a long way in neoepitope discovery. Assays based on sequencing of T-cell receptors (55) may be one such example.
References
- 1.Klein G, Sjorgen HO, Klein E, Hellstrom KE. Demonstration of resistance against methylcholanthrene-induced sarcomas in the primary autochthonous host. Cancer Res. 1960;20:1561–72. [PubMed] [Google Scholar]
- 2.Basombrio MA. Search for common antigenicities among twenty-five sarcomas induced by methylcholanthrene. Cancer Res. 1970;30:2458–62. [PubMed] [Google Scholar]
- 3.Srivastava PK, Old LJ. Individually distinct transplantation antigens of chemically induced mouse tumors. Immunol Today. 1988;9:78–83. doi: 10.1016/0167-5699(88)91269-8. [DOI] [PubMed] [Google Scholar]
- 4.Old LJ. Cancer immunology: the search for specificity--G. H. A. Clowes Memorial lecture. Cancer Res. 1981;41:361–75. [PubMed] [Google Scholar]
- 5.Preuss KD, Zwick C, Bormann C, Neumann F, Pfreundschuh M. Analysis of the B-cell repertoire against antigens expressed by human neoplasms. Immunol Rev. 2002;188:43–50. doi: 10.1034/j.1600-065x.2002.18805.x. [DOI] [PubMed] [Google Scholar]
- 6.Yuasa Y, Srivastava SK, Dunn CY, Rhim JS, Reddy EP, Aaronson SA. Nature. 1983;303:775–9. doi: 10.1038/303775a0. Acquisition of transforming properties by alternative point mutations within c-bas/has human proto-oncogene. [DOI] [PubMed] [Google Scholar]
- 7.Nasti TH, Rudemiller KJ, Cochran JB, Kim HK, Tsuruta Y, Fineberg NS, et al. Immunoprevention of chemical carcinogenesis through early recognition of oncogene mutations. J Immunol. 2015;194:2683–95. doi: 10.4049/jimmunol.1402125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boon T, van der Bruggen P. Human tumor antigens recognized by T lymphocytes. J Exp Med. 1996;183:725–9. doi: 10.1084/jem.183.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramarathinam L, Sarma S, Maric M, Zhao M, Yang G, Chen L, et al. Multiple lineages of tumors express a common tumor antigen, P1A, but they are not cross-protected. J Immunol. 1995;155:5323–9. [PubMed] [Google Scholar]
- 10.GSK MAGE-3 Vaccine Disappointing. http://www.melanoma.org/find-support/patient-community/mpip-melanoma-patients-information-page/gsk-mage-3-vaccine.
- 11.Update on phase III clinical trial of investigational MAGE-A3 antigen-specific cancer immunotherapeutic in non-small cell lung cancer. http://us.gsk.com/en-us/media/press-releases/2014/update-on-phase-iii-clinical-trial-of-investigational-mage-a3-antigen-specific-cancer-immunotherapeutic-in-non-small-cell-lung-cancer/
- 12.Hunder NN, Wallen H, Cao J, Hendricks DW, Reilly JZ, Rodmyre R, et al. Treatment of metastatic melanoma with autologous CD4+ T cells against NY-ESO-1. N Engl J Med. 2008;358:2698–703. doi: 10.1056/NEJMoa0800251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan J, Page DB, Ku GY, Li Y, Mu Z, Ariyan C, et al. Correlation of clinical and immunological data in a metastatic melanoma patient with heterogeneous tumor responses to ipilimumab therapy. Cancer Immun. 2010;10:1. [PMC free article] [PubMed] [Google Scholar]
- 14.Van der Bruggen P, Stroobant V, Vigneron N, Van den Eynde B. Tumor antigens resulting from mutations. 2013 http://cancerimmunity.org/peptide/mutations/ [PMC free article] [PubMed]
- 15.Dranoff G, Jaffee E, Lazenby A, Golumbek P, Levitsky H, Brose K, et al. Vaccination with irradiated tumor cells engineered to secrete murine granulocyte-macrophage colony-stimulating factor stimulates potent, specific, and long-lasting anti-tumor immunity. Proc Natl Acad Sci U S A. 1993;90:3539–43. doi: 10.1073/pnas.90.8.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hege KM, Jooss K, Pardoll D. GM-CSF gene-modifed cancer cell immunotherapies: of mice and men. Int Rev Immunol. 2006;25:321–52. doi: 10.1080/08830180600992498. [DOI] [PubMed] [Google Scholar]
- 17.Srivastava PK. Interaction of Heat shock proteins with peptides and antigen presenting cells : Chaperoning of the innate and adaptive immune responses. Annu Rev Immunol. 2002;20:395–425. doi: 10.1146/annurev.immunol.20.100301.064801. [DOI] [PubMed] [Google Scholar]
- 18.Suto R, Srivastava PK. A mechanism for the specific immunogenicity of heat shock protein - chaperoned peptides. Science. 1995;269:1585–8. doi: 10.1126/science.7545313. [DOI] [PubMed] [Google Scholar]
- 19.Binder RJ, Han DK, Srivastava PK. CD91 is a receptor for heat shock protein gp96. Nat Immunol. 2000;1:151–5. doi: 10.1038/77835. [DOI] [PubMed] [Google Scholar]
