Abstract
Binge eating (BE) is a heritable trait associated with eating disorders and involves episodes of rapid, large amounts of food consumption. We previously identified cytoplasmic FMR1-interacting protein 2 (Cyfip2) as a genetic factor underlying compulsive-like BE in mice. CYFIP2 is a homolog of CYFIP1 which is one of four paternally-deleted genes in patients with Type I Prader-Willi Syndrome (PWS), a neurodevelopmental disorder whereby 70% of cases involve paternal 15q11-q13 deletion. PWS symptoms include hyperphagia, obesity (if untreated), cognitive deficits, and obsessive-compulsive behaviors. We tested whether Cyfip1 haploinsufficiency (+/−) would enhance compulsive-like behavior and palatable food (PF) intake in a parental origin- and sex-dependent manner on two Cyfip2 genetic backgrounds, including the BE-prone C57BL/6N (Cyfip2N/N) background and the BE-resistant C57BL/6J (Cyfip2J/J) background. Cyfip1+/− mice showed increased compulsive-like behavior on both backgrounds and increased PF intake on the Cyfip2N/N background. In contrast, maternal Cyfip1 haploinsufficiency on the BE-resistant Cyfip2J/J background induced a robust escalation in PF intake in wild-type Cyfip1J/J males while having no effect in Cyfip1J/- males. Notably, induction of behavioral phenotypes in wild-type males following maternal Fmr1+/− has previously been reported. In the hypothalamus, there was a paternally-enhanced reduction in CYFIP1 protein whereas in the nucleus accumbens, there was a maternally-enhanced reduction in CYFIP1 protein. Nochange in FMR1 protein (FMRP) was observed in Cyfip1+/− mice, regardless of parental origin. To summarize, Cyfip1 haploinsufficiency increased compulsive-like behavior and induced genetic background-dependent, sex-dependent, and parent-of-origin-dependent effects on PF consumption and CYFIP1 expression that could have relevance for neurodevelopmental and neuropsychiatric disorders.
Keywords: C57BL/6 substrains, binge eating disorder, overeating, anorexia nervosa, Fragile X, FMRP, psychiatric genetics, addiction genetics, neuropsychiatric, Genetics of Sex
Binge eating (BE) refers to the rapid consumption of large quantities of food and is accompanied by feelings of loss of control. Binge eating disorder (BED) is a psychiatric disorder with a lifetime prevalence of 3.5% in women and 2% in men (Hudson et al. 2007). Both BED (Mitchell et al. 2010) and BE are heritable (Bulik et al. 2003). However, genome-wide association studies have yet to identify genetic risk factors associated with BE (Yilmaz et al. 2015). The first genome-wide significant loci were recently identified for anorexia nervosa (restricted eating) (Hinney et al. 2017) and bipolar disorder with BE behavior (PRR5-ARHGAP8) (McElroy et al. 2018). Additional genome-wide significant loci will likely soon be uncovered for BE-associated disorders with increasing sample sizes and power (Huckins et al. 2018).
We used quantitative trait locus (QTL) mapping and gene knockout in C57BL/6 mouse substrains to identify cytoplasmic FMR1-interacting protein 2 (Cyfip2) as a major genetic factor underlying BE and compulsive-like behaviors (Kirkpatrick et al. 2017). The QTL capturing increased palatable food (PF) intake mapped to a single missense mutation in Cyfip2 in the C57BL/6N strain (S968F; “Cyfip2M1N”) that is hypothesized to act as a gain-of-function mutation (Kumar et al. 2013). Accordingly, mice with one copy of a null allele and one copy of the missense allele of Cyfip2 showed a reduction in BE toward the phenotypic direction of the wild-type C57BL/6J level (Kirkpatrick et al. 2017). This same missense SNP in Cyfip2 was first associated with reduced behavioral sensitivity to cocaine (Kumar et al. 2013), which could indicate a common neurobiological mechanism involving synaptic plasticity within the mesocorticolimbic dopamine reward pathway (Bello and Hajnal. 2010; Berridge. 2009) that affects the hedonic component of PF consumption (DiLeone et al. 2012; Lutter and Nestler 2009).
Cyfip2 and the gene homolog Cyfip1 code for proteins that interact with the RNA binding protein Fragile X Mental Retardation Protein (FMRP) and are part of the canonical WAVE regulatory complex and transduce activity-dependent Rac signaling in regulating actin dynamics during neuronal development and synaptic plasticity (Abekhoukh and Bardoni. 2014). CYFIP1 expression is necessary for the maintenance and stabilization of neuronal dendritic arborization and morphological complexity (Pathania et al. 2014). In humans, CYFIP1 resides within a non-imprinted region on chromosome 15 (15q11.2) that contains four genes TUBGCP5, NIPA1, NIPA2, and CYFIP1 (Bittel et al. 2006). The syntenic region in mice is located on chromosome 7C (55.4 Mb - 56 Mb). Preclinical models of Cyfip1 haploinsufficiency demonstrate perturbations in synaptic activity during neural development, activity-dependent plasticity, dendritic morphology, and fear learning (Bozdagi et al. 2012; Chung et al. 2015; Hsiao et al. 2016; Oguro-Ando et al. 2015). Haploinsufficiency of 15q11.2 underlies Microdeletion Syndrome (MDS; a.k.a. Burnside-Butler Syndrome) which can comprise developmental delay (speech, motor), reduced cognitive function, dysmorphic features, intellectual disability, autism, ADHD, obsessive-compulsive disorder, and schizophrenia (Cox and Butler. 2015). One case study of 15q11.2 MDS reported hypotonia, increased food craving and obesity, and obsessive-compulsive disorder (Doornbos et al. 2009). CYFIP1 haploinsufficiency is implicated in multiple symptoms of 15q11.2 MDS and a new study demonstrates parent-of origin effects the microdeletion on the distribution of clinical features (Davis et al. 2019). Converse to microdeletion, microduplication of 15q11.2 containing CYFIP1 was recently associated anorexia nervosa (Chang et al. 2019).
The 15q11.2 region is also paternally-deleted in a subset of individuals with a more severe form (Type I) of Prader-Willi Syndrome (PWS), a neurodevelopmental disorder defined genetically by paternal deletion of 15q11-q13 in a majority of cases (Angulo et al. 2015). Extreme hyperphagia due to lack of satiety is the most defining and debilitating feature of PWS and emerges during childhood, leading to obesity if left untreated. Food-related obsessive-compulsive (OC) behaviors are common in PWS; however, OC symptoms unrelated to food are also frequent (State et al. 1999), and include repetitive, ritualistic behaviors, perseverative speech, counting, adaptive impairment, need to tell, ask, or know, ordering and arranging, repeating rituals, and self-mutilation (Dykens et al. 1996; Feurer et al. 1998; Stein et al. 1994). Genetic deletion in PWS involves either the shorter paternal deletion (Type II) of 15q11-q13 or a larger, paternal Type I deletion that also includes the same 15q11.2 MDS region comprising TUBGCP5, NIPA1, NIPA2, and CYFIP1 (Bittel et al. 2006; Butler et al. 2004). Type I PWS is associated with reduced transcription of these genes and a more severe neurodevelopmental and neuropsychiatric profile, including reduced cognition, increased risk of autism and schizophrenia, and increased severity and lack of control over OC behaviors (e.g., grooming and bathing, arranging objects, object hoarding, checking) that interfere with social functioning (Bittel et al. 2006; Butler et al. 2004; Doornbos et al. 2009; Milner et al. 2005; Zarcone et al. 2007).
Decreased CYFIP1 expression is also implicated in the Prader-Willi Phenotype (PWP) of a subset of individuals with Fragile-X Syndrome (FXS). FXS is the most common genetic cause of intellectual disability and autism and is caused by a CGG trinucleotide repeat expansion within the fragile X mental retardation 1 (FMR1) gene that is located on the X chromosome and codes for FMRP, a major interacting protein of CYFIP proteins (Schenck et al. 2001). Interestingly, 10% of FXS individuals also exhibit a PWP in the absence structural or imprinting differences in 15q11-q13. The PWP includes hallmark hyperphagia, lack of satiation, obesity, and more severe behavioral problems, such as OC behaviors and an increased rate of autism (Muzar et al. 2016; Nowicki et al. 2007). The cause of the PWP is unknown, although one logical candidate gene is CYFIP1, given its association with PWS and its interaction with FMRP (Schenck et al. 2001). PWP-presenting individuals with FXS show a two-to fourfold decrease in CYFIP1 transcription in peripheral blood mononuclear cells compared to FXS individuals without PWP (Nowicki et al. 2007). There was also a twofold decrease in Cyfip1 gene transcription in a mouse model of FXS (Stefan et al. 2005).
We previously mapped the gene homolog Cyfip2 in BE (Kirkpatrick et al. 2017). Because CYFIP1 deletion and reduced CYFIP1 expression are associated with more severe PWS (Type I) and hyperphagia in the PWP (FXS) and conversely, because gene duplication and thus, increased expression of CYFIP1 are associated with restrictive eating (Chang et al. 2019), in this study, we tested the hypothesis that Cyfip1 haploinsufficiency would increase premorbid compulsive-like behavior and increase consumption of palatable food (PF) in our BE paradigm (Babbs et al. 2018; Goldberg et al. 2017; Kirkpatrick et al. 2017). We tested the effect of Cyfip1 haploinsufficiency on two different Cyfip2 genetic backgrounds. Additionally, because a recent preclinical study demonstrated a parental origin (PO) effect of Cyfip1 haploinsufficiency on hippocampal synaptic transmission, learning, and anxiety-like behavior (Chung et al. 2015) and because a recent clinical study indicated an effect of PO on the distribution of clinical features in 15q11.2 MDS (Burnside-Butler Syndrome) (Davis et al. 2019), we tested whether there would be an effect of PO of Cyfip1 deletion on compulsive-like behavior and PF intake.
To gain insight into the molecular mechanisms associated with PO effects of Cyfip1 deletion on PF intake, we examined transcription of Cyfip1, Cyfip2, and Magel2 - a nearby imprinted gene within the syntenic, canonical PWS region that is implicated in hyperphagia and obesity (Tacer and Potts. 2017). Additionally, we examined protein expression of CYFIP1 and its interacting partner FMRP as a function of both Cyfip1 genotype and PO in two brain regions, including the hypothalamus which is critical for homeostatic regulation of feeding and the nucleus accumbens which is critical for hedonic aspects of food intake (DiLeone et al. 2012; Lutter and Nestler. 2009). Finally, because OC behaviors are associated with BE (Kessler et al. 2016; Moore et al. 2017; Wilfley et al. 2016) and hyperphagia in PWS (Griggs et al. 2015), we employed a battery of tests to assess anxiety-like and compulsive-like behaviors and post-BE training behaviors in Cyfip1 haploinsufficient mice, including compulsive-like eating and concomitant behaviors in the light/dark conflict test (Babbs et al. 2018; Kirkpatrick et al. 2017).
Materials And Methods
Mice
All experiments were performed in accordance with the National Institutes of Health Guidelines for the Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee at Boston University. Mice were 50-100 days old at the first day of testing. A minimum sample size of N = 20 per Genotype per Treatment was employed for behavioral studies based on power analysis of PF intake from the Cyfip2 study (Kirkpatrick et al. 2017) (see Supplementary Material for additional details on power analyses). Mice heterozygous for a null deletion in exons 4 through 6 of Cyfip1 (Cyfip1+/−) were generated by the International Knockout Mouse Consortium on C57BL/6N background (Skarnes et al. 2011). We propagated these mice on two different C57BL/6 genetic backgrounds, including the BE-prone isogenic C57BL6/N background or on a mixed background C57BL/6J / C57BL/6N background whereby mice were backcrossed to C57BL/6J to be homozygous for the BE-resistant C57BL6/J allele at the Cyfip2 locus. Additional details regarding mouse breeding and genotyping of Cyfip1 and Cyfip2 are provided in the Supplementary Material.
Premorbid anxiety-like and compulsive-like behavioral battery
Because of the link between anxiety, compulsivity and pathological overeating (Moore et al. 2017) and because OC behavior is associated with eating disorders (Cavallini et al. 2000; Micali et al. 2011), we incorporated a behavioral battery to assess differences in premorbid anxiety-like and compulsive-like behaviors in experimentally naïve, Cyfip1+/− mice. Mice were tested in the behavioral battery and were either killed afterward (all mice on Cyfip1,2N/N background) or were subsequently trained for BE (a subset of mice on the Cyfip1,2J/J background). Mice were assayed in the battery with one test per day over five days in the following order: 1) open field; 2) elevated plus maze; 3) marble burying; 4) hole board; 5) mist-induced grooming. The rationale and procedural details for each behavioral test are provided in the Supplementary Material. Testing was conducted between 0800 and 1300 h. The experimenters responsible for running the mice, video tracking, data curation, and analysis were blinded to Genotype for each cohort.
