Abstract
The generation of induced pluripotent stem cells (iPSCs) from patient’s somatic cells and the subsequent differentiation into desired cell types opens up numerous possibilities in regenerative medicine and tissue engineering. Adult cardiomyocytes have limited self-renewal capacity; thus, the efficient, safe, and clinically applicable generation of autologous cardiomyocytes is of great interest for the treatment of damaged myocardium. In this study, footprint-free iPSCs were successfully generated from urine-derived renal epithelial cells through a single application of self-replicating RNA (srRNA). The expression of pluripotency markers and the in vitro as well as in vivo trilineage differentiation were demonstrated. Furthermore, the resulting iPSCs contained no residual srRNA, and the karyotyping analysis demonstrated no detectable anomalies. The cardiac differentiation of these iPSCs resulted in autologous contracting cardiomyocytes after 10 days. We anticipate that the use of urine as a non-invasive cell source to obtain patient cells and the use of srRNA for reprogramming into iPSCs will greatly improve the future production of clinically applicable cardiomyocytes and other cell types. This could allow the regeneration of tissues by generating sufficient quantities of autologous cells without the risk of immune rejection.
Keywords: self-replicating RNA, reprogramming, iPSCs, urine, renal epithelial cells, cardiomyocytes, regenerative medicine
Introduction
Heart failure is currently one of the leading causes of death and affects 23 million people worldwide.1 In addition to atrial fibrillation and arterial hypertension, myocardial infarction is a common cause of heart failure. The destruction of cardiomyocytes is associated with a reduced pumping capacity of the heart. Because of the extremely low self-renewal capacity of cardiomyocytes with less than 1% per year in adult hearts, a physiologically sufficient regeneration of the myocardium is not feasible.2 In end-stage heart failure, solely organ transplantation can ensure the survival of patients. To overcome the lack of donor organs for transplantation, there is an explicit need for regenerative therapeutic strategies to treat advanced heart failure.
Previous in vivo studies with different species demonstrated that the transplantation of cardiomyocytes can improve the performance of the heart after myocardial infarction.3, 4 However, human implementation is currently failing due to the lack of reliable sources of human cardiomyocytes. To overcome this limitation, various approaches have been developed to generate cardiomyocytes from different cell sources, including bone-marrow-derived stem cells,5 embryonic stem cells (ESCs),6, 7 and induced pluripotent stem cells (iPSCs).8, 9 In particular, iPSCs are a promising cell source to obtain autologous cells. Since iPSCs are produced from a patient’s somatic cells, their generation and application avoid ethical concerns associated with ESCs. Furthermore, rejection reactions are prevented, because the generated cells are autologous. In addition to the application of iPSCs in the field of regenerative medicine, cardiomyocytes derived from iPSCs allow the screening and discovery of drugs, the prediction of cardiotoxicity,10 and the study of cardiovascular diseases.11
Reprogramming of somatic cells into iPSCs was first achieved in murine fibroblasts by Yamanaka and colleagues12, 13 and shortly thereafter in human fibroblasts using retroviral vectors encoding four transcription factors: KLF4, c-MYC, OCT4, and SOX2. Hitherto, human fibroblasts have been the most commonly used cell source for reprogramming studies.14 Fibroblasts are usually obtained from skin biopsies, which represent an invasive procedure and lead to an injury of healthy tissue. This is associated with pain and additional burden for the patient. Naturally, due to physiological self-renewal of the epithelial tissue in the urinary tract, approximately 2,000 to 7,000 human renal proximal tubule epithelial cells are detached daily and excreted with urine.15 Thus, the collection and reprogramming of these urine-derived cells represent a promising simple and non-invasive strategy for obtaining patient’s own somatic cells.
The genome-integrating viral vectors originally used by Yamanaka et al.12, 13 are carrying a certain risk. The random insertion of retroviral vectors into the genome can lead to mutations and, thereby, to the development of tumors. Furthermore, the retroviral introduction of c-MYC in the cells can lead to increased tumorigenesis in mice.16, 17 Thus, these iPSCs generated by retroviral vectors cannot be clinically used. Meanwhile, several non-integrating approaches have been developed that reduce concerns regarding genetic alterations in iPSCs.18, 19, 20, 21, 22 In particular, synthetic mRNA-based approaches are promising.23 Thereby, the exogenous delivery of synthetic mRNAs into somatic cells leads to transient expression of desired proteins under physiological conditions by cells’ translational machinery. In contrast to plasmid DNA, synthetic mRNAs do not need to enter the cell nucleus. This results in an immediate translation of delivered mRNA in the cytosol and eliminates the risk of genomic integration and insertional mutagenesis.24 The incorporation of modified nucleotides during the in vitro transcription (IVT) or codon optimization25 can significantly improve the stability of synthetic mRNAs, drastically reduce immune responses triggered by recognition of foreign RNAs, and increase the recruitment and recycling of ribosomes and, thereby, result in enhanced translation efficiency.26, 27, 28, 29
Due to the transient nature, synthetic mRNAs can be used to express reprogramming factors to obtain footprint-free iPSCs.22 However, the transient presence of the exogenously delivered mRNAs also necessitates the daily transfection of cells with the synthetic mRNAs during the reprogramming process, which is cost intensive, time consuming, and associated with increased cellular stress. To overcome these hurdles, self-replicating RNAs (srRNAs) can be used to express the reprogramming factors for an extended period of time,30 without the need for repeated transfections. The srRNA contains the coding sequences of four transcription factors-OCT4, KLF4, SOX2, and c-MYC-and four non-structural proteins (nsP1–nsP4), which encode the RNA replication complex of Venezuelan equine encephalitis (VEE) virus.30, 31
In this study, using srRNA, footprint-free iPSCs were generated from adult human urine-derived epithelial cells, and the successful generation of beating autologous cardiomyocytes from these iPSCs was demonstrated.
Results
Reprogramming of Urine-Derived RECs into iPSCs Using OCT4, KLF4, SOX2, IRES-cMyc (OKSiM)-GFP srRNA
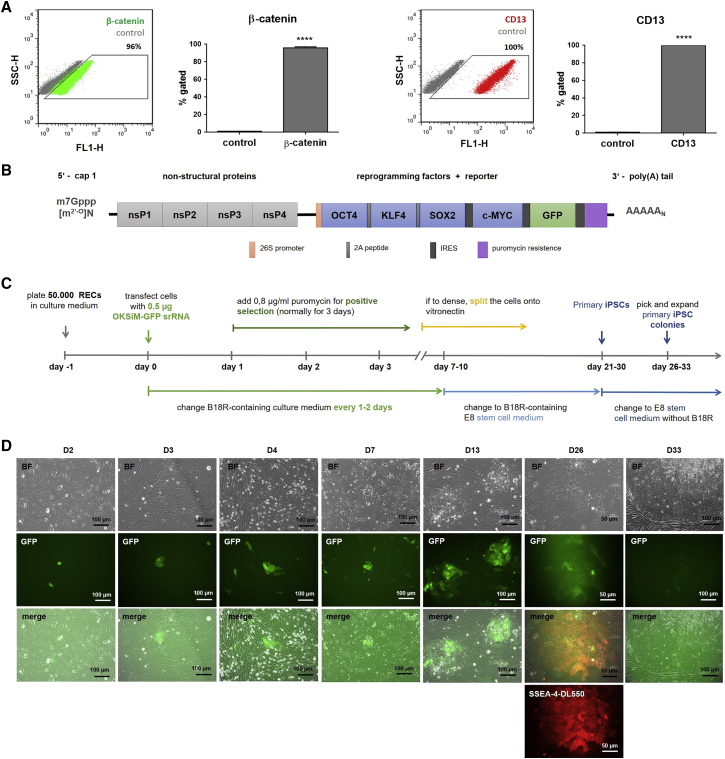
Cells were collected from urine of 4 different donors. The renal epithelial origin of the isolated and expanded cells at passage 2 or 3 was confirmed using flow cytometry. The analysis showed that 95.8% ± 1.3% of the cells positively express epithelial marker β-catenin and that 99.8% ± 0.2% of the cells express the renal proximal tubular marker CD13 (Figure 1A).
Figure 1.
Reprogramming of Urine-Derived RECs into iPSCs Using OKSiM-GFP srRNA
(A) Flow cytometry analysis of RECs after staining with antibodies against epithelial marker β-catenin and renal epithelial marker CD13. Results are shown as mean + SD (n = 4). Statistical differences were determined using paired t test (****p < 0.0001). (B) Schematic overview of srRNA construct. The synthetized srRNA contains the coding sequences for the non-structural proteins (nsP1–nsP4) of VEE to allow self-replication of srRNA, the reprogramming factors (OCT4, KLF4, SOX2, and c-MYC), and the reporter protein GFP. The internal ribosome entry sites (IRES) sequences (linked to c-MYC and GFP) control ribosome entry and, therefore, protein production. The 5′ end was modified with a cap1 structure, and a poly(A) tail was added at the 3′ end. For positive selection, the srRNA also encodes a puromycin resistance. (C) Timeline for the generation of iPSCs from RECs using srRNA. (D) Microscopic images of RECs after the transfection with 0.5 μg srRNA. A few cells expressing the reporter protein GFP were detected 2 days after the transfection (D2). At day 3, puromycin was added for 3 days to eliminate cells, which are not transfected (D3). In the following days, GFP-positive cells proliferated and generated colonies (D4). At day 7, mainly GFP-positive cells had survived, and non-transfected cells were dying (D7). At day 13, an increased size of colonies was observed (D13). After 26 days, iPSC colonies were stained with DL550-labeled live antibody against SSEA-4 (D26). Depending on the colony size, primary iPSCs were picked the next day or further cultivated up to 7 days (D33) until colonies increased in size. The detached iPSCs were transferred into vitronectin-coated wells.
