Abstract
The HOXA gene family is associated with various cancer types. However, the role of HOXA genes in acute myeloid leukemia (AML) have not been comprehensively studied. We compared the transcriptional expression, survival data, and network analysis of HOXA-associated signaling pathways in patients with AML using the ONCOMINE, GEPIA, LinkedOmics, cBioPortal, and Metascape databases. We observed that HOXA2-10 mRNA expression levels were significantly upregulated in AML and that high HOXA1-10 expression was associated with poor AML patient prognosis. The HOXA genes were altered in ~18% of the AML samples, either in terms of amplification, deep deletion, or elevated mRNA expression. The following pathways were modulated by HOXA gene upregulation: GO:0048706: embryonic skeletal system development; R-HSA-5617472: activation of HOX genes in anterior hindbrain development during early embryogenesis; GO:0060216: definitive hemopoiesis; hsa05202: transcriptional mis-regulation in cancer; and GO:0045638: negative regulation of myeloid cell differentiation, and they were significantly regulated due to alterations affecting the HOXA genes. This study identified HOXA3-10 genes as potential AML therapeutic targets and prognostic markers.
Keywords: HOXA gene family, acute myeloid leukemia, transcription factor, prognostic value, molecular functions, bioinformatics integration analysis
1. Introduction
Acute myeloid leukemia (AML) is a heterogeneous hematological malignant cancer, characterized by the clonal expansion of myeloid blasts in the peripheral blood, bone marrow, and/or other tissues. It is the most prevalent acute leukemia amongst adults and the leading cause of leukemia-associated deaths in the United States each year [1]. An estimated 21,450 people will be diagnosed with AML in 2019, and 10,920 will die of this disease [2]. Reports show that the overall 5-year survival rate for AML patients has increased from 16% from 1991–1996 to 23% from 2003–2008 [3], followed by 28.4% from 2009–2015 [4]. In fact, about 45% of AML patients show a normal karyotype detected by conventional cytogenetics at diagnosis, whilst 97.3% display somatic mutations [5]. Therefore, there is an urgent need to identify reliable biomarkers, enabling early diagnosis and improved prognosis prediction of AML, in addition to developing novel targeted therapeutic strategies.
Mammals possess 39 HOX genes, which are arrayed in four linear clusters with 9 to 11 genes per cluster. Based on homologies, the genes are assigned to 13 paralogous groups. The HOXA gene family encodes proteins that contain the DNA-binding homeobox motif and controls the early patterns of embryo segmentation in addition to later developmental events [6]. HOX gene expression is typically inhibited in adults, although controlled reactivation is required for body repair and various homeostatic cellular processes [7,8], including hematopoiesis [9], wound healing [10], vascularization [10], as well as female reproductive tract development and fertility [11].
The expression of HOX genes is a common feature of AML, which is considered a disorder of the HOX pathway, leading to abnormal cell-renewal and leukemia development. In AML patients, the expression of HOX genes correlates most strongly to the translocation of the MLL gene [12]. HOXA9, in particular, has been shown to be a target of MLL fusion oncoproteins [13]. Several studies have shown that certain AML mutations are related to HOX gene expression and that high HOXA gene expression levels are usually accompanied by partial tandem MLL duplications [14]. Conversely, when HOX gene expression is low or absent, it is typically accompanied by PML-RARA and RUNX1-RUNX1T1 gene fusions [15,16], as well as mutations of the CEBPA gene [17].
Therefore, the identification of HOXA-mediated oncogenes or tumor suppressors as potential mechanisms for predicting biomarkers may provide new therapeutic strategies for treating AML. The challenge lays in the fact that the differences in transcriptional levels, prognostic value, molecular functions, and biological processes for the majority of HOXA genes are yet to be elucidated in the context of AML disease. In this study, we expanded the knowledge on AML pathology by interrogating several universally-acknowledged databases to comprehensively analyze the relationship between HOXA subtypes and the pathogenesis and progression of AML.
2. Materials and Methods
2.1. Ethics Statement
This study was approved by the Academic Committee of the Sun Yat-Sen University Cancer Center, and it was carried out in accordance with the principles of the Helsinki Declaration. All the data analyzed within this study were retrieved from published literature, for which we confirm that written informed consent was obtained.
2.2. ONCOMINE Database Analysis
We analyzed HOXA gene transcription levels in different cancers using gene expression array datasets within ONCOMINE (https://www.oncomine.org), an online cancer microarray database [18]. HOXA mRNA expression levels in tumor specimens were compared with the levels in normal controls, and P values were obtained using the Students’ t-test. The cut-off P value and fold change were defined as 0.01 and 2, respectively. Genes co-expressed with HOXA genes were analyzed in the Cancer Genome Atlas (TCGA) for Leukemia, using the ONCOMINE database. Genes with a correlation value >0.4 were selected for further analysis.
2.3. The Gene Expression Profiling Interactive Analysis (GEPIA) Dataset
GEPIA [19] (http://gepia.cancer-pku.cn) is a newly developed interactive web server that enables users to perform expression analyses (such as survival and differential analyses) at the subtype level. GEPIA can be used to analyze gene and isoform expression by performing a comparison to TCGA and GTEx data. As such, we validated the differential expression of HOXA genes in AML and healthy donor samples.
2.4. The LinkedOmics Dataset
LinkedOmics [20] (http://www.linkedomics.orglogin.php) is a publicly-available portal comprising multiomics data from all 32 TCGA cancer types. It can be used to access, analyze, and compare multiomics data within and across tumor types. We performed a prognosis analysis for the HOXA gene family using the LinkedOmics AML databases.
2.5. TCGA Data and the cBioPortal
The cBioPortal [21] (http://www.cbioportal.org/) for cancer genomics is an open-access, open-source resource designed for the interactive exploration of multidimensional cancer genomics datasets. It supports and stores data relating to nonsynonymous mutations, DNA copy-number, mRNA and microRNA expression, protein-level and phosphoprotein level, DNA methylation, and de-identified clinical data. The cBioPortal online tool enabled us to calculate the frequency of gene alterations, mRNA expression z-scores (RNA Seq V2 RSEM), and HOXA family gene correlations.