BE and light/dark conflict test of compulsive-like eating
Mice were trained in an intermittent, limited access, conditioned place preference (CPP) procedure to detect genetic differences in BE (Babbs et al. 2018; Goldberg et al. 2017; Kirkpatrick et al. 2017). All mice on the Cyfip1,2N/N background were experimentally naïve prior to BE training. For mice on the Cyfip1,2J/J background, approximately one-half of the total sample size had previously undergone testing in the five-day behavioral battery (described above) prior to commencement of BE training. Effects of prior battery testing on food intake are described below in the Results section.
Initial locomotor activity was also quantified on Day (D)1 prior to BE training. For details on the BE protocol, see Supplementary Material. Briefly, mice were tested for side preference on D1 and D22. During the intervening three weeks, mice were confined to a food-paired and non-food-paired side on alternating days (Tuesday through Friday). Cages were assigned to either the PF or Chow group in a counterbalanced design in order to ensure equal distribution across Sex, Genotype, Treatment, and PO. On D23, mice were assessed for compulsive eating and associated behaviors, as previously described (Babbs et al. 2018; Kirkpatrick et al. 2017). (Supplementary Material). The experimenters responsible for running the mice, video tracking, data curation, and analysis were blinded to Genotype for each cohort.
Hypothalamus dissections for real-time quantitative PCR (qPCR)
We chose a subset of Chow-trained, PF-naive mice (n = 7-9 per Genotype per PO; both sexes) on the Cyfip1,2N/N background or untrained, naïve mice on a Cyfip1,2J/J background (n = 8-12 per Genotype per PO; both sexes) to examine baseline (PF-naive) gene transcription between Cyfip1N/- vs. Cyfip1N/N mice and PO effects. We examined Cyfip1, Cyfip2, and Magel2 transcript levels in the hypothalamus, a brain region important for hyperphagia in PWS (Griggs et al. 2015).
On D24, brains from Chow-trained mice (Cyfip1,2N/N background) were harvested and the hypothalamus was free form dissected by pinching the entire structure from the ventral surface with forceps while using the anterior commissure and mammillary bodies as landmarks. Tissue was stored in RNAlater Solution (Invitrogen, Carlsbad, CA USA) at 4°. After five days, the tissue was dried and transferred to a -80° freezer.
Real-time quantitative PCR (qPCR)
Total RNA from hypothalamus was extracted and processed for qPCR as described (Goldberg et al. 2017; Kirkpatrick et al. 2017; Yazdani et al. 2015). Briefly, oligo-dT primers were used to synthesize cDNA. PCR reactions were conducted on the StepOne Plus 96-Well Real-Time PCR machine (Life Technologies, Foster City, CA, USA) in technical triplicates and averaged (SD < 0.5). Plates were balanced across Genotype, PO, and Sex. We report the difference in expression in Cyfip1+/− relative to Cyfip1+/+ using the 2-(∆∆CT) method (Schmittgen and Livak. 2008). Primer sequences are provided in the Supplementary Material. All qPCR samples analyzed on the Cyfip1,2N/N background were from Chow-trained mice. All qPCR samples analyzed on the Cyfip1,2J/J genetic background were from experimentally naïve mice.
Immunoblot analysis of CYFIP1 and FMRP From hypothalamus and nucleus accumbens
Hypothalamus was dissected as described above. Nucleus accumbens punches were harvested using 1.2 mm-diameter punches of ventral forebrain centered around anterior commissure from the first 4 mm of brain section in a brain matrix. Samples were processed and analyzed for quantity of CYFIP1 and FMRP proteins. A majority of the tissue samples for immunoblot analysis were collected from PF-trained mice. In addition to the collapsed analysis across PF and Chow samples that we report below, we conducted a separate analysis that excluded the Chow-trained samples and obtained qualitatively the same trending results for CYFIP1 and the same null results for FMRP (also described below). Thus, the addition of Chow samples improved our statistical power without confounding the results. Detailed methods can be found in the Supplementary Material.
Analysis
Statistical analyses were conducted using R (https://www.r.project.org). For the compulsive-like and anxiety-like behavioral tests, two-tailed unpaired t-tests were used to detect genotypic differences for all behaviors except marble burying behaviors which were also analyzed by non-parametric Mann-Whitney U-tests.
Food intake was analyzed using various mixed model ANOVAs that included Genotype, Treatment, Sex, and/or PO as independent variables as indicated in each section below, and Day as a repeated measure using the “aov” function in R. Subsequent follow-up ANOVAs and t-tests were run to determine the source of various interactions among these variables as indicated below. To address issues of non-normality or unequal variance, we included additional, select non-parametric analyses to support key findings that support our conclusions (Supplementary Material).
Slope analyses of food intake across days were calculated to detect escalation in consumption as a measure of BE-like behavior (Babbs et al. 2012; Babbs et al. 2018; Kirkpatrick et al. 2017) using GraphPad Prism 7 (GraphPad Software, La Jolla, CA USA). Linear regression was employed to fit a line for each group, to calculate the slope and y-intercept, and to determine whether each slope was significantly different from zero. Group differences in slopes were detected using Analysis of Covariance (ANCOVA) and post hoc pairwise, Bonferroni-adjusted comparisons. Slope of escalation in food intake was included as an additional analysis in order to represent the degree of escalation of food consumption over time. A positive (non-zero) slope indicates significant escalation, whereas no slope indicates no escalation. The y-intercept is affected by both initial consumption and the degree of stability in average consumption over time. For example, a slope that was not significantly different from zero could be explained by greater initial consumption for the early training trials that was maintained at a similar over time. Significant differences in y-intercepts were cross-referenced with the daily intake values to interpret the results.
Data Availability
The authors affirm that all data necessary for confirming the conclusions of this article are represented fully within the article and its tables and figures. Supplemental material available at Figshare: https://doi.org/10.25387/g3.8316635.
Results
Cyfip1 haploinsufficiency increases compulsive-like behaviors
Sample sizes are listed in Supplementary Table 1. Figure 1 illustrates the breeding scheme for generating Cyfip1+/− mice on two Cyfip1,2 genetic backgrounds as well as the breeding scheme and annotations of the offspring derived from the bidirectional, parent-of-origin crosses. The full set of statistical results, including F statistics, p-values, and slope analyses are provided in the Figure Legends. Specific descriptions of each ANOVA model as well as p-values for main effects, interactions, and post-hoc group comparisons that are relevant to the conclusions are provided in the main text of the Results section below.
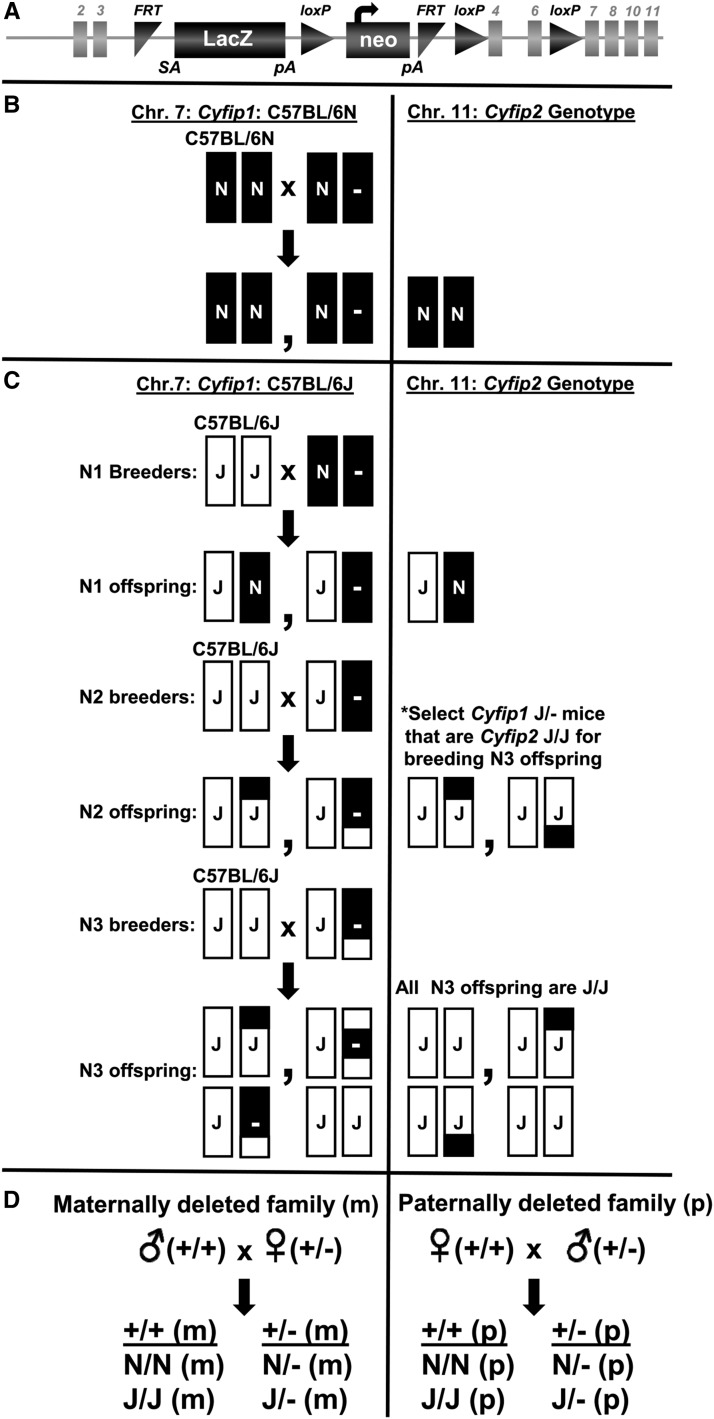
Figure 1.
Generation of the Cyfip1 knockout allele and breeding scheme for Cyfip1 haploinsufficient mice on the Cyfip1,2N/N and Cyfip1,2J/J genetic backgrounds. (A): A schematic of the knockout first allele for KOMP generation of Cyfip1N/- mice was obtained from the International Mouse Phenotyping Consortium (IMPC) website (http://www.mousephenotype.org/data/alleles/MGI:1338801/tm2a(EUCOMM)Wtsi). Mice containing floxed alleles flanking exons 4 through 6 were generated from embryonic stem cells on a C57BL/6N background by the International Knockout Mouse Consortium and were crossed to global Cre-expressing mice, yielding offspring heterozygous for constitutive deletions in exons 4 through 6. Mice heterozygous for the null deletion on a C57BL/6N background were re-derived using sperm obtained from The Jackson Laboratory. (B): Left panel: In the first study, we re-derived Cyfip1N/- and propagated mice on an isogenic C57BL/6N background. Right panel: All mice were homozygous for the N allele (N/N) at Cyfip2 which contains a missense mutation that we previously showed was associated with a marked enhancement of binge eating (BE), accounting for one-third of the genetic variance in parental strain BE (Kirkpatrick et al. 2017). We maintained this colony on an isogenic C57BL/6N background by breeding Cyfip1N/- mice with C57BL/6NJ mice (black bars; N/N) ordered from The Jackson laboratory. (C): In the second study, we generated another colony on a mixed background. The primary goal was to monitor and replace the BE-associated N/N Cyfip2 alleles with C57BL/6J (J/J) alleles via backcrossing Cyfip1N/- mice to C57BL/6J (white bars; J/J) for three and four generations and assess the effect of Cyfip1 deletion on BE on a mixed N3 and N4 background containing a fixed, BE-resistant, homozygous J/J genotype at Cyfip2 (Kirkpatrick et al. 2017). Mixed-color bars illustrate hypothetical recombination events that accumulate through repeated backcrossing to C57BL/6J (white). (D): Schematic of the bidirectional, parent-of-origin crosses for generating wild-type (+/+) and heterozygous (+/−) offspring from either the paternally (p) deleted Cyfip1 families or the maternally (m) deleted Cyfip1 families. There are eight possible annotations, including four on the Cyfip1,2N/N background and four on the Cyfip1,2J/J background. N/N (m): Wild-type offspring from a maternally deleted family on a Cyfip1,2 N/N background; N/- (m): Heterozygous offspring from a maternally deleted family on a Cyfip1,2 N/N background; N/N (p): Wild-type offspring from a paternally deleted family on a Cyfip1,2 N/N background; N/- (p): Heterozygous offspring from paternally deleted family on a Cyfip1,2 N/N background; J/J (m): Wild-type offspring from a maternally deleted family on a Cyfip1,2 J/J background; J/- (m): Heterozygous offspring from a maternally deleted family on a Cyfip1,2 J/J background; J/J (p): Wild-type offspring from paternally deleted family on a Cyfip1,2 J/J background; J/- (p): Heterozygous offspring from a paternally deleted family on a Cyfip1,2 J/J background.