To control the transfection and translation of the srRNA in cells, an IRES-GFP encoding sequence was cloned into the T7-VEE-OKSiM plasmid for the synthesis of OKSiM-GFP srRNA (Figure 1B). For reprogramming, 5 × 104 renal epithelial cells (RECs) were seeded on gelatin-coated wells. Using the established reprogramming protocol (Figure 1C), 21 to 30 days post-transfection, 3 to 25 primary iPSC colonies were obtained per well. After 24 h and reaching approximately 50% confluency (Figure 1D), a single transfection with 0.5 μg OKSiM-GFP srRNA was performed for 4 h. Subsequently, cells were cultivated in B18R-containing medium. Using flow cytometry, a strong GFP reporter protein expression was detected in 7.2% ± 2.8% of the cells 2 days post-transfection (n = 4). On the third day after transfection, 0.8 μg/mL puromycin was added to the medium to select srRNA-transfected cells. After 24 h, only a partial antibiotic-mediated, selective cell death was detected. On day 7 after transfection, a significant reduction in cell numbers was observed, and GFP+ cells carrying the reprogramming srRNAs survived and grew in colonies. On day 7, the cells underwent severe morphological changes, and small round cells with large nuclei appeared (Figure 1D). Afterward, the medium was changed to E8 stem cell medium containing B18R. In the following days, when the cells reached 90% confluency during the reprogramming process, the cells were split at a 1:2 or 1:4 ratio onto vitronectin-coated plates. On day 13, an increase in colony size was detected. After 26 days, iPSC colonies were identified using live-cell staining (antibody against SSEA-4). Thereafter, B18R was withdrawn from the medium to eliminate the reprogramming srRNA from the cells, and iPSCs were either immediately picked or further cultivated for about 4–7 days until primary iPSC colonies increased in size (D33). Manually picked iPSC colonies were transferred onto the vitronectin-coated wells of a 12-well plate.
Characterization of iPSCs Derived from RECs
Expression of Pluripotency Markers
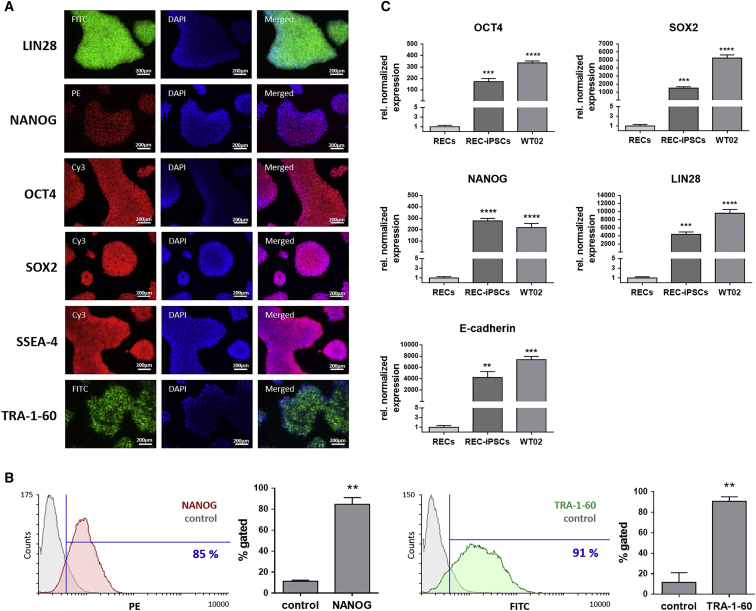
After the reprogramming of adult somatic RECs into iPSCs, the expression of pluripotency markers was analyzed in iPSCs at passage 4 by immunostaining with specific antibodies. Fluorescence microscopy analyses revealed a strong expression of NANOG, OCT4, SOX2, SSEA-4, TRA-1-60, and LIN28 within the entire iPSC colonies (Figure 2A). Furthermore, flow cytometry analyses demonstrated that 85% ± 6% of iPSCs (passages 5–7) were expressing NANOG and that 91% ± 4% of cells were positive for TRA-1-60 (Figure 2B). Further cultivation and expansion of iPSCs until passage 25, corresponding to 4–5 months in culture, had no effect on TRA-1-60 and NANOG expression (Figure S1). Additionally, gene expression of SOX2, OCT4, LIN28, NANOG, and E-cadherin was quantified using qRT-PCR. The obtained REC-iPSCs (passages 3–6) showed—like the commercially available positive control iPSC cell line WT02, which was generated using a Sendai virus vector from dermal fibroblasts—a significantly high expression of SOX2 (1,510-fold), OCT4 (173-fold), LIN28 (4,376-fold), NANOG (733-fold), and E-cadherin (4,230-fold), compared to the initial RECs (Figure 2C).
Figure 2.
Analysis of Pluripotency Marker Expression in iPSCs Generated from RECs
(A) Representative immunofluorescence microscopy images of iPSCs (passage 4) stained with LIN28-, NANOG-, OCT4-, SOX2-, SSEA-4-, and TRA-1-60-specific antibodies. (B) Flow cytometry analysis of iPSCs (passages 5–7) stained with antibodies specific for NANOG and TRA-1-60. Results are shown as mean + SD (n = 3). (C) Expression analysis of SOX2, OCT4, LIN28, NANOG, and E-cadherin transcripts using qRT-PCR. mRNA levels were normalized to GAPDH mRNA levels, and the results are presented relative to the expression levels in RECs. Results are shown as mean + SEM (n = 4). Statistical differences were determined using paired t test (**p < 0.01; ***p < 0.001, ****p<0.0001).
Trilineage Differentiation of iPSCs
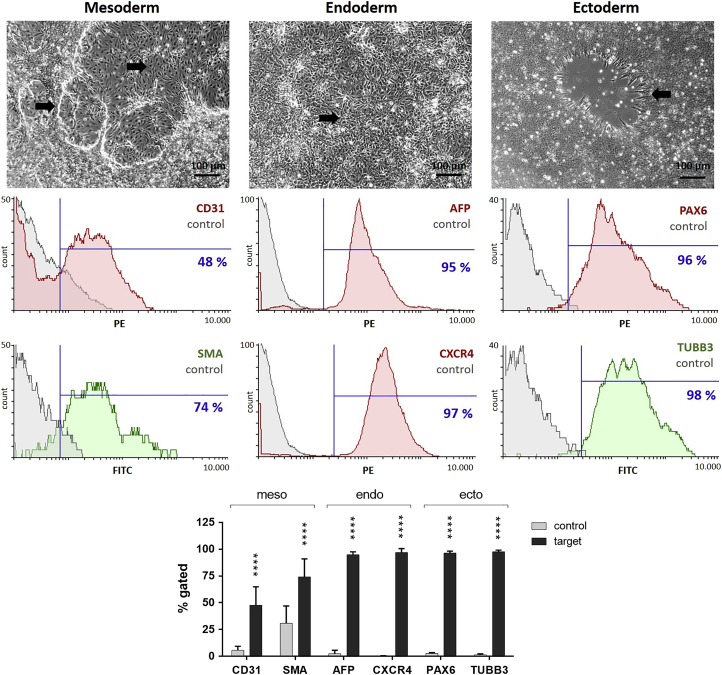
The differentiation potential of the obtained iPSCs (passages 3–7) into all three germ layers was evaluated in vitro using a directed 7-day differentiation protocol (StemMACS Trilineage Differentiation Kit from Miltenyi Biotec) followed by specific antibody staining and quantification using flow cytometry. The mesoderm induction resulted in the generation of elongated endothelial-like cells as well as multilayer accumulations of smooth muscle-like cells (Figure 3). The endoderm induction led to the detection of a dense cell layer similar to that of early hepatocyte-like cells. The ectoderm induction resulted in cells arranged in neural rosettes. The successful mesodermal commitment was shown by detecting 48% ± 17% CD31-positive and 74% ± 17% SMA-positive cells. The endoderm differentiation resulted in 95% ± 3% AFP-positive and 97% ± 4% CXCR4-positive cells. The ectodermal distinction was demonstrated by the detection of 96% ± 2% paired box gene 6 (PAX6)-positive and 98% ± 1% class III β-tubulin (TUBB3)-positive cells.
Figure 3.
In Vitro Differentiation Potential of iPSCs Derived from RECs into the Three Germ Layers: Mesoderm, Endoderm, and Ectoderm
After 7 days of trilineage differentiation, cells with changed phenotypes (indicated with arrows) were observed. Flow cytometry analysis confirmed the differentiation of iPSCs (passages 3–7) into all three germ layers by detection of mesodermal (CD31 and SMA), definitive-endodermal (AFP and CXCR4), and neuroectodermal (PAX6 and TUBB3) cells. Compared to controls, differentiated cells showed significantly increased expression of mesodermal, endodermal, and ectodermal markers. Scale bars of phase contrast microscopic pictures represent 100 μm. Results are shown as mean + SD (n = 4). Statistical differences were determined using one-way ANOVA followed by Bonferroni’s multiple comparison test (****p < 0.0001).
Teratoma Formation of iPSCs
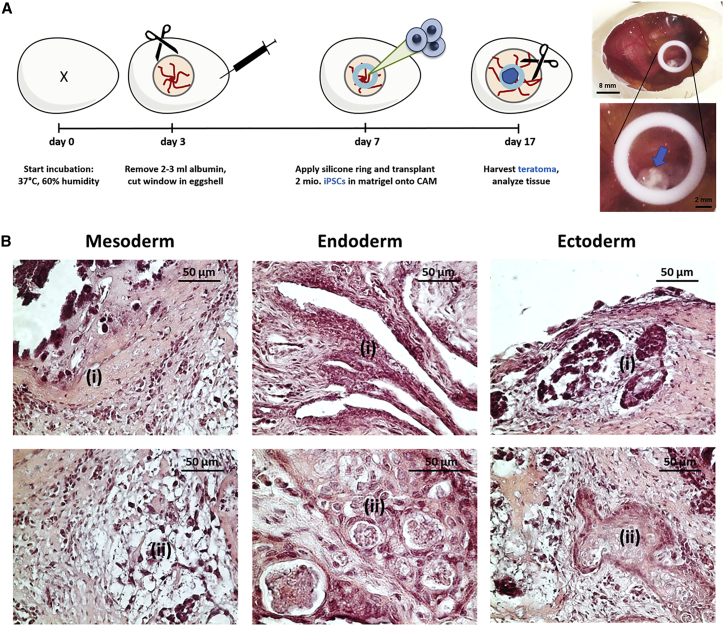
The in vivo differentiation capacity of the generated iPSCs into tissue types of all three germ layers was assessed using chorioallantoic membrane (CAM) assay. Therefore, 2 × 106 iPSCs (passages 8–11) were suspended in Matrigel and applied onto the CAM. After 10 days of incubation, the formed cell mass was excised together with the CAM (Figure 4A). Histological analysis revealed mesodermal, endodermal, and ectodermal differentiation of applied iPSCs within the teratoma tissue mass (Figure 4B). The differentiation into mesodermal tissue was demonstrated by the presence of bone-like and connective tissue structures in teratoma sections. The differentiation toward endodermal tissue was shown by the detection of gland- and gut-like epithelial tissues, and the generation of ectodermal tissue was demonstrated by the presence of squamous epithelial tissue and neural epithelium starting to form rosette-like structures.