2.6. Functional Enrichment and Bioinformatics Analysis
The Metascape [22] (http://metascape.org) online database integrates over forty bioinformatics knowledge bases enabling the extraction of abundant annotations, as well as the identification of enriched pathways and the construction of protein–protein interaction networks from lists of gene and protein identifiers. Using the Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) tools within Metascape, we analyzed a genes list containing HOXA genes to identify the most frequently-altered linked genes.
3. Results
3.1. The Transcriptional Expression of HOXA Genes in Leukemia Patients
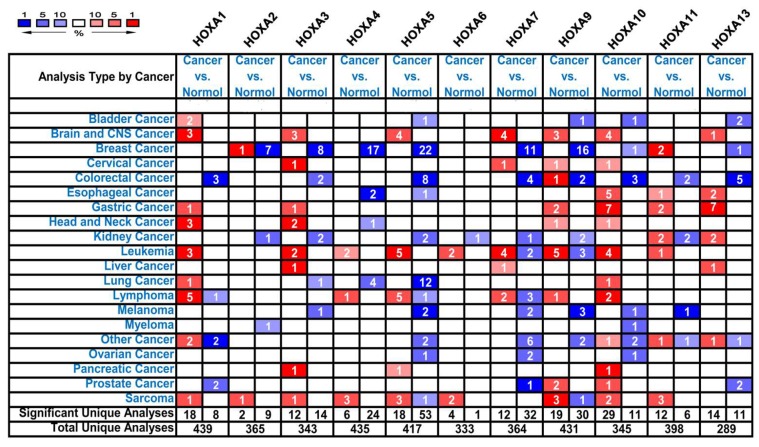
HOXA genes have been identified in the human genome. We used the ONCOMINE databases to compare the transcriptional expression of HOXA genes within tumorigenic and healthy control samples (Figure 1). ONCOMINE analysis revealed that the mRNA expression of HOXA1, HOXA3, HOXA4, HOXA5, HOXA6, HOXA7, HOXA9, and HOXA10 was upregulated in patients with leukemia (Table 1). The analysis revealed that compared to healthy tissues, HOXA1 mRNA was overexpressed 3.45-fold in AML [23] and 10.08-fold in acute adult T-cell leukemia/lymphoma (ATLL) [24]. In the Haferlach et al. dataset, HOXA3 mRNA expression was upregulated in AML (fold change = 2.23) and in pro-B acute lymphoblastic leukemia (ALL) (fold change = 3.98), compared to healthy controls [25]. Moreover, Haferlach and colleagues showed that HOXA4 was elevated in AML (fold change = 2.07) compared to the control samples [25]. Meanwhile, Andersson et al. reported that HOXA4 was overexpressed in pro-B ALL (fold change = 8.08) [26]. In the Haferlach et al. dataset [25], the expression of the following HOXA genes was higher in the cancerous samples compared to the healthy controls: (i) HOXA5 in AML (fold change = 2.52) and pro-B ALL (fold change = 4.36); (ii) HOXA6 expression in pro-B ALL (fold change = 2.01); (iii) HOXA7 expression in AML (fold change = 2.03); (iv) HOXA9 expression in AML (fold change = 2.99) and pro-B ALL (fold change = 7.21); and (v) HOXA10 expression in AML (fold change = 2.17) and pro-B ALL (fold change = 4.80). Stegmaier et al. reported the elevated expression of HOXA5 (fold change = 6.58), HOXA9 (fold change = 53.05), and HOXA10 (fold change = 8.48) in AML [27]. In the Choi et al. dataset, HOXA10 was upregulated in ATLL with a fold change of 3.95 [24]. According to the ONCOMINE analysis, no significant differences were observed in the mRNA expression levels of HOXA2, HOXA11, and HOXA13 between leukemic and healthy control samples.
Figure 1.
The transcription levels of HOXA genes in different types of cancers.
Table 1.
The significant changes in HOXA expression in transcription level between different types of leukemia and normal samples (ONCOMINE Database).
| Gene | Types of leukemia vs. Normal Samples | Fold Change | P Value | t-Test | Reference |
|---|---|---|---|---|---|
| HOXA1 | Acute Myeloid Leukemia vs. Normal | 3.451 | 1.95E-07 | 9.046 | Valk [23] |
| Acute Adult T-Cell Leukemia/Lymphoma vs. Normal | 10.084 | 3.46E-05 | 7.407 | Choi [24] | |
| HOXA2 | NA | NA | NA | NA | NA |
| HOXA3 | Acute Myeloid Leukemia vs. Normal | 2.230 | 2.48E-68 | 19.863 | Haferlach [25] |
| Pro-B Acute Lymphoblastic Leukemia vs. Normal | 3.975 | 8.57E-19 | 11.883 | Haferlach [25] | |
| HOXA4 | Acute Myeloid Leukemia vs. Normal | 8.083 | 4.21E-06 | 6.951 | Andersson [26] |
| Pro-B Acute Lymphoblastic Leukemia vs. Normal | 2.065 | 8.86E-16 | 10.006 | Haferlach [25] | |
| HOXA5 | Acute Myeloid Leukemia vs. Normal | 6.584 | 7.75E-05 | 5.292 | Stegmaier [27] |
| Acute Myeloid Leukemia vs. Normal | 2.522 | 2.78E-38 | 14.237 | Haferlach [25] | |
| Pro-B Acute Lymphoblastic Leukemia vs. Normal | 4.359 | 1.35E-15 | 9.826 | Haferlach [25] | |
| HOXA6 | Pro-B Acute Lymphoblastic Leukemia vs. Normal | 2.012 | 2.30E-14 | 9.174 | Haferlach [25] |
| HOXA7 | Acute Myeloid Leukemia vs. Normal | 2.027 | 1.40E-67 | 19.782 | Haferlach [25] |
| HOXA9 | Acute Myeloid Leukemia vs. Normal | 53.046 | 3.68E-07 | 9.445 | Stegmaier [27] |
| Acute Myeloid Leukemia vs. Normal | 2.483 | 2.88E-33 | 13.493 | Haferlach [25] | |
| Acute Myeloid Leukemia vs. Normal | 2.986 | 9.55E-58 | 17.841 | Haferlach [25] | |
| Pro-B Acute Lymphoblastic Leukemia vs. Normal | 7.209 | 1.32E-20 | 12.798 | Haferlach [25] | |
| HOXA10 | Acute Myeloid Leukemia vs. Normal | 8.483 | 8.82E-06 | 7.123 | Stegmaier [27] |
| Acute Adult T-Cell Leukemia/Lymphoma vs. Normal | 3.946 | 4.70E-05 | 4.888 | Choi [24] | |
| Acute Myeloid Leukemia vs. Normal | 2.174 | 1.30E-40 | 14.641 | Haferlach [25] | |
| Pro-B Acute Lymphoblastic Leukemia vs. Normal | 4.802 | 7.72E-19 | 11.645 | Haferlach [25] | |
| HOXA11 | NA | NA | NA | NA | NA |
| HOXA13 | NA | NA | NA | NA | NA |
Abbreviations: NA, not available.