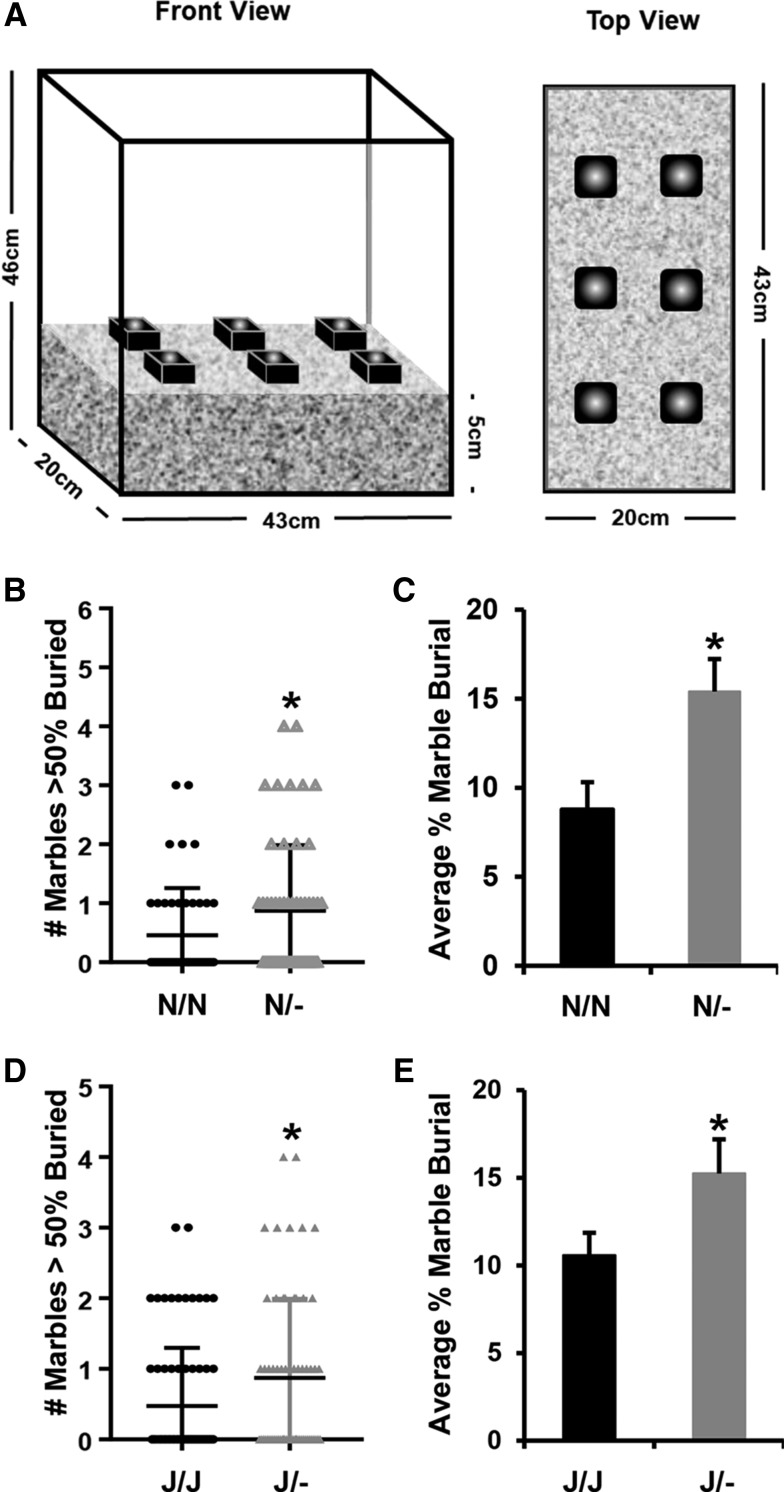
In the marble burying test, Cyfip1N/- mice on the Cyfip1,2N/N genetic background showed a greater number of marbles that were at least 50% buried (Mann-Whitney: *P = 0.031). Furthermore, two-way ANOVA (Genotype, PO) identified a main effect of Genotype (*P = 0.009), indicating a greater average percentage of marbles buried than Cyfip1N/N mice (Figure 2A-C). We replicated these results in Cyfip1J/- on the Cyfip1,2J/J background (Mann-Whitney: *P = 0.019; Effect of Genotype: P = *0.042, respectively; Figure 2D,E). If we collapse across genetic background and run either a Mann-Whitney U-test for number of marbles that were greater than 50% buried or an unpaired t-test for average percent marble burial across the six marbles in Cyfip1+/− vs. Cyfip1+/+ mice and we employ a corrected p-value for statistical significance that accounts for the 14 phenotypes across all five behavioral assays within the compulsive-like battery (P < 0.05/14 = 0.0036), the result is still statistically significant for both phenotypes [U(235) = 5787; P = 0.0014; t(241) = 3.31; P = 0.0011 respectively]. Furthermore, the effect of Genotype in the ANOVA model of the collapsed data for average percent marble burial also survives the multiple correction procedure [F(1,238) = 10.9; P = 0.0011]. Finally, the increase in marble burying in Cyfip1+/− mice cannot be explained by a genotypic difference in locomotor activity as there was no significant difference in distance traveled in Cyfip1+/− vs. Cyfip1+/+ mice [collapsed across genetic backgrounds: t(241) = 0.79; P = 0.43].
Figure 2.
Increase in premorbid, OC-like marble burying in Cyfip1N/- and Cyfip1J/- mice. (A): Schematic of the marble burying apparatus. (B,C): Cyfip1N/- mice buried more marbles with greater than 50% coverage than wild-type Cyfip1N/N mice [B: U(102) = 1050; *P = 0.031; two-tailed], and had a greater average percent marble burial across the six marbles [C: effect of Genotype: F(1,96) = 7.1; *P = 0.009; no effect of PO and no Genotype x PO interaction; p’s > 0.05]. (D,E): Similarly, Cyfip1J/- mice buried more marbles with greater than 50% coverage than Cyfip1J/J mice [D: U(137) = 1884; *P = 0.019, two-tailed], and also had a greater average percent marble burial across the six marbles [E: Effect of Genotype: F(1,134) = 4.2; *P = 0.042; no effect of PO and no Genotype x PO interaction; p’s > 0.05]. Data are presented as the mean ± SEM.
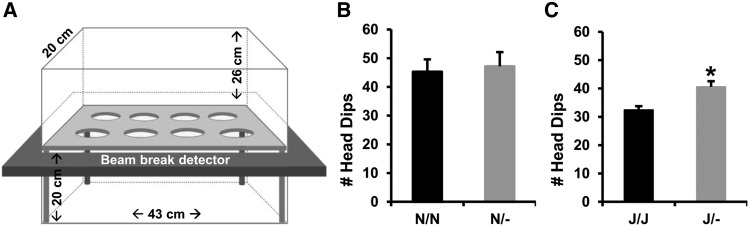
For the most part, Cyfip1 deletion did not induce a statistically significant change in any other behaviors within the battery (Supplementary Table 2, t-tests: all ps > 0.05), including mist spray-induced grooming (p’s > 0.18; Supplementary Table 2). The only exception was that on the Cyfip1,2J/J background, Cyfip1J/- showed a greater number of total head dips in the hole board test compared to Cyfip1J/J (t-test: P = 0.03; Figure 3), further supporting increased compulsive-like behaviors as a result of Cyfip1 haploinsufficiency. The increase in head dips in Cyfip1J/- mice cannot be explained by an overall increase in locomotor activity as an ANOVA model (Genotype and PO as factors) indicated that there was no significant effect of Genotype on distance traveled [F(1,134) = 3.44; P = 0.066] and despite the fact that Cyfip1J/- mice showed a greater number of head dips than their Cyfip1J/J counterparts, they actually tended to show less locomotor activity [8.49 +/− 0.30 m (SEM) vs. 9.23 +/− 0.27 m (SEM), respectively]. Note that the significant increase in head dips in Cyfip1J/- mice is not statistically significant if one employs a Bonferroni-corrected p-value of 0.05/14 (P < 0.0036) to account for the 14 statistical tests across the five behaviors within the compulsive-/anxiety-like battery (12 phenotypes in Supplementary Table 2 plus the two marble burying phenotypes in Figure 2).
Figure 3.
Hole board behavior in Cyfip1N/- and Cyfip1J/- mice. (A): Cartoon and dimensions of the hole board test. The holes (2 × 4) were 2.54 cm in diameter and were spaced center-to-center 10.16 cm apart. Head dips were detected via beam breaks (for additional details, see Supplementary Material). (B): In examining premorbid compulsive-like behavior in the hole board test, for mice on the Cyfip1,2N/N background, Cyfip1N/- mice did not differ from wild-type Cyfip1N/N mice [t(102) = 0.29; P = 0.78]. (C): For mice on the Cyfip1,2J/J background, Cyfip1J/- mice showed a significantly greater number of head dips than Cyfip1J/J mice [t(136) = 2.2; *P = 0.03]. Note that the significant increase in the number of head dips in Cyfip1J/- mice does not survive the cut-off for significance if one corrects for all 14 phenotypes in the five-day behavioral battery (P < 0.0036; 12 phenotypes in Supplementary Table 2 plus two marble burying phenotypes in Fig. 2).
There was no main effect of PO or interaction with Cyfip1 Genotype on marble burying or any other behaviors within the battery (data not shown). Furthermore, there were no significant genotypic differences in any of the other behaviors within the battery (Supplementary Table 2). To summarize, Cyfip1 haploinsufficiency induced a selective increase in compulsive-like marble burying regardless of the genetic background or PO as well as an increase in compulsive-like head-dipping in the hole board test that was observed only on the Cyfip1,2J/J genetic background. The lack of effect of Cyfip1 haploinsufficiency on other OC-related behaviors such as mist spray-induced grooming could reflect differences in specific cell types and neural circuitry underlying complex repetitive action patterns for burying vs., e.g., grooming that are perturbed by Cyfip1 haploinsufficiency (Kim et al. 2016).
