Abstract
The cellulose synthase (Ces) and cellulose synthase-like (Csl) gene families belonging to the cellulose synthase gene superfamily, are responsible for the biosynthesis of cellulose and hemicellulose of the plant cell wall, and play critical roles in plant development, growth and evolution. However, the Ces/Csl gene family remains to be characterized in pineapple, a highly valued and delicious tropical fruit. Here, we carried out genome-wide study and identified a total of seven Ces genes and 25 Csl genes in pineapple. Genomic features and phylogeny analysis of Ces/Csl genes were carried out, including phylogenetic tree, chromosomal locations, gene structures, and conserved motifs identification. In addition, we identified 32 pineapple AcoCes/Csl genes with 31 Arabidopsis AtCes/Csl genes as orthologs by the syntenic and phylogenetic approaches. Furthermore, a RNA-seq investigation exhibited the expression profile of several AcoCes/Csl genes in various tissues and multiple developmental stages. Collectively, we provided comprehensive information of the evolution and function of pineapple Ces/Csl gene superfamily, which would be useful for screening out and characterization of the putative genes responsible for tissue development in pineapple. The present study laid the foundation for future functional characterization of Ces/Csl genes in pineapple.
Keywords: cellulose synthase gene, expression profile, RNA-Seq, Ananas comosus
1. Introduction
The cell wall, as a key component of plant cell, plays vital roles in the whole process of plant growth. The plant cell synthesizes and deposits the wall polymers to adjust the architecture of the cell wall according to requirements and it also manages the physical properties of the cell wall. Generally, cell walls are made of the primary cell wall and the secondary cell wall. The primary cell wall is deposited during cell growth and the secondary cell wall is formed when an expansion ends. Polysaccharides including hemicellulose, cellulose and pectin and proteins are common components of cell walls [1]. Cellulose is the main component of primary and secondary plant cell walls. Cellulose displays huge tensile firmness to the cell wall. In nature, about 180 billion tons of cellulose is generated every year, which is an important renewable resource on the earth [2]. Cellulose synthesis captures a lot of interest and many reviews have highlighted the contributions toward the understanding of cellulose biosynthesis [2,3,4,5,6,7]. The study on the regulation mechanism of plant cell wall synthesis and modification has been one of the most important fields in plant development biology.
Cellulose synthase (CesA) genes encode cellulose which is consists of β-1, 4 linked glucan residue chains [8,9,10]. Cellulose synthesis is coordinated by the strict and complex transcriptional regulation system. Similar to CesA sequences, cellulose synthase-like (Csl) genes also make a gene family. The Csl genes are mainly responsible for the biosynthesis of hemicellulose [11,12] that with cellulose forms a matrix in the cell walls [13,14]. The CesA and Csl gene families constitute the cellulose synthase gene superfamily that is classified as glycoside transferase gene family (GT2) in the Carbohydrate-Active enZYmes Database (CAZy) database based on sequence similarity, GT2 consists of conserved and variable regions [15,16]. GT2 proteins possess a class-specific region (CSR) and a plant-conserved region as well as four conserved motifs (QxxRW, DD, DCD, and TED) to be responsible for binding to the substrate [17].
All CesA and Csl proteins have several transmembrane domains (TMs). At the N-terminal, two TMs are found and the remaining are found near the C-terminal which contains a hydrophilic intracellular region [12]. The maximum homology between CesA and Csl proteins occurs in the intracellular region. Csl and CesA genes have high sequence similarity, and their proteins have glycosyltransferase activity [15]. In addition, they contain an enzyme catalytic site for the motif D-D-D-QxxRW (D, Q, R, and W represent standard amino acid, while x depicts any amino acid) [18]. The main difference between Csl and CesA is that the Csl family lacks a zinc finger structure, which has a very conservative repetitive sequence CXXC (cysteine-X-X-cysteine). The sequence can bind to DNA and play an important role to maintain the stability of cellulose synthase complexes, and be involved in the interaction between the subunits [2]. Since Csl lacks a zinc finger structure, it has been suggested that hemicellulose synthesis probably does not require such a structure, and a single Csl protein may have the catalytic activity to form the main chain of hemicellulose.
The functions of CesAs and Csls have been well studied. In Arabidopsis, AtCesA1, AtCesA3 and AtCesA6, genes function in the cellulose biosynthesis and composition of the primary cell wall [19]. In addition, loss-of-function mutants of AtCesA3 and AtCesA6 leaded to a decrease in cellulose [19,20,21]. The functional loss mutants of AtCesA4, AtCesA7 and AtCesA8 showed reduced cellulose content in the secondary cell wall, which was usually accompanied by changes in xylem structure. Further analysis revealed that AtCesA4, AtCesA7 and AtCesA8 co-expressed, and interacted together [22,23,24]. Csl consists of eight subfamilies (CslA, CslB/HCslC, CslD, CslE, CslF, CslG and CslJ/M), and the CslD subfamily is common in terrestrial plants. The Csl subfamilies show high similarity in sequence with cellulose synthase gene. So far, six, five and five CslD genes have been identified in Arabidopsis, rice and maize (Zea mays), respectively. Several Csl genes have been demonstrated to be directly responsible for the biosynthesis of cellulose in Arabidopsis, poplar (Populus alba) and rice (Oryza sativa). [25,26]. In rice, the CslF gene family plays a crucial role in cell wall formation and growth [26,27]. In Tropaeolum majus, it was found that one TmCsl was related to the synthesis of the 1, 4-glucan skeleton of xyloglucan (XyG), the main component of hemicellulose in the primary cell wall, and this protein is highly homologous to Arabidopsis CslC4. Mutants with the loss-of-function of AtCslD1 and AtCslD4 showed significantly reduced cellulose deposition in the cell wall of pollen tube, and the histological sequence from the cell walls of pollen tube was significantly broken, thus affecting the germination and growth of pollen tube and the transmissibility of male gametophyte [28]. All in all, CesA and Csl proteins play key roles in the plant growth and development.
Gram negative bacterium, Acetobacter xylinus, was first reported to contain a gene encoding cellulose synthase designated as BcsA [29,30,31]. The first plant CesA proteins were identified based on a homology search between bacterial BcsA and expressed sequence tags from cotton [2,29,30,31,32]. Recently, researchers on the CesA and Csl gene superfamily have been reported in various plant species, such as Arabidopsis, maize, wheat (Triticum aestivum), poplar, pine (Pinus), rice, eucalypt (Eucalyptus tereticornis) and alfalfa (Medicago sativa) [12,14,18,25,26,27,33,34,35,36,37,38,39,40,41,42]. However, the CesA and Csl have not been explored in pineapple, a highly valued tropical fruit with a gross production value reaching nine billion dollars annually and having outstanding nutritional and medicinal properties [43]. Recently, the genome of the pineapple was sequenced providing the opportunity to decipher the gene functions during pineapple developmental and stress response [44,45,46,47,48,49].