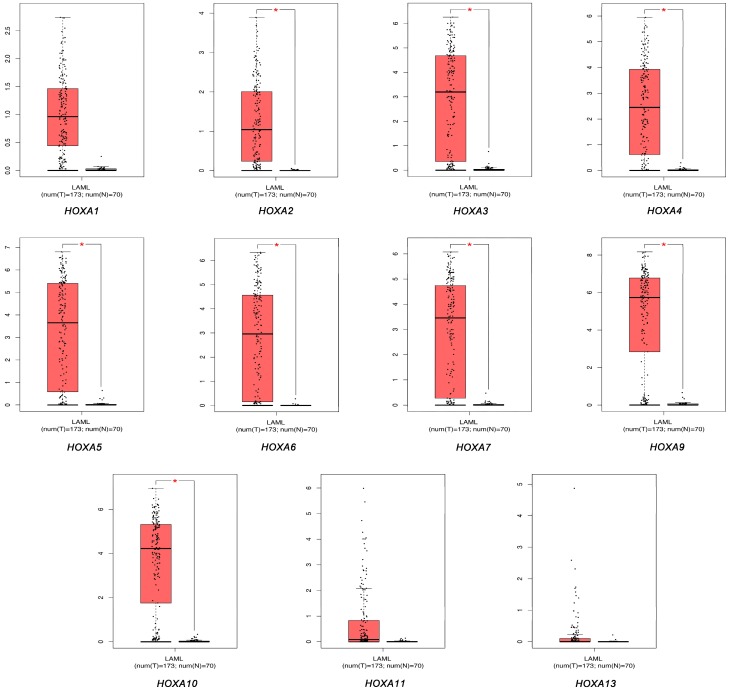
We used the GEPIA dataset to compare the mRNA expression levels of HOXA genes between leukemic and healthy control samples. The results indicated that the expression levels of HOXA2, HOXA3, HOXA4, HOXA5, HOXA6, HOXA7, HOXA9, and HOXA10 were significantly increased in human AML patients compared to those in normal samples (P < 0.05), whilst the expression levels of HOXA1, HOXA11, and HOXA13 were not significantly different (Figure 2).
Figure 2.
The expression of HOXA genes in acute myeloid leukemia (AML) and healthy control samples.
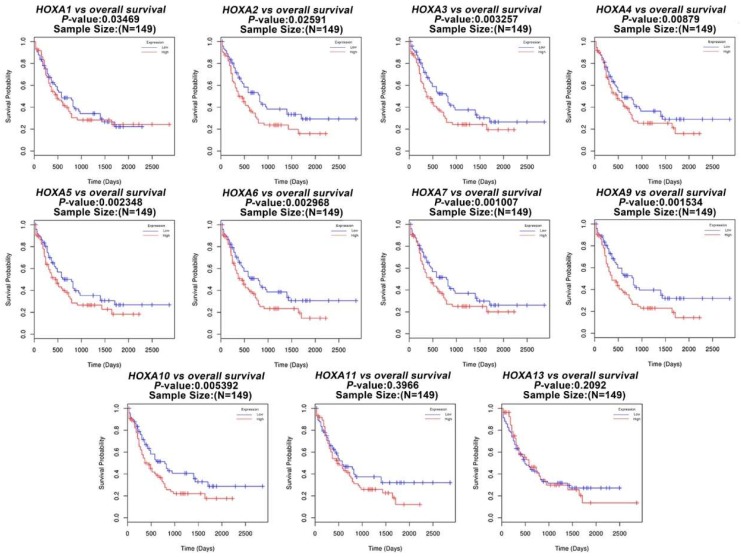
Next, we studied the role of HOXA genes in the survival of patients with AML. Using the LinkedOmics databases, we performed a prognosis analysis of HOXA genes in AML patients. The results revealed that the increased expression levels of HOXA1, HOXA2, HOXA3, HOXA4, HOXA5, HOXA6, HOXA7, HOXA9, and HOXA10 were significantly correlated with poor overall survival (OS, P < 0.05) in all of the AML patients (Figure 3). Therefore, overexpression of HOXA1-10 may represent a poor prognostic factor for AML.
Figure 3.
The prognostic value of the expression levels of HOXA genes in AML patients.
3.2. Genetic Alteration and Correlations of HOXA Genes in AML
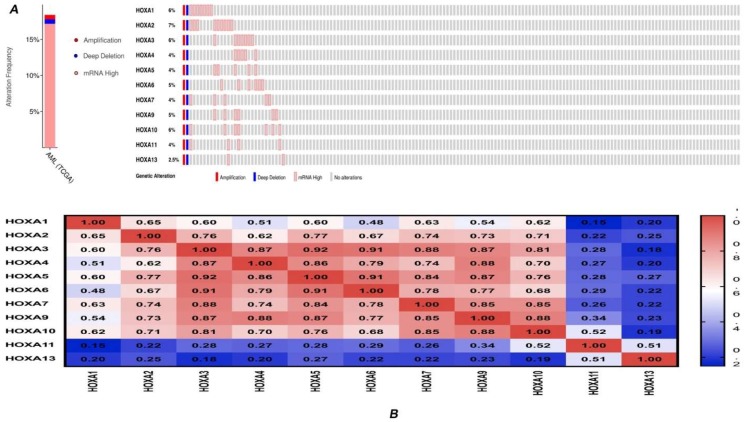
Using the cBioPortal online tool and the ‘TCGA Provisional’ database for AML, we obtained information on genetic alterations in HOXA genes and identified any correlations between genes. HOXA genes were altered in 30/163 (18%) of AML patient samples (Figure 4A). The types of genetic alterations included amplification, deep deletion, and mRNA overexpression. In addition, cBioPortal was employed to analyze the expression of HOXA genes in AML (using mRNA sequencing [RNA-seq] version V2 RSEM), and we calculated the interactions between specific HOXA genes (including the Pearson’s correlation). The results revealed a significant positive correlation between any two HOXA family genes members with the exception of HOXA1 and HOXA11 (Pearson = 0.15, P = 0.0543) (Figure 4B).
