Abstract
Alkaline phosphatase (AP) enables marine phytoplankton to utilize dissolved organic phosphorus (DOP) when dissolved inorganic phosphate (DIP) is depleted in the ocean. Dinoflagellate AP (Dino-AP) represents a newly classified atypical type of AP, PhoAaty. Despite While being a conventional AP, PhoAEC is known to recruit Zn2+ and Mg2+ in the active center, and the cofactors required by PhoAaty have been contended and remain unclear. In this study, we investigated the metal ion requirement of AP in five dinoflagellate species. After AP activity was eliminated by using EDTA to chelate metal ions, the enzymatic activity could be recovered by the supplementation of Ca2+, Mg2+ and Mn2+ in all cases but not by that of Zn2+. Furthermore, the same analysis conducted on the purified recombinant ACAAP (AP of Amphidinium carterae) verified that the enzyme could be activated by Ca2+, Mg2+, and Mn2+ but not Zn2+. We further developed an antiserum against ACAAP, and a western blot analysis using this antibody showed a remarkable up-regulation of ACAAP under a phosphate limitation, consistent with elevated AP activity. The unconventional metal cofactor requirement of Dino-AP may be an adaptation to trace metal limitations in the ocean, which warrants further research to understand the niche differentiation between dinoflagellates and other phytoplankton that use Zn–Mg AP in utilizing DOP.
Keywords: dinoflagellate, alkaline phosphatase, metal cofactor, inducible expression
1. Introduction
In marine ecosystems, the availability of P is critical to the growth of phytoplankton. Thus, P is often found to be the limiting nutrient of the primary productivity in the ocean [1,2]. In the ocean, P is present in both inorganic and organic forms, among which dissolved inorganic phosphorus (DIP) as the preferred form for phytoplankton is quickly consumed and found at a low concentration in the euphotic zone [3]. In contrast, dissolved organic phosphorus (DOP) is relatively abundant in the euphotic zone. DOP mainly occurs in two forms: The major form phosphoesters and the less abundant form phosphonates [4,5]. When facing the DIP limited condition, phytoplankton are able to utilize a broad spectrum of phosphoesters to meet their cellular P requirement and sustain their primary production [6,7].
Enzymatic hydrolysis by alkaline phosphatase (AP) is a well-known mechanism employed by marine phytoplankton to utilize various types of phosphoesters which commonly features a phosphoester (O–P) bond. AP can release Pi from phosphoesters at the characteristic alkaline pH in the ocean [8]. Therefore, the presence and measurement of the bulk AP activity of phytoplankton has been used to indicate the P limitation in the ocean [2,7]. To gain further insights into the DOP utilization conveyed by AP in individual taxa, many studies have been conducted to examine the expression of AP coding genes and to characterize the enzyme properties in both marine bacteria and eukaryotic phytoplankton [9,10,11,12]. Experimental and genomic studies have revealed multiple types of APs which show not only substrate preferences but also different required metal ions as cofactors [13,14,15,16,17].
The conventional AP (PhoAEC) initially identified in Escherichia coli was the best studied and defined as a homodimeric phosphomonoesterase. It mainly hydrolyzes phosphomonoesters with low activity on the phosphate diester and requires 2Zn–Mg as bimetallo activity cores in the protein structure [18,19]. PhoAEC has been documented in marine bacteria, cyanobacteria and diatoms [12,13,20]. The second type of AP with a determined protein folding structure is PhoX, which possesses a unique 3Ca–2Fe core [17] and exhibits a low substrate specificity for DOPs with an O–P bond [21,22]. Besides marine prokaryotes, PhoX homologs have also been identified in green algae, Volvox carteri and Chlamydomonas reinhardtii [23,24,25]. The third, rather common, type of AP in marine prokaryotes is PhoD, which has also been confirmed to be Ca2+-dependent and able to hydrolyze both phosphomonoesters and phosphodiesters [13,26].
In addition, there are several uncategorized types of APs in marine phytoplankton. A Ca2+-dependent putative AP was identified from the pelagophyte Aureoumbra lagunensis through a proteomic analysis [27]. A novel kind of AP, EHAP1, has also been identified in haptophyte Emiliania huxleyi [28]. Besides possessing an up-regulated PhoD under a P limitation [29], the diatom model Phaeodactylum tricornutum has been reported to own another type of AP with some sequence similarity to PhoAEC, but its enzyme activity is enhanced by Mn2+, Mg2+ and Ca2+ instead of Zn2+ [12,30]. Because of their high sequence variability, the structural and functional classification of these APs remains challenging. Though PhoAEC has been documented in marine bacteria, it is less frequently reported than the other two types in marine microbes. Taken together, the multiple types of APs and the prevalence of Ca2+-dependent APs are believed to be an adaptive mechanism which gives different marine microorganisms their niche advantages to utilize available types of phosphoesters under low metal ion availability conditions in the ocean [22].
Dinoflagellates are one of the most important groups of phytoplankton in the ocean and are well known to be capable of utilizing DOP with the aid of an AP under DIP-depleted conditions. Reports showed that the subcellular localization of the Dinoflagellate-AP (Dino-AP) can be both cell surface and intracellular [11,31]. Based on the genomic and transcriptomic data of the several dinoflagellate species currently available, only one type of AP has been identified in dinoflagellates, and we classified it as the PhoAaty type due to a generally high sequence divergence but recognizable conserved domains similar to an atypical PhoA type in cyanobacteria [32]. In these PhoAaty APs, we found conserved aspartic acid (D), glutamic acid (E) and a “proline-aspartic” motif amongst those conserved domains, a motif known to be responsible for Ca2+-binding, thus suggesting that PhoAaty might also be a Ca2+-dependent type of AP [32]. However, this hypothesis has not yet been examined experimentally.
In this study, we investigated the metal ion requirement of APs in five dinoflagellate species (Amphidinium carterae, Alexandrium pacificum, Karenia mikimotoi, Prorocentrum minimum and Fugacium kawagutii) using an EDTA chelating and metal ion resupply approach. To better demonstrate the AP dependency on the metal ion, we overexpressed the AP gene of A. carterae (ACAAP) in E. coli, and enzymatic assays conducted on purified recombinant ACAAP (rACAAP) verified that this AP could be activated by Ca2+, Mg2+, and Mn2+ but not Zn2+. We further developed a polyclonal antiserum against ACAAP, and a western blot analysis showed the remarkable up-regulation of ACAAP expression under a phosphate limitation and a positive correlation between ACAAP and AP activity, confirming the active function and translational regulation of this gene.
2. Materials and Methods
2.1. Algal Strains, Culture Conditions and Sample Collection
Five dinoflagellate strains were used in this study. A. carterae and F. kawagutii (formerly Symbiodinium kawagutii) [33] were provided by the Provasoli-Guillard National Center for Marine Algae and Microbiota (NCMA); A. pacificum (formerly A. catenella) and P. minimum were provided by Center for Collections of Marine Bacteria and Phytoplankton, Xiamen University (CCMBP); and K. mikimotoi was provided by Jinan University. These strains were maintained in sterilized oceanic seawater (filtered through 0.22 µm pore size filters, salinity 30 psu) enriched with the full nutrient regime of the L1 medium with no Si [34] in an incubator with temperature controlled at 20 °C (most examined strains) or 25 °C (F. kawagutii) and illumination under a light dark cycle L:D = 12:12 (most examined strains) or 14:10 (F. kawagutii) with a photon flux of 100 µE m−2 s−1.
