Abstract
Objective
Epigenetic alterations of the malignantly transformed cells have increasingly been regarded as an important event in the carcinogenic development. Induction of some miRNAs such as miR-302/367 cluster has been shown to induce reprogramming of breast cancer cells and exert a tumor suppressive role by induction of mesenchymal to epithelial transition, apoptosis and a lower proliferation rate. Here, we aimed to investigate the impact of miR-302/367 overexpression on transforming growth factor-beta (TGF-β) signaling and how this may contribute to tumor suppressive effects of miR-302/367 cluster.
Materials and Methods
In this experimental study, MDA-MB-231 and SK-BR-3 breast cancer cells were cultured and transfected with miR-302/367 expressing lentivector. The impact of miR-302/367 overexpression on several mediators of TGF-β signaling and cell cycle was assessed by quantitative real-time polymerase chain reaction (qPCR) and flow cytometry.
Results
Ectopic expression of miR-302/367 cluster downregulated expression of some downstream elements of TGF-β pathway in MDA-MB-231 and SK-BR-3 breast cancer cell lines. Overexpression of miR-302/367 cluster inhibited proliferation of the breast cancer cells by suppressing the S-phase of cell cycle which was in accordance with inhibition of TGF-β pathway.
Conclusion
TGF-β signaling is one of the key pathways in tumor progression and a general suppression of TGF-β mediators by the pleiotropically acting miR-302/367 cluster may be one of the important reasons for its anti-tumor effects in breast cancer cells.
Keywords: Breast Cancer, miR-302/367, Reprogramming, Transforming Growth Factor-Beta
Introduction
Despite recent advancements in the treatment of breast cancer, it still remains one of the leading causes of cancer deaths among women (1). Therefore, development of new therapeutic approaches for breast cancer is of the utmost importance. Reprograming of somatic cells and generation of induced pluripotent stem (iPS) cells by some transcription factors including OCT3/4, SOX2, NANOG, KLF4, LIN28 and MYC (2, 3) demonstrated that the cell fate can be manipulated in vitro. Reprogramming is a process accompanied by distinct alterations in chromatin and transcriptional programs.
MiRNAs constitute a class of 17-24 bp small non-coding RNAs, involved in regulation of different biological processes and cancer-related cellular activities such as apoptosis, proliferation and invasion (4, 5). MiR-302/367 cluster possesses a coding sequence located in intron 8 of the LARP7 gene and codes for 5 miRNAs including miR302a, miR302b, miR302c, miR302d, and miR367 which are highly expressed in embryonic stem cells (6-8), but their expression decline rapidly after differentiation (9). It was shown that miR-302/367 cluster can effectively reprogram human and mouse somatic cells to iPS cells (10, 11). miR-302 is also able to reprogram human cancer cells to a human embryonic stem cell-like state with a slow cell cycle rate and dormant cell-like morphology (12, 13). Reprogramming by miR-302/367 cluster has shown tumor suppressive effects on different cancer cells, such as melanoma and colon cancer cells (14), cervical carcinoma cells (15) glioblastoma cells (16), prostate cancer cells (13), endometrial cancer cells (17) and breast cancer (18). The miR-302/367 cluster has been shown to induce reprogramming of somatic cells through multiple pathways, including MECP1/2 and AOF1/2 silencing, repression of suppressor NR2F2 gene expression, and silencing RHOC and TGFBRII (19).
Transforming growth factor-b (TGF-β) signaling pathway is one of the major players in malignant progression through multiple mechanisms which enhance tumor cell invasion, dissemination, and immune evasion (20, 21). In this study we aimed to investigate how overexpression of miR-302/367 cluster in breast cancer cells affects some of the main TGF-β signaling pathway mediators.
Materials and Methods
Cell lines and culture conditions
In this experimental study, human MDA-MB-231 and SK BR-3 breast cancer cell lines were respectively purchased from Pasteur Institute and Iranian Biological Resource Center (IRBC), Iran. Both cell lines were cultured in Dulbecco’s Modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS), 1% L-glutamine and 1% penicillin-streptomycin (all from GibcoTM, Thermo Fisher Scientific, USA) at 5% CO2 and 37°C. The culture medium was renewed every other day.