Figure 4.
Genetic alteration and correlations of HOXA genes in AML. (A) Genetic alteration of HOXA genes in AML. (B) Genetic correlations of HOXA genes in AML.
3.3. Predicted Functions and Pathway Enrichment Analysis of HOXA Genes in AML Patients
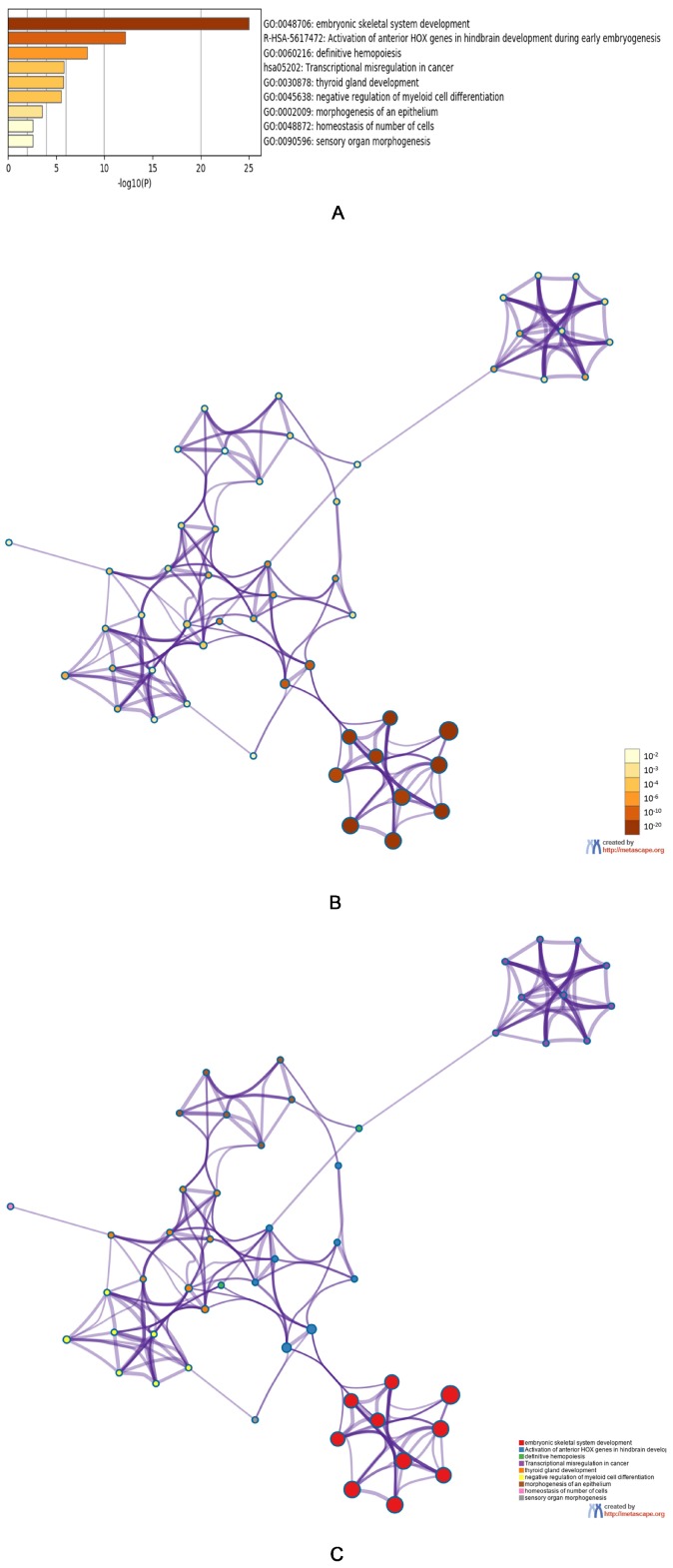
Genes that were co-expressed in conjunction with HOXA genes were analyzed in the ‘TCGA Leukemia’ dataset using the ONCOMINE database. We found that the expression of HOXA genes was positively correlated with the upregulation of the following genes: TSLP, HOXB2, HOXB3, HOXB4, LOC404266, HOXB6, NKX2-3, PBX3, C10orf140, LOC100271722, LOC100289444, MEIS1, CPNE8, LOC400931, SDSL, EMR1, and HOXB7. We subsequently compiled a list of the expressed HOXA genes and the most frequently altered linked genes, prior to analyzing this gene list using the GO and KEGG tools in Metascape (Figure 5A–C). We found that the following processes were affected by HOXA gene alterations: GO:0048706: embryonic skeletal system development; R-HSA-5617472: activation of HOX genes in anterior hindbrain development during early embryogenesis; GO:0060216: definitive hemopoiesis, hsa05202: transcriptional misregulation in cancer; GO:0030878: thyroid gland development; GO:0045638: negative regulation of myeloid cell differentiation; GO:0002009: morphogenesis of epithelium; GO:0048872: homeostasis of several cell types; and GO:0090596: sensory organ morphogenesis.
Figure 5.
The functions of HOXA genes and genes significantly associated with HOXA genes alterations. (A) Heatmap of the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enriched terms colored by P-values. (B) Network of GO and KEGG enriched terms colored by P-value. (C) Network of GO and KEGG enriched terms colored by clusters.
