Abstract
Background
HLA-DRB1 is the strongest susceptibility gene to rheumatoid arthritis (RA). HLA-DRB1 alleles showed significant non-additive and interactive effects on susceptibility to RA in the European population, but these effects on RA susceptibility should vary between populations due to the difference in allelic distribution. Furthermore, non-additive or interactive effects on the phenotypes of RA are not fully known. We evaluated the non-additive and interactive effects of HLA-DRB1 alleles on RA susceptibility and anticitrullinated protein/peptide antibody (ACPA) levels in Japanese patients.
Methods
A total of 5581 ACPA(+) RA and 19 170 controls were genotyped or imputed for HLA-DRB1 alleles. Logistic regression analysis was performed for both allelic non-additive effects and interactive effects of allelic combinations. The significant levels were set by Bonferroni’s correction. A total of 4371 ACPA(+) RA were analysed for ACPA levels.
Results
We obtained evidence of non-additive and interactive effects of HLA-DRB1 on ACPA(+) RA susceptibility (p=2.5×10−5 and 1.5×10−17, respectively). Multiple HLA-DRB1 alleles including HLA-DRB1*04:05, the most common susceptibility allele in the Japanese, showed significant non-additive effects (p≤0.0043). We identified multiple allelic combinations with significant interactive effects including a common combination with the European population as well as novel combinations. Additional variance of ACPA(+) RA susceptibility could be explained substantially by heterozygote dominance or interactive effects. We did not find evidence of non-additive and interactive effects on levels of ACPA.
Conclusion
HLA allelic non-additive and interactive effects on ACPA(+) RA susceptibility were observed in the Japanese population. The allelic non-additive and interactive effects depend on allelic distribution in populations.
INTRODUCTION
Rheumatoid arthritis (RA) is the most frequent autoimmune joint-destructive disease with a prevalence of 0.5%–1.0% worldwide.1,2 Anticitrullinated protein/peptide antibody (ACPA) is a highly specific antibody to RA. Previous studies have revealed involvement of environmental factors such as smoking and genetic components with the onset and disease progression of RA.1,2
The HLA region is the strongest susceptibility locus to RA, and the HLA-DRB1 region shows the strongest susceptibility association in the HLA region.3 ACPA-positive RA and ACPA-negative RA have very different association patterns in the HLA region.4–7 Shared epitope (SE) is a predefined allelic group having common amino acid sequences at positions 70–74 in the HLA-DRB1 protein and has been widely accepted to explain the majority of the association between the HLA region and ACPA-positive RA.8 Recent analyses taking advantages of genome-wide association study (GWAS) combined with imputation revealed that the primary association signal in the HLA region was driven by amino acid positions 11, 71 and 74 in HLA-DRB1 and the remaining associations could be explained by specific amino acid positions in HLA-B, DPB1 and A in the European population.5,9 A following study in the Asian population showed similar genetic architectures to the European population with an Asian-specific association at amino acid position 57.10 In addition to susceptibility to ACPA-positive RA, HLA-DRB1 alleles or amino acid residues are strongly associated with RA phenotypes including levels of ACPA and joint destruction, which is independent of ACPA positivity.11–14
Previous studies have suggested the effect of HLA-DRB1 allelic interactions on RA susceptibility.15,16 A recent genetic study revealed that the effects of HLA-DRB1 allelic dominance or interactions demonstrate a substantial genetic influence on ACPA-positive RA susceptibility in the European population.17 The allelic combinations included DRB1*03:01 and DRB1*04:01, which are frequent alleles in Europeans but rare or uncommon in Asian populations. Since HLA allelic distribution is different between populations, there is a possibility that undiscovered HLA allelic interactions confer RA susceptibility in the Asian population. While our previous study suggested a non-additive effect of Japanese susceptibility alleles,18 detailed interactive analyses have not been conducted to date. Furthermore, HLA allelic interactive effects on RA phenotypes are not yet known. While the original report used assessment of HLA-DRB1 allelic interaction on RA susceptibility in logistic regression model, we can expand this statistical framework into general linear regression model to assume interactive effects on quantitative traits.
Here, we analysed HLA allelic interactive associations with ACPA-positive RA and RA phenotypes by using a total of 24 751 case–control subjects in the Japanese population.
METHODS
Subjects
A total of 5581 patients with RA who were positive for CCP2 and 19 170 control subjects in two data sets were included for ACPA(+) RA susceptibility in the current study. The breakdown of the subjects is shown in table 1. All cases fulfilled the 1987 American College of Rheumatology (ACR) or the 2010 ACR/European League Against Rheumatism RA classification criteria.19,20 As for ACPA levels, 4371 subjects whose data were analysed in our previous genetic association study for ACPA levels were used.11 Most of the subjects in analyses of ACPA levels were included for ACPA(+) RA susceptibility. We also combined data of ACPA(+) RA with that of 931 ACPA(−) RA for analyses of ACPA levels. Written informed consent was obtained from each patient. This study was approved by the ethical review board in each institution.