Transfection with miR-302/367 expressing vector
Transfection of MDA-MB-231 and SK-BR-3 were performed using either a TDH101PA-GP miR-302abcd/367 expressing Lentivector (System Biosciences, SBI, USA) or the same vector without the miR-302/367 cluster as the mock control type, using Lipofectamine® 2000 transfection reagent (Invitrogen, Thermo Fisher Scientific, USA) according to the manufacture’s protocol. 48 hours after transfection, transfected cells were selected by adding 1 mg/ ml puromycin dihydrochloride (Bio Basic Inc., Canada) to the culture medium every other day up to the elimination of untransfected cells. Transfected cells were kept in culture condition for a two-week period.
Analysis of miRNA and gene expression by quantitative real time polymerase chain reaction
For analysis of miRNA expression, total RNA including small RNA, was extracted from the cultured cells using RNX-Plus solution (Sinaclon, Iran) according to the manufacturer’s protocol. Equal amounts of RNA were reverse transcribed into cDNA using BON-miR miRNA 1st-Strand cDNA Synthesis Kit (Stem Cell Technology Co., Iran).
For quantification of mRNAs, total RNA was extracted using the High Pure RNA Isolation Kit (Roche, Germany) according to the manufacturer’s protocol. RNAquality and quantity were assessed using a NanoDropTM 2000/2000c Spectrophotometer (Thermo Fisher Scientific, USA). Equal amount of total RNA from each group was reverse transcribed into cDNA using oligo-dT primers and RevertAid H Minus Reverse Transcriptase (Thermo Fisher Scientific). Assessment of miRNA and mRNA expression was performed, using FastStart SYBR Green Master (Roche, Germany) and specific primers for miR302a, miR-302b, miR-302c, miR-302d, miR-367 and other genes as mentioned in Table 1, on a Rotor-Gene 6000 (Corbett Research, Australia) real-time PCR instrument. SNORD47 was selected as the internal reference gene for quantification of miRNAs. GAPDH and B2M were used as the internal reference genes for quantification of the mRNAs. Comparative analysis of gene expression between different groups was performed using REST 2009 software (Relative Expression Software Tool, Qiagen) based on a Pair Wise Reallocation Randomization Test (22). Four replicates of each group were included in the qPCR reactions.
Table 1.
Primers used for quantitative real-time polymerase chain reaction
| Target | Primer sequence (5ˊ-3ˊ) | Size (bp) | Accession no. |
|---|---|---|---|
| TGFBR2 | F: CCCATCCACTGAGACATATTAAT | 198 | NM_001024847.2 |
| R: CATTCTTTCTCCATACAGCCAC | |||
| BUB1 | F: GAGTCAAATATGGAACGAAGAG | 207 | NM_004336.4 |
| R: GTCTTCATTTACCCATTGCTCA | |||
| RHOC | F: CCTGACAGCCTGGAAAACAT | 153 | NM_175744.4 |
| R: AACGGGCTCCTGCTTCATCT | |||
| AKT1 | F: ACAAACGAGGGGAGTACATCAA | 156 | NM_005163.2 |
| R: TCTTCATCAGCTGGCACTGC | |||
| MAPK1 | F: ATTCCAAGGGCTACACCAAGT | 136 | NM_002745.4 |
| R: GGATCCAAGAATACCCAAAATGT | |||
| MAPK14 | F: TGGCTGTCGACTTGCTGGA | 189 | NM_001315.2 |
| R: CATAGGTCAGGCTTTTCCACT | |||
| SMAD3 | F: CATAATAACTTGGACCTGCAGC | 236 | NM_005902.3 |
| R: ACGCCTCTTCCGATGTGTCT | |||
| B2M | F: TCCAGCGTACTCCAAAGATTCA | 113 | NM_004048.2 |
| R: GTCAACTTCAATGTCGGATGGAT | |||
| GAPDH | F: TCACCATCTTCCAGGAGCGA | 116 | NM_002046.5 |
| R: CAAATGAGCCCCAGCCTTCT | |||
Analysis of cell cycle by flow cytometry
At the end of transfection and cell culture period, the cells were harvested and fixed in 70% cold ethanol and DNA content was stained with propidium iodide (PI) solution. Four replicates of each group were used in this study. Cell cycle analysis was carried out using a FACSCaliburTM flow cytometer (BD Biosciences, USA). FlowJo vX.0.7 software (Tree Star Inc., USA) was used for analysis of the results. Comparison of the cell cycle G1, S and G2/M proportions was performed between the mock and miR-302/367 transfected group of each cell line, using unpaired t test.
