Abstract
Objective
The pathogenetic mechanisms by which HLA-DRB1 alleles are associated with anticitrullinated peptide antibody (ACPA)-positive rheumatoid arthritis (RA) are incompletely understood. RA high-risk HLA-DRB1 alleles are known to share a common motif, the ‘shared susceptibility epitope (SE)’. Here, the electropositive P4 pocket of HLA-DRB1 accommodates self-peptide residues containing citrulline but not arginine. HLA-DRB1 His/Phe13β stratifies with ACPA-positive RA, while His13βSer polymorphisms stratify with ACPA-negative RA and RA protection. Indigenous North American (INA) populations have high risk of early-onset ACPA-positive RA, whereby HLA-DRB1*04:04 and HLA-DRB1*14:02 are implicated as risk factors for RA in INA. However, HLA-DRB1*14:02 has a His13βSer polymorphism. Therefore, we aimed to verify this association and determine its molecular mechanism.
Methods
HLA genotype was compared in 344 INA patients with RA and 352 controls. Structures of HLA-DRB1*1402-class II loaded with vimentin-64Arg59–71, vimentin-64Cit59–71 and fibrinogen β−74Cit69–81 were solved using X-ray crystallography. Vimentin-64Cit59–71-specific and vimentin59–71-specific CD4+ T cells were characterised by flow cytometry using peptide-histocompatibility leukocyte antigen (pHLA) tetramers. After sorting of antigen-specific T cells, TCRα and β-chains were analysed using multiplex, nested PCR and sequencing.
Results
ACPA+ RA in INA was independently associated with HLA-DRB1*14:02. Consequent to the His13βSer polymorphism and altered P4 pocket of HLA-DRB1*14:02, both citrulline and arginine were accommodated in opposite orientations. Oligoclonal autoreactive CD4+ effector T cells reactive with both citrulline and arginine forms of vimentin59–71 were observed in patients with HLA-DRB1*14:02+ RA and at-risk ACPA− first-degree relatives. HLA-DRB1*14:02-vimentin59–71-specific and HLA-DRB1*14:02-vimentin-64Cit59–71-specific CD4+ memory T cells were phenotypically distinct populations.
Conclusion
HLA-DRB1*14:02 broadens the capacity for citrullinated and native self-peptide presentation and T cell expansion, increasing risk of ACPA+ RA.
INTRODUCTION
Rheumatoid arthritis (RA) is an autoimmune disease with peak incidence in the sixth decade and prevalence of 1% in Caucasians, linked to HLA-DRB1. HLA-DRB1 alleles associated with anticitrullinated peptide antibody (ACPA)-positive RA in Caucasians share a common motif in the third hypervariable region, the ‘shared susceptibility epitope (SE)’, which was shown to accommodate citrullinated (Cit) self-epitopes.1–3 Citrulline is post-translationally modified from arginine during inflammation, endoplasmic reticulum (ER) stress and autophagy.4,5 Various RA-associated Cit-autoantigens, recognised by ACPA, are present in inflamed sites, including joint tissues. Low-titre ACPA develops in healthy individuals associated with environmental risk factors including smoking and periodontitis, but are unrelated to HLA-DR SE.6–11 In the immediate pre-RA period, ACPA isotype diversity and titre increase — a process associated with antigen-specific CD4+ T cell help for affinity maturation in germinal centres.8 HLA-DR SE is associated with ACPA+ RA rather than ACPA, implying that presentation of Cit-autoantigens bound to HLA-DR SE molecules to CD4+ T cells is associated with RA development in at-risk individuals carrying HLA-DR SE.
Based on genome-wide studies (GWAS) in patients of predominantly Caucasian and Asian ethnicity, HLA alleles associated with ACPA-positive RA, including HLA-DRB1*04:01, HLA-DRB1*04:05 and HLA-DRB1*01:01 (ORs of 2.17–4.44), were found to share a common motif at amino acid positions 11, 13, 71 and 74, influencing the P4 antigen-binding pocket of DRβ.2 Moreover, Val11βSer and His13βSer polymorphisms within that motif were found in genomic studies to stratify with ACPA-negative RA in Caucasians.3 The discovery that P4-Cit was accommodated but the positively charged Arg was excluded from the electropositive P4 pocket of HLA-DRB1*04:01/04 suggested that preferential presentation of Cit-autoantigens might underpin the association of ACPA+ RA with the SE.12 While HLA-DRB1*04:01-restricted CD4+ memory T cells recognising Cit-autoantigens have been reported in patients with RA,12 13 their role in disease development is unclear. To date it has not been determined how autoreactive T cells respond to Cit-autoantigen in RA and whether they undergo clonal expansion due to antigen experience in patients with RA or in HLA-SE+ at-risk first-degree relatives (FDRs).
Furthermore, in unravelling antigen presentation by HLA-DR molecules in RA, the ethnic mix of the samples included in GWAS may skew interpretations of amino acids contributing to binding motifs made from genomic studies. The Indigenous North American (INA) population has a twofold to threefold higher prevalence of RA than Caucasians, with RA onset peaking earlier in the fourth decade of life.14 Moreover, 90% of INA patients with RA are ACPA+. In this population, FDRs have a high prevalence of joint symptoms and of ACPA positivity, predisposing them to RA.15,16 To date, the HLA association with RA in INA has been sought in studies of <100 patients.17–19 These studies suggested that the HLA-SE alleles also predispose to RA in INA. HLA-DRB1*04:04 is the most frequent SE allele in these populations, followed by HLA-DRB1*14:02. Since HLA-DRB1*14:02 carries the β13Ser residue, which was interpreted to stratify with ACPA-negative RA,3 we investigated the genetic and underlying molecular bases for the increased risk of severe ACPA+ RA in INA.
MATERIALS AND METHODS
Study participants
RA cases, non-RA controls and FDRs were recruited from INA populations in Central Canada (Cree, Ojibway and Ojicree) and Alaska Native people (from Southcentral and Southeast Alaska). DNA for HLA typing, serum and peripheral blood mononuclear cells (PBMC) were isolated.
HLA-DRB1*14:02 expression and purification
Peptide-loaded HLA-DRB1*14:02 molecules were purified, crystallised and structures determined.
Multiplex ACPA assay
Serum levels of antibodies targeting 40 putative RA-associated autoantigens were measured using a custom bead-based immunoassay on a Bio-Plex platform, as previously described.20
Tetramer staining and analysis of TCR repertoire
Tetramer staining and T cell receptor (TCR) repertoire analysis used previously published methods, with some modifications.12,21,22
Details of all methods are available in online supplementary methods, supplementary table 1 and supplementary figure 1.