DIP-depleted and replete cultures (under the same condition as described above) were prepared in triplicates. The DIP-depleted cultures were grown under the same nutrient regime as DIP-replete cultures, except for a decreased phosphate concentration (2 µM). The cell density in the culture was determined daily by the counting of the cells using a Sedgewick–Rafter counting chamber (Phycotech, St. Joseph, MI, USA). The DIP concentration in the culture media was measured using the Molybdate Blue method [35]. Cells from each culture (200 mL) were harvested by filtration onto 3 μm pore size polycarbonate membranes (Millipore, Bedford, MA, USA) and washed off the membrane using an AP buffer (20 mM Tris-HCl, pH 8.0, 25 mM NaCl). Harvested cells were homogenized by bead beating three times separately, using 0.5 mm ceramic beads with the setting of 6 m s−1 for 30 s on an MP FastPrep-24 (MP Biomedicals, CA, USA). After centrifugation at 13,000 g at 4 °C for 10 min, the supernatant that contained extracted crude protein was subjected to enzyme activity measurements.
2.2. Alkaline Phosphatase Activity Measurement on Algal Cells and Protein Extract
The bulk AP activity of cultures was measured by adding 50 μL of 20 mM p-nitro-phenylphosphate (pNPP from Fluka, St. Louis, MO, USA), prepared in 1 M Tris-HCl (pH 8.0) into a 1 mL culture sample [36]. The reaction was carried out in sterile microcentrifuge tubes and incubated in darkness for 2 h at 25 °C. After centrifugation at 10,000 g for 2 min, the supernatant was removed and used for an OD measurement at 405 nm on a NanoDrop ND-2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA).
An in-gel AP activity assay was performed as described previously [27] with minor modifications. The extracted protein of 106 A. carterae cells of both the P-depleted and the P-replete groups were incubated with a non-reducing Laemmli sample buffer at room temperature for 5 min and loaded into (4%–10%) SDS-PAGE gel, with an equal amount of protein in each well (25 μg) and in triplicate for the P-depleted group. After electrophoresis, each gel lane was excised, soaked in an AP buffer (20 mM Tris-HCl, pH 8.0, 25 mM NaCl) containing one of the three different metal ions (10 mM Ca2+, 10 mM Mg2+, 0.1 mM Zn2+), and incubated with ELF97 (1:50 dilution) (ELFTM 97 endogenous phosphatase detection kit, Molecular Probes, OR, USA) for 2 h in the darkness at room temperature. For the P-replete group, each gel slice was subjected to the incubation with all three metal ions included to detect possible AP activity in one (instead of multiple) assay. Fluorescent gel images were acquired on the Bio-Rad Gel Doc XR system using an EtBr filter at 302 nm (Bio-Rad, CA, USA).
2.3. Heterologous Expression of Recombinant ACAAP and Production of Antibody
Based on the AP coding gene (acaap, GenBank: HQ259111.2) identified in A. carterae [36], two different kinds of amplicons were prepared and subjected to heterologous expression. A hydrophilic peptide (pACAAP) comprising 180 amino acid residues (nucleotide site 661–1200) was used to generate a polyclonal antibody, and a recombinant ACAAP (rACAAP) comprising 684 amino acid residues (full ORF region with the exclusion of a signal peptide coding region (nucleotide site 1–60)) was used for an enzyme activity analysis. Both gene fragments were amplified using specific primers (Table 1), and then gel-purified PCR products were cloned into vector pEasy-E1 (TransGen Biotech, Beijing, China) and transformed into E. coli BL21 (DE3). For each amplicon, a single colony containing the gene fragment was grown separately in a 3 mL LB (Luria-Bertani) liquid medium containing 100 µg mL−1 ampicillin on a shaker rotating at 200 rpm at 37 °C for 16 h; each colony was then transferred to 1 L fresh LB with 0.5 mM IPTG (isopropyl-β-D-thiogalactopyranoside) to induce the expression of the inserted gene fragment, grown for 3 h at 37 °C for pACAAP and 10 h at 28 °C for rACAAP, both on a shaker rotating at 200 rpm.
Table 1.
Primers used in this study.
| Applications | Primer Name | Direction | Primer sequence (5′-3′) | PCR Condition |
|---|---|---|---|---|
| pACAAP | PF1 | F | TTGAGCACATACACCGACGA | 94 °C 3min, 35 cycles (94 °C, 30 s, 56 °C, 45s, 72 °C, 1 min), 72 °C, 10 min |
| PR1 | R | AGATGCATTCAGATACATGATG | ||
| rACAAP | PF2 | F | AAGGGACGTAGGCTTGCTAG | |
| PR2 | R | CGCACGCACGGTCAAGAAG |
E. coli cells were harvested by centrifugation at 5000 g, 4 °C for 10 min, resuspended in a 1 mL Tris-HCl buffer (50 mM Tris-HCl, pH 8.0), and then homogenized by ultrasonic treatment. After centrifugation at 15,000 g, 4 °C for 10min, the supernatant containing the heterologously-expressed peptide was purified using an Ni-NTA spin kit column (TransGen Biotech, Beijing, China) following the manufacturer’s protocol. The resultant overexpressed peptide was loaded into a Superdex 75 gel filtration column on an AKTA prime liquid chromatography system (GE Healthcare Bio-Sciences, Uppsala, Sweden) and eluted using a phosphate-buffered saline buffer (PBS buffer, 50 mM NaH2PO4, 150 mM NaCl, pH 8.0) as the mobile phase. All collected fractions were examined by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and fractions containing the target peptide were concentrated using Amicon Ultra centrifugal filter devices (Merck Millipore Ltd., Carrigtwohill, IRL). Purified rACAAP was subjected to further enzymatic characterization, and purified pACAAP was used to immunize a rabbit (Japanese white) to raise a polyclonal antiserum (Proteintech Group Inc., which has the laboratory animal license SYXK2014-0076 issued by Hubei Science Technology Department, Wuhan, China).
2.4. Western Blot Analysis
Crude protein, extracted as described above, was mixed with a reducing Laemmli buffer and incubated at 95 °C for 5 min. The denatured proteins of each sample were loaded into (4%–10%) SDS-PAGE gel (Bio-Rad, CA, USA) at equal amounts in each well. Electrophoresis was carried out at 90 V for 30 min, followed by 120 V for 60 min. The SDS-PAGE gel was then blotted onto a polyvinylidene fluoride (PVDF) membrane (Millipore, Bedford, MA, USA) at 25 V for 30 min using the Trans-Blot SD semi-dry transfer cell (Bio-Rad, CA, USA). The membranes were incubated in a blocking buffer containing 5% non-fat milk for 2 h at room temperature, followed by incubation with the polyclonal antiserum (1:5000 dilution) in TBS (Tris buffered saline) for 1 h at room temperature. After four washes with TBS containing 0.1% Tween 20 (TBST), the membranes were incubated with a biotinylated goat anti-rabbit IgG (TransGen Biotech, Beijing, China) in a 1:10,000 dilution for 1 h at room temperature and then were washed four times in TBST. Meanwhile, the reference protein GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was also examined using the monoclonal antibody (1:5000 dilution, Sangon Biotech, Shanghai, China) in parallel following the same procedure. The immunodetected bands were visualized using the enhanced chemiluminescent (ECL) substrate (Bio-Rad, CA, USA), and chemiluminescene images were acquired on a Bio-Rad Gel Doc XR system (Bio-Rad, CA, USA). The relative quantification of ACAAP expression was calculated in two ways, normalized to the ACAAP expression of P-replete on day 1 and normalized to reference protein GAPDH based on the band intensity estimated by Quantity One software (Bio-Rad, Hercules, CA, USA).