Results
Overexpression of the miR-302/367 members in transfected cells
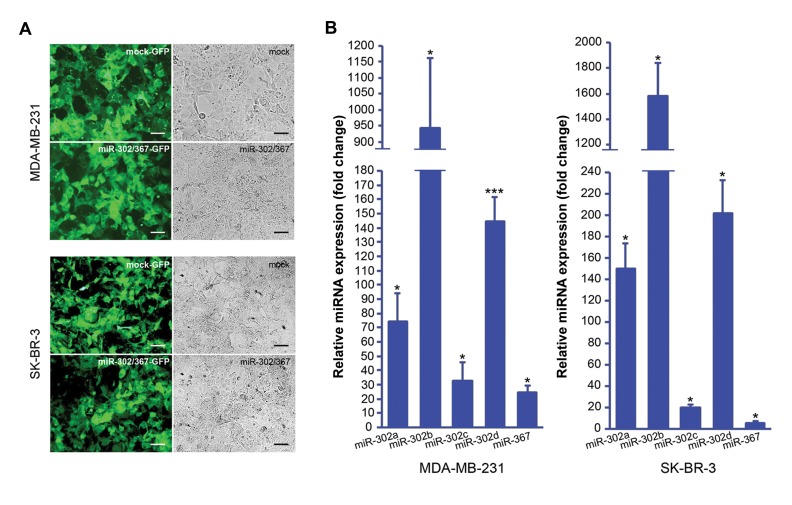
Antibiotic-based selection of the miR-302/367 transfected breast cancer cells caused producing a highly (>90%) GFP-expressing cells population (Fig .1A) which were used for the subsequent experiments. Quantification of the miR-302/367 expression in MDA-MB-231 cells showed upregulation of miR-302a, miR-302b, miR-302c, miR-302d and miR-367 by mean factors of 74, 946, 33, 145 and 25, respectively (Fig .1B). In SK-BR-3 cells, after miR-302/367 transfection, miR-302a, miR-302b, miR-302c, miR-302d and miR-367 were upregulated by mean factors of 145, 1581, 20, 202 and 6, respectively (Fig .1B).
Fig.1.
Ectopic expression of miR-302 cluster in the BC cells. A. Photomicrographs of the MDA-MB-231 and SK-BR-3 cells transfected with either miR-302/367 or mock vector. Transfected cells show GFP expression. Scale bar represents 50 µm and B. Assessment of miR-302/367 expression using quantitative real-time polymerase chain reaction (qPCR) in MDA-MB-231 cells (left) and SK-BR-3 cells (right) transfected with miR-302/367 vector. Fold changes are reflected on the vertical axis compared to the control group (transfected with mock vector) which has been normalized to 1. Analysis performed by REST 2009 software based on a Pair Wise Fixed Reallocation Randomisation Test. and significant P values (*; P<0.05, ***; P<0.001) are indicated on the chart. BC; Breast cancer and GFP; Green fluorescent protein.