RESULTS
HLA-DRB1*14:02 is independently associated with ACPA+ RA in INA
Although very rare in Caucasians and Asians, HLA-DRB1*14:02 has a prevalence of up to 80% in some INA populations, suggesting a particular survival advantage against pathogens.17–19 To determine the HLA-DRB1 association with ACPA+ RA in INA, we genotyped the largest cohort available, comprising 344 INA patients with RA and 352 controls. Rheumatoid factor (RF) and ACPA status was known in 241/344 patients with RA: 90% were seropositive (RF and/or ACPA+). In patients with seropositive RA, 32% carried HLA-DRB1*14:02 and 45% HLA-DRB1*04:04. One-third of patients carrying HLA-DRB1*14:02 carried an additional SE allele. In healthy controls (HCs), 28% carried DRB1*14:02, 22% HLA-DRB1*04:04 and 17% carried an additional SE allele. After stratifying patients with RA according to HLA status, HLA-DRB1*14:02 was a risk factor for seropositive RA (OR 2.38) independent of other SE alleles (table 1). In INA patients with RA, the most commonly associated other SE allele is HLA-DRB1*04:04 (table 1). In INA, RA risk was associated with HLA-DRB1 alleles with a conserved SE motif at 71 and 74 in all RA (OR=2.48, 95% CI 1.70 to 3.60, p<0.0001) and in seropositive patients with RA (OR=2.46, 95% CI 1.58 to 3.81, p = 0.0001) (table 1). We note that without genome-wide genotyping, we cannot rule out the possibility of confounding due to case–control differences in ancestry.
Table 1.
Association of HLA-DRB1*14:02 with all RA and seropositive RA in INA patients with RA and controls
| HC, n (%) | All RA, n (%) | OR (CI) | p | Seropositive RA, n (%) | OR (CI) | p | |
|---|---|---|---|---|---|---|---|
| SE− | 106 (30) | 51 (15) | Ref | 32 (15) | Ref | ||
| 14:02+/Other SE− | 64 (18) | 80 (23) | 2.60 (1.63 to 4.15) | 0.0001 | 46 (21) | 2.38 (1.38 to 4.12) | 0.0019 |
| 14:02−/Other SE+ | 147 (42) | 178 (52) | 2.52 (1.69 to 3.75) | <0.0001 | 117 (54) | 2.64 (1.66 to 4.19) | <0.0001 |
| 14:02+/Other SE+ | 35 (10) | 35 (10) | 2.08 (1.17 to 3.70) | 0.0127 | 23 (11) | 2.18 (1.13 to 4.20) | 0.02 |
| Any SE+ | 246 (70) | 293 (85) | 2.48 (1.70 to 3.60) | <0.0001 | 186 (85) | 2.46 (1.58 to 3.81) | 0.0001 |
Patients and controls were stratified according to the presence of SE. SE-positive individuals were further stratified as shown. Other SE alleles included DRB1*01:01, DRB1*01:02, DRB1*04:01, DRB1*04:04, DRB1*04:05, DRB1*04:08, DRB1*04:10, DRB1*04:13 and DRB1*10:01.
HC, healthy controls; INA, Indigenous North American; RA, rheumatoid arthritis; SE, susceptibility epitope.
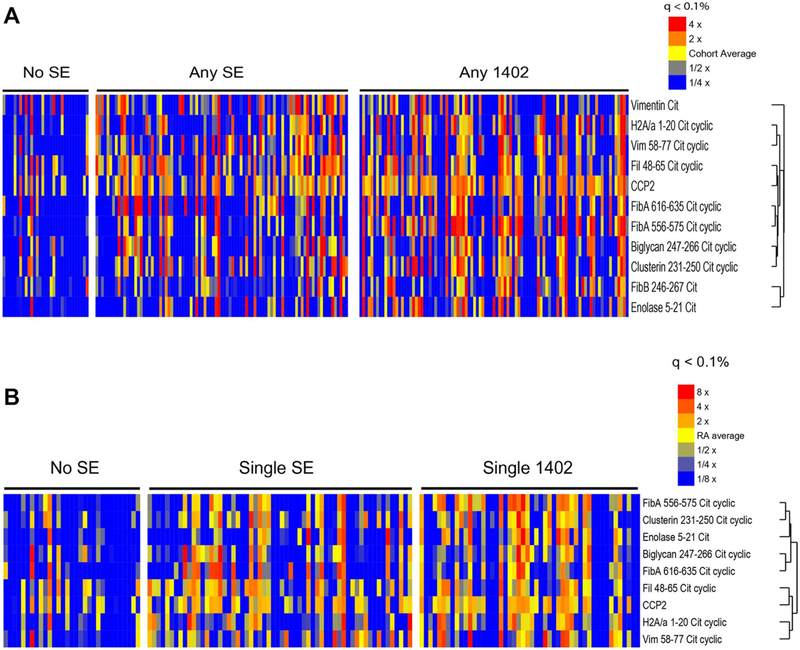
To stratify ACPA response with genotype, sera of 232 INA patients with RA were tested in a multiplex ACPA antigen array and given an ACPA score (sum of all normalised ACPA titres divided by number of epitopes).23 ACPA score was higher in INA patients with RA who were either HLA-DRB1*14:02 homozygotes or HLA-DRB1*14:02/HLA-SE compound heterozygotes than those who were HLA-SE-negative (p<0.05). ACPA scores in HLA-DRB1*14:02 + patients were equivalent to those in HLA-SE+ patients lacking HLA-DRB1*14:02. ACPA specificities increased among HLA-DRB1*14:02 + patients and included vimentin-64Cit58–77, filaggrin-56Cit48–65 and fibrinogen-α−573Cit556–575 (figure 1A,B). Thus, despite polymorphisms of Val11βSer and His13βSer, INA individuals carrying HLA-DRB1*14:02 develop a broad ACPA response whether or not they carry other SE alleles. This implies binding and presentation of a variety of Cit-autoantigens by HLA-DRB1*14:02 to autoreactive T cells.
Figure 1.
HLA-DRB1*14:02 is associated with a broad ACPA response. Serum levels of antibodies targeting RA-associated Cit-autoantigens were measured in serum from 232 INA patients with RA using a custom bead-based fluorescence immunoassay. Fluorescence intensities for 10 Cit-autoantigens and CCP were clustered (A) according to the presence or absence of 1 or 2 HLA-DR-SE alleles or HLA-DRB1*14:02 with any HLA-DR-SE allele, and (B) according to presence or absence of a single HLA-DR-SE or HLA-DRB1*14:02 allele, as shown. Each column represents one patient sample. ACPA, anticitrullinated peptide antibody; CCP, cyclic citrullinated peptide; Cit, citrullinated; RA, rheumatoid arthritis; SE, susceptibility epitope.
