Abstract
A better understanding of the environmental and genetic contribution to migratory behavior and the evolution of traits linked to migration is crucial for fish conservation and fisheries management. Up to date, a few genes with unequivocal influence on the adoption of alternative migration strategies have been identified in salmonids. Here, we used a common garden set-up to measure individual migration distances of generally highly polymorphic brown trout Salmo trutta from two populations. Fish from the assumedly resident population showed clearly shorter migration distances than the fish from the assumed migratory population at the ages of 2 and 3 years. By using two alternative analytical pipelines with 22186 and 18264 SNPs obtained through RAD-sequencing, we searched for associations between individual migration distance, and both called genotypes and genotype probabilities. None of the SNPs showed statistically significant individual effects on migration after correction for multiple testing. By choosing a less stringent threshold, defined as an overlap of the top 0.1% SNPs identified by the analytical pipelines, GAPIT and Angsd, we identified eight candidate genes that are potentially linked to individual migration distance. While our results demonstrate large individual and population level differences in migration distances, the detected genetic associations were weak suggesting that migration traits likely have multigenic control.
Keywords: Life-history strategies, RADseq, GWAS, salmonids
Genome-wide association studies (GWAS) aim to reveal links between genotypes and phenotypes. Originally developed for case-control comparisons in medical sciences (Ku et al. 2010), association mapping has been subsequently adopted for use on other organisms and for addressing agricultural, evolutionary and ecological questions. Recent studies have described genetic determinants for economically and ecologically important traits. For example, vgll3 locus affects maturation age in Atlantic salmon Salmo salar in sex-dependent fashion (Ayllon et al. 2015; Barson et al. 2015) and greb1l affects migration timing in Pacific salmonids Onchorynchus spp. (Prince et al. 2017). These studies have revealed that traits that were traditionally thought to be influenced by tens or hundreds of genes (Waples et al. 2004; Johnston et al. 2014; Gutierrez et al. 2015) can actually be influenced by loci with major effect. Identification of these genotype-phenotype associations have helped to further understand the evolution of these traits in response to both natural and human-induced selection pressures (Czorlich et al. 2018).
Salmonids display a variety of anadromous and potamodromous migratory strategies with populations ranging from fully resident to fully migratory (Klemetsen et al. 2003; Chapman et al. 2012; Dodson et al. 2013). Prior to migration, some juvenile salmonids usually undergo a series of physiological and morphological changes, known as smoltification, that prepare the fish for seawater migration and entry to novel environments (Folmar and Dickhoff 1980; McCormick 2009; McCormick et al. 2013). During recent years, the genetic components underlying the dichotomy between resident and migratory forms have been increasingly studied, particularly in the genus Onchorhynchus (e.g., Hale et al. 2013; Nichols et al. 2016; Veale and Russello 2017). A single genomic region in chromosome 5 has been linked to the migration differences between resident “rainbow trout” and migratory “steelhead” populations of O. mykiss (Hecht et al. 2012; Pearse et al. 2014; Leitwein et al. 2017a), yet not in all (Hale et al. 2013). In addition, an extensive list of candidate genes for migration propensity has been identified for O. mykiss (Hecht et al. 2013; Hess et al. 2016) as well as for other species within the genus (Nichols et al. 2016; Veale and Russello 2017). To obtain a representative view of genetic variants influencing propensity to migrate, genome wide markers with as good coverage as possible should optimally be used. However, any significant indications for genetic control would be important in answering the question whether migration propensity can evolve in response to natural and human-induced selection pressures.
Brown trout Salmo trutta is an ecologically and economically important salmonid native to Europe, Asia and Northern Africa, and occurs currently as migratory, resident and partially migratory populations in all continents except for Antarctica (MacCrimmon and Marshall 1968). Migratory (anadromous and potamodromous) populations are frequently threatened by anthropogenic factors such as dam building and overfishing, while many resident populations are often isolated and occur mainly in small headwaters with lesser human impact. It is crucial to gain knowledge about the underlying causes of migration in brown trout to understand the historical and contemporary interactions between resident and migratory forms, and resolve the evolvability of migration tendency in both hatchery breeding and fisheries that may impose selection on migration traits. Traditionally, brown trout have been considered an extreme example of phenotypic plasticity when it comes to migration, with food availability and conspecific density reportedly driving migratory behavior in empirical field-studies (Olsson et al. 2006; Wysujack et al. 2009; Kendall et al. 2015). However, by comparing migratory and resident brown trout populations using genome-wide genetic markers, we have recently identified a subset of outlier genes that potentially influence migration propensity (Lemopoulos et al. 2018). However, the extent of genetic control over this trait, as well as whether a common basis for migration exists among different populations of brown trout or among salmonids, has yet to be determined (Ferguson et al. 2019).
Studying individual-level migration patterns in wild populations is challenging for a number of reasons, including high cost, intensive workload and potential negative effects of telemetry tags on focal animals (Thorstad et al. 2013). Moreover, natural populations are subject to different environmental pressures and any genetic signatures of selection can reflect processes that are independent of, or completely confounded with, the adaptations directly linked to migration. Experimental common-garden designs can overcome these challenges by providing uniform environmental conditions for all genotypes (Liedvogel et al. 2011; de Villemereuil et al. 2015). Migration tendency consists of overall probability to migrate (migration propensity) and eventual migration distance. Because migration tendency can be a difficult trait to measure per se, experimental studies have often focused on smoltification-related traits such as growth, condition, body coloration, morphology and osmoregulatory enzyme activities (Nichols et al. 2008; Hecht et al. 2012; Baerwald et al. 2016). Despite the potential advantage of using a common-garden design to create uniform environmental conditions and the power of genome-wide markers to reveal genetic determinants of migratory behavior, only few studies to date have successfully combined these two approaches (e.g., Nichols et al. 2008; Narum et al. 2011; Hecht et al. 2014).
Here, we performed a multi-year common garden experiment to investigate the genetic basis of migration tendency, measured as migration distance in brown trout. We artificially propagated brown trout from one predominantly migratory and one predominantly resident population (Lemopoulos et al. 2018) originating from a single water system using a replicated full factorial 3 males × 3 females matrix design, and continuously followed the movements of F1 individuals over two smolt migration seasons at the ages of two and three years. Restriction site associated DNA sequencing (RADseq) was used to genotype 116 individuals from the tails of the estimated distribution of migration distances (i.e., individuals showing the longest and shortest migration distances). We used two complementary association analyses to identify candidate genes for migration tendency in data corrected for population differences. Since brown trout migratory behavior has been recently found to associate with several outlier markers (Lemopoulos et al. 2018), we anticipated that the outliers and/or genomic regions identified by the earlier genome scan would associate with the experimentally determined individual migration distance. Thus, we aimed to a) identify novel candidate genes that would explain individual migration distance, and b) validate the functional links between previously detected and overlapping outlier SNPs and migration tendency.