Table 1.
Characteristics of the study population
| ACPA(+) RA susceptibility | ACPA levels | |
|---|---|---|
| Set 1 | ||
| No of subjects | 3062 vs 2008 | 2488 |
| Institution | Kyoto University, Tokyo Women’s Medical University | Kyoto University, Tokyo Women’s Medical University |
| HLA-DRB1 | Genotyping | Genotyping |
| Set 2 | ||
| No of subjects | 2519 vs 17162 | 1883 |
| Institution | RIKEN | RIKEN |
| HLA-DRB1 | Imputation | Genotyping |
ACPA, anticitrullinated protein/peptide antibody; RA, rheumatoid arthritis.
HLA-DRB1 genotyping
HLA-DRB1 alleles were imputed for subjects in set 2 for ACPA(+) RA susceptibility based on genotypes of single-nucleotide polymorphisms in GWAS by SNP2HLA program21 with the use of the Japanese reference panel.22 The details of the subjects were described in the previous study.18 SNP2HLA gives us integer values of dosages of each HLA allele. The concordance of HLA-DRB1 alleles was 0.972. Since it is not favourable to mix genotyping results and imputation results in the same set, we used only imputation results even when both genotyping and imputation results were available for a subset of subjects in set 2. As for the other subjects, HLA-DRB1 alleles were genotyped by a WAK-flow system or an AlleleSEQR HLA-DRB1 typing kit (Abbott).
CCP2 quantification and detection of its levels
We used the MESACUP CCP ELISA kit (Medical and Biological Laboratories) to quantify positivity or levels of second-generation ACPAs in the patients with RA, based on the manufacturer’s specifications. The details were shown in our previous paper.11
Statistical analysis
Statistical framework to evaluate non-additive effects
The analysis was conducted by the same methods described in the paper reporting HLA allelic interactions on susceptibility to autoimmune diseases.17 Briefly, a heterozygote effect or a homozygote effect of certain HLA-DRB1 allele was assessed in the data set in which we excluded homozygote or heterozygote subjects of the allele, respectively, in logistic regression model with indicator variable of whether each subject belonged to set 2 as a covariate.
Overall non-additive effects of HLA-DRB1 alleles were evaluated by comparing the following two models:
| (model 1) |
| (model 2) |
and the non-additive effect in each HLA-DRB1 allele was evaluated by comparing the following two models:
| (model 3) |
| (model 4) |
where logiti is logit in the th individual; βA,k and βD,k are additive and dominant effect sizes of the kth HLA-DRB1 allele, respectively; γk,i is dosage of the kth HLA-DRB1 allele in the th individual; δk J is an indicator variable whether the th individual is heterozygous for the kth HLA-DRB1 allele; n is number of HLA-DRB1 alleles in data set; β1 is an effect size of set 1 as a covariate; δ1,J is an indicator variable whether the th individual belongs to set 1; and θ represents generalised logistic regression intercept. We compared different models with the χ2 test by calculating deviance statistics, which follow a χ2 distribution with degree of freedom of u, where u is the difference in degree of freedom of the two models. Deviance statistics are equal to product of −2 and log likelihood for the model tested.
Interactive effects were evaluated by the following formula:
| (model 5) |
where βI,k l is an effect size of a combination of the kth and the th alleles, δk,l,i is an indicator variable whether the th individual is heterozygous for the kth and the th alleles.
Since assessment of non-additive effects requires a substantial number of homozygotes of the alleles being evaluated and our data included imputation data in which it is not practical to expect high imputation accuracy for rare alleles, we adopted HLA alleles with more than 10 homozygotes in cases as the full data set or HLA alleles with frequencies of more than 5% in control samples as the common data set. The definition of these two sets was according to the European study.17 We used the full data set as a standard data and confirmed the results in the common data set. Statistical significance levels were determined based on Bonferroni’s method based on numbers of multiple testing in the analyses.
Non-additive effects of amino acid positions
We obtained amino acid sequences of HLA-DRB1 alleles from the HLA-IMGT database.23 We applied the abovementioned statistical framework on amino acid positions. Considering the strong linkage equilibrium between amino acid residues in different positions, it is not practical to analyse interactive effects between amino acid residues. Thus, we sequentially analysed non-additive effects of all amino acid residues at given positions, which showed strong signals in the additive model, namely, amino acid positions 11, 74, 47 and 37 for ACPA(+) RA susceptibility and 74 and 57 for ACPA levels. In this model, we conditioned on each amino acid position sequentially by omnibus test9,12 to assess non-additive effects of the HLA-DRB1 amino acid residues. For example, when we assessed non-additive effects of amino acid residues at position 47 on ACPA(+) susceptibility, we conditioned on amino acid positions 11 and 74.
