Abstract
Long non-coding RNAs (lncRNAs) are transcripts longer than 200 nt that are involved in cardiovascular diseases (CVDs). To determine whether lncRNAs are involved in stable angina pectoris (SAP), we analysed the expression profile of lncRNAs and mRNAs on a genome-wide scale in SAP of Uyghur population. Five pairs of SAP patients and healthy controls were screened by an Agilent microarray (human lncRNA + mRNA Array V4.0). Quantitative real-time polymerase chain reaction (qRT-PCR) was used to validate the lncRNA expression levels in 50 SAP and 50 controls. Data analyses were performed using R and Bioconductor. A total of 1871 up- and 231 down-regulated lncRNAs were identified to be differentially expressed in the peripheral blood mononuclear cells (PBMCs). Microarray analysis results identified the lncRNAs NR_037652.1, ENST00000607654.1, ENST00000589524.1 and uc004bhb.3, which were confirmed by qRT-PCR. Among screened lncRNAs, the annotation result of their co-expressed mRNAs showed that the most significantly related pathways were the NF-κB signalling pathway, apoptosis and the p53 signalling pathway, while the main significantly related diseases were the cholesterol, calcium and coronary disease. Our study indicated that clusters of lncRNAs were significantly differentially expressed between SAP patients and matched controls. These lncRNAs may play a significant role in SAP development and could serve as biomarkers and potential targets for the future treatment of SAP.
Keywords: large intervening non-coding RNA, microarray, stable angina pectoris
Introduction
Stable angina pectoris (SAP) is one of the most common manifestations of coronary heart disease (CHD) and is associated with myocardial ischaemia due to the increased oxygen requirement and decreased diastolic perfusion time [1,2]. Although the annual mortality rate due to SAP is relatively low [3,4], but it seriously affects the prognosis and quality of life for CHD patients. Conventional treatment strategies, such as medication, revascularisation and lifestyle modification, have often been used for SAP patients [5,6], however, these treatments do not completely relieve angina-related symptoms for most patients. Moreover, there is a lack of knowledge regarding multiple conventional risk factors and serum biomarkers for SAP diagnosis, gene expression profiling using microarrays has promoted the development of many novel molecular biomarkers [7,8].
Long non-coding RNAs (lncRNAs) are a class of well-studied non-coding RNAs that are >200 nucleotides long and lack protein-coding capability [9]. Recently, lncRNAs have emerged as powerful regulators of biological processes, including RNA–RNA interactions and epigenetic and post-transcriptional regulation [10]. Functionally, many studies have demonstrated that lncRNAs play indispensable roles in the pathophysiologic process of CHD [11,12]. For example, the expression of the lncRNA ANRIL is associated with the risk of coronary atherosclerosis [13], the expression of the lncRNA MIAT in peripheral blood mononuclear cells (PBMCs) is significantly reduced in patients with ST segment elevation myocardial infarction (STEMI) [14], and the lncRNA LIPCAR is down-regulated after acute myocardial infarction (AMI) but is increased during later stages of heart failure [15]. The Chinese Uyghur population, a Muslim minority, accounts for 48.53% of the population in Xinjiang. The Uyghur population has a unique lifestyle with a high incidence of CHD [16]. To date, the genome-wide expression of lncRNAs and their potential biological functions in Uyghur SAP patients remain unknown.
To further identify the potential lncRNAs markers for SAP, we collected PBMCs from five SAP patients and five matched controls to identify the different expression profiles of lncRNAs and mRNAs among SAP and healthy control. The co-expression relationships among the target genes which confirmed the differentially expressed lncRNAs were analyzes via a regulatory co-expression network. Overall, the present study identified differentially expressed lncRNAs and their potential corresponding mRNAs to predict SAP and to provide a basis for the study of the pathogenesis of SAP.
Materials and methods
Patients and samples collection
All patients were evaluated at the time of diagnostic cardiac catheterisation and coronary angiography. In all cases, at least one haemodynamically significant (≥50%) stenosis was present in a major coronary artery [2]. Coronary angiograms were obtained at the Catheter Laboratory in the Department of Cardiology at the First Affiliated Hospital of Xinjiang Medical University. To weaken the variation between samples as much as possible, blood samples from no coronary stenosis matched with age and sex were used as controls. A history of congestive heart failure, unstable angina pectoris (UAP) and MI, severe hepatic or renal dysfunction, malignant tumour, recent infection or active chronic inflammatory disease during the last 6 weeks was excluded in all participants. Five SAP patients (SAP-U2, SAP-U3, SAP-U6, SAP-U7, SAP-U8) and five matched controls (N-U2, N-U3, N-U4, N-U5, N-U6) were randomly selected for lncRNA chip analysis. Blood samples from the rest of 50 SAP patients and 50 controls were obtained to validate the lncRNA expression level using quantitative real-time polymerase chain reaction (qRT-PCR). Additionally, blood samples (5 ml) were collected from peripheral veins before the administration of any anticoagulants, and put into test tubes containing EDTA.
Collection and purification of PBMCs
Blood samples within 2 h after harvesting were centrifuged at 3000 rpm for 10 min, then divided into two layers. The upper plasma layer was discarded, and the lower layer was used for lysis reaction. Lymphocyte Separation Medium (twice the total blood volume) (TBD, Tianjin Biotechnology) was then added to start the lysis reaction, after mixing gently and then centrifuged at 1500 rpm for 20 min. Thereafter, a white membrane, that is the white cell layer, was transferred to a new tube and centrifuged at 10000 rpm for 5 min. After removal of the supernatant, the PBMCs were left in tubes.
RNA extraction, labelling and hybridisation
Total RNA, including lncRNAs, was extracted using TRIzol reagent (Invitrogen) and then purified according to the manufacturer’s instructions using a mirVana miRNA Isolation Kit (Ambion, Austin, TX, U.S.A.). Total RNA and OD260/280 readings were quantified using a Nanodrop-2000 (Thermo Fisher Scientific, Waltham, MA). The RNA quality and the amount of lncRNAs were measured using an Agilent Bioanalyzer (Agilent Technologies, CA). Sample labelling and array hybridisation were performed according to the Agilent One-Color Microarray-Based Gene Expression Analysis Protocol (Agilent Technology). In brief, Cy5 and Cy3-dCTP were used to label cDNA. Double-stranded cDNAs containing the T7 RNA polymerase promoter were synthesised with T7 Oligo (dT) and T7 Oligo (dN) primers using CbcScript reverse transcriptase (Capitalbio). The dsDNA products were purified using a PCR NucleoSpin Extract II Kit (MN) and eluted with 30 μl of elution buffer for transcription reactions using T7 Enzyme Mix. The amplified cRNA was purified using an RNA Clean-up Kit (MN). Then, a Klenow enzyme labelling strategy [17] was used; 5 μl of Klenow buffer, dNTPs, and Cy5-dCTP or Cy3-dCTP (GE Healthcare) were added, and the mixture was incubated at 37°C for 90 min. Before loading on to a microarray, hybridisation solution including DNA was denatured at 95°C for 3 min. The arrays were hybridised overnight at a rotation speed of 20 rpm in an Agilent Hybridization Oven at 42°C and then washed consecutively with two different solutions (2× saline sodium citrate (SSC) for 5 min at 42°C and 0.2× SSC with 0.2% sodium dodecyl sulphate (SDS) for 5 min at room temperature).
LncRNA expression profiles
Approximately 200 ng of RNA from each sample was applied for the lncRNA microarray analysis. The lncRNA expression profiles were analysed using an Agilent Human lncRNA + mRNA Array V4.0 (4 × 180K format) that contained 41000 human lncRNA probes and approximately 34000 human mRNA probes. The lncRNAs and their mRNA target sequences were obtained from multiple databases, including GENCODE/ENSEMBL, LNCipedia, the Human LincRNA Catalog, the ncRNA Expression Database (NRED), RefSeq, the University of California, Santa Cruz (UCSC), and the Chen Ruisheng laboratory (Institute of Biophysics, Chinese Academy of Science). Each RNA was detected two times by probes.
