Abstract
Background
Plasmodium species feature only four to eight nuclear ribosomal units on different chromosomes, which are assumed to evolve independently according to a birth-and-death model, in which new variants originate by duplication and others are deleted throughout time. Moreover, distinct ribosomal units were shown to be expressed during different developmental stages in the vertebrate and mosquito hosts. Here, the 18S rDNA sequences of 32 species of avian haemosporidian parasites are reported and compared to those of simian and rodent Plasmodium species.
Methods
Almost the entire 18S rDNAs of avian haemosporidians belonging to the genera Plasmodium (7), Haemoproteus (9), and Leucocytozoon (16) were obtained by PCR, molecular cloning, and sequencing ten clones each. Phylogenetic trees were calculated and sequence patterns were analysed and compared to those of simian and rodent malaria species. A section of the mitochondrial CytB was also sequenced.
Results
Sequence patterns in most avian Plasmodium species were similar to those in the mammalian parasites with most species featuring two distinct 18S rDNA sequence clusters. Distinct 18S variants were also found in Haemoproteus tartakovskyi and the three Leucocytozoon species, whereas the other species featured sets of similar haplotypes. The 18S rDNA GC-contents of the Leucocytozoon toddi complex and the subgenus Parahaemoproteus were extremely high with 49.3% and 44.9%, respectively. The 18S sequences of several species from all three genera showed chimeric features, thus indicating recombination.
Conclusion
Gene duplication events leading to two diverged main sequence clusters happened independently in at least six out of seven avian Plasmodium species, thus supporting evolution according to a birth-and-death model like proposed for the ribosomal units of simian and rodent Plasmodium species. Patterns were similar in the 18S rDNAs of the Leucocytozoon toddi complex and Haemoproteus tartakovskyi. However, the 18S rDNAs of the other species seem to evolve in concerted fashion like in most eukaryotes, but the presence of chimeric variants indicates that the ribosomal units rather evolve in a semi-concerted manner. The new data may provide a basis for studies testing whether differential expression of distinct 18S rDNA also occurs in avian Plasmodium species and related haemosporidian parasites.
Keywords: 18S ribosomal RNA, Plasmodium, Haemoproteus, Leucocytozoon, Birth-and-death evolution, Concerted evolution
Background
Ribosomal RNAs of eukaryotes
Ribosomal RNAs (rRNAs) form the core part of the ribosomes and are essential for protein synthesis in all living organisms. Due to functional constraints rRNAs are among the most conserved nucleic acids in nature, and at the same time they constitute also the vast majority of RNA molecules in cells, e.g., the rRNA content (dry weight) of Escherichia coli cells varies from 20% in the early exponential growth phase to 2% in developed cells [1]. These characteristics render rRNAs suitable targets for various molecular genetic approaches. Ribosomal RNAs and their genomic counterparts, the ribosomal DNAs (rDNAs), are the most common targets in hybridization and screenings assays.
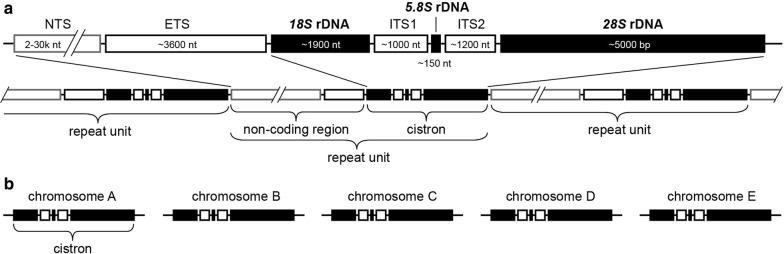
Cytoplasmic ribosomes of eukaryotes are composed of four rRNAs and over 50 proteins, which together form the small ribosomal subunit (SSU) and the large ribosomal subunit (LSU). The 18S rRNA is the rRNA component of the SSU, while 28S rRNA, 5.8S rRNA, and 5S rRNA are the rRNAs of the LSU. Eukaryote nuclear rDNA genes are arranged in units containing 18S rDNA, 5.8S rDNA, and 28S rDNA, separated by the internal transcribed spacers ITS1 and ITS2. The rDNA units of multicellular eukaryotes also include a non-transcribed spacer (NTS) and an external transcribed spacer (ETS), which like ITS1 and ITS2 are not involved in the formation of the ribosomes, but are assumed to play a role in rDNA transcription [2] (Fig. 1a). The 5S rDNAs are usually not directly associated with the other rDNA units but are located in different genomic regions.
Fig. 1.
a Ribosomal units of multicellular eukaryotes and arrangement in ribosomal clusters (NTS: non-transcribed spacer, ETS: external transcribed spacer). b Arrangement of ribosomal units in Plasmodium species. The ribosomal units of Plasmodium spp. do not contain NTS and ETS regions and are not arranged in clusters with multiple units
The rDNA units of most eukaryotes are arranged in clusters of so-called tandem repeats on one or several chromosomes, with each cluster containing up to several hundred copies. The individual rDNA units do not evolve independently, but according to a model of concerted evolution, a process that leads to homogenization [3–5]. Concerted evolution involves mechanisms such as unequal crossing over during recombination, gene duplication, and inter-chromosomal gene conversion, and is assumed to affect the evolution of members of most multigene families [6]. However, there are exceptions to this rule. Multigene families involved in the immune system such as immunoglobulins and the major histocompatibility complex (MHC) do not evolve in a concerted fashion [7], but they are assumed to follow a model of birth-and-death evolution [7, 8]. In this model, new copies originate by gene duplication, whereas others become non-functional and deleted over time. In respect to the evolution of their rDNAs, human, simian and rodent Plasmodium species are considered exceptional within the eukaryote domain because their rDNA units are assumed to evolve according to a birth-and-death model under strong purifying selection. Plasmodium species feature only four to eight rDNA units per haploid genome, which are located on different chromosomes and are assumed to accumulate mutations independently [e.g., 9] (Fig. 1b). Moreover, distinct units were shown to be expressed during different developmental stages of the parasites in the vertebrate and mosquito hosts [10].
The 18S rRNAs of Plasmodium
The first notable studies on 18S rDNAs of Plasmodium were published in the early 1980s when DNA sequencing was still at the very beginning. Although complete sequences were not available at that time, mapping of rDNAs by restriction enzyme analysis and Southern blot hybridization allowed determining the number of distinct units. By using these methods, four nuclear rDNA units were identified in the rodent malaria parasite Plasmodium berghei in different genomic regions, and they were found not to be arranged in tandem arrays like in other eukaryotes [11]. Moreover, the four units could be assigned to two classes, putatively being expressed during different developmental stages [11, 12]. The so-called A-type rDNA units were found to be expressed in asexual blood-stages in the vertebrate host, whereas the expression patterns of the C-type units were still unknown at that time [13]. The naming of rDNA types is quite inconsistent because expression patterns of the C-type rDNAs were unknown at first. Later, McCutchan et al. [14] proposed consistently using the term S-type for the sporozoite specific stages instead of C-type. This recommendation is followed here by using the terms A-type or S-type in the figures and discussion. The first 18S rDNA sequences of P. berghei were published several years later, allowing to directly compare the variants for the first time, and to design oligonucleotide probes specifically targeting the A-type or the C-type [S-type] genes. Hybridization of oligonucleotide probes to rRNA isolated from different life stages lead to the discovery that the C-type [S-type] is expressed in the sporozoite stage in the mosquito vector [15]. Studies on simian malaria species rendered comparable results as those on P. berghei. Restriction mapping of rDNA fragments of the human malaria parasite Plasmodium falciparum also revealed the presence of at least four distinct units belonging to two classes of transcription units [16]. Two structurally distinct 18S rDNA genes were sequenced from P. falciparum and only the A-type was found to be expressed in blood-stage parasites [17]. Of the two C-type [S-type] rDNAs only one type (C2) was highly expressed in the sporozoite stages [18]. The switch from A-type to C-type [S-type] gene expression in P. falciparum was shown to involve the control of rRNA processing [10]. In Plasmodium cynomolgi, a parasite of Old World monkeys, five rDNA units were found to be expressed in tissue and blood stages in the vertebrate host, besides a single one being expressed in sporozoites [19]. A-type and S-type 18S rDNAs were also sequenced and studied in the human malaria species Plasmodium vivax [20–22]. Later a third class of rDNA units, the so-called O-type, was characterized in P. vivax, which is expressed in ookinetes and oocysts developing in the mosquito midgut [23]. A further class, mainly expressed in maturing ookinetes and young oocysts, was also identified in the rodent malaria parasite Plasmodium yoelii. It was termed D-type and characterized as a subtype of the S-type genes [24]. The number of 18S rDNAs was also determined for the avian parasite Plasmodium lophurae using Southern blot analysis and restriction analysis, supporting the presence of six copies, which may be divided into four classes [25].
The temperature was shown to be a major factor involved in transcription of distinct rDNA types. In laboratory cultures of P. falciparum, both A-type genes were preferentially expressed at higher temperatures and the S-type genes at lower temperatures. This pattern might relate to differences in body temperatures between the vertebrate and mosquito hosts [18]. Since parasites entering the mosquito host face a severe drop in glucose concentration compared to the vertebrate host, glucose is another potential regulator of gene transcription. Asexual blood stages of P. falciparum subjected to glucose starvation decreased expression of A-type genes and increased expression of the S-type genes. This effect was shown to be in synergy with temperature changes related to the host shift [26].
For phylogenetic approaches 18S rDNAs expressed in asexual stages were sequenced from a wider range of Plasmodium species, including Plasmodium fragile from macaques [27] and Plasmodium malariae from humans [28], and Plasmodium lophurae [29] and Plasmodium gallinaceum [27] from poultry. The A-type 18S rDNAs of the primate malaria species Plasmodium knowlesi and Plasmodium cynomolgi were sequenced by Waters et al. [30]. The first 18S rDNAs of lizard Plasmodium species were published for Plasmodium mexicanum (accession number L11716) and Plasmodium floridense (L11717) on GenBank only. In a phylogenetic approach, the A-type and S-type 18S rDNAs were analysed of several simian Plasmodium species related to P. vivax: Plasmodium coatneyi, Plasmodium inui, Plasmodium hylobati, Plasmodium fieldi, Plasmodium simiovale, P. fragile, and P. cynomolgi [31]. The latter simian malaria species are closely related to each other and all feature several A-type gene variants, but only a single S-type gene. Comparison of 18S rDNAs strongly indicates that gene duplication events resulting in A-type and S-type genes happened independently at least three times in the genus Plasmodium, which is again supporting evolution according to a model of birth-and-death evolution [31].
In the last two decades, new data on rDNAs of Plasmodium species became available by sequencing complete nuclear genomes of several simian and rodent malaria species. For the first time, not only the numbers and sequences of rDNA units were determined, but they could be assigned to certain chromosomes as well. The first haemosporidian genome published was that of P. falciparum (BioProject: PRJNA148 [32]), followed by those of P. knowlesi (PRJNA33651, [33]), P. vivax (PRJNA20431, [34]), P. cynomolgi (PRJDA49901 [35]), Plasmodium gaboni (PRJNA329100 [36]), Plasmodium reichenowi (PRJNA329100, [36]), P. coatneyi (PRJNA329102 [37]), and the two rodent malaria species P. berghei (PRJNA317456 [38]) and P. yoelii (PRJNA317465 [38]). Annotated nuclear genomes without related publication are available on GenBank from P. ovale curtisi (PRJEB12678) and P. ovale wallikeri (PRJEB12679), P. malariae (PRJEB14392), Plasmodium chabaudi (PRJNA317457) and Plasmodium relictum (PRJEB9074), whereas only raw genome assemblies are available from P. inui (PRJNA257224), Plasmodium vinckei vinckei (PRJNA255250), Plasmodium vinckei petteri (PRJNA257099) and P. fragile (PRJNA282950). The first haemosporidian genome sequenced of a non-Plasmodium species was that of Haemoproteus (Parahaemoproteus) tartakovskyi (PRJNA309868 [39]).