Variance explained
We calculated variance explained in the full data set by additive, dominance (model 1) and interactive (model 5) models, respectively, based on liability threshold model. We assume that all subjects have quantitative risk scores to develop RA with a mean of 0 and a variance of 1 and that subjects whose risk scores exceed a threshold develop RA. We set prevalence of RA as 1%, and based on this, we used OR of HLA haplotypes or combinations as approximation of the relative risk of RA to compute prevalence in groups based on HLA-DRB1 alleles. Prevalence was inversely mapped to normal distribution with a mean of 0 and a variance of 1. Variance explained was defined as follows;
where Vexp, Vbetween and Vwithin are variance explained, variance between groups based on HLA-DRB1 alleles and variance within groups which is equal to 1, respectively.
Expansion of statistical framework to quantitative traits
We expanded the methods written above to a quantitative trait, namely, the levels of ACPA. We used the levels of ACPA as a dependent variable in general linear regression model and the same statistical framework was applied.
Others
To confirm imputation accuracy, we compared allele frequencies of control subjects between sets 1 and 2 by Spearman’s correlation coefficient. Statistical analyses were conducted by R software.
RESULTS
We recruited a total of 5581 RA cases positive for ACPA and 19 171 control subjects in two different sets. At first, we assessed allelic frequencies in control samples. The two data sets showed a very good correlation of allele frequencies (r=0.99, 1; see online supplementary figure 1). We constructed a data set as the full data set in which each subject carried two of the following alleles: HLA-DRB1*01:01, HLA-DRB1*04:05, HLA-DRB1*09:01, HLA-DRB1*15:01, HLA-DRB1*15:02, HLA-DRB1*13:02 and HLA-DRB1*08:03. There were more than 10 homozygotes of all these alleles in case subjects. We also constructed a common data set, another data set composed of only HLA-DRB1*01:01, HLA-DRB1*04:05, HLA-DRB1*09:01, HLA-DRB1*15:01 and HLA-DRB1*15:02, all of which exceeded allele frequencies over 5% in control subjects in each of the two sets to confirm findings in the full data set (for details, see the Methods section). Among alleles in the full data set, DRB1*01:01 and DRB1*15:01 were common between the current data set and the previous European study.
When we assessed an overall non-additive effect, we found that combination of additive and non-additive effects significantly improved fit in comparison with model of additive effects only (p=2.5×10−5). This result indicates that HLA-DRB1 alleles show dominance effects on ACPA(+) RA susceptibility as a whole. When we evaluated each allelic non-additive effect, HLA-DRB1*04:05 showed a significant result (p=3.8×10−4, table 2), compatible with our previous study.18 HLA-DRB1*08:03 and HLA-DRB1*15:02 also showed significant results (p=4.3×10−3 and 2.4×10–3, respectively, table 2). While HLA-DRB1*09:01 is the second strongest susceptibility allele to RA in the Asian population, it did not show an evidence of non-additive effect (p=0.37, table 2). While DRB1*01:01 is a strong susceptibility allele to RA especially in Europeans and showed significant non-additive effects in European population, its effect was not very apparent in the current study (p=0.39, table 2). The common data set confirmed the non-additive effects as a whole (p=4.8×10−7) and in the alleles mentioned above (see online supplementary table 1).
Table 2.
Non-additive HLA-DRB1 allelic effects on ACPA(+) rheumatoid arthritis susceptibility in the full data set
| Non-additive p | Heterozygote OR (95% CI) | Homozygote OR* (95% CI) | |
|---|---|---|---|
| DRB1*01:01 | 0.39 | 1.09 (0.96 to 1.24) | 0.95 (0.72 to 1.27) |
| DRB1*04:05 | 0.00038 | 2.57 (2.33 to 2.85) | 2.07 (1.89 to 2.27) |
| DRB1*08:03 | 0.0043 | 0.47 (0.41 to 0.54) | 0.70 (0.56 to 0.87) |
| DRB1*09:01 | 0.37 | 1.12 (1.01 to 1.23) | 1.18 (1.07 to 1.31) |
| DRB1*13:02 | 0.54 | 0.43 (0.37 to 0.5) | 0.48 (0.35 to 0.66) |
| DRB1*15:01 | 0.11 | 0.70 (0.61 to 0.79) | 0.56 (0.43 to 0.73) |
| DRB1*15:02 | 0.0024 | 0.78 (0.7 to 0.87) | 0.56 (0.46 to 0.68) |
OR per allele.
ACPA, anticitrullinated protein/peptide antibody.
