Abstract
Phlebotomine sand flies are tiny, hairy, blood-sucking nematoceran insects that feed on a wide range of hosts. They are known as a principal vector of parasites, responsible for human and animal leishmaniasis worldwide. In Thailand, human autochthonous leishmaniasis and trypanosomiasis have been reported. However, information on the vectors for Leishmania and Trypanosoma in the country is still limited. Therefore, this study aims to detect Leishmania and Trypanosoma DNA in field-caught sand flies from endemic areas (Songkhla and Phatthalung Provinces) and non-endemic area (Chumphon Province) of leishmaniasis. A total of 439 sand flies (220 females and 219 males) were collected. Head and genitalia dissection of female sandflies were done for morphology identification, and the remaining parts of those sand flies were then used for the detection of Leishmania and Trypanosoma parasites. The DNA was extracted from individual female sand flies. Polymerase chain reaction (PCR) anneal, specific to the ITS1 and SSU rRNA gene regions, was used to detect Leishmania and Trypanosoma DNA, respectively. The positive PCR products were cloned and sequenced. The results showed that the female sand fly species in this study consisted of Sergentomyia khawi (35.9%); Se. anodontis (23.6%); Phlebotomus betisi (18.6%); Ph. kiangsuensis (9.5%); Ph. asperulus (6.4%); Se. barraudi (2.3%); 0.9% of each Se. indica, Ph. stantoni, and Ph. major major; and 0.5% of each Se. sylvatica and Ph. mascomai. The PCR and sequence analysis were able to detect Leishmania and Trypanosoma DNA in sand fly samples, which were identified as L. martiniquensis, 1/220 (0.45%) in Se. khawi, 3/220 (1.36%) of T. noyesi in Se. anodontis, and Ph. asperulus. Fourteen (6.36%) of the unidentified trypanosome species in Se. khawi, Se. indica, Se. anodontis, Ph. asperulus, and Ph. betisi were found in all of the areas of this study. Interestingly, we found a 1/220 (0.45%) co-infection sample of L. martiniquensis and Trypanosoma in Se. khawi from Songkhla Province. These data indicate that several species of sand flies might be potential vectors of Leishmania and Trypanosoma parasites in southern Thailand. However, more extensive study for potential vectors using a larger number of sand flies should be conducted to prove whether these sand flies can be natural vectors of leishmaniasis and trypanosomiasis in both humans and animals. In addition, our study could be useful for the future study of infection prevention, including effective vector control for leishmaniasis and trypanosomiasis in Thailand.
Keywords: Leishmania spp., Trypanosoma sp., sand flies, vector, Thailand
1. Introduction
Phlebotomine sand flies are small dipteran, hairy, and blood-sucking insects belonging to the order Diptera and family Psychodidae [1], which are known as important medical vectors of leishmaniasis worldwide. Leishmaniasis is a neglected tropical disease caused by the flagellate protozoa of the genus Leishmania [2]. Moreover, Phlebotomine sand flies have been reported to be the potential vector of the Trypanosoma species, which can cause trypanosomiasis in mammals, birds, fish, rats, sheep, cows, rabbits, lizards, frogs, and snakes [3,4,5,6,7]. Both parasite genera have been reported in many areas of the world. In Thailand, Leishmania orientalis [8], L. martiniquensis [9], and L. donovani complex [10,11] have been identified as the causative agents of indigenous visceral and cutaneous leishmaniasis. There have been approximately 20 autochthonous leishmaniasis reported in the country from 1996 to the present [12]. The disease is endemic mostly in southern Thailand. Trypanosomiasis is commonly found in animals, for example, T. lewisi and T. evansi. These animals are the major pathogenic trypanosome of domestic animals, such as rodents, cattle, wild animals, elephants, and tigers [13]. Furthermore, T. lewisi [14] and T. lewisi-like infection [15] have been reported in an infant in Thailand. It has been almost 20 years since the first case of autochthonous leishmaniasis was reported in Thailand [10], but data of the sand fly vector for the infectious diseases are still limited. In Thailand, several reports of sand flies in the western, central, northern, southern, and northeastern regions have identified four genera of sand flies, as follows: Sergentomyia, Phlebotomus, Idiophlebotomus, and Chinius [16,17,18,19,20]. The genus Sergentomyia was mostly demonstrated in the Phangnga, Suratthani, and Nakonsitammarat Provinces of southern Thailand, which are considered to be the affected areas of leishmaniasis. Sand fly species, such as Sergentomyia gemmea (81.4%), Se. iyengari, Se. barraudi, Se. indica, Se. sylvatica, and Se. perturbans, as well as another genus, Phlebotomus (Ph. argentipes), are found in the aforementioned affected areas in the country [21]. Moreover, Se. (Neophlebotomus) gemmea collected from an endemic area of leishmaniasis in Thailand were detected for L. martiniquensis DNA; therefore, Se. gemmea was claimed to be a potential vector of L. martiniquensis [22]. Recently, Phumee et al. (2016) reported an unknown Trypanosoma sp. DNA detected in a Ph. stantoni from Thailand [23]. However, the information on other sand fly species, which may transmit leishmaniasis and trypanosomiasis, is also limited. In this study, we focused on the detection of Leishmania and Trypanosoma parasites in sand flies collected from endemic and non-endemic areas of leishmaniasis in southern Thailand. The information from this study might help us to gain a better understanding about the prevalence of parasites in the sand fly population, and about the association among the sand fly species as a potential vector in endemic areas of leishmaniasis in Thailand.
2. Materials and Methods
2.1. Ethics Statement
The study was approved by the animal research ethics committee of Chulalongkorn University Animal Care and Use Protocol (CU-ACUP), Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand (COA No. 019/2561).