The 18S rDNAs were the first nucleotide sequences available for simian Plasmodium species and therefore chosen as targets for parasite screening and detection approaches. Moreover, they feature a succession of extremely conserved and variable sequence regions, which allows detecting and differentiating both high level-taxa (orders, families, genera) and low-level taxa (species, strains). The first approach for distinguishing the human malaria parasites P. falciparum, P. vivax, P. malariae, and P. ovale by hybridizing specific oligonucleotide probes to the parasites’ 18S rRNAs in processed human blood samples followed by autoradiography was developed by Waters and McCutchan [40]. The first comprehensive PCR assay with species-specific primers targeting the 18S rDNAs of the four above-mentioned species was developed by Snounou et al. [41]. Several other PCR protocols for the detection of simian Plasmodium species were published later [42–45].
The 18S rDNAs were the first sequences analysed in human, simian and rodent Plasmodium species, and they are still the targets of choice in molecular screening approaches. Moreover, ribosomal genes of these Plasmodium species are assumed to evolve according to a birth-and-death model and to be differentially expressed in the vertebrate and mosquito hosts, respectively. Despite the strong interest in 18S rDNAs of human, simian and rodent malaria species, ribosomal genes have been neglected widely in studies on avian Plasmodium species and related haemosporidians. For the first time, the 18S rDNAs of a wider range of avian haemosporidians were analysed, including seven Plasmodium, nine Haemoproteus, and 16 Leucocytozoon species. A central question was whether avian haemosporidians feature sequentially distinct ribosomal units like the simian and rodent malaria species, which would be indicative not only for differential expression of distinct rDNA units but also for evolution according to a birth-and-death model. The new data open the way for practical applications in parasite detection (hybridization and PCR assays) but also may facilitate studies on differential gene expression in haemosporidians other than human, simian and rodent Plasmodium species.
Methods
Sample collection and preparation
For the present study, blood and tissue samples of birds containing parasites of three haemosporidian genera, Plasmodium, Haemoproteus, and Leucocytozoon, were collected. The 18S rDNAs were analysed of 17 individual samples containing seven Plasmodium species belonging to four subgenera (see Table 1): Plasmodium (Haemamoeba) relictum SGS1 (3 individuals), Plasmodium (Haemamoeba) matutinum LINN1 (2)/AFTRU5 (1), Plasmodium (Huffia) elongatum GRW06 (3)/ERIRUB01 (1), Plasmodium (Novyella) vaughani SYAT05 (2), Plasmodium (Novyella) delichoni COLL6 (1), Plasmodium (Giovannolaia) homocircumflexum COLL4 (1), and Plasmodium (Giovannolaia) sp. WW12 (2). One sample of Haemoproteus (Haemoproteus) columbae COLIV06 was analysed, besides nine samples belonging to eight species of the subgenus Parahaemoproteus: Haemoproteus (Parahaemoproteus) balmorali ROBIN1, Haemoproteus (Parahaemoproteus) lanii RBS06, Haemoproteus (Parahaemoproteus) minutus TURDUS2, Haemoproteus (Parahaemoproteus) sp. EMCIR01, Haemoproteus (Parahaemoproteus) syrnii CULKIB01, Haemoproteus (Parahaemoproteus) tartakovskyi SISKIN1, Haemoproteus (Parahaemoproteus) sp. LK03, and Haemoproteus (Parahaemoproteus) sp. STAL2 (2). One sample each was analysed in 16 Leucocytozoon species: Leucocytozoon cf. californicus CIAE02, Leucocytozoon sp. SYCON05, Leucocytozoon sp. TUPHI06, Leucocytozoon sp. PARUS20, Leucocytozoon sp. COCOR18, Leucocytozoon sp. STAL05, Leucocytozoon sp. BUBO01, Leucocytozoon sp. ASOT06, Leucocytozoon sp. STAL3, Leucocytozoon sp. MILVUS2, Leucocytozoon sp. COCOR09, Leucocytozoon sp. BT1, Leucocytozoon sp. COCOR13, Leucocytozoon sp. CIAE03, Leucocytozoon sp. ACNI03, and Leucocytozoon sp. BUTBUT03. The latter three species belong to the Leucocytozoon toddi species complex (including Leucocytozoon mathisi and Leucocytozoon buteonis), parasites of accipitriform predators, which form a clade distinct from that comprising the majority of other Leucocytozoon species. The codes following the species names correspond to lineages assigned in the MalAvi database [46], which serves as a reference collection containing most CytB haplotypes isolated from avian haemosporidians. Most samples were collected from birds at the University of Veterinary Medicine Vienna (Austria). Blood spots were taken from living birds at the Bird and Reptile Clinic (Department for Companion Animals and Horses) (19 samples), and tissue samples were obtained from the tissue collections of the Research Institute of Wildlife Ecology (7) and the Institute of Pathology (3). Two samples were collected at the Biological Station Neusiedler See (Illmitz, Burgenland). Blood samples were obtained from seven living birds caught at the Ornithological Station in Ventes Ragas, Lithuania. Three samples were obtained from experimentally infected canaries at the Nature Research Centre in Vilnius (Lithuania). The sample of H. columbae was collected from blood of Columba livia in Cape Town, South Africa. Blood samples of living birds were taken by puncturing the brachial vein and using heparinized microcapillaries. A drop of blood was transferred to filter papers of which DNA was extracted later. Tissue samples were taken from organs (liver and spleen) of dead birds stored at minus 80 °C. DNA was isolated either from blood spots on filter papers or frozen liver tissue using the DNeasy Blood & Tissue Kit (QIAGEN, Venlo, Netherlands). The manufacturer’s protocol for isolation of DNA from tissue was followed, but two eluates of 100 µl each were made from the same column, the first at 8000 rpm, and the second at 13,000 rpm. The second eluate was used for the PCRs.
Table 1.
Samples analysed in the present study for the 18S rDNAs and CytB
| ID | MalAvi lineage | Species | Host species | Origin | 18S clones no. | 18S accession no. | CytB accession no. |
|---|---|---|---|---|---|---|---|
| AH0013P | SGS1 | Plasmodium (Haemamoeba) relictum | Serinus canaria | LT, Vilnius, Nature Research Centre | 11 | MK650473–MK650483 | MK652231 |
| AH0069P | SGS1 | P. (Haemamoeba) relictum | Parus major | AT, Vienna, VetMed (Pathology) | 11 | MK650484–MK650494 | MK652232 |
| AH0392P | SGS1 | P. (Haemamoeba) relictum | Athene noctua | AT, Vienna, VetMed (FIWI) | 10 | MK650495–MK650504 | MK652233 |
| AH0792P | AFTRU5 | P. (Haemamoeba) matutinum | Phoenicurus ochruros | AT, Vienna, VetMed (Bird clinic) | 12 | MK650524–MK650535 | MK652236 |
| AH0524P | LINN1 | P. (Haemamoeba) matutinum | Turdus merula | AT, Vienna, VetMed (Bird clinic) | 9 | MK650515–MK650523 | MK652235 |
| AH0079P | LINN1 | P. (Haemamoeba) matutinum | Turdus merula | AT, Vienna, VetMed (Pathology) | 10 | MK650505–MK650514 | MK652234 |
| AH0846P | GRW06 | P. (Huffia) elongatum | Picus viridis | AT, Vienna, VetMed (Bird clinic) | 9 | MK650564–MK650572 | MK652240 |
| AH0808P | GRW06 | P. (Huffia) elongatum | Parus major | AT, Vienna, VetMed (Bird clinic) | 10 | MK650554–MK650563 | MK652239 |
| AH0420P | GRW06 | P. (Huffia) elongatum | Corvus corone | AT, Vienna, VetMed (FIWI) | 8 | MK650546–MK650553 | MK652238 |
| AH0012P | ERIRUB01 | P. (Huffia) elongatum | Serinus canaria | LT, Vilnius, Nature Research Centre | 10 | MK650536–MK650545 | MK652237 |
| AH0011P | COLL6 | P. (Novyella) delichoni | Carduelis spinus | LT, Ventes Ragas, Ornithological Station | 10 | MK650573–MK650582 | MK652241 |
| AH0551P | SYAT05 | P. (Novyella) vaughani | Turdus merula | AT, Vienna, VetMed (Bird clinic) | 11 | MK650583–MK650593 | MK652242 |
| AH0824P | SYAT05 | P. (Novyella) vaughani | Turdus merula | AT, Vienna, VetMed (Bird clinic) | 6 | MK650594–MK650599 | MK652243 |
| AH0010P | COLL4 | P. (Giovannolaia) homocircumflexum | Serinus canaria | LT, Vilnius, Nature Research Centre | 12 | MK650600–MK650611 | MK652244 |
| AH0017P | WW12 | P. (Giovannolaia) sp. | Carduelis spinus | LT, Vilnius, Nature Research Centre | 11 | MK650612–MK650622 | MK652245 |
| AH0018P | WW12 | P. (Giovannolaia) sp. | Carduelis spinus | LT, Vilnius, Nature Research Centre | 9 | MK650623–MK650631 | MK652246 |
| CL017 | COLIV03 | Haemoproteus (Haemoproteus) columbae | Columba livia | ZA, Cape Town, FitzPatrick Institute | 11 | MK650632–MK650642 | MK652247 |
| AH0004H | ROBIN1 | H. (Parahaemoproteus) balmorali | Luscinia luscinia | LT, Ventes Ragas, Ornithological Station | 10 | MK650643–MK650652 | MK652248 |
| AH0002H | RBS06 | H. (Parahaemoproteus) lanii | Lanius collurio | LT, Ventes Ragas, Ornithological Station | 10 | MK650653–MK650662 | MK652249 |
| AH0014H | TURDUS2 | H. (Parahaemoproteus) minutus | Turdus merula | LT, Ventes Ragas, Ornithological Station | 8 | MK650663–MK650670 | MK652250 |
| AH0775H | CULKIB01 | H. (Parahaemoproteus) syrnii | Strix uralensis | AT, Vienna, VetMed (Bird clinic) | 9 | MK650671–MK650679 | MK652251 |
| AH0005H | SISKIN1 | H. (Parahaemoproteus) tartakovskyi | Loxia curvirostra | LT, Vilnius, Nature Research Centre | 10 | MK650680–MK650689 | MK652252 |
| AH0141H | STAL2 | H. (Parahaemoproteus) sp. | Strix uralensis | AT, Vienna, VetMed (FIWI) | 8 | MK650690–MK650697 | MK652253 |
| AH0460H | LK03 | H. (Parahaemoproteus) sp. | Falco tinnunculus | AT, Vienna, VetMed (Bird clinic) | 9 | MK650698–MK650706 | MK652254 |
| AH0608H | EMCIR01 | H. (Parahaemoproteus) sp. | Emberiza citrinella | AT, Vienna, VetMed (Pathology) | 6 | MK650707–MK650712 | MK652255 |
| AH0776H | STAL2 | H. (Parahaemoproteus) sp. | Strix uralensis | AT, Vienna, VetMed (Bird clinic) | 11 | MK650713–MK650723 | MK652256 |
| AH0255L | CIAE02 | Leucocytozoon cf. californicus | Circus aeruginosus | AT, Vienna, VetMed (FIWI) | 12 | MK650775–MK650786 | MK652257 |
| AH0040L | STAL3 | Leucocytozoon sp. | Strix aluco | AT, Vienna, VetMed (Bird clinic) | 10 | MK650724–MK650733 | MK652258 |
| AH0053L | TUPHI06 | Leucocytozoon sp. | Turdus merula | AT, Vienna, VetMed (Bird clinic) | 2 | MK650741–MK650742 | MK652259 |
| AH0110L | PARUS20 | Leucocytozoon sp. | Corvus corone cornix | AT, Vienna, VetMed (Bird clinic) | 10 | MK650744–MK650753 | MK652260 |
| AH0145L | STAL5 | Leucocytozoon sp. | Strix aluco | AT, Vienna, VetMed (FIWI) | 10 | MK650754–MK650763 | MK652261 |
| AH0232L | MILVUS02 | Leucocytozoon sp. | Haliaeetus albicilla | AT, Vienna, VetMed (FIWI) | 11 | MK650764–MK650774 | MK652262 |
| AH0286L | COCOR09 | Leucocytozoon sp. | Corvus corone | AT, Vienna, VetMed (FIWI) | 10 | MK650787–MK650796 | MK652263 |
| AH0439L | COCOR18 | Leucocytozoon sp. | Corvus corone cornix | AT, Vienna, VetMed (Bird clinic) | 2 | MK650802–MK650803 | MK652264 |
| AH0441L | COCOR13 | Leucocytozoon sp. | Corvus corone corone | AT, Vienna, VetMed (Bird clinic) | 11 | MK650806–MK650816 | MK652265 |
| AH0517L | ASOT06 | Leucocytozoon sp. | Asio otus | AT, Vienna, VetMed (Bird clinic) | 10 | MK650817–MK650826 | MK652266 |