Microarray imaging and data analysis
Data summarisation, normalisation and quality control concerning the lncRNA + mRNA array data were performed in GeneSpring software V13.0 (Agilent). Volcano plot filtering and hierarchical clustering were used to identify the differentially expressed lncRNAs and mRNAs. Differentially expressed genes with statistical significance were identified with a random variance model, and the P-values were determined using paired t tests. Classification of genes as up- and down-regulated required a fold-change (Fc) >2.0 and a P<0.05. A tree was constructed with Java TreeView (Stanford University School of Medicine, Stanford, CA, U.S.A.).
Gene Ontology and Kyoto Encyclopedia of Genes and Genomes analyses
Gene Ontology (GO) analysis was undertaken to illustrate the unique biological significance of the differentially expressed genes [18]. GO categories describe potential functions related to three defined terms: biological processes, molecular functions and cellular components. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was carried out to identify crucial pathways related to gene maps based on the latest KEGG database. The significant GO terms and pathways were identified by Fisher’s exact tests and Chi-square tests, and the significance threshold was defined by false discovery rate (FDR) and P-value [19].
Construction of the regulatory co-expression network
It is important to accurately understand the biological functions of co-expressed proteins, which are important for identifying novel and significant genes. For each pair of genes, a Pearson correlation coefficient was calculated and the significant correlation pairs were selected for construction of the network [8]. The search tool for the open-source bioinformatics software Cytoscape [20] is a reliable online tool that can be used to evaluate many co-expression relationships. In the present study, lncRNAs and mRNAs with Pearson correlation coefficients of at least 0.99 were selected to draw a network with Cytoscape. In a network analysis, a degree is the simplest and most important measure of a gene centrality within a network that determines its relative importance [21].
Validation of lncRNAs by qRT-PCR
The expression of four lncRNAs was validated by qRT-PCR. cDNA was synthesised by reverse transcription from RNA with an RNeasy Mini Kit (QIAGEN, China) in accordance with the manufacturer’s protocol. Then, qRT-PCR was carried out using Power SYBR Green PCR Master Mix (Applied Biosystems, U.S.A.) in a 7900 HT Fast Real-Time PCR system (Applied Biosystems, U.S.A.). Primers for NR_037652.1, ENST00000607654.1, ENST00000589524.1 and uc004bhb.3, and GAPDH were synthesised by Invitrogen (Shanghai, China). All of the primer sequences used are shown in Supplementary Table S1. To quantify the results, the expression of each lncRNA was calculated using the 2−ΔΔct method.
Statistical analysis
The data are presented as the mean ± standard deviation (SD) or standard error (SEM). Continuous variables were compared using either the Mann–Whitney U test or the Kruskal–Wallis test. For categorical variables, either the Chi-square test or Fisher’s test was used as appropriate. GO and KEGG analyses were evaluated using Fisher’s exact test. For all statistical analyses, IBM SPSS Statistics 18.0 software (SPSS Inc, Chicago, IL, U.S.A.) was used, and a two-tailed P<0.05 was considered to be significant.
Results
Patient enrolment
The lncRNA and mRNA expression profiles in five SAP patients and five matched healthy controls were detected using a microarray, and 50 SAP patients and 50 matched controls were used for clinical validation. The clinical characteristics of ten participants for lncRNA chip analysis are shown in Table 1.
Table 1. Clinical characteristics of five Uyghur SAP patients and five Uyghur matched controls.
| Characteristics | Controls (n=5) | SAP patients (n=5) |
|---|---|---|
| Age, years | 51.1 ± 2.14 | 51.6 ± 2.83 |
| Gender (male/female) | 3/2 | 3/2 |
| BMI | 23.7 ± 2.02 | 24.9 ± 2.24 |
| Hypertension (Yes/No) | 0/5 | 0/5 |
| Arrhythmia (Yes/No) | 0/5 | 0/5 |
| Diabetes (Yes/No) | 0/5 | 0/5 |
| Smoking (Yes/No) | 1/4 | 1/4 |
| Drinking (Yes/No) | 1/4 | 1/4 |
| SBP, mmHg | 120.1 ± 7.24 | 120.9 ± 8.35 |
| DBP, mmHg | 82.4 ± 7.25 | 81.6 ± 9.49 |
| Heart rate, beats per minute | 71.6 ± 4.84 | 75.9 ± 6.86 |
| Serum creatinine, μmol/l | 80.7 ± 16.79 | 77.9 ± 17.49 |
| FPG, mmol/l | 4.89 ± 0.41 | 5.56 ± 1.16 |
| TG, mmol/l | 1.79 ± 0.5* | 2.94 ± 1.97* |
| TC mmol/l | 4.57 ± 0.77 | 4.97 ± 0.86 |
| LDL, mmol/l | 3.11 ± 0.91 | 2.97 ± 0.83 |
| Serum calcium, mmol/l | 2.47 ± 0.11 | 1.94 ± 0.65 |
The data are presented as the mean ± standard deviation. Abbreviations: BMI, body mass index; DBP, diastolic blood pressure; FPG, fasting plasma glucose; LDL, low-density lipoprotein; SBP, systolic blood pressure; TC, total cholesterol; TG, triglyceride.
*P<0.05 compared with healthy controls.
Differentially expressed lncRNAs and categorisation between SAP patients and healthy controls
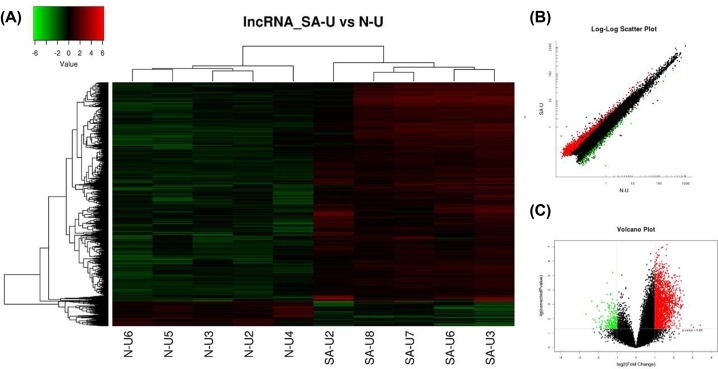
In total, 2102 lncRNAs displayed differential expression between two groups using the criteria of a corrected P<0.05 and an Fc > 2.0, including 1871 up-regulated and 231 down-regulated lncRNAs. A selection of lncRNAs showing distinctive expression in Uyghur SAP patients are presented in Table 2. As hierarchical clustering represents one of the simplest and most widely used techniques, we used this analysis which can then enable the generation of hypotheses about the relationships among samples in our experiment. The results of hierarchical clustering showed distinguishable lncRNA expression profiles between the two groups (Figure 1A). Scatter and volcano plots were also used to assess the variations in gene expression between two groups (Figure 1B,C).
