Abstract
Cancer is the second deadliest disease listed by the WHO. One of the major causes of cancer disease is tobacco and consumption possibly due to its main component, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK). A plethora of studies have been conducted in the past aiming to decipher the association of NNK with other diseases. However, it is strongly linked with cancer development. Despite these studies, a clear molecular mechanism and the impact of NNK on various system-level networks is not known. In the present study, system biology tools were employed to understand the key regulatory mechanisms and the perturbations that will happen in the cellular processes due to NNK. To investigate the system level influence of the carcinogen, NNK rewired protein–protein interaction network (PPIN) was generated from 544 reported proteins drawn out from 1317 articles retrieved from PubMed. The noise was removed from PPIN by the method of modulation. Gene ontology (GO) enrichment was performed on the seed proteins extracted from various modules to find the most affected pathways by the genes/proteins. For the modulation, Molecular COmplex DEtection (MCODE) was used to generate 19 modules containing 115 seed proteins. Further, scrutiny of the targeted biomolecules was done by the graph theory and molecular docking. GO enrichment analysis revealed that mostly cell cycle regulatory proteins were affected by NNK.
Keywords: NNK, cancer, systems biology, protein–protein interaction network, topological analysis, gene ontology
1. Introduction
Cancer is one of the major non-communicable diseases [1] and is accountable for millions of deaths per year worldwide. According to the World Health Organization (WHO), cancer is the second major cause of morbidity, with an estimate of 9.6 billion deaths in 2018 [2]. Cancer is a multistage process caused by aberrations in the cellular processes. Cancer is not only caused by mutation in any single gene but also by the accumulation of mutations in multiple genes, a phenomenon described as ‘oncogene addiction’ [3]. According to the WHO, there are mainly three reasons that lead to these aberrations, with tobacco consumption heading the list, which is single-handedly responsible for around 22% of deaths by cancer globally [4].
Currently, we have immense information on how tobacco consumption has direct implications in cancer, specially lung, head and neck, stomach, liver, and pancreatic cancers [5,6]. 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) is one of the main components in tobacco that plays a major role in the causation of cancer [6]. NNK and its derivative, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL), binds with the DNA and forms DNA adducts, the resultant of which may lead to genetic mutations followed by the deregulation of normal cellular processes [6,7].
NNK is not just responsible for causing cancer but also holds serious implications in other diseases as well. Earlier studies have shown that NNK has significant impact on steatohepatitis [8], Alzheimer’s disease [9], and tuberculosis [10], for example.
In this study, attempts have been made to exploit the system biology approach for an investigation of the overall impact of tobacco-generated carcinogen NNK on molecular systems.
Systems biology is an interdisciplinary field, which is a combination of mathematics, computer science, and biology [11]. Systems biology holds importance as it helps in getting a holistic view of the connections of biomolecules. It provides an anti-reductionist approach towards the involvement of different biomolecular components in a variety of biological systems. It focuses on the development of interactome of the affected targets and then analyzing it by using mathematical models [12]. Graph theory is the core assessment tool for the topological analysis of the interactome that aims to identify hub biomolecular targets based on their clustering coefficients, degrees, and betweenness centralities. Whereas, clustering and gene ontology (GO) enrichment analysis categorize the biomolecules on the basis of functions to procure more promising insight into complex networks. This study aims to find the most probable biomolecular targets of tobacco-associated carcinogen NNK along with its interactions and associated pathways that get perturbated by various cellular mechanisms and lead to cancer development. The most probable key targets of NNK are identified on the basis of their bottleneck scores and based on their thermodynamic interactions with NNK calculated by molecular docking simulations.
2. Materials and Methods
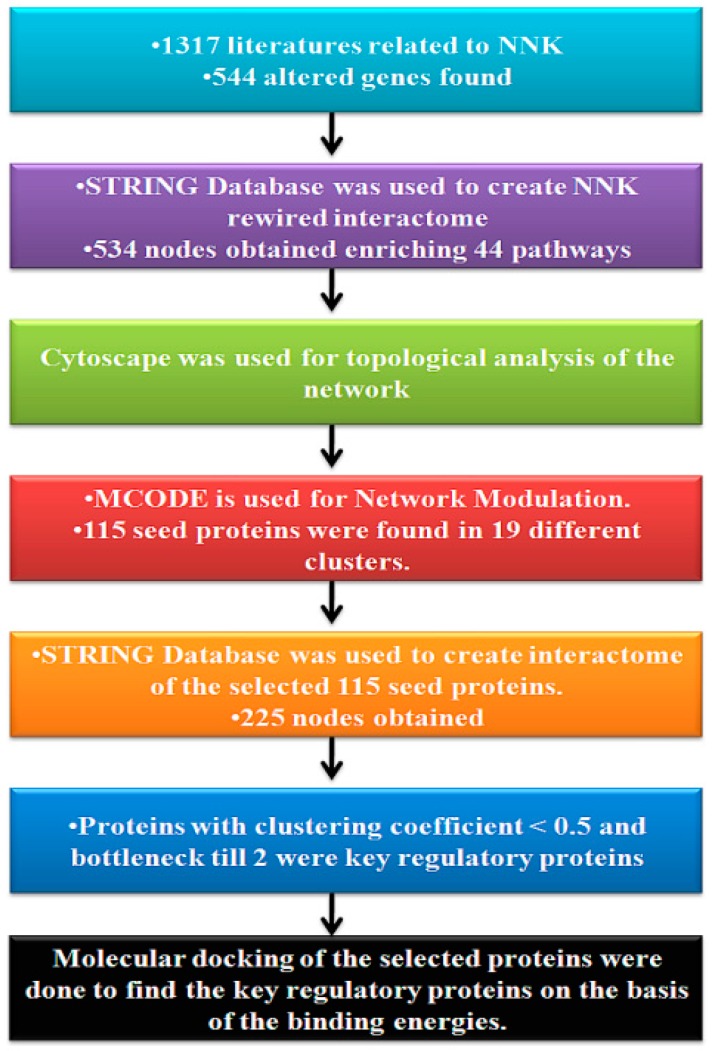
The full methodology scheme is mentioned in Figure 1.
Figure 1.
Schematic diagram of the adopted methodology.
2.1. Construction and Visualization of Protein–Protein Interaction Network
In total, 544 biomolecular targets were found to be affected by NNK in approximately 1320 studies using PubMed. A protein–protein interaction network was developed using the STRING database version 10.5 [13]. The network was evidence-based and developed with the highest confidence level score of 0.9, having 50 interactors in the first as well as second shell.
2.2. Protein–Protein Interaction Network (PPIN) Analysis
The Cytoscape Software (version 3.6.1) program [14] was used for the analysis of the protein–protein interaction network (PPIN) to generate protein interaction networks. Network analyzer, an in-built plugin of Cytoscape, was used to analyze the topological properties of an NNK modulated PPIN [15]. The topological properties of any network provide a deep insight into complex biological networks [16,17]. The topological analysis also helps in reducing noise in the data and offers reliable information regarding the network [18]. Node properties, like degree distribution, shortest path length, average clustering coefficient, betweenness centrality, and closeness centrality, were also analyzed.
2.3. Protein Interaction Network Modular Analysis and Pathway Enrichment
Clusters or modules are closely connected nodes in a network that come together and form a dense sub-network [19]. The analysis of clusters or modules helps in attaining detailed information about PPIN. Molecular COmplex DEtection (MCODE) is a plug-in available in the Cytoscape software program, which was used for the cluster analysis. The clusters are scored on the basis of size and density—a high score means a big and dense cluster—while gene ontology (GO) serves the purpose of validating the cluster that belongs to a specific function [20]. Thus, the enrichment of clusters helps in enriching the pathways by providing an additional number of external genes that are not present in the dataset. GO analysis provides detailed information about the biological process underneath that cluster. For GO functional enrichment analysis, ClueGO (version 2.5.1) plug-in of Cytoscape software was used [21]. The analysis was done using a threshold p value < 0.05. A two-sided hypergeometric test was used for the statistical analysis along with the Bonferroni correction method, in case applied.
2.4. Molecular Docking Analysis
Molecular docking is one of the most preferred methods to find the orientation of two molecules when they form a complex. Docking simulation also explores the thermodynamic stability of the complexes by providing information regarding the binding energy and inhibition constant (Ki) value and helps in finding the best binding modes or orientations of a ligand with its biomolecule. The docking parameters used were based on the studies published by [22]. Autodock 4.0 MGL suite [23] was used for docking simulations. The simulations were performed on AMD E1-6015 APU processor, CPU 1.4 GHz and 4 GB RAM of Hewlett-Packard (HP) machine.