In this study, we identified AcoCes/Csl candidate genes using the reference genome of the pineapple, and analyzed the domain, motif and gene structure of AcoCes/Csl genes. We also studied the evolutionary relationship of AcoCes/Csl genes and analyzed the phylogenetic relationship of the Ces/Csl gene family between Arabidopsis (dicot) and rice (monocot). Furthermore, we studied the expression pattern of Ces/Csl genes in different pineapple tissues and developmental stages. Our results provide the key information for further evaluation and functional characterization of AcoCes/Csl genes in pineapple.
2. Results
2.1. Identification of Ces /Csl Genes in Pineapple
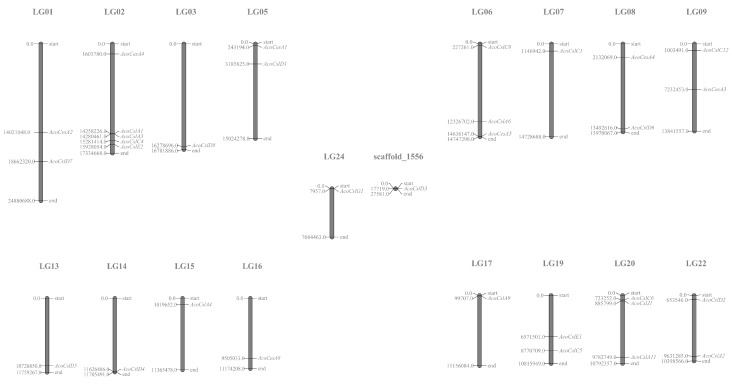
We identified a total of 32 candidate genes from the pineapple genome. The 32 proteins were grouped into seven subfamilies, including AcoCesA, AcoCslA, AcoCslC, AcoCslD, AcoCslE, AcoCslG and AcoCslJ according to the phylogenetic relationships with Arabidopsis and rice (Table 1). CslD had the maximum number of members (with eight genes) among the identified subfamilies. The smallest subfamilies were CslG and CslJ, both containing only one member. The gene distribution in chromosomes was showed in Figure 1. These genes were mapped on 17 pineapple chromosomes and one scaffold. Chr3 possessed five genes, Chr20 contained three genes, seven chromosomes each contained two genes, and eight chromosomes each contained one gene. Moreover, the remaining one gene was located on scaffold 1556.
Table 1.
Properties of the Ces/Csl gene superfamily in pineapple.
| Gene Name | Gene ID | Chr | Length | Amino Acids | Introns | Exons | Isoelectric Point (pI) | Molecular Weight (MW) |
|---|---|---|---|---|---|---|---|---|
| AcoCesA1 | Aco014283 | LG05 | 7376 | 1104 | 13 | 14 | 6.72 | 122.4 |
| AcoCesA2 | Aco018229 | LG01 | 8424 | 1077 | 13 | 14 | 7.92 | 120.7 |
| AcoCesA3 | Aco024230 | LG09 | 12,555 | 1081 | 12 | 13 | 8.31 | 119.6 |
| AcoCesA4 | Aco014585 | LG08 | 7332 | 1070 | 11 | 12 | 7.61 | 119.9 |
| AcoCesA5 | Aco018552 | LG06 | 6358 | 1102 | 13 | 14 | 7.04 | 123.2 |
| AcoCesA7 | Aco012076 | LG02 | 6278 | 1062 | 12 | 13 | 6.33 | 118.9 |
| AcoCesA8 | Aco006039 | LG16 | 6951 | 1003 | 12 | 13 | 5.93 | 111.7 |
| AcoCslA1 | Aco001096 | LG02 | 14,534 | 1126 | 19 | 19 | 8.99 | 127.2 |
| AcoCslA2 | Aco006900 | LG22 | 4546 | 536 | 8 | 9 | 9.19 | 60.9 |
| AcoCslA3 | Aco001095 | LG02 | 5559 | 560 | 9 | 10 | 8.8 | 62.9 |
| AcoCslA4 | Aco004149 | LG15 | 20,041 | 1184 | 20 | 20 | 8.8 | 133.1 |
| AcoCslA6 | Aco002889 | LG06 | 9648 | 555 | 15 | 9 | 8.56 | 63 |
| AcoCslA9 | Aco016682 | LG17 | 9750 | 607 | 10 | 10 | 9.16 | 68.4. |
| AcoCslA11 | Aco014689 | LG20 | 7773 | 568 | 9 | 10 | 7.83 | 64.1 |
| AcoCslC1 | Aco004974 | LG07 | 3939 | 727 | 4 | 5 | 8.4 | 81.1 |
| AcoCslC4 | Aco000968 | LG02 | 2630 | 673 | 4 | 5 | 8.85 | 77.1 |
| AcoCslC5 | Aco008242 | LG19 | 4607 | 747 | 8 | 6 | 8.86 | 84.7 |
| AcoCslC6 | Aco013494 | LG20 | 5582 | 709 | 4 | 5 | 8.67 | 78.7 |
| AcoCslC8 | Aco011603 | LG06 | 3136 | 663 | 4 | 5 | 8.99 | 74.8 |
| AcoCslC12 | Aco008598 | LG09 | 3881 | 773 | 6 | 7 | 6.15 | 85.7 |
| AcoCslD1 | Aco004435 | LG05 | 4309 | 1068 | 4 | 5 | 7.62 | 116.9 |
| AcoCslD2 | Aco015969 | LG22 | 5627 | 1174 | 4 | 3 | 7.49 | 129.2 |
| AcoCslD3 | Aco030607 | scaffold_1556 | 5631 | 1131 | 4 | 3 | 6.74 | 124.7 |
| AcoCslD4 | Aco017129 | LG14 | 6362 | 1159 | 6 | 5 | 6.46 | 127.5 |
| AcoCslD5 | Aco013738 | LG13 | 3953 | 1201 | 2 | 3 | 8.45 | 130.9 |
| AcoCslD6 | Aco016995 | LG08 | 4094 | 1193 | 4 | 5 | 7.9 | 130.5 |
| AcoCslD7 | Aco025070 | LG01 | 7403 | 1170 | 3 | 4 | 6.7 | 128.9 |
| AcoCslD8 | Aco017291 | LG03 | 3892 | 1185 | 2 | 3 | 7.08 | 130.2 |
| AcoCslE1 | Aco026335 | LG19 | 5869 | 680 | 7 | 8 | 6.99 | 76.2 |
| AcoCslE2 | Aco000884 | LG02 | 4750 | 736 | 7 | 8 | 8.02 | 81.9 |
| AcoCslG1 | Aco013153 | LG24 | 47,982 | 1615 | 16 | 17 | 7.92 | 178.6 |
| AcoCslJ1 | Aco013513 | LG20 | 13,869 | 807 | 9 | 10 | 8.91 | 89.2 |
Figure 1.