4. Discussion
HOX genes belong to a family of transcription factor-encoding genes that share a DNA-binding homeobox domain [6]. In addition to their role in determining the characteristics of various body segments during embryogenesis, HOX genes also control the differentiation and regeneration of hematopoietic stem cells and precursor cells. The genes of the HOXA cluster, as well as the smaller HOXB family, are especially highly transcribed in hematopoietic precursor cells, and their expression gradually declines during maturation [28]. Dysregulation of members of the HOXA gene family has been reported in a variety of cancers, including glioblastoma [29], hepatocellular carcinoma [30], prostate cancer [31], gastric cancer [32], epithelial ovarian cancer [33], breast cancer [34], and leukemia [35,36,37,38]. Although the role of HOXA genes in the development of several cancers is well documented, a bioinformatics analysis of this gene family has not been performed in AML patient samples.
Previous research has shown that AML patients displaying favorable cytogenetic characteristics exhibit lower levels of HOX gene expression, whilst poor AML prognosis cases are associated with the upregulation of these genes [39]. HOXA family members were co-expressed with HOXB3, HOXB6, MEIS1, and PBX3 in AML, and the expression levels of these genes were highly correlated [39]. The same results were observed in our study. Furthermore, we found that the expression of HOXA3-10 was elevated in AML samples compared to healthy tissues, and a higher expression of HOXA3-10 was significantly associated with poor OS in AML. The cBioPortal data indicated significant positive correlations between any two HOXA gene members, particularly between any two of the HOXA3-10 genes. In addition, we found that HOXA gene expression was positively correlated with the upregulation of HOXB2, HOXB3, HOXB4, HOXB6, NKX2-3, PBX3, and MEIS1, amongst others. This result was consistent with the aforementioned work of Drabkin et al. [39].
It is worth noting that a previous study identified HOXA4 as the only prognostic gene in AML, the upregulation of which in patients with normal karyotypes was actually associated with longer-term survival [40]. This finding is contrary to our results. However, on closer inspection of the work, we became aware that only 52 of the 126 AML patients (enrolled in the study from 1985 to 1997) were retained for the final analyses. Moreover, the study in question did not consider FLT3 and MLL duplications, stating that the impact of HOXA4 on survival was independent of the white blood cell count and the presence of the FLT3-internal tandem duplication (ITD). In addition to the small sample size, the work of Grubach et al. failed to consider the prognostic stratification of AML due to the limited information available at the time of the study [40]. Moreover, AML treatment regimens have undergone considerable change since the publication of the study in question. In light of this, we adhere to the view that the role of HOXA4 in the survival of AML patients remains unclear and we await the emergence of further large-scale sample studies involving the HOXA gene family.
Numerous studies describe the functional role of the HOXA gene in the pathogenesis of AML. For instance, it has been shown that the KDM3B gene exhibits potential tumor-suppressive activity in AML and it transcriptionally regulates HOXA1 via retinoic acid response elements [41]. A study by Li and colleagues showed that HOXA5 knockout significantly inhibits the proliferation of AML cells, whilst shRNA down-regulation of HOXA5 may induce apoptosis and overcome drug resistance in leukemia cells [42]. In addition, the upregulation of HOXA5 through H3K79 methylation may represent a potential mechanism for leukemia transformation via the CALM-AF10 fusion gene [43].
HOXA7 and HOXA9 appear to be required for efficient in vitro myeloid immortalization via the replacement of leukemia-associated fusion proteins with MLL fusion proteins. The involvement of HOXA7 and HOXA9 in MLL-mediated transformation suggests that the MLL oncoprotein acts as an upstream constitutive activator that promotes myeloid transformation through a HOX-dependent mechanism [44].
Amongst the HOXA genes, HOXA9 is the most studied in AML [35,45,46,47]. Using the RNA interference technique, HOXA9 has been shown to inhibit MLL rearrangement in MLL-rearranged leukemias, suggesting that targeting HOXA9 or its downstream signaling partners may present an effective therapeutic strategy [46]. Similar to HOXA9, HOXA11 has been implicated in leukemia development, exemplified by a patient with juvenile myelomonocytic leukemia (JMML) carrying a NUP98-HOXA11 fusion gene [48]. In addition, a novel NUP98 partner gene for HOXA13, with an expression pattern similar to HOXA9 in leukemic cell lines, implies that the NUP98-HOXA13 fusion protein, like the NUP98-HOXA9 fusion protein, may play a role in leukemia development via the same mechanism [49].
At present, the functional roles of HOXA genes in AML merit further elucidation.
A large body of evidence has shown that AML, like normal hematopoiesis, is initiated and maintained by a subpopulation of leukemic stem cells (LSCs) [50]. Studies have shown that NPM-1, ASXL1, and KAT6A are the most important upstream regulators of LSCs epigenetic marker genes, which mainly regulate HOXA genes, including HOXA5, HOXA6, HOXA7, HOXA9, and HOXA10 [51]. All three of these upstream regulators have been found to be mutated in AML, and may be driving HOXA gene expressions [52]. The work of Jung et al. has implied that the overexpression of HOXA genes may be mediated by DNA hypomethylation, a core mechanism for LSC activity [51]. Another study has demonstrated that NPM1c facilitates HOX/MEIS1 expression and that HOX genes support the leukemic state in NPM1-mutant AML [53].
In this study, we employed GO and KEGG analysis tools to determine how the expression of HOXA genes and the most frequently-altered linked genes were associated with AML initiation and prognosis. We concluded that a few pathways deserved closer attention in the future, including: GO:0048706: embryonic skeletal system development; R-HSA-5617472: activation of HOX genes in anterior hindbrain development during early embryogenesis; GO:0060216: definitive hemopoiesis; hsa05202: transcriptional misregulation in cancer; and GO:0045638: negative regulation of myeloid cell differentiation. The last pathway has attracted our attention.
Bcor, a repressor of HoxA genes in myeloid cells, plays an indispensable role in hematopoiesis by inhibiting myeloid cell proliferation and differentiation [54]. Up-regulation of the HOXA gene in Bcor deletion may at least partially result in prolonged clonal growth of Bcor mutant hematopoietic stem/progenitor cells in replantation experiments [54].
In clinical practice, Venetoclax, a Bcl-2 protein inhibitor of apoptosis, has been approved by the FDA for first-line use in AML, in combination with azacytidine, decitabine, or low-dose cytarabine. Studies have shown that the significant overexpression of specific HOXA gene transcripts was detected in samples with high Bcl-2 inhibitor sensitivity [55]. Bcl-2 inhibition has been shown to be effective in inducing the apoptosis of progenitor cells in patients with high-risk myelodysplastic syndrome and secondary AML [56]. Therefore, the mechanism through which Bcl-2 inhibitors modulate HOXA gene expression deserves further investigation.