2.2. Sand Fly Collection, Sample Preparation, and Identification
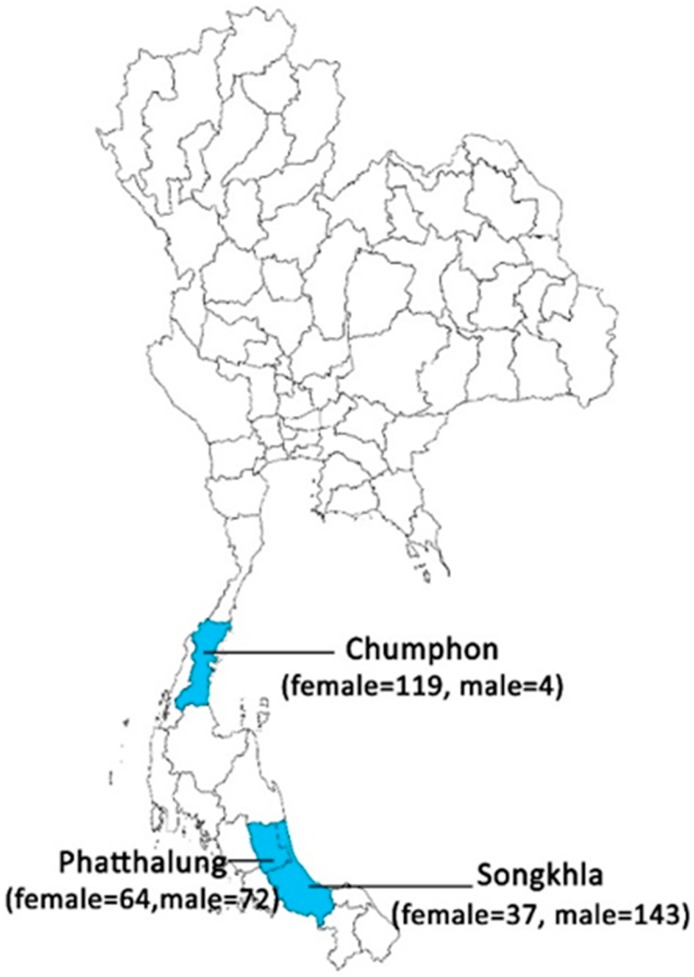
Sand flies were collected from endemic areas, around human settlements (Songkhla and Phatthalung Provinces), and non-endemic areas, in caves (Chumphon Provinces), of leishmaniasis in southern Thailand (Figure 1). The sand fly specimens were collected by Centers for Disease Control and Prevention (CDC) miniature light traps (25 W bulb) without CO2, during September–October 2017. The 10 traps operated for over 12 h, from 18:00 to 06:00, for one night. The collecting bags were kept at −20 °C for 30 min in order to anesthetize the arthropods and separate the sand flies from the other arthropods by morphological features using a stereomicroscope (Olympus, Tokyo, Japan). Each female sand fly was dissected under a stereomicroscope on a sterilized slide using sterile needles. The head and genitalia were mounted on slides with Hoyer’s medium for morphological character identification in all female sand flies, under a light microscope (Olympus, Tokyo, Japan), following the taxonomic keys described by Lewis (1978) [24], Phumee et al. (2016) [23], and Depaquit et al. (2019) [25]. The abdomen and the thorax of each female specimen were transferred to sterile 1.5 mL Eppendorf tubes with a lysis buffer for the detection of Leishmania and Trypanosoma species by the polymerase chain reaction (PCR).
Figure 1.
Map of Thailand showing locations of the sand fly sample-collection sites in the three provinces. Blue denotes the collection locations of sand flies.
2.3. DNA Extraction
DNA was extracted from individual sand fly samples using an Invisorb Spin Tissue Mini Kit (STRATEC Molecular, Berlin, Germany), following the manufacturer’s instructions. The sand flies were lysed in a 200 µL lysis buffer containing 20 µL of proteinase K. At the final step, DNA was eluted in a 40 µL of elution buffer. The extracted DNA samples were kept at −80 °C for long-term storage.
2.4. Detection of Leishmania and Trypanosoma DNA in the Sand Fly Samples
The extracted DNA samples were used to detect the Leishmania DNA using polymerase chain reaction (PCR) amplification annealed specifically to the ITS1 region. The reactions were performed using primers LeR: 5′-CCA-AGT-CAT-CCA-TCG-CGA-CAC-G-3′ and LeF: 5′-TCC-GCC-CGA-AAG-TTC-ACC-GAT-A-3′, under the conditions previously described by Spanakos et al. (2008) [26]. The reaction was carried out in a total volume of 25 μL, containing 2.5 μL of 10× PCR Buffer (NH4SO4), 1.25 μL of 25 mM MgCl2, 2.5 μL of 2.5 mM of each dNTP, 10 mM of each primer, 1 unit of Taq DNA polymerase, and 50 ng of DNA template. After initial denaturation at 95 °C for 5 min, PCR was performed, with 40 cycles consisting of denaturation (95 °C for 1 min), annealing (50 °C for 1 min), extension (72 °C for 1 min), and a final extension at 72 °C for 7 min. The PCR products were analyzed by electrophoresis on a 1.5% agarose gel for 40 min at 100 volts, stained with ethidium bromide, and visualized with Quantity One Quantification Analysis Software Version 4.5.2 (Gel DocEQ System; Bio-Rad, Hercules, CA, USA). For the identification of the Trypanosoma species, the small subunit ribosomal RNA (SSU rRNA) region was amplified from the sand fly using primers designed specifically for trypanosomatids. The sequences primers were TRY927F: 5′-GAA-ACA-AGA-AAC-ACG-GGA-G-3′ and TRY927R: 5′-CTA-CTG-GGC-AGC-TTG-GA-3′ [27]. The reaction and conditions followed the amplification of the ITS1 region described previously.
2.5. Molecular Cloning and Nucleotide Sequencing
The amplified PCR products were cloned into the pGEM-T Easy Vector (Promega, Madison, WI, USA). The ligated vectors were transformed into E. coli DH5∝ competent cells. The recombinant plasmids were screened using the blue-white screening systems. Suspected chimeric colonies were cultured and subjected to plasmid DNA extraction by the Invisorb Spin Plasmid Mini Kit (STRATEC Molecular, Berlin, Germany), following the manufacturer’s instructions. The purified plasmids were sequenced by the sequencing service of Macrogen Inc., Seoul, Korea, using a universal forward T7 primer. The nucleotide sequences were analyzed by comparison with the GenBank database using a BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The sequences were aligned using BioEdit Sequence Alignment Editor Version 7.0.5.3 [28]. The phylogenetic tree was constructed using the maximum likelihood method with Kimura’s two-parameter and bootstrap analysis with 1000 replications in MEGAX version 10.0.1 [29].