3. Results
3.1. Construction of the Network
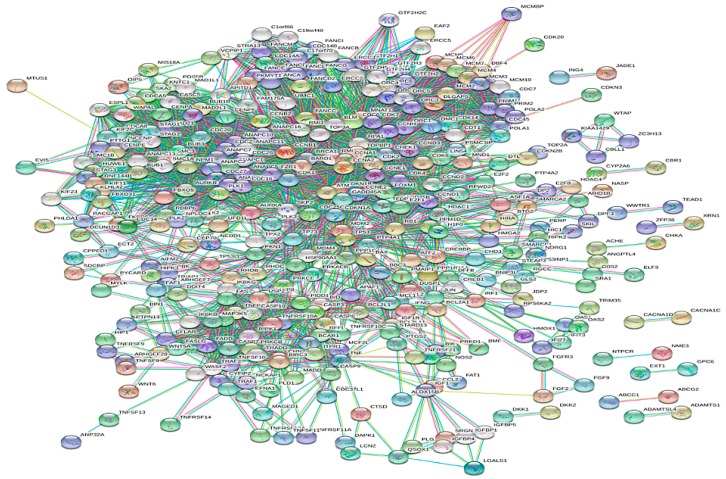
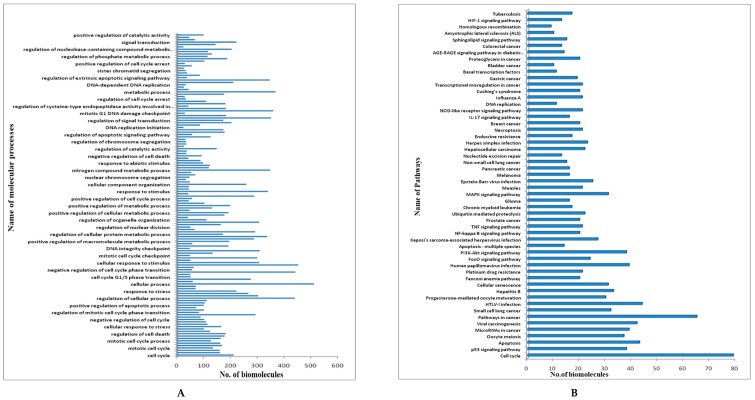
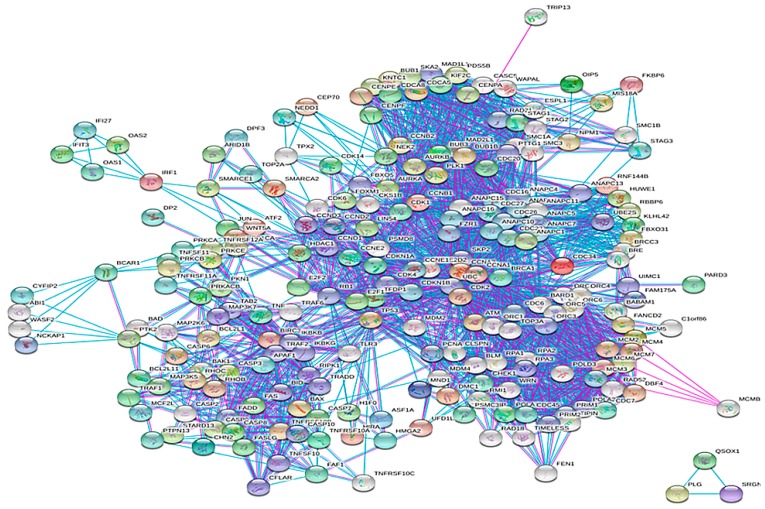
In total, 544 biomolecular targets were found to be affected by NNK interaction through a literature survey. A protein–protein interaction network (PPIN) was developed using the STRING database version 10.5. The network developed at the 0.9 confidence level score and 50–50 interactors in the first and second shell comprised of 534 nodes and 2909 edges. The average node degree was 10.09 and average local clustering coefficient was 0.501. The PPI enrichment p value was <1 × 10−16. Figure 2 represents the NNK rewired protein–protein interaction network having 534 nodes and 2909 edges. Figure 3A,B depicts the number of biomolecular targets involved in various processes and pathways, respectively.
Figure 2.
STRING generated NNK rewired protein–protein interaction network with 534 nodes and 2909 edges.
Figure 3.
(A) Processes enriched by NNK rewired PPIN. (B) Pathways enriched by NNK rewired PPIN.
3.2. Topological Properties of the Network
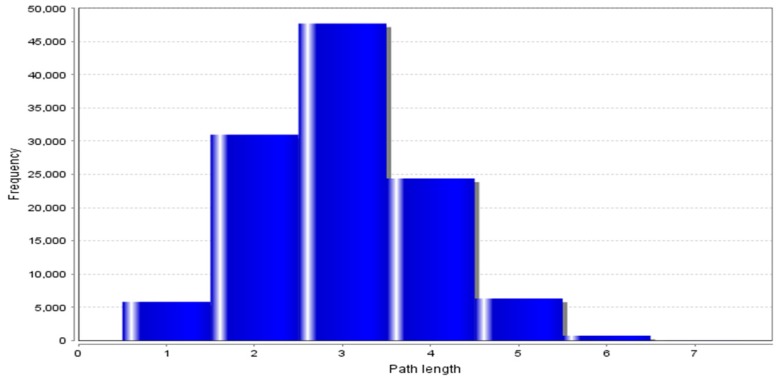
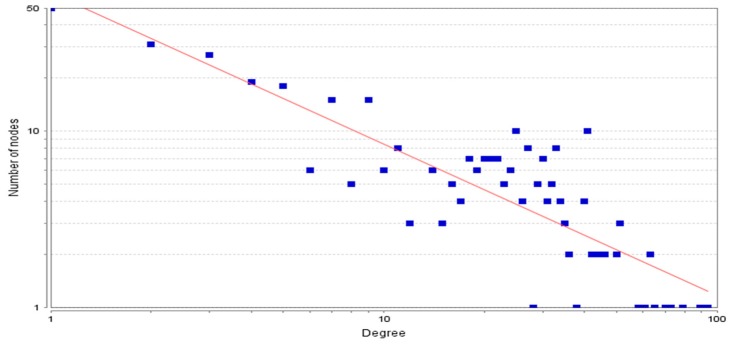
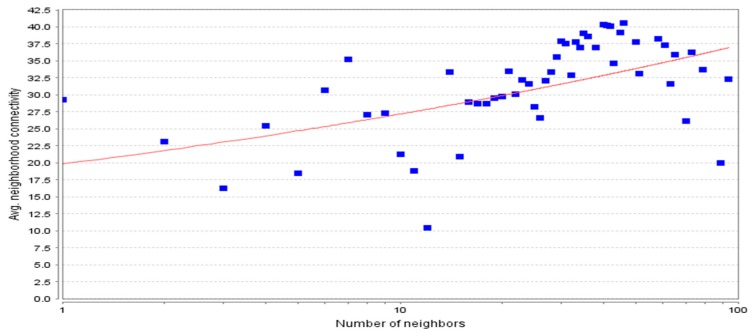
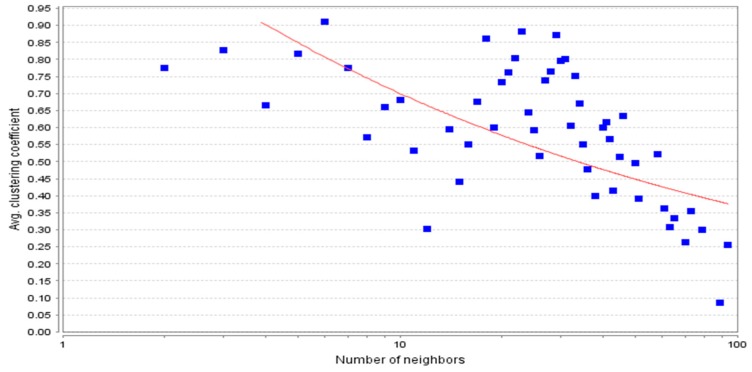
The protein–protein interaction network developed was further analyzed using Cytoscape software. The topological properties of the PPIN calculated with the help of Network Analyzer plug-in were the shortest path length average, neighborhood connectivity distribution, clustering coefficient, node degree distribution, betweenness centrality, closeness centrality, etc. The shortest path length in any network depicts the shortest communication mode between two nodes. Figure 4 shows a graphical representation of path length distribution 2971. This means that at the 2971-unit path length, the information is being passed on at the highest frequency. The degree of a node describes the connectivity of a node with other nodes; it is the total number of links, which are either reaching or starting from that node [24]. The node degree distribution (Figure 5) is one of the most important topological properties of a network. It fits a power law that indicates the presence of hubs in the network [25]. The nodes, which lie close to the power line and have a higher degree, can be considered as the hubs. The average neighborhood connectivity distribution stands for the average number of neighbors, which was observed as 15,940 in this study (Figure 6). Figure 6 shows the average connections of each node with its neighbors. The above parameter helps in understanding the density of a network. The clustering coefficient of a network depicts the tendency of a graph to be divided into clusters [26]. The local clustering coefficient is the number of edges around a particular node, whereas the average clustering coefficient is the clustering coefficient of the whole network [25], and in this study, the average clustering coefficient in this network was found to be 0.597 (Figure 7).
Figure 4.
Characteristic path length distribution: 2971.
Figure 5.
Node degree distribution following power law fitting y = 61,323x−0.861 (R-squared 0.722).
Figure 6.
Average (Avg.) neighborhood connectivity distribution following power law fitting y = 19,838x0.137 (R-squared 0.253). The average number of neighbors is 15,940.
Figure 7.
Average (Avg.) clustering coefficient following power law fitting y = 1.326 x−0.277 (R-squared 0.329). Clustering coefficient distribution of 0.597.
3.3. Clustering and GO Enrichment Analysis
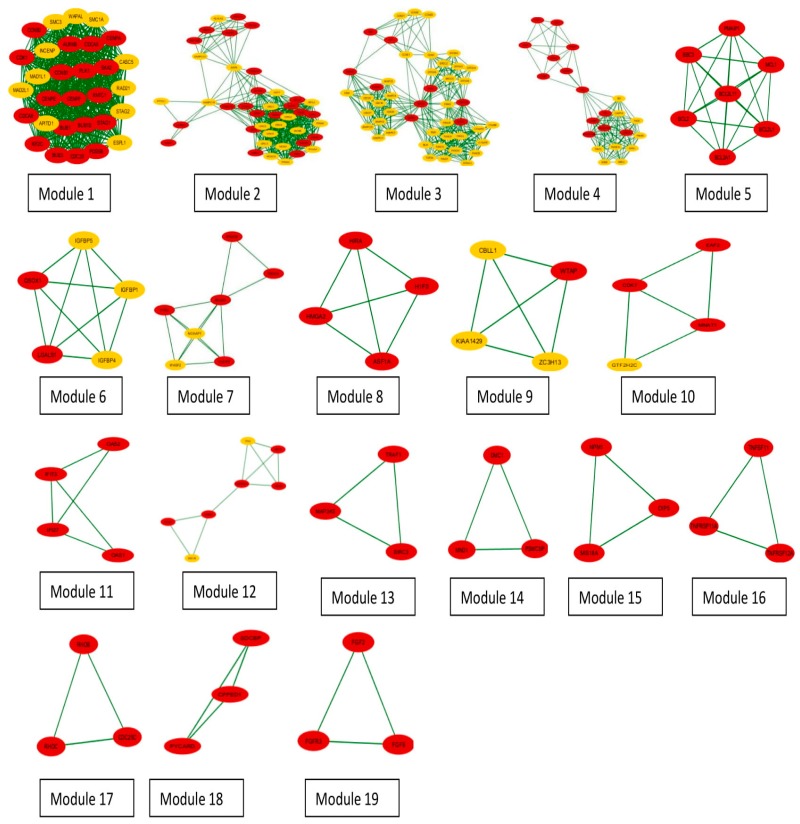
For modular analysis and pathway enrichment, the MCODE plug-in of the Cytoscape was used. Modules or clusters were created from the network and were scored on the basis of their size and density. A high score depicts a denser and tighter cluster. Formation of clusters also helps in the reduction of the noise and getting a better understanding of the genes involved in the clusters. Figure 8 shows the clusters generated by MCODE. The nodes in red color represent the seed proteins, and the yellow nodes are the connectors. The seed proteins are the proteins that were reported earlier to get either upregulated or downregulated by the action of NNK and connector proteins are the proteins that are associated with the seed proteins in the transfer of information, but are not reported to have any direct relation with NNK. The seed proteins were checked in all the clusters and are presented in bold in Table 1, while the remaining proteins (non-highlighted) are the connectors. The cluster that was ranked first had the highest score of 29,862, with 30 nodes and 433 edges. Moreover, during analysis, it was found that cluster 1 had 19 seed proteins and 11 connectors and the overall analysis of the entire cluster found 115 seeds and 88 connectors.