Table 2. Top 20 differentially expressed lncRNAs in SAP patients in Uyghur based on Fc values.
| lncRNA ID | Probe name | Fc (abs) | Regulation | Chrom | Class | Database |
|---|---|---|---|---|---|---|
| ENST00000471935.1 | p15135 | 6.2932 | Down | 7 | Intergenic | ENSEMBL |
| TCONS_00015557 | p24545 | 5.070091 | Down | 9 | Divergent | HumanLincRNACatalog |
| ENST00000443162.1 | p15157 | 4.84097 | Down | 7 | Antisense | ENSEMBL |
| uc001vjh.1 | p25856 | 4.54429 | Down | 13 | Intronic | UCSC |
| ENST00000584157.1 | p6780 | 4.36858 | Down | 17 | Intronic | ENSEMBL |
| XR_158932.2 | p30363 | 4.172306 | Down | 19 | Divergent | RefSeq |
| ENST00000529252.2 | p29496 | 4.092166 | Down | 8 | Antisense | ENSEMBL |
| ENST00000533528.1 | p2878 | 3.868413 | Down | 11 | Divergent | ENSEMBL |
| ENST00000526377.1 | p2591 | 3.836865 | Down | 11 | Intergenic | ENSEMBL |
| ENST00000425554.1 | p913 | 3.818452 | Down | 1 | Divergent | ENSEMBL |
| int-HOXA3-11 | p28119 | 10.6204 | Up | 7 | Intergenic | HOX Loci |
| ENST00000558536.1 | p5543 | 8.313788 | Up | 15 | Antisense | ENSEMBL |
| TCONS_00011823 | p23452 | 7.638344 | Up | 6 | Divergent | HumanLincRNACatalog |
| ENST00000597915.1 | p34744_v4 | 5.823942 | Up | 2 | Antisense | ENSEMBL |
| ENST00000534297.1 | p2546 | 5.668468 | Up | 11 | Antisense | ENSEMBL |
| ENST00000536100.1 | p3485 | 5.616653 | Up | 12 | Antisense | ENSEMBL |
| LIT2106 | p33935 | 5.426726 | Up | 10 | Antisense | RNAdb |
| TCONS_00029064 | p21721 | 5.422044 | Up | 21 | Intergenic | HumanLincRNACatalog |
| TCONS_00022290 | p29659 | 5.31662 | Up | 13 | Intergenic | HumanLincRNACatalog |
| TCONS_00020945 | p29634 | 5.266935 | Up | 12 | Intergenic | HumanLincRNACatalog |
Figure 1. Differential expression of lncRNAs in SAP patients and control individuals of Uyghur ethnicity.
(A) Expression values are represented through hierarchical clustering analysis in red and green, indicating up- and down-regulated lncRNA expressions in SAP patients (SAP-U2, SAP-U3, SAP-U6, SAP-U7, SAP-U8) or control individuals (N-U2, N-U3, N-U4, N-U5, N-U6), respectively. (B) Scatter plot of differential lncRNA expression. x-axis: N-U, y-axis: SAP-U. (C) Volcano plot of differential lncRNA expression. The red and green spots indicate up- and down-regulation, respectively. x-axis: log2 Fc; y-axis: −1 × log10 (corrected P-value) for each probe.
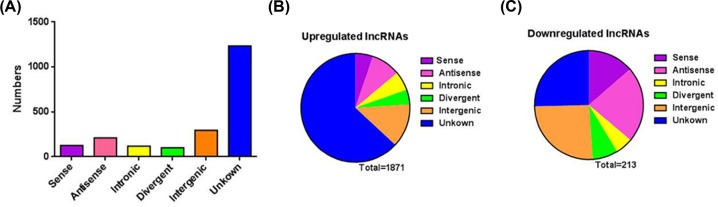
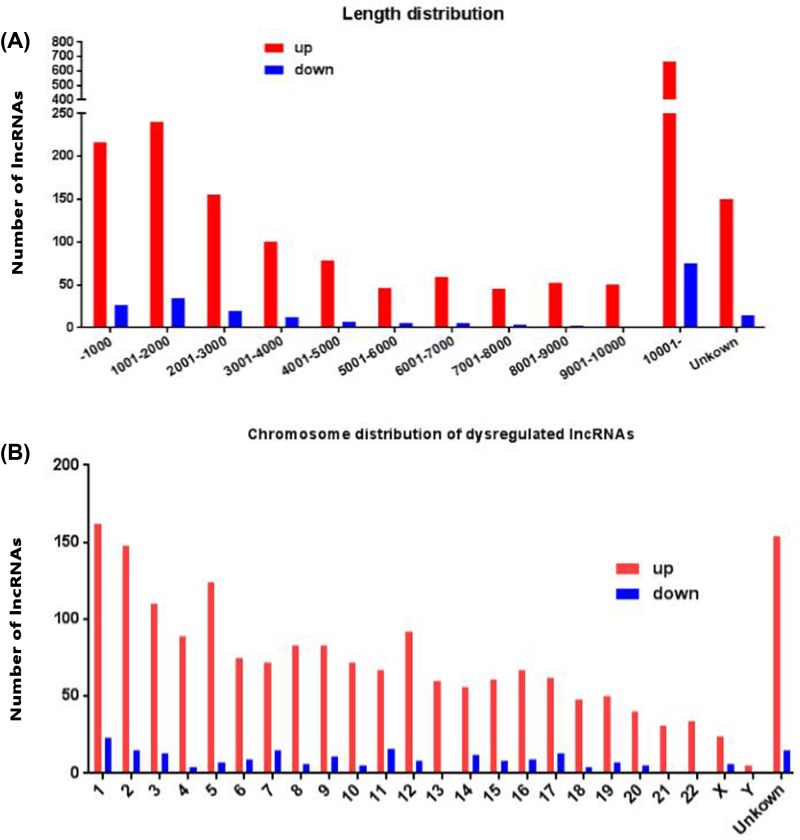
Next, we analysed distinctive lncRNAs based on their categorisations (Figure 2A). Although 59.21% of the lncRNAs were not successfully categorised, the well-annotated lncRNAs were classified into five categories: 14.20% were intergenic, 10.08% were antisense, 5.66% were intronic, 6.09% were sense and 4.75% were bidirectional. This pattern was also present regarding both the up- and down-regulated lncRNAs (Figure 2B,C). The lengths of the dysregulated lncRNAs were mostly between 200 and 3000 bp (Figure 3A). The chromosome distribution shows the number of up- and down-regulated lncRNAs located within each chromosome (Figure 3B). Up-regulated lncRNAs were mainly located in chr1, chr2, chr3, chr5 and unknown chromosome groups, and down-regulated lncRNAs were mainly located in chr1, chr2, chr3, chr7, chr11 and unknown chromosome groups.
Figure 2. The classification of lncRNAs according to their correlations with protein-coding genes.
(A) The numbers of identified lncRNAs in six categories. (B) Pie chart showing the number of up-regulated lncRNAs according to criteria, Fc > 2 and P<0.05 in each category. (C) Pie chart showing the number of down-regulated lncRNAs according to criteria, Fc > 2 and P<0.05 in each category.
Figure 3. The length and chromosomes distribution of dysregulated lncRNAs.
(A) The length distribution of dysregulated lncRNAs. x-axis: length distribution, y-axis: number of lncRNAs. (B) Chromosomes distribution of up- and down-regulated lncRNAs location. x-axis: chromosomes distribution, y-axis: number of lncRNAs.
mRNA expression profiles between SAP patients and healthy controls
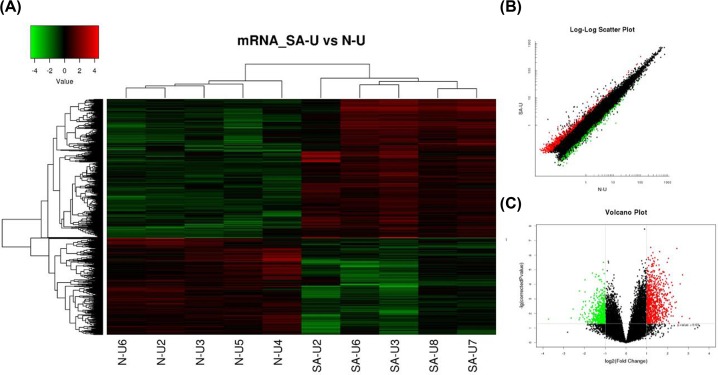
As the function of most of these 2102 differentially expressed lncRNA probes remains unknown, we predicted their potential functions through annotation of their co-expressed mRNAs. Among the 34000 detected mRNA probes, a total of 1349 were found to be significantly differentially expressed between SAP patients and healthy controls (Figure 4A). Of these 1349 probes, 795 were up-regulated and 554 were down-regulated. A partial summary of the distinctively expressed mRNAs in Uyghur SAP patients is presented in Table 3. The scatter and volcano plots generated from these differentially expressed probes are clearly segregated between two groups clusters (Figure 4B,C). This result suggests that these mRNAs were substantially different between the SAP and healthy controls.