Regulation of TGF-ß and MAPK pathway genes by miR-302/367 cluster
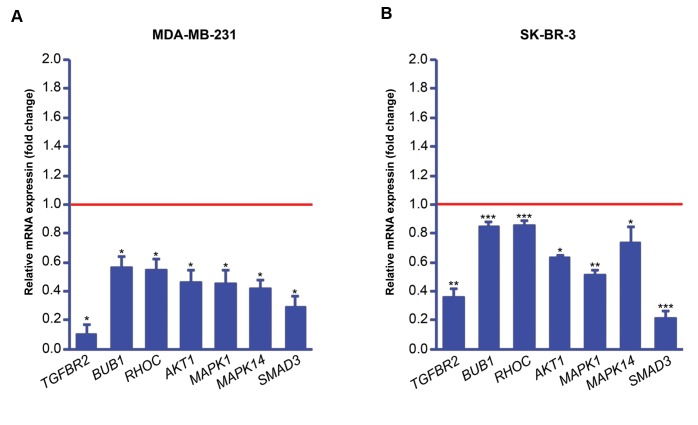
Firstly, we checked how transfection of the breast cancer cells with miR-302/367 cluster affects the expression of some key mediators of TGF-β and mitogenactivated protein kinase (MAPK) pathways at gene level. Quantitative real-time PCR showed that in the both MDA-MB-231 and SK-BR-3 cells, overexpression of miR-302/367 cluster downregulated TGFBR2, BUB1, RHOC, AKT1, MAPK1, MAPK14 and SMAD3 expression compared to the mock transfected cells (Fig .2).
Fig.2.
Expression analysis of some transforming growth factor-beta (TGF-β) mediators at mRNA level after transfection with miR-302/367 vector using quantitative real-time polymerase chain reaction (qPCR). Downregulation of TGF-β-related genes in A. MDA-MB-231 and B. SK-BR-3 cells. Red line represents expression level in the mock transfected group. P values were generated by REST 2009 software based on a Pair Wise Fixed Reallocation Randomisation Test.. Significant P values (*; P<0.05, **; P<0.01, ***; P<0.001) are reflected on the chart.
Cell cycle arrest by overexpression of miR-302/367 cluster
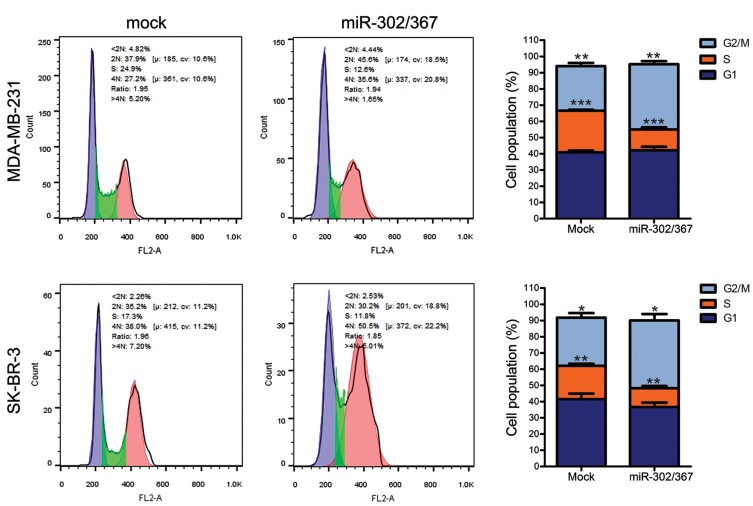
At the end of culture period, transfected breast cancer cells with either miR-302/367 or mock vector were analyzed for the cell cycle phases, using PI staining and flow cytometry. In the miR-302/367 transfected MDA-MB-231 and SK-BR-3 cells, there was a marked decrease in the S-phase population, while the G2/M phase population was partially increased compared to the mock transfected group (Fig .3).
Fig.3.
Flow cytometry analysis of cell cycle. There was a significant decrease in S-phase and a partial increase in G2/M-phase population of both MDA-MB-231 and SK-BR-3 cells, after overexpression of miR-302/367 cluster (unpaired t test, n=4, *; P<0.05, **; P<0.01 and ***; P<0.001).
Discussion
Genetic and epigenetic alterations contribute to cancer initiation and progression through affecting gene expression. While genetic mutations may lead to stable and irreversible alterations, transient and reversible changes are usually caused by epigenetic modifications (23). It has been shown that reprogramming of cancer cells by some pluripotency transcription factors or specific miRNAs, like miR-302/367 cluster, may lead to an embryonic stem cell-like state and less tumorigenicity (13, 24). We previously demonstrated upregulation of some pluripotency factors, including OCT4A, SOX2 and NANOG, by overexpression of miR-302/367 cluster in MDA-MB-231 and SK-BR-3 cells (18).