3. Results
The number of sand flies collected from all of the sites (Songkhla, Phatthalung, and Chumphon Provinces) using a CDC light trap was 439 (220 females and 219 males). For the morphological characteristics, the female sand flies (n = 220) were classified into two genera and 11 species, which were from the Songkhla (Se. anodontis, Se. khawi, Se. barraudi, and Ph. stantoni), Phatthalung (Se. khawi, Se. barraudi, Se. indica, Ph. betisi, and Ph. kiangsuensis), and Chumphon (Se. anodontis, Se. sylvatica, Ph. asperulus, Ph. betisi, Ph. kiangsuensis, Ph. major major, and Ph. mascomai) provinces. The most abundant species in our samples was Se. khawi (35.9%; Table 1). All of the samples of female sand flies were screened for Leishmania and Trypanosoma infection using ITS1-PCR and SSU rRNA-PCR, respectively.
Table 1.
Morphological identification of female sand flies from southern Thailand.
| Species | Provinces | Total | ||
|---|---|---|---|---|
| Songkhla | Phatthalung | Chumphon | ||
| Se. anodontis | 4 | 0 | 48 | 52 |
| Se. khawi | 29 | 50 | 0 | 79 |
| Se. sylvatica | 0 | 0 | 1 | 1 |
| Se. barraudi | 2 | 3 | 0 | 5 |
| Se. indica | 0 | 2 | 0 | 2 |
| Ph. stantoni | 2 | 0 | 0 | 2 |
| Ph. asperulus | 0 | 0 | 14 | 14 |
| Ph. betisi | 0 | 3 | 38 | 41 |
| Ph. kiangsuensis | 0 | 6 | 15 | 21 |
| Ph. major major | 0 | 0 | 2 | 2 |
| Ph. mascomai | 0 | 0 | 1 | 1 |
| Total | 37 | 64 | 119 | 220 |
In the endemic area of leishmaniasis, Songkhla province, one out of thirty-seven female Se. khawi DNA extracts was positive, by PCR, for L. martiniquensis DNA, using ITS1-PCR (Figure S1), and one sample showed co-infection between L. martiniquensis and Trypanosoma parasites (Table 2). An analysis of the L. martiniquensis amplified sequences showed 99% compatibility with L. martiniquensis (accession no. JQ001751; Figure S2). Moreover, PCR targeting the SSU rRNA region of the four positive samples showed 99% identity with Trypanosoma sp. (accession no. AB520638; Figure S3). The female sand flies collected from Phatthalung were 50 sand flies of Se. khawi and two sand flies of Se. indica. One positive sample showed a 99% similarity with Trypanosoma sp. (accession no. AB520638) in each species, whereas Leishmania DNA was not detected in this area (Figure S4).
Table 2.
Molecular detection of Leishmania DNA of ITS1 and Trypanosoma DNA of SSU rRNA from sand fly samples collected in southern Thailand.
| Province | Positive Sand Fly Species | Detection of Leishmania spp. and Trypanosoma sp. | ||
|---|---|---|---|---|
| L. martiniquensis | T. noyesi | Trypanosoma sp. | ||
|
Songkhla
(n = 37) |
Se. khawi | 2 * | ND | 4 |
|
Phatthalung
(n = 64) |
Se. khawi | ND | ND | 1 |
| Se. indica | ND | ND | 1 | |
|
Chumphon
(n = 119) |
Ph. asperulus | ND | 1 | 2 |
| Se. anodontis | ND | 2 | 4 | |
| Ph. betisi | ND | ND | 2 | |
| Total (n = 220) | 1 | 3 | 14 | |
* One specimen was co-infected by L. martiniquensis and Trypanosoma sp.; ND—not detected.
The female sand flies were collected from the non-endemic area of leishmaniasis, Chumphon Province. The results showed that 2/119 of Se. anodontis and 1/119 of Ph. asperulus were detected for T. noyesi DNA, with 98% identity (accession no. KX008320); furthermore, we found positive Trypanosoma sp. in four samples of Se. anodontis and two samples of each Ph. asperulus and Ph. betisi, which showed 96%–99% similarity with Trypanosoma sp. (accession no. AB520638). However, Leishmania DNA was not detected in the non-endemic area of leishmaniasis.