Figure 4.
In Vivo Analysis of Teratoma Formation by Application of iPSCs onto CAM
(A) Schematic representation of the implementation of CAM assay for the analysis of teratoma formation. 2 × 106 iPSCs (passages 8–11) were applied into the inner area of a silicone ring (Ø 8 mm), which was placed onto the CAM on day 7 of incubation. After 10 days of incubation, teratomas (indicated with an arrow) were excised, and histological analyses were performed. (B) A representative microscopic image of H&E-stained teratoma sections showing iPSC-derived tissues of all three germ layers: mesoderm (i, bone-like tissue; ii, connective tissue), endoderm (i, gut-like epithelium; ii, gland-like epithelium), and ectoderm (i, primitive neural rosettes; ii, squamous epithelium).
Downregulation of Oncogenic Transcription Factors and Elimination of srRNA after the Reprogramming of RECs into iPSCs and Analysis of Genomic Stability
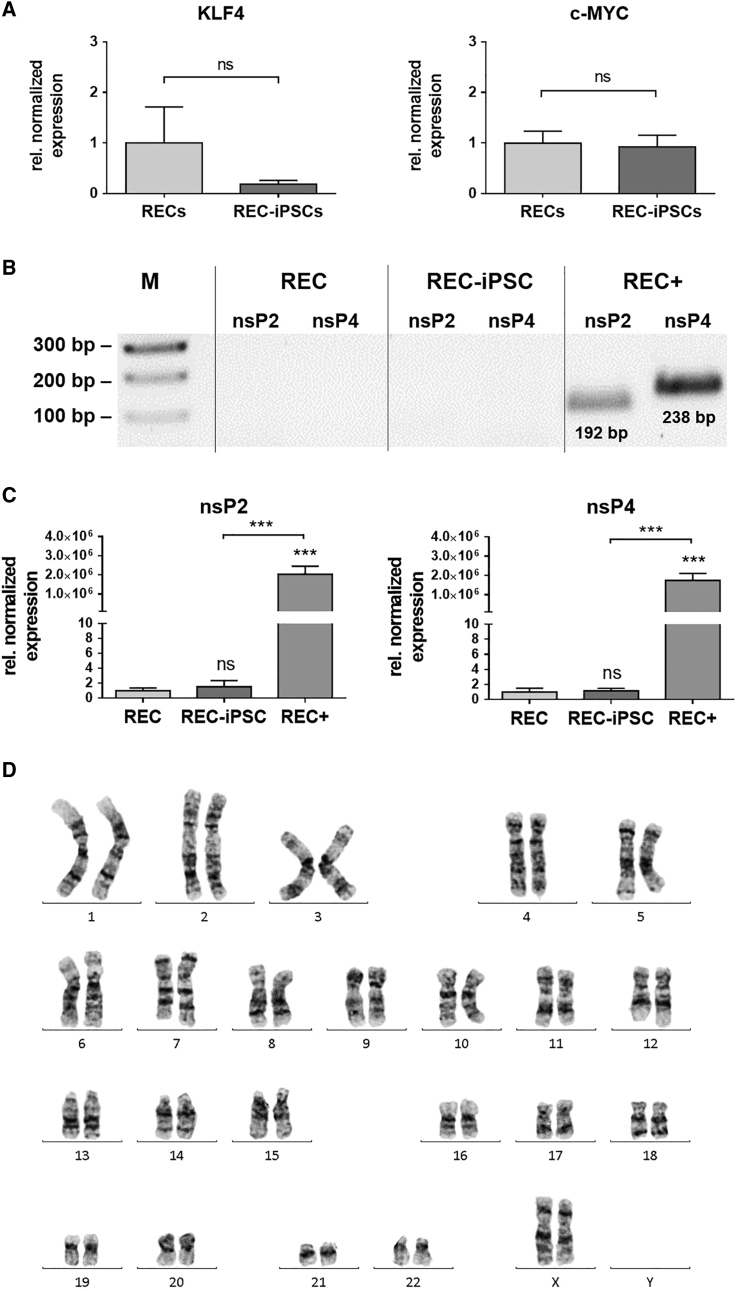
Since a permanently elevated expression of KLF4 and c-MYC is associated with an increased tumorigenesis,16 the expression levels of these transcription factors were determined in obtained iPSCs (passage 3) using qRT-PCR. The analyses revealed that the expression of KLF4 and c-MYC in iPSCs was not significantly different from the expression levels of the initial RECs (Figure 5A). To eliminate the exogenously delivered srRNA in the reprogrammed cells, B18R protein treatment was discontinued between day 21 and day 30 of reprogramming. The elimination of srRNA was analyzed in iPSCs at passage 3 using qRT-PCR. Thereby, the srRNA-specific nsP2 and nsP4 coding regions were detected. RECs transfected with srRNA (RECs+) and cultivated for 2 days demonstrated approximately 2 × 106-fold higher numbers of srRNA transcripts compared to untransfected RECs (Figure 5B). In contrast, no residual srRNA expression was detected in iPSCs. Subsequently, the obtained amplicons were analyzed using agarose gel electrophoresis. As shown in Figure 5C, solely RECs+ showed transcripts with the expected product length of 192 bp for nsP2 and 238 bp for nsP4. To analyze the genomic stability of iPSCs, karyotyping of chromosomes was performed with the initial urine-derived RECs as well as with the generated iPSCs (at passages 4–6). The results revealed no changes in number and appearance of chromosomes, including their length, banding pattern, and centromere position after the reprogramming procedure (Figure 5D).
Figure 5.
Detection of Oncogenic Transcription Factors and srRNA after the Reprogramming of RECs into iPSCs and Analysis of the Genomic Stability of REC-Derived iPSCs
(A) Analysis of KLF4 and c-MYC expression in iPSCs (passage 3) generated by a single transfection with srRNA. The qRT-PCR results are presented relative to the expression levels in the initial RECs. Results are shown as mean + SEM (n = 4). (B) Quantification of srRNA in RECs 2 days after the srRNA transfection (REC+) and in iPSCs (passage 3) using qRT-PCR. Thereby, nsP2- and nsP4-encoding regions of the srRNA were detected using specific primers. The results are presented relative to the expression levels of initial RECs without srRNA transfection. Results are shown as mean + SEM (n = 4). Statistical differences were determined using paired t test (***p < 0.001). (C) Detection of nsP2 and nsP4 specific amplicons in qRT-PCR products using 1% agarose gel electrophoresis. (D) Representative karyotype image of REC-iPSCs (passage 4) obtained from a female subject showing a normal 46XX karyotype by G-banding (n = 3).
Characterization of Generated Cardiomyocytes from iPSCs
iPSCs (passages 4–12) generated from 4 different donors were differentiated within 10–14 days into beating cardiomyocytes (passage 0). Using flow cytometry, the yield of cardiac troponin T (cTNT)-positive cardiomyocytes was determined after the enrichment procedure and resulted in 89.2% ± 1.7% cTNT-positive cells (Figure 6A). The fluorescence microscopic overview of cTNT and α-actinin (ACTN2) double-stained cells confirmed the high amount of cardiac cells compared to a few solely DAPI-stained nuclei (Figure 6B). The differentiated cells also showed the typical elongated rod-like shape of cardiomyocytes. More detailed confocal laser scanning micrographs of the cells in passage 1 demonstrated the structural arrangement of the expressed cardiomyocyte-specific proteins, cTNT, cardiac myosin heavy chain (MYH6), or the muscle-specific marker alpha smooth muscle actin (ACTA2) (Figure 6C). The staining of cells with cTNT-specific antibody revealed the cardiomyocyte-specific sarcomeric structures within the cells. Moreover, using qRT-PCR, the expression of cardiomyocyte-specific markers, atrial natriuretic peptide (ANP), cTNT, α-actin cardiac muscle 1 (ACTC1), and MYH6 was analyzed. The obtained cells (passage 0) expressed 5.4 × 104-fold increased levels of ANP, 2.9 × 104-fold higher cTNT levels, 5.5 × 103-fold higher ACTC1 levels, and 2.2 × 104-fold higher MYH6 levels compared to those in the initial RECs (Figure 6D). Furthermore, the presence of cardiac troponin I (TNNI3) was analyzed in cell culture supernatants 13 to 16 days after starting the iPSC differentiation into cardiomyocytes (Figure 6E). A significantly higher concentration of TNNI3 (0.26 ± 0.11 μg/L) was detected in cardiomyocyte cell culture supernatants compared to iPSC culture medium supernatants (<0.03 μg/L, which represents the lower TNNI3 detection limit). Thereby, the successful differentiation of iPSCs into cardiomyocytes was demonstrated.
Figure 6.
Analysis of Cells Obtained after Cardiac Differentiation of REC-Derived iPSCs
(A) Flow cytometry analysis of cTNT-expressing cells after cardiac differentiation and enrichment. (B) Fluorescence microscopic overview of cTNT- and α-actinin (ACTN2)-positive cells in passage 1 after the cardiac differentiation. Counterstaining of nuclei was performed with DAPI. (C) Confocal-laser-scanning microscopic analysis of cardiomyocyte-specific ACTA2, cTNT, and MYH6 expression. Nuclei were stained with SYTO9. (D) qRT-PCR gene expression analysis of cardiac-specific proteins ANP, cTNT, ACTC1, and MYH6 in passage 1 after the differentiation. (E) Detection of TNNI3 concentration in cardiomyocyte culture supernatants and in iPSC culture supernatants as control. Results are shown as mean + SEM (n = 3). Statistical differences were determined using paired t test (*p < 0.05; **p < 0.01; ***p < 0.001).