Distribution of AcoCes/Csl genes on the pineapple genome. The gene start points are shown on the chromosome. Thirty-two Ces/Csl genes of pineapple were mapped to different chromosomes using MapChart. Only those chromosomes bearing AcoCes/Csl are represented. The prefix ‘Aco’ indicates Ananas comosus.
Characteristics of the 32 Ces/Csl genes were shown in Table 1. Genomic DNA size of genes in this gene superfamily varied from 2630 kb (AcoCslC4) to 20,041 kb (AcoCslA4). The average length of these genes was 8270 kb. Genomic DNA length did not change much in the CesA subfamily rather than CesA3, CslC, CslD and ClsE subfamilies. The numbers of predicted amino acids ranged from 536 aa (AcoCslA2) to 1615 aa (AcoCslG1) with the corresponding molecular weight varied from 60.9 kDa to 178.6 kDa. The CslC subfamily showed great divergence in terms of amino acid length (536 aa–1184 aa), which differed from the other subfamilies. The CesA and CslD subfamilies constituted similar amino acid length. The predicted isoelectric points varied from 5.93 (AcoCesA8) to 9.19 (AcoCslA2). Besides these, the minimum intron number was two that was found in CslD subfamily including AcoCslD2, AcoCslD3, AcoCslD5 and AcoCslD8. The CslA subfamily has the maximum intron number including AcoCslA1 (19) and AcoCslA4 (20). The intron number of CesA, CslC, CslD and CslE subfamily changed little, especially for CslE that two members had the same intron number. The intron number of CslA subfamily varied from eight to 20.
2.2. Phylogenetic Analysis of the Pineapple Ces/Csl Gene Superfamily
Based on the phylogenetic distribution, pineapple Ces/Csl proteins could be divided into five subgroups (Figure S1), including I, II, III, IV and V. Subgroup I possessed all the CslA proteins, Subgroup II contained all the CslC proteins, Subgroup III consisted the CslE, CslG and CslJ proteins, Subgroup IV contained all the CesA proteins and subgroup V consisted all the CslD proteins.
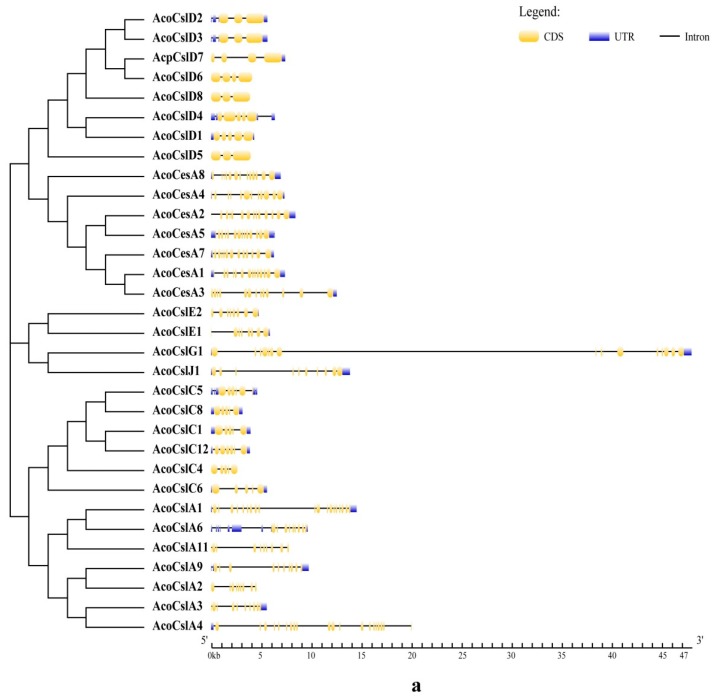
The related sister pairs appeared in the joint phylogenetic tree (such as AcoCslA3 and AcoCslA4) and triplets (such as AcoCslA1, AcoCslA6 and AcoCslA11) [50,51]. We found, seven sister pairs and five triplets among the AcoCes/Csl gene families. The similar intron-exon structure existed in the sister pairs or triplets (Figure 2a) validating the phylogenetic results. The structural diversities among the AcoCes/Csl genes suggested that the gene family could be showing functional divergence.
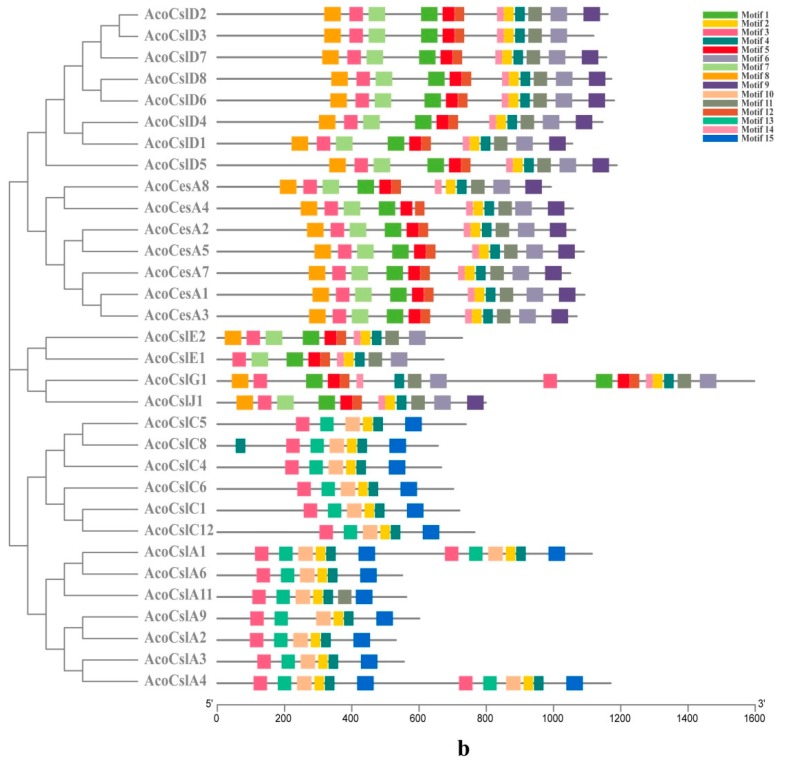
Figure 2.