2.5. Metal Ion Dependency Analysis
The metal ion requirement of APs was investigated in five dinoflagellate species and the purified rACAAP. The five dinoflagellate species were grown under the DIP-depleted condition as described above, and cells were collected at the stationary phase. For each enzymatic reaction, an equal amount of proteins (equal to 104~105 cells dependent on different species) were used. Pre-incubation with EDTA was employed to chelate the cellular metal ions present in the protein. The reaction mixture contained 6 μg protein, 5 μL 100 mM EDTA, and 75 μL AP buffer, and it was incubated at 25 °C in the darkness for 30 min in a 96-well plate. Afterwards, 5 μL 20 mM pNPP was applied to the reaction mix along with 10 μL of one of each of the following: 100 mM Ca2+, 100 mM Mg2+, 100 mM Mn2+, 100 mM Co2+, 100 mM Fe3+, 10 mM Zn2+, and 100 mM EDTA (all prepared in 20 mM Tris-HCl pH 8.0), triplicate wells for each treatment were done separately [13]. After another 2 h incubation, AP activity (APA) was measured on a SpectraMax Paradigm (Molecular Devices, CA, USA), and the spectrum reading of the control treatment (Ctrl) was taken as a base value to normalize the readings of other treatments to calculate the relative fold change of the enzyme activity as follows: (APA–APACtrl)/APACtrl. Meanwhile, a control treatment was maintained in the preserved condition as pre-incubation, and 5 μL (30 U μL−1) commercialized CIAP (calf intestinal alkaline phosphatase, Takara Bio Inc., Kusatsu, Shiga, Japan) were subjected to the same procedure as a positive reference.
3. Results
3.1. Metal Ion Requirement of Dino-AP
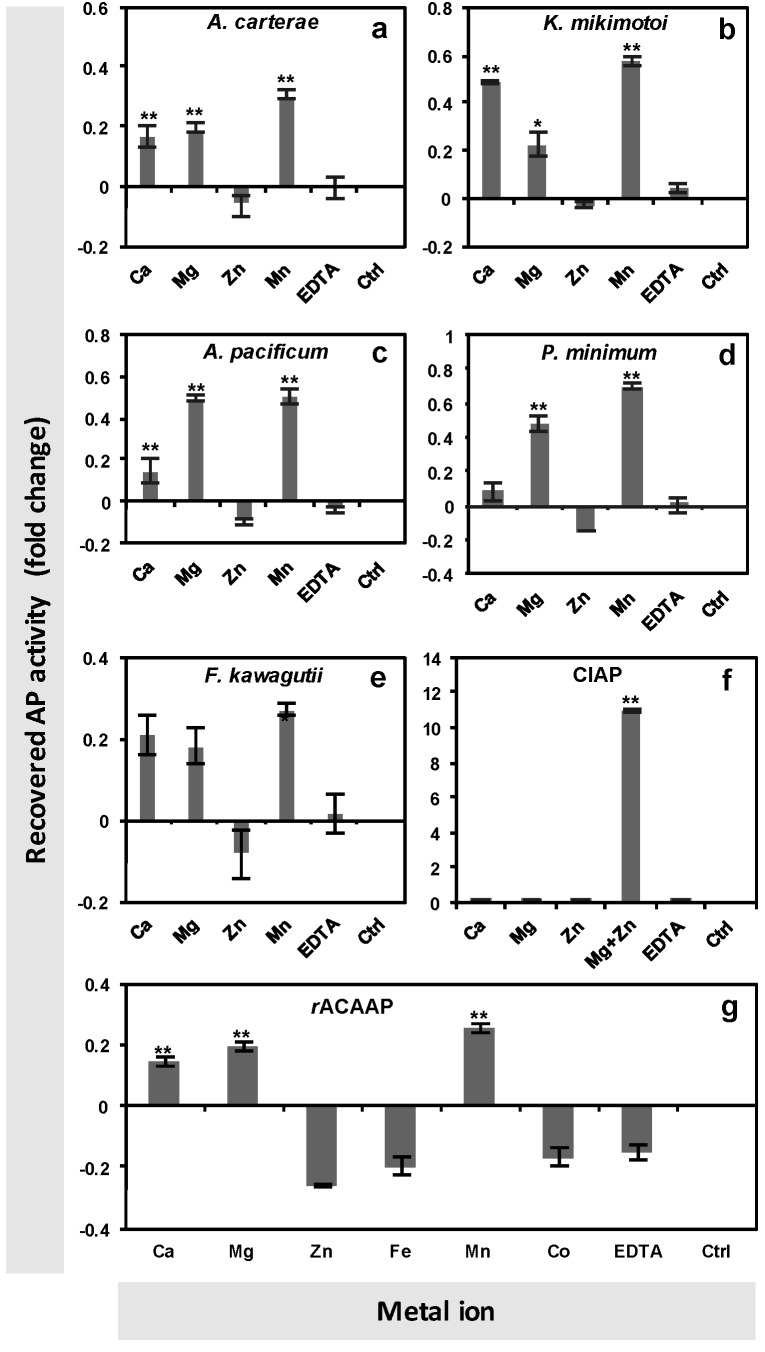
The metal ion requirement analysis was performed on crude protein extracts of A. carterae, K. mikimotoi, A. pacificum, P. minimum and F. kawagutii collected under the P-depleted condition. After pre-incubation with EDTA, AP activity disappeared (Figure 1). The AP activity was recovered significantly with the supplement of Ca2+, Mg2+ and Mn2+ in A. carterae, K. mikimotoi and A. pacificum (Tukey HSD, p < 0.01, Figure 1a–c). In P. minimum, the AP activity was recovered significantly with the supplement of Mg2+ and Mn2+, while in F. kawagutii, the AP activity was recovered by Mn2+ but not significantly by Ca2+ and Mg2+ (Figure 1d,e). To exclude the possibility that the apparent lack of AP activation by Zn2+ was due to a toxic effect of overdose, the commercialized CIAP was used as a control in the assay. In sharp contrast to dinoflagellate APs, the enzymatic activity of the CIAP was significantly recovered by the supply of Mg2+ and Zn2+ (Tukey HSD, p < 0.01,) (Figure 1f), as documented of the conventional AP.
Figure 1.