Next, we assessed whether the significant non-additive effects were driven by interactive effects. We found improvement in model fitting by additive and interactive terms in comparison with additive and non-additive terms (p=1.5×10−17). When we assessed interactive effect in each combination, we found five allelic combinations showing significant interactive effects (p<2.2×10−3, table 3). These combinations include a combination of HLA-DRB1*01:01 and HLA-DRB1*15:01, which showed a significant interactive effect in the European study. We did not observe a significant interactive effect between HLA-DRB1*04:05 and HLA-DRB1*09:01 (p=0.13, table 3), the strongest and the second strongest susceptibility allele to ACPA(+) RA in the Asian populations and whose interactive effect was previously reported in the Korean population.24 These results were confirmed in the common data set (see online supplementary table 2).
Table 3.
Interactive effects of allelic combinations in the full data set
| DRB1*01:01 | DRB1*04:05 | DRB1*08:03 | DRB1*09:01 | DRB1*13:02 | DRB1*15:01 | DRB1*15:02 | |
|---|---|---|---|---|---|---|---|
| DRB1*01:01 | - | 1.17 (0.81 to 1.7) | 1.05 (0.63 to 1.76) | 0.81 (0.55 to 1.21) | 0.37 (0.18 to 0.77) | 2.23 (1.33 to 3.72) | 2.37 (1.55 to 3.62) |
| DRB1*04:05 | 0.40 | - | 0.46 (0.32 to 0.66) | 1.15 (0.96 to 1.38) | 1.47 (0.99 to 2.19) | 1.73 (1.23 to 2.44) | 1.78 (1.36 to 2.32) |
| DRB1*08:03 | 0.85 | 2.9×10−5 | - | 1 (0.72 to 1.39) | 0.45 (0.22 to 0.93) | 0.93 (0.54 to 1.6) | 0.68 (0.42 to 1.08) |
| DRB1*09:01 | 0.31 | 0.13 | 1 | - | 0.87 (0.57 to 1.34) | 0.83 (0.56 to 1.22) | 1.09 (0.81 to 1.46) |
| DRB1*13:02 | 0.0078 | 0.054 | 0.03 | 0.52 | - | 0.75 (0.38 to 1.46) | 0.63 (0.34 to 1.17) |
| DRB1*15:01 | 2.2×10−3 | 1.5×10−3 | 0.79 | 0.34 | 0.39 | - | 1.06 (0.65 to 1.73) |
| DRB1*15:02 | 6.6×10−5 | 2.3×10−5 | 0.10 | 0.57 | 0.14 | 0.82 | - |
p Values and OR (95% CI) are indicated in subdiagonal and superdiagonal, respectively. Red and blue colours indicate significant p values with positive and negative effects, respectively.
We analysed non-additive effects of amino acid residues in the predefined amino acid positions in HLA-DRB1 exhibiting significant associations with ACPA(+) RA susceptibility. We assessed non-additive effects of amino acid residues at positions 11, the most significant amino acid position known for ACPA(+) RA susceptibility. As a result, we found that the amino acid at position 11 showed significant non-additive effects (p=2.2×10−4). These non-additive effects were driven by amino acid residues of valine and serine (p=7.0×10−4 and 2.9×10–3, respectively), both of which were the main drivers of this position for associations with RA. We further sequentially assessed non-additive effects of amino acid positions 74, 37 and 47, showing the sequential significant associations with ACPA(+) RA susceptibility. The full results are shown in online supplementary table 3.
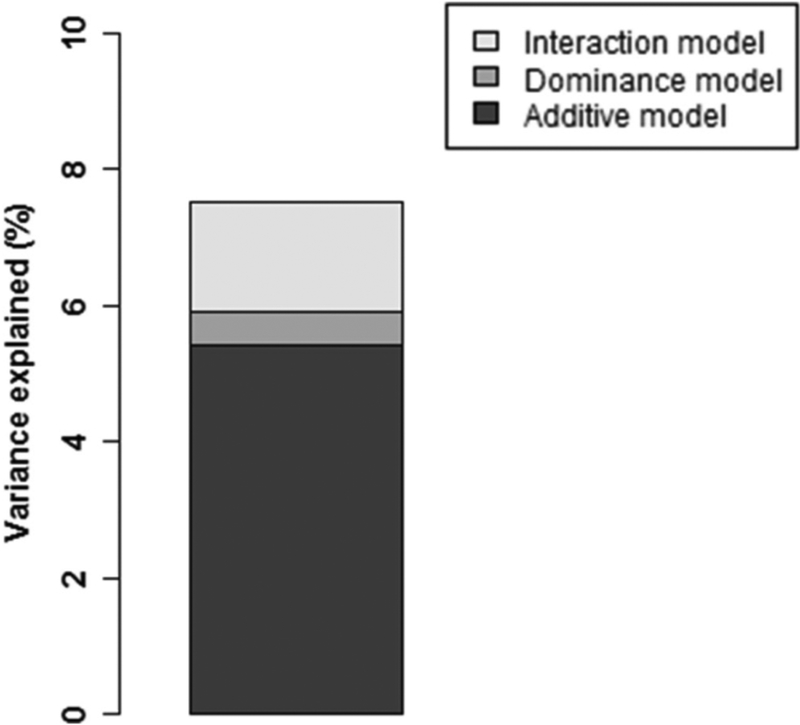
We calculated variance explained by these effects in the full data set. The alleles explained 5.4% of the variance in the subjects (figure 1). The model including non-additive effects of the alleles (dominance model) additionally explained variance by 0.5% (figure 1). The additive and interactive terms (interactive model) explained additionally 1.5% of variance over the dominance model (figure 1).