Material and Methods
Common garden experiment
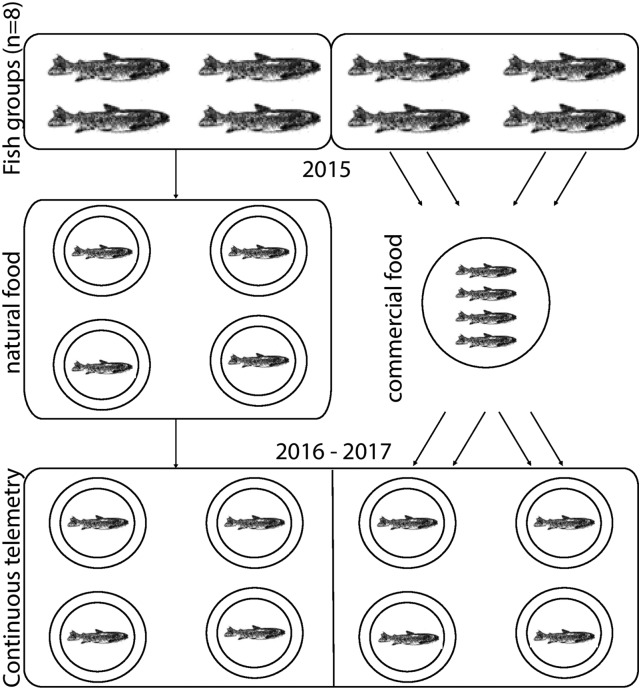
We used two strains of brown trout originating from a single river system discharging to Lake Oulujärvi, Northern Central Finland, for this study: 1) hatchery-bred migratory fish originating from rivers Varisjoki and Kongasjoki (referred as OUV), and 2) wild resident fish collected via electrofishing from a small headwater tributary (Vaarainjoki, VAA; see Lemopoulos et al. 2018). The wild fish were transported and held in seminatural ponds at the Kainuu Fisheries Research Station (Natural Resources Institute Finland (LUKE): www.kfrs.fi; see Lemopoulos et al. 2019) until artificial fertilization on 16th October 2013. The wild (females: n = 9, length 357 mm ± 30.8 mm (mean ± SD), mass 520.3 g ± 126.0 g; males: n = 9, length 400.4 mm ± 82.3 mm, mass 795.8 g ± 412.0 g) and hatchery (n = 9 males and 9 females, year class 2008, individual sizes missing, mean weight 1794 g) fish were crossed in fully factorial 3 × 3 matrices in three replicates (within and between population crosses). The eggs were pooled within strains (equal proportions between families) and incubated in four replicates per strain. After hatching, the progeny was raised in the hatchery in 3.2 m2 fiberglass tanks (four replicate tanks per strain, 1595-2109 fish per tank on 20th February 2014) according to standard methods and fed ad libitum with size-adjusted commercial dry feeds (Raisioagro, www.raisioagro.com) until tagging (Hyvärinen and Rodewald 2013). On 18th September 2015, twenty individuals were randomly dip-netted from each rearing tank (n = 80 per strain including the hybrid strain, n = 240 total) and tagged with a 12 mm half duplex passive integrated transponder (HDX-PIT) tags under benzocaine anesthesia (40 mg L−1). The fish (mean length ± SD 159.9 mm ± 18.1 mm, mean body mass ± SD 47.5 g ± 16.7 g) were randomized evenly into eight circular migration channels (n = 10 per group, n = 30 per channel) (Fig. S1).
The experimental migration channels (width 1.5 m, mean length 26.15 m) were built in 75 m2 circular outdoor concrete ponds with dark green plastic tarpaulin tent covers allowing natural but shaded light conditions inside the tanks (Fig. S1). The water input (∼55 l s-1) was adjacent to the water outlet to create unidirectional flow (average depth 0.3 m, water flow ∼0.11 m s-1). The channels were equipped with four stationary PIT-tag antennae (reading rate 9 s-1) at equal intervals. An antenna was constructed of five coil inductor loops made of PVC-coated multithread copper wire (ø = 4 mm) (1350 mm × 300 mm, length × width), with each antenna connected to one of four computers configured to run and save (TIRIS datalogger program, Citius solutions Oy 2009) ASCII data (for further details see Janhunen et al. (2011)).
Smolt experiment with 2-year old fish
Because smolt migration is expected to be influenced by food availability (Vainikka et al. 2012), we used two feeding regimes: half of the fish groups (n = 4) were transferred to the experimental channels supplied only with natural food on 5th November 2015 (Figure 1), and the rest of the groups (n = 4) were transferred into a 50 m2 concrete rearing pond in order to be fed with commercial fish feeds ad libitum until April 2016. The feed-augmented fish (and one VAA fish tagged on 25th February 2016 replacing one accidental mortality) were recovered on 14th April 2016 and transferred to the four experimental channels the next day. Continuous PIT-telemetry in all eight streams (see also Vainikka et al. 2012 for more details) was started on 16th April 2016 at midnight and continued until the end of the experiment at midnight on 26th June 2016. The fish were measured for body length and body mass and sampled for a piece of caudal fin tissue for DNA extraction under anesthesia on 28th June 2016. After the measurements, four random samples of thirty fish within the natural food treatment (amended with three fish from the spring stocking group to replace mortalities) were returned back to the smolt migration ponds where they were maintained with natural food until the end of the experiment in following summer 2017. The rest of the fish were transferred to a 50 m2 concrete pond to be maintained with commercial dry food until the spring release in 2017 (see below).
Figure 1.
Diagram of the rearing of 8 fish groups used for a 2-year common garden experiment.
Smolt experiment with 3-year-old fish
Brown trout augmented with commercial feed tested in 2016 (n = 115 + five reserve fish with no previous test) were held in a 50 m2 concrete rearing ponds with additional fish from the same cohort (n = 444 in total) over winter. The fish were recovered and measured for length (to 1 mm, mean ± SD 275.2 ± 32.2 mm) and body mass (to 0.1 g, mean ± SD 238.7 ± 91.0 g) under anesthesia on 14th March 2017. These fish were then transferred into four migration channel ponds after one night’s recovery in four 3.2 m2 indoor fiberglass tanks on 15.3.2017 at 10:00 (ten fish per strain, n = 30 per pond). The migration experiment was ended at two occasions: one random pond per feeding regime was emptied on 7th June 2017 to collect lethal physiological samples from killed fish and the rest of the ponds were emptied on 21th June 2017. All recovered fish were measured for length and body mass (within the subset of sequenced fish, OUV: 267.2 mm ± 62.4 mm (average ± SD), 211.7 g ± 139.7 g; VAA: 231.6 mm ± 39.8 mm, 123.2 g ± 60.8 g) under anesthesia, and fish in good condition were transferred back to the rearing ponds under standard rearing and feeding protocols. The fish sampled on 7th June 2017 were analyzed for their migration behavior from 16th March 2017 at midnight to 6th June 2017 at midnight. All the other fish were analyzed for their migration behavior from 16th March 2017 at midnight to 20th June 2017 at midnight.
Quantification of individual migration tendency
PIT detection data were transformed to individual movements per hour by counting the PIT-specific detections at each antenna using application-specific software (by Niko Vuokko, http://pitdata.net/). Based on the order of detections, the movement distances were calculated for each individual as quarter rounds up- and downstream. The hourly data were further analyzed by calculating the total individual distance moved downstream using custom codes in AV Bio-Statistics 5.2 (written by A.V.). The recording computers had to be restarted on a weekly basis, and when calculating migratory distances, the fish were assumed to have maintained their previous position during these maintenance breaks (< 30 min).
To maximize phenotypic variance between the limited number of samples that could be sequenced, we initially chose the four most and least migratory individuals (based on total downstream distance on equal time periods) in each channel (n = 128) and prioritized the presence of data from both years. This selection procedure resulted a subset of 116 individuals (out of 160 in total, n = 58 from both strains) that is not representative of all individuals in the experiment but provided extreme phenotypic values for the GWAS. Because individual migration distances could have been influenced by the test channel (which was confounded with diet treatment), rearing tank, year, sex (see below for the determination method) and feeding treatment, we removed their effects using a linear mixed effects (LME) model including sex, year (also as repeated), test pond and year × pond –interaction as fixed factors and rearing tanks / test pond during the previous year as a random factor. Prior to the analysis, migration distances were ln-transformed to meet normality. The model was fitted in SPSS 23.0.0.2 (IBM Corp.) using restricted maximum likelihood with random terms based on variance components and temporal covariance matrix based on diagonal structure with heterogenous variance. The residuals from this model were used to represent individual tendency to migrate (Fig. S2). The values were comparable between the channels, sexes and years, but included the effect of strain and fish size as these variables could mechanistically explain the originally genetic effect on migration. For the GWAS, the residual values for the two years were averaged; when migration distance was available for one year only (n = 12 fish, including five migratory strain fish that died during 2016 and seven residents that died during 2017), that value was used. This was justified as the inter-year averages were zero and migration residuals showed high individual repeatability between years (ICC = 0.608, 95% C.I. 0.473 – 0.714, ICC package in R environment (Wolak et al. 2012)). To test the effect of strain on the phenotypic migration distance, we also fitted a model with strain as an additional fixed factor.
Library preparation and genotyping
DNA samples were extracted from the 116 individuals using Macherey-Nagel NucleoSpin Tissue kit and quantified by fluorescence using Qubit 2.0. Sex of individuals was obtained through amplification and gel visualization of the sexually dimorphic sdY locus (Quéméré et al. 2014). Sbf1 restriction enzyme was used in the library preparation, and the single-end sequencing was performed on two lanes on an Illumina HiSeq 3500 platform. Final DNA concentration measures, library preparation and sequencing were obtained from a commercial provider, Plateforme MGX - Montpellier GenomiX (Montpellier, France) that delivered the sequences in FastQ format for bioinformatics analyses. Process_radtags function in Stacks v1.40 software (Catchen et al. 2013) was used to demultiplex, quality check and clean the data. Reads of 120 base pairs (average of 1643201 reads per individual, average 13.2X coverage Table S1) were aligned to the Atlantic salmon genome (GenBank: GCA_000233375.4) (Lien et al. 2016) using bowtie2 v2.3 (-p2, -sensitive, other parameters at default) (Langmead and Salzberg 2013).