| AH0856L | BT1 | Leucocytozoon sp. | Phylloscopus trochilus | AT, Burgenland, B.S. Illmitz | 11 | MK650827–MK650837 | MK652267 |
| AH0932L | SYCON05 | Leucocytozoon sp. | Sylvia atricapilla | AT, Burgenland, B.S. Illmitz | 10 | MK650838–MK650847 | MK652268 |
| AH0994L | BUBO01 | Leucocytozoon sp. | Bubo bubo | AT, Vienna, VetMed (Bird clinic) | 11 | MK650848–MK650858 | MK652269 |
| AH0555L | BUTBUT03 | Leucocytozoon sp. (L. toddi complex) | Buteo buteo | AT, Vienna, VetMed (Bird clinic) | 11 | MK650859–MK650869 | MK652270 |
| AH0799L | ACNI03 | Leucocytozoon sp. (L. toddi complex) | Accipiter nisus | AT, Vienna, VetMed (Bird clinic) | 10 | MK650870–MK650879 | MK652271 |
| AH1003L | CIAE03 | Leucocytozoon sp. (L. toddi complex) | Circus aeruginosus | AT, Vienna, VetMed (Bird clinic) | 12 | MK650880–MK650890 | MK652272 |
Indicated are MalAvi lineage names, parasite species, host species, origin of samples, number of clones sequenced, and accession numbers for the 18S rDNA and CytB sequences deposited in NCBI GenBank
18S rDNA primers
Complete 18S rDNA sequences have been published for several Plasmodium species and H. (Parahaemoproteus) tartakovskyi, but no data were available for Leucocytozoon spp. and parasites of other genera and subgenera of the order Haemosporida. The 18S rDNA sequences of Plasmodium and Parahaemoproteus species differ strongly in sequence composition (genetic distances are high even when conserved sections of the 18S are compared) and had to be assessed with two separate primer sets. One primer set was designed based on genome sequences of Plasmodium spp. and allowed amplifying the 18S rDNAs of Plasmodium spp., Leucocytozoon spp., and Haemoproteus columbae. Another primer set was based on two 18S rDNAs isolated from the genome of Haemoproteus (Parahaemoproteus) tartakovskyi (PRJNA309868, [39]) and used for Parahaemoproteus-positive samples only. To identify sequence regions specific to haemosporidians, and to reduce the possibility of unintentional binding to host DNA, 18S rDNAs of various bird species were included in the alignment. At first, a nested PCR approach with two forward and reverse primers each was intended, but most samples featured good quality PCR products in the first round, and the second primer sets were used for some samples only. The primers 18S_P_1F (5′-CAAAGATTAAGCCATGCAAGTGA-3′) and 18S_P_1R (5′-CGGAAACCTTGTTACGACTTCTC-3′), located about 50 base pairs (bp) from the 5′- and 3′-ends of the 18S rDNA, were used to amplify the 18S rDNAs of Plasmodium spp., Leucocytozoon spp., and Haemoproteus columbae. The alternative primers 18S_P_2F (5′-GAACGGCTCATTAAAACAGTTATAATCT-3′) and 18S_P_2R (5′-CGACTTCTCCTTCCTTTAAAAGATAG-3′) are shifted inwards by 100 bp and 20 bp, respectively. The primers 18S_H_1F (5′-TGGTTGATCTTGCCAGTAATATATGT-3′) and 18S_H_1R (5′-CGGAAACCTTGTTACGACTTTTG-3′), located at the 5′-end and 20 bp from the 3′-end, respectively, were used to amplify the 18S rDNAs of species belonging to the subgenus Parahaemoproteus. The alternative primers 18S_H_2F (5′-ACGGCTCCTTAAAACCGTTATAATC-3′ and 18S_H_2R (5′-GCAAAAGGCAGTTACGCATACAG-3′) are shifted inwards by 100 bp and 120 bp, respectively. The primers intended for the nest 1 PCRs allowed for amplification of almost the entire 18S rDNAs (~ 2100 bp) of the haemosporidians under investigation. Based on the 18S rDNAs of Haemoproteus (Haemoproteus) columbae, the primers 18S_Hcol_F2 (5′-ACGGCTCCTTAAAACAGTTATAATCT-3′) and 18S_Hcol_F2 (5′-ACGGCTCCTTAAAACAGTTATAATCT-3′) were designed. They were successfully tested on three H. columbae lineages (COLIV03, COLIV06, and HAECOL1) but not used in the present publication. The latter primers are provided to facilitate future studies on 18S rDNAs of the subgenus Haemoproteus because neither the 18SL_P nor the 18SL_H primer sets allowed for amplification of 18S rDNAs of other lineages in this group.
CytB primers
From all samples, also a section of the mitochondrial Cytochrome B (CytB) was sequenced, which is the standard “DNA-barcode” region for the identification of avian haemosporidian lineages. The common nested PCR approach by Hellgren et al. [47] allows obtaining a 478 bp fragment of the CytB from most avian haemosporidians. PCRs were performed on all samples using the primers HaemNFI and HaemNR3 in the nest 1 PCR, and HaemF/HaemR2 and HaemFL/HaemR2L in the nest 2 PCRs, respectively. The nest 1 PCRs were performed with the original DNA templates and the nest 2 PCRs with each 1 µl product of the first PCRs as template. However, the primers do not allow amplifying all CytB lineages. Samples isolated from birds of the order Accipitriformes were additionally screened with primers specifically targeting parasites of the L. toddi species complex, published by Himmel et al. [48]. These primers allow sequencing a 528 bp section of the CytB. The primers CytB_L2_F (5′-GAGAGTTATGGGCTGGATGGT-3′) and CytB_L2_R (5′-TAGAAAGCCAAGAAATACCATTCTG-3′) were used in the nest 1 PCRs, and CytB_L2_nF (5′-GCTGGATGGTGTTTTAGATAYATGC-3′) and CytB_L2_nR (5′-CCATTCTGGAACAATATGTAAAGGTG-3′) were used in the nest 2 PCRs and for sequencing, respectively. In addition, general primers were designed, which were used for sequencing an 886 bp CytB section in all samples: CytB_HPL_intF1 (5′-GAGAATTATGGAGTGGATGGTG-3′) and CytB_HPL_intR1 (5′-ATGTTTGCTTGGGAGCTGTAATC-3′). The resulting sequences were used for the calculations of the CytB trees.
PCRs and molecular cloning
The PCRs targeting the 18S rDNAs were performed using the GoTaq® Long PCR Master Mix (Promega, Wisconsin, Madison, USA), which has a proof-reading activity and allows for amplification of long PCR products. Master mixes contained 9.5 µl distilled water, 12.5 µl GoTaq® Long PCR Master Mix 2×, 1 µl of each primer (10 pMol/µl), and 1 µl DNA template. PCRs started with an initial denaturation for 2 min at 94 °C, followed by 35 cycles with 30 s at 94 °C, 30 s at 55 °C, 2 min at 68 °C, and a final extension for 10 min at 72 °C. The PCRs for the 886 bp CytB fragment were performed under the same conditions, but with 1 min extension time only. The PCRs targeting all other CytB fragments were performed using the GoTaq® G2 Flexi DNA Polymerase (Promega). The PCRs following the protocol of Hellgren et al. [47] started with an initial denaturation for 2 min at 94 °C, followed by 35 cycles with 30 s at 94 °C, 30 s at 50 °C, 1 min at 72 °C, and a final extension for 10 min at 72 °C. Each 1 µl of nest 1 PCR-product was used as template in the nest 2 PCRs. The nested PCRs targeting the CytB sequences of the L. toddi complex were performed under the same conditions, but at 55 °C in the annealing step. All PCR products were visualized on 1% LB agarose gels using a Gel Doc™ XR + Imager (Biorad, Hercules, California, USA). The CytB PCR-products were sent to Microsynth Austria GmbH (Vienna, Austria) for purification and direct sequencing using the PCR primers (nest 2 primers in case of nested PCRs). The 18S rDNA PCR-products were further processed and subjected to molecular cloning. Each 20 µl of PCR-products were run on 1% LB agarose gels and excised with flamed spatulas. The cut bands were purified using the QIAquick Gel-Extraction Kit (QIAGEN) following the standard protocol and eluted with 20 µl distilled water. Cloning was performed with the TOPO™ TA Cloning™ Kit (Invitrogen, Carlsbad, California, USA) using the pCR™4-TOPO® vector and One Shot® TOP10 competent cells. Following ligation and transformation, E. coli cells were recovered in SOC medium for 1 h at 37 °C, and then plated on LB agar plates and grown for 20 h at 37 °C. From each cloning assay, 15 to 20 individual clones were picked with sterilized tooth sticks and transferred to fresh LB agar plates. The same tooth sticks with remaining E. coli were twisted in PCR-tubes with 25 µl master mix for the colony-PCRs. Colony-PCRs were performed with the GoTaq® Long PCR Master Mix (Promega) under the same conditions as the 18S rDNA PCRs (see above), using the primers M13nF (5′-TGTAAAACGACGGCCAGTGA-3′) and M13nR (5′-GACCATGATTACGCCAAGCTC-3′). The PCR-products of clones carrying inserts of the expected size were sent to Microsynth Austria GmbH (Vienna, Austria) for purification and sequencing (using the colony-PCR primers).
Sequence analyses
The raw forward and reverse sequences (and electropherograms) were carefully checked and aligned in Bioedit v.7.0.8.0 [49]. Following the inspection of individual clones, all sequences were aligned and sorted in MAFFT v.7 [50] applying the default options. In a second control step, all unique positions in the alignment were rechecked in the corresponding electropherograms. For the phylogenetic analyses, GenBank sequences from Plasmodium species for which the whole sets of 18S rDNAs were available (Additional file 1) were included. Sequences of raw genomes were retrieved by performing BLAST searches against the “whole genome shotgun contigs” database of NCBI GenBank. A-type and S-type 18S rDNAs of several simian Plasmodium species published by Nishimoto et al. [31] were also included. The genome of P. malariae (PRJEB14392; Wellcome Trust Sanger Institute WTSI, unpublished) features two similar 18S rDNAs on chromosomes 3 and 10, and an aberrant variant on chromosome 11. The latter variant was too different to be reasonably incorporated in the alignment. Genomic 18S rDNAs were included of P. gallinaceum (PRJEB9073; WTSI, unpublished), but not of the P. relictum (PRJEB9074; WTSI, unpublished), because sequences of the latter were fragmentary only. Moreover, the 18S rDNAs published for Plasmodium cathemerium (AY625607, [51]) and Plasmodium lophurae (X13706, [29]) were not included because these featured only the A-type sequences. The sequence patterns differed strongly between genera, wherefore separate trees were calculated for Plasmodium, Leucocytozoon, and Haemoproteus. The genetic distances between genera were high and even the most conserved sequence sections differed in multiple nucleotide positions. All datasets were aligned with MAFFT v.7 [50] using the option “E-ins-i” for alignments with multiple conserved domains and long gaps. Uncorrected p-distances between 18S rDNA variants were calculated with MEGA v.7.0.21 [52] based on alignments containing all single haplotypes. The alignments were then refined in Bioedit v.7.0.8.0 [49] and trimmed to the size of the shortest sequence(s). For the phylogenetic tree analyses, a subset of clones from the present data set was selected, which included the main sequence variants isolated from each individual. Prior to the tree calculations, all gap positions were removed using trimAl v.1.2 [53]. The 18S alignments produced with the latter approach provided the most reasonable result according to visual inspection by the first author, e.g., because identical sequence regions were correctly aligned throughout the alignment, more reliably than by other programs.