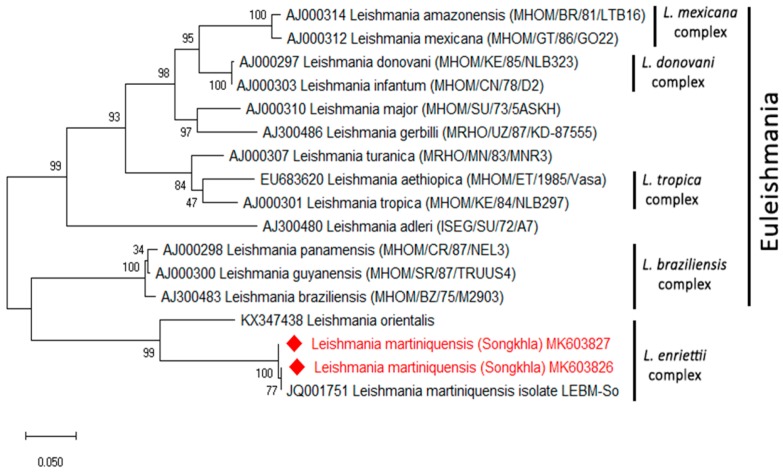
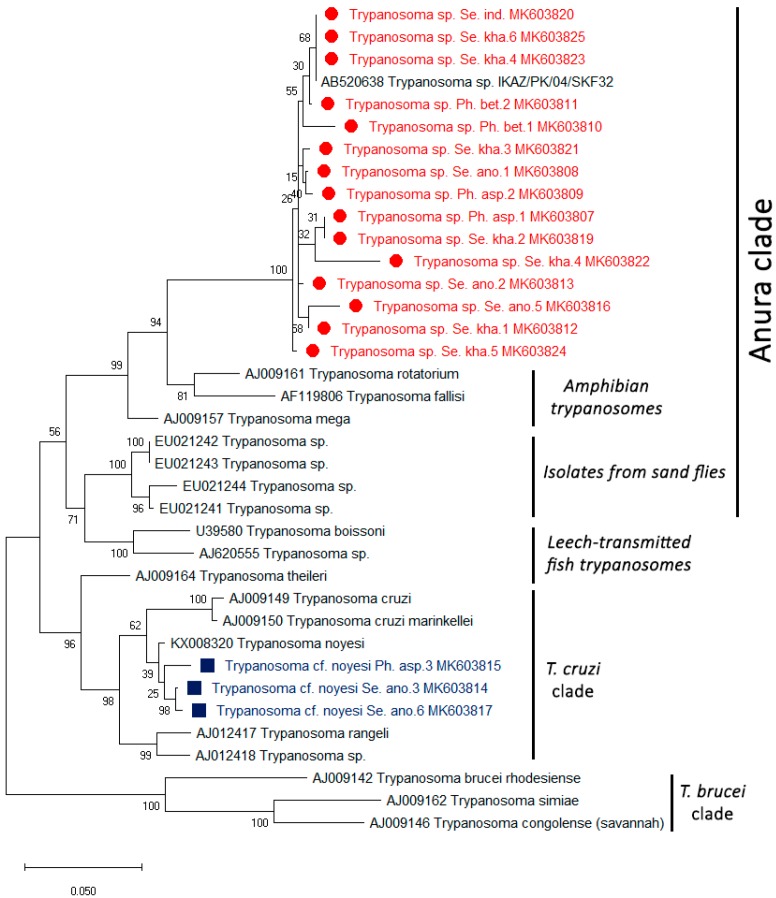
The phylogenetic tree of L. martiniquensis of the ITS1 region was clearly shown in a clade of the L. enriettii complex (Figure 2). The phylogenetic tree of all of the positive Trypanosoma parasites with the SSU rRNA regions was divided into two groups. The first group of 15 samples contained Trypanosoma sp. in the Anura clade described in Pakistan. The second group of three samples was grouped together with T. noyesi with small branch lengths in the T. cruzi clade (Figure 3). The partial nucleotide sequence of the ITS1 and SSU rRNA obtained in this study was deposited in GenBank under the following accession numbers: MK603807–MK603827.
Figure 2.
A maximum likelihood tree was constructed from the partial ITS1 sequences of Leishmania spp. and was compared with the reference sequences using the Kimura two-parameter model with the maximum likelihood method, by testing with 1000 bootstrap values. The sequences from this study are indicated with a red color.
Figure 3.
Phylogenetic tree of the Trypanosoma species in the sand flies constructed from the partial SSU rRNA sequences from all of the regions. The sequences from this study, indicated in red and blue, were compared with the reference sequences obtained from GenBank. The tree was derived using the maximum likelihood method based on the Kimura two-parameter model (bootstrapped 1000 times).
4. Discussion
Autochthonous leishmaniasis in Thailand is caused by two major Leishmania species, namely, L. orientalis and L. martiniquensis (Kinetoplastida: Trypanosomatidae) [8,9,12,30,31,32,33,34,35,36,37,38,39,40]. Studies on the vector of the disease are essential for controlling the disease and for gaining a better understanding of the disease transmission dynamics; however, there are limited data on this disease in Thailand. Ready (2013) has demonstrated that female sand flies are able to transmit several parasites, including Leishmania spp., Trypanosoma spp., and other viruses [41]. In the current study, we described a method for the detection of Leishmania and Trypanosoma protozoans by PCR using primers specific for Leishmania and Trypanosoma DNA in a natural population of sand flies collected from endemic and non-endemic areas of leishmaniasis in southern Thailand. We showed that the only L. martiniquensis-positive sand flies were Se. khawi, by analysing the ITS1 regions. Moreover, one sample showed co-infection between L. martiniquensis and Trypanosoma sp. DNA in Se. khawi (Trypanosoma sp. Se. kha) from the Songkhla Province, an endemic area of leishmaniasis. However, we did not conduct a dissection to demonstrate live parasites in the midgut and did not observe any blood meal in the female sand fly samples; however, we also detected parasite DNA in the samples. These results could infer that there may be some live parasites in the midgut, even after the female sand flies have been defecated. In the next survey study, we will dissect the sand fly midguts for the live parasites. The have been reports of the potential role of Sergentomyia species as a vector of leishmaniasis [42]. L. donovani DNA has been detected in Se. babu from India [43]. L. major DNA has been detected in Se. sintoni from Iran [44], Se. minuta from Portugal [45], and Se. clydei [46] and Se. minuta [47] from Tunisia. In addition, L. infantum DNA has been detected in Se. dubia, Se. magna, and Se. schwetzi from Senegal [48]. In Thailand, L. martiniquensis DNA was detected in both Se. gemmea [12,22] and Se. barraudi [49]. Recently, Siripattanapipong et al. (2018) suggested that L. orientalis was found in Se. iyengari (Diptera: Psychodidae) from southern Thailand [50]. However, some reports have described that the species of Se. gemmea and Se. iyengari sand flies in Thailand may be misidentified based on morphology and molecular techniques [23,25]; moreover, all of the sequences of Se. iyengari deposited in GenBank (accession numbers MG770900, MG770901, MG770913, MG770914, MG770916–MG770918, MG770924, MG770926, and MG770927) and Se. gemmea deposited in GenBank (accession numbers MG770900, MG770901, MG770913, MG770914, MG770916–MG770918, MG770924, MG770926, and MG770927) are grouped as Se. hivernus (Raynal and Gaschen) and Se. khawi (Raynal) by using CytB-PCR [23,25]. In this study, the most abundant species in our samples was Se. khawi, which could be a potential vector of L. martiniquensis infection in both humans and animals in Thailand. However, in order to prove that Se. khawi is a competent vector of leishmaniasis, future studies are needed in order to reveal exact sand fly species in both the morphological and molecular characterization. Additionally, the isolation and cultivation of Leishmania parasites are needed to support the complete life cycle of leishmaniasis in Thailand.