Effect of Cyfip1 haploinsufficiency on PF intake depends on Cyfip2 genetic background
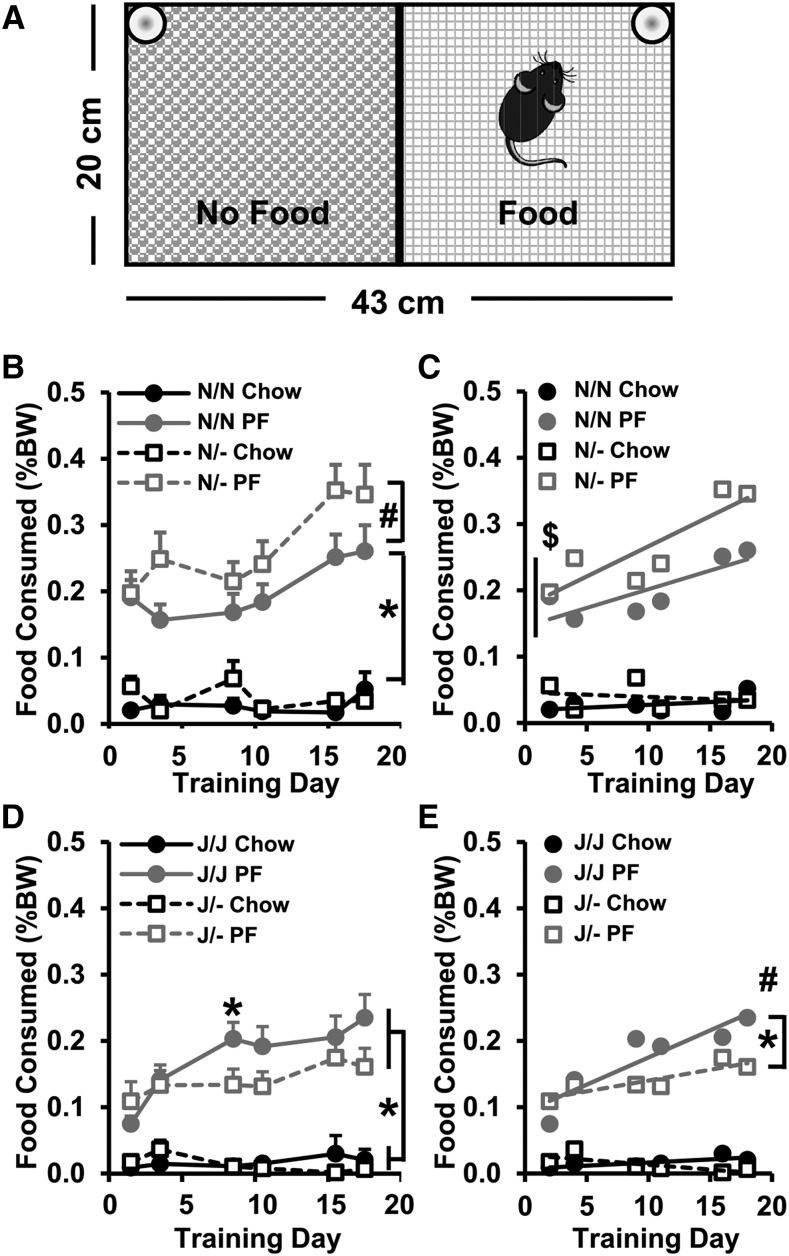
In testing the hypothesis that Cyfip1 haploinsufficiency would increase PF intake in our intermittent, limited access BE and CPP paradigm (Figure 4A), Mixed-effects ANOVA (Factors: Genotype, Treatment, Sex; repeated measure: Day) indicated that PF-trained mice on the Cyfip1,2N/N background consumed significantly more food than Chow-trained mice (Figure 4B; effect of Treatment on intake [PF groups vs. Chow groups: *P < 2x10−16] – the Treatment effect was also reflected by slopes of escalation that were significantly greater than zero in the PF-trained groups (slopes vs. zero: P = 0.024-0.045; Figure 4C) but not in the Chow-trained groups. As predicted, Cyfip1N/- mice consumed more PF than Cyfip1N/N mice (Genotype x Treatment interaction; P = 0.03; t-test for summed PF intake: #P = 0.04; Figure 4B) with no difference in Chow intake [t-test: P = 0.32]. There was also a main effect of Day [P = 6.4 × 10-5] and a Treatment x Day interaction [P = 0.005]. Finally, PF-trained Cyfip1N/- mice showed a greater y-intercept than all three other groups ($p’s < 0.008 vs. each of the three groups; Figure 4C), indicating an initial higher level of consumption during early training days that persisted throughout the study.
Figure 4.
PF consumption in Cyfip1N/- and Cyfip1J/- mice. (A): The conditioned place preference (CPP) chamber that was used for food consumption training had a smooth-textured non-food-paired side (left) and a rough-textured food-paired side. (B): Both wild-type Cyfip1N/N and Cyfip1N/- mice trained with PF in the CPP chamber ate more food over time than Chow-trained mice [main effect of Treatment: F(1,932) = 274.7; *P < 2x10−16]. There was also a main effect of Day [F(5,932) = 16.1; P = 6.4 × 10-5], Sex [F(1,932) = 30.4; P = 4.5 × 10−8], a Treatment x Day interaction [F(5,932) = 7.9; P = 0.005], a Genotype x Treatment interaction [F(1,932) = 4.7; P = 0.03], and a Treatment x Sex interaction [F(1,932) = 22.3; P = 2.7 × 10−6]. Cyfip1N/- mice consumed more PF overall than Cyfip1N/N mice [summed intake across days: t(76) = 2.1; #P = 0.04; Fig.4B) but not Chow [summed intake across days: t(78) = 1.0; P = 0.32]. (C): Both PF-trained genotypes exhibited slopes that were significantly greater than zero (Cyfip1N/N: m = 0.009 ± 0.003, P = 0.024; Cyfip1N/-: m = 0.005 ± 0.002, P = 0.045, respectively, indicating escalation in PF intake over time. Moreover, PF-trained Cyfip1N/- mice showed a significantly greater y-intercept than all three other groups ($p’s < 0.008 vs. each of the three groups), indicating consistently greater overall food consumption throughout the study. (D): When examining the same behaviors in Cyfip1J/- vs. Cyfip1J/J mice, there was a main effect of Treatment [F(1,978) = 191.1; *P = 2 × 10−16], indicating that PF-trained mice consumed more food over time. There was also a main effect of Genotype [F(1,978) = 5.4; P = 0.02], Day [F(5,978) = 4.0; P = 0.001], and a Treatment x Day interaction [F(5,978) = 2.6; P = 0.02] but in contrast to Cyfip1 N/- mice, Cyfip1J/- consumed less food than their wild-type Cyfip1J/J counterparts. There was a significant increase in PF intake on Day (D)9 in Cyfip1J/J vs. Cyfip1J/- mice [t(106) = 2.0; *P = 0.047]. Additionally, there was a main effect of Sex [F(1,978) = 15.7, P = 8.2 × 10−5], a Treatment x Sex interaction [F(1,978) = 6.0; P = 0.01], a Genotype x Sex interaction [F(1,978) = 4.9; P = 0.03], and most importantly, a Genotype x Treatment x Sex interaction [F(1,978) = 10.9; P = 0.001]. Follow-up sex-specific analyses for mice in the Cyfip1,2J/J background are provided in Fig.5. (E): In examining escalation in food intake across days, only PF-trained Cyfip1J/J mice exhibited a slope significantly greater than zero [F(1,328) = 19.7; #P < 0.0001]. The slope value of Cyfip1J/J mice was also greater than Cyfip1J/- mice [F(1,644) = 4.0; *P = 0.046] and further supports reduced food intake in Cyfip1J/- mice. Data are presented as mean ± SEM.
In examining the effect of Cyfip1 haploinsufficiency on food intake on the Cyfip1,2J/J genetic background, as expected (Babbs et al. 2018; Goldberg et al. 2017; Kirkpatrick et al. 2017), there was less overall PF intake in mice on the Cyfip1,2J/J background compared to the Cyfip1,2N/N background [summed PF intake of mice on the Cyfip1,2J/J vs. Cyfip1,2N/N background: t(186) = 3.8; P = 0.001; not shown graphically but compare the two PF groups in Figure 4D vs. Figure 4B]. Furthermore, mixed-effects ANOVA of mice on the Cyfip1,2J/J background (Factors: Genotype, Treatment, Sex; Repeated measure: Day) revealed that PF-trained mice showed greater intake than Chow-trained mice (effect of Treatment: *P = 2 × 10−16; Figure 4D). There was also a main effect of Genotype (P = 0.02), Day [P = 0.001) and a Treatment x Day interaction (P = 0.02) that were in part explained by a significant increase in PF intake on D9 in Cyfip1J/J vs. Cyfip1J/- mice [*P = 0.047; Figure 4D). Slope analysis identified a significant, BE-like slope in escalation of PF intake relative to zero in wildtype Cyfip1J/J mice (#P < 0.0001) that was significantly greater than the slope value of Cyfip1J/- mice (*P = 0.046; Figure 4E).
Approximately one-half of the mice on the Cyfip1,2J/J background had previously undergone prior training in the five-day behavioral battery which had a significant effect on PF intake. Specifically, in an ANOVA model of averaged food intake (collapsed across days) as the dependent measure and Battery, Genotype, and Treatment as factors, there was a significant effect of Battery [F(1,1018) = 18.0; P = 2.38 e-05] and a Battery x Treatment interaction [F(1,1018) = 14.0; P = 0.0002]. Subsequent ANOVA of PF intake alone identified PF treatment as the main source for the effect of Battery on food intake [Effect of Battery with PF-trained mice only: F(1,644) = 33.61; P = 1.06 × 10-8] that was explained by greater average PF intake across days in Battery-exposed mice (0.20 +/− 0.03% body weight consumed) compared to Battery-naïve mice (0.12 +/− 0.02% body weight consumed). Importantly, there was no significant Battery x Genotype interaction in either ANOVA model [F(1,1018) < 1; F(1,644) < 1, respectively], nor was there a significant Battery x Genotype x Treatment interaction [F(1,1018) <1]. Thus, prior training in the Battery increased PF intake and overall phenotypic variance without interacting with Genotype.
PO- and sex-dependent effects of Cyfip1 haploinsufficiency on PF intake
We next investigated the effect of PO of Cyfip1 deletion on food intake in the same data from Figure 4 in light of a recent study demonstrating a PO effect of Cyfip1 deletion on emotional learning and synaptic transmission (Chung et al. 2015). We focused on PF intake rather than Chow intake based on the above results (Figure 4).
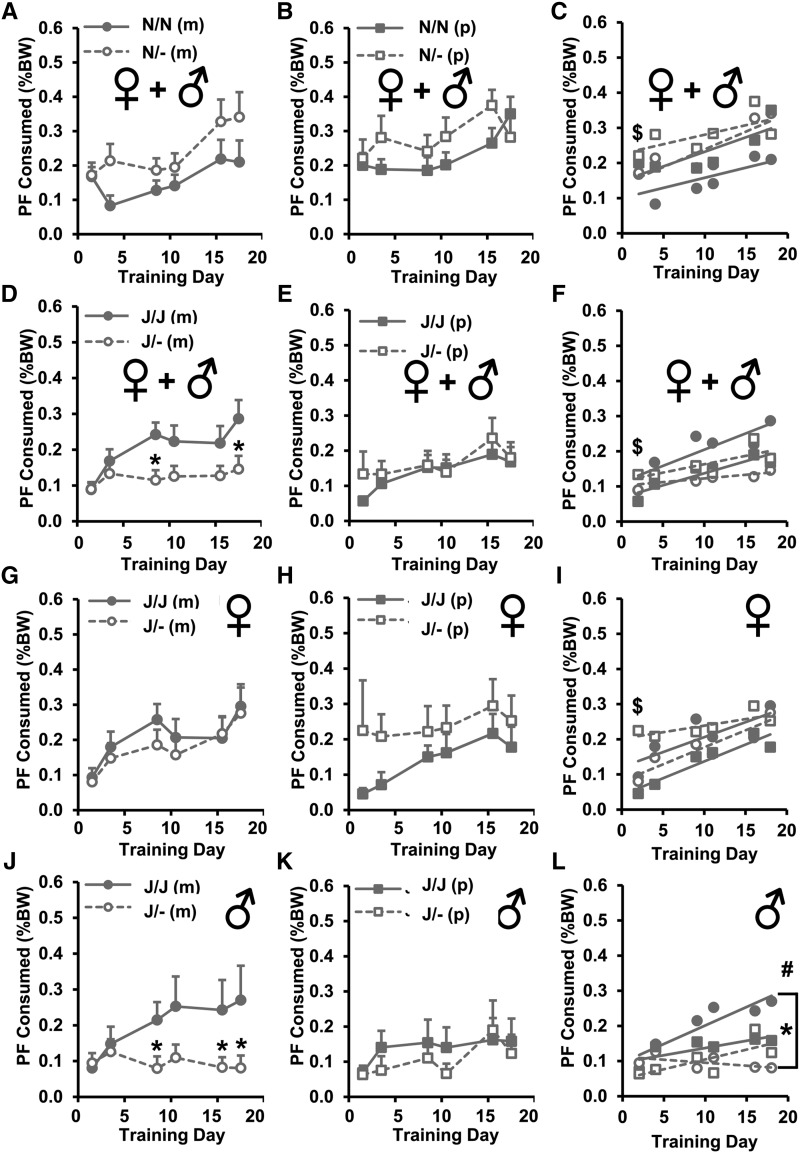
In examining the effect of PO on PF intake on the Cyfip1,2N/N background, mixed-effects ANOVA (Factors: Genotype, PO, Sex; repeated measure: Day) revealed a main effect of PO (P = 0.003) and Day (P = 6.5 × 10−6; Figure 5A,B). Cyfip1N/- (p) mice showed a greater y-intercept compared to their wild-type Cyfip1N/N (p) counterparts and the other wild-type Cyfip1N/N (m) group ($p’s <0.02; Figure 5C). Furthermore, wild-type Cyfip1N/N (p) offspring from paternally deleted families showed a greater y-intercept than wild-type Cyfip1N/N (m) offspring from maternally deleted families (P = 0.046; Figure 5C).
Figure 5.