Hypervariable regions containing long inserts or deletions were generally removed by deleting all gap positions from the alignment before the analyses. These alignments also resulted in trees featuring the highest support values. Alternatively, for the Plasmodium data set only, alignments were also calculated with R-Coffee [54], which uses predicted secondary structures of the ribosomal sequences. Moreover, three different trimming options were applied: (1) only gap positions excluded, (2) “strict” algorithm implemented in trimAl v.1.2 [53], and (3) default options on the G-blocks Server (http://molevol.cmima.csic.es/castresana/Gblocks_server.html) [55]. The GC-contents of sequences were calculated with Microsoft Excel v.2019.
Phylogenetic trees were also calculated with the CytB sequences obtained from the same individuals. The CytB sequences were of same length (886 bp) and aligned manually. Substitution models were evaluated for all individual data sets using IQ-TREE [56]. According to the corrected Akaike information criterion (cAIC), the optimal substitution model for all alignments was GTR + I + G. Maximum Likelihood (ML) trees were calculated with IQ-TREE [56] by performing 1000 bootstrap replicates each. Bayesian Inference (BI) trees were calculated with MrBayes v.3.2 [57], the analyses were run for 10 million generations each (2 runs each with 4 chains, one of which was heated) and every thousandth tree was sampled. The first 25% of trees were discarded as burn-in and 50% majority rule consensus trees were calculated from the remaining 37,500 trees each. The program RDP4 (Recombination Detection Program v.4; [58, 59]) was used to test whether distinct 18S variants isolated from the same species showed chimeric features resulting from recombination events. Prior to the RDP4 analyses, the 18S sequences of each species were separately aligned with MAFFT v.7 [52] using the option “E-ins-i”. Following methods, implemented in RDP4, were used to test for recombination: GENECONV [60], MAXCHI [61], Chimaera [62], SiSscan [63], and 3Seq [64].
Results
Nuclear 18S rDNA sequences were obtained from seven avian Plasmodium species, nine Haemoproteus spp. and 16 Leucocytozoon spp. by PCR, cloning and sequencing. Two to twelve clones were sequenced from each sample (404 in total, 9.6 clones in mean; Table 1). The clones of four samples (not listed in publication) were excluded prior to the analyses, because they contained mixed infections with two different Leucocytozoon species, which was not visible in the sequences amplified with the primers by [47], but in those obtained with the CytB_HPL_intF1 and CytB_HPL_intR1 primers (present study). Another 39 clones featuring sequences of the bird hosts were also excluded. The 18S rDNA sequence patterns varied strongly between species, hampering the classification of haplotypes into groups. Haplotypes of some species were clearly distinct from each other, whereas those of others differed only in a few sites. In the following, similar clones or haplotypes are referred to as “variants”, and groups of substantially different variants as “clusters”. Calculations of uncorrected p-distances between variants, clusters, and species clades were performed on 18S rDNA alignments containing all single clones. The alignments for the calculation of phylogenetic trees contained only a selection of 18S rDNA clones, including the main sequence variants obtained from each specimen. For comparison, already published sequences of simian and rodent malaria species were included in the Plasmodium alignment. It must be noted that alignments of ribosomal genes vary quite strongly depending on the taxa included, and variable sections cannot be aligned with confidence between genetically distant taxa and show high levels of saturation. However, variable sections were kept in the alignments by removing only the positions containing gaps, because these contained phylogenetically informative sites on subgroup level. Plasmodium 18S rDNA trees calculated based on a secondary structure alignment, applying three different trimming methods, are provided in Additional files S2, S4, and S4. The sequences are deposited at NCBI GenBank under the accession numbers MK650473–MK650890 (18S rDNA) and MK652231–MK652272 (CytB), respectively.
18S rDNAs of avian Plasmodium species
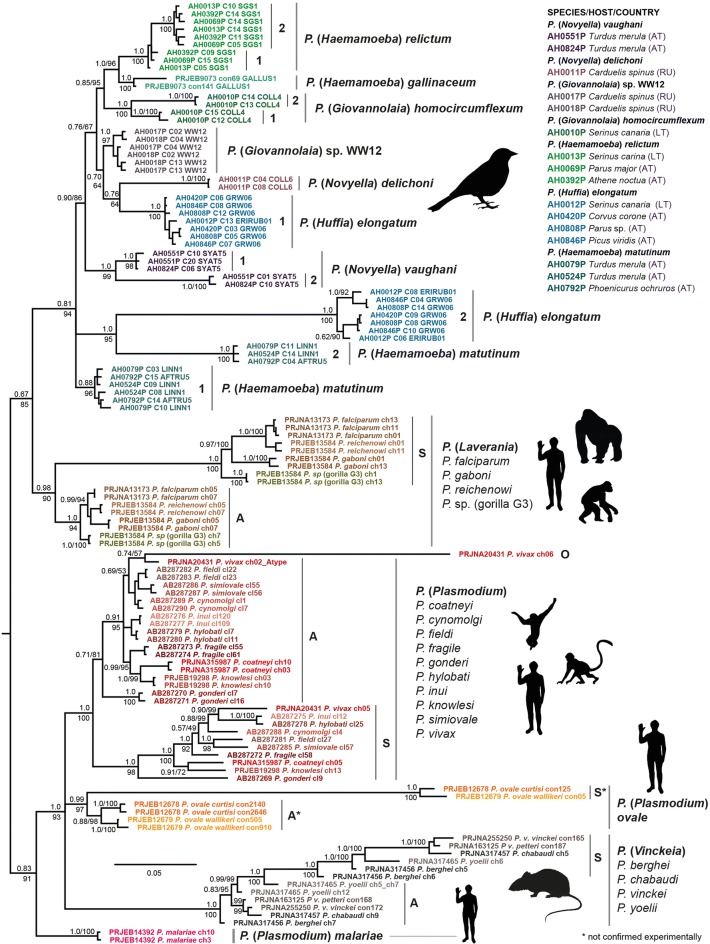
The 18S rDNA data set generated in the present study comprised sequences of seven avian Plasmodium species (16 specimens; 159 clones, 9.9 mean per sample). Multiple samples were analysed of Plasmodium elongatum (4 specimens), Plasmodium matutinum (3), P. relictum SGS1 (3), Plasmodium vaughani (2) and P. sp. WW12 (2), and one sample each of Plasmodium delichoni and Plasmodium homocircumflexum. Conspecifics consistently featured similar sets of sequence variants. Two sequences of the P. gallinaceum genome (PRJEB9073, contigs 69 and 141) and 68 sequences from 20 simian and rodent Plasmodium species were also included. The 18S rDNA alignment was trimmed to the length of the shortest sequence (2992 alignment positions) and contained 1654 sites after all positions with gaps were removed. In the 18S rDNA trees, the avian Plasmodium species grouped within a moderately supported clade (BI posterior probability: 0.90/ML bootstrap support: 86) (Fig. 2). Support values at deeper nodes were low and the topology differed partially from the CytB trees (Additional file 2). Apart from the alignment and trimming methods used for the main tree shown in Fig. 2, trees were calculated also with the Plasmodium 18S sequences aligned with R-Coffee [54], which uses predicted secondary structures of the ribosomal sequences. The tree calculated with the first approach (only gap positions excluded; Additional file 3) shows a similar topology as that presented in Fig. 2, but bootstrap support values are generally lower. Support values were considerably lower in the tree obtained with the second approach (“strict” algorithm implemented in trimAl v.1.2 [53]; Additional file 4). The tree obtained with the third approach, using the most stringent trimming option (default option on the G-blocks Server [55]; Additional file 5), featured the lowest support values, moreover the A-type and S-type clades of the subgenus Laverania and Plasmodium ovale did not group, respectively.
Fig. 2.
Bayesian inference tree of Plasmodium 18S rDNA sequences. Posterior probabilities and maximum likelihood bootstrap values are indicated at most nodes. The scale bar indicates the expected mean number of substitutions per site according to the model of sequence evolution applied. Clades with variants expressed in the vertebrate hosts are labelled with “A”, and clades with sporozoite specific sequences with “S”. As outgroup a sequence of Leucocytozoon sp. (MalAvi lineage SYCON05; AH0932L_c07) was included
Distinct 18S rDNA variants were found in all species, but patterns differed quite strongly between them. The lowest diversity was found in P. delichoni (AH0011P) and Plasmodium sp. WW12 (AH0017P, AH0018P) with distances of up to 0.6% and 1.6% between different haplotypes, respectively (Additional file 6). Plasmodium delichoni featured four similar haplotypes, whereas the Plasmodium sp. WW12 samples featured two main variants each, one of which comprised several similar haplotypes little diverged from each other, and the second of which was represented by two clones (AH0017P c2, AH0018P c4) only. Plasmodium relictum (AH0013P, AH0069P, AH0392P), P. gallinaceum (PRJEB9073), P. homocircumflexum (AH0010P), and P. vaughani (AH0551P, AH0824P) featured two sequence clusters each, whereby different clusters of the same species grouped in all cases. The P. relictum clusters differed from each other by 2.3%, with differences mainly in a short section 650 bp to 850 bp from the 5′-end of the 18S rDNA. The short-branched cluster contained only one or two clones of each sample (AH0013P c5, c7; AH0392 c9; AH0069P c15), the long-branched cluster contained all other haplotypes, being similar to each other, but indicating the presence of three or four additional nuclear 18S rDNA copies. Plasmodium gallinaceum and P. relictum, both members of the subgenus Haemamoeba, presented as sister lineages in the 18S rDNA and the CytB trees, and featured the same distance between the two genomic copies with 2.3%. The distances between sequence clusters were considerably higher in P. (Giovannolaia) homocircumflexum and P. (Novyella) vaughani with 7.2% and 9.3%, respectively. Both species featured two clusters, which contained at least two distinct sequence variants each, suggesting the presence of four or more nuclear 18S rDNA copies. Moreover, two clones of P. homocircumflexum presented with chimeric features, with sequence sections partially matching variants of one of the two main clusters, respectively. The first half of clone 9 (bp 1 to 900) matched clones 5, 10, 12, 15, and 16, whereas the second half (bp 1050 to 2020) matched clones 2, 6, 11, 13, and 14. The first 1620 bp of clone 3 matched those of clones 5, 10, 12, 15, and 16, but the last section (nt. 1650 to 2020) matched clones 2, 6, 11, 13, and 14. All five recombination tests (GENECONV, MAXCHI, Chimaera, SiSscan, and 3Seq) performed with RDP4 [58, 59] were significant for the presence of recombination signals in the latter two clones (Additional file 7). Highest distances between 18S rDNA variants were found in P. (Haemamoeba) matutinum (AH0079P, AH0524P, AH0792P) and P. (Huffia) elongatum (AH0012P, AH0420P, AH0808P, AH0846P) with up to 10.8% and 14.9%, respectively. Both species featured two sequence clusters, each containing at least two distinct variants. However, unlike in the other Plasmodium species, short-branched and long-branched clusters of the two species did not group in reciprocally monophyletic clades [meaning that all lineages of the same species clustered together into “species” clades]. The long-branched cluster of P. elongatum formed a weakly supported clade with P. delichoni (0.76/64), whereas that of P. matutinum branched off from the basal node of the avian Plasmodium clade. The long-branched clusters of P. elongatum and P. matutinum grouped as sister lineages into one clade (1.0/95), although differing from each other by 16.6%. Moreover, several clones of the long-branched P. elongatum cluster presented with chimeric features. Clone 12 of sample AH0012P was largely similar to the clones of the long-branched, but the first 100 bp showed ten mismatches and were identical to sequences of the short-branched cluster, respectively. Other haplotypes of the long-branched cluster showed chimeric features as well, but patterns were complex and not consistent. The RDP4 [58, 59] analyses detected recombinant signals in several 18S clones of P. elongatum, but other than in case of P. homocircumflexum, results were significant only in one or two of the tests each (Additional file 7). Recombinant signals were also detected in some of the tests in clones of P. matutinum (AH0524P), P. relictum (AH0013P, AH0069P, AH0392P) and P. sp. WW12 (AH0017P, AH0018P). The GC-content of the 18S rDNAs ranged from 33.1 to 35.0% (mean 34.0%) and, therefore, was similar in all avian Plasmodium species. The approximate total lengths of the avian Plasmodium 18S rDNAs (extended by sequence sections missing at the 5′- and 3′-ends) ranged from 2100 to 2177 bp (mean 2138 bp).