Alkaline phosphatase activity (APA) recovery of crude proteins of different dinoflagellate strains ((a) A. carterae, (b) K. mikimotoi, (c) A. pacificum, (d) P. minimum, (e) F. kawagutii), the commercialized CIAP (f) and the recombinant AP of A. carterae (rACAAP) (g) with the resupply of different metal ions after EDTA treatment to remove metals and eliminate APA. “Ctrl” represents the EDTA-pre-incubated reaction, which served as a blank control here. “EDTA” represents an experimental control that was resupplied with the same molar amount of EDTA as metal ions in experimental reactions. Asterisks depict the significant recovery of APA by metal ion resupply compared to the EDTA control (Tukey HSD, ** represents p < 0.01, * represents p < 0.05).
The same assay was tested on the purified rACAAP (heterologously expressed ACAAP in E. coli) incubated with the six metal ions separately following the same method as for algal cellular protein extracts. The result was consistent with the observation of crude protein extracts of A. carterae, and the AP activity of rACAAP was recovered significantly with the supplement of Ca2+, Mg2+, and Mn2+ (Tukey HSD, p < 0.01), but not Zn2+, Fe3+ or Co2+ (Figure 1g). Meanwhile, the in-gel AP activity assay conducted on the crude protein extract of A. carterae revealed a unique positive protein band only in the P-depleted group with appended Ca2+, but there was negative in the P-depleted group with Mg2+ and Zn2+ and in the P-replete group (Figure 2).
Figure 2.
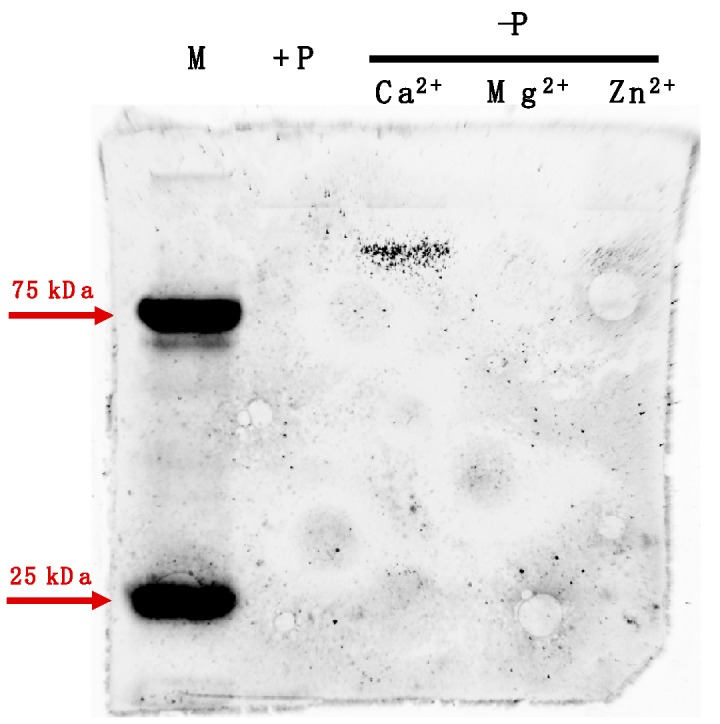
In-gel assay of metal ion specificity required by AP of A. carterae. “M” represents protein marker, “+P” represents extracted protein of the P-replete group, “−P” represents extracted protein of the P-depleted group incubated with specific metal ion respectively.
3.2. Expression Pattern of AP in A. carterae (ACAAP) under Varied P Conditions
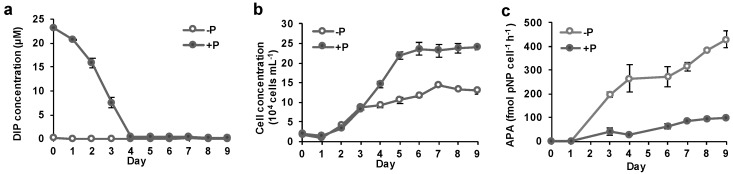
A. carterae was grown in normal phosphate (36 μM) and low phosphate (2 μM ) media. DIP concentration remained scarce (below the detection limit 0.2 μM) in the P-depleted group throughout the experiment. However, in the P-replete group, it was initially high (25 μM) and then decreased quickly, reaching as low as that in the P-depleted group after day four (Figure 3a). Accordingly, markedly different growth curves were observed between two groups of cultures (Figure 3b). During the first three days, the cell densities in both groups increased steadily comparably, from ~1.8 × 104 to ~ 8.4 × 104 cells mL−1. The cells in the DIP-replete group continued the exponential growth until day five and then entered the stationary growth phase in a stable cell density. At the end of this experiment, the cell density of the DIP-replete culture was 2.4 × 105 cells mL−1, a 12-fold increase compared to the starting cell density. In the DIP-depleted group, the cultures grew more slowly after day three, and the cell density was 1.3 × 105 cells mL−1 at the end of this experiment—only about half of that in the DIP-replete group.
Figure 3.
Dissolved inorganic phosphate (DIP) concentration (a), growth curve (b) and AP activity (c) in time course of A. carterae cultures under different phosphate conditions. +P, normal phosphate concentration in an L1 medium; −P, decreased phosphate concentration (2 μM) in an otherwise L1 medium.
By comparison, AP activity started to diverge between the two groups earlier than did the cell density (Figure 3c). At the beginning of the experiment, AP activity was barely detectable in both groups. On day three, when the cell density was about the same between the two groups, AP activity in the P-depleted group (196 fmol pNP cell−1 h−1) increased dramatically to five-fold of that in the P-replete group (37 fmol pNP cell−1 h−1). Since then, the AP activity in the P-depleted group continued to increase quickly, while that in the DIP-replete group only increased slightly despite the significant drop of DIP concentration observed after day three (Figure 3a). At the end of this experiment, the AP activity in the DIP-depleted group (430 fmol pNP cell−1 h−1) was about four-fold of that in the DIP-replete group (99 fmol pNP cell−1 h−1).
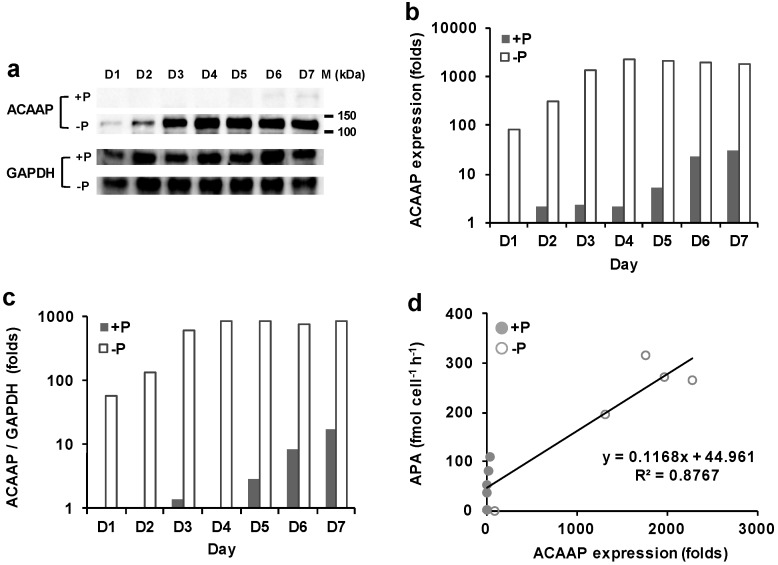
The extracted total proteins of A. carterae from both groups were subjected to SDS-PAGE and a western blot analysis. The result showed that the polyclonal antiserum raised against the pACAAP specifically detected a unique band, which lied between 100 and 150 kDa (Figure 4a), apparently larger than computationally estimated 75 kDa of ACAAP.