Association analyses
To reduce Type I error, association analyses were performed using two complementary pathways based on genotype probabilities (Angsd, Korneliussen et al. 2014) and called genotypes (Stacks, Catchen et al. 2013).
In Angsd, SNP α-value of 1−6 (p-value for being variable) was used for a SNP to be processed. In addition, a minimum mapping quality of ten, minimum base score of 20, a minor allele frequency of 0.05 (Roesti et al. 2012) and a minimum of presence in 87 individuals were required for a SNP to be called. In addition, SNPs were filtered to exclude loci deviating from Hardy-Weinberg equilibrium with a genome-wide threshold accounting for multiple test correction (-do HWE1, -minHWEpval 0.05 / total number of loci). In order to correct for population stratification, we used Pcangsd (Meisner and Albrechtsen 2018) to perform principal component analysis and to obtain a covariance matrix accounting for population structure (one axis was retained). The association study was performed using the -doAsso 2 function (-doMaf 2 -do MajorMinor 1 -do Post 1), corresponding to an association analysis using a quantitative phenotype. Individual LME residuals represented relative migration distances as quantitative trait measures. SNPs that passed all quality filter steps (n = 24330) were ranked based on the likelihood ratio test (LRT) scores. This association test was based on genotype probabilities (see Korneliussen et al. 2014) and used a linear regression to give likelihood scores for each SNP individually (see Skotte et al. 2012 for more details).
In the alternative pathway, Stacks 2.0 (Catchen et al. 2013) and its functions Refmap and populations were used for SNP calling. In addition to the filtering criteria listed above, loci had to present a maximum heterozygosity of 0.5 (Hohenlohe et al. 2011), a minor allele frequency of 0.05 (Roesti et al. 2012), be present in both populations and in at least eighty-seven individuals to be processed. Finally, the dataset obtained in Stacks was tested for Hardy-Weinberg equilibrium in R version 3.4.4 (R Core team 2016) using the hw.test function of adegenet (v2.1.1; Jombart 2008). Loci that deviated from Hardy-Weinberg equilibrium (α = 0.05/number of loci) were excluded. A compressed mixed linear model (MLM) was fitted using the GAPIT package in R environment (Zhang et al. 2010; Lipka et al. 2012). 18 264 SNPs were associated with the phenotype scores that were treated as a continuous variable. We corrected for population stratification by using a covariance matrix based on the genetic structure between the populations (“PC.total =1” option in GAPIT). Correction for kinship was performed by obtaining a kinship matrix based on VanRaden method (VanRaden 2008) in GAPIT and using it as a covariance matrix.
The association method in the first pathway (Angsd) is based on a linear regression while the second (Stacks) relies on a linear mixed model. The linear regression association method takes the uncertainty of the genotypes into account while performing the association and is thus a powerful method than can still control for false positive (Skotte et al. 2012). The linear mixed model used called genotypes and is based on the EMMA algorithm (Kang et al. 2008) which is arguably the mostly used algorithm used in association studies. As such, the two pathways are based on different assumptions inherent to their methods (Zhang et al. 2010; Skotte et al. 2012) and overlapping markers, identified in both pathways, are particularly interesting to examine.
Candidate gene identification
There is a trade-off between identifying true GWAS signals (minimizing false positives) and ignoring more subtle, but real, associations (Type II error) due to low power and multiple testing burden (Kuo 2017). As a consequence, it has been suggested that multiple test corrections are too conservative in a GWAS context (Kuo 2017). For instance, statistically “non-significant” putative candidate genes with sound functional properties have been proposed for different diseases such as essential hypertension (Fowdar et al. 2017) or stroke protection in patients with sickle cell anemia (Flanagan et al. 2013). Moreover, candidate genes, even when not meeting conservative genome-wide significance after multiple test corrections, can a) indicate potential true and functional association with the studied traits, and b) serve as basis for targeted GWAS (e.g., Rivas et al. 2011; Sarzynski et al. 2011; Demontis et al. 2019).
In order to correct statistical significances for multiple testing, we used a false discovery rate correction (Benjamini and Hochberg 1995) separately for both GWAS approaches. The lack of significant associations after correction for multiple testing can be caused by multiple factors such as markers being too far from the causative variant, limited sample size or variant being at too low frequency in a population to reach genome-wide significance. Therefore, we further identified SNPs that showed the strongest genotype-phenotype associations (top 0.1%) based on both Angsd and GAPIT, even if these SNPs did not pass significance in multiple testing. Some of these markers can still represent true genotype-phenotype associations particularly in combination with other markers with small effects. As studied markers likely do not represent causative SNPs affecting migration, we identified the closest genes to the observed SNPs based on the Atlantic salmon reference genome.
Data availability
Supplementary materials are available on Figshare. These include filtered genotypes for both pathways, individual phenotypes, population structure and association results of both analyses. Figure S1 is a picture of the circular channels. Figure S2 is a boxplot of the residuals (i.e., phenotypic scores) between the populations. Figure S3 is a comparison between SNPs obtained for each pipeline. Raw sequence data are deposited on NCBI (PRJNA552287). Movement data and all codes used in this study are available upon request. Supplemental material available at FigShare: https://doi.org/10.25387/g3.7660295.
Results
Phenotypic results
According to the LME added with the strain effect, the assumed migratory OUV strain moved significantly farther downstream than the resident VAA strain (F1, 7.34 = 22.34, P < 0.001, average 160.5 km vs. 69.1 km), indicating the presence of genetic or epigenetic component affecting migration. The type III fixed effect tests indicated that smolt migration was affected by year (F1, 16.59 = 7.08, P = 0.017) and testing pond (F7, 105.56 = 2.98, P = 0.007) but not by sex of the fish (F1, 179.22 = 0.40, P = 0.526) or the testing pond × year interaction (F7, 105.08 = 1.26, P = 0.278). Because the used fish represented a subset of fish in the smolt migration experiment, these results are technical for the purpose of this work, but representative with respect to population differences due to stratified sampling within each test pond.
GWAS - Angsd
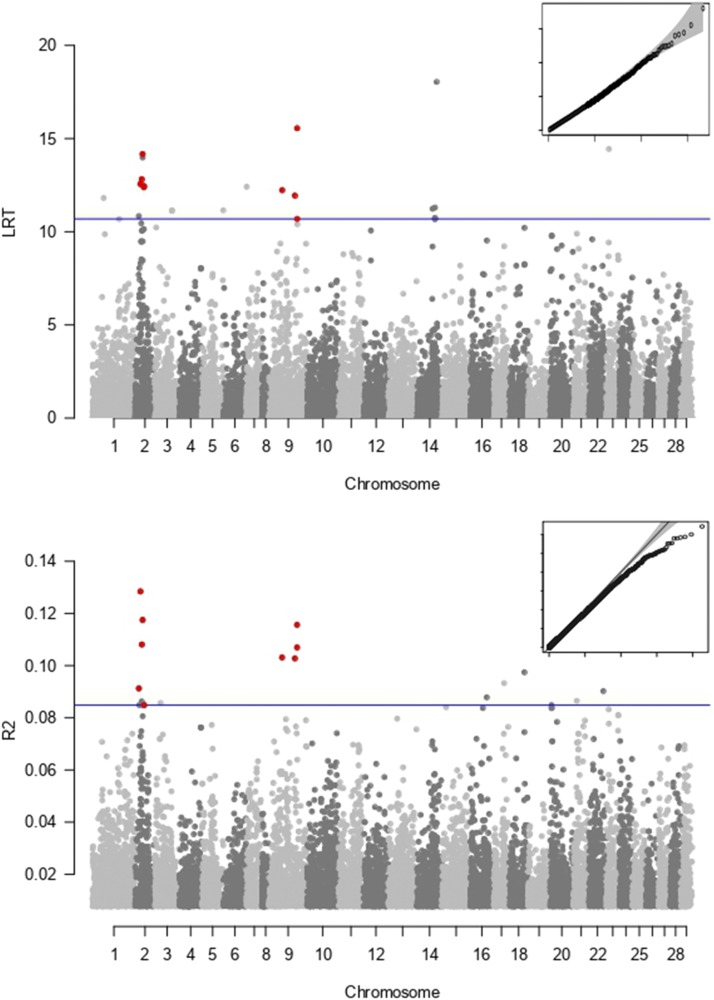
The Q-Q plot showed that there was no inflation in the dataset and the correction for population stratification was adequate (Figure 2). 24430 SNPs were initially obtained in Angsd, but 2114 of them did not pass the filtering criteria of the association function (LRT= -999 in Angsd).