(a) Exon-intron structure of pineapple Ces/Csl genes. Bule boxes indicated untranslated 5’- and 3-regions; yellow boxes indicated exons; black lines indicate introns. The prefix ‘Aco’ indicates Ananas comosus. (b) Motif analysis of the pineapple Ces/Csl protein. Motifs with specific colors can be found on the respective AcoCes/Csl protein. The combined phylogenetic trees of AcoCes/Csl subfamily on the left panel. The motifs of corresponding proteins are shown on the right panel with specific colors on behalf of different motifs using the Multiple Em for Motif Elicitation (MEME). The order of the motifs corresponds to their position within individual protein sequences. Prefix ‘Aco’ indicates Ananas comosus.
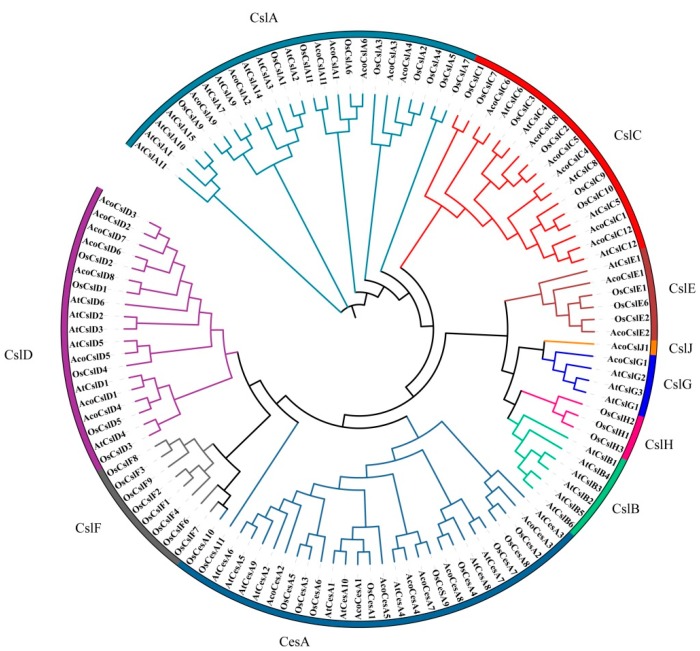
Based on the full-length protein sequences, we constructed a multi-species phylogenetic tree of Ces/Csls from Arabidopsis, pineapple and rice, in order to investigate the functional associations and evolutionary relationships of pineapple Ces/Csl genes (Figure 3). The gene names and IDs of Ces/Csl from Arabidopsis and rice were presented in Table S1. The phylogenetic analysis suggested that Ces/Csl can be grouped into 10 subfamilies: CslD, CslF, CesA, CslB, CslH, CslG, CslJ, CslE, CslC and CslA. The CesA subfamily was the greatest subfamily, with seven pineapple CesA genes, 10 Arabidopsis genes, nine rice genes, accounting for 21% of the total Ces/Csl genes. CslA was the second largest subfamily, having seven genes from pineapple, nine from Arabidopsis, and nine from rice genes. The smallest subfamily was CslJ with only one pineapple gene. No pineapple gene was found in CslB, CslF and CslH. Arabidopsis had six CslB genes, three CslH and eight CslF genes were found in rice.
Figure 3.
Phylogenetic tree depicting the relationships among Ces/Csl proteins from pineapple, Arabidopsis and rice. All the Ces/Csl protein sequences were aligned and phylogenetic tree was constructed using MEGA 7.0. The different colored arcs indicated different subgroups. Prefix ‘Ath’, Osa, and ‘Aco’ indicate Ces/Csl proteins from Arabidopsis, Oryza sativa, and Ananas comosus, respectively.
According to the phylogenetic tree, we identified 10 sister gene pairs between pineapple and rice; AcoCesA7/OsCesA9, AcoCesA8/OsCesA4, AcoCesA1/OsCesA1, AcoCslC5/OsCslC2, AcoCslA6/OsCslA6, AcoCslA11/OsCslA11, AcoCslA9/OsCslA9, AcoCslD6/OsCslD2, AcoCslD8/OsCslD1, AcoCslD4/OsCslD5, six sister gene pairs between pineapple and Arabidopsis; AcoCesA4/AtCesA8, AcoCslC12/AtCslC12, AcoCslC4/AtCslC8, AcoCslC6/AtCslC6, AcoCslD5/AtCslD5, AcoCslD1/AtCslD1 Four triplets were found between pineapple and rice; AcoCesA3/OsCesA2/OsCesA8, AcoCslA4/OsCslA2/OsCslA4, AcoCslC8/AcoCslC5/OsCslC2 and AcoCslA6/OsCslA6/AcoCslA1, and one triplet was identified between pineapple and Arabidopsis; AcoCslC1/AcoCslC12/AtCslC12.
2.3. Gene Structure Analysis and Conserved Motif Identification
The evolutionary aspect and structural diversity of the Ces/Csl genes in pineapple were explored by studying the exon-intron organization. The difference in the gene architecture such as number of exons, introns and the lengths of untranslated region (UTR) among gene pairs suggest that the paralogs could be having separate roles during pineapple growth and development [52]. The number of exons in AcoCes/Csl genes varied from three to 20. AcoCslA4 had the maximum exons, whereas AcoCslD8 and AcoCslD5 had only three exons and three genes (AcoCslD6, AcoCslD8 and AcoCslD5) had no UTR. To further reveal the diversification of AcoCes/Csl gene family in pineapple, we predicted the putative motifs by MEME with the default setting [53]. In total, 15 motifs were identified in the AcoCes/Csl gene family. AcoCslD and AcoCesA subgroup had 12 same motifs, while AcoCslD lacked in the motif 9. Most of the AcoCslA and AcoCslC subgroup contained six same motifs, except for AcoCslC8, AcoCslA1, AcoCslA11 and AcoCslA4 (Figure 2b).