Accumulating evidence supports the function of miR 302 cluster as a tumor suppressor family which can alleviate tumorigenicity of cancer cells through reversal of epithelial to mesenchymal transition (EMT), induction of apoptosis and anti-proliferative effect (13-15). Previously, we demonstrated some anti-tumor effects of miR-302 cluster in melanoma, colon and breast cancer cells (14, 18). Among the pathways promoting EMT, several studies have reported that inhibition of TGF-ß signaling induces somatic cell reprogramming (25, 26).
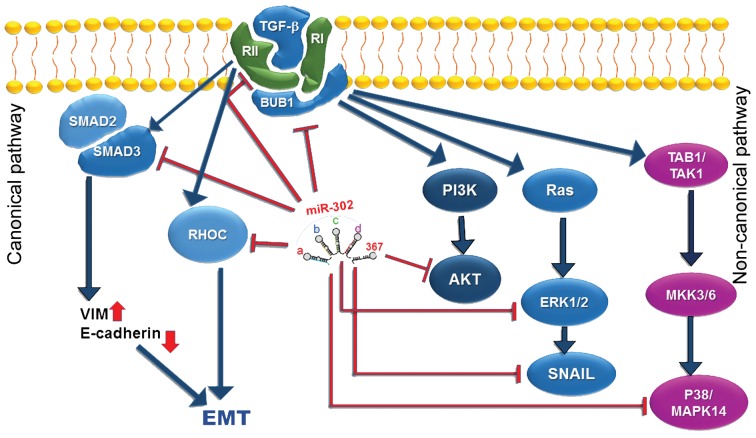
In this study, we investigated how overexpression of miR-302/367 cluster in MDA-MB-231 and SKBR- 3 breast cancer cells affects some mediators of TGF-β/MAPK/AKT signaling pathway at gene level. As shown, TGFBR2 and RHOC are directly targeted by miR-302 cluster, subsequently facilitating human or mouse fibroblast reprogramming towards iPS cells (27, 28). In accordance with these studies, we found that overexpression of miR-302/367 in human breast cancer cells downregulates TGFBR2 and RHOC expressions. TGF-ß signaling has two canonical and non-canonical pathways (Fig .4). In the canonical or SMAD-dependent pathway, SMAD proteins play key regulatory roles among which SMAD2 and SMAD3 proteins are phosphorylated through activity of TGF-ß and activin (29). While, in the non-canonical or SMAD-independent pathway, TGF-ß activates phosphatidylinositol 3kinase (PI3K)/ AKT and MAPK pathways (30). In the current study, expressions of TGFBR2, BUB1, RHOC, AKT1, MAPK1, MAPK14 and SMAD3 were significantly downregulated after overexpression of miR-302/367 cluster in the both cell lines. Previously, Cai et al. (15) reported that miR-302/367 directly targets AKT1 and it suppresses proliferation of HeLa and SiHa cervical carcinoma cells. In the same study, AKT1 protein level was decreased after miR-302/367 transfection, but AKT1 gene expression was not significantly changed. In another study, Li et al. (31) demonstrated that miR-302abcd cluster upregulated OCT4 expression by targeting AKT1 gene at its 3’-UTR. The same report also showed downregulation of AKT1 at both gene and protein levels, following miR-302 transfection. Similarly, we showed that overexpression of miR-302/367 cluster in MDA-MB-231 and SKBR- 3 cells induces expression of OCT4 gene (18) and downregulates expression of AKT1. Therefore, it seems that a mechanism, similar to that of previous reports, is applicable to breast cancer cells.
Fig.4.
Interaction between miR-302/367 cluster and several mediators of transforming growth factor-beta (TGF-β) signaling in both canonical and non-canonical pathways. The inhibitory effect of miR-302/367 cluster is shown by the red lines.
We also detected a significant downregulation of SMAD3 expression following miR-302/367 transfection of the breast cancer cells. Sustained activity of SMAD complexes in the nucleus is one of the key features of TGF-ß signaling in cancer cells. It was reported that an interaction between FOXM1 and SMAD3 is critical for TGF-β-mediated gene expression. Thus, it promotes breast cancer cell invasion and metastasis (32).