Figure 2.
Manhattan plots based on a) 24330 (Angsd) and b) 18264 (Stacks) SNPs. Overlapping SNPs are marked in red. The horizontal significance line corresponding to the top 0.1% of the SNPs is drawn in blue.
None of the individual SNPs showed statistically significant association with migration scores after the multiple test correction (FDR) in the final Angsd dataset of 22186 SNPs. The top 0.1% percent of markers corresponded to 22 SNPs.
GWAS – Stacks/GAPIT
Q-Q plots with the Stacks dataset showed a conservative pattern with slightly right-skewed distribution indicating that the statistical framework used for correcting for various factors was slightly too conservative (Figure 2). In total, 18 264 SNPs were identified. Similar to Angsd, none of the SNPs showed statistically significant individual association with migration distance after the multiple test correction (FDR). The top 0.1% percent of markers corresponded to 18 SNPs.
Overlap
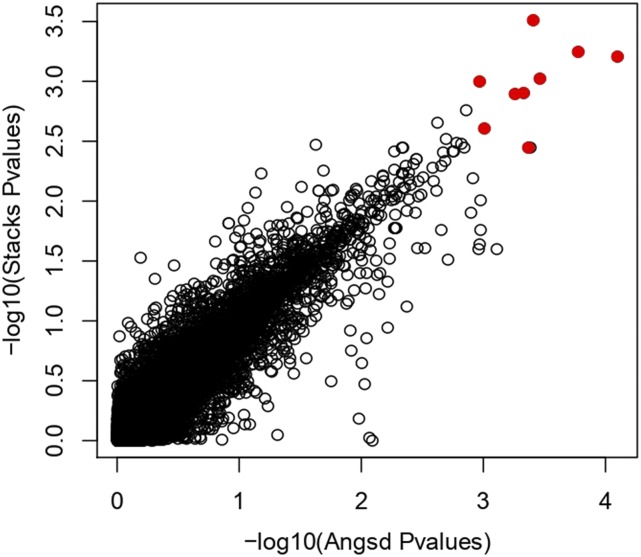
87.4% of the 18264 SNPs identified in Stacks were found among the 24330 SNPs identified by Angsd. The overlap was consistent across chromosomes (Fig. S3). Despite of different analytical approaches, the estimated P-values for Stacks and Angsd showed very high correlation (Pearson’s r = 0.92, Figure 3). Out of the top 0.1% markers of both pipelines (24 and 18 SNPs respectively), nine SNPs were overlapping (Table 1). These SNPs mapped to two chromosomes (Chr 2 and 9) based on the Atlantic salmon reference genome (Figure 2, Table 1). Four SNPs were found within genes, while five SNPs were found from 2565 to 48527 bp from the closest known coding regions.
Figure 3.
Correlation between P-values of two different GWAS based on different pathways (Stacks and Angsd). The nine candidate SNPs identified by both association methods are highlighted in red.
Table 1. Candidate migration genes and protein products identified by both Stacks and Angsd. Chromosome number refers to Atlantic salmon genome.
| Chromosome | SNP position | Distance to closest gene (bp) | Predicted protein | Gene symbol |
|---|---|---|---|---|
| 2 | 13725458 | 48527 - 3′end | FXYD domain containing ion transport regulator 5a precursor | FXYD5A |
| 2 | 19563669 | 2565 – 3′end | flotillin 1 | FLOT1 |
| 2 | 24989435 | 0 | progestin and adipoQ receptor family member 6-like isoform X2 | PAQR6 |
| 2 | 27622691 | 1460 – 3′end | free fatty acid receptor 2-like | FFAR2 |
| 2 | 32550984 | 14430 – 5′end | plectin-like isoform X14 | PLEC |
| 9 | 48392079 | 16589 – 5′end | protocadherin gamma-A11-like | PCDHGA11 |
| 9 | 96081278 | 0 | ATP-binding cassette sub-family F member 3-like | ABCF3 |
| 9 | 103885594 | 0 | limbic system-associated membrane protein-like isoform X | LSAMP |
| 9 | 103988881 | 0 | limbic system-associated membrane protein-like isoform X | LSAMP |
Discussion
This common garden study demonstrated divergent migratory patterns between two populations but did not find genome-wide significant SNPs with strong effect on migratory distance. However, smolt migration distance was individually highly repeatable between the test years and showed strong differences between two populations, which suggests that the trait could be heritable, as repeatable behavioral traits typically show reasonably high heritability (Dochtermann et al. 2015). The two statistical analysis pipelines used for association analysis revealed nine shared SNPs that could potentially affect migratory behavior but none of them overlapped with outlier SNPs identified by Lemopoulos et al. (2018). Together these results suggest that the migration distance in brown trout has underlying genetic components implying evolvability, but the genetic architecture is likely multigenic and the potentially existing regulatory genes with major effects remain to be identified.
Nine migration-associated candidate SNPs were identified by both bioinformatic methods. These markers mapped to two separate chromosomes in Atlantic salmon genome (Chr 2 and 9) and clustered together in relatively narrow genomic regions (Figure 2; Chr 2: 18.8 Mb, Chr 9: 55.6 Mb) indicating that the observed signals most likely reflect real associations, rather than random noise. According to a previous linkage map (Leitwein et al. 2017b), these chromosomes corresponded to brown trout linkage groups 20, 34 and 23. Within the 18.8 Mb region in chromosome 2, 559 genes are annotated in the Atlantic salmon genome. Three of the markers identified in chromosome 9 were mapped within a 7.6 Mb region consisting of 196 genes.
The nine migration-associated candidate SNPs mapped adjacent to eight genes involved in diverse biological functions relevant to migration (Table 1). For example, FYXDa gene, which located 48527 bp from the migration-associated SNP, is an important ion transporter that regulates Na+/K+ ATP-ase pumps. The function of these pumps is well documented in salmonids (e.g., Zaugg 1982; Nielsen et al. 1999; Larsen et al. 2008) as they play significant role for osmoregulation. Similarly, PLEC plays a role in coping with osmotic stress (Osmanagic-Myers et al. 2006) and has been shown to be upregulated in seawater-exposed whitefish Coregonus lavaretus (Papakostas et al. 2012) while ABCF3 is differentially expressed in gills of freshwater or seawater exposed rainbow trout, demonstrating its putative role in acclimation to seawater (Leguen et al. 2015). Among the other candidate genes, LSAMP is part of the limbic system, which itself is important for memory and spatial orientation, via the hypothalamus (Portavella and Vargas 2005; Catani et al. 2013). Nervous system development and memory are key factors in migratory behavior and especially for homing, as shown by differential gene expression and DNA methylation patterns in O. mykiss (McKinney et al. 2015; Baerwald et al. 2016). Moreover, PAQR6 is also expressed in the hypothalamus and other regions of the brain (Thomas and Pang 2012; Morini et al. 2017). The hypothalamus and thalamus regions could play an important role in the migratory behavior of salmonids through different pathways. In particular, circadian rhythms (see Prince et al. 2017; Pritchard et al. 2018), osmoregulatory mechanisms (Prunet et al. 1989; Hourdry 1995) and memory (Carruth et al. 2002; Ebbesson et al. 2003), are all potentially (inter)linked (Kim et al. 2015) within the hypothalamic region. Finally, PCDHGA11 is part of the cadherin gene family that has been identified both in a previous study comparing resident and migratory brown trout (Lemopoulos et al. 2018), and in rainbow trout (Hale et al. 2013; Baerwald et al. 2016). Cadherins play an important role in brain function and our results, together with earlier findings, support the role of cadherins affecting salmonid migratory behavior (Lemopoulos et al. 2018). The functions of the last two candidate genes (FFAR2, FLOT1) are linked to several diverse cellular processes but also exhibit plausible links to migratory behavior. In particular, FFAR2 is involved in lipid metabolism and autoimmune functions (Bjursell et al. 2011; D’Souza et al. 2017), while FLOT1 is involved in signal transduction and vesicular transport processes in the brain (Bickel et al. 1997; Volonté et al. 1999) corroborating earlier studies suggesting that lipid metabolism (Boel et al. 2014), immunity (Sutherland et al. 2014) and brain-related functions (McKinney et al. 2015) may play key roles in affecting migratory strategies.