- 20.Belli R, Testori A, Rivoltini L, Maio M, Andreola G, Sertoli MR, et al. Vaccination of metastatic melanoma patients with autologous Tumor-derived heat shock protein gp96-peptide complexes. J Clin Oncol. 2002;20:4169–80. doi: 10.1200/JCO.2002.09.134. [DOI] [PubMed] [Google Scholar]
- 21.Testori A, Richards J, Whitman E, Mann B, Lutzky J, Camacho L, et al. Comparison of autologous tumor-derived heat shock protein gp96-peptide complex vaccine (vitespen) and physician’s choice in a randomized phase 3 trial in patients with stage IV melanoma. J Clin Oncol. 2008;26:955–62. doi: 10.1200/JCO.2007.11.9941. Erratum in: J Clin Oncol. Aug 1;26:3819. [DOI] [PubMed] [Google Scholar]
- 22.Wood CG, Escudier B, Lacombe L, Wentworth K, Yuh L, Gupta R, et al. A randomized Phase 3 trial comparing adjuvant therapy with autologous tumor-derived heat shock protein gp96 (vitespen) vs. observation in patients at high risk of recurrence of renal cell carcinoma. Lancet. 2008;372:145–54. doi: 10.1016/S0140-6736(08)60697-2. [DOI] [PubMed] [Google Scholar]
- 23.Bloch O, Crane CA, Fuks Y, Kaur R, Aghi MK, Berger MS, et al. Heat-shock protein peptide complex-96 vaccination for recurrent glioblastoma: a phase II, single-arm trial. Neuro Oncol. 2014;16:274–9. doi: 10.1093/neuonc/not203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srivastava PK. (1993) Peptide-binding heat shock proteins in the endoplasmic reticulum: role in immune response to cancer and in antigen presentation. Adv Cancer Res. 1993;62:153–77. doi: 10.1016/s0065-230x(08)60318-8. [DOI] [PubMed] [Google Scholar]
- 25.Segal NH, Parsons DW, Peggs KS, Velculescu V, Kinzler KW, Vogelstein B, et al. Epitope landscapes in breast and colorectal cancer. Cancer Res. 2008;68:889–92. doi: 10.1158/0008-5472.CAN-07-3095. [DOI] [PubMed] [Google Scholar]
- 26.Srivastava N, Srivastava PK. Modeling the repertoire of true tumor-specific epitopes in a human cancer. PLoS ONE. 2009;10:4, e6094. doi: 10.1371/journal.pone.0006094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rajasagi M, Shukla SA, Fritsch EF, Keskin DB, DeLuca D, Carmona E, et al. Systematic identification of personal tumor-specific neoantigens in chronic lymphocytic leukemia. Blood. 2014;124:453–62. doi: 10.1182/blood-2014-04-567933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Al Seesi S, Duan F, Mandoiu II, Srivastava PK, Kueck A. Genomics-guided immunotherapy of human epithelial ovarian cancer. In: Rodriguez-Oquendo Annabelle., editor. Translational Cardiometabolic Genomic Medicine (Textbook) Elsevier Press; 2015. in press. [Google Scholar]
- 29.Lennerz V, Fatho M, Gentilini C, Frye RA, Lifke A, Ferel D, et al. The response of autologous T cells to a human melanoma is dominated by mutated neoantigens. Proc Natl Acad Sci U S A. 2005;102:16013–8. doi: 10.1073/pnas.0500090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown SD, Warren RL, Gibb EA, Martin SD, Spinelli JJ, Nelson BH, et al. Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome Res. 2014;24:743–50. doi: 10.1101/gr.165985.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fritsch EF, Rajasagi M, Ott PA, Brusic V, Harcohen N, Wu CJ. HLA-binding properties in humans. Cancer Immunol Res. 2014;2:522–9. doi: 10.1158/2326-6066.CIR-13-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sette A, Vitiello A, Reherman B, Fowler P, Nayersina R, Kast WM, et al. The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes. J Immunol. 1994;153:5586–92. [PubMed] [Google Scholar]
- 33.Wentworth PA, Vitiello A, Sidney J, Keogh E, Chesnut RW, Grey H, et al. Differences and similarities in the A2.1-restricted cytotoxic T cell repertoire in humans and human leukocyte antigen-transgenic mice. Eur J Immunol. 1996;26:97–101. doi: 10.1002/eji.1830260115. [DOI] [PubMed] [Google Scholar]
- 34.Assarsson E, Sidney J, Oseroff C, Pasquetto V, Bui HH, Frahm N, et al. A quantitative analysis of the variables affecting the repertoire of T cell specificities recognized after vaccinia virus infection. J Immunol. 2007;178:7890–901. doi: 10.4049/jimmunol.178.12.7890. [DOI] [PubMed] [Google Scholar]
- 35.Castle JC, Kreiter S, Diekmann J, Löwer M, van de Roemer N, de Graaf J, et al. Exploiting the mutanome for tumor vaccination. Cancer Res. 2012;72:1081–91. doi: 10.1158/0008-5472.CAN-11-3722. [DOI] [PubMed] [Google Scholar]
