Abstract
In order to gain insight into the genetic architecture of economically important traits in pigs and to derive suitable genetic markers to improve these traits in breeding programs, many studies have been conducted to map quantitative trait loci. Shortcomings of these studies were low mapping resolution, large confidence intervals for quantitative trait loci-positions and large linkage disequilibrium blocks. Here, we overcome these shortcomings by pooling four large F2 designs to produce smaller linkage disequilibrium blocks and by resequencing the founder generation at high coverage and the F1 generation at low coverage for subsequent imputation of the F2 generation to whole genome sequencing marker density. This lead to the discovery of more than 32 million variants, 8 million of which have not been previously reported. The pooling of the four F2 designs enabled us to perform a joint genome-wide association study, which lead to the identification of numerous significantly associated variant clusters on chromosomes 1, 2, 4, 7, 17 and 18 for the growth and carcass traits average daily gain, back fat thickness, meat fat ratio, and carcass length. We could not only confirm previously reported, but also discovered new quantitative trait loci. As a result, several new candidate genes are discussed, among them BMP2 (bone morphogenetic protein 2), which we recently discovered in a related study. Variant effect prediction revealed that 15 high impact variants for the traits back fat thickness, meat fat ratio and carcass length were among the statistically significantly associated variants.
Keywords: Genome wide association study, Whole genome sequencing, Imputation, Meat, carcass, and production traits, Variant calling
Mapping experiments in livestock generally serve two purposes: The first is to understand the genetic architecture of quantitative traits, and to derive and prove new hypotheses of trait expression. The second is the identification of genetic markers that may be useful for livestock breeding. There have been many quantitative trait loci (QTL) mapping experiments carried out over the last decades (see review article by (Rothschild et al. 2007)), mainly in experimental F2 crosses established from two outbred founder pig breeds. In early studies, genotyping was mainly achieved using microsatellite markers and mapping was achieved through linkage analysis (see overview in (Knott 2005)). These designs were set up to enable QTL detection with high power, but they suffered from a low mapping resolution and large confidence intervals for QTL-positions. This was partly due to the limited number of meiosis cycles exploited in these designs in conjunction with typically small numbers of 300 to 500 F2 individuals. Furthermore, this approach assumes the divergent fixation of the QTL alleles in the founder breeds, and highly different gene frequencies and variation within these breeds were not considered (Nagamine et al. 2003). The breed Piétrain, for instance, has been selected for growth and meat yield for many generations and still exhibits a large genetic variation for these traits (Wellmann et al. 2013). More recent QTL-mapping experiments utilized genome-wide association studies (GWAS), which in contrast to linkage analyses, exploit historical meiosis and rely on linkage disequilibrium (LD) requiring high marker densities. The precision of GWAS is then a function of LD block lengths and the number of individuals analyzed, which in turn limits the usefulness of its application in F2 designs (Hayes and Goddard 2001). However, enormous efforts have been made in the establishment of these mapping populations, usually including extensive phenotyping far beyond what would be available in field populations. It would thus be desirable to revisit these resources using current genotyping and sequencing technologies, which would require an increase in the number of individuals and a decrease in the LD block lengths. In a recent simulation study, it was shown to be possible by pooling F2 designs, particularly when founder breeds are closely related and QTL are segregating in one founder breed (Schmid et al. 2018). This approach has already been successfully applied based on medium density SNP chip data (Blaj et al. 2018; Stratz et al. 2018).
With the aim to overcome the aforementioned limits in mapping resolution and to fully exploit the potential of the resource populations, we pooled four well-characterized F2 designs (Table 1), three of them having the founder breed Piètrain in common. Twenty four founder animals were genotyped by high coverage whole genome sequencing (WGS) and 91 of the F1 animals were sequenced at a low coverage for subsequent imputation to a high coverage WGS level. A total of 2,657 F2 animals that were genotyped with the 62K Illumina PorcineSNP60 BeadChip (Ramos et al. 2009) were imputed to WGS levels with pedigree information and analyzed in a joint GWAS (see workflow in Figure 1). As a proof of concept four relevant production traits were analyzed: Average daily gain (ADG), back fat thickness (BFT), meat to fat ratio (MFR), and carcass length (CRCL).
Table 1. Per cross information of the sequenced individuals (F0 and F1) and SNP array genotyped individuals (F2). F0 and F1 animals served as the reference panel for the imputation of the F2 generation to sequence level for subsequent genome wide association analyses.
| Cross/Generation | F0* | F1 | F2 |
|---|---|---|---|
| Piétrain x (Large White x Landrace)/Large White | 13 | 55 | 1750 |
| Meishan x Piétrain | 8 | 19 | 304 |
| Wild Boar x Piétrain | 6 | 17 | 291 |
| Wild Boar x Meishan | 1 | 0 | 312 |
| Total | 24* | 91 | 2657 |
Four founders are common among crosses.
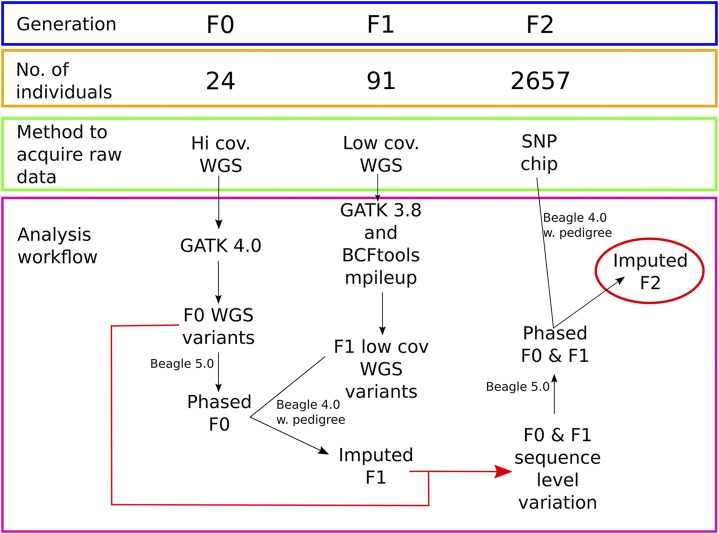
Figure 1.
Genotyping workflow. 24 Founder animals were sequenced with high coverage, variants were called with GATK 4.0 and phased with Beagle 5.0. 91 F1 animals were sequenced with low coverage and variants were called with GATK 3.8 and BCFtools mpileup. The F1 dataset was imputed using Beagle 4.0 and pedigree information with phased Founders as a reference-panel for haplotype structure. The imputed F1 was then merged with the F0 variant call data set and phased with Beagle 5.0. Finally the 2657 chip genotyped F2 individuals were imputed to WGS levels with Beagle 4.0 and pedigree information with the merged and phased Founder/F1-imputed dataset as the reference-panel.
Material and Methods
Description of resource populations and phenotypes
Four well characterized experimental populations were pooled for this study. Detailed descriptions of the resource populations were done by Borchers et al. (Borchers et al. 2000) and Rückert et al. (Rückert and Bennewitz 2010), hence they will only be described briefly. The largest population was obtained from five purebred Piétrain boars and one Large White and six crossbred sows Landrace x Large White. The other three populations stemmed from a Meishan boar or Wild boar crossed with either Piétrain or Meishan females. The Wild boar and three Piétrain females were common founders in three of the crosses. The F2 generation was the result of repeatedly crossing F1 boars with F1 sows in order to obtain large full-sib families. From the crosses, a total number of 2772 animals were chosen (24 F0-generation pigs, 91 F1-generation pigs, 2657 F2-generation pigs) and blood samples were used to extract genomic DNA for genotyping purposes (Table 1). The F0 and F1 animals selected for sequencing were generally chosen according to the number of F2 individuals, i.e., we prioritized individuals from which large families were derived. Four phenotypic traits were considered: ADG, BFT, MFR, and CRCL. The phenotypes were pre-corrected for systematic effects (e.g., stable, slaughter month) and for the effect of RYR1 gene (Fujii et al. 1991) using a general linear model. Trait definition, descriptive statistics and information about the pre-adjustment and the fixed effects used per cross can be found in (Blaj et al. 2018).
Sequencing
A total number of twenty four founder animals were sequenced with an average 19x coverage at the sequencing facility University Hohenheim. Out of 17 F1 families, 91 animals were sequenced with an average 0.9x coverage. All paired-end sequencing (read length 2 × 100 bp) was done on an Illumina HiScan SQ using TruSeq SBS v3 Kits. For the library construction, the DNA samples were fragmented on a Covaris S220 ultrasonicator. Parameters were adjusted to yield 350 bp inserts. Fragment length was measured with High Sensitivity DNA Chips on an Agilent Bioanalyzer. Sequencing adapters and indexes were ligated using Illumina’s TruSeq DNA PCR-Free Library Prep Kits. Quantification of libraries was done by qPCR using KAPA Library Quant Kits. Flow cells were prepared using an Illumina cBot and TruSeq PE v3 Cluster kits. Raw sequencing data were demultiplexed and converted into FASTQ files using Illumina’s CASAVA software.