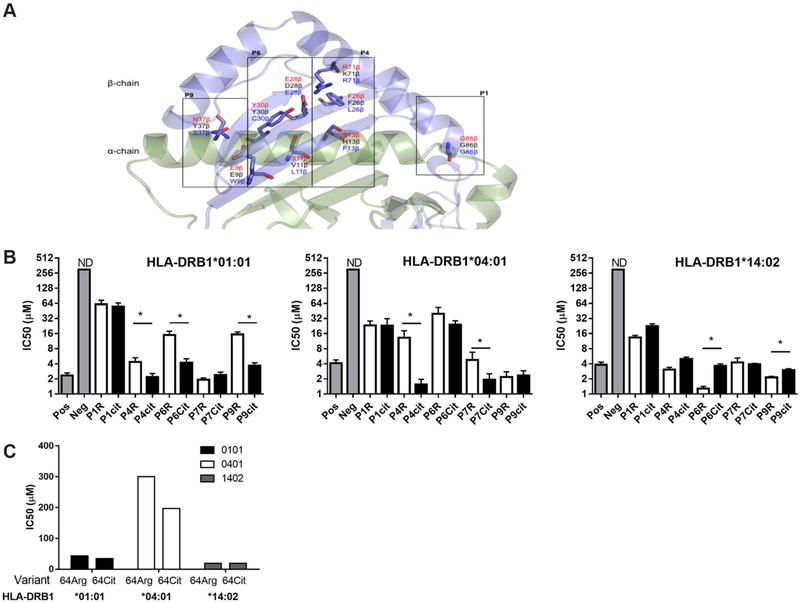
Accommodation of arginine and citrulline residues within the P4 pocket of HLA-DRB1*14:02
The HLA-DRB1 chain contains 12 polymorphic residues that have been directly implicated in peptide binding.24 HLA-DRB1*14:02 differs from HLA-DRB1*04:01, HLA-DRB1*04:04 or HLA-DRB1*01:01 in eight of these residues, which shape P4, P6, P7 and P9 pockets (figure 2A). To test whether HLA-DRB1*14:02 presents autoantigens differently from HLA-DRB1*04:01, HLA-DRB1*04:04 or HLA-DRB1*01:01, we compared the capacity of each HLA binding pocket to accommodate Cit and Arg residues using the influenza-derived HA305–319 peptide.25–28 Conversion of Arg to Cit at peptide positions interacting with P4 enhanced peptide binding affinity to HLA-DRB1*01:01 by twofold and to HLA-DRB1*04:01 by tenfold. Conversion of Arg to Cit at peptide positions interacting with the P6 and P9 pockets enhanced peptide binding affinity to HLA-DRB1*01:01, and P7 to HLA-DRB1*04:01. In contrast, peptides containing an Arg or Cit residue at positions interacting with P4 of HLA-DRB1*14:02 had similar binding affinity, and peptides containing Arg at positions interacting with P6 and P9 had increased affinity relative to Cit peptides (figure 2B). While the IC50 of the self-peptide, vimentin-64Cit59–71, was decreased by 1.3-fold relative to the 64-Arg variant for HLA-DRB1*01:01, and the 64-Arg variant did not bind HLA-DRB1*04:01, HLA-DRB1*14:02 bound both 64-Arg and 64-Cit variants with IC50 of 19 μM (figure 2C), indicating that both residues could be accommodated within P4 of HLA-DRB1*14:02, or that HLA-DRB1*14:02 presented in differing peptide binding registers.
Figure 2.
Accommodation of arginine and citrulline residues by HLA-DRB1*01:01, HLA-DRB1*04:01 and HLA-DRB1*14:02. (A) The peptide binding groove of an HLA-DR molecule is shown in cartoon with polymorphic residues shown as sticks corresponding to the residues present in HLA-DRB1*14:02, which is associated with RA among the INA. Schematic representation of the differences in pockets between HLA-DRB1*14:02 (red font), HLA-DRB1*04:01 (black font) and HLA-DRB1*01:01 (blue font). (B) Competitive binding of a biotin-labelled HA305–319 peptide with an unlabelled HA305–319 peptide or HA305–319 variants with citrulline or arginine residues in P1, P4, P6, P7 and P9 to HLA-DRB1*01:01, HLA-DRB1*04:01 and HLA-DRB1*14:02. ND, non-detectable binding affinity. Pooled binding data from at least three experiments are shown and error bars depict the variation between experiments. (C) Competitive binding of a biotin-labelled HA305–319 peptide with unlabelled vimentin59–71 and vimentin-64Cit59–71 to HLA-DRB1*01:01, HLA-DRB1*04:01 and HLA-DRB1*14:02. INA, Indigenous North American; RA, rheumatoid arthritis.