Effect of Parent-of-Origin (PO) and Sex on PF consumption in Cyfip1N/- and Cyfip1J/- mice. (A,B): For the Cyfip1,2N/N background, there was an effect of Genotype [F(1,464) = 12.3; P = 0.0005], PO [F(1,464) = 9.0; P = 0.003], Sex [F(1,464) = 39.4; P = 8.0 × 10−10], and Day [F(1,464) = 20.8, P = 6.5 × 10−6]. Cyfip1N/- mice consumed more PF than Cyfip1N/N mice on Day (D)4 [A: t(29) = 2.1; *P = 0.046]. Females consumed more PF than males (not shown). (C): No differences were observed among the groups in the slopes of escalation in PF consumption [F(3,16) = 0.7 P = 0.56]; however, paternally-deleted Cyfip1N/- mice (open squares) showed a greater y-intercept than either of the Cyfip1N/N wild-type groups ($: both p’s < 0.02), indicating a greater overall consumption. Furthermore, Cyfip1N/N mice derived from paternal deletion showed a greater y-intercept than Cyfip1N/N mice derived from maternal deletion (P = 0.046). (D,E): For the Cyfip1,2J/J background, there was an effect of Genotype [F(1,600) = 5.2; P = 0.02], Sex [F(1,600) = 14.1; P = 0.0002], Day [F(5,600) = 4.4; P = 0.0006], a Genotype x Sex interaction [F(1,600) = 10.8; P = 0.001], and a Genotype x PO interaction [F(1,600) = 9.3; P = 0.002] that reflected less PF consumption in maternally deleted Cyfip1J/- (m) mice [D: t(59) = 2.9, 2.2; *P = 0.005, 0.03 vs. their Cyfip1J/J counterparts on Day (D)9 and D18, respectively] but no genotypic differences between paternally deleted Cyfip1J/- (p) mice vs. their Cyfip1 J/J (p) counterparts (E). (F): Both maternal and paternal wild-type Cyfip1J/J groups showed significant slope in escalation of intake [Cyfip1J/J (m): F(1,184) = 10.9; P = 0.001; Cyfip1 J/J (p): F(1,142) = 8.4; P = 0.005; closed symbols]. In contrast, neither mutant Cyfip1J/- group showed a significant slope from zero (both ps > 0.15; open symbols). Moreover, paternally deleted Cyfip1J/- (p) mice (open squares) had a greater y-intercept than all three other groups ($ all p’s < 0.0004). (G-I): To understand the source of the above interactions, we next separated PO effects of Cyfip1J/- by Sex. (G-H): In females, there was an effect of Day [F(5,288) = 3.6; P = 0.004] and a Genotype x PO interaction [F(1,288) = 7.1; P = 0.008]. (I): Both Cyfip1J/J wild-type female groups showed a significant slope in escalation (I; both ps < 0.02 vs. zero) as well as maternally-deleted Cyfip1J/- (m) females (P = 0.002 vs. zero). Paternally deleted Cyfip1J/- (p) mice did not show a significant non-zero slope (P = 0.5) but showed the greatest y-intercept compared to all three groups ($P < 0.0002), indicating an initially higher, stable PF consumption across time. (J-L): For males, there was an effect of Genotype [F(1,312) = 13.6; P = 0.0003] and a trending Genotype x PO interaction [F(1,312) = 3.3; P = 0.07]. Maternally deleted Cyfip1J/- (m) males showed significantly less PF intake than their wild-type Cyfip1J/J (m) male counterparts (J: *P < 0.03, 0.04, and 0.04 vs. Cyfip1J/J on D9, D16, and D18, respectively). In contrast, there was no genotypic difference in paternally-deleted Cyfip1J/- (p) males vs. their wild-type Cyfip1J/J (p) male counterparts (panel K). For slope analysis, only the wild-type Cyfip1 J/J (m) males showed a significant slope in escalation of consumption (#P = 0.03 vs. zero) that was also significantly greater than their mutant Cyfip1J/- (m) male counterparts (*P = 0.01; L). Data are presented as mean ± SEM.
For the Cyfip1,2J/J background, mixed effects ANOVA (Factors: Genotype, PO, Sex; repeated measure: Day) revealed a Genotype x PO interaction on PF intake (P = 0.002) and an effect of Day (P = 0.0006). Maternal Cyfip1 deletion [Cyfip1J/- (m)] accounted for the reduced intake in Cyfip1J/- mice that we reported in Figure 4D whereby Cyfip1 J/- (m) mice showed decreased PF intake compared to Cyfip1J/J (m) mice on D9 and D18 (t-tests: P = 0.005, 0.03; Figure 5D). There was no genotypic difference in PF intake in offspring derived from paternal Cyfip1 deletion (Figure 5E). Maternal Cyfip1 deletion induced a significant slope in escalation of PF intake from zero in Cyfip1J/J (m) mice (P = 0.001) but not Cyfip1J/- (m) mice (Figure 5F).
For the Cyfip1,2J/J background, we also observed a Genotype x Sex interaction in PF intake (P = 0.001). To identify the source of this interaction we broke down the maternal and paternal data separately into females and males. Paternally deleted female Cyfip1J/- (p) mice initially showed enhanced PF intake relative to their female wild-type Cyfip1J/J (p) counterparts as hinted by a trending increase in D4 PF intake (t(20) = 2.0; P = 0.06; Figure 5H) that was further supported by a statistically significant increase in y-intercept ($) relative to all three other groups (P = 0.0002 vs. Cyfip1J/J (m) ; P = 0.0002 vs. Cyfip1J/- (m); P < 0.0001 vs. Cyfip1J/J (p); Figure 5I). In contrast, maternally-deleted male Cyfip1J/- (m) mice showed a marked decrease in PF intake relative to male Cyfip1J/J (m) mice on D9, D16, and D18 (t-tests: P = 0.03, 0.04, 0.04) and no significant increase in slope of escalation vs. zero (Figure 5J-L). In contrast, male wild-type Cyfip1 J/J (m) mice showed a significant slope of escalation in PF intake (Cyfip1J/J (m) slope vs. zero: P = 0.03) that was significantly greater than their male Cyfip1 J/- (m) counterparts (vs. J/- (m): P = 0.01) but not greater than paternally deleted male wild-type Cyfip1 J/J (p) (P = 0.29) or mutant Cyfip1J/- (p) mice (P = 0.39; Figure 5L).
To summarize, we observed opposite effects of Cyfip1 deletion on PF intake that depended on Cyfip1,2 genetic background, PO, and Sex. Despite changes in PF intake across Genotype and PO, differences in body weight cannot fully account or the complex interactive effects of Genotype, PO on PF intake (Supplementary Figure 1). I.e., homeostatic mechanisms are unlikely to account for group differences in PF intake.
Conditioned food reward in Cyfip1N/- and Cyfip1J/- haploinsufficient mice
In examining food CPP via the change in preference for the food-paired side (s) between D1 and D22 of training on the Cyfip1,2N/N genetic background, two-way ANOVA (Factors: Genotype, Treatment) revealed no main effect of Cyfip1 Genotype, Treatment, or interaction in mice from either genetic background (p’s > 0.13; Supplementary Figure 2A,B). However, when considering PF treatment alone (as we did for a subset of the above analyses involving PF intake), there was increased PF-CPP in Cyfip1N/- vs. Cyfip1N/N mice that was consistent with increased PF intake (t-test: P = 0.03; Supplementary Figure 2A). For Cyfip1J/- mice on the Cyfip1,2J/J background, there was no genotypic difference in PF-CPP (Supplementary Figure 2B). In considering the effect of PO on PF-CPP, there was no effect of Genotype, PO, or interaction for either Cyfip1,2 genetic background (data not shown).
Compulsive-like eating in the light/dark conflict test in Cyfip1N/- and Cyfip1J/- haploinsufficient mice
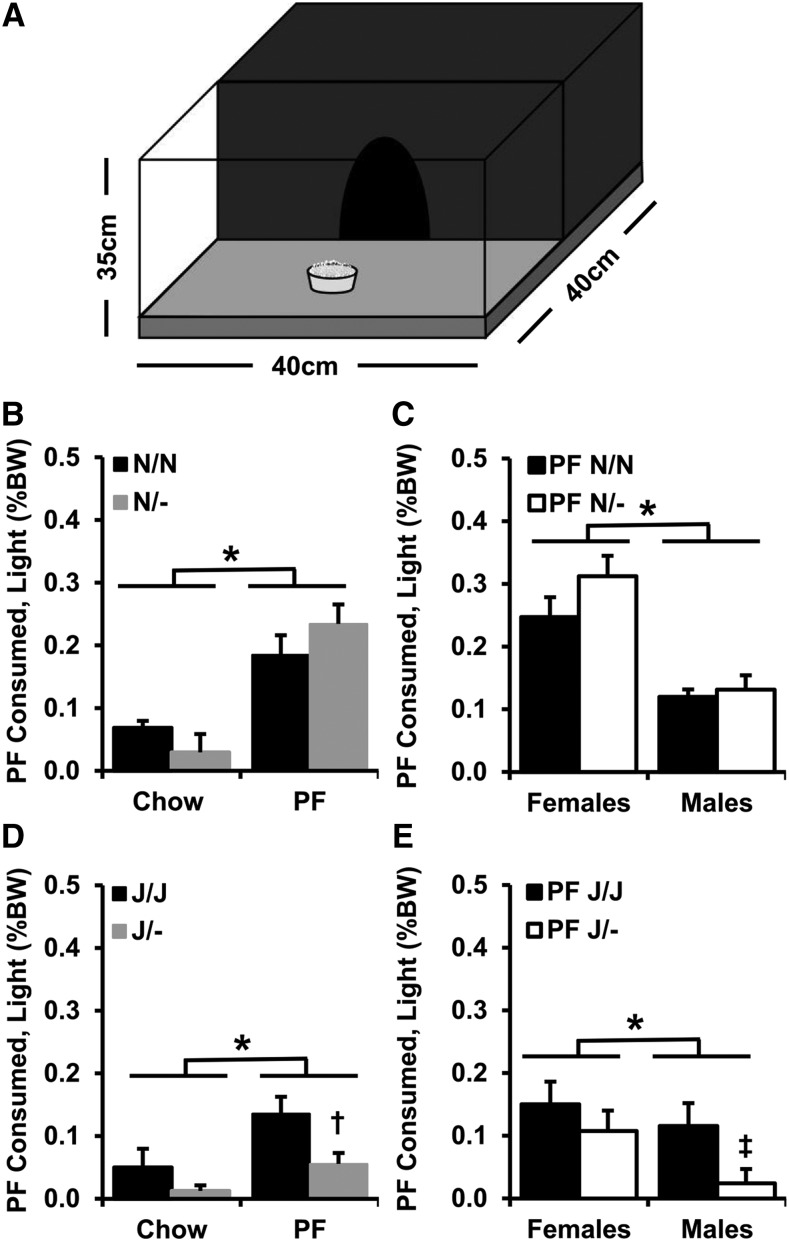
We next examined post-training compulsive-like PF intake using the light/dark conflict test (Figure 6A) (Babbs et al. 2018; Kirkpatrick et al. 2017). Separate, three-way ANOVA (Factors: Genotype, Treatment, Sex) for each of the two genetic backgrounds (Cyfip1,2N//N, Cyfip1,2J/J) revealed that PF-trained mice showed greater PF intake in the light/dark arena than Chow-trained mice (effect of Treatment: *P = 1 × 10−7 and 0.004, respectively; Figure 6B,D). Furthermore, overall, females showed greater PF intake than males on both genetic backgrounds (effect of Sex: P = 0.0004 and 0.05, respectively; Figure 6C,E). For mice on the Cyfip1,2N/N background, there was no genotypic difference in PF consumption in the light/dark arena (Figure 6B,C), regardless of PO (Supplementary Figure 3A-B). For mice on the Cyfip1,2J/J background, Cyfip1J/- mice showed reduced PF consumption (effect of Genotype: P = 0.002; t-test: †P = 0.007; Figure 6D) that was driven primarily by the males (t-test: ‡P = 0.01; Figure 6E).
Figure 6.