Mapping and variant detection
Mapping and variant calling of the F0 generation was performed according to the GATK best practice pipeline using GATK v. 4.0 (McKenna et al. 2010) and genome assembly Sus scrofa 11.1 (GCA_000003025.6 provided by Swine Genome Sequencing Consortium on NCBI). Base quality score recalibration was performed with dbSNP build 150 as the knownSites dataset. Truth datasets used for Variant Quality Score Recalibration (VQSR) were as follows. SNPs: Illumina Infinium PorcineSNP60 v2 BeadChip and Affymetrix Axiom PorcineHD. INDELs: High confidence fraction (filter settings: QD 15.0, FS 200.0, ReadPosRankSum 20.0) of the PigVar database. Training dataset for SNP VQSR was also a high confidence fraction of the PigVar database (filter settings: QD 21.5, FS 60.0, MQ 40.0, MQRankSum 12.5, ReadPosRankSum 8.0, SOR 3.0) (Zhou et al. 2017). A truth sensitivity of 99.0 was chosen for SNPs and INDELs. The known dataset for SNP and INDEL VQSR was dbSNP build 150. Since SNPs were filtered with two truth datasets a Ti/Tv free recalibration according to the GATK best practice guidelines was applied to the data. Low coverage sequencing reads of F1 animals were processed according to the GATK best practice guidelines with the following deviations. SNP Calling was performed using GATK HaplotypeCaller v. 3.8 in joint mode with the settings minPruning 1 and minDanglingBranchLength 1 as well as BCFtools mpileup v 1.9 (Li et al. 2009), respectively. INDELs in the F1 variant call dataset were neglected due to low sequencing depth. An intersection variant call set between HaplotypeCaller, mpileup and the founder SNPs was created and stringently filtered with the following settings: QD 30.0, FS 60.0, MQ 40.0, QUAL 300.0.
Haplotype construction and imputation
To make use of the most recent phasing algorithms Beagle 5.0 was used for all phasing operations (Browning et al. 2018). Beagle 4.0 was applied for genotype imputation since it is the latest version that supports the usage of pedigree information (Browning and Browning 2007). Haplotype phasing of the F0 generation variant call set was done using Beagle 5.0 and subsequent imputation with pedigree information of the F1 low coverage SNPs was achieved with Beagle 4.0. F0 and imputed F1 variants were merged with GATK CombineVariants and phased with Beagle 5.0. F2 generation 60k SNP chip data were imputed with Beagle 4.0 and pedigree information with merged and phased F0 and F1 WGS level variants as the reference panel. Local drops in imputation accuracy were determined by the construction of 24 F0 reference-panels with one animal left out. Genotype data acquired with the 60k SNP chip from each F0 individual was imputed with a reference-panel where the respective individual was missing utilizing Beagle 4.0. The 24 individual datasets were merged and together with the F0 reference dataset converted to additive coding with Plink 1.9 (Chang et al. 2015). Correlation (coefficient of determination, R2) for each variant on QTL harboring chromosomes was calculated with an in house R script.
Genome wide association studies and cluster assignment
Single-trait association analyses were performed with GCTA v. 1.92.4 beta 3 on the F2 population only (Yang et al. 2011). In order to perform a “leave one chromosome out” (LOCO) analysis, multiple genomic relationship matrices (GRMs) were created from the F2 60k SNP chip data by excluding each chromosome once with a minor allele frequency (MAF) cutoff of 1%. Mixed linear model association analyses (MLMAs) were performed with imputed F2 variants for each chromosome separately using the GRM where the respective chromosome was left out and a MAF cutoff of 1%. To account for the pooled population structure, covariates representing the different crosses (4 classes) were included in the MLMA. For further downstream analysis, significance threshold was established by applying Bonferroni correction (i.e., 0.05/number of independent tests). Manhattan plots were created with R, where variants with p-values > 0.001 were excluded due to software limitations. Clusters incorporating potential genomic regions of interest were defined using the Manhattan Harvester (MH) tool (Haller et al. 2019). MH provides quality assignment for each peak via a general quality score (GQS) which can be used as the main parameter for peak assessment. The GQS is generated based on a trained mixed-effects proportional odds model using 16 various parameters (e.g., maximal slope, height to width ratio) and human peak identification data. For this study, the variants with a p-value below 1.0x10−7 (option -inlimit) were included and further the clusters with a GQS > 3.5 (1 is min and 5 is max) were taken into account. Conditional association analyses were performed by including single highly associated variants as fixed effects in a LOCO analysis.
Variant effect prediction and gene enrichment analysis
To predict variant effects the Ensembl Variant Effect Predictor (VEP) release 94 was utilized, which is part of the Ensembl advanced programming interface (API) (McLaren et al. 2016). The vep command using the clusters’ statistically significant variants was executed with the following settings: –merged –force_overwrite –variant_class –symbol –nearest gene. To provide further functional interpretation, the Database for Annotation, Visualization and Integrated Discovery (DAVID) (Huang da et al. 2009) was used for a systematic and integrative analysis. The gene list from the VEP output was the input for DAVID (Huang et al. 2007) and Sus scrofa genes were considered as the background. Gene Ontology (GO) terms (i.e., cellular component, molecular function, and biological process) from the functional annotation chart report which were significantly overrepresented with an EASE Score (i.e., a modified Fisher Exact P-Value) below 0.05 and with a gene count higher or equal to 5 were retained.
Statement on data and reagent availability
Sequencing data, which were used to conduct this study will be made publicly available upon publication of the article in the NCBI Sequence Read Archive (SRA). Supplementary Tables have been uploaded to GSA. Supplementary Table 1 contains the coefficients of determination (R2) for each variant on QTL harboring chromosomes where calculation was possible. Supplementary Table 2 containts the complete list of clusters identified in the GWAS with additional supporting information for cluster assignment. GRMs from F2 60k genotypes (File_S1.zip) were created by a “leave one chromsome out” approach using the program “Genome-wide Complex Trait Analysis (GCTA) version 1.91.4 beta3”. GWAS was conducted with imputed sequence level F2 genotypes (Supplementary File_S2.zip) for each chromosome using the GRM where the respective chromosome was left out(GRM command: gcta64–bfile SG_F2_chip_wo_chrNO–autosome–maf 0.01–make-grm–out SG_F2_chip_wo_chrNO–thread-num 10–autosome-num 18 ; GWAS command: gcta64–mlma–covar Kiel_Hoh_cross.covar–bfile F2_beagle4.0_ped_ChrNO–grm SG_F2_chip_wo_chrNO–pheno TRAIT.pheno–out TRAIT_chrNO–maf 0.01–thread-num 10; replace NO with the respective chromosome number and TRAIT with the respective trait to be analyzed) Phenotype files are located in Supplementary File_S2.zip for the traits ADG, BFT, MFR, and CRCL were used in the GWAS. Crosses were used as covariates in the GWAS and provided as a gcta compatible covar file in Supplemental File_S4.zip. SNP locations can be found in the bim files of the genotype data (Supplementary File_S1.zip and Supplementary File_S2.zip). Population structure information is provided in form of a Beagle 4.0 compatible pedigree file (Supplementary File_S3.zip). Raw sequencing data are accessible via the NCBI Sequence Read Archive (SRA) under BioProject ID PRJNA553106. File_S5 contains the 60k chip genotype data in variant call file (VCF) format. Genomic positions have been lifted to genome assembly Sus scrofa 11.1 (GCA_000003025.6) and annotated with dbSNP build 150. gcta compatible covar files for the conditional association analyses with top variants are provided in File_S6. Supplemental material available at FigShare: https://doi.org/10.25387/g3.8287847.
Results
Whole genome sequencing and variant calling
An average of 592,788,350 (SD = 38,623,216, MIN = 525,921,083, MAX = 649,442,924) sequencing reads per sample were aligned to the reference genome in the F0 generation with an average mapping efficiency of 99.37%. In the F1 generation an average of 26,562,876 (SD = 6,619,568, MIN = 15,915,385, MAX = 59,300,856) reads were mapped to the reference assembly with an average mapping efficiency of 99.33%.
With respect to the number of SNPs detected in the founder population, 22,671,759 were previously reported and 3,950,955 were novel. Furthermore, 1,482,139 of the INDELs were previously reported and 4,335,345 were novel. Per chromosome, average distances among the variants are summarized in Table 2. The Ti/Tv of SNPs in the founder population was 2.39, whereas known SNPs had a Ti/Tv of 2.44 and novel SNPs had a Ti/Tv of 2.08. Due to the low sequencing coverage (average = 0.96 x, min = 0.58 x, max = 2.14 x) only autosomal SNPs were called in the F1 population. The raw output of Haplotypecaller consisted of 20,055,697 known and 3,529,441 novel SNPs and the raw output of mpileup contained 19,932,201 known and 3,291,758 novel SNPs. The intersection of the two datasets resulted in 19,264,662 known and 2,951,058 novel SNPs whereas removing all SNPs that were not present in the founder variant calling dataset lead to a final number of 19,224,132 known and 2,911,780 novel raw SNPs. After the application of a stringent filtering approach (see Material and Methods) 5,753,444 known and 741,155 novel SNPs remained in the variant calling dataset of the F1 population.