Characterization of Contracting Cardiomyocytes
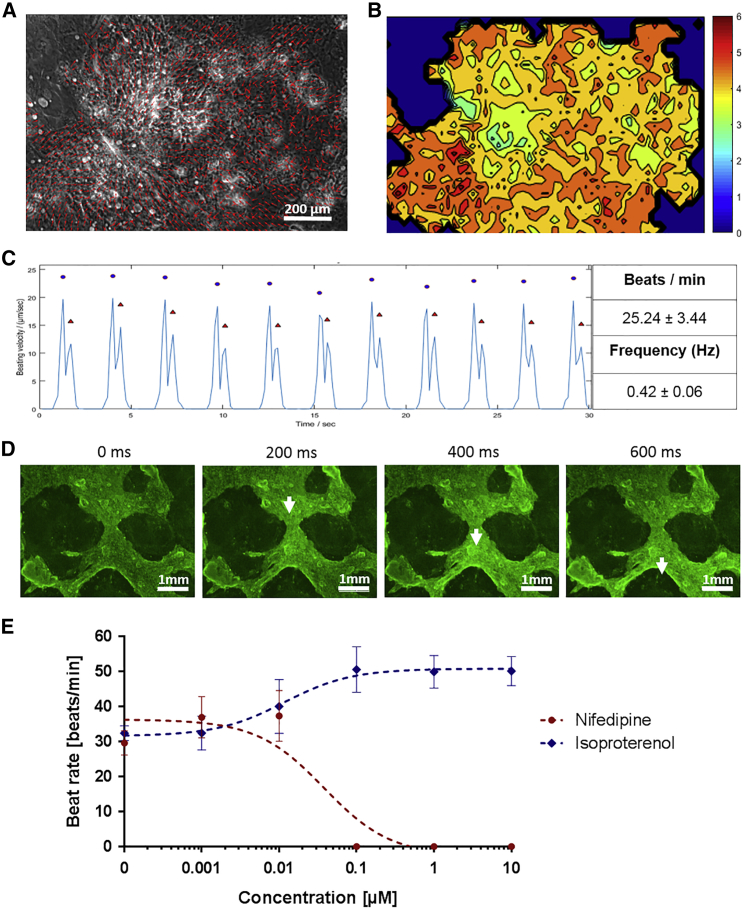
The rhythmic beating is a typical characteristic of fetal and immature stem-cell-derived cardiomyocytes.32 Thus, to show and analyze the contraction of obtained cardiomyocytes (passage 0), short video recordings were made for 30 s, with 7 pictures per second. Using MATLAB application Motion GUI, the motion directions were calculated and indicated by arrows in Figure 7A (Video S1). Thereby, wide ranges of motion and directional synchronous contractions were detected. Furthermore, the areas with the highest movement rate are shown in the heatmap (Figure 7B). Beating rates were calculated by the time shift between contractions. Cardiomyocytes produced from the iPSCs of 4 different REC donors showed uniform motion patterns during the recording time, with a mean beating rate of 25.24 ± 3.44 beats per minute, which corresponds to a beating frequency of 0.42 Hz ± 0.06 Hz (Figure 7C). Fluorescence microscopic recordings of Ca2+ transients in the cardiomyocyte cultures showed Ca2+ oscillations (Figure 7D; Video S2), which proved the electromechanical coupling of obtained cardiomyocytes. These findings revealed that the generated cardiomyocytes were functional in regulating intracellular Ca2+ signaling. In addition, the beating rate was determined in the presence of Ca2+ antagonist nifedipine or β-adrenoceptor agonist isoproterenol (Figure 7E). The treatment of cardiomyocytes with 0.1, 1, or 10 μM nifedipine led to the complete inhibition of cardiomyocyte contractions. In contrast, the treatment of cardiomyocytes with 0.01 μM isoproterenol increased the beating rate from 32.42 ± 2.09 beats per minute to 40.09 ± 7.71 beats per minute. At an isoproterenol concentration of 0.1 μM, the beating rate reached 50.61 ± 6.52 beats per minute. After the removal of nifedipine and isoproterenol and cultivation of cells for a few hours in cardiomyocyte maintenance medium (CMM), the cells recovered and displayed initial beat rates.
Figure 7.
Characterization of Contracting Cardiomyocytes
(A) Arrow-based display of the directed contractions of cardiomyocytes (passage 0). (B) Heatmap depicting the time-averaged magnitude of motion in the x and y directions. An increase of movement is indicated by colors from blue to red. (C) Representative display of beating velocity (micrometers per second) during the recorded time of 30 s. Blue dots indicate contraction, and red triangles indicate the relaxation. Beat rate and frequency (mean ± SD) of iPSC-derived cardiomyocytes were calculated via motion tracking (n = 4). (D) Ca2+ imaging of cardiomyocytes. Representative images of every 200 ms are shown. White arrows indicate the position of Ca2+ transients. (E) Dose-response curves of isoproterenol- and nifedipine-treated cardiomyocytes (mean ± SD; n = 3).
Recordings of 30 s (7 pictures/s) showing wide ranges of motion and directional synchronous contractions. Direction is indicated by arrows using Matlab application Motion GUI.
Fluorescent calcium indicators were used to visualize the intracellular calcium flux during contraction. Recordings of 30 s (7 pictures/s) showing Ca2+ transients in the cardiomyocyte culture.
Discussion
In recent years, the ability to generate iPSCs from somatic cells has led to considerable progress in regenerative medicine, and the reprogramming has become a powerful tool in the field of tissue engineering. In this study, we established a method to generate footprint-free cardiomyocytes by using the autologous cell material from the patient’s urine and srRNA. First, urine-derived human RECs were reprogrammed using srRNA into iPSCs and then differentiated into beating autologous cardiomyocytes. The obtained iPSCs showed self-renewability and the expression of pluripotency-specific markers, while no residual srRNA and genomic abnormalities were detected. The trilineage differentiation potential of REC-derived iPSCs was demonstrated in vitro as well as in vivo. Moreover, the cardiac differentiation of these cells resulted in the generation of contractile cardiomyocytes. Thus, the results clearly demonstrated that autologous footprint-free iPSCs as well as cardiomyocytes can be obtained by using the patient’s urine as a source for somatic cells. Thereby, invasive biopsies for isolation of somatic cells is not required. Since the obtained cells are footprint-free and autologous, they might have a high potential to be used for the regeneration of injured myocardium.
Urine is a naturally excreted material; therefore, the sampling of patients’ urine offers an easy, non-invasive, low-cost, and pain-free method for the collection of sufficient numbers of human somatic cells for reprogramming. Due to natural physiological self-renewal of the epithelial tissue in the urinary tract, approximately 2,000 to 7,000 cells are daily detached and excreted with the urine.15 In urine, 3 types of epithelial cells can be found: renal, transitional, and squamous.33 RECs line the nephron as a single layer. An increased number of these cells in the urine can indicate an infection or kidney disease. Transitional epithelial cells are a multilayer of epithelial cells that line the urinary bladder. A few transitional cells are present in the urine of healthy persons, and increased numbers are associated with infection or transitional cell carcinoma. Most often, these cells are also found in the urine after urethral or ureteral catheterization. Squamous epithelial cells line the urethra and vagina, and this type of epithelial cells is most often found in female urine. Large numbers of squamous cells in female urine generally indicates vaginal contamination. However, during a culturing time of 3 to 5 days in renal epithelial proliferation medium, the squamous epithelial cells do not adhere to the cell culture plate and are removed after the medium change. Typically, 3 to 6 small colonies of RECs appear and grow steadily.
In our study, 100–200 mL urine was enough to obtain a sufficient amount of cells for the reprogramming procedure. However, by using an increased amount of urine, more cells could be obtained; thereby, the expansion time of the cells could be reduced. Furthermore, the proliferation rate of the cells can vary between different donors and ages and might influence the reprogramming efficiency. In this study, the isolated cells from urine samples were positive for the epithelial marker β-catenin and the renal proximal tubular marker CD13. In contrast to urine sampling, the commonly used somatic cells are obtained from skin samples (fibroblasts)14 or blood cells34 by invasive procedures. Cutting out healthy skin pieces is, of course, a painful procedure and is associated with the risk of infection. Furthermore, the reprogramming of blood cells is difficult, and the efficiency is known to be relatively low.35 It has also been shown that iPSCs derived from different cell sources maintain a distinct molecular pattern of epigenetic markers, which is linked to the donor tissue.36 This might result in the improved differentiation potential of iPSCs into the initial somatic cell type. In this study, the successful differentiation of urine-derived REC-iPSCs into cells of all three germline directions was demonstrated.
The reprogramming was performed by a single transfection of RECs with a synthetic srRNA that encodes four reprogramming factors—OCT4, KLF4, SOX2, and c-MYC—as well as the fluorescent reporter protein GFP and contains an open reading frame for puromycin resistance. The puromycin resistance enables the positive selection of srRNA-containing cells, which improves the reprogramming efficiency. In comparison to viral-vector-based methods, the use of srRNA prevents random integration of reprogramming factors into the genome and enables the generation of footprint-free iPSCs. Furthermore, compared to the application of synthetic mRNA for the generation of iPSCs,22 which requires a daily transfection of 5 different mRNAs into the same target cell, a single transfection of cells with srRNA is sufficient for the complete reprogramming period.
The addition of B18R protein into the reprogramming medium suppresses the type-I-interferon-associated immune responses37 to the srRNA; thereby, the premature degradation of the srRNA in the cells can be prevented. After the reprogramming, the termination of B18R addition leads to the degradation of srRNA.30 Furthermore, the incorporation of IRES-GFP into the srRNA allowed the control of successful transfection and translation of srRNA during the reprogramming process. Additionally, in the absence of the immunosuppressive protein B18R, the decrease of the fluorescence intensity indicated the degradation of the srRNA in the cells. Moreover, the implementation of qRT-PCR using specific primers for the nsP2 and nsP4 regions of the srRNA provided the evidence for the srRNA degradation, since, in obtained iPSCs (passage 3), no srRNA could be detected. The proto-oncogenes c-MYC and KLF4 are expressed in different types of cancer,38, 39 and the permanent overexpression of these genes is associated with an increased tumorigenesis.16, 40 Thus, although the expression of these proteins is required during the reprogramming, after the reprogramming, they should be downregulated. This was also demonstrated in our obtained iPSCs; the expression of c-MYC and KLF4 was not significantly different from the expression in precursor RECs. Furthermore, the decrease of c-MYC and KLF4 expression also indicates the degradation of the srRNA construct in the iPSCs.