Figure 8.
Network modules: red circles show the seed proteins and yellow circles are the connectors.
Table 1.
Generated clusters with MCODE scores, seeds (bold letters), and connector (non-bold letters) proteins.
| Cluster | Score | Nodes | Edges | Seeds | Connectors | Node IDs |
|---|---|---|---|---|---|---|
| 1 | 29.862 | 30 | 433 | 19 | 11 | PLK1, CASC5, BUB3, CDCA8, STAG1, BUB1B, CCNB2, CCNB1, BUB1, ESPL1, CENPF, KIF2C, CDCA5, MAD2L1, CDK1, PDS5B, WAPAL, APITD1, SKA2, RAD21, SMC1A, KNTC1, CDC20, CENPA, CENPE, SMC3, STAG2, MAD1L1, AURKB, INCENP |
| 2 | 19.3 | 41 | 386 | 22 | 19 | CDKN1A, FBXO31, CDC6, CDK2, RNF144B, KLHL42, CCNA1, DBF4, CDC7, MCM4, POLA2, PRIM2, CDC34, POLA1, SKP2, ORC3, MCM2, MCM5, ORC2, ORC6, MCM6, MCM3, ORC4, CHEK1, AURKA, PTTG1, CDC45, PRIM1, MCM10, CDT1, ORC1, HUWE1, ORC5, RBBP6, NEK2, MCM7, ANAPC16, ANAPC13, CDKN1B, RPA1, CCNA2 |
| 3 | 14.652 | 47 | 337 | 9 | 38 | ERCC2, E2F1, FBXO5, CDC23, ANAPC5, FANCE, FANCC, FANCG, BLM, ANAPC4, ANAPC1, BRCA1, RB1, FANCF, FANCL, C17orf70, CCNE1, C19orf40, FANCD2, FANCI, ANAPC10, FANCM, FANCB, GTF2H5, CCND2, FZR1, CDC26, RMI1, TOP3A, FANCA, GTF2H4, ANAPC11, CDC27, ANAPC2, GTF2H2, ANAPC7, GTF2H1, STRA13, CDC16, CCNH, C1orf86, ERCC3, ATM, CCND3, BARD1, GTF2H3, CCND1 |
| 4 | 10.762 | 22 | 113 | 13 | 9 | FAS, RIPK1, CASP8, IKBKG, BID, HDAC1, TNFRSF10B, TRADD, TP53, TFDP1, TRAF2, TNFSF10, FADD, FASLG, TNFRSF10A, CASP10, E2F2, TOPBP1, IKBKB, CDK4, CCNE2, CDK6 |
| 5 | 7 | 7 | 21 | 7 | 0 | BBC3, BCL2A1, BCL2L11, BCL2, MCL1, PMAIP1, BCL2L1 |
| 6 | 5 | 5 | 10 | 2 | 3 | IGFBP1, LGALS1, IGFBP4, QSOX1, IGFBP5 |
| 7 | 4.333 | 7 | 13 | 5 | 2 | NCKAP1, WASF2, CYFIP2, PRKCA, PTK2, PRKCB, BCAR1 |
| 8 | 4 | 4 | 6 | 4 | 0 | H1F0, HMGA2, ASF1A, HIRA |
| 9 | 4 | 4 | 6 | 1 | 3 | KIAA1429, WTAP, CBLL1, ZC3H13 |
| 10 | 3.333 | 4 | 5 | 3 | 1 | EAF2, GTF2H2C, CDK7, MNAT1 |
| 11 | 3.333 | 4 | 5 | 4 | 0 | OAS1, IFI27, OAS2, IFIT3 |
| 12 | 3.333 | 7 | 10 | 5 | 2 | STAG3, CEP70, FKBP6, SMC1B, NEDD1, HSP90AA1, TPX2 |
| 13 | 3 | 3 | 3 | 3 | 0 | MAP3K5, TRAF1, BIRC3 |
| 14 | 3 | 3 | 3 | 3 | 0 | PSMC3IP, MND1, DMC1 |
| 15 | 3 | 3 | 3 | 3 | 0 | OIP5, MIS18A, NPM1 |
| 16 | 3 | 3 | 3 | 3 | 0 | TNFRSF11A, TNFRSF12A, TNFSF11 |
| 17 | 3 | 3 | 3 | 3 | 0 | RHOB, RHOC, CDC25C |
| 18 | 3 | 3 | 3 | 3 | 0 | SDCBP, PYCARD, CPPED1 |
| 19 | 3 | 3 | 3 | 3 | 0 | FGF2, FGFR3, FGF9 |
Nodes in “bold text” represent the seed proteins and the nodes in “normal” text represent the connector proteins.
The seed proteins were identified in each cluster. Thereafter, the PPIN was generated using the STRING database with 0.9 confidence level, where 50 interactors in the first and another 50 in the second shell were recorded. The PPIN generated now was further enriched using the ClueGO plug-in.
3.4. PIN Construction and Topological and GO Analysis of Final Selected Seed Proteins
After the modularization process, 115 seed proteins were obtained, and used to further create a PPIN (Figure 9) with 100 connectors using the highest confidence level score of 0.9. The PPIN generated had 213 nodes and 2509 edges, with an average node degree 23.6 and average clustering coefficient of 0.761. The PPI enrichment p-value was less than 1 × 10−16.
Figure 9.
Protein–protein interaction network (PPIN) of final seed proteins.
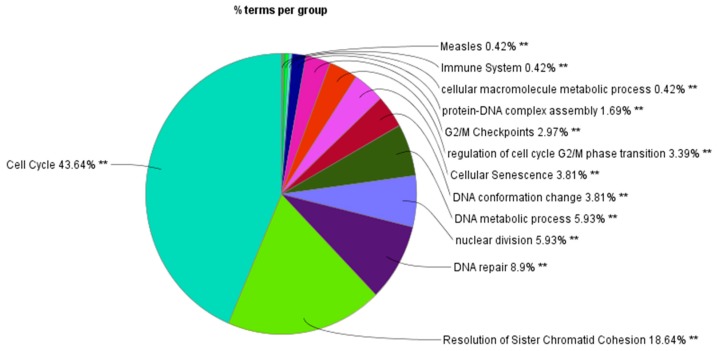
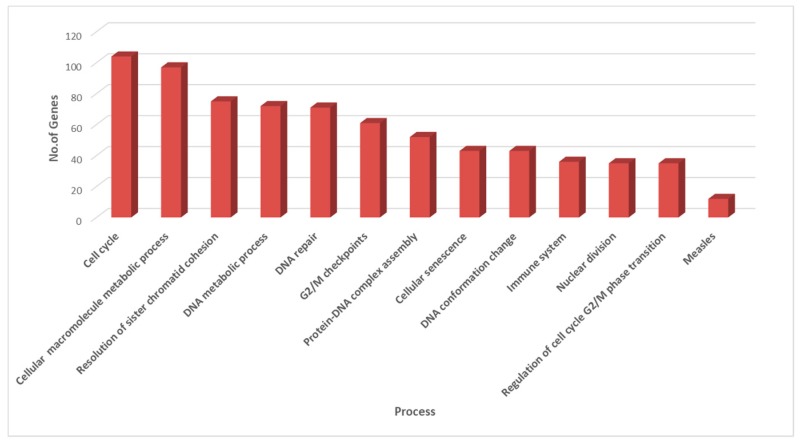
The network generated was analyzed using Cytohubba, a plug-in of Cytoscape [27]. Moreover, cytohubba analysis of each node of the network was performed and scored based on various parameters, like the degree, closeness centrality, clustering coefficients, betweenness, bottleneck, and stress. The proteins were first sorted on the basis of the clustering coefficient and then on the basis of bottleneck. The proteins with a clustering coefficient less than 0.5 were selected. The proteins having a clustering coefficient more than 0.5 were rejected as this depicts that these proteins were highly clustered and have no further spaces for the attachment of other molecules. The nodes with a clustering coefficient less than 0.5 and high degrees were found to be highly significant. A high degree and clustering coefficient less than 0.5 depict that the nodes are important in various connecting networks and they also have binding spaces available on their surface for other molecules to bind to. Once the nodes were sorted on the basis of clustering coefficients, the next most important parameter was the bottleneck. The proteins with high bottleneck were considered the most critical proteins in any PPIN. The median of the bottleneck scores of selected proteins was calculated which came out to be 3. All the proteins with bottleneck more than or equal to 2 were finally selected. Table 2 enlists the final selected proteins, which were sorted on the basis of bottleneck, with clustering coefficients less than 0.5. CHEK1, showing the highest bottleneck score of 29, was ranked in first position followed by TP53 with a bottleneck of 27. These were also analyzed with ClueGO for the pathway enrichment analysis. Figure 10 shows the pie chart representation of the enriched pathways in the form of groups. In Figure 10, the cell cycle is occupying the maximum area (43.64%) of the pie chart. From this, we can say that the cell cycle is the most enriched pathway, having the maximum number of biomolecules (a detailed graph displaying the sub-pathways in each group along with the number of genes present in it and the proteins involved in each group is enlisted in Table 3, also depicted in Supplementary Figure S1). Figure 11 shows that the maximum number of genes are associated with the cell cycle followed by the cellular macromolecule metabolic process.
Table 2.