For Trypanosoma DNA detection, we found 1.81% of T. noyesi in Se. anodontis (Trypanosoma cf. noyesi Se. ano) and Ph. asperulus (Trypanosoma cf. noyesi Se. asp) and 6.36% of Trypanosoma sp. Se. kha, Se. indica (Trypanosoma sp. Se. ind), Se. anodontis (Trypanosoma sp. Se. ano), Ph. asperulus (Trypanosoma sp. Ph. asp), and Ph. betisi (Trypanosoma sp. Ph. bet) from both endemic and non-endemic regions of leishmaniasis. Three positive samples were clustered together with T. noyesi (accession no. KX008320) with small branch lengths and in the T. cruzi clade. Botero et al. (2016) determined that their study provided novel information on the morphological, behavioral observations in vitro, and genetic variability of an Australian trypanosome of T. noyesi within the T. cruzi clade isolated from the critically endangered woylie (Bettongia pencillata) [51]. A previous report in Thailand was the first study on Trypanosoma sp. DNA detection in a Ph. stantoni collected from southern Thailand. It was suspected that the detected sequences belonged to a novel species of the genus Trypanosoma [23]. In this study, we found that 15 samples were grouped to Trypanosoma sp. from Ph. Kazeruni, and Theodor and Mesghali described this in Pakistan (accession no. AB520638), in an Anura clade [52]. In contrast, we have not found unknown Trypanosoma sp. in sand flies, as described by Phumee et al., (2016) in Thailand. This might be because of the low number of samples, and the vector for unknown Trypanosoma sp. may be other sand fly species. Many reports also described the detection of Trypanosome DNA in sand flies. For example, Nzelu et al. (2014) revealed that L. tropica and L. major DNA, and Trypanosoma DNA can be detected in Sergentomyia sand flies in Ghana [53]. However, the information of Leishmania and Trypanosoma parasites in sand flies associated with their hosts, geographic distribution, and human or animal diseases are quite limited. Therefore, more extensive and systematic epidemiological surveys on potential vectors by using a larger number of sand flies should be conducted for proving whether these sand flies can be natural vectors of leishmaniasis and trypanosomiasis in both humans and animals. In addition, a better understanding of the epidemiological and ecological relationships of the disease could be used for studying the prevention, as well as effective control for both Leishmania and Trypanosoma vectors in Thailand.
5. Conclusions
The natural infection of sand flies by Leishmania or Trypanosoma alone and the co-infection of both parasites were detected in both the Sergentomyia and Phlebotomus genera of sand flies. The detection and identification of natural infections in sand flies by these parasites in endemic and non-endemic areas of Thailand are key factors in assessing the disease transmission risk and potential vectors, designing prevention and control measures, and predicting disease epidemics in leishmaniasis and trypanosomiasis in Thailand.
Acknowledgments
We would like to express my sincere thanks to the staff of the staff of Vector Biology and Vector Borne Disease Research Unit, Department of Parasitology, Faculty of Medicine, Chulalongkorn University, Thailand.
Supplementary Materials
The following are available online at https://www.mdpi.com/2075-4450/10/8/238/s1. Figure S1: ITS1-PCR amplification for Leishmania parasites. Lane M: molecular mass marker (100 base pairs [bp]); lane P: L. martiniquensis culture, lane N: negative control, lanes 1–10 female sand fly samples. Figure S2: Sequence alignment of Leishmania parasites based on the partial ITS1 gene. Figure S3: SSU rRNA-PCRs for Trypanosoma parasites. Lane M: molecular mass marker (100 base pairs [bp]); lane P: Trypanosoma sp. culture, lane N: negative control, lanes 1–10 female sand fly samples. Figure S4: Sequence alignment of Trypanosoma parasites base on the partial SSU rRNA gene.
Author Contributions
Conceptualization, P.S. (Padet Siriyasatien); methodology, P.S. (Pimpilad Srisuton); software, P.S. (Pimpilad Srisuton), S.S. and A.P.; validation, P.S. (Pimpilad Srisuton), P.S. (Padet Siriyasatien), and A.P.; formal analysis, P.S. (Padet Siriyasatien); investigation, R.B., S.S.-s. and T.P.; resources, P.S. (Padet Siriyasatien); data curation, P.S. (Padet Siriyasatien); writing—original draft preparation, P.S. (Pimpilad Srisuton); writing—review and editing, P.S. (Padet Siriyasatien), A.P. and N.B.; visualization, P.S. (Padet Siriyasatien); supervision, P.S. (Padet Siriyasatien); project administration, P.S. (Pimpilad Srisuton); funding acquisition, P.S. (Padet Siriyasatien).