In addition to expressing certain proteins responsible for maintaining the pluripotency and the self-renewal capacity of iPSCs, another key feature of iPSCs is the ability to differentiate into each of the three germ layers: mesoderm, endoderm, and ectoderm. We successfully demonstrated the ability of the obtained iPSCs to form each of the germ layers after directed differentiation in vitro as well as in vivo after the application of iPSCs on CAM. Compared to the teratoma formation in mice, which is incubated for about 4 weeks, the application of 2 × 106 iPSCs on CAM and the incubation time of 10 days were sufficient to form all three germ layers. The cardiac differentiation of renal-epithelial-cell-derived iPSCs resulted already after 7 days in beating cardiomyocytes with a beating rate of approximately 25 beats per minute. The treatment of these cells with 0.1 μM nifidipine resulted in complete inhibition of beating. In contrast, the pharmacological modulation of REC-iPSC-derived cardiomyocytes with isoproterenol led to an increased beating rate.
The future application of these cells for the repair of damaged heart tissues requires the production of pure cardiomyocyte cultures in a large-scale format and the selection of the appropriate subtype-specific cardiomyocytes—nodal, atrial, or ventricular cardiomyocytes.41, 42, 43 Furthermore, the complete differentiation of iPSCs into mature cardiomyocytes should be ensured.44, 45 Dubois et al.46 demonstrated by using an anti-SIRPA antibody and fluorescence-activated cell sorting (FACS) that cardiomyocytes can be enriched from human pluripotent stem cells. In our study, we used the lactate-based method to enrich cardiomyocytes.47 Due to differences in energy substrate metabolism, compared to other mammalian cells, cardiomyocytes are able to produce energy also from lactate or fatty acids instead of glucose.48 Thus, the cultivation of cells in glucose-depleted and lactate-supplemented medium leads to the survival of cardiomyocytes and the elimination of undifferentiated cells. Using this method, 89.2% ± 1.7% cTNT-positive cells were obtained.
Heart failure and myocardial infarction mainly affect the ventricles of the heart; thus, to prevent arrhythmias after transplantation of the cells into cardiac ventricles, the ventricular subtype of cardiomyocytes should be applied. Therefore, cell sorting can be performed to obtain pure cardiomyocyte subtypes for different applications. The commonly used marker specific for ventricular cardiomyocytes is the myosin light chain 2v (MLC-2v), and the myosin light chain 2a (MLC-2a) is considered as a specific marker for atrial cardiomyocytes.
Conclusions
The non-invasive collection of somatic cells from urine and the one-off application of srRNA could allow the easy and efficient generation of sufficient and unlimited numbers of patient-specific iPSCs, which can then be differentiated besides cardiomyocytes also into other desired cell types without any genomic integration. Thereby, personalized cell therapy of different diseases can be enabled, which prevents rejection reactions and the use of immunosuppressive drugs with their long-term complications. Thereby, the outcome of various cell therapy approaches can be greatly improved, and the generated cells can also serve as cell models for studying specific genetic diseases and treatment methods.
Materials and Methods
Production of Synthetic srRNA
The T7-VEE-OKSiM plasmid30 containing the VEE non-structural protein coding sequences (nsP1 to nsP4) to enable the RNA replication and the coding sequences of OCT4, KLF4, SOX2, and c-MYC was purchased from Addgene (LGC Standards, Teddington, UK). To monitor the transfection and reprogramming efficiency, an additional sequence encoding an IRES (internal ribosome entry site) and the reporter protein GFP were cloned by Aldevron (Fargo, ND, USA) into the plasmid, which is then called OKSiM-GFP plasmid.
To amplify the OKSiM-GFP plasmid, E. coli competent cells (α-Select Chemically Competent Cells, Bioline, Luckenwalde, Germany) were transformed with 100 ng plasmid and cultivated in lysogeny broth (LB) medium containing 50 μg/mL ampicillin. Plasmid isolation was performed using the QIAprep Spin Miniprep Kit (QIAGEN, Hilden, Germany). Afterward, 36 μg OKSiM-GFP plasmid was linearized using 5 μL FastDigest MluI restriction enzyme (Thermo Fisher Scientific, Waltham, MA, USA) and 20 μL 1× reaction buffer in a total volume of 200 μL for 3 h at 37°C. The linearized DNA was purified using the Isolate II PCR and Gel Kit (Bioline) and analyzed using 1% agarose gel electrophoresis.
For the synthesis of srRNA, IVT was performed for 2 h at 37°C using the RiboMAX Large Scale Production System—T7 Kit (Promega, Madison, WI, USA) according to the manufacturer’s instructions. The IVT reaction mixture contained 10 μg linearized OKSiM-GFP plasmid DNA and 40 U RiboLock RNase Inhibitor (Thermo Fisher Scientific) in 100 μL. Afterward, DNA templates were removed by adding 1 μL TURBO DNase for 15 min at 37°C. Next, 5′-end capping was performed using the ScriptCap Cap1 Capping System followed by 3′-end polyadenylation with the A-Plus Poly(A) Polymerase Tailing Kit (both from Cellscript, Madison, WI, USA) according to manufacturer’s instructions. Following each reaction step, the srRNA was purified using the RNeasy Kit (QIAGEN) and eluted in nuclease-free water. The purity and specific length of generated srRNA products were analyzed using 1% agarose gel containing 2.2 M formaldehyde and 1× GelRed (Biotium, Fremont, CA, USA) in 1× MOPS (3-(N-morpholino)propanesulfonic acid) buffer. Electrophoresis was performed at 100 V for 60 min in 1× MOPS buffer.
Isolation and Cultivation of RECs from Urine
RECs were isolated from 100–200 mL urine from healthy donors (men and women between 25 and 35 years of age) by centrifugation at 400 × g for 10 min. Afterward, cells were washed with 50 μg/mL gentamicin and 250 μg/mL amphotericin B (Sigma-Aldrich) containing Dulbecco’s PBS (DPBS; Thermo Fisher Scientific) and centrifuged at 200 × g for 10 min. Then, cells were suspended in 1 mL primary medium consisting of DMEM/F12 high glucose supplemented with 10% fetal bovine serum (FBS), REGM Renal Epithelial Growth Medium SingleQuots Kit (Lonza, Basel, Switzerland), 50 μg/mL gentamicin, and 250 μg/mL amphotericin B and plated in one well of a 0.1% gelatin-coated 12-well plate. For the next 3 days, 1 mL primary medium was added each day. The obtained cells were then cultivated in proliferation medium (REMC) consisting of 50% renal epithelial (RE) basal medium with REGM Bullet Kit supplements (Lonza, Basel, Switzerland) and 50% mesenchymal cell (MC) proliferation medium (DMEM high glucose supplemented with 10% FBS, 1× GlutaMax, 1× MEM (minimum essential medium) non-essential amino acids (NEAA), 50 μg/mL gentamicin, and 250 μg/mL amphotericin B, 5 ng/mL basic fibroblast growth factor (bFGF), 5 ng/mL platelet-derived growth factor (PDGF)-AB, and 5 ng/mL epidermal growth factor (EGF). Cell culture reagents were obtained from Thermo Fisher Scientific and recombinant human growth factors were obtained from Peprotech (Hamburg, Germany). Cells were cultivated at 37°C with 5% CO2, and the medium was changed every 2–3 days. After reaching 80% confluency, RECs were detached using 0.04% trypsin/0.03% EDTA, and the reaction was stopped using trypsin-neutralizing solution (TNS; 0.05% trypsin inhibitor in 0.1% BSA, PromoCell, Heidelberg, Germany). Afterward, cells were centrifuged for 5 min at 300 × g and seeded on 0.1% gelatin (Sigma-Aldrich Chemie, Steinheim, Germany)-coated cell culture plates. The characterization of RECs was performed by flow cytometry using antibodies specific for epithelial marker β-catenin and renal proximal tubular marker CD13 (Miltenyi Biotec, Bergisch Gladbach, Germany).
Reprogramming of RECs into iPSCs Using srRNA
To perform reprogramming, 5 × 104 RECs (from 4 different donors, passages 2–3) were seeded per well of a 12-well plate coated with 0.1% gelatin and incubated overnight at 37°C in proliferation medium. Next day, the cells were incubated for 45–60 min with proliferation medium containing 200 ng/mL B18R interferon inhibitor (Thermo Fisher Scientific) at hypoxia (37°C, 5% CO2, 5% O2). For the transfection, lipoplexes were generated by the incubation of 0.5 μg OKSiM-GFP srRNA and 1.5 μL Lipofectamine Messenger Max (Thermo Fisher Scientific) in 0.5 mL Opti-MEM I reduced serum medium (Opti-MEM, Thermo Fisher Scientific) for 15 min at room temperature (RT). RECs were washed with DPBS, and lipoplexes were added. After 4 h of incubation, lipoplexes were discarded, and 1 mL fresh proliferation medium containing 200 ng/mL B18R was added for further incubation at 37°C with 5% CO2 and 5% O2 for 24h. Until day 3, the medium was replaced daily, and at day 3, 0.8 μg/mL puromycin (Sigma-Aldrich) was added to the proliferation medium to select srRNA-containing transfected cells. After 2–3 days of incubation, untransfected cells were eliminated. Thereafter, B18R-containing proliferation medium was changed every day. At day 7 of reprogramming, medium was changed to E8 stem cell medium (Essential 8, Thermo Fisher Scientific) supplemented with 200 ng/mL B18R. After the first appearance of iPSC colonies (beginning on day 21), B18R was withdrawn from the medium. iPSC colonies, which were positively stained with DyLight 550-labeled mouse anti-human StainAlive SSEA-4 antibody (Stemgent) were picked manually and seeded on 0.5 μg/cm2 vitronectin (Thermo Fisher Scientific)-coated tissue culture plates and expanded in E8 medium.
Cultivation of iPSCs Derived from RECs
After reaching confluence, iPSCs were washed once with DPBS and incubated for 5–10 min at RT with DPBS containing 0.5 mM EDTA (Sigma-Aldrich). After the detachment, cells were suspended in E8 medium containing 10 μg/mL ROCK inhibitor Y-27632 (Enzo Life Sciences, Lausen, Switzerland) and passaged at a split ratio of 1:10 or 5 × 105 cells per vitronectin-coated well of a 6-well plate or in T25 culture flasks. iPSCs were cultivated at 37°C and 5% CO2, and E8 medium was changed daily.