Key proteins with their bottleneck, clustering coefficient, and degree scores.
| Name | Betweenness | Bottleneck | Closeness | Clustering Coefficient | Degree |
|---|---|---|---|---|---|
| CHEK1 | 835.45994 | 29 | 116.61667 | 0.36501 | 52 |
| TP53 | 8007.14223 | 27 | 126.41667 | 0.19394 | 55 |
| BRCA1 | 2686.48081 | 23 | 127.45 | 0.26346 | 65 |
| CDK1 | 3705.28949 | 19 | 140.41667 | 0.32157 | 85 |
| CDK4 | 935.26901 | 14 | 112.66667 | 0.42521 | 35 |
| HSP90AA1 | 3657.32532 | 13 | 101.66667 | 0.27273 | 22 |
| RPA2 | 1523.40238 | 9 | 125.78333 | 0.33978 | 64 |
| ATM | 1803.73545 | 8 | 115.36667 | 0.33718 | 40 |
| TFDP1 | 426.33754 | 6 | 111.78333 | 0.46702 | 34 |
| CDKN1B | 1218.04331 | 4 | 120.41667 | 0.44245 | 50 |
| CASP8 | 1842.98595 | 4 | 90.11667 | 0.44853 | 17 |
| PYCARD | 796 | 3 | 62.18333 | 0.33333 | 3 |
| CCNA1 | 894.78122 | 3 | 125.2 | 0.39548 | 60 |
| CCNB1 | 1208.39103 | 3 | 127.26667 | 0.40665 | 69 |
| RPA1 | 1657.28981 | 2 | 126.45 | 0.33269 | 65 |
| CDK2 | 1887.40725 | 2 | 134.2 | 0.34035 | 76 |
| CHEK2 | 126.13416 | 2 | 95.95 | 0.3619 | 15 |
| BID | 577.03969 | 2 | 90.78333 | 0.4269 | 19 |
| RB1 | 608.84911 | 2 | 107.66667 | 0.44138 | 30 |
| PLK1 | 1284.13385 | 2 | 121.25 | 0.49545 | 56 |
| CDK7 | 488.15015 | 2 | 108.78333 | 0.49733 | 34 |
Figure 10.
ClueGO results of gene ontology (GO) functional enrichment of key proteins.
Table 3.
Pathway enrichment of the modulated seed proteins using ClueGO.
| Function | Groups | Group Genes |
|---|---|---|
| Cell cycle | Group12 | ANAPC1|ASF1A|ATM|AURKA|AURKB|BARD1|BBC3|BCAR1|BCL2|BCL2A1|BCL2L1|BCL2L11| BIRC3|BRCA1|BUB1|BUB1B|BUB3|CASP8|CCNA1|CCNA2|CCNB1|CCNB2|CCNE2|CDC20|CDC25C| CDC34|CDC45|CDC6|CDC7|CDCA5|CDCA8|CDK1|CDK2|CDK6|CDKN1A|CENPA|CENPE|CENPF|CEP70|CHEK1|CYFIP2|DBF4|DMC1|E2F1|E2F2|EAF2|FANCC|FANCD2|FAS|FASLG|FBXO31|FBXO5|FGF2| FGF9|FGFR3|FKBP6|H1F0|HDAC1|HIRA|HMGA2|HSP90AA1|HUWE1|KIF2C|KNTC1|LGALS1|MCL1| MCM2|MCM4|MCM5|MCM6|MCM7|MDM2|MIS18A|MNAT1|MND1|NEDD1|NEK2|NPM1|OIP5| PDS5B|PLK1|PMAIP1|POLA1|PRKCA|PRKCB|PSMC3IP|PTK2|PYCARD|RBBP6|RHOB|RHOC|RNF144B|SDCBP|SKA2|STAG1|STAG3|TFDP1|TNFRSF10A|TNFRSF10B|TNFRSF10C|TNFRSF12A|TOPBP1|TP53|TRAF1 |
| Cellular senescence | Group06 | ANAPC1|ASF1A|ATM|BBC3|BCL2|BCL2L1|BCL2L11|BIRC3|BRCA1|CASP8|CCNA1| CCNA2|CCNE2|CDC20|CDC25C|CDCA5|CDK1|CDK2|CDK6|CDKN1A|CHEK1|E2F1|E2F2|FAS| FASLG|FGF2|FGF9|FGFR3|H1F0|HDAC1|HIRA|HMGA2|HSP90AA1|MAP3K5|MCL1|MDM2|PMAIP1| PRKCA|PRKCB|PTK2|TFDP1|TP53|TRAF1 |
| DNA conformation change | Group07 | ASF1A|ATM|BBC3|BCL2L11|BIRC3|CASP8|CCNB1|CDC45|CDCA5|CDK1|CENPA|CENPE|CENPF| DMC1|FANCC|FAS|FBXO5|H1F0|HDAC1|HIRA|HMGA2|HSP90AA1|KNTC1|MCM2|MCM4|MCM5| MCM6|MCM7|MDM2|MIS18A|MNAT1|NPM1|OAS1|OIP5|PMAIP1|POLA1|PRKCA|PTK2|PYCARD| RHOC|TNFSF11|TP53|TRAF1 |
| DNA metabolic process | Group08 | ASF1A|ATM|AURKA|AURKB|BARD1|BBC3|BCL2|BCL2A1|BCL2L1|BCL2L11|BRCA1|BUB1|BUB1B|BUB3|CCNA1|CCNA2|CCNB1|CCNE2|CDC25C|CDC34|CDC45|CDC6|CDC7|CDCA5|CDK1|CDK2|CDK6|CDKN1A|CENPF|CHEK1|DBF4|DMC1|E2F1|EAF2|FANCC|FANCD2|FAS|FBXO31|FBXO5|FKBP6|H1F0|HDAC1|HMGA2|HSP90AA1|HUWE1|KNTC1|MCL1|MCM2|MCM4|MCM5|MCM6|MCM7|MDM2|MIS18A|MNAT1|MND1|NEK2|NPM1|PDS5B|PLK1|PMAIP1|POLA1|PRKCB|PSMC3IP|PYCARD|RBBP6|TFDP1|TNFRSF10A|TNFRSF10B|TNFRSF10C|TOPBP1|TP53 |
| DNA repair | Group10 | ANAPC1|ASF1A|ATM|AURKA|AURKB|BARD1|BBC3|BCL2|BCL2L11|BRCA1|CASP8|CCNA1|CCNA2| CCNB1|CCNB2|CCNE2|CDC20|CDC25C|CDC34|CDC45|CDC6|CDC7|CDCA5|CDK1|CDK2|CDK6|CDKN1A|CENPF|CEP70|CHEK1|DBF4|DMC1|E2F1|E2F2|EAF2|FANCC|FANCD2|FAS|FASLG|H1F0|HDAC1| HIRA|HMGA2|HSP90AA1|HUWE1|MAP3K5|MCM2|MCM4|MCM5|MCM6|MCM7|MDM2|MNAT1| MND1|NEDD1|NEK2|NPM1|PDS5B|PLK1|PMAIP1|POLA1|PRKCA|PRKCB|RBBP6|TFDP1|TNFRSF10A|TNFRSF10B|TNFRSF10C|TOPBP1|TP53|TRAF1 |
| G2/M checkpoints | Group04 | ASF1A|ATM|AURKA|AURKB|BARD1|BBC3|BCL2|BCL2A1|BCL2L1|BCL2L11|BRCA1|CCNA1|CCNA2| CCNB1|CCNB2|CCNE2|CDC25C|CDC34|CDC45|CDC6|CDC7|CDCA5|CDK1|CDK2|CDKN1A|CENPF| CHEK1|DBF4|DMC1|E2F1|FANCC|FANCD2|FBXO31|FKBP6|H1F0|HDAC1|HMGA2|HSP90AA1|HUWE1|MCL1|MCM2|MCM4|MCM5|MCM6|MCM7|MDM2|MIS18A|MNAT1|MND1|NEK2|NPM1|PDS5B|PLK1|PMAIP1|POLA1|PSMC3IP|PYCARD|RBBP6|TFDP1|TOPBP1|TP53 |
| Immune system | Group01 | ANAPC1|BCL2|BCL2L1|BIRC3|CASP8|CDC20|CDC34|CDK1|CDKN1A|CENPE|CPPED1|CYFIP2|FASLG| FBXO31|FGF2|FGF9|FGFR3|HSP90AA1|HUWE1|IFI27|IFIT3|KIF2C|MCL1|OAS1|OAS2|PRKCB|PTK2| PYCARD|QSOX1|RBBP6|RNF144B|SDCBP|TNFRSF11A|TNFRSF12A|TNFSF11|TP53 |
| Measles | Group00 | BBC3|CCNE2|CDK2|CDK6|FAS|FASLG|OAS1|OAS2|TNFRSF10A|TNFRSF10B|TNFRSF10C|TP53 |
| Resolution of sister chromatid cohesion | Group11 | ANAPC1|ATM|AURKA|AURKB|BARD1|BBC3|BCL2|BCL2L11|BIRC3|BRCA1|BUB1|BUB1B|BUB3|CASP8|CCNA1|CCNA2|CCNB1|CCNB2|CCNE2|CDC20|CDC25C|CDC34|CDC6|CDC7|CDCA5|CDCA8|CDK1|CDK2|CDKN1A|CENPA|CENPE|CENPF|CEP70|CHEK1|CYFIP2|DMC1|E2F1|FANCD2|FAS|FASLG|FBXO31|FBXO5|FKBP6|HMGA2|HSP90AA1|HUWE1|IFI27|KIF2C|KNTC1|MAP3K5|MDM2|MNAT1|MND1|NEDD1|NEK2|NPM1|PDS5B|PLK1|PMAIP1|PRKCA|PRKCB|PSMC3IP|PTK2|PYCARD|RBBP6|RNF144B|SDCBP|SKA2|STAG1|STAG3|TFDP1|TNFRSF10A|TNFRSF10B|TOPBP1|TP53 |
| Cellular macromolecule metabolic process | Group02 | ANAPC1|ASF1A|ATM|AURKA|AURKB|BARD1|BBC3|BCL2|BCL2L11|BIRC3|BRCA1|BUB1|BUB1B|BUB3|CASP8|CCNA1|CCNA2|CCNB1|CCNE2|CDC20|CDC25C|CDC34|CDC45|CDC6|CDC7|CDCA5|CDCA8|CDK1|CDK2|CDK6|CDKN1A|CENPE|CENPF|CHEK1|CPPED1|CYFIP2|DBF4|DMC1|E2F1|E2F2|EAF2|FANCC|FANCD2|FAS|FASLG|FBXO31|FBXO5|FGF2|FGF9|FGFR3|FKBP6|H1F0|HDAC1|HIRA|HMGA2|HSP90AA1|HUWE1|IFI27|LGALS1|MAP3K5|MCM2|MCM4|MCM5|MCM6|MCM7|MDM2|MIS18A|MNAT1|MND1|NEK2|NPM1|OAS1|OAS2|PDS5B|PLK1|PMAIP1|POLA1|PRKCA|PRKCB|PSMC3IP|PTK2|PYCARD|QSOX1|RBBP6|RNF144B|SDCBP|STAG1|TFDP1|TNFRSF10A|TNFRSF10B|TNFRSF10C|TNFRSF11A|TNFSF11|TOPBP1|TP53|TRAF1|WTAP |
| Nuclear division | Group09 | ANAPC1|ATM|AURKA|AURKB|BRCA1|BUB1|BUB1B|BUB3|CCNA1|CCNA2|CCNB1|CCNE2|CDC20| CDC25C|CDC6|CDCA5|CDCA8|CDK1|CDK2|CENPE|CENPF|CHEK1|DMC1|FANCD2|FBXO5|FKBP6| KIF2C|KNTC1|MND1|NEK2|PDS5B|PLK1|PSMC3IP|STAG1|STAG3 |
| Protein–DNA complex assembly | Group03 | ASF1A|ATM|AURKA|AURKB|BBC3|BCL2|BCL2L11|BIRC3|BRCA1|CASP8|CCNB1|CDC20|CDC34|CDC45|CDK1|CDK2|CENPA|CENPE|CENPF|CEP70|DMC1|FANCC|FAS|FBXO5|H1F0|HDAC1|HIRA|HMGA2|HSP90AA1|KNTC1|MCM2|MCM7|MDM2|MIS18A|MNAT1|NEDD1|NEK2|NPM1|OAS1|OIP5|PLK1|PMAIP1|PRKCA|PTK2|PYCARD|RHOC|SDCBP|STAG1|STAG3|TNFSF11|TP53|TRAF1 |
| Regulation of cell cycle G2/M phase transition | Group05 | ANAPC1|ATM|AURKA|AURKB|BARD1|BRCA1|BUB1B|BUB3|CCNA1|CCNA2|CCNB1|CCNB2|CDC20| CDC25C|CDC7|CDK1|CDK2|CDK6|CDKN1A|CENPF|CEP70|CHEK1|FBXO5|HMGA2|HSP90AA1|MDM2|MNAT1|NEDD1|NEK2|NPM1|PLK1|PRKCA|PRKCB|TOPBP1|TP53 |
Figure 11.