BUB1 was also downregulated in the breast cancer cells after ectopic expression of miR-302/367 cluster. BUB1 is a serine/threonine kinase playing a significant role in cell cycle regulation, chromosome cohesion (33), and it is a key mediator of TGF-β signaling. It has been shown that BUB1 promotes canonical and non-canonical TGF-β signaling and mediates TGF-β-dependent EMT, cell migration and invasion through interaction with both TGFBRI and TGFBRII (34). Here, for the first time, we are reporting downregulation of BUB1 expression in breast cancer cells following overexpression of miR302/ 367 cluster. This provides further evidence regarding the significance of miR-302/367 suppressive impact on TGF-β signaling through inhibition of both canonical and non-canonical pathways.
MAPK pathway is part of the non-SMAD pathways, activated by the TGF-ß receptors (35). MAPK1, also known as ERK2, is encoded by MAPK1 gene. The ERK1/2 pathway plays a pivotal role in regulation of cell proliferation, and it is known as a master regulator of G1 to S-phase progression (36, 37). Another player of the non- canonical TGF-β pathway, MAPK14/p38α is encoded by MAPK14 gene. MAPK14/p38α is 50% identical to ERK2 and generally expressed in cell lines and tissues (36). There has been controversy regarding the role of p38 MAPKs in regulating cell proliferation and survival (38). This primarily depends on the cell type determining whether p38 MAPK induces progression or inhibition of G1/S transition through differential regulation of cyclin A or D levels, phosphorylation of RB protein (39, 40), and phosphorylation of p53 (40). In our study, ectopic expression of miR-302/367 cluster in the breast cancer cells downregulated expression of all of the investigated TGF-β mediators, including TGFBR2, BUB1, RHOC, AKT1, MAPK1, MAPK14 and SMAD3. These findings indicate a strong suppressive effect of miR-302/367 cluster on the TGF-β signaling (Fig .4). In this study we report a lower proliferation rate and S-phase suppression of the breast cancer cells by overexpression of miR-302/367, confirming our previous report (18). This can be explained by suppression of MAPK1 and MAPK14 to some extent. Therefore, suppression of TGF-β mediators may provide a good reason behind the partial cell cycle arrest observedin the breast cancers, following overexpression of miR302/ 367 cluster.
Conclusion
Overexpression of miR-302/367 cluster in human breast cancer cells resulted in a general suppressive effect on multiple mediators of TGF-β signaling and BUB1. This finding was accompanied by inhibition of cell proliferation. Previously, we reported anti-tumor Ahmadalizadeh Khanehsar et al. effects of either miR-302bcad or miR-302bcad/367 clusters on melanoma, colon and breast cancer cells due to induction of apoptosis and suppression of proliferation and invasion. Current results are providing new evidence that suppression of TGF-β signaling at gene level may be one of the important reasons for anti-tumor effects of miR-302/367 cluster in breast cancer cells.
Acknowledgments
There is no financial support and conflict of interest in this study.