Table 2. Average distance between variants discovered in the founder population. A number of 24 F0 animals were sequenced at high coverage and the average distances between variants (SNPs and INDELs) were calculated per chromosome.
| Chromosome | Avg. distance (bp) | SD |
|---|---|---|
| 1 | 105,78729 | 196,8571 |
| 2 | 84,83889 | 201,4813 |
| 3 | 79,13779 | 183,627 |
| 4 | 80,16339 | 174,2767 |
| 5 | 73,37176 | 177,8913 |
| 6 | 85,34639 | 214,5176 |
| 7 | 79,12655 | 175,2067 |
| 8 | 78,90639 | 164,8448 |
| 9 | 79,61324 | 166,9157 |
| 10 | 56,5826 | 141,8648 |
| 11 | 67,6446 | 139,1412 |
| 12 | 65,41473 | 190,7484 |
| 13 | 102,70483 | 209,4908 |
| 14 | 83,58366 | 158,1296 |
| 15 | 93,02928 | 187,2321 |
| 16 | 71,18928 | 155,2467 |
| 17 | 65,91122 | 163,1281 |
| 18 | 76,95204 | 149,7542 |
| Mean | 82,21004 | 180,6988 |
Identification of local drops in imputation accuracy
To detect local inaccuracies in the imputed data, we imputed chip data from each founder with the remaining 23 founders as a reference panel. The data does not provide information about the imputation accuracy of the experiment since pedigree information could not be used. The coefficient of determination for each variant located on a chromosome harboring relevant QTL was determined where feasible. The average coefficients of determination for each chromosome analyzed are summarized in Table 3 (complete analysis results in Supplementary Table 1).
Table 3. Identification of local imputation inaccuracies. Chip data from each of the 24 founders was imputed using the remaining 23 founder animals as the reference panel. Coefficients of determination (R2) were calculated for each variant in order to calculate average R2 for SSC1, SSC2, SSC4, SSC7, SSC17, and SSC18.
| Chromosome | Average R2 | SD |
|---|---|---|
| 1 | 0.28 | 0.32 |
| 2 | 0.22 | 0.29 |
| 4 | 0.25 | 0.30 |
| 7 | 0.25 | 0.31 |
| 17 | 0.18 | 0.25 |
| 18 | 0.29 | 0.32 |
GWAS results and clusters
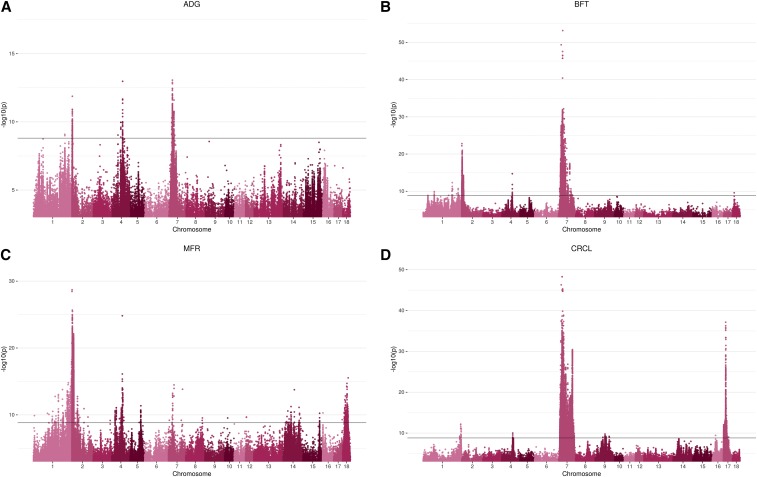
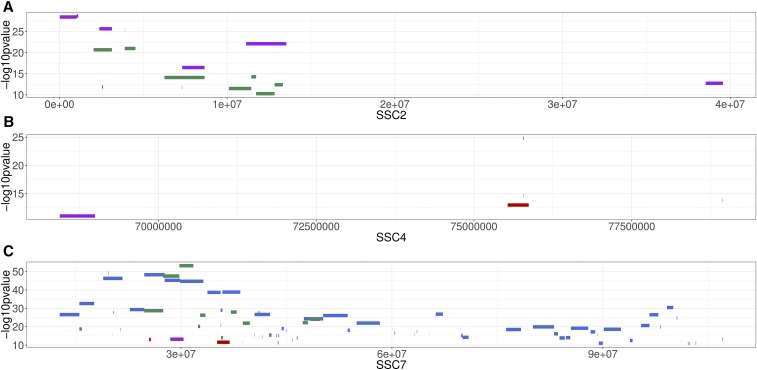
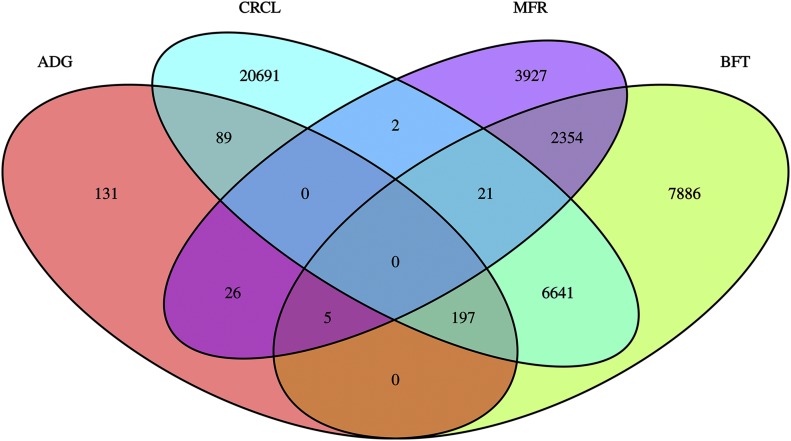
From the genome-wide association study conducted in the pooled F2 population, the following number of variants exceeded the genome-wide significance threshold: 448, 17,105, 6635, and 27,641 for ADG, BFT, MFR, and CRCL, respectively. Manhattan plots of the GWAS for the four phenotypic traits are shown in Figure 2. A total of 120 clusters were designated by the MH tool (i.e., 4 for ADG, 33 for BFT, 22 for MFR and 61 for CRCL) and they were located on the following Sus Scrofa chromosomes (SSC): 1, 2, 4, 5, 7, 17, and 18. The complete cluster list with additional supporting information for cluster assignment can be found in Supplementary Table 2. From each of the defined clusters, the top 5 variants were retained. The genes incorporating or lying nearby these highly significant associations are presented in Table 4. The clusters associated with the traits overlapped on several chromosomes, specifically on SSC2, SSC4, and SSC7. The location and the extent of the overlapping clusters is depicted in Figure 3. Particular chromosomes had exclusive clusters assigned, e.g., SSC17 for CRCL and SSC18 for MFR. To evaluate all possible relations among the variants exceeding the significance threshold for each trait, a Venn diagram was used (Figure 4). The highest number of common variants (i.e., 6,859) was between BFT and CRCL and the second highest was between BFT and MFR (i.e., 2,380). To get an estimate of systemic bias, quantile-quantile plots were generated for all p-values from each GWAS (Supplementary Figure 2). As a measure of association between observed and expected p-values, lambda values were calculated for all four traits: λADG = 1.282319, λBFT = 1.333425, λCRCL = 1.422044, and λMFR = 1.35587.
Figure 2.
Manhattan plots of the −log10 p-values for association of variants with the traits (A) average daily gain (ADG), (B) back fat thickness (BFT), (C) meat to fat ratio (MFR), and (D) carcass length (CRCL). P-values > 0.001 were excluded from the plots.