Figure 1.
Variance additionally explained by dominance and interaction model. Variance explained in the full data set by each model is indicated.
Most ACPA(+) patients are known to be positive for rheumatoid factor (RF).25 When we conducted the same analyses for ACPA(+) RF(+) subjects, we obtained similar results (data not shown).
Next, we applied this statistical framework to the levels of ACPA, a quantitative trait, by using generalised linear regression model. As a result, we did not find evidence of improvement in fit brought about by the model including additive and non-additive effects (p=0.68). Since there is a possibility that significant results of a subset of alleles are cancelled out by non-significant results of the other alleles, we further analysed the non-additive effects of individual haplotypes. As a result, none of the haplotypes, including DRB1*09:01, showing the strongest association with ACPA levels in our previous study, did not show evidence of non-additive effects (see online supplementary table 4). When we included interactive effects of haplotypes instead of non-additive effects, we did not observe strong improvement of fit in this model in comparison with models with additive and non-additive effects (p=0.18). In fact, none of the allelic combinations showed significant associations (table 4). These results were confirmed in the common data set (see online supplementary tables 5 and 6). In addition, when we further analysed non-additive effects of amino acid positions on ACPA levels, we did not find significant results (see online supplementary table 7). Collectively, we did not obtain any evidence of non-additive or interactive effects of HLA-DRB1 alleles on the levels of ACPA.
Table 4.
Lack of evidence of interactive effects on anticitrullinated protein/peptide antibody levels in the full data set
| DRB1*01:01 | DRB1*04:05 | DRB1*08:03 | DRB1*09:01 | DRB1*13:02 | DRB1*15:01 | DRB1*15:02 | |
|---|---|---|---|---|---|---|---|
| DRB1*01:01 | - | 0.103 (0.093) | 0.133 (0.147) | 0.11 (0.102) | 0.298 (0.22) | 0.291 (0.142) | −0.007 (0.115) |
| DRB1*04:05 | 0.27 | - | 0.076 (0.104) | 0.041 (0.042) | 0.206 (0.112) | −0.07 (0.094) | 0.061 (0.074) |
| DRB1*08:03 | 0.37 | 0.46 | - | −0.036 (0.09) | 0.036 (0.127) | 0.032 (0.114) | 0 (0.085) |
| DRB1*09:01 | 0.28 | 0.32 | 0.69 | - | 0.107 (0.21) | 0.206 (0.178) | 0.035 (0.148) |
| DRB1*13:02 | 0.18 | 0.07 | 0.61 | 0.78 | - | −0.089 (0.229) | 0.019 (0.199) |
| DRB1*15:01 | 0.04 | 0.46 | 0.25 | 0.78 | 0.70 | - | −0.028 (0.144) |
| DRB1*15:02 | 0.95 | 0.41 | 0.81 | 1 | 0.93 | 0.85 | - |
p Values and beta (SE) are indicated in subdiagonal and superdiagonal, respectively.
When we expanded the analyses to include a total of 913 ACPA(−) subjects for potential increase of statistical power, we obtained similar results (see online supplementary table 8).
DISCUSSION
In the current study, we found substantial evidence of non-additive and interactive effects on susceptibility to ACPA(+) RA in the Japanese population. While the presence of non-additive and interactive effects are in common with European populations, we found population-specific haplotypes showing significant non-additive or interactive effects. We also identified a lack of evidence for non-additive or interactive effects of HLA-DRB1 alleles on ACPA levels, a quantitative trait of RA.
We limited HLA-DRB1 alleles analysed in the current study to those with frequencies of more than 5% or more than 10 homozygotes in case subjects, which allowed us to limit imputation errors and appropriate estimation of non-additive effects. In fact, the alleles in the common allele or the full data sets have imputation accuracy of more than 98.8%. Taking into account the accuracy of imputation and the strong correlation of allele frequencies between the two sets, we concluded that influence of imputation failure on the analyses would be minimal. Furthermore, the data using only genotyped data revealed the same significant results of non-additive and interactive effects as a whole (p<4.2×10−4, data not shown), again indicating that the significant results were not driven by imputation artefact.
Our previous study18 showed preliminary results of the non-additive effects of HLA alleles, but it used only GWAS imputation data and did not construct full or common data set.