Compulsive-like PF intake in the light/dark conflict test in Cyfip1N/- and Cyfip1J/- mice. (A): A cartoon of the apparatus for the light/dark conflict test of compulsive-like PF consumption is shown. (B): For the Cyfip1,2N/N background, there was a main effect of Training Treatment [F(1,148) = 31.1; *P = 1 × 10−7], indicating increased PF intake. However, there was no effect of Genotype [F(1,148) = 0.1; P = 0.7] or Genotype x Training Treatment interaction in consumption [F(1,148) = 2.3; P = 0.13]. (C): In examining PF-trained mice alone, females showed increased intake [effect of Sex: F(1,72) = 13.6; *P = 0.0004]; however, there was no effect of Genotype [F(1,72) = 1.2; P = 0.3] or Genotype x Sex interaction [F(1,72) = 0.7; P = 0.4]. (D): For mice on the Cyfip1,2J/J genetic background, there was a main effect of Training Treatment [F(1,163) = 8.5; *P = 0.004], indicating greater PF intake in PF-trained mice. There was also a main effect of Genotype [F(1,163) = 9.8; P = 0.002] that was explained primarily by less PF intake in Cyfip1J/- mice vs. Cyfip1J/J mice [t(92) = 2.8; †P = 0.007]. (E): In considering only PF-trained mice, females trended toward greater overall intake [effect of Sex: F(1,100) = 3.9; *P = 0.05] and Cyfip1J/- mice showed overall less intake than Cyfip1J/J mice [effect of Genotype: F(1,100) = 7.6; P = 0.007] that was explained primarily by males [‡: t(54) = 2.7; P = 0.01]. Data are presented as the mean ± SEM.
Reduced transcription of Cyfip1 but not Cyfip2 or Magel2 in the hypothalamus of Cyfip1+/− mice
We hypothesized that the PO- and genetic background-dependent effects of Cyfip1 deletion on PF intake could involve differences in hypothalamic gene transcription of Cyfip1 and other genes, including Cyfip2 and Magel2. Supplementary Table 3 lists the qPCR results as a function of both Cyfip1 haploinsufficiency and PO. For the Cyfip1,2N/N background, there was a reduction in Cyfip1 transcription in Cyfip1N/- mice following either maternal (t-test: P = 0.04) or paternal Cyfip1 deletion (t-test: P = 0.04; Supplementary Table 3A) as these two groups did not differ from one another [t(14) < 1]. In contrast, when the effect of Cyfip1 haploinsufficiency on Cyfip1 transcription was assessed on the Cyfip1,2J/J background, maternally deleted Cyfip1 J/- (m) mice showed a significant decrease in Cyfip1 transcription relative to wild-type Cyfip1J/J (t-test: *P = 0.02) and relative to paternally deleted Cyfip1J/- (p) mice (t-test: P = 0.0061; Supplementary Table 3B). There were no genotypic differences in hypothalamic transcription of Cyfip2 or Magel2 (Supplementary Table 3).
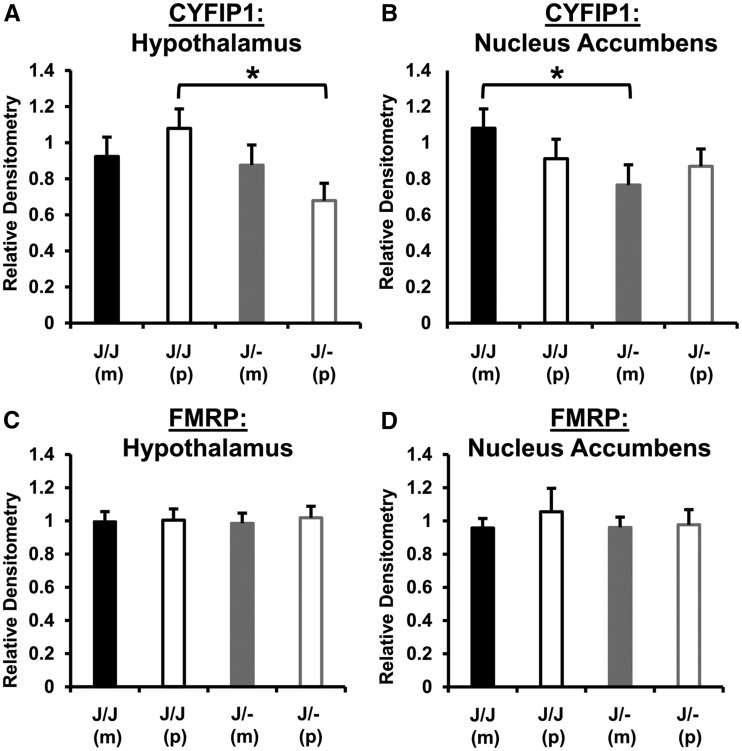
Reduced CYFIP1 protein expression in Cyfip1+/− mice depends on PO
Because preliminary evidence indicated possible effects of PO on Cyfip1 expression at the mRNA level in Cyfip1J/- mice on a Cyfip2J/J background (Supplementary Table 3), we next investigated CYFIP1 expression at the protein level on the same genetic background. We examined the hypothalamus as well as nucleus accumbens. In the hypothalamus, paternally deleted Cyfip1J/- (p) mice showed a significant decrease in CYFIP1 protein relative to their respective Cyfip1 J/J (p) wild-type mice as measured via immunoblot (t-test: P = 0.0055; Figure 7A). In the nucleus accumbens, maternally-deleted Cyfip1J/- (m) mice showed a significant decrease in CYFIP1 protein expression relative to their respective Cyfip1 J/J (m) wild-type mice (t-test: P = 0.049; Figure 7B; Supplementary Figure 4). Thus, either maternal or paternal Cyfip1+/− can result in an enhanced reduction in CYFIP1 protein expression, depending on the brain region.
Figure 7.
CYFIP1 and FMRP protein expression in the hypothalamus and nucleus accumbens. For CYFIP1 and FMRP, there was no effect of Sex (ps > 0.1) or interaction of Sex with other factors (ps > 0.30). There was also no effect of prior Treatment or interaction of Treatment with other factors (ps > 0.10). Therefore, we collapsed across Sex and Treatment. The majority of samples used in CYFIP1 analysis were from PF-trained mice (60 to 61 PF-trained samples per brain region; 16 to 18 Chow-trained samples per brain region) and trending results for the same statistics reported below were observed when analyzing only the PF-trained samples (p’s = 0.065-0.15). Similarly, the majority of samples used for FMRP analysis were from PF-trained mice for hypothalamus (31 to 48 PF-trained samples per brain region; 0 to 8 Chow-trained samples per brain region) and for the nucleus accumbens, all samples were PF-trained (0 Chow-trained). The same null results were observed in the hypothalamus for the effect of Genotype (P = 0.43), PO (P = 0.86), and interaction (P = 0.26). (A): In examining CYFIP1 protein levels in the hypothalamus via immunoblotting, there was a main effect of Genotype [F(1,73) = 7.1; P = 0.009] and a Genotype x PO interaction [F(1,73) = 4.; P = 0.039]. Paternally deleted Cyfip1J/- (p) mice showed lower protein expression than their wild-type Cyfip1J/J (p) counterparts [t(36) = 3.0; *P = 0.0055]. (B): In examining CYFIP1 protein levels in the nucleus accumbens, the effect of Genotype was not significant [F(1,72) = 2.9; P = 0.09]. Maternally deleted Cyfip1J/-(m) mice showed a lower level of immunostaining for CYFIP1 protein than their wild-type Cyfip1J/J (m) littermates [t(37) = 2.0; *P = 0.049]. (C): In examining FMRP levels in the hypothalamus, there was no effect of Genotype, PO, or Genotype x PO interaction [F(1,71) < 1]. (D): In examining FMRP levels in the nucleus accumbens, all samples were from PF-trained mice. Densitometry analysis revealed no effect of Sex or interaction of Sex with any other factors [F(1,23) <`1]. Analysis of FMRP immunoblots from nucleus accumbens revealed no effect of Genotype, PO, or Genotype x PO interaction [F(1,27) < 1].
No effect of CYFIP1 haploinsufficiency on FMRP protein expression
Fmr1 could potentially explain Sex- and PO-specific effects of Cyfip1+/− on PF intake because 1) it is located on the X chromosome; 2) its protein product FMRP interacts with CYFIP1 (Abekhoukh and Bardoni. 2014); and 3) similar maternal effects of Fmr1 haploinsufficiency on behavior in wild-type male mice have been reported on locomotor activity in male mice (Gleason et al. 2011; Zupan and Toth. 2008). We examined locomotor activity in offspring derived from Cyfip1J/-(m) and Cyfip1J/- (p) during initial preference assessment on D1 in the CPP apparatus and again, observed a selective effect of Cyfip1 haploinsufficiency on behavior in maternally deleted male Cyfip1J/- (m) mice that showed increased locomotor activity relative to their wild-type Cyfip1J/J (m) counterparts (Supplementary Figure 5). Therefore, we examined FMRP expression in Cyfip1J/- vs. Cyfip1J/J mice but found no effect of Cyfip1 Genotype or PO on FMRP protein levels in the hypothalamus or nucleus accumbens via immunoblot (Figure 7C,D; Supplementary Figure 6). These null results do not support the hypothesis that FMRP is involved in the downstream mechanisms underlying Sex- and PO-specific effects of Cyfip1J/- haploinsufficiency on behavior.
Discussion
Cyfip1 haploinsufficiency increased OC-like behavior on two different Cyfip2 genetic backgrounds (Figures 1-3) and altered PF consumption and Cyfip1 gene expression at the RNA and protein level, depending on genetic background (Cyfip1,2), PO, and Sex (Figures 4,5,7; Supplementary Table 3). These findings identify a significant contribution of CYFIP1 haploinsufficiency to OC-like behaviors and PF intake that could have relevance for neurodevelopmental disorders (e.g., Type I PWS and FXS) and for neuropsychiatric disorders (e.g., OCD, eating disorders). Sex differences in PWS hyperphagia have not been widely reported (Irizarry et al. 2015). Our findings suggest the possibility of sex differences in PWS hyperphagia, specifically with Type I PWS, which could have implications for developing sex-specific pharmacotherapeutic treatments.
The relatively selective increase in OC-like but not anxiety-like behavior following Cyfip1 deletion (Figure 2; Supplementary Table 2) is consistent with a lack of genetic correlation between marble burying and anxiety and supports marble burying as a repetitive, perseverative-like behavior (Thomas et al. 2009). Nevertheless, there is likely an anxiety-like component to marble burying (Albelda and Joel. 2012) as there is with OC behaviors in humans. For the Cyfip1,2J/J genetic background, the increase in head-dipping behavior in the hole board task in Cyfip1J/- mice (Figure 3) further supports an increase in OC-like/anxiety-like behaviors (Takeda et al. 1998) following Cyfip1 haploinsufficiency, although it should be noted that in contrast to the marble burying behavior (see Results), the p-value for statistical significance for head dipping in the hole board does not survive correction for the 14 statistical tests across the battery of five behavioral assays (P < 0.0036).
Cyfip1+/− mice showed an increase in marble burying which has been shown to predict BE (Freund et al. 2015; Satta et al. 2016). However, in our studies, there was no clear relationship between OC-like behaviors and PF intake because Cyfip1 haploinsufficiency increased marble burying on both Cyfip1,2 genetic backgrounds (Figure 2) yet had opposite effects on PF consumption, depending on the background (Figures 4-5). These results effectively dissociate increased OC-like behavior from increased PF intake following Cyfip1 haploinsufficiency. This dissociation is also evident in patients with PWS who show an increase in OC behaviors that is unrelated to food and is exacerbated in Type I PWS (with CYFIP1 deletion) (Bittel et al. 2006; Butler et al. 2004; Doornbos et al. 2009; Milner et al. 2005; Zarcone et al. 2007). Thus, CYFIP1 deletion could increase the severity of OC symptoms in Type I PWS without modulating eating behavior. Furthermore, multiple types of CYFIP1 variants (structural, coding, intronic, upstream, intergenic) could act more broadly within the general population to associate with OC symptoms (Figures 2-3) or eating behavior in a manner that depends on genetic background (Figures 4-5).
The selective modulation of sweetened PF intake as evidenced during training (Figure 4) and during assessment of compulsive-like eating (Figure 6) combined with the selective demonstration of conditioned reward for sweetened PF (Supplementary Figure 2A) are observations that are consistent with increased preference for sweetened PF as a consequence of Cyfip1 haploinsufficiency and are consistent with a role of Cyfip genes in modulating the hedonic aspects of food intake. In support, the Cyfip2N/N S968F missense mutation in the closely related Cyfip2 gene was associated with both cocaine neurobehavioral sensitivity and plasticity (Kumar et al. 2013) and increased compulsive-like BE (Kirkpatrick et al. 2017). Furthermore, differences in in Cyfip2 mRNA expression genetically correlate with differences in cocaine self-administration in the BXD recombinant inbred strain panel (Dickson et al. 2015). We observed PO-dependent decreases in CYFIP1 protein in both the hypothalamus and nucleus accumbens (Figure 7), a brain region critical for the hedonic aspects of palatable food intake (Lutter and Nestler. 2009). Finally, previous transcriptome analysis of the striatum from Cyfip2N/- vs. Cyfip2N/N genotypes identified “morphine addiction” and “cocaine addiction” as two of the top five KEGG enrichment terms (Kirkpatrick et al. 2017). Together, these findings indicate that both Cyfip1 and Cyfip2 could alter the rewarding/hedonic response to PF consumption to affect food intake.