Table 4. Top associated genes for average daily gain (ADG), back fat thickness (BFT), meat to fat ratio (MFR), and carcass length (CRCL) identified in the GWAS. Genes incorporating or nearby the top 5 variants in the clusters are listed with chromosome and cluster numbers.
| Trait | SSC | Cluster no./SSC | Genes |
|---|---|---|---|
| ADG | 2 | 1 | SHANK2 |
| 4 | 1 | RPS20, LYN, PLAG1 | |
| 7 | 2 | HMGCLL1, TFEB | |
| BFT | 1 | 1 | ZNF462, ENSSSCG00000005432 |
| 2 | 7 | LOC102158414, PGA5, MRPL16, ENSSSCG00000013151, ZFP91, CTNND1, ENSSSCG00000024984, OR9Q2, OR10Q1 | |
| 4 | 1 | RPS20 | |
| 7 | 24 | SCGN, LRFN2, DAAM2, C7H6orf223, C7H6orf132, RIPOR2, CARMIL1, BMP5, ENSSSCG00000001500, KIFC1, C6orf106, PPARD, FKBP5, CPNE5, ENSSSCG00000001574, TMEM217, LRFN2, MRPS10, TRERF1, RUNX2, RCAN2, MEP1A, ADGRF5, PTCHD4, ENSSSCG00000001734, PGK2, IREB2, ABHD17C, GSTA2, CRABP1, CRISP3, PRSS16, TBC1D2B, ENSSSCG00000038708, BCL2A1, E2F3 | |
| MFR | 1 | 3 | LOC106507123, TMEM245, SCAI, ABL1, RAPGEF1, CFAP77, DDX31, MAPKAP1 |
| 2 | 8 | LOC102158414, LOC110259166, LOC110259708, TMEM80, DEAF1, EHD1, MACROD1, ATL3, NAV2, DHCR7, ENSSSCG00000028537, CTTN, SHANK2, ENSSSCG00000036180 (KRTAP5-5-like), NELL1 | |
| 4 | 3 | PDE7A, SNTG1, RPS20 | |
| 5 | 1 | ENSSSCG00000034097 | |
| 7 | 1 | ENSSSCG00000001500 | |
| 18 | 6 | PPP1R3A, IMMP2L, LRRC4, EXOC4, SND1, ELMO1, MDFIC, TFEC | |
| CRCL | 1 | 1 | FNBP1 |
| 7 | 52 | VEGFA, FLRT2, LRFN2, MCTP2, DAAM2, PGF, SV2B, MAX, COL21A1, KLHL25, NPAS3, LOC110261756, NHLRC1, TPMT, CDKAL1, GMNN, RIPOR2, MDC1, DDX39B, HMGCLL1, ENSSSCG00000001500, C6orf106, KCTD20, SRSF3, ENSSSCG00000001612, FOXP4, TFEB, RCAN2, ADGRF1, MUT, CRISP1, TFAP2D, PKHD1, BNC1, ENSSSCG00000001827, TMEM266, NKX2-1, PRKD1, LPCAT4, NR2F2, MCTP2, SLCO3A1, ENSSSCG00000002270, FUT8, ENSSSCG00000002317, DPF3, PTGR2, ZNF410, FAM161B, EIF2B2, MLH3, VIPAS39, SPTLC2, ENSSSCG00000010328, RF01299, RF00100, HMGN4, NRXN3, ID4, SYNJ2BP, ZFP36L1, RAD51B, AVEN, ANG, GCM1, FOXG1, ENSSSCG00000033840, ENSSSCG00000035274, RSL24D1, NSSSCG00000036697,ENSSSCG00000037115, ENSSSCG00000038445, CEMIP, SLC25A21, SPTSSA, ENSSSCG00000039877, DIO2, ENSSSCG00000040930 | |
| 17 | 8 | BMP2, JAG1, SPTLC3, TMX4 |
Figure 3.
Cluster overlap for (A) SSC2, (B) SSC4 and (C) SSC7 for all traits (average daily gain (ADG) – red, back fat thickness (BFT) – green, meat to fat ratio (MFR) – purple, and carcass length (CRCL) - blue). The heights of the clusters are according to the top variant (– log10 p-value) within each given cluster.
Figure 4.
Variants concordance and discordance between the traits average daily gain (ADG), back fat thickness (BFT), meat to fat ratio (MFR), and carcass length (CRCL). The Venn diagram contains statistically significant variants. Intersections between traits include the number of common variants. Numbers of variants that were exclusively found in the single traits are outside of intersections.
VEP and high impact variants
To predict functional consequences on genes the ensembl VEP tool was employed. Multiple transcripts per gene resulted in larger numbers of annotations that are reflected in the higher number of predicted effects as compared to the actual number of identified variants per trait. All inferred consequences for bonferroni corrected variants per trait and their percentage breakdown are summarized in Table 5. The large majority (over 70%) of the consequences were classified as intron variants. According to the severity of the variant consequence, intron variants are assigned to having a modifier impact, which means that predictions are difficult to be made or there is no solid evidence of impact. Variants inferred to have a disruptive impact on the protein, leading to protein truncation, loss of function or causing nonsense-mediated decay were of further interest. These significant high impact variants (Table 6) were mostly located on SSC7, with the exception of SSC2:rs1110687780 (splice donor variant) affecting TCN1 for the trait MFR. For the BFT, the most severe consequences were located in the genes C6orf89, PI16, DST, and PRIM2, while for the CRCL disruptive impact variants were found in NEU1, four novel genes, ABCD4, DST, PRIM2, and LPCAT4. Notably, the same two splice donor variants affect the common genes for BFT and CRCL: DST and PRIM2. Sorting Intolerant From Tolerant (SIFT) scores were determined for all significant missense variants and are summarized in Supplementary Table 3 (Ng and Henikoff 2003).
Table 5. Results of variant effect prediction for the production traits average daily gain (ADG), back fat thickness (BFT), meat to fat ratio (MFR), and carcass length (CRCL). Bonferroni-corrected variants were analyzed.
| Predicted effect | ADG | ADG % | BFT | BFT % | MFR | MFR % | CRCL | CRCL % |
|---|---|---|---|---|---|---|---|---|
| Missense variant | 2 | 0.1580 | 962* | 0.6523* | 58 | 0.1893 | 787* | 0.4750* |
| Frameshift variant | 0 | 0 | 0 | 0 | 0* | 0* | 6* | 0.0036* |
| Start lost | 0 | 0 | 1 | 0.0007 | 0 | 0 | 0 | 0 |
| Stop gained | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0.0006 |
| Inframe deletion | 0 | 0 | 1* | 0.0007* | 0 | 0 | 2 | 0.0012 |
| Intron variant | 936 | 73.9336 | 116815 | 79.2090 | 21556* | 70.3525* | 131590 | 79.4275 |
| 5 prime UTR variant | 0 | 0 | 229* | 0.1553* | 89 | 0.2905 | 277* | 0.1672* |
| 3 prime UTR variant | 8 | 0.6319 | 1160* | 0.7866* | 1242 | 4.0535 | 1543* | 0.9314* |
| Upstream gene variant | 50 | 3.9494 | 5680* | 3.8514* | 2195* | 7.1638* | 5300* | 3.1991* |
| Downstream gene variant | 44* | 3.4755* | 5791* | 3.9267* | 3442 | 11.2337 | 6893* | 4.1606* |
| Frameshift variant, splice region variant | 0 | 0 | 2 | 0.0014 | 0 | 0 | 0 | 0 |
| Missense variant, splice region variant | 0 | 0 | 41* | 0.0278* | 0 | 0 | 75* | 0.0453* |
| Splice region variant, non coding transcript exon variant | 0 | 0 | 2 | 0.0014 | 3* | 0.0098* | 5 | 0.0030 |
| Splice region variant, 3 prime UTR variant | 0 | 0 | 3* | 0.0020* | 3* | 0.0098* | 0 | 0 |
| Splice region variant, intron variant, non coding transcript variant | 0 | 0 | 2* | 0.0014* | 4 | 0.0131 | 20* | 0.0121* |
| Splice region variant, intron variant | 0 | 0 | 426* | 0.2889* | 41* | 0.1338* | 489* | 0.2952* |
| Splice region variant, synonymous variant | 0 | 0 | 21 | 0.0142 | 22* | 0.0718* | 28* | 0.0169* |
| Splice donor variant | 0 | 0 | 36 | 0.0244* | 1* | 0.0033 | 37 | 0.0223 |
| Intergenic variant | 109 | 8.6098 | 3318 | 2.2498 | 644 | 2.1018 | 9909 | 5.9811 |
| Synonymous variant | 0 | 0 | 2837 | 1.9237 | 214* | 0.6984* | 2751 | 1.6605 |
| Intron variant, non coding transcript variant | 117* | 9.2417* | 9636 | 6.5339 | 1060* | 3.4595* | 5759* | 3.4761* |
| Non coding transcript exon variant | 0 | 0 | 514* | 0.3485* | 66 | 0.2154 | 200* | 0.1207* |
| Start lost, start retained variant, 5 prime UTR variant | 0 | 0 | 0 | 0 | 0 | 0 | 1* | 0.0006* |
| Total | 1266 | 147477 | 30640 | 165673 |
Table 6. Statistically significant high impact variants that were discovered in the genome wide association studies for the production traits average daily gain (ADG), back fat thickness (BFT), meat to fat ratio (MFR), and carcass length (CRCL).