Author’s Contributions
M.A.K.; Did the gene expression analysis and writing the draft. M.H.; Performed cell culture and transfection of the cells. M.F.T.; Gave technical assistance on the reprogramming process and contributed to writing the draft. A.J.; Designed the study, supervised the project, performed the cell cycle analysis and edited the manuscript draft. All authors read and approved the final manuscript.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet‐Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 3.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Ma L, Weinberg RA. MicroRNAs in malignant progression. Cell Cycle. 2008;7(5):570–572. doi: 10.4161/cc.7.5.5547. [DOI] [PubMed] [Google Scholar]
- 6.Pi J, Tao T, Zhuang T, Sun H, Chen X, Liu J, et al. A microRNA302- 367-Erk1/2-Klf2-S1pr1 Pathway prevents tumor growth via restricting angiogenesis and improving vascular stability. Circ Res. 2017;120(1):85–98. doi: 10.1161/CIRCRESAHA.116.309757. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Z, Hong Y, Xiang D, Zhu P, Wu E, Li W, et al. MicroRNA- 302/367 cluster governs hESC self-renewal by dually regulating cell cycle and apoptosis pathways. Stem Cell Reports. 2015;4(4):645–657. doi: 10.1016/j.stemcr.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suh M-R, Lee Y, Kim JY, Kim S-K, Moon S-H, Lee JY, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270(2):488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 9.Kuo CH, Ying SY. Advances in microRNA-mediated reprogramming technology. Stem Cells Int. 2012;2012:823709–823709. doi: 10.1155/2012/823709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8(4):376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galoian K, Qureshi A, D’Ippolito G, Schiller PC, Molinari M, Johnstone AL, et al. Epigenetic regulation of embryonic stem cell marker miR302C in human chondrosarcoma as determinant of antiproliferative activity of proline-rich polypeptide 1. Int J Oncol. 2015;47(2):465–472. doi: 10.3892/ijo.2015.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin SL, Chang DC, Ying SY, Leu D, Wu DT. MicroRNA miR-302 inhibits the tumorigenecity of human pluripotent stem cells by coordinate suppression of the CDK2 and CDK4/6 cell cycle pathways. Cancer Res. 2010;70(22):9473–9482. doi: 10.1158/0008-5472.CAN-10-2746. [DOI] [PubMed] [Google Scholar]
- 13.Lin SL, Chang DC, Chang-Lin S, Lin CH, Wu DT, Chen DT, et al. Mir-302 reprograms human skin cancer cells into a pluripotent EScell- like state. RNA. 2008;14(10):2115–2124. doi: 10.1261/rna.1162708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maadi H, Moshtaghian A, Taha MF, Mowla SJ, Kazeroonian A, Haass NK, et al. Multimodal tumor suppression by miR-302 cluster in melanoma and colon cancer. Int J Biochem Cell Biol. 2016;81(Pt A):121–132. doi: 10.1016/j.biocel.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Cai N, Wang YD, Zheng PS. The microRNA-302-367 cluster suppresses the proliferation of cervical carcinoma cells through the novel target AKT1. RNA. 2013;19(1):85–95. doi: 10.1261/rna.035295.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang CM, Chiba T, Brill B, Delis N, von Manstein V, Vafaizadeh V, et al. Expression of the miR‐302/367 cluster in glioblastoma cells suppresses tumorigenic gene expression patterns and abolishes transformation related phenotypes. Int J Cancer. 2015;137(10):2296–2309. doi: 10.1002/ijc.29606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan GJ, Yu F, Wang B, Zhou HJ, Ge QY, Su J, et al. MicroRNA miR- 302 inhibits the tumorigenicity of endometrial cancer cells by suppression of Cyclin D1 and CDK1. Cancer Lett. 2014;345(1):39–47. doi: 10.1016/j.canlet.2013.11.023. [DOI] [PubMed] [Google Scholar]
- 18.Ramezankhani B, Taha MF, Javeri A. Vitamin C counteracts miR-302/367-induced reprogramming of human breast cancer cells and restores their invasive and proliferative capacity. J Cell Physiol. 2019;234(3):2672–2682. doi: 10.1002/jcp.27081. [DOI] [PubMed] [Google Scholar]
- 19.Kuo CH, Deng JH, Deng Q, Ying SY. A novel role of miR-302/367 in reprogramming. Biochem Biophys Res Commun. 2012;417(1):11–16. doi: 10.1016/j.bbrc.2011.11.058. [DOI] [PubMed] [Google Scholar]