5. Conclusions
Our results collectively suggest that the increased expression of HOXA3-10 genes may be associated with the occurrence of AML, and that the increased expression of HOXA3-10 may serve as a molecular marker to identify AML patients with an adverse prognosis. Thus, HOXA3-10 may represent a prognostic marker and potential therapeutic target to improve the diagnosis and treatment of AML.
Author Contributions
Conceptualization, S.-L.C., Z.-Y.Q., and Y.L.; methodology, S.-L.C., Z.-Y.Q., and F.H.; formal analysis, S.-L.C., Z.-Y.Q., and Y.W.; investigation, S.-L.C., Z.-Y.Q., and Y.-J.D.; writing—original draft preparation, S.-L.C. and Z.-Y.Q.; writing—review and editing, S.-L.C., Z.-Y.Q., and Y.L.; supervision, Y.L.; project administration, Y.L.; funding acquisition, Y.L.
Funding
This study was funded by the National Natural Science Foundation of China (NSFC Grant No. 81873428) to Yang Liang.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Tallman M.S., Wang E.S., Altman J.K., Appelbaum F.R., Bhatt V.R., Bixby D., Coutre S.E., De Lima M., Fathi A.T., Fiorella M., et al. Acute Myeloid Leukemia, Version 3.2019, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2019;17:721–749. doi: 10.6004/jnccn.2019.0028. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Pulte D., Redaniel M.T., Jansen L., Brenner H., Jeffreys M. Recent trends in survival of adult patients with acute leukemia: Overall improvements, but persistent and partly increasing disparity in survival of patients from minority groups. Haematologica. 2013;98:222–229. doi: 10.3324/haematol.2012.063602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howlader A.M.N., Krapcho D., Miller A., Brest M., Yu J., Ruhl Z., Tatalovich A., Mariotto D.R., Lewis H.S., Chen E.J., et al. SEER Cancer Statistics Review, 1975–2016. National Cancer Institute; Bethesda, MD, USA: 2019. [(accessed on 16 July 2019)]. Available online: https://seer.cancer.gov/csr/1975_2016. [Google Scholar]
- 5.Patel J.P., Gonen M., Figueroa M.E., Fernandez H., Sun Z., Racevskis J., Van Vlierberghe P., Dolgalev I., Thomas S., Aminova O., et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N. Engl. J. Med. 2012;366:1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rezsohazy R., Saurin A.J., Maurel-Zaffran C., Graba Y. Cellular and molecular insights into HOX protein action. Development. 2015;142:1212–1227. doi: 10.1242/dev.109785. [DOI] [PubMed] [Google Scholar]
- 7.Dunwell T.L., Holland P.W. Diversity of human and mouse homeobox gene expression in development and adult tissues. BMC Dev. Biol. 2016;16:40. doi: 10.1186/s12861-016-0140-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rux D.R., Wellik D.M. HOX genes in the adult skeleton: Novel functions beyond embryonic development. Dev. Dyn. 2017;246:310–317. doi: 10.1002/dvdy.24482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alharbi R.A., Pandha H.S., Simpson G.R., Pettengell R., Poterlowicz K., Thompson A., Harrington K., El-Tanani M., Morgan R. Inhibition of HOX/PBX dimer formation leads to necroptosis in acute myeloid leukemia cells. Oncotarget. 2017;8:89566–89579. doi: 10.18632/oncotarget.20023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kachgal S., Mace K.A., Boudreau N.J. The dual roles of homeobox genes in vascularization and wound healing. Cell Adhes. Migr. 2012;6:457–470. doi: 10.4161/cam.22164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du H., Taylor H.S. The Role of HOX Genes in Female Reproductive Tract Development, Adult Function, and Fertility. Cold Spring Harb. Perspect. Med. 2015;6:a023002. doi: 10.1101/cshperspect.a023002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ernst P., Mabon M., Davidson A.J., Zon L.I., Korsmeyer S.J. An Mll-dependent HOX program drives hematopoietic progenitor expansion. Curr. Biol. 2004;14:2063–2069. doi: 10.1016/j.cub.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 13.Milne T.A., Martin M.E., Brock H.W., Slany R.K., Hess J.L. Leukemogenic MLL fusion proteins bind across a broad region of the HOX a9 locus, promoting transcription and multiple histone modifications. Cancer Res. 2005;65:11367–11374. doi: 10.1158/0008-5472.CAN-05-1041. [DOI] [PubMed] [Google Scholar]
- 14.Dorrance A.M., Liu S., Yuan W., Becknell B., Arnoczky K.J., Guimond M., Strout M.P., Feng L., Nakamura T., Yu L., et al. Mll partial tandem duplication induces aberrant HOX expression in vivo via specific epigenetic alterations. J. Clin. Investig. 2006;116:2707–2716. doi: 10.1172/JCI25546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andreeff M., Ruvolo V., Gadgil S., Zeng C., Coombes K., Chen W., Kornblau S., Baron A.E., Drabkin H.A. HOX expression patterns identify a common signature for favorable AML. Leukemia. 2008;22:2041–2047. doi: 10.1038/leu.2008.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson A., Quinn M.F., Grimwade D., O’Neill C.M., Ahmed M.R., Grimes S., McMullin M.F., Cotter F., Lappin T.R. Global down-regulation of HOX gene expression in PML-RARalpha acute promyelocytic leukemia identified by small-array real-time PCR. Blood. 2003;101:1558–1565. doi: 10.1182/blood.V101.4.1558. [DOI] [PubMed] [Google Scholar]
- 17.Dufour A., Schneider F., Metzeler K.H., Hoster E., Schneider S., Zellmeier E., Benthaus T., Sauerland M.C., Berdel W.E., Buchner T., et al. Acute myeloid leukemia with biallelic CEBPA gene mutations and normal karyotype represents a distinct genetic entity associated with a favorable clinical outcome. J. Clin. Oncol. 2010;28:570–577. doi: 10.1200/JCO.2008.21.6010. [DOI] [PubMed] [Google Scholar]