Figure 4. Differential expression of mRNAs in SAP patients and control individuals of Uyghur ethnicity.
(A) Expression values are represented through hierarchical clustering analysis in red and green, indicating up- and down-regulated mRNA expressions in SAP patients (SAP-U2, SAP-U3, SAP-U6, SAP-U7, SAP-U8) or control individuals (N-U2, N-U3, N-U4, N-U5, N-U6), respectively. (B) Scatter plot of differential mRNA expression. x-axis: N-U, y-axis: SAP-U. (C) Volcano plot of differential mRNA expression. The red and green spots indicate up- and down-regulation, respectively. x-axis: log2 Fc; y-axis: −1 × log10 (corrected P-value) for each probe.
Table 3. Top 20 differentially expressed mRNAs in SAP patients in Uyghur based on Fc values.
| Probe name | P | FC (abs) | Regulation | Gene symbol | Ensembl ID |
|---|---|---|---|---|---|
| A_21_P0004887 | 0.022326836 | 8.341828283 | Up | lnc-CD2AP-2 | Unknown |
| A_23_P216556 | 2.24801E-05 | 6.589254801 | Up | EPB41L4B | ENST00000374557 |
| A_21_P0003860 | 0.01624716 | 6.498708617 | Up | LOC101928223 | ENST00000504509 |
| A_33_P3251896 | 9.39614E-05 | 6.088366922 | Up | APBB2 | ENST00000543538 |
| A_33_P3280561 | 0.044000295 | 5.807894799 | Up | KRTAP16-1 | ENST00000391352 |
| A_23_P74609 | 0.041502611 | 5.779144021 | Up | G0S2 | ENST00000367029 |
| A_21_P0014105 | 3.50089E-07 | 5.46040203 | Up | LOC101927206 | Unknown |
| A_23_P415541 | 0.001620401 | 5.376366065 | Up | GPR26 | ENST00000284674 |
| A_23_P6596 | 0.004156197 | 5.246830347 | Up | HES1 | ENST00000476918 |
| A_21_P0008258 | 0.001797978 | 5.140095227 | Up | LINC00376 | ENST00000439454 |
| A_33_P3294720 | 0.024651055 | 13.271854 | Down | LOC100130865 | Unknown |
| A_23_P217917 | 0.020856044 | 6.223089456 | Down | GSTM4 | ENST00000336075 |
| A_33_P3238579 | 0.005048665 | 5.830537859 | Down | Unknown | ENST00000611787 |
| A_33_P3424067 | 0.026767724 | 4.778261734 | Down | SUV420H1 | ENST00000615954 |
| A_23_P319874 | 0.000290347 | 4.725299189 | Down | TCAIM | ENST00000396078 |
| A_23_P407565 | 0.020509796 | 4.675297736 | Down | CX3CR1 | ENST00000541347 |
| A_24_P79617 | 0.011647027 | 4.560089019 | Down | KIAA0040 | ENST00000423313 |
| A_24_P930111 | 0.005143622 | 4.327681889 | Down | SLC4A10 | ENST00000446997 |
| A_24_P276490 | 0.013011988 | 4.290633557 | Down | LYPLA2 | ENST00000420982 |
| A_24_P6903 | 0.00363308 | 4.218718187 | Down | ACTBL2 | ENST00000423391 |
Identification of potentially functional mRNAs in SAP patients
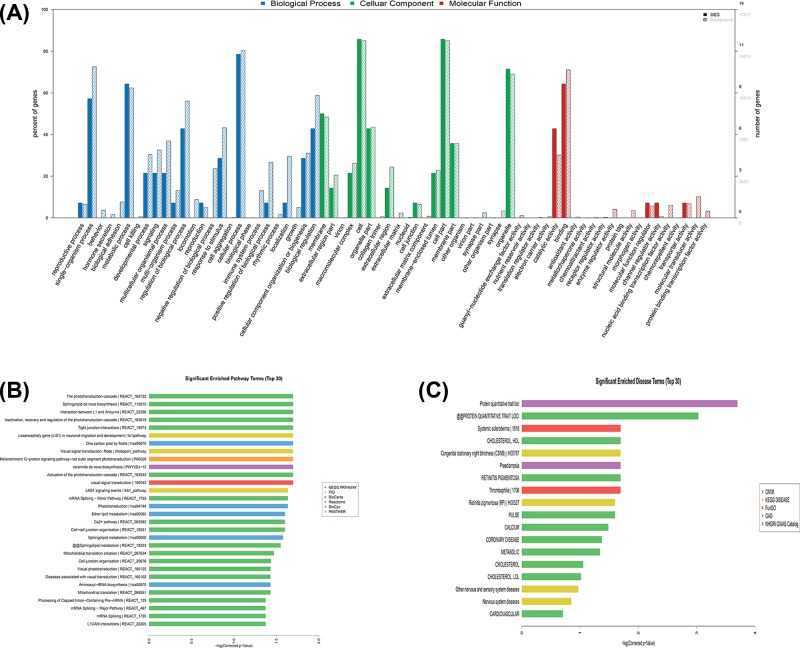
Among the GO terms enriched among the differentially expressed mRNAs in SAP patients, the top ten terms associated with biological processes included: (1) single-organism processes, (2) metabolic processes, (3) regulation of biological processes, (4) responses to stimuli, (5) cellular processes, (6) cellular component organisation or biogenesis, (7) biological regulation, (8) developmental processes, (9) signalling and (10) multicellular organismal processes. The top ten GO terms related to cellular components in the SAP patients included the following: (1) membrane, (2) cell, (3) organelle, (4) membrane-enclosed lumen, (5) organelle part, (6) extracellular region, (7) cell part, (8) extracellular region part, (9) macromolecular complex and (10) membrane part. The top ten GO terms related to molecular functions in SAP patients included: (1) antioxidant activity, (2) enzyme regulator activity, (3) structural molecule activity, (4) morphogen activity, (5) molecular function regulator, (6) channel regulator activity, (7) nucleic acid binding transcription factor activity, (8) transporter activity, (9) molecular transducer activity and (10) protein-binding transcription factor activity (Figure 5A).
Figure 5. GO, KEGG pathways and Disease analyses of differentially expressed mRNAs.
(A) GO analysis of differentially expressed mRNAs. The top 30 significantly enriched GO categories were determined and were plotted as −1 × log10 (P-value). (B) KEGG pathways of differentially expressed mRNAs. The top 30 significantly enriched KEGG categories were determined and were plotted as −1 × log10 (P-value). (C) Disease analysis of differentially expressed mRNAs. The top 30 significantly enriched disease categories and pathways were determined and were plotted as −1 × log10 (P-value).
Based on KEGG analysis, the most enriched pathways corresponding to the dysregulation of mRNAs related to SAP were the phototransduction cascade; inactivation, recovery and regulation of the phototransduction cascade; heterotrimeric G-protein signalling pathway-rod outer segment phototransduction; activation of the phototransduction cascade; and visual phototransduction (Figure 5B). The top ten significantly enriched disease terms included: (1) protein quantitative trait loci, (2) systemic scleroderma, (3) cholesterol and HDL, (4) congenital stationary night blindness, (5) preeclampsia, (6) retinitis pigmentosa, (7) thrombophilia, (8) pulse, (9) calcium and (10) coronary disease (Figure 5C).