The present study aligns with the previous ones by suggesting that the migration propensity is likely a polygenic trait in brown trout (Lemopoulos et al. 2018; Ferguson et al. 2019). This is plausible given that smolt migration is associated with changes in the expression of multitude of genes (e.g., Giger et al. 2006; Seear et al. 2010; Hecht et al. 2014) with multiple physiological functions, such as osmoregulation (Hecht et al. 2014), immunity (Sutherland et al. 2014), and growth (Boel et al. 2014). Surprisingly, while fish ascending toward spawning grounds often have female-biased sex-ratio (Dodson et al. 2013), individual sex was not found to be significant with regard to migration distance in our experiment. This could potentially reflect population specific patterns or suggest that rather demographic effects than true biological differences between the sexes drive the patterns observed in nature. As we included individuals originating from both resident and migratory populations, population structure was inherently confounded with the individual traits inducing migration. While the analysis was performed at the individual level, the removal of population stratification from the GWAS could have led to discarding functionally relevant population-specific variation. Thus, further experiments using multiple populations or populations with mixed origin should be performed to increase the likelihood of finding a signal inherent to species-specific rather than population-specific genes.
Similar to many other studies on non-model species, our experimental design likely suffered from non-optimal statistical power due to limited sample size and limited number of markers. On the other hand, it plausible that the multiple testing correction was too conservative to single out loci with real but small biological effects (e.g., Lantieri et al. 2010; Fowdar et al. 2017). Genomic data typically consist of thousands of markers tested for potential association and in most cases, very few - if any - markers reach the required level of statistical likelihood for a significant effect (e.g., Correa et al. 2017; Barría et al. 2018). This is particularly true when a) the studied traits are under the control of multiple loci of small effects (i.e., a polygenic trait, see Boyle et al. 2017) and b) when the studied populations are structured in terms of the trait of interest (Zhao et al. 2011), as in our case. As an alternative strategy, we searched for overlapping candidate genes that were identified as potentially interesting by two analytical approaches (Stacks and Angsd). Depending on data structure and method assumptions different approaches can perform differently (Wojcik et al. 2015; Zhu et al. 2018), and thus combining methods can potentially reduce both Type I and Type II error, analogously to genome-scans (Vasemägi and Primmer 2005). In this regard, the association results from Stacks and Angsd were similar, but not identical, demonstrating the usefulness of complementary statistical approaches in GWAS (Figure 3).
Compared to previous comparative work (Lemopoulos et al. 2018), none of the top migration-associated SNPs were overlapping between studies. Given that different enzymes were used in the RAD protocol (Sbf1 vs. PstI-BamHI respectively) this is hardly surprising, as these studies analyzed to large extent non-overlapping parts of brown trout genome. Yet, the lack of statistically significant individual SNPs after FDR correction is not valid counterevidence for migration being genetically influenced, because SNPs identified using RADSeq cover only a small proportion of the whole genome. Thus, it is possible that many causative variants affecting migration were not captured, given the low level of linkage disequilibrium in brown trout (Ahmad et al. 2018). To address this issue, the use of hundreds of thousands rather than tens of thousands of markers screened using genome-wide SNP arrays, high frequency RADseq (e.g., using four cutters instead of six or eight cutter enzymes) or whole-genome sequencing would be necessary.
Our study shows how brown trout individuals and populations differing in their migration strategies could bear genetic signatures associated with their life-history. This alone is a valuable result for management and conservation purposes, as it indicates that ecotypes should be managed differently in order to maintain the life-history diversity (Waples and Lindley 2018). These results are also valuable in gene-targeted conservation plans perspective. While clear-cut diagnostic candidate genes are still needed for sound conservation plans and application for conservation practitioners (Shafer et al. 2015; Kardos and Shafer 2018), these results expand the existing knowledge of brown trout migration and can thus serve as a basis for future conservation-oriented studies.
To conclude, our study demonstrates that migration in brown trout has a genetic or epigenetic component but does not fully resolve the mechanistic and causal pathways for variation in migration tendency. By linking telemetry in common garden with genomic data, we identified two promising genomic regions and eight candidate genes potentially associated with migratory behavior. However, additional testing using higher number of SNPs and analysis of inter-population hybrids is still needed for validation of the putative association signals and for better understanding of the molecular function and adaptive significance of the identified candidate genes.
Acknowledgments
We thank Dr. Robin Cristofari for fruitful discussions and help with Unix and R scripts. Aurora Hatanpää is acknowledged for helping during fieldwork and for helpful discussions. We also thank the Kainuu Fisheries Research Station staff for support during the common garden experiment. Dr. Hugues Parrinello and Plateforme MGX - Montpellier GenomiX are acknowledged for smooth collaboration and advice during the genotyping process. We also thank the two anonymous reviewers for the critical comments that helped improve this manuscript. Academy of Finland (decision #286261) funded the study.
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25387/g3.7660295.
Communicating editor: R. Houston
Literature Cited
- Ahmad F., Debes P. V., Palomar G., and Vasemägi A., 2018. Association mapping reveals candidate loci for resistance and anaemic response to an emerging temperature-driven parasitic disease in a wild salmonid fish. Mol. Ecol. 27: 1385–1401. 10.1111/mec.14509 [DOI] [PubMed] [Google Scholar]
- Ayllon F., Kjærner-semb E., Furmanek T., Wennevik V., Solberg F. et al. , 2015. The vgll3 locus controls age at maturity in wild and domesticated Atlantic Salmon (Salmo salar L. ) males. PLoS Genet. 11: e1005628 10.1371/journal.pgen.1005628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerwald M. R., Meek M. H., Stephens M. R., Nagarajan R. P., Goodbla A. M. et al. , 2016. Migration-related phenotypic divergence is associated with epigenetic modifications in rainbow trout. Mol. Ecol. 25: 1785–1800. 10.1111/mec.13231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barría A., Christensen K. A., Yoshida G. M., Correa K., Jedlicki A. et al. , 2018. Genomic Predictions and Genome-Wide Association Study of Resistance Against Piscirickettsia salmonis in Coho Salmon (Oncorhynchus kisutch) Using ddRAD Sequencing. G3: (Bethesda) 8: 1183–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barson, N. J., T. Aykanat, K. Hindar, M. Baranski, G. H. Bolstad et al., 2015 Sex-dependent dominance at a single locusmaintains variation in age at maturity in salmon. Nature 000: 405–408. [DOI] [PubMed]
- Benjamini Y., and Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Bickel P. E., Scherer P. E., Schnitzer J. E., Oh P., Lisanti M. P. et al. , 1997. Flotillin and Epidermal Surface Antigen Define a New Family of Caveolae-associated Integral Membrane Proteins *. J. Biol. Chem. 272: 13793–13802. 10.1074/jbc.272.21.13793 [DOI] [PubMed] [Google Scholar]