| Trait | High impact consequence | Variant | Position bp | Gene | Gene name |
|---|---|---|---|---|---|
| BFT | Start lost | SSC7:rs319855624 | 32544657 | C6orf89 | chromosome 7 C6orf89 homolog |
| Frameshift variant, splice region variant | SSC7:._504514 | 32606375 | PI16 | peptidase inhibitor 16 | |
| SSC7:._504513 | 32606373 | PI16 | peptidase inhibitor 16 | ||
| Splice donor variant | SSC7:rs80834233 | 29157904 | DST | dystonin | |
| SSC7:rs327743463 | 28571665 | PRIM2 | DNA primase subunit 2 | ||
| MFR | Splice donor variant | SSC2:rs1110687780 | 11630410 | TCN1 | transcobalamin 1 |
| CRCL | Start lost, start retained variant, 5 prime UTR variant | SSC7:rs793752812 | 23958518 | NEU1 | neuraminidase 1 |
| Stop gained | SSC7:rs334442580 | 87783592 | novel gene | ||
| Frameshift variant | SSC7:._1165873 | 97574140 | ABCD4 | ATP binding cassette subfamily D member 4 | |
| SSC7:rs693811701 | 48561663 | novel gene | aurora kinase A-like | ||
| SSC7:._1068730 | 87783712 | novel gene | |||
| SSC7:._1068731 | 87783718 | novel gene | |||
| Splice donor variant | SSC7:rs80834233 | 29157904 | DST | dystonin | |
| SSC7:rs327743463 | 28571665 | PRIM2 | DNA primase subunit 2 | ||
| SSC7:rs331245426 | 80150975 | LPCAT4 | lysophosphatidylcholine acyltransferase 4 |
Gene set analysis
GO functional enrichment analysis revealed eleven significantly overrepresented GO terms including molecular functions (MF), biological processes (BP), and cellular components (CC). A list containing the GO terms and the associated list of genes is presented in Table 7. For BFT a GO-MF term was overrepresented and related to calcium ion binding (GO:0005509). Several olfactory receptor genes were prevalent for the GO terms assigned to MFR (e.g., GO-BP GO:0007186 G-protein coupled receptor signaling pathway, GO-MF GO:0005549 odorant binding). The gene set for the CRCL trait was associated with two BP terms (GO:0001666 response to hypoxia and GO:0008283 cell proliferation) and two CC terms (GO:0045177 apical part of the cell and GO:0031410 cytoplasmic vesicle).
Table 7. Most significant Gene Ontology (GO) terms from DAVID for the top associated genes that were identified in genome wide association studies for the the traits back fat thickness (BFT), meat to fat ratio (MFR), and carcass length (CRCL).
| Trait | Category | Term | Genes |
|---|---|---|---|
| BFT | MF | GO:0005509 | DST, LOC100152993, SCGN, GUCA1B, ITPR3, CIB2, GUCA1A, RASGRP2 |
| calcium ion binding | |||
| MFR | BP | GO:0007186 | OR5B3, LOC100623017, LOC106509349, LOC100512519, LOC100513457, OR9Q2, LOC100628183, LOC100511243, LOC100512154, LOC100514032, LOC100521066, LOC100519351, OR10Q1, LOC100511620, LOC106509346 |
| G-protein coupled receptor signaling pathway | |||
| CC | GO:0016021 | ANO9, OR5B3, LOC100512519, LOC100519082, LOC100513457, LOC100628183, SIGIRR, LOC100512154, BET1L, LOC100521066, TMX2, OR10Q1, TMEM80, LOC100623017, LOC106509349, OR9Q2, LOC100511243, ZDHHC5, ATL3, LOC100514032, LRRC4, PPP1R3A, LOC100519351, LRRN3, LOC100511620, STX3, LOC100521938, CCDC136, LOC106509346, NRXN2 | |
| integral component of membrane | |||
| CC | GO:0005886 | OR5B3, EHD1, LOC100623017, LOC106509349, LOC100512519, OR9Q2, LOC100513457, LOC100628183, CTNND1, LOC100511243, ELMO1, LOC100512154, ZDHHC5, LOC100514032, LOC100521066, LOC100519351, STX3, LOC100511620, RABEPK, LOC106509346, RASGRP2 | |
| plasma membrane | |||
| MF | GO:0004930 | OR5B3, LOC100623017, LOC106509349, LOC100512519, LOC100513457, OR9Q2, LOC100628183, LOC100511243, LOC100512154, LOC100514032, LOC100521066, LOC100519351, OR10Q1, GPR141, LOC100511620, LOC106509346 | |
| G-protein coupled receptor activity | |||
| MF | GO:0004984 | OR5B3, LOC100623017, LOC106509349, LOC100512519, LOC100513457, OR9Q2, LOC100628183, LOC100511243, LOC100512154, LOC100514032, LOC100521066, LOC100519351, OR10Q1, LOC100511620, LOC106509346 | |
| olfactory receptor activity | |||
| MF | GO:0005549 | OR5B3, LOC100623017, LOC106509349, LOC100513457, OR9Q2, LOC100628183, LOC100512154, LOC100514032, LOC100521066, LOC100519351, OR10Q1, LOC100511620, LOC106509346 | |
| odorant binding | |||
| CRCL | BP | GO:0001666 | ANG, TGFB3, PGF, PLAT, VEGFA |
| response to hypoxia | |||
| BP | GO:0008283 | FURIN, FAM83B, ZFP36L1, MORF4L1, BYSL, RASGRF1 | |
| cell proliferation | |||
| CC | GO:0045177 | ADGRF5, VASH1, PLAT, HOMER2, BYSL | |
| apical part of cell | |||
| CC | GO:0031410 | ANG, ADGRF5, FES, NEU1, GRM4, RHGC | |
| cytoplasmic vesicle |
Discussion
Genotyping strategy
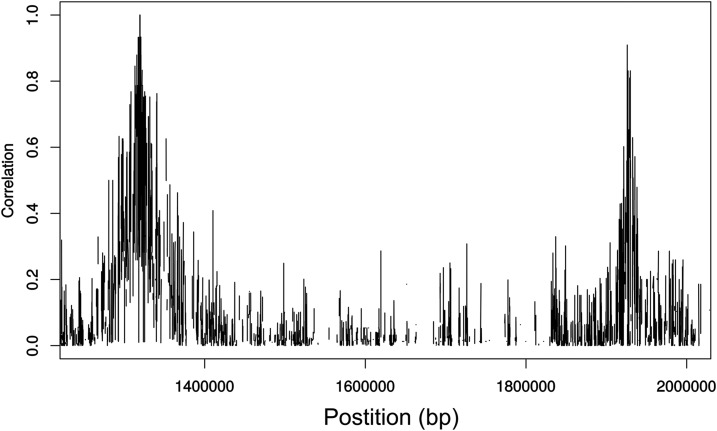
The genotyping strategy that we developed for this study is outlined in Figure 1. Briefly: 24 F0 pigs were subjected to high coverage Illumina short read sequencing and in addition 91 F1 animals were sequenced at low coverage and imputed to high coverage WGS levels in order to allow phasing. 2657 F2 animals were chip genotyped and imputed using a merged dataset of F0 and imputed F1 as reference-panel. All imputation steps involved pedigree information. Opposed to a population-based strategy this approach does not rely on a large reference-panel but on the relatedness of individuals. In general, the genotyping strategy can be considered reliable since the majority of the QTL identified were already described for the four traits analyzed in this study (cross-reference with Pig QTL database (Hu et al. 2019)). Nevertheless, we expected to identify a variant that was associated with muscle mass and fat deposition in exon 2 of IGF2, which has been extensively described to influence muscle development (Nezer et al. 1999). The absence of IGF2 associated variants can be explained by a local drop in coefficients of determination from an average of R2 = 0.22 to R2 = 0.03 in the genomic region where IGF2 resides (SSC2 1469183 – 1496417 bp, Figure 5). It must be pointed out that those coefficients of determination cannot be used to draw conclusions about the actual accuracy of the imputation. Since no pedigree information was included in the simulation, it can solely be used to identify local inaccuracies, which were most likely due to assembly errors in the reference genome.
Figure 5.
Imputation accuracy on SSC2 between positions 1,250,000 and 2,000,000. IGF2 is located between bp 1,469,183 and 1,496,417.
The genotyping approach presented in this study can be considered a reasonable strategy to radically increase the marker density of large F2 populations to WGS levels. By sequencing the founder individuals with high coverage and the F1 with low coverage, which are only a fracture of the number of F2 animals, the approach provides an affordable opportunity to improve the power and potential of otherwise obsolete datasets. Due to the relatedness of the animals deep sequencing of only a few animals is necessary, rendering it economically attractive.
Cluster identification and exploratory analysis
To fully exploit the potential of the four resource populations, the crosses were pooled and further used for conducting GWAS. The increased sample size together with the increased marker density ensures a high resolution that might allow the pinpointing of more specific causative genes and mutations. Further experiments, e.g., Sanger sequencing of promising regions could elaborate on that. Designing F2 populations implies that the LD-blocks are longer, a fact that is counteracted to some extent by jointly analyzing the four designs. Lambda values of 1.282319 to 1.422044 point to a moderate degree of p-value inflation in the GWAS, which is most likely caused by the usage of WGS data and a LOCO GWAS approach. However, to exploit the whole depth and power of the dataset we chose a LOCO analysis approach. To further comprehend the closely linked association signals from GWAS, the following approach was employed: i) clusters incorporating strong evidence for trait-associated chromosomal regions were defined, ii) the effect of the significant variants was predicted, and iii) a gene set analysis was employed to identify sets of genes jointly associated with the traits of interest.