Representation of the pathways enriched and the total number of genes associated with them.
3.5. Molecular Docking Simulation of NNK with Key Proteins
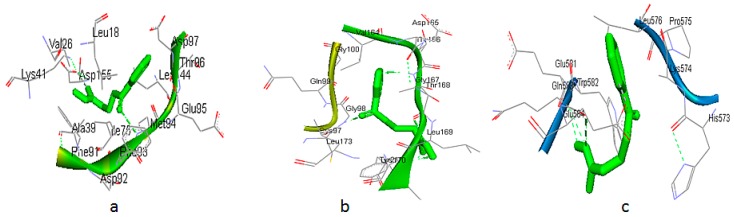
Autodock 4.0 was used for docking the final 30 target proteins with NNK. CDK7 showed the highest binding energy of −5.93 Kcal/Mol followed by CCNA1 (−5.6 Kcal/Mol). The binding energies of the proteins with NNK will further help in refining the results in the selection of the best-suited target proteins of NNK. The more negative the binding energy, the stronger the interaction between the ligand and the protein. The binding energies, Ki values, and the H-bonds formed along with their distances for all the 20 target proteins are listed in Table 4. Figure 12 shows the top three interactions of NNK with its target biomolecules, namely CDK7, CCNA1, and CDKN1B. Table 5 shows the key biomolecular targets of NNK and their role in cell cycle regulation.
Table 4.
Final 22 target proteins docked with NNK.
| S.No | Protein | Ligand | Binding Energy (Kcal/Mol) | Ki | Binding Residues | H-Bond | Distance |
|---|---|---|---|---|---|---|---|
| 1. | CDK7 | NNK | −5.93 | 45.31 μM | Leu18, Val26, Ala39, Lys41, Ile75, Phe91, Asp92, Phe93, Met94, Glu95, Thr96, Asp97, Leu144, Asp155 | CDK7:MET94:N - :NNK:O7 | 2.86224 |
| 2. | CCNA1 (connector) | NNK | −5.60 | 79.09 μM | Cys97, Gly98, Gln99, Gly100, Val164, Asp165, Thr166, Gly167, Thr168, Leu169, Lys170, Leu173, Tyr218 | :GLY98:H - :NNK:O7 :GLY167:H - :NNK:N10 :THR168:H - :ASP165:O :LYS170:H - :NNK:N14 :LYS170:H - :NNK:O15 :NNK:N14 - :THR168:O |
1.83311 1.98057 2.11393 2.36928 1.99807 3.02246 |
| 3. | CDKN1B | NNK | −5.42 | 106.27 μM | His573, Lys574, Pro575, Leu576, Glu581, Trp582, Gln583, Glu584 | CDKN1B:GLN583:N - :NNK:N14 CDKN1B:GLN583:N - :NNK:O15 :NNK:N14 - CDKN1B:GLN583:O |
2.84311 2.79833 2.96186 |
| 4. | CASP8 | NNK | −5.35 | 119.75 μM | Lys2224, Tyr2226, Gln2227, Asp2308, Gly2350, Lys2351, Pro2352, Asp2398, Arg2471, Lys2472 | CASP8:LYS2351:HZ1 - :NNK:N10 CASP8:ARG2471:HH21 - :NNK:N2 CASP8:ARG2471:HH21 - :NNK:N14 CASP8:ARG2471:HH21 - :NNK:O15 CASP8:LYS2472:HZ1 - :NNK:O7 |
1.9527 2.48624 2.19123 2.25672 1.82255 |
| 5. | CHEK2 (connector) | NNK | −5.35 | 120.25 μM | Ser228, Gly229, Ala230, Cys231, Gly232, Val234, Lys249, Leu301, Thr367, Asp368 | CHEK2:CYS231:N - :NNK:N10 CHEK2:GLY232:N - :NNK:N10 CHEK2:LYS249:NZ - :NNK:N14 CHEK2:ASP368:N - :NNK:O15 |
3.11295 3.11412 3.20168 2.86381 |
| 6. | PLK1 | NNK | −5.21 | 152.98 μM | Lys413, Trp414, Val415, Asp416, Leu490, Asn533, Lys540 | PLK1:TRP414:N - :NNK:O7 PLK1:ASP416:N - :NNK:N14 PLK1:ASP416:N - :NNK:O15 PLK1:ASN533:ND2 - :NNK:N10 |
2.97424 2.94596 2.71702 3.10323 |
| 7. | BID (connector) | NNK | −5.13 | 174.27 μM | Leu21, Phe24, Gly25, Gln28, Leu39, Asp40, Leu42, Gly43, Arg86, Ala89, Arg90, Phe173 | :NNK:N14 - BID:GLN28:OE1 BID:PHE24:HA - :NNK:O15 BID:ARG86:HA - :NNK:N10 :NNK:C3 - BID:GLN28:OE1 :NNK:C1 - BID:LEU39:O :NNK:C11 - BID:ARG86:O :NNK:O15 - BID:PHE24 :NNK:N14 - BID:GLN28:OE1 BID:PHE24:HA - :NNK:O15 BID:ARG86:HA - :NNK:N10 :NNK:C3 - BID:GLN28:OE1 :NNK:C1 - BID:LEU39:O :NNK:C11 - BID:ARG86:O :NNK:O15 - BID:PHE24 |
3.29101 2.94776 2.82375 3.41587 3.00214 3.32997 3.70686 3.29101 2.94776 2.82375 3.41587 3.00214 3.32997 3.70686 |
| 8. | HSP90AA1 | NNK | −5.10 | 183.46 μM | Leu48, Asn51, Ser52, Ala55, Asp93, Ile96, Gly97, Met98, Asn106, Phe138, Thr184, Val186 | HSP90AA1:ASN51:ND2 - :NNK:N14 HSP90AA1:ASN51:ND2 - :NNK:O15 HSP90AA1:THR184:OG1 - :NNK:O7 |
3.12471 2.99653 2.69827 |
| 9. | BRCA1 | NNK | −5.08 | 187.68 μM | Val1654, Ser1655, Gly1656, Leu1657, Thr1658, Pro1659, Phe1662, Thr1700, Leu1701, Lys1702 | BRCA1:SER1655:OG - :NNK:N14 BRCA1:GLY1656:N - :NNK:O7 BRCA1:LEU1657:N - :NNK:O7 BRCA1:LEU1701:N - :NNK:O15 BRCA1:LYS1702:N - :NNK:O15 :NNK:N14 - BRCA1:SER1655:OG |
3.19252 2.75464 2.77131 3.00037 2.75018 3.19252 |
| 10. | CDK1 | NNK | −5.00 | 217.06 μM | Lys88, Leu91, Asp92, Ile94, Pro95, Pro96, Glu196, Lys200 | CDK1:LYS200:NZ - :NNK:N14 CDK1:LYS200:NZ - :NNK:O15 :NNK:N14 - CDK1:ILE94:O |
3.0124 2.96559 2.89466 |
| 11. | CDK2 | NNK | −4.90 | 255.02 μM | Val29, Glu81, Phe82, Leu83, His84, Ile135, Asn136, Thr137 | CDK2:PHE82:N - :NNK:N10 CDK2:HIS84:N - :NNK:N14 CDK2:HIS84:N - :NNK:O15 CDK2:HIS84:ND1 - :NNK:N14 :NNK:O15 - CDK2:ILE135:O |
2.88593 2.95506 2.76842 3.18956 2.91961 |
| 12. | CCNB1 | NNK | −4.86 | 274.97 μM | Ile253, Lys256, Tyr257, Glu285, Leu289, Phe294, Gly295, Leu296, Gly297 |
CCNB1:TYR257:N - :NNK:N10 CCNB1:LEU296:N - : NNK:N14 CCNB1:LEU296:N - : NNK:O15 CCNB1:GLY297:N - : NNK:O15 : NNK:N14 - CCNB1:PHE294:O |
3.14377 2.86319 2.81795 2.91169 3.10697 |
| 13. | CHEK1 | NNK | −4.80 | 303.50 μM | Val23, Val37, Ile39, Glu55, Asn59, Leu82, Phe149 | CHEK1:ILE39:N - :NNK:N10 CHEK1:ASN59:ND2 - :NNK:N14 CHEK1:ASN59:ND2 - :NNK:O15 CHEK1:PHE149:N - :NNK:O15 :NNK:N14 - CHEK1:GLU55:OE2 :NNK:O15 - CHEK1:GLU55:OE2 :NNK:O15 - CHEK1:PHE149:N |
2.8265 2.71952 3.02158 3.15867 3.05383 2.67183 3.15867 |
| 14. | RPA2 (connector) | NNK | −4.70 | 358.90 μM | Cys49, Thr50, Ile76, Val77, Asp96, Met97, Tyr125, Phe155, His158, Ile159 | RPA2:VAL77:N - :NNK:O15 :NNK:O15 - RPA2:CYS49:O :NNK:O15 - RPA2:VAL77:O :NNK:O15 - RPA2:HIS158:NE2 |
3.09031 3.06724 3.18087 2.98064 |
| 15. | ATM | NNK | −4.48 | 523.49 μM | Thr2059, Ala2062, Gly2063, Ile2065, Gln2066, Gln2069, Leu2077, Tyr2080, Leu2081, Leu2084, Glu2094, Leu2095, Leu2098 | ATM:GLN2066:N - :NNK:O7 | 3.06063 |
| 16. | CDK4 | NNK | −4.35 | 650.98 μM | Val44, Leu54, Pro55, Thr58, Val59, Val62, Ala63, Arg66, Val82, Ile92, Val94 | CDK4:PRO55:CD - :NNK:O15 CDK4:VAL59:CA - :NNK:O7 |
2.9883 2.92285 |
| 17. | TFDP1 | NNK | −4.20 | 833.96 μM | Val264, Phe285, Asn286, Phe287, Phe291 | TFDP1:PHE287:N - :NNK:O15 :NNK:O15 - TFDP1:PHE287:O |
2.78538 3.08744 |
| 18. | TP53 | NNK | −4.14 | 927.72 μM | Gln136, Leu137, Ala138, His179, Cys182, Asp184, Asn239, Cys275, Ala276 | TP53:LEU137:N - :NNK:O15 TP53:ASN239:ND2 - :NNK:N14 TP53:ASN239:ND2 - :NNK:O15 :NNK:O15 - TP53:CYS275:O |
2.93164 3.07617 2.87469 2.82627 |
| 19. | RB1 | NNK | −4.02 | 1.14 mM | Val434, Gly435, Gln436, Cys438, Asn505, Leu506, Asp507, Ser508, Gly509, Thr510 | RB1:GLN436:HN - :NNK:O15 RB1:SER508:HN - :NNK:O7 RB1:GLY509:HN - :NNK:O7 |
1.93726 2.36625 1.90514 |
| 20. | RPA1 (connector) | NNK | −3.67 | 2.03 mM | Val375, Asn402, Pro403, Ala408, Tyr409, Arg412, Gly413 | RPA1:ARG412:NH1 - :NNK:N14 :NNK:O15 - RPA1:ALA408:O |
3.06161 2.78707 |
Figure 12.