- Bjursell M., Admyre T., Göransson M., Marley A. E., Smith D. M. et al. , 2011. Improved glucose control and reduced body fat mass in free fatty acid receptor 2-deficient mice fed a high-fat diet. Am. J. Physiol. Endocrinol. Metab. 300: E211–E220. 10.1152/ajpendo.00229.2010 [DOI] [PubMed] [Google Scholar]
- Boel M., Aarestrup K., Baktoft H., Madsen S. S., Skov C. et al. , 2014. The Physiological Basis of the Migration Continuum in Brown Trout (Salmo trutta). Physiol. Biochem. Zool. 87: 334–345. 10.1086/674869 [DOI] [PubMed] [Google Scholar]
- Boyle E. A., Li Y. I., and Pritchard J. K., 2017. An expanded view of complex traits: from polygenic to omnigenic. Cell 169: 1177–1186. 10.1016/j.cell.2017.05.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carruth L. L., Jones R. E., and Norris D. O., 2002. Cortisol and Pacific Salmon: a new look at the role of stress hormones in Olfaction and home-stream migration. Integr. Comp. Biol. 42: 574–581. 10.1093/icb/42.3.574 [DOI] [PubMed] [Google Scholar]
- Catani M., Dell’acqua F., and Thiebaut de Schotten M., 2013. A revised limbic system model for memory, emotion and behaviour. Neurosci. Biobehav. Rev. 37: 1724–1737. 10.1016/j.neubiorev.2013.07.001 [DOI] [PubMed] [Google Scholar]
- Catchen J., Hohenlohe P. A., Bassham S., Amores A., and Cresko W. A., 2013. Stacks: An analysis tool set for population genomics. Mol. Ecol. 22: 3124–3140. 10.1111/mec.12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman B. B., Skov C., Hulthén K., Brodersen J., Nilsson P. A. et al. , 2012. Partial migration in fishes: Definitions, methodologies and taxonomic distribution. J. Fish Biol. 81: 479–499. 10.1111/j.1095-8649.2012.03349.x [DOI] [PubMed] [Google Scholar]
- Correa K., Lhorente J. P., Bassini L., López M. E., Di Genova A. et al. , 2017. Genome wide association study for resistance to Caligus rogercresseyi in Atlantic salmon (Salmo salar L.) using a 50K SNP genotyping array. Aquaculture 472: 61–65. 10.1016/j.aquaculture.2016.04.008 [DOI] [Google Scholar]
- Czorlich Y., Aykanat T., Erkinaro J., Orell P., and Robert C., 2018. Rapid sex-specific evolution of age at maturity is shaped by genetic architecture in Atlantic salmon. Nat. Ecol. Evol. 2: 1800–1807. 10.1038/s41559-018-0681-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis D., Walters R. K., Martin J., Mattheisen M., Als T. D. et al. , 2019. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet. 51: 63–75. 10.1038/s41588-018-0269-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dochtermann N. A., Schwab T., Sih A., and Dochtermann N. A., 2015. The contribution of additive genetic variation to personality variation : heritability of personality. Proceedings of the Royal Society B 282. [DOI] [PMC free article] [PubMed]
- Dodson J. J., Aubin-Horth N., Thériault V., and Páez D. J., 2013. The evolutionary ecology of alternative migratory tactics in salmonid fishes. Biol. Rev. Camb. Philos. Soc. 88: 602–625. 10.1111/brv.12019 [DOI] [PubMed] [Google Scholar]
- D’Souza W. N., Douangpanya J., Mu S., Jaeckel P., Zhang M. et al. , 2017. Differing roles for short chain fatty acids and GPR43 agonism in the regulation of intestinal barrier function and immune responses. PLoS One 12: e0180190 10.1371/journal.pone.0180190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbesson L. O. E., Ekstro P., Ebbesson S. O. E., Stefansson S. O., and Holmqvist B., 2003. Neural circuits and their structural and chemical reorganization in the light – brain – pituitary axis during parr – smolt transformation in salmon. Aquaculture 222: 59–70. 10.1016/S0044-8486(03)00102-9 [DOI] [Google Scholar]
- Ferguson A., Reed T. E., Cross T. F., Mcginnity P., and Prodöhl P. A., 2019. Anadromy, potamodromy and residency in brown trout Salmo trutta: the role of genes and the environment. J. Fish Biol.: 1–27. 10.1111/jfb.14005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan J. M., Sheehan V., Linder H., Howard T. A., Wang Y. et al. , 2013. Genetic mapping and exome sequencing identify 2 mutations associated with stroke protection in pediatric patients with sickle cell anemia. Blood 121: 3237–3245. 10.1182/blood-2012-10-464156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmar C., and Dickhoff W., 1980. The Parr-Smolt Transformation (Smoltification) and Seawater Adaptation in Salmonids. A Review of Selected Literature. Aquaculture 21, 1–37. [Google Scholar]
- Fowdar J. Y., Grealy R., Lu Y., and Griffiths L. R., 2017. A genome-wide association study of essential hypertension in an Australian population using a DNA pooling approach. Mol. Genet. Genomics 292: 307–324. 10.1007/s00438-016-1274-0 [DOI] [PubMed] [Google Scholar]
- Giger T., Excoffier L., Day P. J., Champigneulle A., Hansen M. M. et al. , 2006. Life history shapes gene expression in salmonids. Curr. Biol. 16: R281–R282. 10.1016/j.cub.2006.03.053 [DOI] [PubMed] [Google Scholar]
- Gutierrez A. P., Yáñ Ez J. M., Fukui S., Swift B., and Davidson W. S., 2015. Genome-Wide association study (GWAS) for growth rate and age at sexual maturation in atlantic salmon (Salmo salar). PLoS One 10: e0119730 10.1371/journal.pone.0119730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale M. C., Thrower F. P., Berntson E. A., Miller M. R., and Nichols K. M., 2013. Evaluating adaptive divergence between migratory and nonmigratory ecotypes of a Salmonid fish, Oncorhynchus mykiss. G3 (Bethesda) 3: 1273–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht B. C., Campbell N. R., Holeced D. E., and Narum S. R., 2013. Genome-wide association reveals genetic basis for the propensity to migrate in wild populations of rainbow and steelhead trout. Mol. Ecol. 18: 1199–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht B. C., Thrower F. P., Hale M. C., Miller M. R., and Nichols K. M., 2012. Genetic architecture of migration-related traits in rainbow and steelhead trout, Oncorhynchus mykiss. G3 (Bethesda) 2: 1113–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht B. C., Valle M. E., Thrower F. P., and Nichols K. M., 2014. Divergence in Expression of Candidate Genes for the Smoltification Process Between Juvenile Resident Rainbow and Anadromous Steelhead Trout. Mar. Biotechnol. (NY) 16: 638–656. 10.1007/s10126-014-9579-7 [DOI] [PubMed] [Google Scholar]
- Hess J. E., Zendt J. S., Matala A. R., Narum S. R., and Hess J. E., 2016. Genetic basis of adult migration timing in anadromous steelhead discovered through multivariate association testing. Proceedings of the Royal Society B 283. 10.1098/rspb.2015.3064 [DOI] [PMC free article] [PubMed]
- Hohenlohe P. A., Amish S. J., Catchen J. M., Allendorf F. W., and Luikart G., 2011. Next-generation RAD sequencing identifies thousands of SNPs for assessing hybridization between rainbow and westslope cutthroat trout. Mol. Ecol. Resour. 11: 117–122. 10.1111/j.1755-0998.2010.02967.x [DOI] [PubMed] [Google Scholar]
- Hourdry J., 1995. Fish and cydostome migrations between fresh water and sea water : Osmoregulatory modifications between fresh water and sea water. Ital. J. Zool. (Modena) 62: 97–108. [Google Scholar]
- Hyvärinen P., and Rodewald P., 2013. Enriched rearing improves survival of hatchery-reared Atlantic salmon smolts during migration in the River Tornionjoki. Can. J. Fish. Aquat. Sci. 70: 1386–1395. 10.1139/cjfas-2013-0147 [DOI] [Google Scholar]
- Janhunen M., Kekalainen J., Kortet R., Hyvarinen P., and Piironen J., 2011. No evidence for an indirect benefit from female mate preference in Arctic charr Salvelinus alpinus, but female ornamentation decreases offspring viability. Biological Journal of the Linnean Society, 103: 602–611. 10.1111/j.1095-8312.2011.01659.x [DOI] [Google Scholar]
- Johnston S. E., Orell P., Pritchard V. L., Kent M. P., Lien S. et al. , 2014. Genome-wide SNP analysis reveals a genetic basis for sea-age variation in a wild population of Atlantic salmon (Salmo salar). Mol. Ecol. 23: 3452–3468. 10.1111/mec.12832 [DOI] [PubMed] [Google Scholar]
- Jombart T., 2008. Adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 24: 1403–1405. 10.1093/bioinformatics/btn129 [DOI] [PubMed] [Google Scholar]