Funding
This study was supported by Rachadapisaek Sompote Fund (Grant No. RA 62/016), Faculty of Medicine, Chulalongkorn University and Thailand research fund (Grant No. RSA 5780024).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Killick-Kendrick R. The biology and control of phlebotomine sand flies. Clin. Dermatol. 1999;17:279–289. doi: 10.1016/S0738-081X(99)00046-2. [DOI] [PubMed] [Google Scholar]
- 2.Akhoundi M., Kuhls K., Cannet A., Votýpka J., Marty P., Delaunay P., Sereno D. A Historical Overview of the Classification, Evolution, and Dispersion of Leishmania Parasites and Sandflies. PLoS Negl. Trop. Dis. 2016;10:e0004349. doi: 10.1371/journal.pntd.0004349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooper C., Clode P.L., Peacock C., Thompson R.C.A. Chapter Two—Host–Parasite Relationships and Life Histories of Trypanosomes in Australia. In: Rollinson D., Stothard J.R., editors. Advances in Parasitology. Volume 97. Academic Press; Cambridge, MA, USA: 2017. pp. 47–109. [DOI] [PubMed] [Google Scholar]
- 4.Njagu Z., Mihok S., Kokwaro E., Verloo D. Isolation of Trypanosoma brucei from the monitor lizard (Varanus niloticus) in an endemic focus of rhodesian sleeping sickness in Kenya. Acta Trop. 1999;72:137–148. doi: 10.1016/S0001-706X(98)00092-8. [DOI] [PubMed] [Google Scholar]
- 5.Ferreira R.C., De Souza A.A., Freitas R.A., Campaner M., Takata C.S., Barrett T.V., Shaw J.J., Teixeira M.M. A phylogenetic lineage of closely related trypanosomes (Trypanosomatidae, Kinetoplastida) of anurans and sand flies (Psychodidae, Diptera) sharing the same ecotopes in brazilian amazonia. J. Eukaryot. Microbiol. 2008;55:427–435. doi: 10.1111/j.1550-7408.2008.00342.x. [DOI] [PubMed] [Google Scholar]
- 6.Lemos M., Morais D.H., Carvalho V.T., D’Agosto M. First record of Trypanosoma chattoni in Brazil and occurrence of other Trypanosoma species in Brazilian frogs (Anura, Leptodactylidae) J. Parasitol. 2008;94:148–151. doi: 10.1645/GE-1095.1. [DOI] [PubMed] [Google Scholar]
- 7.Viola L.B., Campaner M., Takata C.S., Ferreira R.C., Rodrigues A.C., Freitas R.A., Duarte M.R., Grego K.F., Barrett T.V., Camargo E.P., et al. Phylogeny of snake trypanosomes inferred by SSU rDNA sequences, their possible transmission by phlebotomines, and taxonomic appraisal by molecular, cross-infection and morphological analysis. Parasitology. 2008;135:595–605. doi: 10.1017/S0031182008004253. [DOI] [PubMed] [Google Scholar]
- 8.Jariyapan N., Daroontum T., Jaiwong K., Chanmol W., Intakhan N., Sor-Suwan S., Siriyasatien P., Somboon P., Bates M.D., Bates P.A. Leishmania (Mundinia) orientalis n. sp. (Trypanosomatidae), a parasite from Thailand responsible for localised cutaneous leishmaniasis. Parasites Vectors. 2018;11:351. doi: 10.1186/s13071-018-2908-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiewchanvit S., Tovanabutra N., Jariyapan N., Bates M.D., Mahanupab P., Chuamanochan M., Tantiworawit A., Bates P.A. Chronic generalized fibrotic skin lesions from disseminated leishmaniasis caused by Leishmania martiniquensis in two patients from northern Thailand infected with HIV. Br. J. Dermatol. 2015;173:663–670. doi: 10.1111/bjd.13812. [DOI] [PubMed] [Google Scholar]
- 10.Thisyakorn U., Jongwutiwes S., Vanichsetakul P., Lertsapcharoen P. Visceral leishmaniasis: The first indigenous case report in Thailand. Trans. R. Soc. Trop. Med. Hyg. 1999;93:23–24. doi: 10.1016/S0035-9203(99)90166-9. [DOI] [PubMed] [Google Scholar]
- 11.Maharom P., Siripattanapipong S., Mungthin M., Naaglor T., Sukkawee R., Pudkorn R., Wattana W., Wanachiwanawin D., Areechokchai D., Leelayoova S. Visceral leishmaniasis caused by Leishmania infantum in Thailand. Southeast Asian J. Trop. Med. Public Health. 2008;39:988–990. [PubMed] [Google Scholar]
- 12.Leelayoova S., Siripattanapipong S., Manomat J., Piyaraj P., Tan-Ariya P., Bualert L., Mungthin M. Leishmaniasis in Thailand: A Review of Causative Agents and Situations. Am. J. Trop. Med. Hyg. 2017;96:534–542. doi: 10.4269/ajtmh.16-0604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milocco C., Kamyingkird K., Desquesnes M., Jittapalapong S., Herbreteau V., Chaval Y., Douangboupha B., Morand S. Molecular demonstration of Trypanosoma evansi and Trypanosoma lewisi DNA in wild rodents from Cambodia, Lao PDR and Thailand. Transbound. Emerg. Dis. 2013;60:17–26. doi: 10.1111/j.1865-1682.2012.01314.x. [DOI] [PubMed] [Google Scholar]
- 14.Sim S.T., Wiwanitkit V. Emerging human trypanosomiasis in Southeast Asia: A coming new threat? J. Nat. Sci. 2015;1:e29. [Google Scholar]
- 15.Sarataphan N., Vongpakorn M., Nuansrichay B., Autarkool N., Keowkarnkah T., Rodtian P., Stich R.W., Jittapalapong S. Diagnosis of a Trypanosoma lewisi-like (Herpetosoma) infection in a sick infant from Thailand. J. Med. Microbiol. 2007;56:1118–1121. doi: 10.1099/jmm.0.47222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Apiwathnasorn C., Sucharit S., Rongsriyam Y., Leemingsawat S., Kerdpibule V., Deesin T., Surathin K., Vutikes S., Punavuthi N. A brief survey of phlebotomine sandflies in Thailand. Southeast Asian J. Trop. Med. Public Health. 1989;20:429–432. [PubMed] [Google Scholar]
- 17.Apiwathnasorn C., Samung Y., Prummongkol S., Phayakaphon A., Panasopolkul C. Cavernicolous species of phlebotomine sand flies from Kanchanaburi Province, with an updated species list for Thailand. Southeast Asian J. Trop. Med. Public Health. 2011;42:1405–1409. [PubMed] [Google Scholar]