Binding interactions of the top three proteins with NNK: (a) CDK7 (−5.93 Kcal/Mol), (b) CCNA1 (−5.6 Kcal/Mol), (c) CDKN1B (−5.42 Kcal/Mol).
Table 5.
Key proteins and their roles in cell cycle regulation.
| S.No | Protein | Role in Cell Cycle | References |
|---|---|---|---|
| 1. | CDK7 | Catalytic subunit of CAK complex which activates the cyclin-associated kinases CDK1, CDK2, CDK4, and CDK6 by threonine phosphorylation, thus regulating cell cycle progression | [28,29] |
| 2. | CCNA1 | Controls the cell cycle at the G1/S (start) and G2/M (mitosis) transitions | [30] |
| 3. | CDKN1B | Important regulator of cell cycle progression. Acts either as an inhibitor or an activator of cyclin type D-CDK4 complexes depending on its phosphorylation state and/or stoichometry. | [31] |
| 4. | CASP8 | Responsible for the positive and negative regulation of apoptosis and inflammation | [32] |
| 5. | CHEK2 | required for checkpoint-mediated cell cycle arrest in response to p53 defects | [33] |
| 6. | PLK1 | Regulates entry and exit from mitosis and cytokinesis. Regulates DNA replication | [34] |
| 7. | BID | induces apoptosis on interaction with BCl2 family proteins | [35] |
| 8. | HSP90AA1 | regulates the function of ATM in sensing and repairing the DNA damages | [36] |
| 9. | BRCA1 | Has role in DNA damage repair, cell cycle control and transcriptional regulation | [37] |
| 10. | CDK1 | Promotes G2-M transition and regulates G1 progress and G1-S transition via association with multiple interphase cyclins. | [38] |
| 11. | CDK2 | Essential for transition of cell cycle from G1 to S phase and then from S to G2 phase. | [39,40] |
| 12. | CCNB1 | Essential for the control of the cell cycle at the G2/M (mitosis) transition | [41] |
| 13. | CHEK1 | required for smooth cellular proliferation by inducing the degradation of cdc25a | [42] |
| 14. | RPA2 | Binds and stabilizes single-stranded DNA intermediates. Controls DNA repair and DNA damage checkpoint activation. | [43,44] |
| 15. | ATM | Activates checkpoint signaling upon double strand breaks (DSBs), apoptosis. May function as a tumor suppressor. | [45,46] |
| 16. | CDK4 | Hypophosphorylates RB1 in early G1 phase. Cyclin D-CDK4 complexes are major integrators of various mitogenic and antimitogenic signals. | [47] |
| 17. | TFDP1 | Can stimulate E2F-dependent transcription induces growth arrest. | [48] |
| 18. | TP53 | Prevents CDK7 kinase activity when associated to CAK complex in response to DNA damage, thus stopping cell cycle progression. | [49,50] |
| 19. | RB1 | Key regulator of entry into cell division that acts as a tumor suppressor. Promotes G0-G1 transition when phosphorylated by CDK3/cyclin-C. | [51] |
| 20. | RPA1 | controls DNA repair and DNA damage checkpoint activation | [52] |
4. Discussion
Cancer is a global inflammable problem, which pathologically occurs due to the accumulation of mutations in one or many genes, a phenomenon known as “oncogene addiction”. The loss of fidelity during the replication of DNA or the repair of damaged DNA leads to a cell becoming cancerous. Tobacco consumption has a major influence on these aberrations due to its main component, NNK, which has been strongly related with various cancers, mainly lung cancer [53,54]. NNK binds with various proteins involved in several cellular processes and leads to cancer. Hence, it is vital to fully understand the precise mechanism of the disease development caused by NNK and the interactome analysis of biomolecular target genes/proteins for attaining an overview of the highly complex functional interdependency of these targets for the efficient performance of the cell.
Systems biology is an interdisciplinary field that facilitates an understanding of complex biological network systems. These networks can be metabolic networks, gene regulation network, cell signaling network, or protein–protein interaction network [55]. With the help of system biology, we can analyze huge real-time networks precisely. Significantly, the huge amount of data from research being conducted globally were collected and statistically analyzed to uncover the functions of individual genes and proteins. The results may then be integrated to provide higher-level information [56]. Mathematical modeling and computational simulations help in understanding the internal dynamics of the system or the process and thus help in predicting the future of the process [57]. The networks created represent the genes or proteins with the nodes and their relations or physical connections with the edges [58]. The biological systems are immensely huge and complex networks depicting how one protein is connected with other proteins and how a slight impact on a gene or protein will affect the whole interactome [59]. These highly complicated and tightly packed networks, which govern all biological processes, are also referred to as protein–protein interaction networks (PPINs). All these graphs and networks work on the principle of graph theory. In the current study, we focused on NNK rewired PPIN to identify its bio-molecular targets that regulate the various cellular processes and cause cancer.
To construct PPIN, a literature survey was performed on PubMed database using keywords, like NNK, and around 544 genes extracted from 1317 research articles. The STRING database was used for the generation of the PPIN as it gives freedom to the users to choose the active interaction sources for their network. For this study, we used experimental data sources, like BIND, IntAct, etc., and pathway database interaction sources, like KEGG and reactome. The network generated had 534 nodes, including 100 connectors, at the highest confidence level score of 0.9. It contained 2909 edges that depicted that all the reported nodes were well connected with each other, which were further confirmed by the topological parameter, i.e., the average node degree, which was found to be 10.9. The degree of a node gives the information about the connection of the node with other nodes or simply the number of edges that either enter a node or leave it. All the biological systems are undirected and scale-free so the node degree depicts the average connectivity of each node with other nodes present in the network. The identification of key nodes in any network is not possible just by calculating its degree, hence various other parameters require evaluation, like the betweenness centrality, closeness centrality, and bottleneck scores [60]. Nodes with clustering coefficients less than 0.5 are key nodes because lesser clustering coefficients depict that the protein still have space for interaction with other molecules. The average neighborhood connectivity details provide the information about the density of the network. The shortest path length is important as it gives details about the minimum number of the edges between the two nodes and how fast information can pass on from one node to another node.
The network generated was then divided into modules. Modularization is an important step as it helps in reducing the noise of the data. On modularization, 19 clusters were obtained and each cluster contained seed proteins. Finally, 115 seed proteins were procured from the initial list of 534 proteins. Again, a PPIN was generated using these 115 seed proteins and 100 connectors at the first and second shells, and we obtained 213 nodes and 2509 edges. In addition, 100 connectors were done in order to include some more relevant proteins, which were possibly missed in the above procedure. The topological analysis was done on the basis of clustering coefficient and bottleneck scores. We used the bottleneck score as our final selection criteria as bottlenecks are the significant proteins, which have great functional and dynamic significance in a network [61]. To find the statistically significant cut-off for the proteins to be selected as key targets, the median was calculated on the bottleneck score, which came out to be 2. The enrichment analysis was done to find the pathways and processes that are enriched in the process of the PPIN generation. On the GO enrichment analysis, it was found that most of the seed proteins fall under the pathways that are related to cell cycle regulation and these results were also reflected on the final selected seed proteins that had a bottleneck score of 2 or more than 2. The cell cycle is a tightly regulated and a precisely timed event that is divided into phases and is monitored by checkpoints [62]. It is a highly orchestrated event, which ensures the integrity and stability of the genome. NNK binds to various receptors and hampers various pathways, leading to the cell becoming cancerous [63]. It not only binds with the receptors but also targets many important genes and proteins, like CDKs and cyclins, that are crucial for cell cycle processes [6]. Table 3 and Figure 10, Supplementary Figure S1, and Figure 11 clearly depict that most of the pathways that were enriched fall under cell cycle regulation at different phases.