2.4. Synteny Analysis of Pineapple Ces/Csl Genes
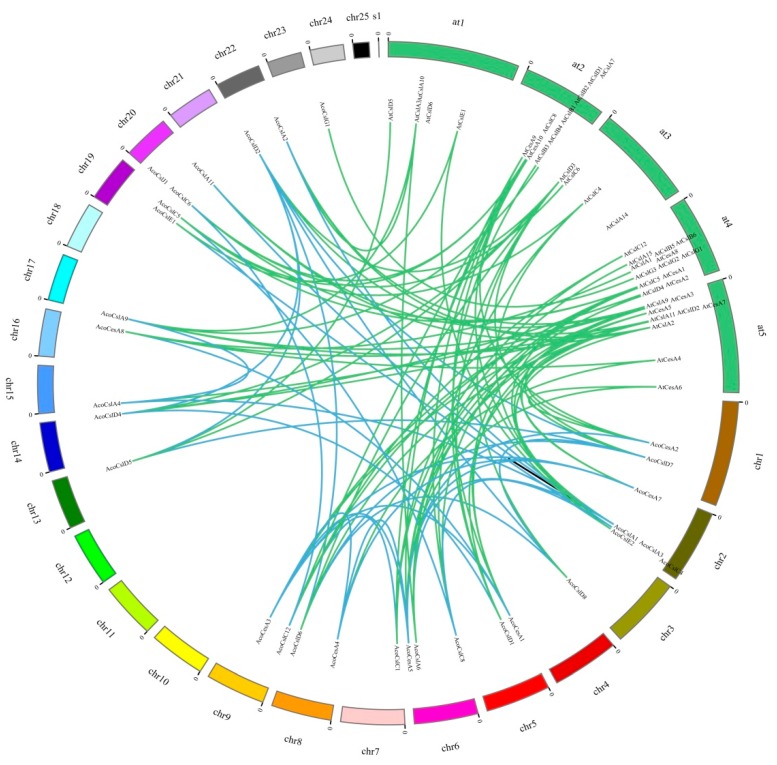
Segmental and tandem duplication gene pairs (identity ≥ 50%) of the Ces/Csl gene family were studied to test the duplication effect. One tandem duplication pair (AcoCesA3 and AcoCesA7), which showed a high coding sequence similarity, was distributed closely on the chromosome 2. In addition, we identified 34 pairs of segmental duplication events where each pair of gene is situated at separate chromosomes in pineapple Ces/Csl genes, such as AcoCesA8/AcoCesA1, AcoCslD1/AcoCslD4, AcoCslC5/AcoCslC8 (Figure 4). Overall, our results showed that tandem and segmental duplication resulted in the expansion of the pineapple Ces/Csl gene family.
Figure 4.
Synteny analysis between pineapple and Arabidopsis Ces/Csl genes. Chromosomes and scaffolds of pineapple and Arabidopsis are shown in different colors and in partial circles. Colored curves indicate the syntenic relationships between pineapple and Arabidopsis Ces/Csl genes. The prefix ‘Aco’ indicates Ananas comosus.
Additionally, the syntenic relationship between pineapple and Arabidopsis was also investigated to study the evolution of pineapple Ces/Csl genes. Two types of pineapple Ces/Csl genes were found in synteny analysis. The first type of pineapple Ces/Csl genes was that a pineapple Ces/Csl gene related with a single Arabidopsis gene viz., AcoCslA4-AtCslA9 and AcoCesA4-AtCesA4. The second type is that a pineapple Ces/Csl gene associated with multiple Arabidopsis genes, for example, AcoCslC8-AtCslC8, AtCslC5, AtCslC4; AcoCesA1-AtCesA1, AtCesA3, AtCesA10. The more elaborated information is given in Supplemental Table S2.
2.5. Expression Patterns of AcoCes/Csl Genes in Four Different Tissues
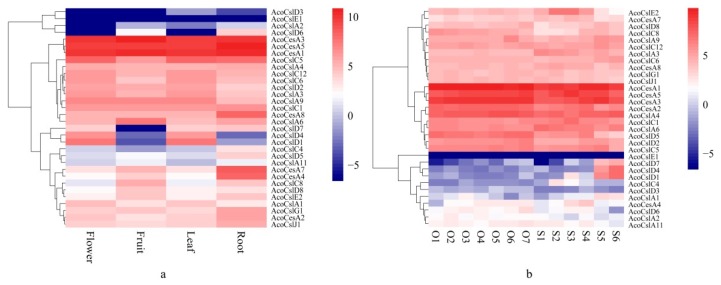
The transcriptome analysis was carried out to understand tissue-specific expression patterns of AcoCes/Csl genes. From the RNA-seq data the expressions of 32 AcoCes/Csl genes in flower, fruit, leaf and root were studied using their fragments per kilobase of exon model per million mapped reads (FPKM) values [44].
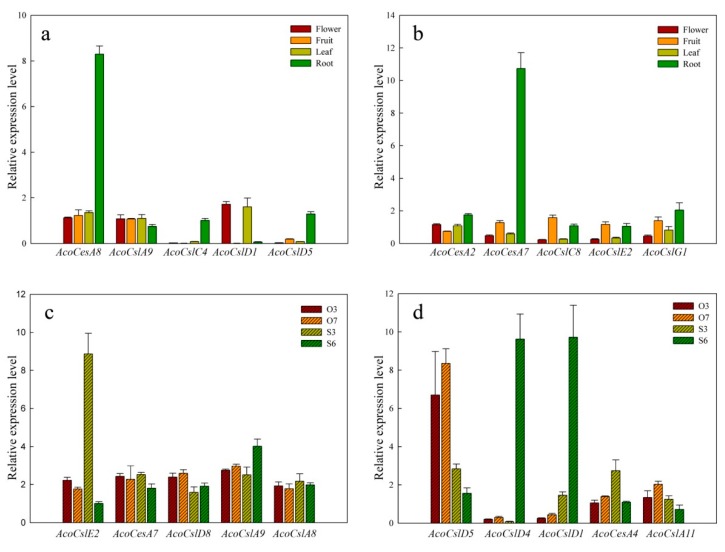
As showed in Figure 5a, AcoCslD3, AcoCslE1, AcoCslA2 and AcoCslD2 were expressed in all sampled tissues at very low levels, implying that those genes might be expressed under special conditions or in other non-sampled pineapple tissues. Transcript levels of AcoCesA3, AcoCesA5, AcoCesA1 and AcoCslC5 were similar, showing very high expression in all the tested tissues, which suggested that these genes might be playing a crucial role in the plant development. Similar expression profiles were found in AcoCslA4, AcoCslC12, AcoCslC6, AcoCslD2, AcoCslA3 and AcoCslA3 with moderate and even expression level in four tissues. The expression level of AcoCslD4 and AcoCslD1 were higher in flower and leaf than fruit and root, indicating that the two genes may be responding in the growth of flower and leaf. Furthermore, AcoCesA4, AcoCesA7 and AcoCesA8, showed high root-specific expression, suggesting these genes may be working during the root development. The remaining genes showed similar expression pattern. The RNA-seq data was further verified using qRT-PCR. For qRT-PCR, 10 genes (AcoCesA8, AcoCslA9, AcoCslC4, AcoCslD1, AcoCslD5, AcoCesA2, AcoCesA7, AcoCslC8, AcoCslE2 and AcoCslG1) were selected. AcoCesA8 and AcoCesA7 showed higher expression in root and relative lower expression than the other three tissues. AcoCesC4 and AcoCesD5 also exhibited root-specific expression, but barely expressed it in flower, fruit and leaf (Figure 6a,b). However, for AcoCesD5, the higher expression was found in flower and leaf, but did not express it in the root and fruit. The other five candidate genes expressed showed no significant difference between four tissues which were consistent with the results from RNA seq data.