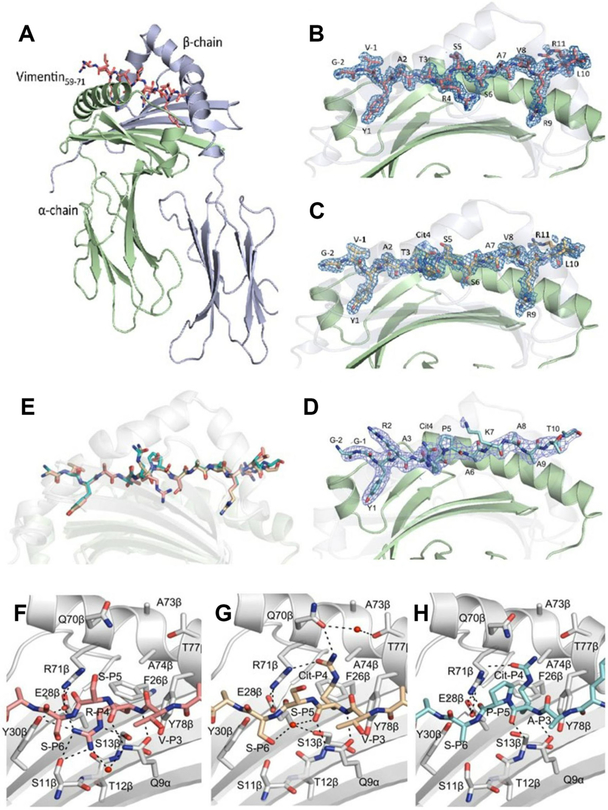
Structural basis of peptide presentation by HLA-DRB1*14:02
We solved the structure of HLA-DRB1*14:02 in complex with vimentin-64Arg59–71, vimentin-64Cit59–71 and fibrinogen β74Cit69–81 (online supplementary table 3, figure 3A–D). These HLA-DRB1*14:02 structures overlaid closely (figure 3E), ruling out markedly differing binding modes to accommodate these differing epitopes. Within the HLA-DRB1*14:02-vimentin-64Cit59–71 and HLA-DRB1*14:02-vimentin-64Arg59–71 structures, Tyr, Ser and Arg occupied the P1, P6 and P9 pockets of HLA-DRB1*14:02, respectively (figure 3B,C). In the HLA-DRB1*14:02-fibrinogen β74Cit69–81 complex, Tyr, Ala and Ala occupied the P1, P6 and P9 pockets of HLA-DRB1*14:02, respectively (figure 3D). The largest structural differences between the peptides in each binary complex centred on the residue occupying the P4 pocket of HLA-DRB1*14:02, namely P4-Cit and P4-Arg in vimentin-64Cit59–71, fibrinogen β74Cit69–81 and vimentin-64Arg59–71, respectively. These structures clearly show that P4-Arg and P4-Cit are presented in alternative orientations, whereby the P4-Arg projects inwards, whereas the P4-Cit projects outwards from the HLA-DRB1*14:02 Ag-binding cleft (figure 3F–H). The P4-Arg is buried in the pocket to avoid interactions with the positively charged β71Arg residue. This orientation is promoted by β11Ser and β13Ser, whereupon these two small polar residues allow the accommodation of Arg by providing the necessary space and H-bonding partners (figure 3F). Larger residues in HLA-DRB1*04:01 (β11Val and β13His) and HLA-DRB1*01:01 (β11Leu and β13Phe) as well as the charge repulsion of β13His in HLA-DRB1*04:01 would prevent the accommodation of Arg at P4.12 The P4-Arg residue is stabilised by H-bonds with Ser11β, Ser13 β and Tyr30β, and a salt bridge with Glu28β (figure 3F). In contrast, the P4-Cit sits upright in both Cit-epitopes, similar to its orientation in P4-Cit from Cit epitopes presented by HLA-DRB1*04:01/04:04 (figure 3G,H).12 The P4-Cit is stabilised by H-bonds with Arg71β and Gln70β (figure 3G,H). Accordingly, we demonstrate a conserved positioning of the citrulline residue in two distinct epitopes that contrast the orientation of the non-Cit residue within the P4 pocket. Similar to its orientation in HLA-DRB1*04:01 and HLA-DRB1*04:04,12,29 P4-Cit was solvent-exposed in HLA-DRB1*14:02 and could potentially interact with TCR.
Figure 3.
Crystal structure of HLA-DRB1*1402 in complex with vimentin59–71, vimentin-64Cit59–71 and fibrinogen β 74Cit69–81. The α-chains and β-chains of (A) the HLA-DRB1*1402 in complex with vimentin59–71 are shown in cartoon representation and coloured in light green and light blue, respectively, while peptide is displayed as sticks. Side view of (B) HLA-DRB1*1402-vimentin59–71, (C) HLA-DRB1*1402-vimentin-64Cit59–71 and (D) HLA-DRB1*1402-fibrinogen β74Cit69–81. The carbons are coloured deep salmon, light orange and teal for vimentin59–71, vimentin-64Cit59–71 and fibrinogen β74Cit69–81, respectively; nitrogens are coloured in blue and oxygens are coloured in red. The β-chain is transparent to help visualise the peptide. The unbiased 2Fo-Fc electron density map of the peptides is shown in blue and contoured to 1 σ. (E) Superposition of the three peptides bound to HLA-DRB1*1402. The P4 pocket of HLA-DRB1*1402 bound to (F) vimentin59–71, (G) vimentin-64Cit59–71 and (H) fibrinogenβ74Cit69–81. The P4-Arg in the vimentin59–71 peptide is buried in the P4 pocket. The P4-Cit of the vimentin-64Cit59–71 and fibrinogenβ74Cit69–81 peptide adopts an upright conformation.
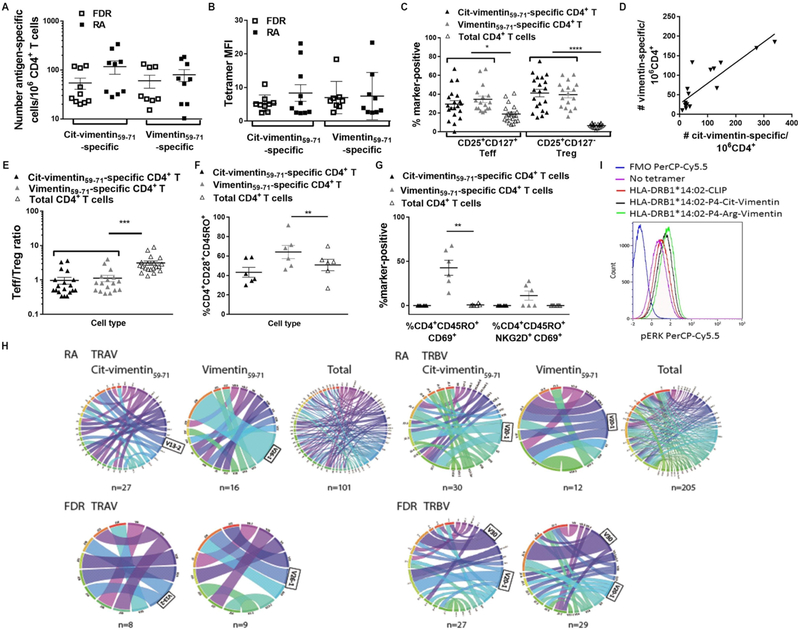
Antigen-experienced, oligoclonally expanded T cells recognise Arg and Cit variants of vimentin59–71 presented by HLA-DRB1*14:02
Although both vimentin-64-Arg and 64-Cit variant peptides bound HLA-DRB1*14:02, only P4-Cit sits upright, and thereby potentially able to interact with the TCR. Thus, we addressed whether autoreactive T cells with TCRs recognising one or both epitopes were present in the periphery and displayed evidence of in vivo expansion in response to antigen presentation. We analysed T cells recognising HLA-DRB1*14:02-vimentin-64Cit59–71 and HLA-DRB1*1402-vimentin59–71 tetramers in 10 HLA-DRB1*14:02+ INA patients with RA, 10 HLA-DRB1*14:02+ ACPA− FDRs and 6 HLA-DRB1*14:02+ non-INA HC subjects. FDRs in the INA population have a high burden of environmental risk factors for RA, a high level of background HLA-DR SE genes, high inflammatory C-reactive protein (CRP) and a high prevalence of joint symptoms.16 Among the 13 FDRs studied (online supplementary table 3), 85% were past smokers and 31% had an abnormal CRP >8. Therefore we could compare T cells recognising vimentin or Cit-vimentin in individuals both with RA and with high risk of future RA. Similar frequencies of CD4+ T cells recognised vimentin 64-Arg and 64-Cit variant peptides in HLA-DRB1*14:02+ FDRs and patients with RA, and with similar tetramer staining intensity (mean fluorescence intensity (MFI)) (figure 4A,B). In all individuals, HLA-DRB1*14:02-vimentin-64Cit59–71-reactive and HLA-DRB1*14:02-vimentin59–71-reactive CD4+ T cells were significantly enriched in CD25+CD127+ effector (Teff) and CD25+ CD127−regulatory T cells (Treg)30 compared with the total CD4+ PB T cell pool (Treg p<0.0001, Teff p<0.05; figure 4C), consistent with antigen experience in vivo. This enrichment did not differ between patients with RA and FDRs, indicating that antigen experience and formation of memory develop before the onset of ACPA in INA. The numbers of circulating HLA-DRB1*14:02-vimentin-64Cit59–71-reactive and HLA-DRB1*14:02-vimentin59–71-reactive CD4+ T cells were correlated in each individual patient with RA and each FDR (r2 = 0.74, p<0.0001; figure 4D).