Finally, docking studies were performed for the target proteins using NNK as a ligand. Molecular docking is the key tool to check whether the binding of ligand and protein is thermodynamically possible or not. The binding energies of these docking results further helped in refining the targets for NNK. The major targets of NNK found were CDK7, CCNA1, CDKN1B, CASP8, CHEK2, PLK1, BID, HSP90AA1, BRCA1, CDK1, CDK2, CCNB1, CHEK1, RPA2, ATM, CDK4, TFDP1, TP53, RB1, and RPA1, with their binding energies ranging from −5.93 to −3.67 Kcal/Mol. PYCARD and QXOS1 were excluded from this screening because of their low degree and 0 clustering coefficient. In addition to the above key proteins, our present study also revealed five connector proteins, which can be proven as putative biomolecular targets of NNK. These proteins are CHEK2, CCNA1, BID, RPA1, and RPA2. From these docking results, we can easily conclude that a majority of the key targets of NNK belong to the cell cycle regulatory proteome. CDKs and cyclins are the key regulatory proteins of the cell cycle that control the various phases of the cell. Different CDK/cyclin complexes are required for a smooth transition of the cell cycle. Other proteins, like ATM, RB1, and TP53, may act as tumor suppressor proteins and signaling proteins for DNA damage. Table 5 enlists the final key biomolecular targets of NNK and their role in the cell cycle.
5. Conclusions
In conclusion, a NNK rewired PPIN, with the help of various systems biology tools, was explored for the identification of potential targets involved in tobacco induced cancer development. The biomolecules that were suspected for being the most probable targets of NNK were further screened using functional enrichment techniques and then using molecular docking techniques. The results of this study show that the maximum proteins that are the most probable targets of NNK are involved in cell cycle regulation. With these, we can conclude that NNK has a major impact on the biomolecules of the cell cycle regulatory proteome. The present study opens future possibilities to identify new biomarkers for the cancers associated with NNK. This can also help in the pre-symptomatic diagnosis of diseases and in the development of precision medicines.
Acknowledgments
The authors are grateful to Vijay Dhasmana, Honorable Vice Chancellor Swami Rama Himalayan University, India, Sunil Saini Chairperson Research Committee, Head of Department of Biosciences, other faculty members of Biosciences and all members of Research Committee—Himalayan Institute of Medical Sciences, Swami Rama Himalayan University, India, for providing the motivation and necessary facilities for this study.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/10/8/564/s1, Figure S1: Graph of GO enriched pathways depicting the involvement of genes under various sub-graphs.
Author Contributions
Conceptualization, A.D., U.B., M.G., and K.K.K.; methodology, A.D., A.N.; software, A.N., M.L.; validation, A.D., A.N., D.K.; formal analysis, A.D., S.U., P.S., U.B., M.G., S.H. and K.K.K.; investigation, A.N., A.D., S.H.; resources, A.D., K.K.K.; data curation, A.D., A.N., S.U. and P.S.; writing—original draft preparation, A.D., A.N., K.K.K.; writing—review and editing, D.K., M.L., J.R.; visualization, A.N.; supervision, A.D.; project administration, A.D.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Pavlopoulou A., Spandidos D.A., Michalopoulos I. Human cancer databases (Review) Oncol. Rep. 2015;33:3–18. doi: 10.3892/or.2014.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F., Colombet M., Mery L., Piñeros M., Znaor A., Zanetti R., Ferlay J. Cancer Incidence in Five Continents. Volume XI IARC; Lyon, France: 2017. [Google Scholar]
- 3.Weinstein I.B., Joe A.K. Mechanisms of Disease: Oncogene addiction—A rationale for molecular targeting in cancer therapy. Nat. Rev. Clin. Oncol. 2006;3:448–457. doi: 10.1038/ncponc0558. [DOI] [PubMed] [Google Scholar]
- 4.Forouzanfar M.H., Afshin A., Alexander L.T., Anderson H.R., Bhutta Z.A., Biryukov S., Brauer M., Burnett R., Cercy K., Charlson F.J., et al. GBD 2015 Risk Factors Collaborators. Global, regional, and national comparative risk assessment of 79 behavioural, environmental and occupational, and metabolic risks or clusters of risks, 1990–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388:1659–1724. doi: 10.1016/S0140-6736(16)31679-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hecht S.S. Tobacco smoke carcinogens and breast cancer. Environ. Mol. Mutagen. 2002;39:119–126. doi: 10.1002/em.10071. [DOI] [PubMed] [Google Scholar]
- 6.Haneef M., Lohani M., Dhasmana A., Jamal Q.M., Shahid S.M., Firdaus S. Molecular Docking of Known Carcinogen 4-(Methyl-nitrosamino)-1-(3-pyridyl)-1-butanone (NNK) with Cyclin Dependent Kinases towards Its Potential Role in Cell Cycle Perturbation. Bioinformation. 2014;10:526–532. doi: 10.6026/97320630010526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xue J., Yang S., Seng S. Mechanisms of Cancer Induction by Tobacco-Specific NNK and NNN. Cancers. 2014;6:1138–1156. doi: 10.3390/cancers6021138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zabala V., Tong M., Yu R., Ramirez T., Yalcin E.B., Balbo S., Silbermann E., Deochand C., Nunez K., Hecht S., et al. Potential Contributions of the Tobacco Nicotine-Derived Nitrosamine Ketone (NNK) in the Pathogenesis of Steatohepatitis in a Chronic Plus Binge Rat Model of Alcoholic Liver Disease. Alcohol Alcohol. 2015;50:118–131. doi: 10.1093/alcalc/agu083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nunez K., Kay J., Krotow A., Tong M., Agarwal A.R. Cigarette Smoke-Induced Alterations in Frontal White Matter Lipid Profiles Demonstrated by MALDI-Imaging Mass Spectrometry: Relevance to Alzheimer’s Disease. J. Alzheimers Dis. 2016;51:151–163. doi: 10.3233/JAD-150916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pai M., Mohan A., Dheda K., Leung C.C., Yew W.W., Christopher D.J., Sharma S.K. Lethal Interaction: The colliding epidemics of tobacco and tuberculosis. Expert Rev. Anti Infect. Ther. 2007;5:385–391. doi: 10.1586/14787210.5.3.385. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y., Zhang X.S., Chen L. Modelling biological systems from molecules to dynamical networks. BMC Syst. Biol. 2012;6(Suppl. 1):2–5. doi: 10.1186/1752-0509-6-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Villaverde A.F., Ross J., Banga J.R. Reverse engineering cellular networks with information theoretic methods. Cells. 2013;2:306–329. doi: 10.3390/cells2020306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P., et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doncheva N.T., Assenov Y., Domingues F.S., Albrecht M. Topological analysis and interactive visualization of biological networks and protein structures. Nat. Protoc. 2012;7:670–685. doi: 10.1038/nprot.2012.004. [DOI] [PubMed] [Google Scholar]
- 16.Barabasi A.L., Oltvai Z.N. Network biology: Understanding the cell’s functional organization. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 17.Almaas E. Biological impacts and context of network theory. J. Exp. Biol. 2007;210:1548–1558. doi: 10.1242/jeb.003731. [DOI] [PubMed] [Google Scholar]
- 18.Klemm K., Bornholdt S. Topology of biological networks and reliability of information processing. Proc. Natl. Acad. Sci. USA. 2005;102:18414–18419. doi: 10.1073/pnas.0509132102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cingovska I., Kovachev A., Gmbh A.T., Trivodaliev K., Kalajdziski S. Protein Function Prediction by Clustering of Protein-Protein Interaction Network. In: Kocarev L., editor. Proceedings of the ICT Innovations 2011. Volume 150. Springer; Berlin, Germany: 2012. Advances in Intelligent and Soft Computing. [DOI] [Google Scholar]
- 20.Rajendran P., Thangavel D. Clustering of Microarray Data to Identify Enriched Go Terms of Genes in Severe Asthma Dataset using Gene Enrichment Analysis. Ind. J. Sci. Tech. 2016;9:1–11. doi: 10.17485/ijst/2016/v9i8/86068. [DOI] [Google Scholar]
- 21.Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W.H., Pagès F., Trajanoski Z., Galon J., et al. Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dhasmana A., Jamal Q.M., Gupta R., Siddiqui M.H., Kesari K.K., Wadhwa G., Khan S., Haque S., Lohani M. Titanium dioxide nanoparticles provide protection against polycyclic aromatic hydrocarbon BaP and chrysene-induced perturbation of DNA repair machinery: A computational biology approach. Biotechnol. Appl. Biochem. 2016;63:497–513. doi: 10.1002/bab.1388. [DOI] [PubMed] [Google Scholar]
- 23.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huberman B., Alto P., Kaneko K., Ouyang Q. Handbook on Biological Networks. Volume 10. World Scientific Publishing Co. Pte. Ltd. 5 Toh Tuck Link; Singapore: 2009. [Google Scholar]
- 25.Junker B.H., Schreiber F. Analysis of Biological Networks. A Wiley Interscience Publication; New York, NY, USA: 2008. [Google Scholar]