- 18.Artemiev M.A. Classification of the subfamily Phlebotominae. Parassitologia. 1991;33:69–77. [PubMed] [Google Scholar]
- 19.Marcondes C.B. A proposal of generic and subgeneric abbreviations for Phlebotomine sandflies (Diptera: Psychodidae: Phlebotominae) of the World. Entomol. News. 2007;118:351–356. doi: 10.3157/0013-872X(2007)118[351:APOGAS]2.0.CO;2. [DOI] [Google Scholar]
- 20.Galati E.A.B. In: Classificação de Phlebotominae, in Flebotomíneos do Brasil. Rangel E.F., Lainson R., editors. Fiocruz; Rio de Janeiro, Brazil: 2003. pp. 23–51. [Google Scholar]
- 21.Sukra K., Kanjanopas K., Amsakul S., Rittaton V., Mungthin M., Leelayoova S. A survey of sandflies in the affected areas of leishmaniasis, southern Thailand. Parasitol. Res. 2013;112:297–302. doi: 10.1007/s00436-012-3137-x. [DOI] [PubMed] [Google Scholar]
- 22.Kanjanopas K., Siripattanapipong S., Ninsaeng U., Hitakarun A., Jitkaew S., Kaewtaphaya P., Tan-ariya P., Mungthin M., Charoenwong C., Leelayoova S. Sergentomyia (Neophlebotomus) gemmea, a potential vector of Leishmania siamensis in southern Thailand. BMC Infect. Dis. 2013;13:333. doi: 10.1186/1471-2334-13-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phumee A., Tawatsin A., Thavara U., Pengsakul T., Thammapalo S., Depaquit J., Gay F., Siriyasatien P. Detection of an Unknown Trypanosoma DNA in a Phlebotomus stantoni (Diptera: Psychodidae) Collected From Southern Thailand and Records of New Sand Flies With Reinstatement of Sergentomyia hivernus Raynal & Gaschen, 1935 (Diptera: Psychodidae) J. Med. Entomol. 2017;54:429–434. doi: 10.1093/jme/tjw161. [DOI] [PubMed] [Google Scholar]
- 24.Lewis D.J. The phlebotomine sandflies (Diptera: Psychodidae) of the oriental region. Bull. Br. Mus. (Nat. Hist.) B. Entomol. 1978;37:217–343. doi: 10.1111/j.1365-3113.1987.tb00194.x. [DOI] [Google Scholar]
- 25.Depaquit J., Vongphayloth K., Siriyasatien P., Polseela R., Phumee A., Loyer M., Vol A., Varlot G., Rahola N., Brey P.T., et al. On the true identity of Sergentomyia gemmea and description of a closely related species: Se. raynali n. sp. Med. Vet. Entomol. 2019 doi: 10.1111/mve.12393. [DOI] [PubMed] [Google Scholar]
- 26.Spanakos G., Piperaki E.T., Menounos P.G., Tegos N., Flemetakis A., Vakalis N.C. Detection and species identification of old world Leishmania in clinical samples using a PCR-based method. Trans. R. Soc. Trop. Med. Hyg. 2008;102:46–53. doi: 10.1016/j.trstmh.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 27.Noyes H.A., Stevens J.R., Teixeira M., Phelan J., Holz P. A nested PCR for the ssrRNA gene detects Trypanosoma binneyi in the platypus and Trypanosoma sp. in wombats and kangaroos in Australia. Int. J. Parasitol. 1999;29:331–339. doi: 10.1016/S0020-7519(98)00167-2. [DOI] [PubMed] [Google Scholar]
- 28.Hall T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 1999;41:95–98. [Google Scholar]
- 29.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bualert L., Charungkiattikul W., Thongsuksai P., Mungthin M., Siripattanapipong S., Khositnithikul R., Naaglor T., Ravel C., El Baidouri F., Leelayoova S. Case Report: Autochthonous disseminated dermal and visceral leishmaniasis in an AIDS patient, southern Thailand, caused by Leishmania siamensis. Am. J. Trop. Med. Hyg. 2012;86:821–824. doi: 10.4269/ajtmh.2012.11-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chusri S., Hortiwakul T., Silpapojakul K., Siriyasatien P. Case Report: Consecutive cutaneous and visceral leishmaniasis manifestations involving a novel Leishmania species in two HIV patients in Thailand. Am. J. Trop. Med. Hyg. 2012;87:76–80. doi: 10.4269/ajtmh.2012.11-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jungudomjaroen S., Phumee A., Chusri S., Kraivichian K., Jariyapan N., Payungporn S., Siriyasatien P. Detection of Leishmania martiniquensis DNA in various clinical samples by quantitative PCR. Trop. Biomed. 2015;32:736–744. [PubMed] [Google Scholar]
- 33.Noppakun N., Kraivichian K., Siriyasatien P. Disseminated dermal leishmaniasis caused by Leishmania siamensis in a systemic steroid therapy patient. Am. J. Trop. Med. Hyg. 2014;91:869–870. doi: 10.4269/ajtmh.13-0711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Osatakul S., Mungthin M., Siripattanapipong S., Hitakarun A., Kositnitikul R., Naaglor T., Leelayoova S. Recurrences of visceral leishmaniasis caused by Leishmania siamensis after treatment with amphotericin B in a seronegative child. Am. J. Trop. Med. Hyg. 2014;90:40–42. doi: 10.4269/ajtmh.13-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phumee A., Kraivichian K., Chusri S., Noppakun N., Vibhagool A., Sanprasert V., Tampanya V., Wilde H., Siriyasatien P. Detection of Leishmania siamensis DNA in saliva by polymerase chain reaction. Am. J. Trop. Med. Hyg. 2013;89:899–905. doi: 10.4269/ajtmh.12-0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phumee A., Chusri S., Kraivichian K., Wititsuwannakul J., Hortiwakul T., Thavara U., Silpapojakul K., Siriyasatien P. Multiple Cutaneous Nodules in an HIV-Infected Patient. PLoS Negl. Trop. Dis. 2014;8:e3291. doi: 10.1371/journal.pntd.0003291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pothirat T., Tantiworawit A., Chaiwarith R., Jariyapan N., Wannasan A., Siriyasatien