Figure 4.
Presentation of vimentin59–71 and vimentin-64Cit59–71 self-antigens in context of HLA-DRB1*14:02 to CD4+ T cells in individuals with and at high risk of ACPA+ RA. (A–D) PBMC from HLA-DRB1*14:02+ RA patients W1:1–10 and HLA-DRB1*14:02+ FDR W1:1–10 were stained with PE-labelled HLA-DRB1*1402-vimentin64Cit59–71 or HLADRB1*14:02-vimentin59–71 tetramer, CD4-APC, CD25-PE/Cyt, and CD127-BV421. Live CD4+ FITC-lineage-negative tetramer+ T cells were gated based on an FMO sample stained with unlabelled primary and conjugated secondary antibodies. The number calculated relative to the CD4+ T cell count, (A) and mean fluorescence intensity (MFI) (B) of tetramer+ cells within the total CD4+ population is shown for RA patients and FDR; (C) the proportion of CD4+CD25+CD127+ Teff and CD4+CD25+CD127− Treg within vimentin64Cit59–71-reactive and vimentin59–71-reactive CD4+ T cell gates and the total CD4+ gate is shown for pooled RA patients and FDR. (D) The numbers of vimentin64Cit59–71-reactive and vimentin59–71-reactive CD4+ T cells were correlated in pooled RA patients and FDR (R2 0.74, p<0.0001). (E) The Teff/Treg ratio of vimentin64Cit59–71 and vimentin59–71-reactive CD4+ T cells is plotted relative to total CD4+ T cell ratio in pooled RA patients and FDR; (F) CD45RO+CD28+, (G) CD45RO+CD69+ and CD45RO+CD69+ NKG2D+ vimentin59–71-reactive CD4+, vimentin64Cit59–71-reactive CD4+ and total CD4+ T cells are plotted in pooled RA patients W2:1–3 and FDR W2:1–3. (H) Frequencies of V-J pairing for TCRα and TCRβ among vimentin64Cit59–71-specific and vimentin59–71-specific CD4+ T cells from HLADRB1*14:02+ RA patients (n=5) and FDR (n=5), and tetramer-negative CD3+ T cells sorted from DRB1*14:02+ RA patients (n=3). The width of the bands is proportional to the frequency of TCR sequences with a particular V-J pairing. Details of productive TCRα and TCRβ sequences in online supplementary table 2 and 4. The figures were generated using the Circos online Table Viewer software (http://mkweb.bcgsc.ca/tableviewer/). (I) 106 P2F3 SKW3 cells were stimulated with 5 ng/mL unlabelled HLA-DRB1*14:02-vimentin59–71 or HLA-DRB1*14:02-vimentin-64Cit59–71 tetramer for 4 hours at 37°C. Cells were fixed and permeabilised, then stained with PerCP/Cy5.5-ERK1/2 and PB-CD3. Live CD3+ cells expressing the GFP marker gene were gated. Representative of 2 experiments.
T cell effector function is balanced by the suppressive activity of Treg.31 The ratio of HLA-DRB1*14:02-vimentin59–71Cit-64-reactive and HLA-DRB1*14:02-vimentin59–71-reactive Teff/Treg was significantly lower than that of the total CD4+ T cell pool in patients with RA and FDRs (p<0.001, figure 4E), consistent with active regulation of the autoreactive T cells. To understand the particular role of CD4+ T cells of each vimentin specificity further, we analysed an additional six HLA-DRB1*14:02+ INA individuals (three with RA, three FDRs; clinical details in online supplementary table 3) and six HLA-DRB1*14:02+ HC for markers of memory T cell activation and differentiation relative to total CD4+ T cells. In INA patients with RA and FDRs, the proportion of CD28+ memory T cells was significantly higher among vimentin59–71-reactive CD4+ T cells (p<0.05; figure 4F). Moreover, cells expressing CD69 were significantly enriched among vimentin59–71-reactive CD4+ T cells in these individuals. Some CD69+ vimentin59–71-reactive memory CD4+ T cells also expressed NKG2D (figure 4G). In contrast, HC vimentin-64-Cit59–71-reactive and vimentin59–71-reactive CD4+ T cells did not differ in phenotype from total CD4+ T cells (online Supplementary figure 2). Thus, in INA FDRs and patients with RA but not HC, vimentin-64-Cit59–71-reactive and vimentin59–71-reactive CD4+ T cells reflect antigen-driven activation and differentiation.