Figure 4.
AP expression in A. carterae cultures grown under DIP-replete (+P) and depleted (−P) conditions. (a) ACAAP and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) western blot. (b) Relatively quantified ACAAP expression. (c) ACAAP expression normalized to GAPDH. (d) Pearson correlation between ACAAP expression and APA (p < 0.05).
The immunodetected protein exhibited a significant difference in abundance between P-replete and P-depleted groups. A clear band was detected on day two and became thicker in the course of the experiment, indicating the inducible expression of ACCAP (Figure 4a). A remarkable up-regulation in the expression of ACCAP was observed in the DIP-depleted group compared to that in the DIP-replete group, regardless which normalization method used to evaluate the fold change of expression level (T-test, p < 0.05) (Figure 4b,c). The expression of ACAAP in the P-depleted group reached the peak on day four and maintained at a stable level until day seven, which was about 80% of that on day four, 21-fold of that on day one, and 57-fold of that in the P-replete group on day seven (Figure 4b). In contrast, the expression of ACAAP in the DIP-replete group generally maintained at low levels (shown as faint detected bands in gel, Figure 4a) throughout the experiment, despite the slight increase after the cultures entered the stationary phase (Figure 4b,c). This result was consistent with the trend observed in AP activity (Figure 1c). Overall the protein expression of ACAAP was positively correlated with enzyme activity (R2 = 0.88, n = 10, Figure 4d).
4. Discussion
4.1. Inducible Expression of Dinoflagellate APs Through Transcriptional and Translational Regulation
AP activities have been shown to be induced by P limitations in various dinoflagellate species in previous work [36,37,38], with which the results of the current study are consistent. Previously, the induction has been attributed to the transcriptional up-regulation of the AP gene under P limitations in dinoflagellate A. carterae, K. brevis and P. donghaiense [36,37,39]. In this study, using the antiserum developed against ACAAP in a western blot analysis, we observed the marked stimulation of AP protein expression in the DIP-depleted A. carterae, which was positively correlated with the AP activity increase. However, the magnitude of the increase in ACAAP abundance was much higher than in its activity. All these results suggest that the regulation of ACAAP expression and enzymatic activity in response to phosphate limitation lies at both the transcriptional level and the translational level, which is in agreement with the expression observed in EH-PhoAaty of E. huxleyi [40]. Furthermore, the increase of both AP activity and AP expression appeared earlier than growth arrest in the P-depleted culture of A. carterae, and we noted a slight increase of both in the stationary growth phase of the P-replete group when there was a sharp drop in DIP concentration. Our previous study showed that AP of A. carterae was localized at both cell surface and intracellular compartment [11]. Taken together, these observations suggest that A. carterae might employ AP to utilize the intracellular DOP storage to maintain population growth when both DIP and DOP were deficient in the medium. Therefore, the expression of AP in A. carterae, and likely in other dinoflagellates as well, can be triggered by a low ambient Pi concentration, but it is really regulated by the internal P availability, similar to the case of EH-PhoAaty [40].
4.2. Evidence of Dinoflagellate AP Protein Modification from Observation on ACAAP
The generation of ACAAP antiserum has enabled us to estimate the molecular size, besides abundance, of a dinoflagellate AP based on a western blot analysis. We noticed that there was a discrepancy in molecular mass between the ACAAP detected in SDS-PAGE (~130 kDa) and the in silico prediction based on the amino acid sequence of ACAAP (~75 kDa). A striking coincidence occurred in PhoAaty of E. huxleyi (EH-PhoAaty), in which while the amino acid sequence gave a predicted molecular mass of 75 kDa, the detected band in the western blot lied between 100 and 120 kDa, ~110kDa [40]. Our subsequent mass spectroscopic analysis showed that the ~110 kDa band was truly an EH- PhoAaty protein. One possibility is that the mature PhoAaty type AP, of which both ACAAP and EH-PhoAaty are, may function in a dimer-hinge conformation like the typical AP, PhoAEC [19]. Though protein complexes are usually broken in the protein denaturing buffer and SDS-PAGE, it cannot be ruled out that this AP holozyme may stay intact under these conditions. As documented, some stable dimers can remain un-dissociated in SDS-PAGE gel and able to perform enzyme activity [41,42]. Another possibility is that the post-translational modifications (PTMs) may occur and alter the molecular mass of the AP monomer or dimer, as has been reported previously for other proteins [43,44]. PTMs have been reported in algal APs. APs in the diatom P. tricornutum and the coccolithophorid E. huxleyi undergo proteolytic modification while released from the cell surface [12,28]. Our previous study proposed that the PhoAaty-type AP represents a phylogenetically distinct type of AP, sharing little sequence similarity to any other classified type of AP, and, as such, their substrate preferences and protein structure still remain undetermined [32]. PTMs are of great importance to the biological function of proteins, including the enzyme activity state, subcellular localization, turnover and interactions with other proteins [43]. Thus, further efforts are required to investigate possible PTMs of PhoAaty and their effects on enzymatic function.
4.3. Metal Ion Requirement of Dino-AP
Metal ions are indispensable for many enzymes to form and stabilize the conformational structure of folded proteins, providing the specific function in substrates binding and hydrolysis [45,46]. Besides transition metals (Fe, Cu and Mn) previously found to be required particularly in biochemical redox process, the metalloproteins containing Mg, Ca and Zn have also been extensively studied [45]. For instance, the archetypal PhoAEC has been well characterized and shown to require 2Zn–Mg in each subunit to coordinate the active center, while another major characterized type of AP, PhoX, owns a complex active-site comprising 2Fe–3Ca [17,19,46]. To implement the catalytic function, bimetallo cores require the transition metal Fe3+/Zn2+ to interact with the oxygen atoms of the O–P bond of the substrate molecule, the same as Ca2+ in PhoX [19,46]. Even though no available active site and structural model could be applied to PhoAaty, we have identified conserved motifs in PhoAaty, including residues of aspartic acid (D) and glutamic acid (E), that feature a high affinity to Ca2+ [47] and a highly conserved Ca2+-binding “proline-aspartic acid (PD)” motif shared by PhoX and phosphotriesterase [17,21,32,48].