Figure 5.
Organ-specific expression profiles of the pineapple AcoCes/Csl genes. (a) Expression profiles of AcoCes/Csl genes in flower, fruit, root and leaf of pineapple. (b) Expression patterns of AcoCes/Csl genes in developing ovule and stamen of pineapple. Hierarchical clustering of expression profiles of pineapple Ces/Csl genes in different organs and developmental stages. Red colors indicate high levels of transcript abundance, and blue colors indicate low transcript abundance. The color scale is shown at right side of the figure. Sample details are mentioned at the bottom of each lane: Ovule O1–O7, stamen S1–S6. The number after different tissues name refers to the different stage. Prefix ‘Aco’ indicates Ananas comosus.
Figure 6.
Expression profiles of the pineapple Ces/Csl genes in different tissues. (a,b) qPCR expression profiles of AcoCes/Csl genes in flower, fruit, root and leaf of pineapple. (c,d) qPCR expression pattern of AcoCes/Csl genes in developing ovule and stamen of pineapple. Sample details are mentioned at the top right corner: Ovule O3 and O7, stamen S3 and S6. The number after the different samples name refers to the different developmental stages. The gene details are mentioned at the bottom of each lane. Data were normalized to the EF1a gene. Vertical bars indicate standard error (SE). All experiments were performed with three technical and three biological repeats. The prefix ‘Aco’ indicates Ananas comosus.
2.6. Expression of AcoCes/Csl Genes during Gametophyte Development
The roles of AcoCes/Csl genes in pineapple were further studied to understand their roles in reproductive development. The expression patterns of 32 AcoCes/Csl in ovules and stamens were investigated using transcriptome data. As showed in Figure 5b, the expression profiles revealed that 10 AcoCes/Csl genes (AcoCesA1, AcoCesA5, AcoCesA3, AcoCesA2, AcoCslA4, AcoCslC1, AcoCslA5, AcoCslD2, AcoCslD2, AcoCslD5 and AcoCslC5) were expressed highly in various stages of ovule and six stages of stamens, implying they may be performing crucial role in the formation reproductive organs. Eleven AcoCes/Csl genes were expressed moderately and evenly in all tested tissues. AcoCslD7, AcoCslD4 and AcoCslD1 showed similar expression level that had higher expression in the stage 5 and stage 6 of stamens than other tissues. Seven AcoCes/Csl genes showed low expression levels in every tissue. AcoCslE1 had the lowest expression in all the tissues. The different stages in pineapple reproductive organs were selected as reported earlier [54]. To further validate these results, 10 genes were selected (AcoCslE2, AcoCesA7, AcoCslD8, AcoCslA9, AcoCslA8, AcoCslD5, AcoCesD4, AcoCesD1, AcoCesA4 and AcoCslA12) to perform qRT-PCR analysis. AcoCslE2 expressed higher in stage 3 stamens than other tissues. AcoCslD5 showed higher expression in stamens than ovules. AcoCesD4 and AcoCesD1 exhibited highest expression in stage 6 of stamens, but barely expressed in ovules and stage 3 of stamens (Figure 6c,d). The other 6 candidate genes expressed showed no significant difference between four tissues which were also coincided the results from RNA seq data.
3. Discussion
Based on the Arabidopsis database, the cellulose synthase superfamily was initially divided into the CesA family and six Csl families including A, B, C, D, E and G [12]. They belong to the integral membrane proteins, CesA proteins are located in the plasma membrane, however CslB, CslG and CslE are believed to locate in the Golgi [2]. The conservation of intron-exon structure exists in CesA, CslB, CslG and CslE, but not in other three families [2]. The CesA is responsible for the synthesis of cellulose, and the Csl participates in the synthesis of hemicellulose [12]. Three specific lineages including CslF [26], CslH [26] and CslJ [55] have been identified in the Poaceae. All of them have functions in the biosynthesis of the cell wall, and the three lineages have a wide distribution in the Poaceae but a narrow distribution in other angiosperms [55,56,57]. Base on the available genome sequence CslF, which was phylogenetically originated from the oldest family, CslD is presented in the graminid and restiid families [13]. However, no CslF genes were found in pineapple. CslH, which showed the monocot-specific sister branch to CslB, was not found in our study. While the CslH genes are involved in the synthesis of (1,3; 1,4)-β-glucan [57], the function of the CslB genes were not found. The CslJ was reported in barley, mediating the synthesis of the cell wall polysaccharide [13,55]. Even the CslJ genes were widely found in monocots, but only one was identified in our study. The CslM was discovered to form a reciprocally monophyletic eudicot-monocot grouping with the CslJ clade. However, heterologous expression of the grape VvCslM (Vitis vinifera) is unable to produce any detectable signs, as shown in Table 1, 4-β-glucan [13]. The CslM and CslJ branches families were different in evolutionary histories, therefore CslJ lineage should be monocot-specific and CslM lineage is eudicot-specific [13].
Based on the Arabidopsis database, the cellulose synthase superfamily was initially divided into the CesA family and six Csl families including A, B, C, D, E and G [12]. They belong to the integral membrane proteins, CesA proteins are located in the plasma membrane, however CslB, CslG and CslE are believed to locate in the Golgi [2]. The conservation of intron-exon structure exists in CesA, CslB, CslG and CslE, but not in other three families [2]. The CesA is responsible for the synthesis of cellulose, and the Csl participates in the synthesis of hemicellulose [12]. Three specific lineages including CslF [26], CslH [26] and CslJ [55] have been identified in the Poaceae. All of them have functions in the biosynthesis of the cell wall, and the three lineages have a wide distribution in the Poaceae but a narrow distribution in other angiosperms [55,56,57]. Base on the available genome sequence CslF, which was phylogenetically originated from the oldest family, CslD is presented in the graminid and restiid families [13]. However, no CslF genes were found in pineapple. CslH, which showed the monocot-specific sister branch to CslB, was not found in our study. While the CslH genes are involved in the synthesis of (1,3; 1,4)-β-glucan [57], the function of the CslB genes were not found. The CslJ was reported in barley, mediating the synthesis of the cell wall polysaccharide [13,55]. Even the CslJ genes were widely found in monocots, but only one was identified in our study. The CslM was discovered to form a reciprocally monophyletic eudicot-monocot grouping with the CslJ clade. However, heterologous expression of the grape VvCslM (Vitis vinifera) is unable to produce any detectable signs, as shown in Table 1, 4-β-glucan [13]. The CslM and CslJ branches families were different in evolutionary histories, therefore CslJ lineage should be monocot-specific and CslM lineage is eudicot-specific [13].