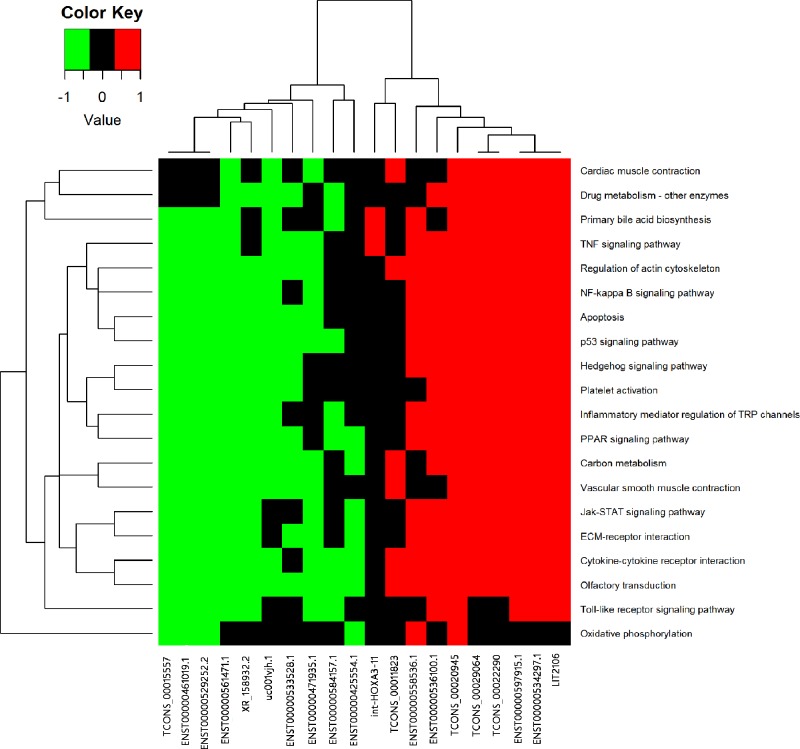
In the fact, the GO terms and KEGG analysis do not refer to lncRNAs directly, so it is necessary to draw the relationship intuitively. In the Figure 6, the hierarchical clustering figure presents a more intuitive tool to see the relationship between the top 20 lncRNAs and pathways through enrichment analysis of the co-expression of mRNA in microarray analysis, which were related to cardiac muscle contraction, drug metabolism-other enzymes, primary bile acid biosynthesis, the TNF signalling pathway, regulation of the actin cytoskeleton, the NF-κB signalling pathway, apoptosis, the p53 signalling pathway, the hedgehog signalling pathway, platelet activation, inflammatory mediator regulation of TRP channels, the PPAR signalling pathway, carbon metabolism, vascular smooth muscle contraction, the Jak-STAT signalling pathway, ECM–receptor interaction, cytokine–cytokine receptor interaction, olfactory transduction, the Toll-like receptor signalling pathway and oxidative phosphorylation (Figure 6).
Figure 6. KEGG annotation for the top 20 lncRNAs via co-expression of their mRNA functions.
Red shows higher expression, green shows lower expression, and the black indicates no relationship.
LncRNA–mRNA network analysis
To determine which lncRNAs and mRNAs play critical roles in SAP progression, we constructed co-expression networks of the differentially expressed correlated lncRNAs and mRNAs. As shown in Figure 7, there were three source lncRNAs (NR_024505.1, ENST00000448425.1, and TCONS_00008970) indicated by yellow cycles) that were correlated with differential expression (down- or up-regulation) of corresponding genes by the criterion that coefficients obtained with no less than 0.99. The details of the source lncRNAs and their corresponding genes are described in Table 4. According to the correlated references and other studies, ROS1 [22] is a protein coding gene which related to transferase activity, transferring phosphorus-containing groups and protein tyrosine kinase activity, and is associated with ERK signalling and Akt signalling pathways. CD276 [23] is related to signalling receptor binding and NF-κB signalling pathway.
Figure 7. LncRNA–mRNA network was constructed to present the correlation analysis between the differentially expressed lncRNAs and mRNAs.
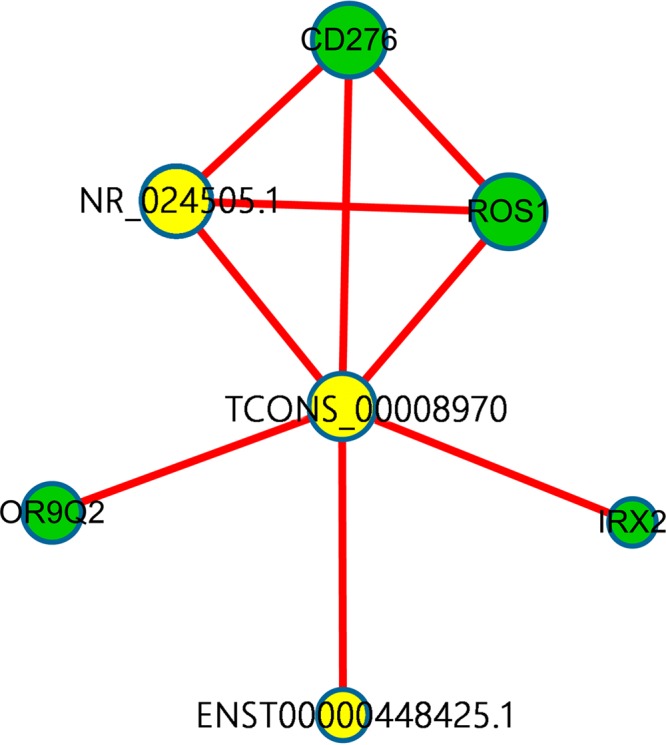
Yellow node represents the lncRNAs and green node represents the target mRNAs. The red outgoing link represents up-regulation, the size of each circle represents the significance of the correlation.
Table 4. LncRNA–mRNA network analysis.
| Source | Target | Correlation | P | Target. Gene symbol |
|---|---|---|---|---|
| NR_024505.1 | A_23_P70278 | 0.993 | 8.252e-09 | ROS1 |
| A_33_P3222917 | 0.995 | 3.9524e-09 | CD276 | |
| ENST00000448425.1 | p22630 | 0.997 | 5.101e-10 | - |
| TCONS_00008970 | A_33_P3222917 | 0.995 | 1.8592e-09 | CD276 |
| A_33_P3297562 | 0.990 | 3.9582e-08 | IRX2 | |
| A_33_P3358779 | 0.990 | 3.9284e-08 | OR9Q2 | |
| A_23_P70278 | 0.992 | 2.1285e-08 | ROS1 | |
| p42653_v4 | 0.995 | 2.7186e-09 | - |
qRT-PCR validation
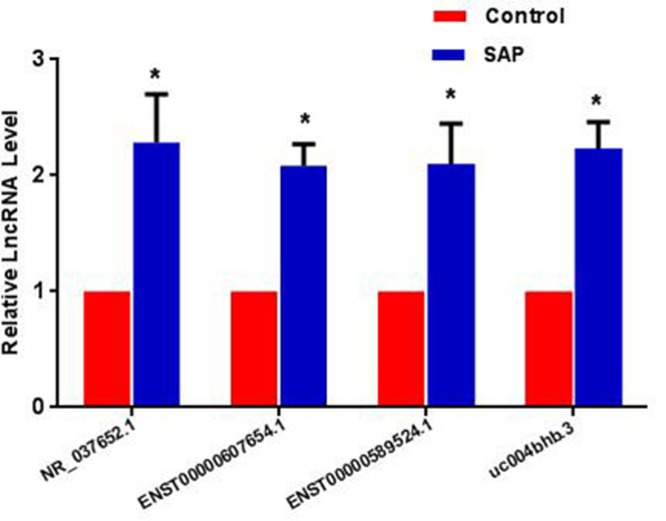
To validate the microarray data, four up-regulated lncRNAs (NR_037652.1, ENST00000607654.1, ENST00000589524.1 and uc004bhb.3) were selected based on the functional co-expression networks, the signal value and the validation of designed primers. A description of the four strictly screened lncRNAs is presented in Table 5. The four lncRNAs were all up-regulated between 50 Uyghur SAP patients and 50 matched controls, which were consistent with the microarray data (Figure 8). The characteristics of the 100 validated participants are shown in Table 6. For an in-depth understanding of the four lncRNAs, the target genes are mainly based on correlation of at least 0.8, not only the possible function (Supplementary Figure S1).