Consistent with the prediction, experiments in this study showed that the AP activity eliminated by metal iron depletion could be restored by the resupply of Ca2+, Mn2+ or Mg2+ in all five examined dinoflagellate strains but not by that of Zn2+. It is worth noting that E. huxleyi was able to express two different types of AP (EH-PhoAaty and EHAP1) at the same time, and the AP activity of cell lysate (excluding most EHAP1 because it is secretory) could be restored more by Ca2+ than Zn2+ [40]. This result is comparable to the previous study carried out in the AP of other algae and PhoX of bacteria. Besides Ca2+ and Fe3+, the activity of PhoX can also be recovered with the presence of Mn2+ (shown in supplementary data in [17]) and Mg2+ [48]. A cell-surface AP isolated from P. minimum also exhibited the activity with the presence of both Ca2+ and Mg2+ [49,50], and the AP in dinoflagellate Pyrocystis noctiluca has previously been described as Mg2+-dependent [51]. A similar cofactor requirement was also observed in the diatom P. tricornutum, in which PhoA activity could be enhanced by Mn2+, Mg2+ and Ca2+, and the AP of the brown tide alga A. lagunensis was found to be Ca2+-dependent [12,27]. This compatibility may be due to the similar chemical characteristics shared by Ca2+ and Mg2+, as the Mg2+ binding residues also show preference for “D” as well as the Ca2+ binding site [52]. Nevertheless, there was a difference between the in-gel assay and the algal cell assay, in this study in that the resupply of Mg2+ and Mn2+ failed to recover AP activity (data not shown) in the in-gel assay, which can be explained as SDS-PAGE may disable protein conformational changes needed to accommodate these bivalent cations [53].
The prevalence of the Ca2+-dependent AP reported in marine microorganisms has been attributed to the relative low availability of Zn2+ in the ocean and the high abundance of Ca2+ and Mg2+ in the ocean [1,14,16,22,27]. Therefore, the adoption of Ca2+ in an AP may be an adaptive strategy that has evolved in the marine microorganisms in the Zn–P co-limited environment. However, the identification of the Ca2+/Fe3+ active site of PhoX poses a possible Fe-deficiency based constrain of AP activity and a nutrient limitation-driven environmental selection of microorganism in the ocean [17,54]. We also noticed that the activity of the Dino-AP could also be recovered by the addition of Mn2+, one common transition metal found in metalloproteins, which, like Ca2+, is more abundant in the sea than Zn2+ [1].
In the PhoAEC family, the structural flexibility of the bimetallic site has been examined to interpret the promiscuous catalytic activity [46]. Structural comparisons of AP superfamily members showed distinct structure features outside of the conserved bimetallo site, which may not only impact specific substrate binding but may also modulate the properties of the bimetallo core to recognize substrates [19]. However, it is still unclear how Ca2+ and Mn2+ function in the active center of Dino-AP. Our results showed negative effect of Zn2+ on AP activity recovery in dinoflagellate, we also found resupply of Zn2++Mg2+, like resupply of Zn2+ alone, did not recover AP (data not shown). However, without a further high-resolution structural observation of the AP proteins with metal ions inside, caution is required to interpret it as the dispensability or even antagonism of Zn2+ for dinoflagellate APs. Though we do not have crystallographic data to directly show the metal ion conformation of the Dino-AP protein, the possibility of an experimental artifact—for instance the addition of Zn2+, albeit only 1% dosage of other metal ions, caused toxic effects to the Dino-AP in our assays—is unequivocally excluded by the result of exactly the same assay for the calf intestinal alkaline phosphatase (CIAP), which exhibited total recovery of AP activity by the resupply of Zn2++Mg2+, as expected of a conventional AP.
5. Conclusions
Dinoflagellate AP belongs to a newly classified type of AP, PhoAaty, and its enzyme expression and characterization has remained largely unexplored. This study is the first attempt to examine the metal ion cofactor requirement of the AP in core dinoflagellate species and develop an antibody to quantify AP abundance. Our results from five dinoflagellate species revealed that Dino-AP was activated by Ca2+, Mg2+ and Mn2+ but not by Zn2+. Additionally, the AP activity could be restored to different degrees by above-mentioned divalent cation among different species, in contrast to the conventional AP which requires the pair of Zn2++Mg2+ as cofactors. Furthermore, our results showed for the first time that the mature enzyme of dinoflagellate APaty might function in a dimer-hinge conformation or undergo a post-translational modification that alters the molecular mass dramatically. These results, albeit a bit surprising, are not entirely unprecedented considering the similar behavior of the PhoAaty AP (EH-PhoAaty) in the haptophyte E. huxelyi and of other proteins in other organisms. Therefore, we propose here, with caution, that PhoAaty may require unconventional metal ions such as Ca2+ as cofactors, which is likely a result of adaptive evolution under the selection pressure of limited or unpredictable trace metal availability in the ocean. Further study is required to illuminate the protein structure of PhoAaty and to determine the exact cofactor conformation in order to gain further understanding of the regulation of growth and the distribution of dinoflagellates from the perspective of P nutrient strategy in the trace metal variable marine environment.
Acknowledgments
We wish to thank Zhiyong Lin from Xiamen University for his technical assistance in protein purification using AKTA purifier. We thank Meizhen Li for her help with statistical analysis.
Author Contributions
Conceptualization, X.L. and S.L.; methodology, X.L., C.G. and L.L.; validation, C.G. and T.L.; formal analysis, C.G.; investigation, X.L. and C.G.; writing—original draft preparation, X.L., C.G. and S.L.; writing—review and editing, X.L. and S.L.; supervision, X.L. and S.L.; project administration, X.L. and S.L.; funding acquisition, X.L. and S.L.
Funding
This research was funded by the National Key Research and Development Program of China grant # 2017YFC1404302 (S.L., X.L.), the National Natural Science Foundation of China grants #41776116, #31661143029 (S.L.) and #41706116 (X.L.), Fundamental Research Funds for the Central Universities 20720180101 (X.L.).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design, execution, interpretation, or writing of this study.