Early publications revealed that the CslD genes mediated functions in tip growth [25,58,59]. In this study, AcoCslD1 and AcoCslD4 had very high expression levels in the developing stamen of pineapple, suggesting that AcoCslDs may regulate stamen development. The plant Ces/Csl superfamily perhaps comes from cyanobacteria by endosymbiotic transferring. The putative CesA genes in cyanobacteria Anabaena spp. exhibited homology to that shown from the previously reported plants [60]. The CesA lineage in a marine cyanobacterium (Synechoccus spp.) existed monophyletic to the embryophyte CesA clades. At present, the phylogenetic analysis divided the superfamily into two distinct evolutionary branches, the CslA and CslC clades and the CesA and CslB/D/E/F/G/H/M lineages [13]. The CslA/C genes represented an independent lineage to CesA and CslB/D/E/F/G/H/J lineage, probably being originated from a different endosymbiotic transfer. The CslA was more similar to CslC than bacterial CesA, and the CslA/C proteins were smaller than bacterial CesA protein [60,61]. Unlike CslCs, some CslAs showed mannan synthase activity [62]. In pineapple, the CslA/CslC lineages had seven and six members, respectively. It is suggested that CslA genes mediate the biosynthesis of mannan [63] and CslC genes are responsible for the biosynthesis of xyloglucan [62,64,65]. However, not all the genes from the CslA and CslC clades were in participation with the biosynthesis of mannan or xyloglucan. Two AcoCslE and one AcoCslG were identified in the pineapple, but no clear functions with respect to the types of synthesized polysaccharides have been assigned to these genes. The gene architecture differences lead to functional diversification or functional redundancy. It is possible that these functional redundant genes are marching to pseudogenes due to lack of selective stress. We did observe that one gene (AcoCslE1) had extremely low expression in all of the tested samples, indicating that this gene is likely a peseudogene without evidence of function after divergence with other family members. In addition, we found that 26 members of the superfamily aggregate in sub-telomeric regions of the chromosomes, however, the reason is not clear so far.
The pineapple is one of the nutrient-rich tropical fruits, containing lots of nutrients including vitamin C, vitamin B6 and folate, as well as dietary fiber. The fiber is divided into soluble type and insoluble type. The soluble fiber comes from the inside of plant cells, reducing the blood sugar and decreasing cholesterol levels by binding to the cholesterol. The insoluble fiber originates from the cell walls of plant cells and can bind to water, making the stool softer, speeding up its movement through the digestive tract and decreases the risks for hemorrhoids, diverticulosis and constipation. The content and quality of fiber is regulated by Ces/Csl genes. The research on this gene family is very useful for biotechnology to improve the quality and yield of pineapple.
In summary, our identifications of the AcoCes/Csl gene families provide useful information to understand the biosynthetic mechanisms of (1,3; 1,4)-β-glucan in pineapple and lay the foundation for studying the origin of cell wall polysaccharides. Furthermore, the AcoCes/Csl genes pave the way to further functional identification and can be candidate genes for quality improvement of pineapple in future works.
4. Materials and Methods
4.1. Identification of Ces/Csl in Pineapple
The Ces/Csl amino acid sequences of Arabidopsis and Oryza sativa were downloaded from The Arabidopsis Information Resource (TAIR) (http://www.Arabidopsis.org) and the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/index.shtml). To identify the pineapple Ces/Csl genes, we used the Arabidopsis Ces/Csl amino acid sequences to search pineapple proteome with Basic Local Alignment Search Tool (BLAST-P) and we downloaded the hidden Markov model (HMM) profiles of the cellulose synthase (PF03552) domain from the pfam database (http://pfam.xfam.org/). Then we used the HMM profiles to search the pineapple proteome database through the hmm search program with the e-value set 0.01. We used Simple Modular Architecture Research Tool (SMART) [66] to verify these sequences we obtained last step, and deleted the redundant sequences. The rest of the Ces/Csl sequences were subjected to the phylogenetic analysis. We used Multiple Sequence Comparison by Log-Expectation (MUSCLE) 3.7 [67] with default setting and performed multiple alignments with Ces/Csls sequences from pineapple, Arabidopsis and rice.
4.2. Physicochemical Properties and Phylogenetic Analysis
To further understand the physicochemical properties, we used ExPASy (http://web.expasy.org/compute_pi/) to predict the isoelectric point (PI) and molecular weight (MW) of pineapple Ces/Csl amino acid sequences. We constructed the phylogenetic tree by MEGA 7 [68] through the maximum likelihood (ML) method with a bootstrap option of n = 1000 and the pairwise deletion of gaps.
4.3. Conserved Motifs Analysis of Pineapple Ces/Csl Protein
The MEME program (http://meme-suite.org/) was used to find the conserved motifs of pineapple Ces/Csl proteins with the motifs number set 15, and other options were default.
4.4. Chromosome Localization and Gene Structural Analysis of Pineapple Ces/Csl Genes
We downloaded the information of chromosome localization of AcoCes/Csl genes from Phytozome. The information was visualized by MapChart, including the localization and the length in corresponding chromosomes. In addition, the online gene structure display server (http://gsds.cbi.pku.edu.cn/) [69] was used to visualize the Ces/Csl genes structure information about the quantity and distribution of exon and intron.
4.5. Synteny Analysis of Pineapple Ces/Csl Genes
We first used blastp program to search homolog pairs between pineapple and Arabidopsis. After that, MCSCANX was used to identify synteny block with default parameter, which means that at least 5 genes should be preserved in a collinear block [70].