Table 5. The description of the four strictly screened lncRNAs.
| lncRNA ID | P | Fc | Regulation | Probe | Chrom | Start | End | Class |
|---|---|---|---|---|---|---|---|---|
| NR_037652.1 | 0.018 | 2.286 | Up | p33678 | 15 | 80191181 | 80216096 | Antisense |
| ENST00000607654.1 | 0.015 | 2.083 | Up | p37346 | 2 | 220550102 | 220552067 | Unkown |
| ENST00000589524.1 | 0.000 | 2.101 | Up | p8706 | 19 | 36440385 | 36442421 | Intergenic |
| uc004bhb.3 | 0.001 | 2.124 | Up | p26507 | 9 | 116075501 | 116077893 | Divergent |
Figure 8. Validation data of the four strictly screened up-regulated lncRNAs by qRT-PCR in 50 SAP patients and 50 controls.
*P<0.05 compared with controls.
Table 6. Clinical characteristics of 50 Uyghur SAP patients and 50 Uyghur controls.
| Characteristics | Controls (n=50) | SAP patients (n=50) |
|---|---|---|
| Age, years | 52.1 ± 6.14 | 51.6 ± 5.83 |
| Gender (male/female) | 38/12 | 38/12 |
| BMI | 24.7 ± 3.02 | 25.4 ± 3.26 |
| Hypertension (Yes/No) | 3/47* | 10/40* |
| Diabetes (Yes/No) | 7/43 | 11/39 |
| Arrhythmia (Yes/No) | 2/48 | 5/45 |
| Medication (Yes/No) | 2/48 | 8/42 |
| Smoking (Yes/No) | 8/42* | 20/30* |
| Drinking (Yes/No) | 3/47 | 6/44 |
| SBP, mmHg | 123.6 ± 14.31 | 116.9 ± 20.35 |
| DBP, mmHg | 83 ± 9.75 | 79.6 ± 16.19 |
| Heart rate, beats per minute | 71 ± 7.14 | 81 ± 9.51 |
| Serum creatinine, μmol/l | 80.4 ± 10.41 | 77.44 ± 17.05 |
| FPG, mmol/l | 4.94 ± 0.26 | 5.36 ± 1.17 |
| TG, mmol/l | 1.79 ± 0.5* | 2.74 ± 2.59* |
| TC, mmol/l | 4.86 ± 0.82 | 4.17 ± 0.69 |
| LDL, mmol/l | 3.49 ± 0.91 | 2.97 ± 0.58 |
| Serum calcium, mmol/l | 2.24 ± 0.07 | 1.94 ± 0.68 |
The data are presented as the mean ± SD. Abbreviations: BMI, body mass index; DBP, diastolic blood pressure; FPG, fasting plasma glucose; LDL, low-density lipoprotein; SBP, systolic blood pressure; TC, total cholesterol; TG, triglyceride.
*P<0.05 compared with controls.
Discussion
Cardiovascular disease (CVD) is the leading cause of death and disability worldwide, and SAP is the most common form of heart disease. Patients may have chest pain or breathlessness on exertion, but not at rest. It is important to reduce the occurrence of SAP, due to such reductions that have been shown to reduce heart attack and death rates [24]. Meanwhile, it is critical to detect SAP early and select appropriate treatments. Notably, recent genetic studies have demonstrated that the genetic component plays an important role in the development of CVD.
LncRNAs have been found to be an important genetic component of the genome regulatory network and to play important roles in disease processes [25]. Many efforts have been made to identify diagnostic and prognostic lncRNA biomarkers in various human cancers [26–28]. To date, there has been a lack of investigation into the utility of lncRNAs as biomarkers for the early diagnosis of SAP. Several profile-based studies have identified altered lncRNA expression during the initiation and progression of MI. For example, Zangrando et al. [29] used microarray analysis on MI mice to investigate the roles of differentially expressed lncRNAs in left ventricular remodelling, and Qu et al. [30] identified 545 deregulated lncRNAs involved in cardiac fibrogenesis induced by MI using microarray analysis. Furthermore, two other studies constructed dysregulated lncRNA–mRNA co-expression networks to investigate the functional roles of lncRNAs in MI and identified some key lncRNA candidates [31,32], emphasising the potential of lncRNAs to be used as biomarkers for the early diagnosis of MI.
Following the exploration in expression pattern of lncRNAs in MI [8], we investigated the genome-wide expression profile of lncRNAs in Uyghur ethnicity SAP in Xinjiang. A total of 1871 up- and 231 down-regulated lncRNAs were identified to be significantly and differentially expressed in SAP patients, accordingly in mRNAs level, 795 mRNAs were up-regulated and 554 mRNAs were down-regulated when compared with the healthy matched controls. To predict the potential functions of the lncRNAs, their co-expressed mRNAs were subjected to GO annotation and KEGG analysis. The most enriched GO annotation corresponding to the dysregulation of mRNAs related to SAP was the regulation of antioxidant activity. The significantly enriched disease terms included cholesterol (HDL), thrombophilia, pulse, calcium, coronary disease and metabolism. Among the top 20 aberrantly expressed lncRNAs, the annotation results of their co-expressed mRNAs showed that the most significantly related pathways were the NF-κB signalling pathway, apoptosis and the p53 signalling pathway. According to previous study, it has been proved that NF-κB signalling pathway [33,34] and p53 signalling pathway [34,35] serve not only an essential role in CHD, but also a significant effect on inflammatory response, oxidative stress and apoptosis. In order to reveal the co-expressions, we specifically picked out three lncRNAs (NR_024505.1, ENST00000448425.1 and TCONS_00008970), mainly due to the potential function of their co-expressed mRNAs (ROS1, CD276, OR9Q2 and IRX2). So far, several researches [36,37] have presented that ROS1 is correlated with CVDs, especially MI and sudden cardiac death. Anzalone et al. [38] has proved that CD276 is associated with heart failure, and IRX2 [39] undertakes an essential role in the development of heart. Importantly, previous studies [22,23] have revealed that the potential mechanisms of the mRNAs are related to NF-κB signalling pathway, ERK signalling pathway and Akt signalling pathways in cancers. These findings suggest that coordinated patterns of lncRNAs and their co-expressed mRNAs might be involved in the development of SAP. We speculated that these mRNAs may participate in the occurrence and development of SAP in the Uyghur population by regulating the three selected lncRNAs in the present study. However, this hypothesis needs to be further investigated.
Some lncRNAs have been reported to be biomarkers for the diagnosis of CVD; for example the lncRNA PCA3 has been reported to be a biomarker for severe left ventricular remodelling after MI [40], the circulating levels of lincRNA-P21 [41] are markedly increased in atherosclerosis and may be important in its pathogenesis, and the lncRNA OTTHUMT00000387022 [42] has been reported as a biomarker in coronary artery disease. However, these previous studies did not involve different ethnicities. Compared with these reports, our previous research [8] aimed to identify novel biological biomarkers which may have a potential in better management and stratification of Uyghur AMI patients. The result not only presented the expression profile of lncRNA using the same microarray, but also identified that three non-reported novel lncRNAs (ENST00000416860.2, ENST00000421157.1 and TCONS_00025701) were decreased in AMI patients, which had served as potential biomarkers. In order to illustrate whether lncRNAs can provide new biomarkers for Uyghur SAP patients, we have searched some lncRNAs as follows: firstly, we explored the primary data, and selected the functionally related mRNA through GO, KEGG and Disease enrichment (the main function of CVDs, the level of lipid and inflammation pathway). Secondly, we focussed on the co-expression network and chose the closely related lncRNAs. Finally, we selected according to the signal value and primers which must be designed and validated successfully. As a result, we singled out the four lncRNAs (NR_037652.1, ENST00000607654.1, ENST00000589524.1 and uc004bhb.3), and found that the levels of the four lncRNAs were elevated in Uyghur SAP patients. These results indicate that these differentially expressed lncRNAs may be potential biomarkers for the diagnosis of SAP in the Uyghur population. Our results provide a supplementary data in this field.