18S rDNAs of human, simian and rodent Plasmodium species
The 18S rDNAs of species belonging to subgenus Laverania grouped with those of avian haemosporidians with low support (0.81/85). Plasmodium malariae formed the sister lineage to a clade containing all other simian and rodent Plasmodium species groups except for Laverania. This clade presented as a trichotomy with three well supported subclades, the first comprising species of the subgenus Vinckeia (P. berghei, P. chabaudi, P. berghei, and P. yoelii) (1.0/100), the second P. ovale wallikeri and P. ovale curtisi (0.99/97), and the third P. vivax, P. coatneyi, P. cynomolgi, P. fieldi, P. fragile, P. gonderi, P. hylobati, P. inui, P. knowlesi, and P. simiovale (1.0/100) (Fig. 2). Plasmodium species of these clades and the Laverania clade featured two sequence clusters with A-type and S-type variants each, indicating that ribosomal variants with differential expression patterns originated independently at least in four Plasmodium species groups. In P. vivax and related simian Plasmodium species, the distances between A-type and S-type variants ranged between 7.8% and 10.9% (mean 9.5%) (Additional file 1). Distances between variants were considerably higher in P. ovale curtisi and P. ovale wallikeri with 17.0% and 17.6%, respectively. In the subgenus Laverania, distances between A-type and S-type clusters ranged from 11.4% to 12.9% (mean 12.1%), whereas in the rodent malaria subgenus Vinckeia they ranged from 8.0% to 11.3% (mean 9.9%). The RDP4 [58, 59] analyses detected recombinant signals in 18S rDNA variants of all four Plasmodium species belonging to the subgenus Laverania, two species belonging to the subgenus Vinckeia (P. berghei and P. yoelii), and five species belonging to the subgenus Plasmodium (P. coatneyi, P. cynomolgi, P. knowlesi, P. vivax, and P. ovale curtisi) (Additional file 7). The four Laverania species featured the lowest GC-contents ranging from 32.3% to 35.7% (mean 33.9%). The values were higher in the four species of the subgenus Vinckeia with 35.9% to 38.8% (mean 37.4%). The GC-contents of the species group related to P. vivax ranged from 34.7% to 40.2% (mean 37.3%), whereby the O-type variant of P. vivax showed the highest value. The two P. ovale subspecies featured 18S rDNAs with GC-contents from 36.0% to 37.5% (mean 36.6%), and P. malariae showed GC-contents ranging from 33.2 to 34.2 (mean 33.8%). The approximate total lengths of the 18S rDNAs ranged from 2039 to 2676 bp (mean 2121 bp). The longest sequences were those of the S-type variants of P. ovale wallikeri and P. ovale curtisi with 2559 bp and 2676 bp, respectively, followed by the P. vivax O-type variant with 2216 bp. The P. malaria genome (PRJEB14392) features a unique 3350 bp long 18S rDNA copy on chromosome 11, which was not included in the alignment because it aligned only partially with the other Plasmodium sequences. It remains unclear whether this aberrant variant is functional, because 18S rDNA expression patterns have not been studied yet in P. malariae.
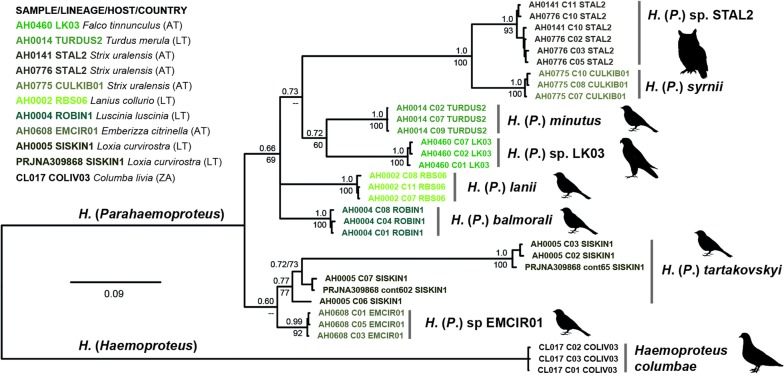
18S rDNAs of Haemoproteus species
The Haemoproteus data set comprised sequences of eight species of the subgenus Parahaemoproteus and one sample of the subgenus Haemoproteus, Haemoproteus columbae (92 clones, 9.2 mean per sample). For taxonomic reasons, the Parahaemoproteus trees were rooted with H. columbae, although the 18S rDNAs differed strongly between parasites of the two subgenera. The Parahaemoproteus 18S rDNAs differed from those of other haemosporidians (including H. columbae) by 31% to 32% and featured clearly distinct sequence patterns. Most Parahaemoproteus species clades were well supported in the 18S rDNA trees, except for the Haemoproteus (P.) tartakovskyi clade (0.7/70), but relationships between species-clades were mostly not resolved (Fig. 3). The 18S rDNA variants of the Parahaemoproteus parasites were similar within, but distinct between most species. The p-distances between clones were low in Haemoproteus (P.) minutus (AH0014H), Haemoproteus (P.) lanii (AH0002H), Haemoproteus (P.) sp. (AH0608H), and Haemoproteus (P.) balmorali (AH0004H) with up to 0.1%, 0.4%, 0.6%, and 0.9%, respectively. The latter four samples featured several different haplotypes each, but these did not form distinct clusters with unique sequence patterns. The intraspecific p-distances were only slightly higher in Haemoproteus (P.) sp. LK03 (AH0640H), Haemoproteus (P.) sp. STAL2 (AH0141H, AH0776H), and Haemoproteus (P.) syrnii (AH0775H) with 1.0%, 1.2%, and 1.3%, respectively. Each of the latter taxa featured at least three variants, indicating the presence of several distinct genomic 18S rDNA copies. The Haemoproteus (P.) tartakovskyi sample (AH0005H) was exceptional, because the clones grouped into three clearly distinct sequence clusters. The haplotypes of the first cluster (clones 1, 2, 3, 4, 5, 8 and 9) differed from that of the second (clones 7 and 11) and the third cluster (clone 6) by 21.5% and 18.7%, respectively. The sequences of the latter two clusters differed from each other by 6.3%. The RDP4 [58, 59] analyses detected recombinant signals in several clones belonging to Haemoproteus (P.) tartakovskyi (AH0005H), Haemoproteus (P.) syrnii (AH0775H), and Haemoproteus (P.) sp. STAL2 (AH0141H, AH0776H), whereby at least two of five tests showed significant results for all of the latter. For three clones (AH0005H_C06, AH0775_C02, and AH0775_C10), significant results were obtained in four of five recombination tests (Additional file 7). The GC-content of the Parahaemoproteus 18S rDNAs ranged from 37.4% to 47.4% (mean 44.9%), and the approximate total lengths ranged from 1939 to 2176 bp. The GC-content of the Haemoproteus columbae (CL017) 18S rDNAs was lower (36.5%), the approximate total length of the 18S rDNAs was greater with 2280 bp. The Haemoproteus columbae haplotypes differed only in 0.4% from each other and no subdivision into distinct variants was evident.
Fig. 3.
Bayesian inference tree of Haemoproteus 18S rDNA sequences. Posterior probabilities and maximum likelihood bootstrap values are indicated at most nodes. The scale bar indicates the expected mean number of substitutions per site according to the model of sequence evolution applied. The tree was midpoint-rooted, no outgroup was used
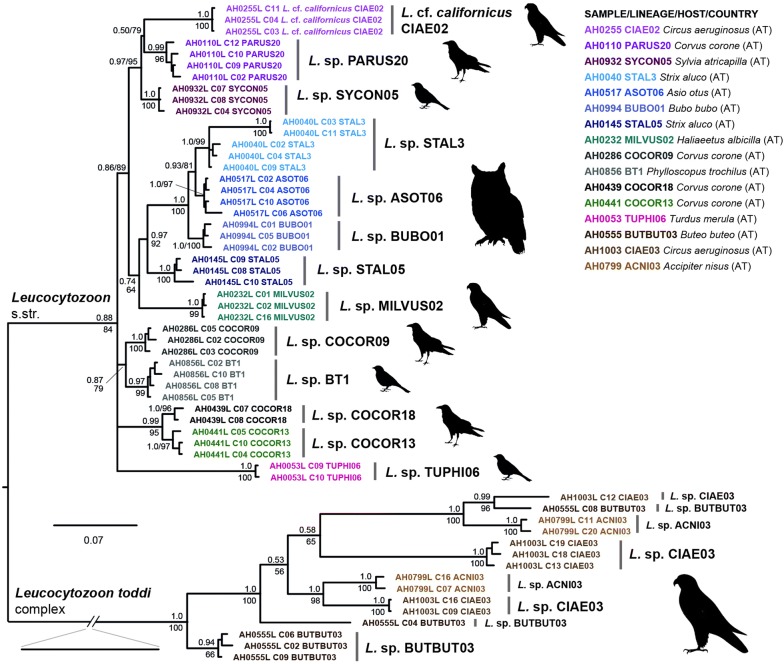
18S rDNAs of Leucocytozoon species
The Leucocytozoon data set comprised 18S rDNAs of 16 species (167 clones, 10.4 mean per sample). The trees were rooted with a sequence of P. relictum and featured to main clades, one clade with sequences of 13 Leucocytozoon species, and a second one with sequences of the three species belonging to the Leucocytozoon toddi species complex. The 18S rDNAs of most Leucocytozoon species in the first clade were similar and grouped into reciprocally monophyletic clades (Fig. 4).
Fig. 4.
Bayesian inference tree of Leucocytozoon 18S rDNA sequences. Posterior probabilities and maximum likelihood bootstrap values are indicated at most nodes. The scale bar indicates the expected mean number of substitutions per site according to the model of sequence evolution applied. As outgroup a sequence of P. relictum (MalAvi lineage SGS1; AH0013P_c05) was included
The lowest distances between intraspecific 18S rDNA variants were found in Leucocytozoon sp. TUPHI06 (AH0053L), Leucocytozoon sp. SYCON5 (AH0932L), Leucocytozoon sp. MILVUS2 (AH0232L), and Leucocytozoon sp. COCOR18 (AH0439L) with up to 0.3%, 0.4%, 0.5%, 0.8%, respectively. The maximum distance between variants was only slightly higher in Leucocytozoon cf. californicus CIAE02 (AH0255L) and Leucocytozoon sp. BUBO01 (AH0994L) with 0.9% each. No structuring of haplotypes into different clusters was evident in the latter samples. The 18S rDNAs of Leucocytozoon sp. COCOR09 (AH0286L), Leucocytozoon sp. BT1 (AH0856L), Leucocytozoon sp. PARUS20 (AH0110L), and Leucocytozoon sp. COCOR13 (AH0441L) differed by up to 1.3%, 1.6%, 1.8%, and 2.0%, respectively. The first two samples featured at least three, the latter two four distinct variants. The distances were slightly higher in Leucocytozoon sp. STAL5 (AH0145L), Leucocytozoon sp. ASOTO6 (AH0517L) and Leucocytozoon sp. STAL3 (AH0040L) with up to 2.2%, 2.3% and 5.7%, respectively. The first two species featured at least four distinct variants. Clone 6 of Leucocytozoon sp. ASOTO6 (AH0517L) differed considerably from the others, but only in a short sequence section (bp 1400 to 1600). The clones of Leucocytozoon sp. STAL3 (AH0040L) formed two clusters, with three similar haplotypes in the first, and clones 3 and 11 in the second cluster, both of the latter featuring clearly distinct sequence variants. The 18S rDNA GC-content of the 13 Leucocytozoon species ranged from 34.0 to 38.6% (mean 36.9%), and the approximate total lengths ranged from 2094 to 2138 bp.