P., Supparatpinyo K., Bates M.D., Kwakye-Nuako G., Bates P.A. First isolation of Leishmania from Northern Thailand: Case report, identification as Leishmania martiniquensis and phylogenetic position within the Leishmania enriettii complex. PLoS Negl. Trop. Dis. 2014;8:e3339. doi: 10.1371/journal.pntd.0003339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siriyasatien P., Chusri S., Kraivichian K., Jariyapan N., Hortiwakul T., Silpapojakul K., Pym A.M., Phumee A. Early detection of novel Leishmania species DNA in the saliva of two HIV-infected patients. BMC Infect. Dis. 2016;16:89. doi: 10.1186/s12879-016-1433-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suankratay C., Suwanpimolkul G., Wilde H., Siriyasatien P. Case Report: Autochthonous visceral leishmaniasis in a human immunodeficiency virus (HIV)-infected patient: The first in Thailand and review of the literature. Am. J. Trop. Med. Hyg. 2010;82:4–8. doi: 10.4269/ajtmh.2010.09-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sukmee T., Siripattanapipong S., Mungthin M., Worapong J., Rangsin R., Samung Y., Kongkaew W., Bumrungsana K., Chanachai K., Apiwathanasorn C., et al. A suspected new species of Leishmania, the causative agent of visceral leishmaniasis in a Thai patient. Int. J. Parasitol. 2008;38:617–622. doi: 10.1016/j.ijpara.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 41.Ready P. Biology of phlebotomine sand flies as vectors of disease agents. Annu. Rev. Entomol. 2013;58:227–250. doi: 10.1146/annurev-ento-120811-153557. [DOI] [PubMed] [Google Scholar]
- 42.Maia C., Depaquit J. Can Sergentomyia (Diptera, Psychodidae) play a role in the transmission of mammal-infecting Leishmania? Parasite. 2016;23:55. doi: 10.1051/parasite/2016062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mukherjee S., Hassan M.Q., Ghosh A., Ghosh K.N., Bhattacharya A., Adhya S. Leishmania DNA in Phlebotomus and Sergentomyia species during a kala-azar epidemic. Am. J. Trop. Med. Hyg. 1997;57:423–425. doi: 10.4269/ajtmh.1997.57.423. [DOI] [PubMed] [Google Scholar]
- 44.Arvizi P., Amirkhani A. Mitochondrial DNA characterization of Sergentomyia sintoni populations and finding mammalian Leishmania infections in this sandfly by using ITS-rDNA gene. Iran. J. Vet. Res. 2008;9:9–18. [Google Scholar]
- 45.Campino L., Cortes S., Dionísio L., Neto L., Afonso M.O., Maia C. The first detection of Leishmania major in naturally infected Sergentomyia minuta in Portugal. Mem. Inst. Oswaldo Cruz. 2013;108:516–518. doi: 10.1590/S0074-02762013000400020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ayari C., Ben Othman S., Chemkhi J., Tabbabi A., Fisa R., Ben Salah A., BenAbderrazak S. First detection of Leishmania major DNA in Sergentomyia (Sintonius) clydei (Sinton, 1928, Psychodidae: Phlebotominae), from an outbreak area of cutaneous leishmaniasis in Tunisia. Infect. Genet. Evol. 2016;39:241–248. doi: 10.1016/j.meegid.2015.10.030. [DOI] [PubMed] [Google Scholar]
- 47.Aouadi K., Ghawar W., Salem S., Gharbi M., Bettaieb J., Yazidi R., Harrabi M., Hamarsheh O., Ben Salah A. First report of naturally infected Sergentomyia minuta with Leishmania major in Tunisia. Parasites Vectors. 2015;8:649. doi: 10.1186/s13071-015-1269-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Senghor M., Niang A., Depaquit J., Faye M., Ferté H., Elguero E., Gaye O., Alten B., Perktas U., Cassan C., et al. Transmission of Leishmania infantum in the canine leishmaniasis focus of Mont-Rolland, Senegal: Ecological, parasitological and molecular evidence for a possible role of Sergentomyia sand flies. PLoS Negl. Trop. Dis. 2016;10:e0004940. doi: 10.1371/journal.pntd.0004940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chusri S., Thammapalo S., Chusri S., Thammapalo S., Silpapojakul K., Siriyasatien P. Animal reservoirs and potential vectors of Leishmania siamensis in southern Thailand. Southeast Asian J. Trop. Med. Public Health. 2014;45:13–19. [PubMed] [Google Scholar]
- 50.Siripattanapipong S., Leelayoova S., Ninsaeng U., Mungthin M. Detection of DNA of Leishmania siamensis in Sergentomyia (Neophlebotomus) iyengari (Diptera: Psychodidae) and Molecular Identification of Blood Meals of Sand Flies in an Affected Area, Southern Thailand. J. Med. Entomol. 2018;55:1277–1283. doi: 10.1093/jme/tjy069. [DOI] [PubMed] [Google Scholar]
- 51.Botero A., Cooper C., Thompson C.K., Clode P.L., Rose K., Thompson R.C. Morphological and Phylogenetic Description of Trypanosoma noyesi sp. nov.: An Australian Wildlife Trypanosome within the T. cruzi Clade. Protist. 2016;167:425–439. doi: 10.1016/j.protis.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 52.Kato H., Uezato H., Sato H., Bhutto A.M., Soomro F.R., Baloch J.H., Iwata H., Hashiguchi Y. Natural infection of the sand fly Phlebotomus kazeruni by Trypanosoma species in Pakistan. Parasites Vectors. 2010;3:10. doi: 10.1186/1756-3305-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nzelu C.O., Kato H., Puplampu N., Desewu K., Odoom S., Wilson M.D., Sakurai T., Katakura K., Boakye D.A. First detection of Leishmania tropica DNA and Trypanosoma species in Sergentomyia sand flies (Diptera: Psychodidae) from an outbreak area of cutaneous leishmaniasis in Ghana. PLoS Negl. Trop. Dis. 2014;8:e2630. doi: 10.1371/journal.pntd.0002630. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.