The quantitative traits considered for this study have been investigated in the past and are mostly well represented in the Pig QTL database (Hu et al. 2019), except for MFR. The clusters assigned to each trait were compared with the QTL regions from the database. For MFR, additional fat-related traits (e.g., fat percentage in the carcass and fat-cuts percentage) were considered in order to allow an adequate comparison given that the trait has few records in the database and the trait definition can be country dependent. Most of the clusters overlapped or were in the vicinity of the previously reported QTL. This was expected as the database has been recently updated and also includes our previous results (Blaj et al. 2018) using SNP chip data and three out of the four pig populations which were taken into account here. Some of the earlier reported QTL in the database spread over large genomic regions (e.g., > 5Mb). It is assumed that many of these large QTL regions might in fact not be due to a single mutation, thus representing haplotype effects caused by several causative variants (Andersson 2009). In the current study, we were able to assign numerous clusters within these regions, which implies that a higher genomic resolution was achieved and that it may be possible to disentangle distinct quantitative trait nucleotides.
Conditional association analyses by including the top variant as a fixed effect in the MLMA were carried out in order to gather statistical evidence for putative causality (Cohen-Zinder et al. 2005) and was specifically applied to CRCL and BFT on SSC7. This chromosome exhibits the highest number of clusters (SM with Clusters) and the highest association signals. By including the top variant (rs81228492) for BFT, only one well-supported peak was above the significance threshold (Supplementary Figure 1) meaning that there is additional genetic variation within this region. Similarly, for CRCL the two top variants (rs333021601 and rs319044994) representing the two different significant genomic regions were included alternatively in the model. After fixing the effect of the latter variant, the surrounding significant region disappeared, pointing to the possibility that there could be only one QTL responsible for CRCL on SSC7 around the 99 Mb region. An alternative or additional explanation could be the presence of long LD blocks, long-range LD and/or various epistatic interactions among the loci. The overlap among the BFT and CRCL significant variants (see Figure 3 and Figure 4) localized mostly in the genomic region 24-32 Mb indicate the existence of pleiotropic loci for the two traits. When conditioned on the top BFT variant (rs81228492) as a fixed effect for a MLMA on CRCL and the top CRCL variant (rs333021601) for MLMA on BFT, the initially associated clusters and those nearby dropped in the intensity of the association signals (Supplementary Figure 1), supporting the presence of pleiotropic loci. It is also noteworthy that CRCL might be influenced by the number of thoracolumbar vertebrae (Rohrer et al. 2015). Since the variant that has been associated with a higher number of vertebrae is a large Indel in intron 1 of the VRTN gene (Fan et al. 2013) we were not able to discover this variant since the genotyping pipeline applied in this study does only cover small INDELs.
In order to gain insight into the possible genetic mechanisms that control the traits, an enrichment analysis of the gene function was performed with DAVID, prioritizing on the GO terms. The GO-MF calcium ion binding term found for BFT supports the relationship between the calcium ion, food intake and lipid metabolism previously described in the literature (Cui et al. 2017). Furthermore, one of the genes in this group is DST, a strong candidate gene for which high impact variants were found via VEP, which is discussed in detail below. A GO-BP term related to cell proliferation comprised the FAM83B gene, which is the gene incorporating the top variant found for CRCL. Interestingly, the majority of the genes included in the over-represented terms for MFR were olfactory receptors. This enrichment is a consequence of the MFR-identified clusters overlapping regions that are rich in various olfactory receptor genes. This particular gene family is known to have significant expansion throughout time within the Sus Scrofa genome (Nguyen et al. 2012).
Variant effect prediction
ADG:
A QTL for ADG found on SSC7 comprises 115 statistically significant intron variants and 83 variants upstream (min. p-value 8.71 × 10−14) of the HMGCLL1 gene, which was shown by Comuzzie et al. to be associated with childhood obesity in the Hispanic population and to influence creatinine levels. Another QTL on SSC2 contains 112 intron variants in SHANK2 (min. p-value 1.33 × 10−12). SHANK2 was also shown to be associated with childhood obesity in the same study and to have an influence on estradiol blood concentrations (Comuzzie et al. 2012). A third QTL on SSC4 harbors 2 intron and 12 downstream variants (min p-value 1.06 × 10−13) affecting LYN, which encodes for the LYN proto-oncogene, which was also identified by Comuzzie et al. and correlated with the amount of fat mass in obese children (Comuzzie et al. 2012). Six additional variants in the QTL on SSC4 (min. p-value 2.44 × 10−12) lie in an intergenic region 13,463 – 14,460 bp downstream of RPS20, a gene which in interplay with GNL1 is critical for cell growth (Krishnan et al. 2018). Another likely candidate SNP to influence ADG is an intron variant in the PLAG1 transcription factor (p-value 1.32 × 10−11), which is a regulator of IGF2 expression (Zatkova et al. 2004).
BFT:
A QTL for BFT with a very prominent peak was detected on SSC7. The SNP with the lowest p-value (6.63 × 10−54) is an intron variant in gene C6orf106. C6orf106 is a target of the human miRNA has-miR-192, which has been identified to have regulatory functions in type 2 diabetes mellitus (Cui et al. 2016). The second top scoring SNP is an intron variant in the RIPOR2 gene (p-value 4.34 × 10−50). RIPOR2 expression and protein levels are upregulated during muscle cell differentiation in human fetal muscle cells (Yoon et al. 2007). Another gene containing top scoring variants on SSC7 is KIFC1 (7 intron variants, min p-value 3.12 × 10−47). Overexpression of KIFC1 promotes cell proliferation in non-small cell lung cancer (Liu et al. 2016). 21 intron and 8 downstream variants in BMP5 (min. p-value 1.91 × 10−29), which induces cartilage and bone formation (Wozney et al. 1988), are also located in a cluster on SSC7. 6 variants downstream of the aforementioned RPS20 (min. p-value 1.90 × 10−15) were found in the cluster on SSC4.
MFR:
GWAS for the MFR trait revealed a strong QTL on SSC2 with variant rs81327136 upstream of KRTAP5-5-like being the most significant (p-value 1.59 × 10−23). Of 72 variants 6 were located in KRTAP5-5-like introns and 66 in the vicinity of the gene. KRTAP5-5 was shown to control cytoskeletal function and cancer cell vascular invasion (Berens et al. 2017). Other variants found in clusters on SSC2 are located in or adjacenct to DEAF1 (8 intron variants, min p-value 3.47 × 10−29), which is a transcription factor that regulates proliferation of epithelial cells (Barker et al. 2008) and that forms a dominant-negative splice isoform in type 1 diabetes, which correlates with disease severity (Yip et al. 2015). Clusters on SSC2 also harbor variants associated with SHANK2 (1,714 intron variants, 3 5′ UTR variants, min p-value 2.18 × 10−26) and CTTN (188 up- and downstream variants, min p-value 1.53 × 10−25). CTTN’s protein product Cortactin binds to and is indirectly phosphorylated by obesity factor PTP1B (Stuible et al. 2008). A noteworthy intron variant is located in the vitamin D pathway gene DHCR7 (p-value 3.06 × 10−25), which has been associated with obesity traits in humans (Vimaleswaran et al. 2013). A total of 14 DHCR7 intron variants were above the significance threshold. A less prominent QTL on SSC4 harbors variants in or close to the aforementioned genes RPS20 (17 downstream variants, min p-value 1.51 × 10−25) and in SNTG1 (19 intron variants, min p-value 1.48 × 10−14), which has been associated with type 2 diabetes (Ban et al. 2010). A third, rather minor QTL on SSC18, contains 21 variants downstream of MDFIC (min p-value 1.97 × 10−15), a gene which has been linked to improved piglet birth weight (Zhang et al. 2014). 25 intron and 42 downstream variants were found for the PPP1R3A gene (min p-value 6.92 × 10−15), which in a whole exome sequencing study was found to be associated with type 2 diabetes in a Mayan population (Sánchez-Pozos et al. 2018).