Flow Cytometry
Cells were washed with 1 mL DPBS and detached using 0.04% trypsin/0.03% EDTA, and the reaction was stopped by adding TNS (PromoCell). The cells were then centrifuged (5 min at 400 × g), washed with DPBS, and fixed for 10 min at RT in 0.5 mL fixation solution (R&D Systems). After washing with DPBS, cells were suspended in washing buffer (Permeabilization/Wash Buffer I, R&D Systems), and 5 μL fluorescently labeled antibody was added and incubated for 45 min at RT. Afterward, cells were washed with 0.5 mL washing buffer, suspended in 200 μL 1× BD CellFIX solution (Becton Dickinson, Heidelberg, Germany), and measured using a BD FACScan flow cytometer (Becton Dickinson) and Flowing Software (Turku Centre for Biotechnology, Turku, Finland).
qRT-PCR
To perform qRT-PCR analysis, 300 ng RNA was reverse transcribed into complementa DNA (cDNA) using the iScript Kit (Bio-Rad). The primers used for the specific amplification of transcripts are listed in Table S1, and they were ordered from Ella Biotech (Martinsried, Germany) and used at a final concentration of 300 nM. Real-time qRT-PCR reactions were performed in a CFX Connect Real-Time PCR Detection System (Bio-Rad) using IQ SYBR Green Supermix (Bio-Rad). Expression of constitutively expressed gene GAPDH (glyceraldehyde 3-phosphate dehydrogenase) was used as an internal control for the amount of RNA input. Primers were designed by using the Primer-Blast tool from NCBI.49 Melting temperatures and self-complementarities were checked using the Oligonucleotide Properties Calculator from Northwestern University Medical School.50
The qRT-PCR amplification of cDNA was performed under the following conditions: 3 min at 95°C for one cycle, followed by 40 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 10 s. After 40 cycles, melt curve analysis was performed to ensure the specificity of the products. The qRT-PCR reactions were run in triplicate with a total volume of 15 μL per well. Levels of mRNA for each gene were normalized to GAPDH, and the results are shown relative to control mRNA levels.
Characterization of iPSCs Derived from RECs
Detection of Pluripotency Markers
Immunocytochemistry of iPSCs
5 × 105 iPSCs (passages 4–5) were seeded on vitronectin-coated glass slides in 12-well plates and cultured for 2–3 days in cell culture medium until reaching 50%–70% confluency. Cells were washed 2× with 1 mL DPBS and fixed for 10 min at RT with 0.5 mL fixation solution (R&D Systems, Minneapolis, MN, USA). After washing with 0.5 mL washing buffer, the cells were incubated for 1.5 h at RT in washing buffer containing 5% BSA. Then, cells were incubated 3 h at RT with fluorescently labeled antibodies in washing buffer containing 1% BSA or overnight at 4°C with primary antibodies. After washing 3× with 0.5 mL washing buffer, the staining of the cells with fluorescently labeled secondary antibodies was performed for 1 h at RT in washing buffer containing 1% BSA. Afterward, the cells were washed 3× with washing buffer, DPBS, and then water. Subsequently, the coverslips were mounted using Fluoroshield mounting medium with DAPI (Abcam, Cambridge, UK). Rabbit anti-human POU5F1 (OCT4) (Sigma-Aldrich Chemie), rabbit anti-human SOX2 (Stemgent, Cambridge, MA, USA), and mouse anti-human LIN28A (6D1F9) (Thermo Fisher Scientific) antibodies were used as primary antibodies. Fluorescein isothiocyanate (FITC)-labeled sheep anti-mouse immunoglobulin G (IgG) (whole molecule; Sigma-Aldrich) and Cy3-labeled goat anti-rabbit IgG cross-adsorbed secondary antibody (Thermo Fisher Scientific) were used according to the manufacturer’s instructions. Furthermore, phycoerythrin (PE)-labeled mouse anti-human NANOG antibody (BD, Franklin Lakes, NJ, USA), DyLight 488-labeled mouse anti-human StainAlive TRA-1-60 antibody (Stemgent), and DyLight 550-labeled mouse anti-human StainAlive SSEA-4 antibody (Stemgent) were used. Fluorescence images were taken using an Axiovert 135 microscope and AxioVision 4.8.2 software (Carl Zeiss, Oberkochen, Germany).
Gene Expression Analysis of iPSCs
RNA from 1 × 106 iPSCs (passages 3–6) was isolated using the Aurum Total RNA Mini Kit (Bio-Rad, Munich, Germany) according to manufacturer’s instructions, and qRT-PCR analysis was performed to detect the expression of OCT4, SOX2, NANOG, LIN28, E-cadherin, KLF4, and c-MYC. Levels of mRNA for each gene were normalized to that of GAPDH, and the results are shown relative to control mRNA levels in RECs. The commercially available iPSC line WTSli020-A (referred to as WT02 and generated from dermal fibroblasts using Sendai virus vector by the delivery of OCT4, SOX2, KLF4, and c-MYC; European Bank for induced pluripotent Stem Cells; Babraham, UK]) was used as iPSC control.
Trilineage Differentiation of iPSCs
To analyze the ability of the obtained iPSCs to differentiate into the three embryonal germ layers the directed differentiation of iPSCs (at passages 3–7) into meso-, endo-, and ectoderm was tested using the human StemMACS Trilineage Differentiation Kit (Miltenyi Biotec, Bergisch Gladbach, Germany) according the manufacturer’s instructions. Therefore, optimized cell numbers were seeded per well of a vitronectin-coated 12-well plate: mesoderm differentiation (1 × 105 iPSCs), endoderm differentiation (2 × 105 iPSCs), and ectoderm differentiation (1.5 × 105 iPSCs). At day 7, the differentiated cells were analyzed using flow cytometry.
The mesodermal differentiation capacity was analyzed by the formation of endothelial cells, using PE-labeled mouse anti-human CD31 antibody (BD Biosciences, Franklin Lakes, NJ, USA), and the formation of smooth muscle cells, using Alexa Fluor 488-labeled anti-human α-smooth muscle actin (SMA) antibody (R&D Systems). The endodermal differentiation capacity is characterized by the presence of definitive endoderm cells using PE-labeled anti-human α-fetoprotein (AFP) antibody (R&D Systems) and PE-labeled anti-human C-X-C chemokine receptor type 4 (CXCR4) antibody (R&D Systems). Ectodermal differentiation potential was assessed through the presence of neuroectoderm cells using PE-labeled anti-PAX6 antibody (Miltenyi Biotec) and Alexa Fluor 488-labeled anti-human neuron-specific TUBB3 antibody (BD Biosciences).
Teratoma Formation of iPSCs Using Chicken Embryo CAM Assay
To confirm the trilineage differentiation potential of iPSCs, the in vivo formation of teratomas was analyzed using CAM assay. Fresh fertilized chicken eggs of the Lohmann White × White Rock breed chicken variety were obtained from the breeding facility Matthias Sittig (Buchholz, Germany). The eggs were incubated for 3 days at 37°C and 60% relative humidity in an egg incubator (Heka-Brutgeräte, Rietberg-Varensell, Germany) and completely rotated twice a day. At day 3 of incubation, 2–3 mL albumin was aspirated by inserting an 18G needle at the tip of the egg without harming the yolk. Subsequently, a semi-permeable adhesive tape, Suprasorb F (Lohmann & Rauscher, Rengsdorf, Germany), was stuck to the eggshell. A circular window (Ø 1–1.5 cm) was cut into the shell. Unfertilized eggs showing no vasculature or heart beating were removed. Then, using the adhesive tape, the window was sealed to prevent dehydration and to minimize the risk of infection. Afterward, the eggs were incubated without rotation. At day 7, 2 × 106 iPSCs (passages 8–11) were suspended in 50 μL cell culture medium and mixed with 50 μL Matrigel (hECS qualified, Corning). A silicone ring with an inner diameter of 0.8 cm (neoLab, Leonberg, Germany) was carefully placed onto the CAM, and 100 μL Matrigel-containing cells was applied into the inner circle of the ring. The eggs were then sealed and further incubated. At day 17, the CAMs were excised around the application area and fixed overnight at 4°C with 4% paraformaldehyde (Merck, Darmstadt, Germany). After washing with DPBS, the specimens were dehydrated using ascending ethanol series and embedded in paraffin for sectioning. Sections were cut at 8-μm thickness and stained with H&E (Morphisto, Frankfurt, Germany).
Genomic Stability of iPSCs
The genomic stability of iPSCs was analyzed by karyotyping at the Institute of Medical Genetics and Applied Genomics, University of Tübingen, Tübingen, Germany. Therefore, RECs and RE-derived iPSCs (at passages 4–7) were treated for 1 h with Colcemid (Biochrom), incubated with 0.075 M KCl for 30 min at 37°C, and fixed with 1:3 acidic acid:methanol. Karyotyping was performed on G-banded metaphase chromosomes (banding quality of 400–500 bp) using standard cytogenetic procedures. An intact genome was demonstrated by karyotyping (numerical analysis of 15 mitoses and structural analysis of at least 5 mitoses).
Detection of srRNA Elimination after the Reprogramming of Cells
RNA from 1 × 106 iPSCs (passage 3) was isolated using the Aurum Total RNA Mini Kit according to manufacturer’s instructions. Using nsP2- and nsP4-specific primers (Table S1) and qRT-PCR, the presence of srRNA in generated iPSCs was analyzed. RNA levels were normalized to GAPDH, and the results are shown relative to control RNA levels in RECs. In addition, to obtain a positive control, cells were transfected with 1 μg srRNA, and the RNA was isolated after 2 days of cultivation.
Differentiation of iPSCs into Cardiomyocytes
The differentiation of iPSCs from 4 different donors into cardiomyocytes was performed using the PSC Cardiomyocyte Differentiation Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Therefore, 2 × 105 iPSCs (passages 4–12) were seeded on Geltrex LDEV-Free hESC-Qualified Reduced Growth Factor Basement Membrane Matrix (Thermo Fisher Scientific) (1:100 diluted in CMM) or on 0.5 μg/cm2 vitronectin-coated wells of a 6-well plate. After 3–4 days cultivation in E8 medium, the differentiation protocol started. The cells were incubated for 2 days with cardiomyocyte differentiation medium A and then for 2 days with cardiomyocyte differentiation medium B. On the fifth day of differentiation, CMM was added to the cells for the following days of differentiation.