TCR bias and oligoclonal TCR reactive with vimentin-64-Cit59–71 and vimentin59–71
To obtain evidence of in vivo expansion in HLA-DRB1*14:02+ INA, we used multiplex PCR to sequence the TCRs from a total of 53 single vimentin59–71-reactive and 71 vimentin-64Cit59–71-reactive tetramer-positive CD4+ T cells derived from five patients with HLA-DRB1*14:02+ RA and five HLA-DRB1*14:02+ FDRs. TRAV and TRBV gene usage among CD4+ T cells reactive to each vimentin epitope was generally diverse in RA and FDR (online supplementary table 4). However, TRBV20–1 and TRBV30 were preferentially used variable gene segments for recognition of vimentin 64-Arg and 64-Cit variant peptides bound to HLA-DRB1*14:02 (figure 4H). TRAV13–2 and TRAV26–1 were preferentially used variable gene segments among both vimentin59–71-reactive and vimentin-64Cit59–71-reactive TCRs. Use of preferential variable gene segments in the repertoires of T cells recognising each epitope was observed in both patients with RA and FDRs and suggested underlying oligoclonality of the autoreactive T cell populations when compared with the total CD3+ T cell population (figure 4H). Indeed, multiple vimentin-64Cit59–71-reactive and vimentin59–71-reactive CD4+ T cells bearing the same CDR3α and/or CDR3β sequences were identified among the single cells sorted from two of the patients with RA and two FDRs (table 2). In two FDRs, CD4+ T cells bearing the same TRBV30 CDR3 sequences were identified multiple times among single cells reactive for vimentin 64-Arg and 64-Cit variant peptides bound to HLA-DRB1*14:02. In one patient with RA, the same TRBV2 CDR3 sequences were identified multiple times among single vimentin-64Cit59–71-reactive CD4+ T cells, and in another patient the same TRBV10–3 CDR3 sequences were identified multiple times among single vimentin59–71-reactive CD4+ T cells. These repeated CDR3α and CDR3β sequences indicate antigen-reactive clonal expansion within the blood of these HLA-DRB1*14:02+ patients and at-risk FDRs (table 2). In all cases, clonally expanded tetramer+ T cells were CD4+CD25+CD127+ Teff, as determined by index sorting. Individuals in whom any oligoclonal sequences were detected in peripheral blood (PB) were more likely to be HLA-DRB1*14:02 homozygous or HLA-DRB1*14:02/*04:04 compound heterozygous than individuals without oligoclonal expansion (p<0.05, X2 test). Remarkably, a common TRBV30 CDR3 sequence (SI/VGAGNQPQ) was expanded in the blood of two individual FDRs, which in each case encoded TCRs recognising vimentin59–71 as well as vimentin-64Cit59–71. Of 34 sorted antigen-reactive T cells yielding productive TRBV gene sequences from FDRs, 23.5% contained this CDR3β sequence. We used retroviral vectors encoding HLA-DR14:02-vimentin59–71-restricted TCR P2F3 (online supplementary table 4, bold) to transduce the αβ TCR-deficient SKW-3 cell line.32,33 When stimulated with HLA-DRB1*14:02-vimentin59–71 or vimentin-64Cit59–71 tetramers, P2F3 SKW-3 cells upregulated phospho- extracellular signal-related kinases (pERK) relative to HLA-DRB1*14:02-CLIP tetramers (figure 4I), confirming that TCR identified from cells with HLA-DRB1*14:02-vimentin59–71 tetramer reactivity recognises vimentin59–71 and vimentin-64Cit59–71 in the context of HLA-DRB1*14:02.
Table 2.
Repeated TCR TRAV, TRBV and TRAV/TRBV clonotypes used by HLA-DRB1*14:02-restricted vimentin59–71 or vimentin-64Cit59–71-reactive CD4+ T cells
| Subject | Tetramer | TRAV | CDR3α | TRAJ | Frequency number/Total, (%) | HLA-DRB1 |
| RA W1:1 | Cit-vimentin | 13–2 | SQPGTAL | 15 | 2/17 (11.8%) | 14:02, 14:02 |
| RA W1:2 | Vimentin | 26–1 | SGAGSYQL | 28 | 5/13 (38%) | 04:04, 14:02 |
| Subject | Tetramer | TRBV | CDR3β | TRBJ | Frequency* (%) | |
| RA W1:1 | Cit-vimentin | 2 | SEAADNEQ | 2–1 | 2/14 (14.2%) | 14:02, 14:02 |
| RA W1:2 | Vimentin | 10–3 | GGTRTESSYEQ | 2–7 | 2/5 (40%) | 04:04, 14:02 |
| FDR W1:3* | Cit-vimentin | 30 | SIGAGNQPQ | 1–5 | 1/19 (5.2%) | 04:04, 14:02 |
| FDR W1:3* | Vimentin | 30 | SIGAGNQPQ | 1–5 | 3/7 (42.8%) | |
| FDR W1:5† | Cit-vimentin | 30 | SVGAGNQPQ | 1–5 | 2/2 (100%) | 13:02, 14:02 |
| FDR W1:5† | Vimentin | 30 | SVGAGNQPQ | 1–5 | 2/6 (33%) |
| Subject | Tetramer | TRAV/TRBV | CDR3a/CDR3β | TRAJ/TRBJ | Frequency* (%) | ||||
| RA W1:1 | Cit-vimentin | 13–2/2 | SQPGTAL/SEAADNEQ | 15/2–1 | 2/12 (16.6%) | 14:02, 14:02 | |||
| Subject | TRBV | ||||||||
| FDR W1:3† | S | I | G | A | G | N | Q | P | Q |
| agt | atc | ggg | gcg | ggc | aat | cag | ccc | cag | |
| FDR W1:5† | S | V | G | A | G | N | Q | P | Q |
| agt | gtg | ggg | gca | ggc | aat | cag | ccc | cag |
TCR repertoire analysis was undertaken in RA patients W1:1–5 and FDR W1:1–5. Productive single TRAV and TRBV clonotypes detected from two patients with RA and three FDRs are shown.
Frequency reflects the frequency of the repeated TRAV or TRBV clonotype divided by the total number of tetramer+ cells with productive TRAV or TRBV sequence, for each individual.
Nucleotide sequences encoding each of the public CDR3β amino acid sequences, which require a minimal number of N or P additions to be produced. Nucleotides attributed by the germline Vβ, Dβ and Jβ genes are shown in blue, red and green, respectively. N-additions are in black and P-additions in purple text. The nucleotides at the D–J junction encoding the same amino acid are underlined in each case.
FDR, first-degree relatives; RA, rheumatoid arthritis.