References
- 1.Moore C.M., Mills M.M., Arrigo K.R., Bermanfrank I., Bopp L., Boyd P.W., Galbraith E.D., Geider R.J., Guieu C., Jaccard S.L., et al. Processes and patterns of oceanic nutrient limitation. Nat. Geosci. 2013;6:701–710. doi: 10.1038/ngeo1765. [DOI] [Google Scholar]
- 2.Karl D.M. Microbially mediated transformations of phosphorus in the sea: New views of an old cycle. Annu. Rev. Mar. Sci. 2014;6:279–337. doi: 10.1146/annurev-marine-010213-135046. [DOI] [PubMed] [Google Scholar]
- 3.Benitez-Nelson C.R. The biogeochemical cycling of phosphorus in marine systems. Earth-Sci. Rev. 2000;51:109–135. doi: 10.1016/S0012-8252(00)00018-0. [DOI] [Google Scholar]
- 4.Kolowith L.C., Ingall E.D., Benner R. Composition and cycling of marine organic phosphorus. Limnol. Oceanogr. 2001;46:309–321. doi: 10.4319/lo.2001.46.2.0309. [DOI] [Google Scholar]
- 5.Paytan A., McLaughlin K. The oceanic phosphorus cycle. Chem. Rev. 2007;107:563–576. doi: 10.1021/cr0503613. [DOI] [PubMed] [Google Scholar]
- 6.Mather R.L., Reynolds S.E., Wolff G.A., Williams R.G., Torresvaldes S., Woodward E.M.S., Landolfi A., Pan X., Sanders R., Achterberg E.P. Phosphorus cycling in the north and south atlantic ocean subtropical gyres. Nat. Geosci. 2008;1:439–443. doi: 10.1038/ngeo232. [DOI] [Google Scholar]
- 7.Lin S., Litaker R.W., Sunda W.G. Phosphorus physiological ecology and molecular mechanisms in marine phytoplankton. J. Phycol. 2016;52:10–36. doi: 10.1111/jpy.12365. [DOI] [PubMed] [Google Scholar]
- 8.Dyhrman S.T., Ammerman J.W., Mooy B.A.S.V. Microbes and the marine phosphorus cycle. Oceanography. 2007;20:110–116. doi: 10.5670/oceanog.2007.54. [DOI] [Google Scholar]
- 9.Armbrust E.V., Berges J.A., Bowler C., Green B.R., Martinez D., Putnam N.H., Zhou S., Allen A.E., Apt K.E., Bechner M., et al. The genome of the diatom Thalassiosira pseudonana: Ecology, evolution, and metabolism. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- 10.Dyhrman S.T., Jenkins B.D., Rynearson T.A., Saito M.A., Mercier M.L., Alexander H., Whitney L.P., Drzewianowski A., Bulygin V.V., Bertrand E.M., et al. The transcriptome and proteome of the diatom Thalassiosira pseudonana reveal a diverse phosphorus stress response. PLoS ONE. 2012;7:e33768. doi: 10.1371/journal.pone.0033768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin X., Zhang H., Cui Y., Lin S. High sequence variability, diverse subcellular localizations, and ecological implications of alkaline phosphatase in dinoflagellates and other eukaryotic phytoplankton. Front. Microbiol. 2012;3:235. doi: 10.3389/fmicb.2012.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin H.Y., Shih C.Y., Liu H.C., Chang J., Chen Y.L., Chen Y.R., Lin H.T., Chang Y.Y., Hsu C.H., Lin H.J. Identification and characterization of an extracellular alkaline phosphatase in the marine diatom Phaeodactylum tricornutum. Mar. Biotechnol. 2013;15:425–436. doi: 10.1007/s10126-013-9494-3. [DOI] [PubMed] [Google Scholar]
- 13.Luo H., Benner R., Long R.A., Hu J. Subcellular localization of marine bacterial alkaline phosphatases. Proc. Natl. Acad. Sci. USA. 2009;106:21219–21223. doi: 10.1073/pnas.0907586106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sebastian M., Ammerman J.W. The Alkaline Phosphatase PhoX is more widely distributed in marine bacteria than the classical PhoA. ISME J. 2009;3:563–572. doi: 10.1038/ismej.2009.10. [DOI] [PubMed] [Google Scholar]
- 15.White A.E. New insights into bacterial acquisition of phosphorus in the surface ocean. Proc. Natl. Acad. Sci. USA. 2009;106:21013–21014. doi: 10.1073/pnas.0912475107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kathuria S., Martiny A.C. Prevalence of a calcium-based alkaline phosphatase associated with the marine cyanobacterium Prochlorococcus and other ocean bacteria. Environ. Microbiol. 2011;13:74–83. doi: 10.1111/j.1462-2920.2010.02310.x. [DOI] [PubMed] [Google Scholar]
- 17.Yong S.C., Roversi P., Lillington J., Rodriguez F., Krehenbrink M., Zeldin O.B., Garman E.F., Lea S.M., Berks B.C. A complex iron-calcium cofactor catalyzing phosphotransfer chemistry. Science. 2014;345:1170–1173. doi: 10.1126/science.1254237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coleman J.E. Structure and mechanism of alkaline phosphatase. Annu. Rev. Biophys. Biomol. Struct. 1992;21:441–483. doi: 10.1146/annurev.bb.21.060192.002301. [DOI] [PubMed] [Google Scholar]
- 19.Zalatan J.G., Fenn T.D., Brunger A.T., Herschlag D. Structural and functional comparisons of nucleotide pyrophosphatase/phosphodiesterase and alkaline phosphatase: Implications for mechanism and evolution. Biochemistry. 2006;45:9788–9803. doi: 10.1021/bi060847t. [DOI] [PubMed] [Google Scholar]
- 20.Luo M., Guo Y.C., Deng J.Y., Wei H.P., Zhang Z.P., Leng Y., Men D., Song L.R., Zhang X.E., Zhou Y.F. Characterization of a monomeric heat-labile classical alkaline phosphatase from Anabaena sp. PCC7120. Biochemistry. 2010;75:655. doi: 10.1134/S0006297910050172. [DOI] [PubMed] [Google Scholar]
- 21.Zaheer R., Morton R., Proudfoot M., Yakunin A., Finan T.M. Genetic and biochemical properties of an alkaline phosphatase PhoX family protein found in many bacteria. Environ. Microbiol. 2009;11:1572–1587. doi: 10.1111/j.1462-2920.2009.01885.x. [DOI] [PubMed] [Google Scholar]
- 22.Sebastian M., Ammerman J.W. Role of the phosphatase PhoX in the phosphorus metabolism of the marine bacterium Ruegeria pomeroyi DSS-3. Environ. Microbiol. Rep. 2011;3:535–542. doi: 10.1111/j.1758-2229.2011.00253.x. [DOI] [PubMed] [Google Scholar]
- 23.Quisel J.D., Wykoff D.D., Grossman A.R. Biochemical characterization of the extracellular phosphatases produced by phosphorus-deprived Chlamydomonas reinhardtii. Plant Physiol. 1996;111:839–848. doi: 10.1104/pp.111.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hallmann A. Enzymes in the extracellular matrix of Volvox: An inducible, calcium-dependent phosphatase with a modular composition. J. Biol. Chem. 1999;274:1691–1697. doi: 10.1074/jbc.274.3.1691. [DOI] [PubMed] [Google Scholar]
- 25.Moseley J.L., Chang C.W., Grossman A.R. Genome-based approaches to understanding phosphorus deprivation responses and PSR1 control in Chlamydomonas reinhardtii. Eukaryot. Cell. 2006;5:26–44. doi: 10.1128/EC.5.1.26-44.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kageyama H., Tripathi K., Rai A.K., Chaum S., Waditeesirisattha R., Takabe T. An alkaline phosphatase/phosphodiesterase, PhoD, induced by salt stress and secreted out of the cells of Aphanothece halophytica, a halotolerant cyanobacterium. Appl. Environ. Microbiol. 2011;77:5178–5183. doi: 10.1128/AEM.00667-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun M.M., Sun J., Qiu J.W., Jing H., Liu H. Characterization of the proteomic profiles of the brown tide alga Aureoumbra lagunensis under phosphate- and nitrogen-limiting conditions and of its phosphate limitation-specific protein with alkaline phosphatase activity. Appl. Environ. Microbiol. 2012;78:2025–2033. doi: 10.1128/AEM.05755-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu Y., Wahlund T.M., Feng L., Shaked Y., Morel F.M.M. A novel alkaline phosphatase in the Coccolithophore Emiliania huxleyi (Prymnesiophyceae) and its regulation by phosphorus. J. Phycol. 2006;42:835–844. doi: 10.1111/j.1529-8817.2006.00243.x. [DOI] [Google Scholar]