CRCL:
In the GWAS for CRCL 52 clusters were identified on SSC7. Although not located in one of the clusters, the two lowest p-values (min p-value 5.40 × 10−49) were found in the intron and coding region (silent mutation) of FAM83B (or C6orf143) respectively. A total of 62 significant variants in FAM83B were discovered comprising of 60 intron variants, 1 silent mutation, and 1 missense mutation. Cipriano et al. demonstrated that overexpression or mutation of FAM83B leads to EGFR hyperactivation by direct interaction and consequent hyperactivation of the EGFR downstream effector phospholipase D1, which was previously associated with BMI in humans (Davenport et al. 2015). An intron variant in the RIPOR2 gene with a p-value of 5.08 × 10−47 is the same SNP, which was found in the GWAS for BFT. A total of 85 mostly intronic RIPOR2 variants were found for the CRCL trait. A second, less prominent QTL on SSC7 harbors 9 intron, 12 downstream and 317 upstream variants (min p-value 3.62 × 10−31), which have been assigned to the RSL24D1 gene. RSL24D1 has been identified as a potential target in familial hypercholesterolemia (Li et al. 2015). One of the clusters identified for CRCL on SSC17 contains 230 variants 122,416-126,520 bp downstream of BMP2 (min p-value 7.21 × 10−38), a bone formation inducing factor (Wang et al. 2013). In addition, 18 intron and 114 variants upstream of TMX4 were discovered. TMX4 was associated with feed conversion ratios in chickens (Shah et al. 2016).
High impact variants:
Various high impact variants were discovered by variant effect prediction. A splice donor variant (rs80834233) in DST, the gene encoding Dystonin, is associated with BFT (p-value 1.98 × 10−19) and CRCL (p-value 1.25 × 10−19). Knockout of DST leads to intrinsic muscle weakness and instability of skeletal muscle cytoarchitecture in mice (Dalpé et al. 1999). Variant rs793752812 leads to a probable start codon loss in NEU1 and is associated with CRCL (p-value 1.49 × 10−12). A deficiency of the NEU1 gene product Neuraminidase 1 leads to vertebral deformities in humans (Sphranger et al. 1977), which is reasonable considering CRCL is largely determined by the number of vertebrae. Furthermore one frameshift variant in AURKA (rs693811701, p-value 2.95 × 10−12) and one splice donor variant in NUTM1 (rs331245426, p-value 1.38 × 10−9), both oncogenes (Umene et al. 2015) (Schaefer et al. 2018), are associated with CRCL. The splice donor variant rs1110687780, which affects the gene coding for placenta-specific protein 1-like, was detected in the GWAS for MFR. In humans, PLAC1 has been found to be highly expressed in various types of tumors (Koslowski et al. 2007).
Application of results in breeding programs and follow up studies
Functional validation studies based on appointed candidate genes and genetic variants will be considered in follow-up studies. Besides understanding the underlying molecular mechanisms of ADG, BFT, MFR and CRCL, the results of GWAS can render a substantial increase in the reliability of genomic predictions in breeding programs. This concept was demonstrated in several studies in cattle (Brøndum et al. 2015; Porto-Neto et al. 2015; van den Berg et al. 2016) and in Drosophila melanogaster (Ober et al. 2015) by including pre-selected variants from GWAS results in the prediction models. Even though implementing genomic selection is becoming a common practice, the usage of marker-assisted selection or genomic screening is not obsolete pointing out that the identification of relevant genetic markers via GWAS and post-GWAS analyses is still of practical importance in pig breeding.
Conclusion
Putting the results of previous simulation studies to test, we conducted GWAS in four pooled F2 designs, which have been imputed to sequence level based on high coverage founder and low coverage F1 sequencing. We found that by pooling the designs the sequence level marker density can be exploited efficiently. QTL for four well-characterized traits were identified in agreement with previous mapping studies and candidate genes and pathways were unraveled, that should be subject to further studies. Thus, the approach applied herein is a feasible strategy to efficiently utilize extremely well phenotyped experimental designs that have been established in the past.
Acknowledgments
We acknowledge support by the German Research Foundation and the Open Access Publication Funds of the Göttingen University. We also express our gratitude to Dr. Alexander Charles Mott for proof reading and language advice.
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25387/g3.8287847.
Communicating editor: D. J. de Koning
Literature Cited
- Andersson L., 2009. Genome-wide association analysis in domestic animals: a powerful approach for genetic dissection of trait loci. Genetica 136: 341–349. 10.1007/s10709-008-9312-4 [DOI] [PubMed] [Google Scholar]
- Ban H.-J., Heo J. Y., Oh K.-S., and Park K.-J., 2010. Identification of Type 2 Diabetes-associated combination of SNPs using Support Vector Machine. BMC Genet. 11: 26 10.1186/1471-2156-11-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker H. E., Smyth G. K., Wettenhall J., Ward T. A., Bath M. L. et al. , 2008. Deaf-1 regulates epithelial cell proliferation and side-branching in the mammary gland. BMC Dev. Biol. 8: 94 10.1186/1471-213X-8-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berens E. B., Sharif G. M., Schmidt M. O., Yan G., Wellstein A. et al. , 2017. Keratin-associated protein 5-5 controls cytoskeletal function and cancer cell vascular invasion. Oncogene 36: 593–605. 10.1038/onc.2016.234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaj I., Tetens J., Preuß S., Bennewitz J., and Thaller G., 2018. Genome-wide association studies and meta-analysis uncovers new candidate genes for growth and carcass traits in pigs. PLoS One 13: e0205576 10.1371/journal.pone.0205576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchers N., Reinsch N., and Kalm E., 2000. Familial cases of coat colour-change in a Piétrain cross. J. Anim. Breed. Genet. 117: 285–287. 10.1046/j.1439-0388.2000.00255.x [DOI] [Google Scholar]
- Brøndum R. F., Su G., Janss L., Sahana G., Guldbrandtsen B. et al. , 2015. Quantitative trait loci markers derived from whole genome sequence data increases the reliability of genomic prediction. J. Dairy Sci. 98: 4107–4116. 10.3168/jds.2014-9005 [DOI] [PubMed] [Google Scholar]
- Browning B. L., Zhou Y., and Browning S. R., 2018. A One-Penny Imputed Genome from Next-Generation Reference Panels. Am. J. Hum. Genet. 103: 338–348. 10.1016/j.ajhg.2018.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning S. R., and Browning B. L., 2007. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 81: 1084–1097. 10.1086/521987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. C., Chow C. C., Tellier L. C., Vattikuti S., Purcell S. M. et al. , 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4: 7 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Zinder M., Seroussi E., Larkin D. M., Loor J. J., Wind A. E. et al. , 2005. Identification of a missense mutation in the bovine ABCG2 gene with a major effect on the QTL on chromosome 6 affecting milk yield and composition in Holstein cattle. Genome Res. 15: 936–944. 10.1101/gr.3806705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comuzzie A. G., Cole S. A., Laston S. L., Voruganti V. S., Haack K. et al. , 2012. Novel genetic loci identified for the pathophysiology of childhood obesity in the Hispanic population. PLoS One 7: e51954 10.1371/journal.pone.0051954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H., Yang S., Zheng M., Liu R., Zhao G. et al. , 2017. High-salt intake negatively regulates fat deposition in mouse. Sci. Rep. 7: 2053 10.1038/s41598-017-01560-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Chen W., Chi J., and Wang L., 2016. Comparison of Transcriptome Between Type 2 Diabetes Mellitus and Impaired Fasting Glucose. Med. Sci. Monit. 22: 4699–4706. 10.12659/MSM.896772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalpé G., Mathieu M., Comtois A., Zhu E., Wasiak S. et al. , 1999. Dystonin-deficient mice exhibit an intrinsic muscle weakness and an instability of skeletal muscle cytoarchitecture. Dev. Biol. 210: 367–380. 10.1006/dbio.1999.9263 [DOI] [PubMed] [Google Scholar]
- Davenport E. R., Cusanovich D. A., Michelini K., Barreiro L. B., Ober C. et al. , 2015. Genome-Wide Association Studies of the Human Gut Microbiota. PLoS One 10: e0140301 10.1371/journal.pone.0140301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., Xing Y., Zhang Z., Ai H., Ouyang Z. et al. , 2013. A further look at porcine chromosome 7 reveals VRTN variants associated with vertebral number in Chinese and Western pigs. PLoS One 8: e62534 10.1371/journal.pone.0062534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii J., Otsu K., Zorzato F., de Leon S., Khanna V. et al. , 1991. Identification of a mutation in porcine ryanodine receptor associated with malignant hyperthermia. Science 253: 448–451. 10.1126/science.1862346 [DOI] [PubMed] [Google Scholar]