After 7–12 days, when contracting cardiomyocytes were observed (passage 0), the medium was changed to cardiomyocyte enrichment medium (CEM): RPMI 1640 medium without glucose (Thermo Fisher) containing 0.25% BSA (Fraction V, Sigma-Aldrich), 4 mM sodium lactate (Fisher Chemicals), 4 mM HEPES (Thermo Fisher), and 6.5 μM ascorbic acid (Acros Organics, Geel, Belgium). After 4–6 days, cardiomyocytes were passaged using 1 mL TrypLE solution (Thermo Fisher), and the reaction was stopped by adding an equal volume of TNS (PromoCell). Cell suspension was filtered through a 100-μm cell strainer (Greiner Bio-One, Frickenhausen, Germany) and centrifuged at 200 × g for 5 min. The cells were suspended in CMM, seeded onto Geltrex- or vitronectin-coated cell culture plates with a density of 1 × 105 cells per square centimeter (passage 1), and cultivated in a humidified atmosphere at 37°C and 5% CO2. After 4–6 days of enrichment procedure, the yield of differentiated cardiomyocytes was determined by flow cytometry using FITC-labeled mouse anti-human cTNT antibody from Miltenyi Biotec.
Detection of Cardiomyocyte Markers
Immunocytochemistry of Cardiomyocytes Derived from iPSCs
1 × 106 iPSC-derived cardiomyocytes (passage 1) were plated on Geltrex-coated glass slides in 12-well plates and cultivated for 2 days in CMM. Rabbit anti-human cTNT antibody, mouse anti-human ACTA2 (both from R&D Systems), and mouse anti-human MYH6 antibody (GeneTex, Irvine, CA, USA) were used as cardiomyocyte-specific primary antibodies. NL637-conjugated donkey anti-mouse IgG (R&D Systems) and Cy3-labeled goat anti-rabbit IgG (Thermo Fisher Scientific) were used as secondary antibodies. Primary and secondary antibodies were applied as recommended by the manufacturer. Nuclei were stained using a final concentration of 5 μM SYTO 9 Green Fluorescent Nucleic Acid Stain (Thermo Fisher Scientific) in DPBS. Fluorescence images were taken after washing with DBPS using a Leica TCS SP5 confocal laser scanning microscope and the Leica Application Suite Advanced Fluorescence (2.7.3.9723) software (Leica, Wetzlar, Germany). To obtain fluorescence microscopic overview images of cardiomyocytes derived from iPSCs, cells were stained using FITC-labeled anti-human cTNT and PE-labeled anti-human (sarcomeric) α-actinin (ACTN2) antibodies from Miltenyi Biotech and Fluoroshield Mounting Medium with DAPI (Abcam). Fluorescence images were taken using an Axiovert 135 microscope and AxioVision 4.8.2 software (Carl Zeiss).
Gene Expression Analysis of Cardiomyocytes Derived from iPSCs
RNA from 1 × 106 iPSC-derived cardiomyocytes (12–16 days after starting the differentiation; passage 0) was isolated using the Aurum Total RNA Mini Kit according to manufacturer’s instructions, and qRT-PCR analysis was performed to detect the expression of ANP, cTNT, MYH6, and α-actinin, cardiac muscle 1 (ACTC1). Levels of mRNA for each gene were normalized to GAPDH, and the results are shown relative to control mRNA levels in RECs.
Detection of Cardiac Troponin I
1 × 106 iPSCs (passages 4–8) were seeded per well on Geltrex-coated 6-well plates and differentiated for 12–16 days into cardiomyocytes. The supernatants (CMM) of beating cardiomyocyte cultures (passage 0) were analyzed to measure the TNNI3 content using ADVIA Centaur XPT (TnI-Ultra chemiluminescent immunoassay, Siemens Healthcare Diagnostics, Eschborn, Germany) according to the manufacturer’s instructions. Additionally, the TNNI3 concentration was measured in cell culture supernatants of RECs (REMC medium) and REC-derived iPSCs (E8 medium) as well as in fresh CMM.
Characterization of Contracting Cardiomyocytes
Video Microscopy
Video recordings of beating cells were performed to analyze the mechanical beating behavior of obtained cardiomyocytes from 4 different donors. Therefore, 7 images per second were taken for 30 s using an Axiovert 135 microscope and AxioVision 4.8.2 software (Carl Zeiss). The beating center(s) and the beat rate of obtained cardiomyocytes were determined using the MATLAB application Motion GUI.51
Analysis of Electromechanical Coupling Using Ca2+ Imaging
To analyze the electromechanical coupling of the obtained cardiomyocytes, calcium imaging was performed. Ca2+ oscillations are an indication of a fully differentiated cardiac phenotype and key regulator in controlling cardiomyocyte relaxation and contraction. To evaluate the intracellular Ca2+ behavior, Ca2+ transients were measured from spontaneously contracting cardiomyocytes (passage 0, 4 donors) using the Fluo-4 Direct Calcium Assay Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Beating cardiomyocytes were obtained 12 days after the differentiation of iPSCs on Geltrex-coated wells of a 6-well plate. The obtained cells were incubated for 30 min at 37°C with 1 mL 1× Fluo-4 Direct Calcium Assay Reagent Solution and 1 mL CMM. Using a fluorescence microscope (Axiovert 135), Ca2+ transients were recorded within the next 30 min at RT with 5 pictures per second and a 30 ms exposure time at 494 nm excitation wavelength.
Response of Cardiomyocytes to Pharmacological Modulation
To analyze the reaction of the generated REC-iPSC derived cardiomyocytes on pharmaceutical drugs, the cells were treated with 0.0001, 0.001, 0.01, 0.1, 1.0, or 10 μM Ca2+ channel blocker nifedipine or β-adrenoceptor agonist isoproterenol. Stock solutions of 100 mM nifedipine or 100 mM isoproterenol (both from Sigma Aldrich) were prepared in DMSO and diluted in CMM. Cardiomyocyte culture medium was replaced 11 or 13 days after starting the differentiation process (passage 0) by preheated (37°C) drug dilutions in CMM and incubated for 5 min at 37°C. Video recordings of 20 s each were performed using AxioCam and an Axiovert 135 microscope, and the beat rate was analyzed using Motion GUI. After recordings, the medium was changed to CMM.
Statistical Analysis
Data are shown as mean ± SD or SEM. Paired t test or one-way ANOVA for repeated measurements followed by Bonferroni’s multiple comparison test was performed to compare the means. Two-tailed statistical analyses were performed using GraphPad Prism 6.01 (GraphPad Software, La Jolla, CA, USA). Differences of p < 0.05 were considered significant.
Author Contributions
H.S. and M.A.-A. conceived and designed the experiments. H.S. and M.W. performed the experiments with support from A.B., A.-F.P., H.P.W., and M.A.-A., and analyzed the data. U.M.-H. performed karytotyping analysis and interpretation. C.v.O. performed CLSM investigations. H.P.W. and C.S. contributed reagents, materials, and analysis tools. H.S., M.W., and M.A.-A. wrote the paper. M.A.-A. supervised the project.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
For TNNI3 detection in supernatants, the authors would like to thank Dr. Ingo Rettig at the Division of Endocrinology, Diabetology, Vascular Medicine, Nephrology and Clinical Chemistry, Department of Internal Medicine IV, University of Tübingen, Germany. We would also like to thank Jeannette Schöne and Elisa Kächele for excellent cytogenetic technical assistance and Annika Hechler for assistance with the collection of CLSM data. This study was funded by the German Research Foundation (Deutsche Forschungsgemeinschaft; DFG) through AV 133/7-1.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.07.016.