Some limitations of our study should be acknowledged. First, the present study used relatively few lncRNA probes compared with the number of known lncRNAs in some databases because the lncRNA expression profiles were obtained based on the Agilent Human lncRNA + mRNA Array V4.0. Second, the patients in the present study were from one hospital, not a multi-centre and larger scale study. Further verification from different areas and different races should be carried out to study the functional roles of these candidate lncRNA biomarkers in SAP. Finally, we forecast the function of lncRNA through high-throughput microarray and complex bioinformatics analysis. In the follow-up stage, we need to investigate the biological significances in model systems or cell lines which may contribute to understanding of the pathological mechanism of SAP.
In conclusion, our study examines the expression profile of lncRNA and mRNA in PBMCs from Uyghur SAP patients in comparison with matched controls using microarray. The results provide previously unreported bioinformation on Uyghur SAP patients, based on genomic-wide lncRNA expression and corresponding mRNA expression. These findings, on the one hand, provide useful bioinformation which may have a potential role in the development of SAP; on the other hand, emphasised the potential of lncRNAs to be used as biomarkers for the early diagnosis of SAP.
Supporting information
Supplementary Figure S1. (A) LncRNA-mRNA network was constructed between the NR_037652.1 and mRNAs. (B) LncRNA-mRNA network was constructed between the ENST00000607654.1 and mRNAs. (C) LncRNA-mRNA network was constructed between the ENST00000589524.1 and mRNAs. (D) LncRNA-mRNA network was constructed between the uc004bhb.3 and mRNAs.
Supplementary Material.
Abbreviations
- AMI
acute myocardial infarction
- CHD
coronary heart disease
- CVD
cardiovascular disease
- Fc
fold-change
- GO
Gene Ontology
- HDL
high density lipoprotein
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LDL
low density lipoprotein
- lncRNA
long non-coding RNA
- PBMC
peripheral blood mononuclear cell
- qRT-PCR
quantitative real-time polymerase chain reaction
- SAP
stable angina pectoris
- SSC
saline sodium citrate
Ethical approval
The present study was approved by the Research Ethics Committee of Xinjiang Medical University. All participants from the First Affiliated Hospital of Xinjiang Medical University were enrolled between December 2014 and September 2015. All patients involved in the present study received oral and written information regarding the objectives of the study and provided written informed consent.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Author Contribution
Conceived and designed the experiments: Yi-Ning Yang and Xiao-Mei Li. Performed the experiments: Xin-Rong Zhou and Ning Song. Analysed the data: Jun-Yi Luo and Hui Zhai. Contributed reagents/materials/analysis tools: Xiang-Mei Li and Qian Zhao. Quality control the study and revision: Fen Liu and Ning Song. Wrote the paper: Xin-Rong Zhou.
Funding
This work was supported by the National Natural Science Foundation of China [grant numbers 81660058, 81770363, U1503322]; the Special Funds for the Key Laboratory of Xinjiang Autonomous Region in China [grant number 2018D04029]; the Graduate Research Innovation Project of Xinjiang Autonomous region [grant number CXCY2018026]; and the Major Disease Medical Key Laboratory Open Subject of Xinjiang in China [grant number SKLIB-XJMDR-2016-Y4].
References
- 1.Liu B., Du Y., Cong L., Jia X. and Yang G. (2016) Danshen (Salvia miltiorrhiza) compounds improve the biochemical indices of the patients with coronary heart disease. Evid. Based Complement. Alternat. Med. 2016, 9781715 10.1155/2016/9781715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fihn S.D., Gardin J.M., Abrams J., Berra K., Blankenship J.C., Dallas A.P.. et al. (2012) 2012 ACCF/AHA/ACP/AATS/PCNA/SCAI/STS Guideline for the diagnosis and management of patients with stable ischemic heart disease: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines, and the American College of Physicians, American Association for Thoracic Surgery, Preventive Cardiovascular Nurses Association, Society for Cardiovascular Angiography and Interventions, and Society of Thoracic Surgeons. J. Am. Coll. Cardiol. 60, e44–e164 10.1016/j.jacc.2012.07.013 [DOI] [PubMed] [Google Scholar]
- 3.Pepine C.J., Handberg E.M., Cooper-DeHoff R.M., Marks R.G., Kowey P., Messerli F.H.. et al. (2003) A calcium antagonist vs a non-calcium antagonist hypertension treatment strategy for patients with coronary artery disease. The International Verapamil-Trandolapril Study (INVEST): a randomized controlled trial. JAMA 290, 2805–2816 [DOI] [PubMed] [Google Scholar]
- 4.Dargie H.J., Ford I. and Fox K.M. (1996) Total Ischaemic Burden European Trial (TIBET). Effects of ischaemia and treatment with atenolol, nifedipine SR and their combination on outcome in patients with chronic stable angina. The TIBET Study Group. Eur. Heart J. 17, 104–112 [DOI] [PubMed] [Google Scholar]
- 5.Ren Y., Li D., Zheng H., Lv J., Leng J., Zhang L.. et al. (2014) Acupoint application in patients with chronic stable angina pectoris: study protocol of a randomized, double-blind, controlled trial. Evid. Based Complement. Alternat. Med. 2014, 619706 10.1155/2014/619706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wee Y., Burns K. and Bett N. (2015) Medical management of chronic stable angina. Aust. Prescr. 38, 131–136 10.18773/austprescr.2015.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li L., Wang L., Li H., Han X., Chen S., Yang B.. et al. (2018) Characterization of LncRNA expression profile and identification of novel LncRNA biomarkers to diagnose coronary artery disease. Atherosclerosis 275, 359–367 10.1016/j.atherosclerosis.2018.06.866 [DOI] [PubMed] [Google Scholar]
- 8.Zhai H., Li X.M., Liu F., Chen B.D., Zheng H., Wang X.M.. et al. (2017) Expression pattern of genome-scale long noncoding RNA following acute myocardial infarction in Chinese Uyghur patients. Oncotarget 8, 31449–31464 10.18632/oncotarget.16355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ponting C.P. and Belgard T.G. (2010) Transcribed dark matter: meaning or myth? Hum. Mol. Genet. 19, R162–R168 10.1093/hmg/ddq362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prensner J.R. and Chinnaiyan A.M. (2011) The emergence of lncRNAs in cancer biology. Cancer Discov. 1, 391–407 10.1158/2159-8290.CD-11-0209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uchida S. and Dimmeler S. (2015) Long noncoding RNAs in cardiovascular diseases. Circ. Res. 116, 737–750 10.1161/CIRCRESAHA.116.302521 [DOI] [PubMed] [Google Scholar]
- 12.Boon R.A., Jae N., Holdt L. and Dimmeler S. (2016) Long noncoding RNAs: from clinical genetics to therapeutic targets? J. Am. Coll. Cardiol. 67, 1214–1226 10.1016/j.jacc.2015.12.051 [DOI] [PubMed] [Google Scholar]
- 13.Holdt L.M., Beutner F., Scholz M., Gielen S., Gabel G., Bergert H.. et al. (2010) ANRIL expression is associated with atherosclerosis risk at chromosome 9p21. Arteriosclerosis Thromb. Vasc. Biol. 30, 620–627 10.1161/ATVBAHA.109.196832 [DOI] [PubMed] [Google Scholar]
- 14.Vausort M., Wagner D.R. and Devaux Y. (2014) Long noncoding RNAs in patients with acute myocardial infarction. Circ. Res. 115, 668–677 10.1161/CIRCRESAHA.115.303836 [DOI] [PubMed] [Google Scholar]