The biased TCR usage suggests a structural requirement for conserved amino acid sequences to recognise vimentin59–71 and vimentin-64Cit59–71. Nucleotide sequences encoding TRBV30 CDR3 reveal that although the second N region at the D–J junction in each TCR is different, they encode the same amino acid sequence (table 2). These data implicate convergent recombination events in the selection of this sequence during TCR gene rearrangement.34
DISCUSSION
HLA-DRB1*14:02 and HLA-DRB1*04:04 are shown to be independent risk alleles for ACPA+ RA in the INA population. Analysis of the structures and T cell responses to Cit and non-Cit epitopes shows that, consequent to the His13βSer polymorphism and altered P4 pocket, HLA-DRB1*14:02 can present both variant peptides. This contrasts with structures of HLA-DRB1*04:01 and *04:04, in which Cit but not Arg can be accommodated in P4.12,29 Presentation of both 64-Cit and 64-Arg vimentin59–71 variants promoted autoreactive CD4+ T cell activation and differentiation to Teff and Treg, and clonal expansion of Teff in patients with HLA-DRB1*14:02+ RA and at-risk FDRs. In HLA-DRB1*14:02+ HC, we observed no activation or differentiation of antigen-specific T cells above the background total CD4+ T cells. Previous studies of CD4+ T cells in individuals carrying Caucasian SE alleles show that T cell responses to Cit-autoantigenic peptides are increased relative to Arg-variant peptides,12,29,35,36 reinforcing that T cell function aligns with HLA-DR-peptide structure.
We show preferential variable gene segments and clonally expanded TCR among vimentin59–71-reactive and vimentin-64Cit59–71-reactive CD4+CD25+CD127+ Teff sorted from patients with RA and FDRs, including a public TRBV CDR3. These data suggest that presentation of vimentin self-epitopes in vivo continues in genetically predisposed individuals before and after onset of RA, and selects T cells making productive TCR rearrangements, as identified by the nucleotide sequences, for antigen recognition and T cell expansion.
Although limited by small sample numbers, the phenotypic profiles of 64-Cit and 64-Arg variant-specific autoreactive T cells appeared to be different. The fibroblast antigen, vimentin, has widespread tissue expression.37 Vimentin 64-Arg-specific memory CD4+ T cells specifically expressed CD69. CD69 is a marker of recent activation or tissue residency and exposure to cytokines such as tumour necrosis factor (TNF),30 suggesting that vimentin59–71-reactive T cells are activated in tissue inflammatory sites. NKG2D signifies Teff costimulatory function and is TNF-activated.31 Intriguingly, our data suggest cross-reactivity of some TCRs for the 64-Cit and 64-Arg variants: cell frequency of each specificity was correlated, the public clonotype occurred in T cells reactive with each variant, and T cells expressing TCR P2F3 were activated with both tetramers. In general, coexpansion of T cells recognising Cit and Arg variants might be advantageous, for example, for cross-protection against infectious antigens requiring rapid immunity.38–40 This suggests a hypothesis for persistence of HLA-DRB1*14:02 in the INA population, even though its molecular structure permits presentation of multiple self-antigens driving ACPA+ RA.
Low-titre ACPA develops in healthy individuals independent of HLA-DR SE, particularly in inflammatory contexts.6–8,10,11 In HLA-DR SE+ individuals, memory T cells driven by antigen experience would provide required B cell help for increased titres and epitope spreading in patients developing RA.8,41,42 Since HLA-DRB1*14:02 broadens capacity for autoantigen presentation and T cell expansion, our study provides a mechanism for enhanced risk of early onset of ACPA+ RA in INA.
Supplementary Material
Acknowledgements
We thank the staff at the National Australian Synchrotron for assistance with data collection, and staff at the Monash Macromolecular Crystallisation Facility and Nishta Ramnoruth for assistance with flow cytometric staining.
Funding This work was supported by the Australian National Health and Medical Research Council (NHMRC), Australian Research Council (ARC) and Canadian Institutes of Health Research (CIHR: MOP77700) funding. JR is an ARC Australian Laureate Fellow and RT an NHMRC Fellow.
Footnotes
Competing interests There was no commercial support for this work. However we wish to declare that RT is a director of a spin-off company that is commercialising antigen-specific immunotherapy for rheumatoid arthritis.
Ethics approval University of Manitoba, Alaska Area Institutional Review Board, Metro South HREC, University of Queensland HREC.
Provenance and peer review Not commissioned; externally peer reviewed.
Data sharing statement No additional data are available.
REFERENCES
- 1.Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis. an approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum 1987;30:1205–13. [DOI] [PubMed] [Google Scholar]
- 2.Raychaudhuri S, Sandor C, Stahl EA, et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet 2012;44:291–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han B, Diogo D, Eyre S, et al. Fine mapping seronegative and seropositive rheumatoid arthritis to shared and distinct HLA alleles by adjusting for the effects of heterogeneity. Am J Hum Genet 2014;94:522–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ireland JM, Unanue ER. Autophagy in antigen-presenting cells results in presentation of citrullinated peptides to CD4 T cells. J Exp Med 2011;208:2625–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ireland JM, Unanue ER. Processing of proteins in autophagy vesicles of antigen-presenting cells generates citrullinated peptides recognized by the immune system. Autophagy 2012;8:429–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Padyukov L, Silva C, Stolt P, et al. A gene-environment interaction between smoking and shared epitope genes in HLA-DR provides a high risk of seropositive rheumatoid arthritis. Arthritis Rheum 2004;50:3085–92. [DOI] [PubMed] [Google Scholar]
- 7.Quirke AM, Perry E, Cartwright A, et al. Bronchiectasis is a Model for chronic bacterial infection inducing autoimmunity in rheumatoid Arthritis. Arthritis Rheumatol 2015;67:2335–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koning F, Thomas R, Rossjohn J, et al. Coeliac disease and rheumatoid arthritis: similar mechanisms, different antigens. Nat Rev Rheumatol 2015;11:450–61. [DOI] [PubMed] [Google Scholar]
- 9.Alpizar-Rodriguez D, Brulhart L, Mueller RB, et al. The prevalence of anticitrullinated protein antibodies increases with age in healthy individuals at risk for rheumatoid arthritis. Clin Rheumatol 2017;36:677–82. [DOI] [PubMed] [Google Scholar]
- 10.Fisher BA, Cartwright AJ, Quirke AM, et al. Smoking, Porphyromonas gingivalis and the immune response to citrullinated autoantigens before the clinical onset of rheumatoid arthritis in a southern european nested case-control study. BMC Musculoskelet Disord 2015;16:331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Terao C, Asai K, Hashimoto M, et al. Significant association of periodontal disease with anti-citrullinated peptide antibody in a japanese healthy population - The Nagahama study. JAutoimmun 2015;59:85–90. [DOI] [PubMed] [Google Scholar]