HLA-DRB1*04:05, the most common susceptibility allele to ACPA(+) RA in the Asian population, drove the signal of non-additive effects. On the contrary, HLA-DRB1*09:01, a non-SE allele and the second strongest susceptibility allele to ACPA(+) RA in the Asian population, did not show a significant signal. Since the previous European study showed that SE alleles and non-SE alleles showed non-additive effects to ACPA(+) RA, the non-additive effects seemed different across the alleles. It would be interesting to clarify the common features underlying HLA-DRB1 alleles with significant non-additive effects.
While DRB1*01:01 showed a strong non-additive effect in the European study (p=1.3×10−8), we did not find such evidence in the current study. This is also true for DRB1*15:01 (p=1.2×10−3 in the European population). The difference between the populations may be explained by the difference in frequencies of allelic combinations between the populations. In fact, the non-additive effect of DRB1*01:01 in the European population was driven by interactive effects of DRB1*01:01 and DRB1*15:01, DRB1*07:01, DRB1*04:01 or DRB1*03:01, but three out of the four alleles were absent in the current data sets. As for DRB1*15:01, the non-additive effect of DRB1*15:01 was driven by interactive effects of DRB1*15:01 and DRB1*01:01 or DRB1*04:01. DRB1*04:01 was not included in the current data sets. Importantly, the interactive effect between DRB1*01:01 and DRB1*15:01 was observed in the current data set, and the direction of the interactive effect was shared between the two populations. This result indicates common genetic interactive architecture underlying ACPA(+) RA susceptibility.
The non-additive effect of amino acid position 11 was driven by valine and serine residues. These interactive effects matched the results of allelic non-additive effects. The non-additive effect of valine could be explained by that of DRB1*04:05. The effect of serine could be explained by that of DRB1*08:03. Non-additive effects of amino acid positions of 37 and 47 could also be explained by DRB1*04:05 and DRB1*08:03.
The interactive effects in the current study identified novel allelic combinations with significant results. Since DRB1*15:02, DRB1*04:05 and DRB1*08:03 are relatively rare alleles in European populations, we could not say that the interactive effects were population specific. We need expanded subjects in the European population to address this point.
While the previous Korean study showed interactive effect between DRB1*04:05 and DRB1*09:01,24 we did not find a significant interactive effect between the two alleles. Since the Korean study did not use the same model as the current study, it would be reasonable to assess the interactive effect of the two alleles in the current model in the Korean population.
We did not find significant results of non-additive and interactive effects on ACPA levels. It is compatible that we did not find significant results of non-additive effects in amino acid positions since neither DRB1*09:01 nor DRB1*04:05, the main drivers of ACPA levels, showed non-additive effects.
Since the analyses for non-additive and interactive effects require a substantial number of homozygotes of the alleles being evaluated, we need thousands of subjects to assess conclusive effects. This makes it difficult to assess non-additive and interactive effects on phenotypes of RA especially joint destruction since most of the data sets contain less than 1000 individuals due to the intensive burden of evaluating joint X-rays.12 However, it would be quite interesting to analyse the presence of non-additive effects of HLA haplotypes on joint destruction since we previously found that DRB1*04:05 showed a specific association with joint destruction independently of ACPA.12
Supplementary Material
Key messages.
HLA-DRB1 shows non-additive and interactive effects on anticitrullinated protein/peptide antibody (ACPA) (+) rheumatoid arthritis beyond population.
Allelic combinations seem dependent on allelic frequencies in the population.
The additive and interactive effects are not observed in ACPA levels.
Acknowledgements
We appreciate the cooperators in the GARNET consortium.
Funding This study was supported by JSPS KAKENHI (grant nos JP16H06251 and JP16K15513), KANAE foundation for the promotion of medical science, The Uehara Memorial Foundation, The John Mung Advanced Program, Kyoto University.
Footnotes
Competing interests
None declared.
Patient consent
Detail has been removed from this case description/these case descriptions to ensure anonymity. The editors and reviewers have seen the detailed information available and are satisfied that the information backs up the case the authors are making.
Ethics approval
This study was approved by the ethical review board in each institution.
Provenance and peer review
Not commissioned; externally peer reviewed.
Additional material is published online only. To view please visit the journal online (http://dx.doi.org/10.1136/jmedgenet-2017-104779).