- 20.Miyazono K. Transforming growth factor-beta signaling in epithelial- mesenchymal transition and progression of cancer. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85(8):314–323. doi: 10.2183/pjab.85.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meulmeester E, Ten Dijke P. The dynamic roles of TGF-βeta in cancer. J Pathol. 2011;223(2):205–218. doi: 10.1002/path.2785. [DOI] [PubMed] [Google Scholar]
- 22.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30(9):e36–e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishikawa S, Ishii H, Haraguchi N, Kano Y, Fukusumi T, Ohta K, et al. microRNA-based cancer cell reprogramming technology. Exp Ther Med. 2012;4(1):8–14. doi: 10.3892/etm.2012.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyoshi N, Ishii H, Nagai K, Hoshino H, Mimori K, Tanaka F, et al. Defined factors induce reprogramming of gastrointestinal cancer cells. Proc Natl Acad Sci USA. 2010;107(1):40–45. doi: 10.1073/pnas.0912407107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maherali N, Hochedlinger K. Tgfbeta signal inhibition cooperates in the induction of iPSCs and replaces Sox2 and cMyc. Curr Biol. 2009;19(20):1718–1723. doi: 10.1016/j.cub.2009.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin T, Ambasudhan R, Yuan X, Li W, Hilcove S, Abujarour R, et al. A chemical platform for improved induction of human iPSCs. Nat Methods. 2009;6(11):805–808. doi: 10.1038/nmeth.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao B, Bao X, Liu L, Feng S, Zovoilis A, Liu W, et al. MicroRNA cluster 302-367 enhances somatic cell reprogramming by accelerating a mesenchymal-to-epithelial transition. J Biol Chem. 2011;286(19):17359–17364. doi: 10.1074/jbc.C111.235960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanyam D, Lamouille S, Judson RL, Liu JY, Bucay N, Derynck R, et al. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol. 2011;29(5):443–448. doi: 10.1038/nbt.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaudhury A, Howe PH. The tale of transforming growth factorbeta (TGFbeta) signaling: a soigne enigma. IUBMB Life. 2009;61(10):929–939. doi: 10.1002/iub.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chapnick DA, Warner L, Bernet J, Rao T, Liu X. Partners in crime: the TGFβ and MAPK pathways in cancer progression. Cell Biosci. 2011;1:42–42. doi: 10.1186/2045-3701-1-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li HL, Wei JF, Fan LY, Wang SH, Zhu L, Li TP, et al. miR-302 regulates pluripotency, teratoma formation and differentiation in stem cells via an AKT1/OCT4-dependent manner. Cell Death Dis. 2016;7:e2078–e2078. doi: 10.1038/cddis.2015.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xue J, Lin X, Chiu WT, Chen YH, Yu G, Liu M, et al. Sustained activation of SMAD3/SMAD4 by FOXM1 promotes TGF-βeta-dependent cancer metastasis. J Clin Invest. 2014;124(2):564–579. doi: 10.1172/JCI71104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson VL, Scott MI, Holt SV, Hussein D, Taylor SS. Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J Cell Sci. 2004;117(Pt 8):1577–1589. doi: 10.1242/jcs.01006. [DOI] [PubMed] [Google Scholar]
- 34.Nyati S, Schinske-Sebolt K, Pitchiaya S, Chekhovskiy K, Chator A, Chaudhry N, et al. The kinase activity of the Ser/Thr kinase BUB1 promotes TGF-β signaling. Sci Signal. 2015;8(358):ra1–ra1. doi: 10.1126/scisignal.2005379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang YE. Non-smad signaling pathways of the TGF-βeta family. Cold Spring Harb Perspect Biol. 2017;9(2) doi: 10.1101/cshperspect.a022129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev. 2011;75(1):50–83. doi: 10.1128/MMBR.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meloche S, Pouysségur J. The ERK1/2 mitogen-activated protein kinase pathway as a master regulator of the G1- to S-phase transition. Oncogene. 2007;26(22):3227–3239. doi: 10.1038/sj.onc.1210414. [DOI] [PubMed] [Google Scholar]
- 38.Cuenda A, Rousseau S. p38 MAP-kinases pathway regulation, function and role in human diseases. Biochim Biophys Acta. 2007;1773(8):1358–1375. doi: 10.1016/j.bbamcr.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 39.Brancho D, Tanaka N, Jaeschke A, Ventura JJ, Kelkar N, Tanaka Y, et al. Mechanism of p38 MAP kinase activation in vivo. Genes Dev. 2003;17(16):1969–1978. doi: 10.1101/gad.1107303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ambrosino C, Nebreda AR. Cell cycle regulation by p38 MAP kinases. Biol Cell. 2001;93(1-2):47–51. doi: 10.1016/s0248-4900(01)01124-8. [DOI] [PubMed] [Google Scholar]