- 18.Rhodes D.R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D., Barrette T., Pandey A., Chinnaiyan A.M. Oncomine: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. Gepia: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vasaikar S.V., Straub P., Wang J., Zhang B. LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46:D956–D963. doi: 10.1093/nar/gkx1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valk P.J., Verhaak R.G., Beijen M.A., Erpelinck C.A., Barjesteh van Waalwijk van Doorn-Khosrovani S., Boer J.M., Beverloo H.B., Moorhouse M.J., van der Spek P.J., Lowenberg B., et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N. Engl. J. Med. 2004;350:1617–1628. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 24.Choi Y.L., Tsukasaki K., O’Neill M.C., Yamada Y., Onimaru Y., Matsumoto K., Ohashi J., Yamashita Y., Tsutsumi S., Kaneda R., et al. A genomic analysis of adult T-cell leukemia. Oncogene. 2007;26:1245–1255. doi: 10.1038/sj.onc.1209898. [DOI] [PubMed] [Google Scholar]
- 25.Haferlach T., Kohlmann A., Wieczorek L., Basso G., Kronnie G.T., Bene M.C., De Vos J., Hernandez J.M., Hofmann W.K., Mills K.I., et al. Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: Report from the International Microarray Innovations in Leukemia Study Group. J. Clin. Oncol. 2010;28:2529–2537. doi: 10.1200/JCO.2009.23.4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andersson A., Ritz C., Lindgren D., Eden P., Lassen C., Heldrup J., Olofsson T., Rade J., Fontes M., Porwit-Macdonald A., et al. Microarray-based classification of a consecutive series of 121 childhood acute leukemias: Prediction of leukemic and genetic subtype as well as of minimal residual disease status. Leukemia. 2007;21:1198–1203. doi: 10.1038/sj.leu.2404688. [DOI] [PubMed] [Google Scholar]
- 27.Stegmaier K., Ross K.N., Colavito S.A., O’Malley S., Stockwell B.R., Golub T.R. Gene expression-based high-throughput screening (GE-HTS) and application to leukemia differentiation. Nat. Genet. 2004;36:257–263. doi: 10.1038/ng1305. [DOI] [PubMed] [Google Scholar]
- 28.Argiropoulos B., Humphries R.K. HOX genes in hematopoiesis and leukemogenesis. Oncogene. 2007;26:6766–6776. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- 29.Kurscheid S., Bady P., Sciuscio D., Samarzija I., Shay T., Vassallo I., Criekinge W.V., Daniel R.T., van den Bent M.J., Marosi C., et al. Chromosome 7 gain and DNA hypermethylation at the HOXA10 locus are associated with expression of a stem cell related HOX-signature in glioblastoma. Genome Biol. 2015;16:16. doi: 10.1186/s13059-015-0583-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quagliata L., Matter M.S., Piscuoglio S., Arabi L., Ruiz C., Procino A., Kovac M., Moretti F., Makowska Z., Boldanova T., et al. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59:911–923. doi: 10.1002/hep.26740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo Z., Rhie S.K., Lay F.D., Farnham P.J. A Prostate Cancer Risk Element Functions as a Repressive Loop that Regulates HOXA13. Cell Rep. 2017;21:1411–1417. doi: 10.1016/j.celrep.2017.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu D.C., Wang S.S.W., Liu C.J., Wuputra K., Kato K., Lee Y.L., Lin Y.C., Tsai M.H., Ku C.C., Lin W.H., et al. Reprogramming Antagonizes the Oncogenicity of HOXA13-Long Noncoding RNA HOTTIP Axis in Gastric Cancer Cells. Stem Cells. 2017;35:2115–2128. doi: 10.1002/stem.2674. [DOI] [PubMed] [Google Scholar]
- 33.Richards E.J., Permuth-Wey J., Li Y., Chen Y.A., Coppola D., Reid B.M., Lin H.Y., Teer J.K., Berchuck A., Birrer M.J., et al. A functional variant in HOXA11-AS, a novel long non-coding RNA, inhibits the oncogenic phenotype of epithelial ovarian cancer. Oncotarget. 2015;6:34745–34757. doi: 10.18632/oncotarget.5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun M., Song C.X., Huang H., Frankenberger C.A., Sankarasharma D., Gomes S., Chen P., Chen J., Chada K.K., He C., et al. HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis. Proc. Natl. Acad. Sci. USA. 2013;110:9920–9925. doi: 10.1073/pnas.1305172110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shima Y., Yumoto M., Katsumoto T., Kitabayashi I. MLL is essential for NUP98-HOXA9-induced leukemia. Leukemia. 2017;31:2200–2210. doi: 10.1038/leu.2017.62. [DOI] [PubMed] [Google Scholar]
- 36.Spencer D.H., Young M.A., Lamprecht T.L., Helton N.M., Fulton R., O’Laughlin M., Fronick C., Magrini V., Demeter R.T., Miller C.A., et al. Epigenomic analysis of the HOX gene loci reveals mechanisms that may control canonical expression patterns in AML and normal hematopoietic cells. Leukemia. 2015;29:1279–1289. doi: 10.1038/leu.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z., Huang H., Li Y., Jiang X., Chen P., Arnovitz S., Radmacher M.D., Maharry K., Elkahloun A., Yang X., et al. Up-regulation of a HOXA-PBX3 homeobox-gene signature following down-regulation of miR-181 is associated with adverse prognosis in patients with cytogenetically abnormal AML. Blood. 2012;119:2314–2324. doi: 10.1182/blood-2011-10-386235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kok C.H., Brown A.L., Ekert P.G., D’Andrea R.J. Gene expression analysis reveals HOX gene upregulation in trisomy 8 AML. Leukemia. 2010;24:1239–1243. doi: 10.1038/leu.2010.85. [DOI] [PubMed] [Google Scholar]