REFERENCES
- 1.Terao C, Raychaudhuri S, Gregersen PK. Recent advances in defining the genetic basis of rheumatoid arthritis. Annu Rev Genomics Hum Genet 2016;17:273–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Terao C, Ikari K, Nakayamada S, Takahashi Y, Yamada R, Ohmura K, Hashimoto M, Furu M, Ito H, Fujii T, Yoshida S, Saito K, Taniguchi A, Momohara S, Yamanaka H, Mimori T, Matsuda F. A twin study of rheumatoid arthritis in the Japanese population. Mod Rheumatol 2016;26:685–9. [DOI] [PubMed] [Google Scholar]
- 3.Okada Y, Terao C, Ikari K, Kochi Y, Ohmura K, Suzuki A, Kawaguchi T, Stahl EA, Kurreeman FA, Nishida N, Ohmiya H, Myouzen K, Takahashi M, Sawada T, Nishioka Y, Yukioka M, Matsubara T, Wakitani S, Teshima R, Tohma S, Takasugi K, Shimada K, Murasawa A, Honjo S, Matsuo K, Tanaka H, Tajima K, Suzuki T, Iwamoto T, Kawamura Y, Tanii H, Okazaki Y, Sasaki T, Gregersen PK, Padyukov L, Worthington J, Siminovitch KA, Lathrop M, Taniguchi A, Takahashi A, Tokunaga K, Kubo M, Nakamura Y, Kamatani N, Mimori T, Plenge RM, Yamanaka H, Momohara S, Yamada R, Matsuda F, Yamamoto K. Meta-analysis identifies nine new loci associated with rheumatoid arthritis in the Japanese population. Nat Genet 2012;44:511–6. [DOI] [PubMed] [Google Scholar]
- 4.Terao C, Ohmura K, Kochi Y, Ikari K, Okada Y, Shimizu M, Nishina N, Suzuki A, Myouzen K, Kawaguchi T, Takahashi M, Takasugi K, Murasawa A, Mizuki S, Iwahashi M, Funahashi K, Natsumeda M, Furu M, Hashimoto M, Ito H, Fujii T, Ezawa K, Matsubara T, Takeuchi T, Kubo M, Yamada R, Taniguchi A, Yamanaka H, Momohara S, Yamamoto K, Mimori T, Matsuda F. Anti-citrullinated peptide/protein antibody (ACPA)-negative RA shares a large proportion of susceptibility loci with ACPA-positive RA: a meta-analysis of genome-wide association study in a Japanese population. Arthritis Res Ther 2015;17:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han B, Diogo D, Eyre S, Kallberg H, Zhernakova A, Bowes J, Padyukov L, Okada Y, González-Gay MA, Rantapää-Dahlqvist S, Martin J, Huizinga TW, Plenge RM, Worthington J, Gregersen PK, Klareskog L, de Bakker PI, Raychaudhuri S. Fine mapping seronegative and seropositive rheumatoid arthritis to shared and distinct HLA alleles by adjusting for the effects of heterogeneity. Am J Hum Genet 2014;94:522–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohmura K, Terao C, Maruya E, Katayama M, Matoba K, Shimada K, Murasawa A, Honjo S, Takasugi K, Tohma S, Matsuo K, Tajima K, Yukawa N, Kawabata D, Nojima T, Fujii T, Yamada R, Saji H, Matsuda F, Mimori T. Anti-citrullinated peptide antibody-negative RA is a genetically distinct subset: a definitive study using only bone-erosive ACPA-negative rheumatoid arthritis. Rheumatology 2010;49:2298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Terao C, Ohmura K, Kochi Y, Ikari K, Maruya E, Katayama M, Shimada K, Murasawa A, Honjo S, Takasugi K, Matsuo K, Tajima K, Suzuki A, Yamamoto K, Momohara S, Yamanaka H, Yamada R, Saji H, Matsuda F, Mimori T. A large-scale association study identified multiple HLA-DRB1 alleles associated with ACPA-negative rheumatoid arthritis in Japanese subjects. Ann Rheum Dis 2011;70:2134–9. [DOI] [PubMed] [Google Scholar]
- 8.Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum 1987;30:1205–13. [DOI] [PubMed] [Google Scholar]
- 9.Raychaudhuri S, Sandor C, Stahl EA, et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet 2012;44:291–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okada Y, Kim K, Han B, et al. Risk for ACPA-positive rheumatoid arthritis is driven by shared HLA amino acid polymorphisms in Asian and European populations. Hum Mol Genet 2014;23:6916–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Terao C, Suzuki A, Ikari K. An association between the 74th amino acid position of HLA-DRB1 and ACPA levels of Japanese ACPA-positive RA. Arthritis Rheumatol 2015. [DOI] [PubMed] [Google Scholar]
- 12.Terao C, Yano K, Ikari K, Furu M, Yamakawa N, Yoshida S, Hashimoto M, Ito H, Fujii T, Ohmura K, Yurugi K, Miura Y, Maekawa T, Taniguchi A, Momohara S, Yamanaka H, Mimori T, Matsuda F. Brief report: main contribution of DRB1*04:05 among the shared epitope alleles and involvement of DRB1 amino acid position 57 in association with joint destruction in anti-citrullinated protein antibody-positive rheumatoid arthritis. Arthritis Rheumatol 2015;67:1744–50. [DOI] [PubMed] [Google Scholar]