- 29.Feng T.Y., Yang Z.K., Zheng J.W., Xie Y., Li D.W., Murugan S.B., Yang W.D., Liu J.S., Li H.Y. Examination of metabolic responses to phosphorus limitation via proteomic analyses in the marine diatom Phaeodactylum tricornutum. Sci. Rep. 2015;5:10373. doi: 10.1038/srep10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bowler C., Allen A.E., Badger J.H., Grimwood J., Jabbari K., Kuo A., Maheswari U., Martens C., Maumus F., Otillar R.P., et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;456:239–244. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- 31.Ou L., Qin X., Shi X., Feng Q., Zhang S., Lu S., Qi Y. Alkaline phosphatase activities and regulation in three harmful Prorocentrum species from the coastal waters of the East China Sea. Microb. Ecol. 2019:1–13. doi: 10.1007/s00248-019-01399-3. [DOI] [PubMed] [Google Scholar]
- 32.Lin X., Wang L., Shi X., Lin S. Rapidly diverging evolution of an atypical alkaline phosphatase (PhoAaty) in marine phytoplankton: Insights from dinoflagellate alkaline phosphatases. Front. Microbiol. 2015;6:868. doi: 10.3389/fmicb.2015.00868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.LaJeunesse T.C., Parkinson J.E., Gabrielson P.W., Jeong H.J., Reimer J.D., Voolstra C.R., Santos S.R. Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr. Biol. 2018;28:2570–2580. doi: 10.1016/j.cub.2018.07.008. [DOI] [PubMed] [Google Scholar]
- 34.Guillard R.R.L., Hargraves P.E. Stichochrysis immobilis is a diatom, not a chrysophyte. Phycologia. 1993;32:234–236. doi: 10.2216/i0031-8884-32-3-234.1. [DOI] [Google Scholar]
- 35.Parsons T.R., Marita Y., Lalli C.M. A Manual of Chemical and Biological Methods for Seawater Analysis. 1st ed. Pergamon Press; Oxford, UK: 1984. pp. 22–25. [Google Scholar]
- 36.Lin X., Zhang H., Cui Y., Lin S. Alkaline phosphatase gene sequence and transcriptional regulation by phosphate limitation in Amphidinium carterae (Dinophyceae) J. Phycol. 2011;47:1110–1120. doi: 10.1111/j.1529-8817.2011.01038.x. [DOI] [PubMed] [Google Scholar]
- 37.Lin X., Zhang H., Cui Y., Lin S. Alkaline phosphatase gene sequence characteristics and transcriptional regulation by phosphate limitation in Karenia brevis (Dinophyceae) Harmful Algae. 2012;17:14–24. doi: 10.1016/j.hal.2012.02.005. [DOI] [Google Scholar]
- 38.Zhang C., Lin S., Huang L., Lu W., Li M., Liu S. Suppression subtraction hybridization analysis revealed regulation of some cell cycle and toxin genes in Alexandrium catenella by phosphate limitation. Harmful Algae. 2014;39:26–39. doi: 10.1016/j.hal.2014.06.005. [DOI] [Google Scholar]
- 39.Shi X., Lin X., Li L., Palenik B., Lin S. Transcriptomic and microRNAomic profiling reveals multi-faceted mechanisms to cope with phosphate stress in a dinoflagellate. ISME J. 2017;11:2209. doi: 10.1038/ismej.2017.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li T., Guo C., Zhang Y., Wang C., Lin X., Lin S. Identification and expression analysis of an atypical alkaline phosphatase in Emiliania huxleyi. Front. Microbiol. 2018;9:2156. doi: 10.3389/fmicb.2018.02156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gentile F., Amodeo P., Febbraio F., Picaro F., Motta A., Formisano S., Nucci R. SDS-resistant active and thermostable dimers are obtained from the dissociation of homotetrameric β-Glycosidase from hyperthermophilic Sulfolobus solfataricus in SDS stabilizing role of the A-C intermonomeric interface. J. Biol. Chem. 2002;277:44050–44060. doi: 10.1074/jbc.M206761200. [DOI] [PubMed] [Google Scholar]
- 42.Rosen R.F., Tomidokoro Y., Ghiso J.A., Walker L.C. SDS-PAGE/immunoblot detection of Aβ multimers in human cortical tissue homogenates using antigen-epitope retrieval. J. Vis. Exp. 2010;38:e1916. doi: 10.3791/1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mann M., Jensen O.N. Proteomic analysis of post-translational modifications. Nat. Biotechnol. 2003;21:255–261. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- 44.Khoury G.A., Baliban R.C., Floudas C.A. Proteome-wide post-translational modification statistics: Frequency analysis and curation of the swiss-prot database. Sci. Rep. 2011;1:1161–1166. doi: 10.1038/srep00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dudev M., Lim C. Principles governing Mg, Ca, and Zn binding and selectivity in proteins. Chem. Rev. 2003;103:773–788. doi: 10.1021/cr020467n. [DOI] [PubMed] [Google Scholar]
- 46.Hou G., Cui Q. Stabilization of different types of transition states in a single enzyme active site: QM/MM analysis of enzymes in the alkaline phosphatase superfamily. J. Am. Chem. Soc. 2013;135:10457–10469. doi: 10.1021/ja403293d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jing Z., Liu C., Qi R., Ren P. Many-body effect determines the selectivity for Ca2+ and Mg2+ in proteins. Proc. Natl. Acad. Sci. USA. 2018;115:E7495–E7501. doi: 10.1073/pnas.1805049115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu J.R., Shien J.H., Shieh H.K., Hu C.C., Gong S.R., Chen L.Y., Chang P.C. Cloning of the gene and characterization of the enzymatic properties of the monomeric alkaline phosphatase (PhoX) from Pasteurella multocida strain X-73. FEMS Microbiol. Lett. 2007;267:113–120. doi: 10.1111/j.1574-6968.2006.00542.x. [DOI] [PubMed] [Google Scholar]
- 49.Dyhrman S.T., Palenik B.P. The identification and purification of a cell-surface alkaline phosphatase from the dinoflagellate Prorocentrum minimum (Dinophyceae) J. Phycol. 1997;33:602–612. doi: 10.1111/j.0022-3646.1997.00602.x. [DOI] [Google Scholar]
- 50.Dyhrman S.T. Ectoenzymes in Prorocentrum minimum. Harmful Algae. 2005;4:619–627. doi: 10.1016/j.hal.2004.08.011. [DOI] [Google Scholar]
- 51.Rivkin R., Swift E. Characterization of alkaline phosphatase and organic phosphorous utilization in the oceanic dinoflagellate Pyrocystis noctiluca. Mar. Biol. 1980;61:1–8. doi: 10.1007/BF00410336. [DOI] [Google Scholar]
- 52.Dudev M., Lim C. Discovering structural motifs using a structural alphabet: Application to magnesium-binding sites. BMC Bioinform. 2007;8:106. doi: 10.1186/1471-2105-8-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dudev M., Lim C. Competition among metal ions for protein binding sites: Determinants of metal ion selectivity in proteins. Chem. Rev. 2013;114:538–556. doi: 10.1021/cr4004665. [DOI] [PubMed] [Google Scholar]
- 54.Moore C.M. Microbial proteins and oceanic nutrient cycles. Science. 2014;345:1120–1121. doi: 10.1126/science.1258133. [DOI] [PubMed] [Google Scholar]