- 12.Scally SW, Petersen J, Law SC, et al. A molecular basis for the association of the HLA-DRB1 locus, citrullination, and rheumatoid arthritis. J Exp Med 2013;210:2569–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.James EA, Rieck M, Pieper J, et al. Citrulline-specific Th1 cells are increased in rheumatoid arthritis and their frequency is influenced by disease duration and therapy. Arthritis Rheumatol 2014;66:1712–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peschken CA, Esdaile JM. Rheumatic diseases in North America’s indigenous peoples. Semin Arthritis Rheum 1999;28:368–91. [DOI] [PubMed] [Google Scholar]
- 15.Ferucci ED, Darrah E, Smolik I, et al. Prevalence of anti-peptidylarginine deiminase type 4 antibodies in rheumatoid arthritis and unaffected first-degree relatives in indigenous North American Populations. J Rheumatol 2013;40:1523–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smolik I, Robinson DB, Bernstein CN, et al. First-degree relatives of patients with rheumatoid arthritis exhibit high prevalence of joint symptoms. J Rheumatol 2013;40:818–24. [DOI] [PubMed] [Google Scholar]
- 17.Willkens RF, Nepom GT, Marks CR, et al. Association of HLA-Dw16 with rheumatoid arthritis in Yakima Indians. further evidence for the “shared epitope” hypothesis. Arthritis Rheum 1991;34:43–7. [DOI] [PubMed] [Google Scholar]
- 18.Nelson JL, Boyer G, Templin D, et al. HLA antigens in Tlingit Indians with rheumatoid arthritis. Tissue Antigens 1992;40:57–63. [DOI] [PubMed] [Google Scholar]
- 19.Williams RC, Jacobsson LT, Knowler WC, et al. Meta-analysis reveals association between most common class II haplotype in full-heritage native Americans and rheumatoid arthritis. Hum Immunol 1995;42:90–4. [DOI] [PubMed] [Google Scholar]
- 20.Sokolove J, Bromberg R, Deane KD, et al. Autoantibody epitope spreading in the pre-clinical phase predicts progression to rheumatoid arthritis. PLoS One 2012;7:e35296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tungatt K, Bianchi V, Crowther MD, et al. Antibody stabilization of peptide-MHC multimers reveals functional T cells bearing extremely low-affinity TCRs. J Immunol 2015;194:463–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang GC, Dash P, McCullers JA, et al. T cell receptor αβ diversity inversely correlates with pathogen-specific antibody levels in human cytomegalovirus infection. Sci Transl Med 2012;4:128ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagner CA, Sokolove J, Lahey LJ, et al. Identification of anticitrullinated protein antibody reactivities in a subset of anti-CCP-negative rheumatoid arthritis: association with cigarette smoking and HLA-DRB1 ‘shared epitope’ alleles. Ann Rheum Dis 2015;74:579–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geluk A, van Meijgaarden KE, Southwood S, et al. HLA-DR3 molecules can bind peptides carrying two alternative specific submotifs. J Immunol 1994;152:5742–8. [PubMed] [Google Scholar]
- 25.Hennecke J, Wiley DC. Structure of a complex of the human alpha/beta T cell receptor (TCR) HA1.7, influenza hemagglutinin peptide, and major histocompatibility complex class II molecule, HLA-DR4 (DRA*0101 and DRB1*0401): insight into TCR cross-restriction and alloreactivity. J Exp Med 2002;195:571–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stern LJ, Brown JH, Jardetzky TS, et al. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature 1994;368:215–21. [DOI] [PubMed] [Google Scholar]
- 27.James EA, Moustakas AK, Bui J, et al. HLA-DR1001 presents “altered-self” peptides derived from joint-associated proteins by accepting citrulline in three of its binding pockets. Arthritis Rheum 2010;62:2909–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.James EA, Moustakas AK, Bui J, et al. The binding of antigenic peptides to HLA-DR is influenced by interactions between pocket 6 and pocket 9. J Immunol 2009;183:3249–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerstner C, Dubnovitsky A, Sandin C, et al. Functional and Structural Characterization of a Novel HLA-DRB1*04:01-Restricted α-Enolase T Cell Epitope in Rheumatoid Arthritis. Front Immunol 2016;7:494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seddiki N, Santner-Nanan B, Martinson J, et al. Expression of interleukin (IL)-2 and IL-7 receptors discriminates between human regulatory and activated T cells. J Exp Med 2006;203:1693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakaguchi S Regulatory T cells: key controllers of immunologic self-tolerance. Cell 2000;101:455–8. [DOI] [PubMed] [Google Scholar]
- 32.Gras S, Chen Z, Miles JJ, et al. Allelic polymorphism in the T cell receptor and its impact on immune responses. J Exp Med 2010;207:1555–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beringer DX, Kleijwegt FS, Wiede F, et al. T cell receptor reversed polarity recognition of a self-antigen Major histocompatibility complex. Nat Immunol 2015;16:1153–61. [DOI] [PubMed] [Google Scholar]
- 34.Kedzierska K, Thomas PG, Venturi V, et al. Terminal deoxynucleotidyltransferase is required for the establishment of private virus-specific CD8+ TCR repertoires and facilitates optimal CTL responses. J Immunol 2008;181:2556–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Law SC, Street S, Yu CH, Ch Y, et al. T-cell autoreactivity to citrullinated autoantigenic peptides in rheumatoid arthritis patients carrying HLA-DRB1 shared epitope alleles. Arthritis Res Ther 2012;14:R118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.James EA, Rieck M, Pieper J, et al. Citrulline-specific Th1 cells are increased in rheumatoid arthritis and their frequency is influenced by disease duration and therapy. Arthritis Rheumatol 2014;66:1712–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kishaba Y, Matsubara D, Niki T. Heterogeneous expression of nestin in myofibroblasts of various human tissues. Pathol Int 2010;60:378–85. [DOI] [PubMed] [Google Scholar]
- 38.Furman D, Jojic V, Sharma S, et al. Cytomegalovirus infection enhances the immune response to influenza. Sci Transl Med 2015;7:281ra43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang MH, Moonesinghe R, Athar HM, et al. Trends in disparity by sex and race/Ethnicity for the leading causes of death in the United States-1999–2010. J Public Health Manag Pract 2016;22 Suppl 1(Suppl 1):S13–S24. [DOI] [PubMed] [Google Scholar]
- 40.Foote EM, Singleton RJ, Holman RC, et al. Lower respiratory tract infection hospitalizations among american indian/Alaska native children and the general United States child population. Int J Circumpolar Health 2015;74:29256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hunt L, Hensor EM, Nam J, et al. T cell subsets: an immunological biomarker to predict progression to clinical arthritis in ACPA-positive individuals. Ann Rheum Dis 2016;75:1884–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Janssen KM, Westra J, Chalan P, et al. Regulatory CD4+ T-Cell subsets and Anti-Citrullinated protein antibody repertoire: potential biomarkers for Arthritis Development in Seropositive Arthralgia Patients? PLoS One 2016;11:e0162101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.