- 26.Pavlopoulos G.A., Maria Secrier M., Moschopoulos C.N., Soldatos T.G., Kossida S., Aerts J., Schneider R., Bagos P.G. Using graph theory to analyze biological networks. BioData Min. 2011;4:10. doi: 10.1186/1756-0381-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chin C.H., Chen S.H., Wu H.H., Ho C.W., Ko M.T., Lin C.Y. Cytohubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014;8(Suppl. 4):S11. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fisher R.P. Secrets of a double agent: CDK7 in cell-cycle control and transcription. J. Cell Sci. 2005;118:5171–5180. doi: 10.1242/jcs.02718. [DOI] [PubMed] [Google Scholar]
- 29.Bartkova J., Zemanova M., Bartek J. Expression of CDK7/CAK in normal and tumor cells of diverse histogenesis, cell-cycleposition and differentiation. Int. J. Cancer. 1996;66:732–737. doi: 10.1002/(SICI)1097-0215(19960611)66:6<732::AID-IJC4>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 30.Yang R., Müller C., Huynh V., Fung Y.K., Yee A.S., Koeffler H.P. Functions of cyclin A1 in the cell cycle and its interactions with transcription factor E2F-1 and the Rb family of proteins. Mol. Cell. Biol. 1999;19:2400–2407. doi: 10.1128/MCB.19.3.2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ray A., James M.K., Larochelle S., Fisher R.P., Blain S.W. P27kip1 inhibits cyclin D-cyclin-dependent kinase 4 by two independent modes. Mol. Cell. Biol. 2009;29:986–999. doi: 10.1128/MCB.00898-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liccardi G., Ramos G.L., Tenev T., Annibaldi A., Legrand A.J., Robertson D., Feltham R., Anderton H., Darding M., Peltzer N., et al. RIPK1 and Caspase-8 Ensure Chromosome Stability Independently of Their Role in Cell Death and Inflammation. Mol. Cell. 2019;73:413–428. doi: 10.1016/j.molcel.2018.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Furusawa Y., Yamanouchi Y., Iizumi T., Zhao Q., Mitsuhashi Y., Morita A., Enomoto A., Tabuchi Y., Kondo T. Checkpoint kinase 2 is dispensable for regulation of the p53 response but is required for G2/M arrest and cell survival in cells with p53 defects under heat stress. Apoptosis. 2017;22:1225–1234. doi: 10.1007/s10495-017-1402-2. [DOI] [PubMed] [Google Scholar]
- 34.Giráldez S., Galindo-Moreno M., Limón-Mortés M.C., Rivas A.C., Herrero-Ruiz J., Mora-Santos M., Sáez C., Japón M.Á., Tortolero M., Romero F. G1/S phase progression is regulated by PLK1 degradation through the CDK1/βTrCP axis. FASEB J. 2017;31:2925–2936. doi: 10.1096/fj.201601108R. [DOI] [PubMed] [Google Scholar]
- 35.Renshaw S.A., Dempsey C.E., Barnes F.A., Bagstaff S.M., Dower S.K., Bingle C.D., Whyte M.K. Three novel Bid proteins generated by alternative splicing of the human Bid gene. J. Biol. Chem. 2004;279:2846–2855. doi: 10.1074/jbc.M309769200. [DOI] [PubMed] [Google Scholar]
- 36.Pennisi R., Antoccia A., Leone S., Ascenzi P., di Masi A. Hsp90α regulates ATM and NBN functions in sensing and repair of DNA double-strand breaks. FEBS J. 2017;284:2378–2395. doi: 10.1111/febs.14145. [DOI] [PubMed] [Google Scholar]
- 37.Mullan P.B., Quinn J.E., Harkin D.P. The role of BRCA1 in transcriptional regulation and cell cycle control. Oncogene. 2006;25:5854–5863. doi: 10.1038/sj.onc.1209872. [DOI] [PubMed] [Google Scholar]
- 38.Timofeev O., Cizmecioglu O., Settele F., Kempf T., Hoffmann I. Cdc25 phosphatases are required for timely assembly of CDK1-cyclin B at the G2/M transition. J. Biol. Chem. 2010;285:16978–16990. doi: 10.1074/jbc.M109.096552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neganova I., Vilella F., Atkinson S.P., Lloret M., Passos J.F., von Zglinicki T., O’Connor J.E., Burks D., Jones R., Armstrong L., et al. An important role for CDK2 in G1 to S checkpoint activation and DNA damage response in human embryonic stem cells. Stem Cells. 2011;29:651–659. doi: 10.1002/stem.620. [DOI] [PubMed] [Google Scholar]
- 40.De Boer L., Oakes V., Beamish H., Giles N., Stevens F., Somodevilla-Torres M., Desouza C., Gabrielli B. Cyclin A/cdk2 coordinates centrosomal and nuclear mitotic events. Oncogene. 2008;27:4261–4268. doi: 10.1038/onc.2008.74. [DOI] [PubMed] [Google Scholar]
- 41.Brown N.R., Lowe E.D., Petri E., Skamnaki V., Antrobus R., Johnson L.N. Cyclin B and cyclin A confer different substrate recognition properties on CDK2. Cell Cycle. 2007;6:1350–1359. doi: 10.4161/cc.6.11.4278. [DOI] [PubMed] [Google Scholar]
- 42.Goto H., Natsume T., Kanemaki M.T., Kaito A., Wang S., Gabazza E.C., Inagaki M., Mizoguchi A. Chk1-mediated Cdc25A degradation as a critical mechanism for normal cell cycle progression. J. Cell Sci. 2019;132:jcs223123. doi: 10.1242/jcs.223123. [DOI] [PubMed] [Google Scholar]
- 43.Sleeth K.M., Sørensen C.S., Issaeva N., Dziegielewski J., Bartek J., Helleday T. RPA mediates recombination repair during replication stress and is displaced from DNA by checkpoint signalling in human cells. J. Mol. Biol. 2007;373:38–47. doi: 10.1016/j.jmb.2007.07.068. [DOI] [PubMed] [Google Scholar]
- 44.Grudic A., Jul-Larsen A., Haring S.J., Wold M.S., Lønning P.E., Bjerkvig R., Bøe S.O. Replication protein A prevents accumulation of single-stranded telomeric DNA in cells that use alternative lengthening of telomeres. Nucleic Acids Res. 2007;35:7267–7278. doi: 10.1093/nar/gkm738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bakkenist C.J., Kastan M.B. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 46.Taira N., Yamamoto H., Yamaguchi T., Miki Y., Yoshida K. ATM augments nuclear stabilization of DYRK2 by inhibiting MDM2 in the apoptotic response to DNA damage. J. Biol. Chem. 2010;285:4909–4919. doi: 10.1074/jbc.M109.042341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Z., Xie Y., Zhang L., Zhang H., An X., Wang T., Meng A. Migratory localization of cyclin D2-Cdk4 complex suggests a spatial regulation of the G1-S transition. Cell Struct. Funct. 2008;33:171–183. doi: 10.1247/csf.08019. [DOI] [PubMed] [Google Scholar]
- 48.Zaragoza K., Bégay V., Schuetz A., Heinemann U., Leutz A. Repression of transcriptional activity of C/EBPalpha by E2F-dimerization partner complexes. Mol. Cell. Biol. 2010;30:2293–2304. doi: 10.1128/MCB.01619-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo A., Salomoni P., Luo J., Shih A., Zhong S., Gu W., Pandolfi P.P. The function of PML in p53-dependent apoptosis. Nat. Cell Biol. 2000;2:730–736. doi: 10.1038/35036365. [DOI] [PubMed] [Google Scholar]
- 50.Huarte M., Guttman M., Feldser D., Garber M., Koziol M.J., Kenzelmann-Broz D., Khalil A.M., Zuk O., Amit I., Rabani M., et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ren S., Rollins B.J. Cyclin C/cdk3 promotes Rb-dependent G0 exit. Cell. 2004;117:239–251. doi: 10.1016/S0092-8674(04)00300-9. [DOI] [PubMed] [Google Scholar]
- 52.Maréchal A., Li J.M., Ji X.Y., Wu C.S., Yazinski S.A., Nguyen H.D., Liu S., Jiménez A.E., Jin J., Zou L. PRP19 transforms into a sensor of RPA-ssDNA after DNA damage and drives ATR activation via a ubiquitin-mediated circuitry. Mol. Cell. 2014;53:235–246. doi: 10.1016/j.molcel.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jamal Q.M.S., Dhasmana A., Lohani M., Firdaus S., Ansari M.Y., Sahoo G.C., Haque S. Binding pattern elucidation of NNK and NNAL cigarette smoke carcinogens with NER pathway enzymes: An onco-informatics study. Asian Pac. J. Cancer Prev. 2015;16:5311–5317. doi: 10.7314/APJCP.2015.16.13.5311. [DOI] [PubMed] [Google Scholar]
- 54.Huang R.Y., Li M.Y., Hsin M.K., Underwood M.J., Ma L.T., Mok T.S., Warner T.D., Chen G.G. 4-Methylnitrosamino-1-3-pyridyl-1-butanone (NNK) promotes lung cancer cell survival by stimulating thromboxane A 2 and its receptor. Oncogene. 2011;30:106–116. doi: 10.1038/onc.2010.390. [DOI] [PubMed] [Google Scholar]
- 55.Boccaletti S., Latora V., Moreno Y., Chavez M., Hwang D.U. Complex networks: Structure and dynamics. Phys. Rep. 2006;424:175–308. doi: 10.1016/j.physrep.2005.10.009. [DOI] [Google Scholar]
- 56.Koutsogiannouli E., Papavassiliou A.G., Papanikolaou N.A. Complexity in cancer biology: Is systems biology the answer? Cancer Med. 2013;2:164–177. doi: 10.1002/cam4.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kitano H. Systems Biology: A brief overview. Science. 2002;295:162–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- 58.Bosley A.D., Das S., Andresson T. A Role for Protein–Protein Interaction Networks in the Identification and Characterization of Potential Biomarkers. In: Issaq H.J., editor. Proteomic and Metabolomic Approaches to Biomarker Discovery. Academic Press; London, UK: 2013. pp. 333–347. Chapter 21. [Google Scholar]
- 59.Klipp E., Liebermeister W., Wierling C., Kowald A., Lehrach H., Herwig R. Systems Biology: A Textbook. Wiley-VCH; Weinheim, Germany: 2009. [Google Scholar]
- 60.Liu J., Xiong Q., Shi W., Shi X., Wang K. Evaluating the importance of nodes in complex networks. Phys. A Stat. Mech. Appl. 2016;452:209–219. doi: 10.1016/j.physa.2016.02.049. [DOI] [Google Scholar]
- 61.Yu H., Kim P.M., Sprecher E., Trifonov V., Gerstein M. The importance of bottlenecks in protein networks: Correlation with gene essentiality and expression dynamics. PLoS Comput. Biol. 2007;3:713–720. doi: 10.1371/journal.pcbi.0030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hustedt N., Durocher D. The control of DNA repair by the cell cycle. Nat. Cell Boil. 2017;19:1–9. doi: 10.1038/ncb3452. [DOI] [PubMed] [Google Scholar]
- 63.Huang R.Y., Chen G.G. Cigarette smoking, cyclooxygenase-2 pathway and cancer. Biochim. Biophys. Acta Rev. Cancer. 2011;1815:158–169. doi: 10.1016/j.bbcan.2010.11.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.