- 36.Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–4. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duan F, Duitama J, AlSeesi S, Ayres CM, Corcelli SA, Pawashe AP, et al. Genomic and bioinformatic profiling of mutational neoepitopes reveals new rules to predict anticancer immunogenicity. J Exp Med. 2014;211:2231–48. doi: 10.1084/jem.20141308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gubin MM, Zhang X, Schuster H, Caron E, Ward JP, Noguchi T, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515:577–81. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yadav M, Jhunjhunwala S, Phung QT, Lupardus P, Tanguay J, Bumbaca S, et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature. 2014;515:572–6. doi: 10.1038/nature14001. [DOI] [PubMed] [Google Scholar]
- 40.van Rooij N, van Buuren MM, Philips D, Velds A, Toebes M, Heemskerk B, et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J Clin Oncol. 2013;31:e439–e442. doi: 10.1200/JCO.2012.47.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blum JS, Wearsch PA, Cresswell P. Pathways of antigen processing. Annu Rev Immunol. 2013;31:443–73. doi: 10.1146/annurev-immunol-032712-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stroobant V, Demotte N, Luiten RM, Leonhardt RM, Cresswell P, Bonehill A, et al. Inefficient exogenous loading of a tapasin-dependent peptide onto HLA-B*44:02 can be improved by acid treatment or fixation of target cells. Eur J Immunol. 2012;42:1417–28. doi: 10.1002/eji.201141954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Callahan MK, Wolchok JD, Allison JP. Anti-CTLA4 antibody therapy: immune monitoring during clinical development of a novel immunotherapy. Semin Oncol. 2010;37:473–484. doi: 10.1053/j.seminoncol.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic Basis for Clinical Response to CTLA-4 Blockade in Melanoma. N Engl J Med. 2014;371:2189–99. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Postow MA, Chesney J, Pavlick AC, Robert C, Grossmann K, McDermott D, et al. Nivolumab and ipilimumab versus ipilimumab in untreated melanoma. N Engl J Med. 2015;372:2006–17. doi: 10.1056/NEJMoa1414428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang RF, Wang X, Atwood AC, Topalian SL, Rosenberg SA. Cloning genes encodingMHCclass II-restricted antigens: mutated CDC27 as a tumor antigen. Science. 1999;284:1351–4. doi: 10.1126/science.284.5418.1351. [DOI] [PubMed] [Google Scholar]
- 47.Monach PA, Meredith SC, Siegel CT, Schreiber H. A unique tumor antigen produced by a single amino acid substitution. Immunity. 1995;2:45–59. doi: 10.1016/1074-7613(95)90078-0. [DOI] [PubMed] [Google Scholar]
- 48.Matsutake T, Srivastava PK. The immunoprotective MHC II epitope of a chemically-induced tumor harbors a unique mutation in a ribosomal protein. Proc Natl Acad Sci U S A. 2001;98:3992–7. doi: 10.1073/pnas.071523398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kreiter S, Vormehr M, van de Roemer N, Diken M, Löwer M, Diekmann J, et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature. 2015;520:692–6. doi: 10.1038/nature14426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O’Rawe J, Jiang T, Sun G, Wu Y, Wang W, Hu J, et al. Low concordance of multiple variant-calling pipelines: practical implications for exome and genome sequencing. Genome Med. 2013;5:28. doi: 10.1186/gm432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim SY, Speed TP. Comparing somatic mutation-callers: beyond Venn diagrams. BMC Bioinformatics. 2013;14:189. doi: 10.1186/1471-2105-14-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Butros P, Ewing A, Ellrott K, Norman T, Dang K, Hu Y, et al. Global optimization of somatic variant identification in cancer genomes with a global community challenge. Nat Genetics. 2014;46:318–9. doi: 10.1038/ng.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y, Waters J, Leung ML, Unruh A, Roh W, Shi X, et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014;512:155–60. doi: 10.1038/nature13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klinger M, Kong K, Moorhead M, Weng L, Zheng J, Faham M. Combining next-generation sequencing and immune assays: a novel method for identification of antigen-specific T cells. PLoS One. 2013;8:e74231. doi: 10.1371/journal.pone.0074231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]