In contrast to our prediction, Cyfip1J/- mice on the BE-resistant Cyfip1,2J/J background did not show an escalation of PF intake (Figure 4D,E). Instead, we observed what appeared to be a decrease in PF intake in Cyfip1J/- mice that was explained by the surprising induction of a BE phenotype (escalated PF intake) in wild-type mice on a Cyfip1,2J/J background (Figure 4E). This observation was puzzling, given that we have repeatedly shown that mice (especially males) on a mixed F2 background with a homozygous Cyfip2J/J genotype or on an isogenic C57BL/6J background do not show BE (Babbs et al. 2018; Goldberg et al. 2017; Kirkpatrick et al. 2017). Closer inspection revealed that the induction of BE in genetically unaffected wild-type mice was completely accounted for by wild-type male offspring derived from maternal Cyfip1 deletion (Cyfip1J/J (m); Figure 5J,L). Interestingly, although we failed to provide evidence for an association of FMRP expression with behavior (Figure 7C,D; Supplementary Figure 6), a similar pattern of results was observed with maternal haploinsufficiency of Fmr1 (coding for FMRP) whereby genetically unaffected wild-type males demonstrated constitutive locomotor hyperactivity (Zupan and Toth. 2008) relative to wild-type males derived from wild-type dams. Thus, while our results do not support a link between Cyfip1 haploinsufficiency, FMRP, and behavior, in the context of the prior Fmr1 literature, they illustrate the importance of both Sex and PO as a biological variables when investigating the phenotypic effects of gene haploinsufficiency.
Genetic interactions with the social environment can contribute significantly to behavioral variance (Baud et al. 2017). Maternal vs. paternal Cyfip1 deletion could affect social interactions with the dam and sire or with the maternal/paternal care of the pups. As an example, both genetically affected and unaffected male offspring derived from maternal Fmr1 haploinsufficiency showed increased social approach behaviors toward conspecific strangers and neurobiological evidence supporting social aversion (Zupan et al. 2016). In addition to the maternal effects of gene deletion on neurobehavioral phenotypes of genetically unaffected offspring (Gleason et al. 2011), males can demonstrate paternal pup retrieval (Liu et al. 2013) and thus, paternal Cyfip1 deletion could also affect sire-pup contact and behavior in the offspring. For example, selective effects of paternal deletion of neuregulin 1 on multiple behaviors of genetically affected male offspring have also been reported, including decreased fear learning and increased social interactions (Shang et al. 2017). Given the association between CYFIP1 deletion and social deficits in neurodevelopmental disorders (Abekhoukh and Bardoni. 2014), Cyfip1+/− in the dam or sire could affect the social dynamics in the offspring in a PO-dependent, genotype (offspring)-dependent, and sex-dependent manner, leading to long-term neurobehavioral effects. One hypothesis is that wild-type Cyfip1J/J males are particularly susceptible to social influences of maternal-pup and/or pup-pup interactions in the maternally-deleted Cyfip1J/- environment (whereas the Cyfip1J/- mice are resistant), ultimately explaining the selective induction of escalated PF intake.
What is the mechanism underlying sex-dependent, PO-specific effects of Cyfip1 deletion on behavior and gene expression? There is no published evidence that Cyfip1 is imprinted and while our analysis of Cyfip1 transcript and protein expression indicate PO-dependent effects as previously reported (Chung et al. 2015), the direction was not always consistent with maternal imprinting and was dependent on the particular brain region (Supplementary Table 3; Figure 7). A recent study of nearly 100 phenotypes showed that most complex traits exhibit PO effects and that non-imprinted KO alleles (e.g., Cyfip1) can induce extensive PO effects by interacting in trans with imprinted loci throughout the genome to affect gene networks (Mott et al. 2014). If a trans-acting genomic mechanism underlies the effects of Cyfip1 haploinsufficiency on behavior, the trans-acting factor(s) must be faithfully co-inherited with the maternal or paternal deletion to explain the specific PO effects. One such mechanism could involve inheritance of sex-dependent gene expression patterns originating from sex chromosomes that interact with Cyfip1 deletion to affect neurobehavioral phenotypes. Fmr1 is located on the X chromosome and codes for FMRP, an RNA-binding protein that interacts with CYFIP and regulates mRNA translation (Abekhoukh and Bardoni. 2014). However, despite observing Fmr1-like PO effects of maternal Cyfip1 haploinsufficiency on initial locomotor activity prior to BE training (Supplementary Figure 5), we did not observe any evidence for a relationship between FMRP protein expression and the complex interactive effects of Cyfip1 haploinsufficiency on PF intake (Figure 7C, D), nor did we observe any difference in FMRP expression between females and males [t(73)=1.68; P = 0.10]. These null results are consistent with FMR1 undergoing X-inactivation (Kirchgessner et al. 1995) and fail to support a mechanistic role for FMRP, although we should note that differential FMRP expression could still be involved at some stage of neurodevelopment in the underlying mechanisms. Residual heterozygosity of B6NJ alleles on the X chromosome could also affect the expression of X-linked genes that act as modifiers of Cyfip1 transcription or that skew X-inactivation and account for the background-dependent PO effects of Cyfip1 deletion on behavior. The use of the four core genotypes model (XX, XY, XX-male, XY-female) could test the involvement of sex chromosomes in PO- and Sex-dependent effects of Cyfip1 deletion on PF intake (Arnold and Chen. 2009). Notably, a recent study using this genetic model identified a contribution of sex chromosomes to operant reinforcement for PF (Seu et al. 2014).
Our preclinical findings provide evidence that reduced CYFIP1 expression could contribute to OC behaviors and disordered eating (Chang et al. 2019). Future genomic studies of brain regions, cell types, and neurodevelopmental time points could inform molecular mechanisms of eating behaviors on different genetic backgrounds and the potential interaction of Cyfip1 deletion with gene expression on sex chromosomes.
Acknowledgments
These studies were funded by NIH/NIDA R21DA038738 (C.D.B.), NIH/NIDA R01DA039168, NIDA Diversity Scholars Network (F.R.), Burroughs-Wellcome Fund Transformative Training Program in Addiction Science (TTPAS; 1011479), NIH/NIGMS T32GM008541, and NIH/NIDA U01DA041668 (V.K.). We thank Dr. Rachel Wevrick for providing us with the primer sequences used for gene expression analysis of Magel2. We would also like to acknowledge Dr. Lynn Deng and Matthew Au of the Boston University Analytical Instrumentation Core Facility (S10OD023663) for their support in conducting the qPCR studies.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.8316635.
Communicating editor: D. Threadgill
Literature Cited
- Abekhoukh S., and Bardoni B., 2014. CYFIP family proteins between autism and intellectual disability: Links with fragile X syndrome. Front. Cell. Neurosci. 8: 81 10.3389/fncel.2014.00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albelda N., and Joel D., 2012. Animal models of obsessive-compulsive disorder: Exploring pharmacology and neural substrates. Neurosci. Biobehav. Rev. 36: 47–63. 10.1016/j.neubiorev.2011.04.006 [DOI] [PubMed] [Google Scholar]
- Angulo M. A., Butler M. G., and Cataletto M. E., 2015. Prader-willi syndrome: A review of clinical, genetic, and endocrine findings. J. Endocrinol. Invest. 38: 1249–1263. 10.1007/s40618-015-0312-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold A. P., and Chen X., 2009. What does the “four core genotypes” mouse model tell us about sex differences in the brain and other tissues? Front. Neuroendocrinol. 30: 1–9. 10.1016/j.yfrne.2008.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babbs R. K., Wojnicki F. H., and Corwin R. L., 2012. Assessing binge eating. an analysis of data previously collected in bingeing rats. Appetite 59: 478–482. 10.1016/j.appet.2012.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babbs R. K., Kelliher J. C., Scotellaro J. L., Luttik K. P., Mulligan M. K.. et al., 2018. Genetic differences in the behavioral organization of binge eating, conditioned food reward, and compulsive-like eating in C57BL/6J and DBA/2J strains. Physiol. Behav. 197: 51–66. 10.1016/j.physbeh.2018.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud A., Mulligan M. K., Casale F. P., Ingels J. F., Bohl C. J.. et al., 2017. Genetic variation in the social environment contributes to health and disease. PLoS Genet. 13: e1006498 10.1371/journal.pgen.1006498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello N. T., and Hajnal A., 2010. Dopamine and binge eating behaviors. Pharmacol. Biochem. Behav. 97: 25–33. 10.1016/j.pbb.2010.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge K. C., 2009. Wanting and liking: Observations from the neuroscience and psychology laboratory. Inquiry (Oslo) 52: 378–398. 10.1080/00201740903087359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittel D. C., Kibiryeva N., and Butler M. G., 2006. Expression of 4 genes between chromosome 15 breakpoints 1 and 2 and behavioral outcomes in prader-willi syndrome. Pediatrics 118: e1276–e1283. 10.1542/peds.2006-0424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdagi O., Sakurai T., Dorr N., Pilorge M., Takahashi N.. et al., 2012. Haploinsufficiency of Cyfip1 produces fragile X-like phenotypes in mice. PLoS One 7: e42422 10.1371/journal.pone.0042422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik C. M., Sullivan P. F., and Kendler K. S., 2003. Genetic and environmental contributions to obesity and binge eating. Int. J. Eat. Disord. 33: 293–298. 10.1002/eat.10140 [DOI] [PubMed] [Google Scholar]
- Butler M. G., Bittel D. C., Kibiryeva N., Talebizadeh Z., and Thompson T., 2004. Behavioral differences among subjects with prader-willi syndrome and type I or type II deletion and maternal disomy. Pediatrics 113: 565–573. 10.1542/peds.113.3.565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallini M. C., Bertelli S., Chiapparino D., Riboldi S., and Bellodi L., 2000. Complex segregation analysis of obsessive-compulsive disorder in 141 families of eating disorder probands, with and without obsessive-compulsive disorder. Am. J. Med. Genet. 96: 384–391. [DOI] [PubMed] [Google Scholar]
- Chang X., Qu H., Liu Y., Glessner J., Hou C.. et al., 2019. Microduplications at the 15q11.2 BP1–BP2 locus are enriched in patients with anorexia nervosa. J. Psychiatr. Res. 113: 34–38. 10.1016/j.jpsychires.2019.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung L., Wang X., Zhu L., Towers A. J., Cao X.. et al., 2015. Parental origin impairment of synaptic functions and behaviors in cytoplasmic FMRP interacting protein 1 (Cyfip1) deficient mice. Brain Res. 1629: 340–350. 10.1016/j.brainres.2015.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox D. M., and Butler M. G., 2015. The 15q11.2 BP1–BP2 microdeletion syndrome: A review. Int. J. Mol. Sci. 16: 4068–4082. 10.3390/ijms16024068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis K. W., Serrano M., Loddo S., Robinson C., Alesi V.. et al., 2019. Parent-of-origin effects in 15q11.2 BP1–BP2 microdeletion (burnside-butler) syndrome. Int. J. Mol. Sci. 20: 1459 10.3390/ijms20061459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson P. E., Miller M. M., Calton M. A., Bubier J. A., Cook M. N.. et al., 2015. Systems genetics of intravenous cocaine self-administration in the BXD recombinant inbred mouse panel. Psychopharmacology (Berl) 233: 701–14. 10.1007/s00213-015-4147-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLeone R. J., Taylor J. R., and Picciotto M. R., 2012. The drive to eat: Comparisons and distinctions between mechanisms of food reward and drug addiction. Nat. Neurosci. 15: 1330–1335. 10.1038/nn.3202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doornbos M., Sikkema-Raddatz B., Ruijvenkamp C. A., Dijkhuizen T., Bijlsma E. K.. et al., 2009. Nine patients with a microdeletion 15q11.2 between breakpoints 1 and 2 of the prader-willi critical region, possibly associated with behavioural disturbances. Eur. J. Med. Genet. 52: 108–115. 10.1016/j.ejmg.2009.03.010 [DOI] [PubMed] [Google Scholar]