- 15.Kumarswamy R., Bauters C., Volkmann I., Maury F., Fetisch J., Holzmann A.. et al. (2014) Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ. Res. 114, 1569–1575 10.1161/CIRCRESAHA.114.303915 [DOI] [PubMed] [Google Scholar]
- 16.Yan Y.Z., Ma R.L., Ding Y.S., Guo H., Zhang J.Y., Mu L.T.. et al. (2015) Association of inflammation with metabolic syndrome among low-income rural Kazakh and Uyghur adults in far Western China. Mediators Inflamm. 2015, 706768 10.1155/2015/706768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H., Li H., Yue H., Wang W., Yu L., Wang S.. et al. (2017) Comparison between smaller ruptured intracranial aneurysm and larger un-ruptured intracranial aneurysm: gene expression profile analysis. Neurosurg. Rev. 40, 419–425 10.1007/s10143-016-0799-3 [DOI] [PubMed] [Google Scholar]
- 18.Huang da W., Sherman B.T. and Lempicki R.A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 19.Rinn J.L. and Chang H.Y. (2012) Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166 10.1146/annurev-biochem-051410-092902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smoot M.E., Ono K., Ruscheinski J., Wang P.L. and Ideker T. (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27, 431–432 10.1093/bioinformatics/btq675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barabasi A.L. and Oltvai Z.N. (2004) Network biology: understanding the cell’s functional organization. Nat. Rev. Genet. 5, 101–113 10.1038/nrg1272 [DOI] [PubMed] [Google Scholar]
- 22.Yasuda H., de Figueiredo-Pontes L.L., Kobayashi S. and Costa D.B. (2012) Preclinical rationale for use of the clinically available multitargeted tyrosine kinase inhibitor crizotinib in ROS1-translocated lung cancer. J. Thorac. Oncol. 7, 1086–1090 10.1097/JTO.0b013e3182570919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen X., Meng X., Foley N.M., Shi X., Liu M., Chai Y.. et al. (2017) Activation of the TLR2-mediated downstream signaling pathways NF-kappaB and MAPK is responsible for B7-H3-augmented inflammatory response during S. pneumoniae infection. J. Neuroimmunol. 310, 82–90 10.1016/j.jneuroim.2017.07.002 [DOI] [PubMed] [Google Scholar]
- 24.Chwojnicki K., Wierucki L., Zagozdzon P., Wojtyniak B., Nyka W.M. and Zdrojewski T. (2016) Long-term mortality after stroke is higher than after myocardial infarction. Neurol. Sci. 37, 891–898 10.1007/s10072-016-2502-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X., Wu Z., Fu X. and Han W. (2014) lncRNAs: insights into their function and mechanics in underlying disorders. Mutat. Res. Rev. Mutat. Res. 762, 1–21 10.1016/j.mrrev.2014.04.002 [DOI] [PubMed] [Google Scholar]
- 26.Zhou M., Wang X., Shi H., Cheng L., Wang Z., Zhao H.. et al. (2016) Characterization of long non-coding RNA-associated ceRNA network to reveal potential prognostic lncRNA biomarkers in human ovarian cancer. Oncotarget 7, 12598–12611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou M., Zhang Z., Zhao H., Bao S., Cheng L. and Sun J. (2018) An immune-related six-lncRNA signature to improve prognosis prediction of glioblastoma multiforme. Mol. Neurobiol. 55, 3684–3697 [DOI] [PubMed] [Google Scholar]
- 28.Li J., Chen Z., Tian L., Zhou C., He M.Y., Gao Y.. et al. (2014) LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut 63, 1700–1710 10.1136/gutjnl-2013-305806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zangrando J., Zhang L., Vausort M., Maskali F., Marie P.Y., Wagner D.R.. et al. (2014) Identification of candidate long non-coding RNAs in response to myocardial infarction. BMC Genomics 15, 460 10.1186/1471-2164-15-460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qu X., Song X., Yuan W., Shu Y., Wang Y., Zhao X.. et al. (2016) Expression signature of lncRNAs and their potential roles in cardiac fibrosis of post-infarct mice. Biosci. Rep. 36, e00337 10.1042/BSR20150278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun C., Jiang H., Sun Z., Gui Y. and Xia H. (2016) Identification of long non-coding RNAs biomarkers for early diagnosis of myocardial infarction from the dysregulated coding-non-coding co-expression network. Oncotarget 7, 73541–73551 10.18632/oncotarget.11999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang P., Fu H., Cui J. and Chen X. (2016) Differential lncRNAmRNA coexpression network analysis revealing the potential regulatory roles of lncRNAs in myocardial infarction. Mol. Med. Rep. 13, 1195–1203 10.3892/mmr.2015.4669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X., Zhou J. and Huang K. (2017) Inhibition of the lncRNA Mirt1 attenuates acute myocardial infarction by suppressing NF-kappaB activation. Cell. Physiol. Biochem. 42, 1153–1164 10.1159/000478870 [DOI] [PubMed] [Google Scholar]
- 34.Chatterjee A., Mir S.A., Dutta D., Mitra A., Pathak K. and Sarkar S. (2011) Analysis of p53 and NF-kappaB signaling in modulating the cardiomyocyte fate during hypertrophy. J. Cell. Physiol. 226, 2543–2554 10.1002/jcp.22599 [DOI] [PubMed] [Google Scholar]
- 35.Ma H., Wang X., Ha T., Gao M., Liu L., Wang R.. et al. (2016) MicroRNA-125b prevents cardiac dysfunction in polymicrobial sepsis by targeting TRAF6-mediated nuclear factor kappaB activation and p53-mediated apoptotic signaling. J. Infect. Dis. 214, 1773–1783 10.1093/infdis/jiw449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horne B.D., Carlquist J.F., Muhlestein J.B., Nicholas Z.P. and Anderson J.L. (2007) Associations with myocardial infarction of six polymorphisms selected from a three-stage genome-wide association study. Am. Heart J. 154, 969–975 10.1016/j.ahj.2007.06.032 [DOI] [PubMed] [Google Scholar]
- 37.Ivanova A.A., Maksimov V.N., Orlov P.S., Ivanoshchuk D.E., Savchenko S.V., Voevoda M.I.. et al. (2016) Association of the genetic markers for myocardial infarction with sudden cardiac death. Indian Heart J. 69, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anzalone R., Corrao S., Lo I.M., Loria T., Corsello T., Cappello F.. et al. (2013) Isolation and characterization of CD276+/HLA-E+ human subendocardial mesenchymal stem cells from chronic heart failure patients: analysis of differentiative potential and immunomodulatory markers expression. Stem Cells Dev. 22, 1. [DOI] [PubMed] [Google Scholar]
- 39.Christoffels V.M., Keijser A.G.M., Houweling A.C., Clout D.E.W. and Moorman A.F.M. (2000) Patterning the embryonic heart: identification of five mouse iroquois homeobox genes in the developing heart. Dev. Biol. 224, –274 10.1006/dbio.2000.9801 [DOI] [PubMed] [Google Scholar]
- 40.Ren S., Wang F., Shen J., Sun Y., Xu W., Lu J.. et al. (2013) Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur. J. Cancer 49, 2949–2959 10.1016/j.ejca.2013.04.026 [DOI] [PubMed] [Google Scholar]
- 41.Tang S.S., Cheng J., Cai M.Y., Yang X.L., Liu X.G., Zheng B.Y.. et al. (2016) Association of lincRNA-p21 haplotype with coronary artery disease in a Chinese Han population. Dis. Markers 2016, 9109743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cai Y., Yang Y., Chen X., Wu G., Zhang X., Liu Y.. et al. (2016) Circulating ‘lncRNA OTTHUMT00000387022’ from monocytes as a novel biomarker for coronary artery disease. Cardiovasc. Res. 112, 714–724 10.1093/cvr/cvw022 [DOI] [PubMed] [Google Scholar]