Supplemental Information
References
- 1.Roger V.L. Epidemiology of heart failure. Circ. Res. 2013;113:646–659. doi: 10.1161/CIRCRESAHA.113.300268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergmann O., Zdunek S., Felker A., Salehpour M., Alkass K., Bernard S., Sjostrom S.L., Szewczykowska M., Jackowska T., Dos Remedios C. Dynamics of cell generation and turnover in the human heart. Cell. 2015;161:1566–1575. doi: 10.1016/j.cell.2015.05.026. [DOI] [PubMed] [Google Scholar]
- 3.Li R.K., Jia Z.Q., Weisel R.D., Mickle D.A.G., Zhang J., Mohabeer M.K., Rao V., Ivanov J. Cardiomyocyte transplantation improves heart function. Ann. Thorac. Surg. 1996;62:654–660. doi: 10.1016/s0003-4975(96)00389-x. [DOI] [PubMed] [Google Scholar]
- 4.Watanabe E., Smith D.M., Jr., Delcarpio J.B., Sun J., Smart F.W., Van Meter C.H., Jr., Claycomb W.C. Cardiomyocyte transplantation in a porcine myocardial infarction model. Cell Transplant. 1998;7:239–246. doi: 10.1177/096368979800700302. [DOI] [PubMed] [Google Scholar]
- 5.Valarmathi M.T., Fuseler J.W., Goodwin R.L., Davis J.M., Potts J.D. The mechanical coupling of adult marrow stromal stem cells during cardiac regeneration assessed in a 2-D co-culture model. Biomaterials. 2011;32:2834–2850. doi: 10.1016/j.biomaterials.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caspi O., Itzhaki I., Kehat I., Gepstein A., Arbel G., Huber I., Satin J., Gepstein L. In vitro electrophysiological drug testing using human embryonic stem cell derived cardiomyocytes. Stem Cells Dev. 2009;18:161–172. doi: 10.1089/scd.2007.0280. [DOI] [PubMed] [Google Scholar]
- 7.Mummery C., Ward-van Oostwaard D., Doevendans P., Spijker R., van den Brink S., Hassink R., van der Heyden M., Opthof T., Pera M., de la Riviere A.B. Differentiation of human embryonic stem cells to cardiomyocytes: role of coculture with visceral endoderm-like cells. Circulation. 2003;107:2733–2740. doi: 10.1161/01.CIR.0000068356.38592.68. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J., Wilson G.F., Soerens A.G., Koonce C.H., Yu J., Palecek S.P., Thomson J.A., Kamp T.J. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ. Res. 2009;104:e30–e41. doi: 10.1161/CIRCRESAHA.108.192237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burridge P.W., Zambidis E.T. Highly efficient directed differentiation of human induced pluripotent stem cells into cardiomyocytes. Methods Mol. Biol. 2013;997:149–161. doi: 10.1007/978-1-62703-348-0_12. [DOI] [PubMed] [Google Scholar]
- 10.Mathur A., Loskill P., Shao K., Huebsch N., Hong S., Marcus S.G., Marks N., Mandegar M., Conklin B.R., Lee L.P., Healy K.E. Human iPSC-based cardiac microphysiological system for drug screening applications. Sci. Rep. 2015;5:8883. doi: 10.1038/srep08883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moretti A., Bellin M., Welling A., Jung C.B., Lam J.T., Bott-Flügel L., Dorn T., Goedel A., Höhnke C., Hofmann F. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N. Engl. J. Med. 2010;363:1397–1409. doi: 10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 13.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 14.Raab S., Klingenstein M., Liebau S., Linta L. A comparative view on human somatic cell sources for iPSC generation. Stem Cells Int. 2014;2014:768391. doi: 10.1155/2014/768391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ingelfinger J.R. Nephrogenic adenomas as renal tubular outposts. N. Engl. J. Med. 2002;347:684–686. doi: 10.1056/NEJMe020084. [DOI] [PubMed] [Google Scholar]
- 16.Okita K., Ichisaka T., Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 17.Wernig M., Meissner A., Cassady J.P., Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell. 2008;2:10–12. doi: 10.1016/j.stem.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Stadtfeld M., Nagaya M., Utikal J., Weir G., Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008;322:945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaji K., Norrby K., Paca A., Mileikovsky M., Mohseni P., Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu J., Hu K., Smuga-Otto K., Tian S., Stewart R., Slukvin I.I., Thomson J.A. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou H., Wu S., Joo J.Y., Zhu S., Han D.W., Lin T., Trauger S., Bien G., Yao S., Zhu Y. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warren L., Manos P.D., Ahfeldt T., Loh Y.H., Li H., Lau F., Ebina W., Mandal P.K., Smith Z.D., Meissner A. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steinle H., Behring A., Schlensak C., Wendel H.P., Avci-Adali M. Concise review: application of in vitro transcribed messenger RNA for cellular engineering and reprogramming: progress and challenges. Stem Cells. 2017;35:68–79. doi: 10.1002/stem.2402. [DOI] [PubMed] [Google Scholar]
- 24.Rabinovich P.M., Weissman S.M. Cell engineering with synthetic messenger RNA. Methods Mol. Biol. 2013;969:3–28. doi: 10.1007/978-1-62703-260-5_1. [DOI] [PubMed] [Google Scholar]
- 25.Thess A., Grund S., Mui B.L., Hope M.J., Baumhof P., Fotin-Mleczek M., Schlake T. Sequence-engineered mRNA without chemical nucleoside modifications enables an effective protein therapy in large animals. Mol. Ther. 2015;23:1456–1464. doi: 10.1038/mt.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson B.R., Muramatsu H., Nallagatla S.R., Bevilacqua P.C., Sansing L.H., Weissman D., Karikó K. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010;38:5884–5892. doi: 10.1093/nar/gkq347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karikó K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 28.Karikó K., Muramatsu H., Welsh F.A., Ludwig J., Kato H., Akira S., Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Svitkin Y.V., Cheng Y.M., Chakraborty T., Presnyak V., John M., Sonenberg N. N1-methyl-pseudouridine in mRNA enhances translation through eIF2α-dependent and independent mechanisms by increasing ribosome density. Nucleic Acids Res. 2017;45:6023–6036. doi: 10.1093/nar/gkx135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshioka N., Gros E., Li H.R., Kumar S., Deacon D.C., Maron C., Muotri A.R., Chi N.C., Fu X.D., Yu B.D., Dowdy S.F. Efficient generation of human iPSCs by a synthetic self-replicative RNA. Cell Stem Cell. 2013;13:246–254. doi: 10.1016/j.stem.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petrakova O., Volkova E., Gorchakov R., Paessler S., Kinney R.M., Frolov I. Noncytopathic replication of Venezuelan equine encephalitis virus and eastern equine encephalitis virus replicons in Mammalian cells. J. Virol. 2005;79:7597–7608. doi: 10.1128/JVI.79.12.7597-7608.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mummery C.L., Zhang J., Ng E.S., Elliott D.A., Elefanty A.G., Kamp T.J. Differentiation of human embryonic stem cells and induced pluripotent stem cells to cardiomyocytes: a methods overview. Circ. Res. 2012;111:344–358. doi: 10.1161/CIRCRESAHA.110.227512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ringsrud K.M. Cells in the urine sediment. Lab. Med. 2001;32:153–155. [Google Scholar]
- 34.Loh Y.H., Agarwal S., Park I.H., Urbach A., Huo H., Heffner G.C., Kim K., Miller J.D., Ng K., Daley G.Q. Generation of induced pluripotent stem cells from human blood. Blood. 2009;113:5476–5479. doi: 10.1182/blood-2009-02-204800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim Y., Rim Y.A., Yi H., Park N., Park S.H., Ju J.H. The generation of human induced pluripotent stem cells from blood cells: an efficient protocol using serial plating of reprogrammed cells by centrifugation. Stem Cells Int. 2016;2016:1329459. doi: 10.1155/2016/1329459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim K., Doi A., Wen B., Ng K., Zhao R., Cahan P., Kim J., Aryee M.J., Ji H., Ehrlich L.I. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim Y.G., Baltabekova A.Z., Zhiyenbay E.E., Aksambayeva A.S., Shagyrova Z.S., Khannanov R., Ramanculov E.M., Shustov A.V. Recombinant Vaccinia virus-coded interferon inhibitor B18R: Expression, refolding and a use in a mammalian expression system with a RNA-vector. PLoS ONE. 2017;12:e0189308. doi: 10.1371/journal.pone.0189308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foster K.W., Ren S., Louro I.D., Lobo-Ruppert S.M., McKie-Bell P., Grizzle W., Hayes M.R., Broker T.R., Chow L.T., Ruppert J.M. Oncogene expression cloning by retroviral transduction of adenovirus E1A-immortalized rat kidney RK3E cells: transformation of a host with epithelial features by c-MYC and the zinc finger protein GKLF. Cell Growth Differ. 1999;10:423–434. [PubMed] [Google Scholar]
- 39.Foster K.W., Frost A.R., McKie-Bell P., Lin C.Y., Engler J.A., Grizzle W.E., Ruppert J.M. Increase of GKLF messenger RNA and protein expression during progression of breast cancer. Cancer Res. 2000;60:6488–6495. [PubMed] [Google Scholar]
- 40.Yu F., Li J., Chen H., Fu J., Ray S., Huang S., Zheng H., Ai W. Kruppel-like factor 4 (KLF4) is required for maintenance of breast cancer stem cells and for cell migration and invasion. Oncogene. 2011;30:2161–2172. doi: 10.1038/onc.2010.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Später D., Hansson E.M., Zangi L., Chien K.R. How to make a cardiomyocyte. Development. 2014;141:4418–4431. doi: 10.1242/dev.091538. [DOI] [PubMed] [Google Scholar]
- 42.Cyganek L., Tiburcy M., Sekeres K., Gerstenberg K., Bohnenberger H., Lenz C., Henze S., Stauske M., Salinas G., Zimmermann W.H. Deep phenotyping of human induced pluripotent stem cell-derived atrial and ventricular cardiomyocytes. JCI Insight. 2018;3:99941. doi: 10.1172/jci.insight.99941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schweizer P.A., Darche F.F., Ullrich N.D., Geschwill P., Greber B., Rivinius R., Seyler C., Müller-Decker K., Draguhn A., Utikal J. Subtype-specific differentiation of cardiac pacemaker cell clusters from human induced pluripotent stem cells. Stem Cell Res. Ther. 2017;8:229. doi: 10.1186/s13287-017-0681-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Besser R.R., Ishahak M., Mayo V., Carbonero D., Claure I., Agarwal A. Engineered microenvironments for maturation of stem cell derived cardiac myocytes. Theranostics. 2018;8:124–140. doi: 10.7150/thno.19441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Di Baldassarre A., Cimetta E., Bollini S., Gaggi G., Ghinassi B. Human-induced pluripotent stem cell technology and cardiomyocyte generation: progress and clinical applications. Cells. 2018;7:E48. doi: 10.3390/cells7060048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dubois N.C., Craft A.M., Sharma P., Elliott D.A., Stanley E.G., Elefanty A.G., Gramolini A., Keller G. SIRPA is a specific cell-surface marker for isolating cardiomyocytes derived from human pluripotent stem cells. Nat. Biotechnol. 2011;29:1011–1018. doi: 10.1038/nbt.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tohyama S., Hattori F., Sano M., Hishiki T., Nagahata Y., Matsuura T., Hashimoto H., Suzuki T., Yamashita H., Satoh Y. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell. 2013;12:127–137. doi: 10.1016/j.stem.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 48.Lopaschuk G.D., Jaswal J.S. Energy metabolic phenotype of the cardiomyocyte during development, differentiation, and postnatal maturation. J. Cardiovasc. Pharmacol. 2010;56:130–140. doi: 10.1097/FJC.0b013e3181e74a14. [DOI] [PubMed] [Google Scholar]
- 49.Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T.L. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kibbe W.A. OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res. 2007;35:W43–W46. doi: 10.1093/nar/gkm234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huebsch N., Loskill P., Mandegar M.A., Marks N.C., Sheehan A.S., Ma Z., Mathur A., Nguyen T.N., Yoo J.C., Judge L.M. Automated video-based analysis of contractility and calcium flux in human-induced pluripotent stem cell-derived cardiomyocytes cultured over different spatial scales. Tissue Eng. Part C Methods. 2015;21:467–479. doi: 10.1089/ten.tec.2014.0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Recordings of 30 s (7 pictures/s) showing wide ranges of motion and directional synchronous contractions. Direction is indicated by arrows using Matlab application Motion GUI.
Fluorescent calcium indicators were used to visualize the intracellular calcium flux during contraction. Recordings of 30 s (7 pictures/s) showing Ca2+ transients in the cardiomyocyte culture.