- Dykens E. M., Leckman J. F., and Cassidy S. B., 1996. Obsessions and compulsions in prader-willi syndrome. J. Child Psychol. Psychiatry 37: 995–1002. 10.1111/j.1469-7610.1996.tb01496.x [DOI] [PubMed] [Google Scholar]
- Feurer I. D., Dimitropoulos A., Stone W. L., Roof E., Butler M. G.. et al., 1998. The latent variable structure of the compulsive behaviour checklist in people with prader-willi syndrome. J. Intellect. Disabil. Res. 42: 472–480. 10.1046/j.1365-2788.1998.4260472.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund N., Thompson B. S., Norman K. J., Einhorn P., and Andersen S. L., 2015. Developmental emergence of an obsessive-compulsive phenotype and binge behavior in rats. Psychopharmacology (Berl.) 232: 3173–3181. 10.1007/s00213-015-3967-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason G., Zupan B., and Toth M., 2011. Maternal genetic mutations as gestational and early life influences in producing psychiatric disease-like phenotypes in mice. Front. Psychiatry 2: 25 10.3389/fpsyt.2011.00025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg, L. R., S. L. Kirkpatrick, N. Yazdani, K. P. Luttik, O. A. Lacki et al, 2017 Casein kinase 1-epsilon deletion increases mu opioid receptor-dependent behaviors and binge eating. Genes Brain Behav 16: 725–738. 10.1111/gbb.12397. Erratum: 2017 16: 823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griggs J. L., Sinnayah P., and Mathai M. L., 2015. Prader-willi syndrome: From genetics to behaviour, with special focus on appetite treatments. Neurosci. Biobehav. Rev. 59: 155–172. 10.1016/j.neubiorev.2015.10.003 [DOI] [PubMed] [Google Scholar]
- Hinney A., Kesselmeier M., Jall S., Volckmar A. L., Focker M.. et al., 2017. Evidence for three genetic loci involved in both anorexia nervosa risk and variation of body mass index. Mol. Psychiatry 22: 192–201. Erratum: 22: 321–322. 10.1038/mp.2016.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao K., Harony-Nicolas H., Buxbaum J. D., Bozdagi-Gunal O., and Benson D. L., 2016. Cyfip1 regulates presynaptic activity during development. J. Neurosci. 36: 1564–1576. 10.1523/JNEUROSCI.0511-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huckins L. M., Hatzikotoulas K., Southam L., Thornton L. M., Steinberg J.. et al., 2018. Investigation of common, low-frequency and rare genome-wide variation in anorexia nervosa. Mol. Psychiatry 23: 1169–1180. Erratum: 23: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson J. I., Hiripi E., Pope H. G. Jr., and Kessler R. C., 2007. The prevalence and correlates of eating disorders in the national comorbidity survey replication. Biol. Psychiatry 61: 348–358. 10.1016/j.biopsych.2006.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry K. A., Bain J., Butler M. G., Ilkayeva O., Muehlbauer M.. et al., 2015. Metabolic profiling in prader-willi syndrome and nonsyndromic obesity: Sex differences and the role of growth hormone. Clin. Endocrinol. (Oxf.) 83: 797–805. 10.1111/cen.12766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler R. M., Hutson P. H., Herman B. K., and Potenza M. N., 2016. The neurobiological basis of binge-eating disorder. Neurosci. Biobehav. Rev. 63: 223–238. 10.1016/j.neubiorev.2016.01.013 [DOI] [PubMed] [Google Scholar]
- Kim, H., C. S. Lim and B. K. Kaang, 2016 Neuronal mechanisms and circuits underlying repetitive behaviors in mouse models of autism spectrum disorder. Behav. Brain Funct. 12: 3–016–0087-y. 10.1186/s12993-016-0087-y [DOI] [PMC free article] [PubMed]
- Kirchgessner C. U., Warren S. T., and Willard H. F., 1995. X inactivation of the FMR1 fragile X mental retardation gene. J. Med. Genet. 32: 925–929. 10.1136/jmg.32.12.925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick S. L., Goldberg L. R., Yazdani N., Babbs R. K., Wu J.. et al., 2017. Cytoplasmic FMR1-interacting protein 2 is a major genetic factor underlying binge eating. Biol. Psychiatry 81: 757–769. 10.1016/j.biopsych.2016.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V., Kim K., Joseph C., Kourrich S., Yoo S. H.. et al., 2013. C57BL/6N mutation in cytoplasmic FMRP interacting protein 2 regulates cocaine response. Science 342: 1508–1512. 10.1126/science.1245503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. X., Lopatina O., Higashida C., Fujimoto H., Akther S.. et al., 2013. Displays of paternal mouse pup retrieval following communicative interaction with maternal mates. Nat. Commun. 4: 1346 10.1038/ncomms2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutter M., and Nestler E. J., 2009. Homeostatic and hedonic signals interact in the regulation of food intake. J. Nutr. 139: 629–632. 10.3945/jn.108.097618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy, S. L., S. J. Winham, A. B. Cuellar-Barboza, C. L. Colby, A. M. Ho et al, 2018 Bipolar disorder with binge eating behavior: A genome-wide association study implicates PRR5-ARHGAP8. Transl. Psychiatry. 8: 40–017–0085–3. 10.1038/s41398-017-0085-3 [DOI] [PMC free article] [PubMed]
- Micali N., Hilton K., Nakatani E., Heyman I., Turner C.. et al., 2011. Is childhood OCD a risk factor for eating disorders later in life? A longitudinal study. Psychol. Med. 41: 2507–2513. 10.1017/S003329171100078X [DOI] [PubMed] [Google Scholar]
- Milner K. M., Craig E. E., Thompson R. J., Veltman M. W., Thomas N. S.. et al., 2005. Prader-willi syndrome: Intellectual abilities and behavioural features by genetic subtype. J. Child Psychol. Psychiatry 46: 1089–1096. 10.1111/j.1469-7610.2005.01520.x [DOI] [PubMed] [Google Scholar]
- Mitchell K. S., Neale M. C., Bulik C. M., Aggen S. H., Kendler K. S.. et al., 2010. Binge eating disorder: A symptom-level investigation of genetic and environmental influences on liability. Psychol. Med. 40: 1899–1906. 10.1017/S0033291710000139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C. F., Sabino V., Koob G. F., and Cottone P., 2017. Pathological overeating: Emerging evidence for a compulsivity construct. Neuropsychopharmacology 42: 1375–1389. 10.1038/npp.2016.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott R., Yuan W., Kaisaki P., Gan X., Cleak J.. et al., 2014. The architecture of parent-of-origin effects in mice. Cell 156: 332–342. 10.1016/j.cell.2013.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzar Z., Lozano R., Kolevzon A., and Hagerman R. J., 2016. The neurobiology of the prader-willi phenotype of fragile X syndrome. Intractable Rare Dis. Res. 5: 255–261. 10.5582/irdr.2016.01082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowicki S. T., Tassone F., Ono M. Y., Ferranti J., Croquette M. F.. et al., 2007. The prader-willi phenotype of fragile X syndrome. J. Dev. Behav. Pediatr. 28: 133–138. 10.1097/01.DBP.0000267563.18952.c9 [DOI] [PubMed] [Google Scholar]
- Oguro-Ando A., Rosensweig C., Herman E., Nishimura Y., Werling D.. et al., 2015. Increased CYFIP1 dosage alters cellular and dendritic morphology and dysregulates mTOR. Mol. Psychiatry 20: 1069–1078. 10.1038/mp.2014.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathania M., Davenport E. C., Muir J., Sheehan D. F., Lopez-Domenech G.. et al., 2014. The autism and schizophrenia associated gene CYFIP1 is critical for the maintenance of dendritic complexity and the stabilization of mature spines. Transl. Psychiatry 4: e374 Erratum: 4: e423. 10.1038/tp.2014.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satta V., Scherma M., Giunti E., Collu R., Fattore L.. et al., 2016. Emotional profile of female rats showing binge eating behavior. Physiol. Behav. 163: 136–143. 10.1016/j.physbeh.2016.05.013 [DOI] [PubMed] [Google Scholar]
- Schenck A., Bardoni B., Moro A., Bagni C., and Mandel J. L., 2001. A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P. Proc. Natl. Acad. Sci. USA 98: 8844–8849. 10.1073/pnas.151231598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T. D., and Livak K. J., 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3: 1101–1108. 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- Seu E., Groman S. M., Arnold A. P., and Jentsch J. D., 2014. Sex chromosome complement influences operant responding for a palatable food in mice. Genes Brain Behav. 13: 527–534. 10.1111/gbb.12143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang K., Talmage D. A., and Karl T., 2017. Parent-of-origin effects on schizophrenia-relevant behaviours of type III neuregulin 1 mutant mice. Behav. Brain Res. 332: 250–258. 10.1016/j.bbr.2017.05.057 [DOI] [PubMed] [Google Scholar]
- Skarnes W. C., Rosen B., West A. P., Koutsourakis M., Bushell W.. et al., 2011. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474: 337–342. 10.1038/nature10163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- State M. W., Dykens E. M., Rosner B., Martin A., and King B. H., 1999. Obsessive-compulsive symptoms in prader-willi and “prader-willi-like” patients. J. Am. Acad. Child Adolesc. Psychiatry 38: 329–334. 10.1097/00004583-199903000-00021 [DOI] [PubMed] [Google Scholar]
- Stefan M., Portis T., Longnecker R., and Nicholls R. D., 2005. A nonimprinted prader-willi syndrome (PWS)-region gene regulates a different chromosomal domain in trans but the imprinted pws loci do not alter genome-wide mRNA levels. Genomics 85: 630–640. 10.1016/j.ygeno.2005.02.004 [DOI] [PubMed] [Google Scholar]
- Stein D. J., Keating J., Zar H. J., and Hollander E., 1994. A survey of the phenomenology and pharmacotherapy of compulsive and impulsive-aggressive symptoms in prader-willi syndrome. J. Neuropsychiatry Clin. Neurosci. 6: 23–29. 10.1176/jnp.6.1.23 [DOI] [PubMed] [Google Scholar]
- Tacer K. F., and Potts P. R., 2017. Cellular and disease functions of the prader-willi syndrome gene MAGEL2. Biochem. J. 474: 2177–2190. 10.1042/BCJ20160616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda H., Tsuji M., and Matsumiya T., 1998. Changes in head-dipping behavior in the hole-board test reflect the anxiogenic and/or anxiolytic state in mice. Eur. J. Pharmacol. 350: 21–29. 10.1016/S0014-2999(98)00223-4 [DOI] [PubMed] [Google Scholar]
- Thomas A., Burant A., Bui N., Graham D., Yuva-Paylor L. A.. et al., 2009. Marble burying reflects a repetitive and perseverative behavior more than novelty-induced anxiety. Psychopharmacology (Berl.) 204: 361–373. 10.1007/s00213-009-1466-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfley D. E., Citrome L., and Herman B. K., 2016. Characteristics of binge eating disorder in relation to diagnostic criteria. Neuropsychiatr. Dis. Treat. 12: 2213–2223. 10.2147/NDT.S107777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdani N., Parker C. C., Shen Y., Reed E. R., Guido M. A.. et al., 2015. Hnrnph1 is A quantitative trait gene for methamphetamine sensitivity. PLoS Genet. 11: e1005713 10.1371/journal.pgen.1005713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz Z., Hardaway J. A., and Bulik C. M., 2015. Genetics and epigenetics of eating disorders. Adv. Genomics Genet. 5: 131–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarcone J., Napolitano D., Peterson C., Breidbord J., Ferraioli S.. et al., 2007. The relationship between compulsive behaviour and academic achievement across the three genetic subtypes of prader-willi syndrome. J. Intellect. Disabil. Res. 51: 478–487. 10.1111/j.1365-2788.2006.00916.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupan B., and Toth M., 2008. Wild-type male offspring of fmr-1+/− mothers exhibit characteristics of the fragile X phenotype. Neuropsychopharmacology 33: 2667–2675. 10.1038/sj.npp.1301651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupan B., Sharma A., Frazier A., Klein S., and Toth M., 2016. Programming social behavior by the maternal fragile X protein. Genes Brain Behav. 15: 578–587. 10.1111/gbb.12298 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors affirm that all data necessary for confirming the conclusions of this article are represented fully within the article and its tables and figures. Supplemental material available at Figshare: https://doi.org/10.25387/g3.8316635.