- 13.Terao C, Ikari K, Ohmura K, Suzuki T, Iwamoto T, Takasugi K, Saji H, Taniguchi A, Momohara S, Yamanaka H, Matsuda F, Mimori T. Quantitative effect of HLA-DRB1 alleles to ACPA levels in Japanese rheumatoid arthritis: no strong genetic impact of shared epitope to ACPA levels after stratification of HLA-DRB1*09:01. Ann Rheum Dis 2012;71:1095–7. [DOI] [PubMed] [Google Scholar]
- 14.Okada Y, Suzuki A, Yamada R, Kochi Y, Shimane K, Myouzen K, Kubo M, Nakamura Y, Yamamoto K. HLA-DRB1*0901 lowers anti-cyclic citrullinated peptide antibody levels in Japanese patients with rheumatoid arthritis. Ann Rheum Dis 2010;69:1569–70. [DOI] [PubMed] [Google Scholar]
- 15.Balandraud N, Picard C, Reviron D, Landais C, Toussirot E, Lambert N, Telle E, Charpin C, Wendling D, Pardoux E, Auger I, Roudier J. HLA-DRB1 genotypes and the risk of developing anti citrullinated protein antibody (ACPA) positive rheumatoid arthritis. PLoS One 2013;8:e64108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall FC, Weeks DE, Camilleri JP, Williams LA, Amos N, Darke C, Gibson K, Pile K, Wordsworth BP, Jessop JD. Influence of the HLA-DRB1 locus on susceptibility and severity in rheumatoid arthritis. QJM 1996;89:821–30. [DOI] [PubMed] [Google Scholar]
- 17.Lenz TL, Deutsch AJ, Han B, Hu X, Okada Y, Eyre S, Knapp M, Zhernakova A, Huizinga TW, Abecasis G, Becker J, Boeckxstaens GE, Chen WM, Franke A, Gladman DD, Gockel I, Gutierrez-Achury J, Martin J, Nair RP, Nöthen MM, Onengut-Gumuscu S, Rahman P, Rantapää-Dahlqvist S, Stuart PE, Tsoi LC, van Heel DA, Worthington J, Wouters MM, Klareskog L, Elder JT, Gregersen PK, Schumacher J, Rich SS, Wijmenga C, Sunyaev SR, de Bakker PI, Raychaudhuri S. Widespread non-additive and interaction effects within HLA loci modulate the risk of autoimmune diseases. Nat Genet 2015;47:1085–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okada Y, Suzuki A, Ikari K, Terao C, Kochi Y, Ohmura K, Higasa K, Akiyama M, Ashikawa K, Kanai M, Hirata J, Suita N, Teo YY, Xu H, Bae SC, Takahashi A, Momozawa Y, Matsuda K, Momohara S, Taniguchi A, Yamada R, Mimori T, Kubo M, Brown MA, Raychaudhuri S, Matsuda F, Yamanaka H, Kamatani Y, Yamamoto K. Contribution of a non-classical HLA gene, HLA-DOA, to the risk of rheumatoid arthritis. Am J Hum Genet 2016;99:366–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, Healey LA, Kaplan SR, Liang MH, Luthra HS. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum 1988;31:315–24. [DOI] [PubMed] [Google Scholar]
- 20.Aletaha D, Neogi T, Silman AJ. 2010 Rheumatoid Arthritis Classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Ann Rheum Dis 2010;62:2569–81. [DOI] [PubMed] [Google Scholar]
- 21.Jia X, Han B, Onengut-Gumuscu S, Chen WM, Concannon PJ, Rich SS, Raychaudhuri S, de Bakker PI. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One 2013;8:64683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okada Y, Momozawa Y, Ashikawa K, Kanai M, Matsuda K, Kamatani Y, Takahashi A, Kubo M. Construction of a population-specific HLA imputation reference panel and its application to Graves’ disease risk in Japanese. Nat Genet 2015;47:798–802. [DOI] [PubMed] [Google Scholar]
- 23.Robinson J, Waller MJ, Parham P, de Groot N, Bontrop R, Kennedy LJ, Stoehr P, Marsh SG. IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res 2003;31:311–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee HS, Lee KW, Song GG, Kim HA, Kim SY, Bae SC. Increased susceptibility to rheumatoid arthritis in Koreans heterozygous for HLA-DRB1*0405 and *0901. Arthritis Rheum 2004;50:3468–75. [DOI] [PubMed] [Google Scholar]
- 25.Terao C, Ohmura K, Ikari K, Kochi Y, Maruya E, Katayama M, Yurugi K, Shimada K, Murasawa A, Honjo S, Takasugi K, Matsuo K, Tajima K, Suzuki A, Yamamoto K, Momohara S, Yamanaka H, Yamada R, Saji H, Matsuda F, Mimori T. ACPA-negative RA consists of two genetically distinct subsets based on RF positivity in Japanese. PLoS One 2012;7:e40067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.