The sequences of the three species belonging to the Leucocytozoon toddi species complex grouped into two or three distinct clusters each. The clusters did not form reciprocally monophyletic clades, but clusters of different species grouped. The p-distances between 18S rDNA variants of Leucocytozoon sp. BUTBUT03 (AH0555L), Leucocytozoon sp. ACNI03 (AH0799L) and Leucocytozoon sp. CIAE03 (AH1003L) were high with up to 16.9%, 17.4%, and 18.5%, respectively. One sequence cluster of Leucocytozoon sp. BUTBUT03 contained several distinct haplotypes with chimeric features (some haplotypes only partially matched two other clones, respectively), whereas the other two clusters contained only sequences of clones 4 and 8, respectively. Three sequence clusters were also identified in Leucocytozoon sp. CIAE03, two clusters of which together contained four distinct variants, and a third cluster which was represented by clone 12 only. The sequences of Leucocytozoon sp. ACNI03 formed two clusters, each containing at least three distinct variants. The RDP4 [58, 59] analyses detected recombinant signals in multiple 18S clones of all three species belonging to the Leucocytozoon toddi complex. For some of the clones (AH0555L_C10, AH0555L_C12, AH0799_C16, AH1003L_C01, and AH1003L_C02) significant results were obtained in at least four of five recombination tests (Additional file 7). The species of the Leucocytozoon toddi species complex featured 18S rDNAs with exceptionally high GC-contents, ranging between 47.0 and 50.5% (mean 49.3%). The approximate total lengths of the 18S rDNAs ranged from 2125 to 2308 bp.
Discussion
In the present study, the 18S rDNAs of seven Plasmodium, nine Haemoproteus, and 16 Leucocytozoon species were analysed. Ten clones per lineage were sequenced in mean, whereby multiple samples were analysed of most avian Plasmodium species. The data might be more complete for the latter taxa, but it is possible that not all 18S rDNA variants of the taxa investigated were obtained. The actual number of distinct 18S rDNA variants could be determined in future studies by mapping rDNAs by restriction enzyme analysis and Southern blot hybridization as it was done in earlier studies on P. berghei [11], P. falciparum [16], and P. cynomolgi [19]. Another approach would be high-coverage sequencing, assembling the whole genomes of the parasites, and identifying the number and location of ribosomal units on the respective chromosomes. Therefore, further studies are required to determine the total number of ribosomal units in the species studied, and correctly assign them to specific chromosomes. In that context, the data cannot be used to formally test for a birth-and-death model.
Phylogenetic analyses were performed particularly to show the diversity of 18S rDNA variants of the species studied, rather than aiming to resolve the relationships between those taxa. Generally, multi-copy genes, especially those of Plasmodium species, are not a good choice as phylogenetic markers for studying relationships between species. The human, simian and rodent Plasmodium species studied so far feature distinct ribosomal clusters, being differentially expressed in the vertebrate and mosquito hosts, and therefore subjected to different selective pressure [65]. Moreover, gene conversion between non-homologous genes also may generate misleading results [19]. Haemosporidian parasites feature the most variable nc ribosomal genes of all eukaryotes analysed to date, showing by far the highest intraspecific variability. Extremely variable sections cannot be aligned with confidence between distantly related taxa because of the presence of numerous insertion/deletions, even if secondary structure models are applied. Thus, more recent phylogenetic approaches for human, simian and rodent Plasmodium species rather tended to use the mt CytB [65] or nc coding genes [e.g., 66], whereas nc ribosomal genes were almost entirely neglected in avian haemosporidian research.
Although the species number of avian haemosporidians exceeds by far those in mammalian hosts, 18S rDNAs have been published only from four avian Plasmodium species and Haemoproteus (P.) tartakovskyi [39]. Only the 18S rDNA variants expressed in the vertebrate host (A-type) were sequenced from P. lophurae (X13706, [29]) and P. cathemerium (AY625607, [51]), and the 18S rDNAs of the P. relictum genome (PRJEB9074; WTSI, unpublished) are fragmentary. The present data show that the patterns with several distinct 18S rDNA variants (or ribosomal units) are not unique to human, simian and rodent malaria parasites, but are similar in most of the avian Plasmodium species studied belonging to three different subgenera. Plasmodium (Haemamoeba) relictum, P. (Haemamoeba) gallinaceum, P. (Giovannolaia) homocircumflexum, and P. (Novyella) vaughani each featured two sequence clusters, whereby the two clusters of each species grouped together in well-supported reciprocally monophyletic clades, suggesting that new variants originated independently in malaria parasites of different subgenera in the course of their evolution or that of related ancestral species. Differently, the two sequence clusters of P. (Huffia) elongatum and P. (Haemamoeba) matutinum were extremely diverged and did not group into reciprocally monophyletic clades, suggesting that they originated before the two species emerged. With exception of P. (Novyella) delichoni and P. gallinaceum, all Plasmodium species under investigation featured at least four clearly distinct sequence variants, indicating the presence of four or more genomic rDNA units. Apart from the Plasmodium species, the three species of the Leucocytozoon toddi complex also featured clearly distinct 18S rDNA variants showing up in two or three clusters. Distinct variants of the latter species did not group in reciprocally monophyletic clades, but with those of the other species, suggesting that the last recent common ancestor of the L. toddi species complex already featured a set of different 18S rDNA variants. Since only about ten clones per sample were sequenced and intraspecific diversity of haplotypes was extremely high, probably not all 18S rDNA variants in the species studied were recovered. This species group was also peculiar regarding the high GC-content of their 18S rDNAs (47.0% to 50.5%), exceeding that of Parahaemoproteus (37.4% to 47.4%), the other Leucocytozoon species (34.9% to 38.6%), Haemoproteus (36.5%), and Plasmodium (33.2% to 38.8%).
The patterns were different in the other Leucocytozoon species, with most species featuring similar haplotypes, which grouped in reciprocally monophyletic clades. Most Parahaemoproteus species and Haemoproteus columbae contained sets of similar clones, only Haemoproteus (P.) tartakovskyi featured three clearly distinct sequence variants, which grouped into a reciprocally monophyletic clade (Fig. 3). It cannot be excluded that the other seven Parahaemoproteus species contained additional, undetected 18S rDNA copies because the primers were designed based on sequences of the Haemoproteus (P.) tartakovskyi genome only. However, it seems unlikely that the primers would not have recovered additional variants in at least some of the other seven species.
Expression of 18S rDNA genes has not been studied in avian haemosporidians yet (except for that of the A-type variants in P. lophurae; [25]), but the present data indicate that differential expression may also occur in avian Plasmodium species and some other haemosporidians. The simian and rodent Plasmodium species each featured at least two distinct 18S rDNAs clusters, the less diverged (short-branched) clusters contain the A-type variants expressed in the vertebrate host, and the more diverged (long-branched) clusters contain the S-type variants expressed in the mosquito hosts. A similar pattern is also evident in most avian Plasmodium species studied, because these also featured short-branched and long-branched clusters each (Fig. 2). However, additional experiments are required to clarify whether 18S rDNAs of avian Plasmodium species (and species of related haemosporidian genera) show similar expression patterns as the human, simian and rodent Plasmodium species. The new data may provide the basis for studying 18S rDNA expression of different developmental stages in the avian Plasmodium species by targeting rRNAs of distinct clusters with specific oligonucleotide probes in both the bird and mosquito hosts. Molecular hybridization assays could also be performed on the species of the Leucocytozoon toddi species complex and Haemoproteus (P.) tartakovskyi, to study if their rDNA units are expressed differentially as well. Due to the high number of considerably distinct 18S rDNA clusters, these species would be promising candidates to test stage-specific expression of distinct ribosomal units in haemosporidian parasites belonging to genera other than Plasmodium.
The results of the recombination tests indicate that at least some species in all three genera studied have 18S rDNA variants with chimeric features to a varying extent, potentially resulting from recombination events. This finding might relate to the general evolution and maintenance of ribosomal units in apicomplexan parasites. Whereas ribosomal units of most eukaryotes are assumed to evolve in a concerted fashion involving mechanisms such as unequal crossing-over during recombination, gene duplication, and inter-chromosomal gene conversion [6], the evolution of ribosomal units in malaria parasites was explained with a birth-and-death model, assuming that new variants originate by gene duplication and others are inactivated or deleted from the genome [9, 67]. However, the evolution of ribosomal units in apicomplexans is probably more complicated and involves mechanisms other than gene duplication and deletion only. Corredor & Enea [19] analysed the 18S rDNA sequence patterns of P. cynomolgi, P. berghei, and P. falciparum and concluded that the two main gene types do not evolve independently but interact through gene conversion or some form of recombination [19, 68]. They found that the extent of divergence between sequence regions of isomorphs varies erratically and that the same regions show varying degrees of divergence depending on the species analysed. In this context, they point out that the ribosomal RNAs of Plasmodium species rather evolve in a semi-concerted manner [19]. The present data generally support this assumption, and the recombination tests detected chimeric features not only in 18S rDNA alignments of human, simian and rodent Plasmodium species, but also in several avian Plasmodium, Leucocytozoon, and Haemoproteus species (Additional file 7). The finding that recombinant features were not detected in all species investigated, does not necessarily mean that their ribosomal genes were not affected by recombination, because not all 18S rDNA variants might have been recovered (particularly in the genus Haemoproteus), or recombinant variants might have been deleted throughout evolution, thus obscuring past events. A general problem in avian haemosporidian research is that assembled and annotated genomes of Leucocytozoon and Haemoproteus parasites are still missing, and neither has the number and location of distinct ribosomal units been determined in any species by mapping rDNA by restriction enzyme analysis and Southern blot hybridization like in the first studies on 18S rDNAs of rodent, human, and simian Plasmodium species (e.g., [11, 16]). Nonetheless, the fundamental reasons for the maintenance of sequentially distinct ribosomal genes in haemosporidian parasites generally remain insufficiently investigated. Many non-haemosporidian parasites of the Apicomplexa (e.g., Theileria spp., Babesia spp., and Hepatozoon spp.) also show complex life cycles with arthropod vectors and vertebrate hosts, but they do not feature considerably distinct ribosomal variants.
Malaria parasites and related haemosporidians cause diseases in many bird species. Recent molecular studies show that widespread Haemoproteus parasites, which have been formerly considered to be relatively benign [69], are actually virulent and cause severe pathology and even mortality in non-adapted birds due to damage of organs by tissue stages [70]. However, diagnostics of these parasites remain insufficiently developed in avian hosts during tissue stage of development. Ribosomal genes constitute the most frequent RNA molecules in cells and therefore are the most common targets in hybridization assays targeting parasites in host tissue. In situ hybridization techniques are particularly important in pathological studies, because they allow studying the effects of parasite infections in host tissues before the development of parasitemia. Additionally, this sensitive approach provides opportunities to distinguish development stages of haemosporidians belonging to different genera even during co-infections, which predominate in wildlife. The new data allow developing oligonucleotide probes specifically targeting and differentiating genera or species in host tissue. A chromogenic in situ hybridization protocol targeting the 18S rRNAs of Plasmodium species in paraffin wax-embedded tissue samples of penguins was already developed [71]. The protocol was successfully applied to a wider range of parasite species [72, 73] and should allow for identification of all avian Plasmodium species belonging to different subgenera [personal comment based on the sequence data of the present study, JH]. Based on the 18S rDNA data of the present study, Himmel et al. [48] developed chromogenic in situ hybridization protocols, which reliably allowed detecting blood and tissue stages of parasites belonging to Haemoproteus (Parahaemoproteus) species and Leucocytozoon spp. in paraffin wax-embedded tissue samples of birds by targeting the parasites’ ribosomes with genus specific probes.
In human malaria research, 18S rDNAs are also the most common targets in PCR screening approaches. The first comprehensive PCR assay with species-specific primers targeting the 18S rDNAs of the four human malaria species P. falciparum, P. malariae, P. vivax and P. ovale was published by [41]. Several other PCR protocols for the detection of simian Plasmodium species were developed later [43–45]. Differently, PCR assays developed for avian haemosporidians almost exclusively target mitochondrial genes [47, 74, 75]. In contrast to mitochondrial coding genes, the 18S rDNAs feature multiple sequence sections being entirely conserved within haemosporidian parasites of certain genera but differing considerably between species of different genera. Based on the new 18S rDNA data, PCR screening assays could be developed to reliably detect and differentiate between avian haemosporidians belonging to different genera.