- 39.Drabkin H.A., Parsy C., Ferguson K., Guilhot F., Lacotte L., Roy L., Zeng C., Baron A., Hunger S.P., Varella-Garcia M., et al. Quantitative HOX expression in chromosomally defined subsets of acute myelogenous leukemia. Leukemia. 2002;16:186–195. doi: 10.1038/sj.leu.2402354. [DOI] [PubMed] [Google Scholar]
- 40.Grubach L., Juhl-Christensen C., Rethmeier A., Olesen L.H., Aggerholm A., Hokland P., Ostergaard M. Gene expression profiling of Polycomb, HOX and Meis genes in patients with acute myeloid leukaemia. Eur. J. Haematol. 2008;81:112–122. doi: 10.1111/j.1600-0609.2008.01083.x. [DOI] [PubMed] [Google Scholar]
- 41.Xu X., Nagel S., Quentmeier H., Wang Z., Pommerenke C., Dirks W.G., Macleod R.A.F., Drexler H.G., Hu Z. KDM3B shows tumor-suppressive activity and transcriptionally regulates HOXA1 through retinoic acid response elements in acute myeloid leukemia. Leuk. Lymphoma. 2018;59:204–213. doi: 10.1080/10428194.2017.1324156. [DOI] [PubMed] [Google Scholar]
- 42.Li N., Jia X., Wang J., Li Y., Xie S. Knockdown of homeobox A5 by small hairpin RNA inhibits proliferation and enhances cytarabine chemosensitivity of acute myeloid leukemia cells. Mol. Med. Rep. 2015;12:6861–6866. doi: 10.3892/mmr.2015.4331. [DOI] [PubMed] [Google Scholar]
- 43.Okada Y., Jiang Q., Lemieux M., Jeannotte L., Su L., Zhang Y. Leukaemic transformation by CALM-AF10 involves upregulation of Hoxa5 by hDOT1L. Nat. Cell Biol. 2006;8:1017–1024. doi: 10.1038/ncb1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dickson G.J., Liberante F.G., Kettyle L.M., O’Hagan K.A., Finnegan D.P., Bullinger L., Geerts D., McMullin M.F., Lappin T.R., Mills K.I., et al. HOXA/PBX3 knockdown impairs growth and sensitizes cytogenetically normal acute myeloid leukemia cells to chemotherapy. Haematologica. 2013;98:1216–1225. doi: 10.3324/haematol.2012.079012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mohr S., Doebele C., Comoglio F., Berg T., Beck J., Bohnenberger H., Alexe G., Corso J., Strobel P., Wachter A., et al. Hoxa9 and Meis1 Cooperatively Induce Addiction to Syk Signaling by Suppressing miR-146a in Acute Myeloid Leukemia. Cancer Cell. 2017;31:549–562. doi: 10.1016/j.ccell.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Faber J., Krivtsov A.V., Stubbs M.C., Wright R., Davis T.N., van den Heuvel-Eibrink M., Zwaan C.M., Kung A.L., Armstrong S.A. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood. 2009;113:2375–2385. doi: 10.1182/blood-2007-09-113597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ogawara Y., Katsumoto T., Aikawa Y., Shima Y., Kagiyama Y., Soga T., Matsunaga H., Seki T., Araki K., Kitabayashi I. IDH2 and NPM1 Mutations Cooperate to Activate Hoxa9/Meis1 and Hypoxia Pathways in Acute Myeloid Leukemia. Cancer Res. 2015;75:2005–2016. doi: 10.1158/0008-5472.CAN-14-2200. [DOI] [PubMed] [Google Scholar]
- 48.Mizoguchi Y., Fujita N., Taki T., Hayashi Y., Hamamoto K. Juvenile myelomonocytic leukemia with t(7;11)(p15;p15) and NUP98-HOXA11 fusion. Am. J. Hematol. 2009;84:295–297. doi: 10.1002/ajh.21373. [DOI] [PubMed] [Google Scholar]
- 49.Taketani T., Taki T., Ono R., Kobayashi Y., Ida K., Hayashi Y. The chromosome translocation t(7;11)(p15;p15) in acute myeloid leukemia results in fusion of the NUP98 gene with a HOXA cluster gene, HOXA13, but not HOXA9. Genes Chromosomes Cancer. 2002;34:437–443. doi: 10.1002/gcc.10077. [DOI] [PubMed] [Google Scholar]
- 50.Majeti R. Monoclonal antibody therapy directed against human acute myeloid leukemia stem cells. Oncogene. 2011;30:1009–1019. doi: 10.1038/onc.2010.511. [DOI] [PubMed] [Google Scholar]
- 51.Jung N., Dai B., Gentles A.J., Majeti R., Feinberg A.P. An LSC epigenetic signature is largely mutation independent and implicates the HOXA cluster in AML pathogenesis. Nat. Commun. 2015;6:8489. doi: 10.1038/ncomms9489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cancer Genome Atlas Research Network. Ley T.J., Miller C., Ding L., Raphael B.J., Mungall A.J., Robertson A., Hoadley K., Triche T.J., Jr., Laird P.W., et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brunetti L., Gundry M.C., Sorcini D., Guzman A.G., Huang Y.H., Ramabadran R., Gionfriddo I., Mezzasoma F., Milano F., Nabet B., et al. Mutant NPM1 Maintains the Leukemic State through HOX Expression. Cancer Cell. 2018;34:499–512. doi: 10.1016/j.ccell.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cao Q., Gearhart M.D., Gery S., Shojaee S., Yang H., Sun H., Lin D.C., Bai J.W., Mead M., Zhao Z., et al. BCOR regulates myeloid cell proliferation and differentiation. Leukemia. 2016;30:1155–1165. doi: 10.1038/leu.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kontro M., Kumar A., Majumder M.M., Eldfors S., Parsons A., Pemovska T., Saarela J., Yadav B., Malani D., Floisand Y., et al. HOX gene expression predicts response to BCL-2 inhibition in acute myeloid leukemia. Leukemia. 2017;31:301–309. doi: 10.1038/leu.2016.222. [DOI] [PubMed] [Google Scholar]
- 56.Jilg S., Reidel V., Muller-Thomas C., Konig J., Schauwecker J., Hockendorf U., Huberle C., Gorka O., Schmidt B., Burgkart R., et al. Blockade of BCL-2 proteins efficiently induces apoptosis in progenitor cells of high-risk myelodysplastic syndromes patients. Leukemia. 2016;30:112–123. doi: 10.1038/leu.2015.179. [DOI] [PubMed] [Google Scholar]