- Kang H. M., Zaitlen N. A., Wade C. M., Kirby A., Heckerman D. et al. , 2008. Efficient control of population structure in model organism association mapping. Genetics 178: 1709–1723. 10.1534/genetics.107.080101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardos M., and Shafer A. B. A., 2018. The Peril of Gene-Targeted Conservation. Trends Ecol. Evol. 33: 827–839. 10.1016/j.tree.2018.08.011 [DOI] [PubMed] [Google Scholar]
- Kendall N. W., Mcmillan J. R., Sloat M. R., Buehrens T. W., Quinn T. P. et al. , 2015. Anadromy and residency in steelhead and rainbow trout (Oncorhynchus mykiss): a review of the processes and patterns. Can. J. Fish. Aquat. Sci. 72: 319–342. 10.1139/cjfas-2014-0192 [DOI] [Google Scholar]
- Kim N. N., Young J. C., Lim S., Jeong M., Jin D. et al. , 2015. Effect of salinity changes on olfactory memory-related genes and hormones in adult chum salmon Oncorhynchus keta. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 187: 40–47. 10.1016/j.cbpa.2015.04.011 [DOI] [PubMed] [Google Scholar]
- Klemetsen A., Amundsen P. A., Dempson J. B., Jonsson B., Jonsson N. et al. , 2003. Atlantic salmon Salmo salar L., brown trout Salmo trutta L. and Arctic charr Salvelinus alpinus L.: a review of aspects of their life histories. Ecol. Freshwat. Fish 12: 1–59. 10.1034/j.1600-0633.2003.00010.x [DOI] [Google Scholar]
- Korneliussen T. S., Albrechtsen A., and Nielsen R., 2014. ANGSD : analysis of next generation sequencing data. BMC Bioinformatics 15: 356 10.1186/s12859-014-0356-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku C. S., Loy E. Y., Pawitan Y., and Chia K. S., 2010. The pursuit of genome-wide association studies : where are we now? J. Hum. Genet. 55: 195–206. 10.1038/jhg.2010.19 [DOI] [PubMed] [Google Scholar]
- Kuo K. H. M., 2017. Multiple testing in the context of gene discovery in sickle cell disease using genome-wide association studies. Genomic Insights 10: 1178631017721178 10.1177/1178631017721178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., and Salzberg S. L., 2013. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantieri F., Glessner J. T., Hakonarson H., Elia J., and Devoto M., 2010. Analysis of GWAS top hits in ADHD suggests association to two polymorphisms located in genes expressed in the cerebellum. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 153: 1127–1133. [DOI] [PubMed] [Google Scholar]
- Larsen P. F., Nielsen E. E., Koed A., Thomsen D. S., Olsvik P. A. et al. , 2008. Interpopulation differences in expression of candidate genes for salinity tolerance in winter migrating anadromous brown trout (Salmo trutta L.). BMC Genet. 9: 12 10.1186/1471-2156-9-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leguen I., Le Cam A., Montfort J., Peron S., and Fautrel A., 2015. Transcriptomic analysis of trout gill ionocytes in fresh water and sea water using laser capture microdissection combined with microarray analysis. PLoS One 10: e0139938 10.1371/journal.pone.0139938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitwein M., Garza J. C., and Pearse D. E., 2017a Ancestry and adaptive evolution of anadromous, resident, and adfluvial rainbow trout (Oncorhynchus mykiss) in the San Francisco bay area: application of adaptive genomic variation to conservation in a highly impacted landscape. Evol. Appl. 10: 56–67. 10.1111/eva.12416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitwein M., Guinant B., Pouzadoux J., Desmarais E., Berrebi P. et al. , 2017b A Dense Brown Trout (Salmo trutta) Linkage Map Reveals Recent Chromosomal Rearrangements in the Salmo Genus and the Impact of Selection on Linked Neutral Diversity. G3 (Bethesda) 7: 1365–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemopoulos A., Prokkola J. M., Huusko A., Hyvärinen P., Koljonen M.-L. et al. , 2019. Comparing RADseq and microsatellites for estimating genetic diversity and relatedness — Implications for brown trout conservation. Ecol. Evol. 9: 2106–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemopoulos A., Uusi-Heikkila S., Huusko A., Vasemägi A., and Vainikka A., 2018. Comparison of migratory and resident populations of brown trout reveals candidate genes for migration tendency. Genome Biol. Evol. 10: 1493–1503. 10.1093/gbe/evy102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liedvogel M., Åkesson S., and Bensch S., 2011. The genetics of migration on the move. Trends Ecol. Evol. 26: 561–569. 10.1016/j.tree.2011.07.009 [DOI] [PubMed] [Google Scholar]
- Lien S., Koop B. F., Sandve S. R., Miller J. R., Matthew P. et al. , 2016. The Atlantic salmon genome provides insights into rediploidization. Nature 533: 200–205. 10.1038/nature17164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka A. E., Tian F., Wang Q., Peiffer J., Li M. et al. , 2012. GAPIT: Genome association and prediction integrated tool. Bioinformatics 28: 2397–2399. 10.1093/bioinformatics/bts444 [DOI] [PubMed] [Google Scholar]
- MacCrimmon H. R., and Marshall T., 1968. World Distribution of Brown Trout. J. Fish. Res. Board Can. 25: 2527–2548. 10.1139/f68-225 [DOI] [Google Scholar]
- McCormick S. D., 2009. Evolution of the hormonal control of animal performance: Insights from the seaward migration of salmon. Integr. Comp. Biol. 49: 408–422 Crossref, Medline, Google Scholar. [Google Scholar]
- McCormick S. D., 2013. Smolt physiology and endrocrinology. In S. D. McCormick, A. P. Farrell, and C. J. Brauner (Eds.), Euryhaline fishes (pp. 199–251). Amsterdam, Netherlands: Academic Press. Google Scholar. [Google Scholar]
- McKinney G. J., Hale M. C., Goetz G., Gribskov M., Thrower F. P. et al. , 2015. Ontogenetic changes in embryonic and brain gene expression in progeny produced from migratory and resident Oncorhynchus mykiss. Mol. Ecol. 24: 1792–1809. 10.1111/mec.13143 [DOI] [PubMed] [Google Scholar]
- Meisner J., and Albrechtsen A., 2018. Inferring population structure and admixture proportions in low-depth NGS data. Genetics 210: 719–731. 10.1534/genetics.118.301336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morini M., Peñaranda D. S., Vílchez M. C., Nourizadeh-lillabadi R., Lafont A. et al. , 2017. Nuclear and membrane progestin receptors in the European eel: Characterization and expression in vivo through spermatogenesis. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 207: 79–92. 10.1016/j.cbpa.2017.02.009 [DOI] [PubMed] [Google Scholar]
- Narum S. R., Zendt J. S., Frederiksen C., Campbell N., Matala A. et al. , 2011. Candidate genetic markers associated with anadromy in Oncorhynchus mykiss of the Klickitat river. Trans. Am. Fish. Soc. 140: 843–854. 10.1080/00028487.2011.588131 [DOI] [Google Scholar]
- Nichols K. M., Edo A. F., Wheeler P. A., and Thorgaard G. H., 2008. The genetic basis of smoltification-related traits in Oncorhynchus mykiss. Genetics 179: 1559–1575. 10.1534/genetics.107.084251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols K. M., Kozfkay C. C., and Narum S. R., 2016. Genomic signatures among Oncorhynchus nerka ecotypes to inform conservation and management of endangered Sockeye Salmon. Evol. Appl. 9: 1285–1300. 10.1111/eva.12412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen C., Madsen S. S., and Bjoernsson B. T., 1999. Changes in branchial and intestinal osmoregulatory mechanisms and growth hormone levels during smolting in hatchery-reared and wild brown trout. J. Fish Biol. 54: 799–818. 10.1111/j.1095-8649.1999.tb02034.x [DOI] [Google Scholar]
- Olsson I. C., Greenberg L. A., Bergman E., and Wysujack K., 2006. Environmentally induced migration: the importance of food. Ecol. Lett. 9: 645–651. 10.1111/j.1461-0248.2006.00909.x [DOI] [PubMed] [Google Scholar]
- Osmanagic-Myers S., Gregor M., Walko G., Burgstaller G., Reipert S. et al. , 2006. Plectin-controlled keratin cytoarchitecture affects MAP kinases involved in cellular stress response and migration. J. Cell Biol. 174: 557–568. 10.1083/jcb.200605172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papakostas S., Vasemägi A., Vähä J.-P., Himberg M., Peil L. et al. , 2012. A proteomics approach reveals divergent molecular responses to salinity in populations of European whitefish (Coregonus lavaretus). Mol. Ecol. 21: 3516–3530. 10.1111/j.1365-294X.2012.05553.x [DOI] [PubMed] [Google Scholar]