Conclusion
In the present study, 18S rDNA sequences of 32 avian haemosporidian species were sequenced, which more than doubles the number of haemosporidian taxa for which 18S sequences are available. Moreover, literature on 18S rDNAs of human, simian and rodent Plasmodium species was reviewed, and already published data was included in the analyses to compare patterns with those found in the avian haemosporidians. The present data show that gene duplication events giving rise to two diverged main sequence clusters happened independently in at least six out of seven avian Plasmodium species, thus supporting evolution according to a birth-and-death model like proposed for the ribosomal units of human, simian and rodent Plasmodium species. The 18S rDNA patterns of the three species of the Leucocytozoon toddi complex and Haemoproteus tartakovskyi also support evolution according to the latter model. However, the other species featured sets of similar 18S rDNA haplotypes, raising the question of whether their ribosomal units rather evolve in concerted fashion like in other eukaryotes (including most non-haemosporidian apicomplexans). Moreover, the presence of chimeric 18S rDNA variants in some Plasmodium and Leucocytozoon species indicates that the distinct ribosomal units are not evolving entirely independently, but rather in a semi-concerted evolution as suggested by [19]. The new data may facilitate studying differential expression of distinct ribosomal units in haemosporidians other than human, simian and rodent Plasmodium species. In a study published recently [48], using the 18S rDNA data of the present study, hybridization assays with genus-specific probes for Leucocytozoon and Haemoproteus (subgenus Parahaemoproteus), targeting the parasites’ ribosomes in host tissue, were already developed.
Supplementary information
Additional file 1. Human, simian and rodent malaria species included in the Plasmodium dataset.
Additional file 2. Bayesian inference trees calculated from CytB sequences of Plasmodium species (A), Haemoproteus spp. (B) and Leucocytozoon spp. (C). The trees were midpoint-rooted, no outgroups were used. Posterior probabilities and maximum likelihood bootstrap values are indicated at most nodes. The scale bar indicates the expected mean number of substitutions per site according to the model of sequence evolution applied.
Additional file 3. Maximum-likelihood tree of Plasmodium 18S rDNA sequences based on a secondary structure alignment calculated with R-Coffee and only gaps trimmed.
Additional file 4. Maximum-likelihood tree of Plasmodium 18S rDNA sequences based on a secondary structure alignment calculated with R-Coffee applying the “strict” algorithm implemented in trimAl v.1.2 [53] for trimming.
Additional file 5. Maximum-likelihood tree of Plasmodium 18S rDNA sequences based on a secondary structure alignment calculated with R-Coffee applying the default option on the G-blocks Server [55] for trimming.
Additional file 6. Sequence features of the 18S rDNAs analysed.
Additional file 7. Results of the recombination tests.
Acknowledgements
We would like to thank Alexandra Scope and Cornelia Konicek for collecting blood samples from living birds and Anna Kübber-Heiss for providing tissue samples of dead birds. We thank Petra Sumasgutner for the samples of H. columbae from South Africa, Ivan Maggini for catching birds at the Biological Station Illmitz, and Thomas Zechmeister for providing access to the latter. The staff of P.B. Šivickis Laboratory of Parasitology, Nature Research Centre Vilnius is acknowledged for participation in field and experimental work during sample collection, and Tatjana Iezhova and Mikas Ilgūnas are acknowledged for assistance in the laboratory during parasite species identification and selection of blood samples. We also feel indebted to Hans-Peter Fuehrer for valuable discussions and comments on the manuscript. Finally, we want to thank the three anonymous reviewers for their critical comments and valuable suggestions, which helped to improve the manuscript.
Abbreviations
- nc
nuclear
- mt
mitochondrial
- rDNA
ribosomal DNA
- rRNA
ribosomal RNA
- Cytb
cytochrome b
- SSU
small ribosomal subunit
- LSU
large ribosomal subunit
- NTS
non-transcribed spacer
- ETS
external transcribed spacer
- ITS
internal transcribed spacer
- MHC
major histocompatibility complex
Authors’ contributions
JH, HW, TH: study conception and design; JH: drafting of manuscript; JH, GV, TH: sample collection and processing; GV, TH: microscopy; JH: primer design, molecular lab work, sequence analyses, and illustrations; all authors read and critically revised the manuscript. All authors read and approved the final manuscript.
Funding
The present study was funded by the Austrian Science Fund FWF (P 29628).
Availability of data and materials
All DNA, blood and tissue samples are deposited in the collection of the Institute of Pathology (Department of Pathobiology, University of Veterinary Medicine Vienna). All sequence data were uploaded to NCBI GenBank. DNA sequence alignments are available from J.H. upon request.
Ethics approval and consent to participate
The study was part of the FWF-project P 29628 and approved by the institutional ethics and animal welfare committee and the national authority according to §§ 26ff. of Animal Experiments Acts, TVG 2012, Austria (BMWFW-68.205/0036-WF/V/3b/2017). The collection of bird blood samples in Lithuania was approved by the Environmental Protection Agency Lithuania (permit 2018-04-13, no. 24).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12936-019-2940-6.
References
- 1.Failmezger J, Ludwig J, Nieß A, Siemann-Herzberg M. Quantifying ribosome dynamics in Escherichia coli using fluorescence. FEMS Microbiol Lett. 2017;364:1–6. doi: 10.1093/femsle/fnx055. [DOI] [PubMed] [Google Scholar]
- 2.Hillis DM, Dixon MT. Ribosomal DNA: molecular evolution and phylogenetic inference. Q Rev Biol. 1991;66:411–453. doi: 10.1086/417338. [DOI] [PubMed] [Google Scholar]
- 3.Elder JF, Jr, Turner BJ. Concerted evolution of repetitive DNA sequences in eukaryotes. Q Rev Biol. 1995;70:297–320. doi: 10.1086/419073. [DOI] [PubMed] [Google Scholar]
- 4.Arnheim N, Krystal M, Schmickel R, Wilson G, Ryder O, Zimmer E. Molecular evidence for genetic exchanges among ribosomal genes on nonhomologous chromosomes in man and apes. Proc Natl Acad Sci USA. 1980;77:7323–7327. doi: 10.1073/pnas.77.12.7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schlötterer C, Tautz D. Chromosomal homogeneity of Drosophila ribosomal DNA arrays suggests intrachromosomal exchanges drive concerted evolution. Curr Biol. 1994;4:777–783. doi: 10.1016/S0960-9822(00)00175-5. [DOI] [PubMed] [Google Scholar]
- 6.Liao D. Concerted evolution: molecular mechanism and biological implications. Am J Hum Genet. 1999;64:24–30. doi: 10.1086/302221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nei M, Gu X, Sitnikova T. Evolution by the birth-and-death process in multigene families of the vertebrate immune system. Proc Natl Acad Sci USA. 1997;94:7799–7806. doi: 10.1073/pnas.94.15.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nei M, Hughes AL. Balanced polymorphism and evolution by the birth-and-death process in the MHC loci. In: Tsuji K, Aizawa M, Sasazuki T, editors. 11th histocompatibility workshop and conference. Oxford: Oxford University Press; 1992. [Google Scholar]
- 9.Rooney AP. Mechanisms underlying the evolution and maintenance of functionally heterogeneous 18S rRNA genes in apicomplexans. Mol Biol Evol. 2004;21:1704–1711. doi: 10.1093/molbev/msh178. [DOI] [PubMed] [Google Scholar]
- 10.Waters AP, Syin C, McCutchan TF. Developmental regulation of stage-specific ribosome populations in Plasmodium. Nature. 1989;342:438–440. doi: 10.1038/342438a0. [DOI] [PubMed] [Google Scholar]
- 11.Dame JB, McCutchan TF. The four ribosomal DNA units of the malaria parasite Plasmodium berghei. Identification, restriction map, and copy number analysis. J Biol Chem. 1983;258:6984–6990. [PubMed] [Google Scholar]
- 12.Dame JB, McCutchan TF. Cloning and characterization of a ribosomal RNA gene from Plasmodium berghei. Mol Biochem Parasitol. 1983;8:263–279. doi: 10.1016/0166-6851(83)90048-8. [DOI] [PubMed] [Google Scholar]
- 13.Dame JB, Sullivan M, McCutchan TF. Two major sequence classes of ribosomal RNA genes in Plasmodium berghei. Nucleic Acids Res. 1984;12:5943–5952. doi: 10.1093/nar/12.14.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCutchan TF, Li J, McConkey GA, Rogers MJ, Waters AP. The cytoplasmic ribosomal RNAs of Plasmodium spp. Parasitol Today. 1995;11:134–138. doi: 10.1016/0169-4758(95)80132-4. [DOI] [PubMed] [Google Scholar]
- 15.Gunderson JH, Sogin ML, Wollett G, Hollingdale M, De La Cruz VF, Waters AP, et al. Structurally distinct, stage-specific ribosomes occur in Plasmodium. Science. 1987;238:933–937. doi: 10.1126/science.3672135. [DOI] [PubMed] [Google Scholar]
- 16.Langsley G, Hyde JE, Goman M, Scaife JG. Cloning and characterisation of the rRNA genes from the human malaria parasite Plasmodium falciparum. Nucleic Acids Res. 1983;11:8703–8717. doi: 10.1093/nar/11.24.8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCutchan TF, de la Cruz VF, Lal AA, Gunderson JH, Elwood HJ, Sogin ML. Primary sequences of two small subunit ribosomal RNA genes from Plasmodium falciparum. Mol Biochem Parasitol. 1988;28:63–68. doi: 10.1016/0166-6851(88)90181-8. [DOI] [PubMed] [Google Scholar]
- 18.Fang J, McCutchan TF. Malaria: thermoregulation in a parasite’s life cycle. Nature. 2002;418:742. doi: 10.1038/418742a. [DOI] [PubMed] [Google Scholar]
- 19.Corredor V, Enea V. The small ribosomal subunit RNA isoforms in Plasmodium cynomolgi. Genetics. 1994;136:857–865. doi: 10.1093/genetics/136.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qari SH, Goldman IF, Pieniazek NJ, Collins WE, Lal AA. Blood and sporozoite stage-specific small subunit ribosomal RNA-encoding genes of the human malaria parasite Plasmodium vivax. Gene. 1994;150:43–49. doi: 10.1016/0378-1119(94)90855-9. [DOI] [PubMed] [Google Scholar]
- 21.Li J, Wirtz RA, McConkey GA, Sattabongkot J, McCutchan TF. Transition of Plasmodium vivax ribosome types corresponds to sporozoite differentiation in the mosquito. Mol Biochem Parasitol. 1994;65:283–289. doi: 10.1016/0166-6851(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 22.Waters AP, McCutchan TF. Partial sequence of the asexually expressed SU rRNA gene of Plasmodium vivax. Nucleic Acids Res. 1989;17:2135. doi: 10.1093/nar/17.5.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Gutell RR, Damberger SH, Wirtz RA, Kissinger JC, Rogers MJ, et al. Regulation and trafficking of three distinct 18S ribosomal RNAs during development of the malaria parasite. J Mol Biol. 1997;269:203–213. doi: 10.1006/jmbi.1997.1038. [DOI] [PubMed] [Google Scholar]
- 24.Qi Y, Zhu F, Eastman RT, Fu Y, Zilversmit M, Pattaradilokrat S, et al. Regulation of Plasmodium yoelii oocyst development by strain- and stage-specific small-subunit rRNA. MBio. 2015;6:e00117. doi: 10.1128/mBio.00117-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Unnasch TR, Wirth DF. The avian malaria Plasmodium lophurae has a small number of heterogeneous ribosomal RNA genes. Nucleic Acids Res. 1983;11:8443–8459. doi: 10.1093/nar/11.23.8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fang J, Sullivan M, McCutchan TF. The effects of glucose concentration on the reciprocal regulation of rRNA promoters in Plasmodium falciparum. J Biol Chem. 2004;279:720–725. doi: 10.1074/jbc.M308284200. [DOI] [PubMed] [Google Scholar]