- Haller T., Tasa T., and Metspalu A., 2019. Manhattan Harvester and Cropper: a system for GWAS peak detection. BMC Bioinformatics 20: 22 10.1186/s12859-019-2600-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes B., and Goddard M. E., 2001. The distribution of the effects of genes affecting quantitative traits in livestock. Genetics, Selection. Evolution GSE 33: 209–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Park C. A., and Reecy J. M., 2019. Building a livestock genetic and genomic information knowledgebase through integrative developments of Animal QTLdb and CorrDB. Nucleic Acids Res. 47: D701–D710. 10.1093/nar/gky1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T., and Lempicki R. A., 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Tan Q., Collins J. R., and Alvord W. G. et al. , 2007. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 8: R183 10.1186/gb-2007-8-9-r183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott S. A., 2005. Regression-based quantitative trait loci mapping: robust, efficient and effective. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360: 1435–1442. 10.1098/rstb.2005.1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koslowski M., Sahin U., Mitnacht-Kraus R., Seitz G., Huber C. et al. , 2007. A placenta-specific gene ectopically activated in many human cancers is essentially involved in malignant cell processes. Cancer Res. 67: 9528–9534. 10.1158/0008-5472.CAN-07-1350 [DOI] [PubMed] [Google Scholar]
- Krishnan R., Boddapati N., and Mahalingam S., 2018. Interplay between human nucleolar GNL1 and RPS20 is critical to modulate cell proliferation. Sci. Rep. 8: 11421 10.1038/s41598-018-29802-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Wu X.-J., Kong X.-Q., Wang L., and Jin X., 2015. Cytochrome c oxidase subunit VIIb as a potential target in familial hypercholesterolemia by bioinformatical analysis. Eur. Rev. Med. Pharmacol. Sci. 19: 4139–4145. [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J. et al. , 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Zhan P., Zhou Z., Xing Z., Zhu S. et al. , 2016. The overexpression of KIFC1 was associated with the proliferation and prognosis of non-small cell lung cancer. J. Thorac. Dis. 8: 2911–2923. 10.21037/jtd.2016.10.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K. et al. , 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W., Gil L., Hunt S. E., Riat H. S., Ritchie G. R. S. et al. , 2016. The Ensembl Variant Effect Predictor. Genome Biol. 17: 122 10.1186/s13059-016-0974-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamine Y., Haley C. S., Sewalem A., and Visscher P. M., 2003. Quantitative trait loci variation for growth and obesity between and within lines of pigs (Sus scrofa). Genetics 164: 629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezer C., Moreau L., Brouwers B., Coppieters W., Detilleux J. et al. , 1999. An imprinted QTL with major effect on muscle mass and fat deposition maps to the IGF2 locus in pigs. Nat. Genet. 21: 155–156. 10.1038/5935 [DOI] [PubMed] [Google Scholar]
- Ng P. C., and Henikoff S., 2003. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 31: 3812–3814. 10.1093/nar/gkg509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D. Truong, Lee K., Choi H., Choi M.-k., Thong Le M. et al. , 2012. The complete swine olfactory subgenome: expansion of the olfactory gene repertoire in the pig genome. BMC genomics 13: 584 10.1186/1471-2164-13-584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober U., Huang W., Magwire M., Schlather M., Simianer H. et al. , 2015. Accounting for genetic architecture improves sequence based genomic prediction for a Drosophila fitness trait. PLoS One 10: e0126880 10.1371/journal.pone.0126880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porto-Neto L. R., Barendse W., Henshall J. M., McWilliam S. M., Lehnert S. A. et al. , 2015. Genomic correlation: harnessing the benefit of combining two unrelated populations for genomic selection. Genetics, Selection. Evolution GSE 47: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos A. M., Crooijmans R. P. M. A., Affara N. A., Amaral A. J., Archibald A. L. et al. , 2009. Design of a high density SNP genotyping assay in the pig using SNPs identified and characterized by next generation sequencing technology. PLoS One 4: e6524 10.1371/journal.pone.0006524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrer G. A., Nonneman D. J., Wiedmann R. T., and Schneider J. F., 2015. A study of vertebra number in pigs confirms the association of vertnin and reveals additional QTL. BMC Genet. 16: 129 10.1186/s12863-015-0286-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothschild M. F., Hu Z., and Jiang Z., 2007. Advances in QTL mapping in pigs. Int. J. Biol. Sci. 3: 192–197. 10.7150/ijbs.3.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rückert C., and Bennewitz J., 2010. Joint QTL analysis of three connected F2-crosses in pigs. Genetics, Selection. Evolution GSE 42: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Pozos K., Ortíz-López M. G., Peña-Espinoza B. I., de Los Ángeles Granados-Silvestre M., Jiménez-Jacinto V. et al. , 2018. Whole-exome sequencing in maya indigenous families: variant in PPP1R3A is associated with type 2 diabetes. Molecular genetics and genomics MGG 293: 1205–1216. 10.1007/s00438-018-1453-2 [DOI] [PubMed]
- Schaefer I.-M., Dal Cin P., Landry L. M., Fletcher C. D. M., Hanna G. J. et al. , 2018. CIC-NUTM1 fusion: A case which expands the spectrum of NUT-rearranged epithelioid malignancies. Genes Chromosomes Cancer 57: 446–451. 10.1002/gcc.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Wellmann R., and Bennewitz J., 2018. Power and precision of QTL mapping in simulated multiple porcine F2 crosses using whole-genome sequence information. BMC Genet. 19: 22 10.1186/s12863-018-0604-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah T. M., Patel N. V., Patel A. B., Upadhyay M. R., Mohapatra A. et al. , 2016. A genome-wide approach to screen for genetic variants in broilers (Gallus gallus) with divergent feed conversion ratio. Molecular genetics and genomics MGG 291: 1715–1725. [DOI] [PubMed]
- Sphranger J., Gehler J., and Cantz M., 1977. Mucolipidosis I–a sialidosis. Am. J. Med. Genet. 1: 21–29. 10.1002/ajmg.1320010104 [DOI] [PubMed] [Google Scholar]
- Stratz P., Schmid M., Wellmann R., Preuß S., Blaj I. et al. , 2018. Linkage disequilibrium pattern and genome-wide association mapping for meat traits in multiple porcine F2 crosses. Anim. Genet. 49: 403–412. 10.1111/age.12684 [DOI] [PubMed] [Google Scholar]
- Stuible M., Dubé N., and Tremblay M. L., 2008. PTP1B Regulates Cortactin Tyrosine Phosphorylation by Targeting Tyr446*S. J. Biol. Chem. 283: 15740–15746. 10.1074/jbc.M710534200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umene K., Yanokura M., Banno K., Irie H., and Adachi M. et al. , 2015. Aurora kinase A has a significant role as a therapeutic target and clinical biomarker in endometrial cancer. Int. J. Oncol. 46: 1498–1506. 10.3892/ijo.2015.2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg I., Boichard D., and Lund M. S., 2016. Sequence variants selected from a multi-breed GWAS can improve the reliability of genomic predictions in dairy cattle. Genetics, Selection. Evolution GSE 48: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vimaleswaran K. S., Cavadino A., Berry D. J., Whittaker J. C., Power C. et al. , 2013. Genetic association analysis of vitamin D pathway with obesity traits. Int. J. Obes. 37: 1399–1406. 10.1038/ijo.2013.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Park P., La Marca F., Than K., Rahman S. et al. , 2013. Bone formation induced by BMP-2 in human osteosarcoma cells. Int. J. Oncol. 43: 1095–1102. 10.3892/ijo.2013.2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellmann R., Preuß S., Tholen E., Heinkel J., Wimmers K. et al. , 2013. Genomic selection using low density marker panels with application to a sire line in pigs. Genetics, Selection. Evolution GSE 45: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozney J. M., Rosen V., Celeste A. J., Mitsock L. M., Whitters M. J. et al. , 1988. Novel regulators of bone formation: molecular clones and activities. Science 242: 1528–1534. 10.1126/science.3201241 [DOI] [PubMed] [Google Scholar]
- Yang J., Lee S. H., Goddard M. E., and Visscher P. M., 2011. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88: 76–82. 10.1016/j.ajhg.2010.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip L., Fuhlbrigge R., Taylor C., Creusot R. J., Nishikawa-Matsumura T. et al. , 2015. Inflammation and hyperglycemia mediate Deaf1 splicing in the pancreatic lymph nodes via distinct pathways during type 1 diabetes. Diabetes 64: 604–617. 10.2337/db14-0803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S., Molloy M. J., Wu M. P., Cowan D. B., and Gussoni E., 2007. C6ORF32 is upregulated during muscle cell differentiation and induces the formation of cellular filopodia. Dev. Biol. 301: 70–81. 10.1016/j.ydbio.2006.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatkova A., Rouillard J.-M., Hartmann W., Lamb B. J., Kuick R. et al. , 2004. Amplification and overexpression of the IGF2 regulator PLAG1 in hepatoblastoma. Genes Chromosomes Cancer 39: 126–137. 10.1002/gcc.10307 [DOI] [PubMed] [Google Scholar]
- Zhang L., Zhou X., Michal J. J., Ding B., Li R. et al. , 2014. Genome Wide Screening of Candidate Genes for Improving Piglet Birth Weight Using High and Low Estimated Breeding Value Populations. Int. J. Biol. Sci. 10: 236–244. 10.7150/ijbs.7744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z.-Y., Li A., Otecko N. O., Liu Y.-H., Irwin D. M. et al. . 2017. PigVar: a database of pig variations and positive selection signatures. Database the journal of biological databases and curation. [DOI] [PMC free article] [PubMed]