- Pearse D. E., Miller M. R., Abadı A., and Garza J. C., 2014. Rapid parallel evolution of standing variation in a single, complex, genomic region is associated with life history in steelhead / rainbow trout. Proceedings of the Royal Society B 281. 10.1098/rspb.2014.0012 [DOI] [PMC free article] [PubMed]
- Portavella M., and Vargas J. P., 2005. Emotional and spatial learning in goldfish is dependent on different telencephalic pallial systems. Eur. J. Neurosci. 21: 2800–2806. 10.1111/j.1460-9568.2005.04114.x [DOI] [PubMed] [Google Scholar]
- Prince D. J., O’Rourke S. M., Thompson T. Q., Ali O. A., Lyman H. S. et al. , 2017. The evolutionary basis of premature migration in Pacific salmon highlights the utility of genomics for informing conservation. Sci. Adv. 3: e1603198 10.1126/sciadv.1603198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard V. L., Mäkinen H., Vähä J., Erkinaro J., Orell P. et al. , 2018. Genomic signatures of fine-scale local selection in Atlantic salmon suggest involvement of sexual maturation, energy homeostasis, and immune defence-related genes. Mol. Ecol. 27: 2560–2575. 10.1111/mec.14705 [DOI] [PubMed] [Google Scholar]
- Prunet P., Boeuf G., Bolton J. P., and Young G., 1989. Smoltification and seawater adaptation in Atlantic Salmon (Salmo salar): plasma prolactin, growth hormone, and thyroid hormones. Gen. Comp. Endocrinol. 74: 355–364. 10.1016/S0016-6480(89)80031-0 [DOI] [PubMed] [Google Scholar]
- Quéméré E., Perrier C., Besnard A.-L., Evanno G., Baglinière J. et al. , 2014. An improved PCR-based method for faster sex determination in brown trout (Salmo trutta) and Atlantic salmon (Salmo salar). Conserv. Genet. Resour. 6: 825–827. 10.1007/s12686-014-0259-8 [DOI] [Google Scholar]
- Rivas M. A., Beaudoin M., Gardet A., Stevens C., Sharma Y. et al. , 2011. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat. Genet. 43: 1066–1073. 10.1038/ng.952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesti M., Salzburger W., and Berner D., 2012. Uninformative polymorphisms bias genome scans for signatures of selection. BMC Evol. Biol. 12: 94 10.1186/1471-2148-12-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarzynski M. A., Jacobson P., Rankinen T., Carlsson B., Sjöström L. et al. , 2011. Association of GWAS-based candidates genes with HDL-Cholesterol levels before and after Bariatric Surgery in the Swedish Obese Subjects Study. J. Clin. Endocrinol. Metab. 96: E953–E957. 10.1210/jc.2010-2227 [DOI] [PubMed] [Google Scholar]
- Seear P. J., Carmichael S. N., Talbot R., Taggart J. B., Bron J. E. et al. , 2010. Differential gene expression during smoltification of Atlantic salmon (Salmo salar L.): A first large-scale microarray study. Mar. Biotechnol. (NY) 12: 126–140. 10.1007/s10126-009-9218-x [DOI] [PubMed] [Google Scholar]
- Shafer A. B. A., Wolf J. B. W., Alves P. C., Bergstro L., De Meester L. et al. , 2015. Genomics and the challenging translation into conservation practice. Trends Ecol. Evol. 30: 78–87. 10.1016/j.tree.2014.11.009 [DOI] [PubMed] [Google Scholar]
- Skotte L., Korneliussen T. S., and Albrechtsen A., 2012. Association Testing for Next-Generation Sequencing Data Using Score Statistics. Genet. Epidemiol. 36: 430–437. 10.1002/gepi.21636 [DOI] [PubMed] [Google Scholar]
- Sutherland B. J. G., Hanson K. C., Jantzen J. R., Koop B. F., and Smith C. T., 2014. Divergent immunity and energetic programs in the gills of migratory and resident Oncorhynchus mykiss. Mol. Ecol. 23: 1952–1964. 10.1111/mec.12713 [DOI] [PubMed] [Google Scholar]
- Thomas P., and Pang Y., 2012. Membrane progesterone receptors (mPRs): Evidence for neuroprotective, neurosteroid signaling and neuroendocrine functions in neuronal cells. Neuroendocrinology 96: 162–171. 10.1159/000339822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorstad E. B., Rikardsen A. H., Alp A., and Økland F., 2013. The Use of Electronic Tags in Fish Research – An Overview of Fish Telemetry Methods. Turk. J. Fish. Aquat. Sci. 896: 881–896. [Google Scholar]
- Vainikka A., Huusko R., Hyvärinen P., Korhonen P. K., Laaksonen T. et al. , 2012. Food restriction prior to release reduces precocious maturity and improves migration tendency of Atlantic salmon (Salmo salar) smolts. Can. J. Fish. Aquat. Sci. 69: 1981–1993. 10.1139/f2012-119 [DOI] [Google Scholar]
- VanRaden P. M., 2008. Efficient methods to compute genomic predictions. J. Dairy Sci. 91: 4414–4423. 10.3168/jds.2007-0980 [DOI] [PubMed] [Google Scholar]
- Vasemägi A., and Primmer C. R., 2005. Challenges for identifying functionally important genetic variation: The promise of combining complementary research strategies. Mol. Ecol. 14: 3623–3642. 10.1111/j.1365-294X.2005.02690.x [DOI] [PubMed] [Google Scholar]
- Veale A. J., and Russello M. A., 2017. Genomic changes associated with reproductive and migratory ecotypes in sockeye salmon (Oncorhynchus nerka). Genome Biol. Evol. 9: 2921– 2939 10.1093/gbe/evx215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Villemereuil P., Gaggiotti O. E., Mouterde M., and Till-Bottraud I., 2015. Common garden experiments in the genomic era : new perspectives and opportunities. Heredity 116: 249–254. 10.1038/hdy.2015.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volonté D., Galbiati F., Li S., Nishiyama K., Okamoto T. et al. , 1999. Flotillins / Cavatellins Are Differentially Expressed in Cells and Tissues and Form a Hetero-oligomeric Complex with Caveolins in Vivo. J. Biol. Chem. 274: 12702–12709. 10.1074/jbc.274.18.12702 [DOI] [PubMed] [Google Scholar]
- Waples R. S., and Lindley S. T., 2018. Genomics and conservation units : The genetic basis of adult migration timing in Pacific salmonids. Evol. Appl. 11: 1518–1526. 10.1111/eva.12687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waples R. S., Teel D. J., Myers J. M., and Marshall A. R., 2004. Life-History Divergence in Chinook Salmon : Historic Contingency and Parallel Evolution. Evolution 58: 386–403. 10.1111/j.0014-3820.2004.tb01654.x [DOI] [PubMed] [Google Scholar]
- Wojcik G. L., Kao W. H. L., and Duggal P., 2015. Relative performance of gene- and pathway-level methods as secondary analyses for genome-wide association studies. BMC Genet. 16: 34 10.1186/s12863-015-0191-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolak M. E., Fairbairn D. J., and Paulsen Y. R., 2012. Guidelines for Estimating Repeatability. Methods Ecol. Evol. 3: 129–137. 10.1111/j.2041-210X.2011.00125.x [DOI] [Google Scholar]
- Wysujack K., Greenberg L. A., Bergman E., and Olsson I. C., 2009. The role of the environment in partial migration: Food availability affects the adoption of a migratory tactic in brown trout Salmo trutta. Ecol. Freshwat. Fish 18: 52–59. 10.1111/j.1600-0633.2008.00322.x [DOI] [Google Scholar]
- Zaugg W. S., 1982. Some changes in smoltification and seawater adaptability of salmonids resulting from environmental and other factors. Aquaculture 28: 143–151. 10.1016/0044-8486(82)90017-5 [DOI] [Google Scholar]
- Zhang Z., Ersoz E., Lai C., Todhunter R. J., Tiwari H. K. et al. , 2010. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 42: 355–360. 10.1038/ng.546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., Tung C., Eizenga G. C., Wright M. H., Ali M. L. et al. , 2011. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat. Commun. 2: 467 10.1038/ncomms1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Anttila V., Smoller J. W., and Lee P. H., 2018. Statistical power and utility of meta-analysis methods for cross-phenotype genome-wide association studies. PLoS One 13: e0193256 10.1371/journal.pone.0193256 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Supplementary materials are available on Figshare. These include filtered genotypes for both pathways, individual phenotypes, population structure and association results of both analyses. Figure S1 is a picture of the circular channels. Figure S2 is a boxplot of the residuals (i.e., phenotypic scores) between the populations. Figure S3 is a comparison between SNPs obtained for each pipeline. Raw sequence data are deposited on NCBI (PRJNA552287). Movement data and all codes used in this study are available upon request. Supplemental material available at FigShare: https://doi.org/10.25387/g3.7660295.