- 27.Waters AP, Higgins DG, McCutchan TF. Plasmodium falciparum appears to have arisen as a result of lateral transfer between avian and human hosts. Proc Natl Acad Sci USA. 1991;88:3140–3144. doi: 10.1073/pnas.88.8.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goman M, Mons B, Scaife J. The complete sequence of a Plasmodium malariae SSUrRNA gene and its comparison to other plasmodial SSUrRNA genes. Mol Biochem Parasitol. 1991;45:281–288. doi: 10.1016/0166-6851(91)90096-O. [DOI] [PubMed] [Google Scholar]
- 29.Waters AP, Unnasch TR, Wirth DF, McCutchan TF. Sequence of a small ribosomal RNA gene from Plasmodium lophurae. Nucleic Acids Res. 1989;17:1763. doi: 10.1093/nar/17.4.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waters AP, Higgins DG, McCutchan TF. Evolutionary relatedness of some primate models of Plasmodium. Mol Biol Evol. 1993;10:914–923. doi: 10.1093/oxfordjournals.molbev.a040038. [DOI] [PubMed] [Google Scholar]
- 31.Nishimoto Y, Arisue N, Kawai S, Escalante AA, Horii T, Tanabe K, et al. Evolution and phylogeny of the heterogeneous cytosolic SSU rRNA genes in the genus Plasmodium. Mol Phylogenet Evol. 2008;47:45–53. doi: 10.1016/j.ympev.2008.01.031. [DOI] [PubMed] [Google Scholar]
- 32.Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pain A, Böhme U, Berry AE, Mungall K, Finn RD, Jackson AP, et al. The genome of the simian and human malaria parasite Plasmodium knowlesi. Nature. 2008;455:799–804. doi: 10.1038/nature07306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, Caler E, et al. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008;455:757–763. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tachibana S-I, Sullivan SA, Kawai S, Nakamura S, Kim HR, Goto N, et al. Plasmodium cynomolgi genome sequences provide insight into Plasmodium vivax and the monkey malaria clade. Nat Genet. 2012;44:1051–1057. doi: 10.1038/ng.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sundararaman SA, Plenderleith LJ, Liu W, Loy DE, Learn GH, Li Y, et al. Genomes of cryptic chimpanzee Plasmodium species reveal key evolutionary events leading to human malaria. Nat Commun. 2016;7:1–14. doi: 10.1038/ncomms11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chien J-T, Pakala SB, Geraldo JA, Lapp SA, Humphrey JC, Barnwell JW, et al. High-quality genome assembly and annotation for Plasmodium coatneyi, generated using single-molecule real-time PacBio technology. Genome Announc. 2016;4:e00883. doi: 10.1128/genomeA.00883-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Otto TD, Böhme U, Jackson AP, Hunt M, Franke-Fayard B, Hoeijmakers WAM, et al. A comprehensive evaluation of rodent malaria parasite genomes and gene expression. BMC Biol. 2014;12(86):1–18. doi: 10.1186/s12915-014-0086-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bensch S, Canbäck B, DeBarry JD, Johansson T, Hellgren O, Kissinger JC, et al. The Genome of Haemoproteus tartakovskyi and its relationship to human malaria parasites. Genome Biol Evol. 2016;8:1361–1373. doi: 10.1093/gbe/evw081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waters A, Mccutchan T. Rapid, sensitive diagnosis of malaria based on ribosomal RNA. Lancet. 1989;333:1343–1346. doi: 10.1016/S0140-6736(89)92800-6. [DOI] [PubMed] [Google Scholar]
- 41.Snounou G, Viriyakosol S, Jarra W, Thaithong S, Brown KN. Identification of the four human malaria parasite species in field samples by the polymerase chain reaction and detection of a high prevalence of mixed infections. Mol Biochem Parasitol. 1993;58:283–292. doi: 10.1016/0166-6851(93)90050-8. [DOI] [PubMed] [Google Scholar]
- 42.Snounou G, Viriyakosol S, Xin PZ, Jarra W, Pinheiro L, do Rosario VE, et al. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol. 1993;61:315–320. doi: 10.1016/0166-6851(93)90077-B. [DOI] [PubMed] [Google Scholar]
- 43.Kimura M, Kaneko O, Liu Q, Zhou M, Kawamoto F, Wataya Y, et al. Identification of the four species of human malaria parasites by nested PCR that targets variant sequences in the small subunit rRNA gene. Parasitol Int. 1997;46:91–95. doi: 10.1016/S1383-5769(97)00013-5. [DOI] [Google Scholar]
- 44.Singh B, Bobogare A, Cox-Singh J, Snounou G, Abdullah MS, Rahman HA. A genus-and species-specific nested polymerase chain reaction malaria detection assay for epidemiologic studies. Am J Trop Med Hyg. 1999;60:687–692. doi: 10.4269/ajtmh.1999.60.687. [DOI] [PubMed] [Google Scholar]
- 45.Rougemont M, Van Saanen M, Sahli R, Hinrikson HP, Bille J, Jaton K. Detection of four Plasmodium species in blood from humans by 18S rRNA gene subunit-based and species-specific real-time PCR assays. J Clin Microbiol. 2004;42:5636–5643. doi: 10.1128/JCM.42.12.5636-5643.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bensch S, Hellgren O, Pérez-Tris J. MalAvi: a public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Mol Ecol Resour. 2009;9:1353–1358. doi: 10.1111/j.1755-0998.2009.02692.x. [DOI] [PubMed] [Google Scholar]
- 47.Hellgren O, Waldenström J, Bensch S. A new PCR assay for simultaneous studies of Leucocytozoon, Plasmodium, and Haemoproteus from avian blood. J Parasitol. 2004;90:797–802. doi: 10.1645/GE-184R1. [DOI] [PubMed] [Google Scholar]
- 48.Himmel T, Harl J, Kübber-Heiss A, Konicek C, Juan-Sallés C, Ilgūnas M, et al. Molecular probes for the identification of avian Haemoproteus and Leucocytozoon parasites in tissue sections by chromogenic in situ hybridization. Parasit Vectors. 2019;12:282. doi: 10.1186/s13071-019-3536-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hall TA. BioEdit: a user-friendly biological sequences alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 50.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wiersch SC, Maier WA, Kampen H. Plasmodium (Haemamoeba) cathemerium gene sequences for phylogenetic analysis of malaria parasites. Parasitol Res. 2005;96:90–94. doi: 10.1007/s00436-005-1324-8. [DOI] [PubMed] [Google Scholar]
- 52.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilm A, Higgins DG, Notredame C. R-Coffee: a method for multiple alignment of non-coding RNA. Nucleic Acids Res. 2008;36:e52. doi: 10.1093/nar/gkn174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 56.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics. 2010;26:2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015;1:vev003. doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Padidam M, Sawyer S, Fauquet CM. Possible emergence of new geminiviruses by frequent recombination. Virology. 1999;265:218–225. doi: 10.1006/viro.1999.0056. [DOI] [PubMed] [Google Scholar]
- 61.Smith JM. Analyzing the mosaic structure of genes. J Mol Evol. 1992;34:126–129. doi: 10.1007/BF00182389. [DOI] [PubMed] [Google Scholar]
- 62.Posada D, Crandall KA. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc Natl Acad Sci USA. 2001;98:13757–13762. doi: 10.1073/pnas.241370698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gibbs MJ, Armstrong JS, Gibbs AJ. Sister-scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics. 2000;16:573–582. doi: 10.1093/bioinformatics/16.7.573. [DOI] [PubMed] [Google Scholar]
- 64.Boni MF, Posada D, Feldman MW. An exact nonparametric method for inferring mosaic structure in sequence triplets. Genetics. 2007;176:1035–1047. doi: 10.1534/genetics.106.068874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Escalante AA, Freeland DE, Collins WE, Lal AA. The evolution of primate malaria parasites based on the gene encoding cytochrome b from the linear mitochondrial genome. Proc Natl Acad Sci USA. 1998;95:8124–8129. doi: 10.1073/pnas.95.14.8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Escalante AA, Cornejo OE, Freeland DE, Poe AC, Durrego E, Collins WE, et al. A monkey’s tale: the origin of Plasmodium vivax as a human malaria parasite. Proc Natl Acad Sci USA. 2005;102:1980–1985. doi: 10.1073/pnas.0409652102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Enea V, Corredor V. The evolution of plasmodial stage-specific rRNA genes is dominated by gene conversion. J Mol Evol. 1991;32:183–186. doi: 10.1007/BF02515391. [DOI] [PubMed] [Google Scholar]
- 69.Bennett GF, Peirce MA, Ashford RW. Avian haematozoa: mortality and pathogenicity. J Nat Hist. 1993;27:993–1001. doi: 10.1080/00222939300770621. [DOI] [Google Scholar]
- 70.Ortiz-Catedral L, Brunton D, Stidworthy MF, Elsheikha HM, Pennycott T, Schulze C, et al. Haemoproteus minutus is highly virulent for Australasian and South American parrots. Parasit Vectors. 2019;12(40):1–10. doi: 10.1186/s13071-018-3255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dinhopl N, Mostegl MM, Richter B, Nedorost N, Maderner A, Fragner K, et al. Application of in situ hybridization for the detection and identification of avian malaria parasites in paraffin wax-embedded tissues from captive penguins. Avian Pathol. 2011;40:315–320. doi: 10.1080/03079457.2011.569533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dinhopl N, Nedorost N, Mostegl MM, Weissenbacher-Lang C, Weissenböck H. In situ hybridization and sequence analysis reveal an association of Plasmodium spp. with mortalities in wild passerine birds in Austria. Parasitol Res. 2015;114:1455–1462. doi: 10.1007/s00436-015-4328-z. [DOI] [PubMed] [Google Scholar]
- 73.Ilgūnas M, Bukauskaitė D, Palinauskas V, Iezhova TA, Dinhopl N, Nedorost N, et al. Mortality and pathology in birds due to Plasmodium (Giovannolaia) homocircumflexum infection, with emphasis on the exoerythrocytic development of avian malaria parasites. Malar J. 2016;15:256. doi: 10.1186/s12936-016-1310-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ciloglu A, Ellis VA, Bernotienė R, Valkiūnas G, Bensch S. A new one-step multiplex PCR assay for simultaneous detection and identification of avian haemosporidian parasites. Parasitol Res. 2019;118:191–201. doi: 10.1007/s00436-018-6153-7. [DOI] [PubMed] [Google Scholar]
- 75.Pacheco MA, Cepeda AS, Bernotienė R, Lotta IA, Matta NE, Valkiūnas G, et al. Primers targeting mitochondrial genes of avian haemosporidians: PCR detection and differential DNA amplification of parasites belonging to different genera. Int J Parasitol. 2018;48:657–670. doi: 10.1016/j.ijpara.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Human, simian and rodent malaria species included in the Plasmodium dataset.
Additional file 2. Bayesian inference trees calculated from CytB sequences of Plasmodium species (A), Haemoproteus spp. (B) and Leucocytozoon spp. (C). The trees were midpoint-rooted, no outgroups were used. Posterior probabilities and maximum likelihood bootstrap values are indicated at most nodes. The scale bar indicates the expected mean number of substitutions per site according to the model of sequence evolution applied.
Additional file 3. Maximum-likelihood tree of Plasmodium 18S rDNA sequences based on a secondary structure alignment calculated with R-Coffee and only gaps trimmed.
Additional file 4. Maximum-likelihood tree of Plasmodium 18S rDNA sequences based on a secondary structure alignment calculated with R-Coffee applying the “strict” algorithm implemented in trimAl v.1.2 [53] for trimming.
Additional file 5. Maximum-likelihood tree of Plasmodium 18S rDNA sequences based on a secondary structure alignment calculated with R-Coffee applying the default option on the G-blocks Server [55] for trimming.
Additional file 6. Sequence features of the 18S rDNAs analysed.
Additional file 7. Results of the recombination tests.
Data Availability Statement
All DNA, blood and tissue samples are deposited in the collection of the Institute of Pathology (Department of Pathobiology, University of Veterinary Medicine Vienna). All sequence data were uploaded to NCBI GenBank. DNA sequence alignments are available from J.H. upon request.