4.6. RNA-Seq and qRT-PCR
The different pineapple tissues including flower, fruits, leaf and root, and ovule and stamen from different development stages (MD2) were selected according to the previous method [71]. The total RNA was isolated using RNA extraction kit (Omega Bio-Tek, Shanghai, China). The cDNA libraries were established using the NEBNext Ultra™ RNA Library Prep Kit for Illumina (NEB) according to the manufacturer’s protocol. The qualified libraries were sequenced on the Hiseq2500 machine (NEBNext RNA-Seq data (SRA315090) of different tissues were downloaded from the National Center for Biotechnology Information (NCBI) database [44]. The trimmed pair-end reads of all tissues were aligned to pineapple genome by using TopHat v2.1.1 with default parameter settings. The FPKM values were estimated and further processed by Cufflinks v2.2.1 software. qRT-PCR was employed using the SYBR Taq II (TakaRa, China) and the program was as follows: 94 °C for 25 s; 39 cycles of 94 °C for 5 s and 60 °C for 40 s; 94 °C for 20 s. All the experiments were carried out with three technical and three biological replicates.
5. Conclusions
The Ces/Csl gene superfamily plays a critical role in the biosynthesis of cellulose and hemicellulos, however, information about the pineapple Ces/Csl gene family remains elusive in pineapple. Here, we identified 32 AcoCes/Csl genes in the pineapple which could be divided into five groups. We also studied the basic features including isoelectric point, molecular weight, transmembrane domains, gene structure, chromosome location, phylogenetic analysis and the syntenic relationship of the 32 pineapple Ces/Csl genes compared to Arabidopsis. Gene expression profiles showed they could be playing necessary roles in the development of reproductive organs. Overall, the studies of pineapple AcoCes/Csl genes present important information for functional study and future pineapple research.
Abbreviations
| Aco | Ananas comosus |
| At | Arabidopsis thaliana |
| Ces | Cellulose synthase |
| Csl | Cellulose synthase-like |
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/2223-7747/8/8/275/s1. Figure S1: Phylogenetic tree depicting the relation between pineapple Ces/Csl genes, Table S1: The genes of Ces/Csl in Arabidopsis and rice, Table S2: Synteny block of Ces/Csl genes between pineapple and Arabidopsis genomes.
Author Contributions
S.C. and H.C. performed experiments, data analysis and manuscript writing; J.Z., M.Y. worked on qRT-PCR; S.P.O. and M.A. revised and edited the manuscript; H.Z., S.V.G.N.P., A.H. and L.L. helped to perform experiment. Y.Y., G.C. and Y.Q. conceived the study and revised manuscript. All authors read and approved the final manuscript.
Funding
This work was funded by National Natural Science Foundation of China grant number KAE16010A to S.C.; U1605212, 31761130074 to Y.Q., and the fund from Fujian Agriculture and Forestry University Forestry peak discipline construction project grant number 71201800739.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1.Griffiths J.S., North H.M. Sticking to cellulose: exploiting Arabidopsis seed coat mucilage to understand cellulose biosynthesis and cell wall polysaccharide interactions. New Phytol. 2017;214:959–966. doi: 10.1111/nph.14468. [DOI] [PubMed] [Google Scholar]
- 2.Delmer D.P. CELLULOSE BIOSYNTHESIS: Exciting Times for A Difficult Field of Study. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999;50:245–276. doi: 10.1146/annurev.arplant.50.1.245. [DOI] [PubMed] [Google Scholar]
- 3.Williamson R.E., Burn J.E., Hocart C.H. Towards the mechanism of cellulose synthesis. Trends Plant Sci. 2002;7:461–467. doi: 10.1016/S1360-1385(02)02335-X. [DOI] [PubMed] [Google Scholar]
- 4.Saxena I.M., Brown R.M. Cellulose biosynthesis: Current views and evolving concepts. Ann. Bot.-London. 2005;96:9–21. doi: 10.1093/aob/mci155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Somerville C. Cellulose synthesis in higher plants. Annu. Rev. Cell. Dev. Biol. 2006;22:53–78. doi: 10.1146/annurev.cellbio.22.022206.160206. [DOI] [PubMed] [Google Scholar]
- 6.Crowell E.F., Gonneau M., Stierhof Y.D., Höfte H., Vernhettes S. Regulated trafficking of cellulose synthases. Curr. Opin. Plant Biol. 2010;13:700–705. doi: 10.1016/j.pbi.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 7.Lei L., Li S.D., Gu Y. Cellulose synthase complexes: composition and regulation. Front. Plant Sci. 2012;3:75. doi: 10.3389/fpls.2012.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li S.D., Bashline L., Zheng Y.Z., Xin X.R., Huang S.X., Kong Z.S., Kim S.H., Cosgrove D.J., Gu Y. Cellulose synthase complexes act in a concerted fashion to synthesize highly aggregated cellulose in secondary cell walls of plants. Proc. Natl. Acad. Sci. USA. 2016;113:11348–11353. doi: 10.1073/pnas.1613273113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bessueille L., Bulone V. A survey of cellulose biosynthesis in higher plants. Plant Biotechnol. 2008;25:315–322. doi: 10.5511/plantbiotechnology.25.315. [DOI] [Google Scholar]
- 10.Guerriero G., Fugelstad J., Bulone V. What Do We Really Know about Cellulose Biosynthesis in Higher Plants? J. Integr. Plant Biol. 2010;52:161–175. doi: 10.1111/j.1744-7909.2010.00935.x. [DOI] [PubMed] [Google Scholar]
- 11.Lerouxel O., Cavalier D.M., Liepman A.H., Keegstra K. Biosynthesis of plant cell wall polysaccharides - a complex process. Curr. Opin. Plant Biol. 2006;9:621–630. doi: 10.1016/j.pbi.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 12.Richmond T.A., Somerville C.R. The Cellulose Synthase Superfamily. Plant Physiol. 2000;124:495–498. doi: 10.1104/pp.124.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Little A., Schwerdt J.G., Shirley N.J., Khor S.F., Neumann K., O’Donovan L.A., Lahnstein J., Collins H.M., Henderson M., Fincher G.B., et al. Revised Phylogeny of the Cellulose Synthase Gene Superfamily: Insights into Cell Wall Evolution. Plant Physiol. 2018;177:1124–1141. doi: 10.1104/pp.17.01718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin Y.B., Huang J.L., Xu Y. The cellulose synthase superfamily in fully sequenced plants and algae. Bmc Plant Biol. 2009;9:99. doi: 10.1186/1471-2229-9-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lairson L.L., Henrissat B., Davies G.J., Withers S.G. Glycosyltransferases: Structures, functions, and mechanisms. Annu. Rev. Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 16.Cantarel B.L., Coutinho P.M., Rancurel C., Bernard T., Lombard V., Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morgan J.L.W., Strumillo J., Zimmer J. Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature. 2013;493:U181–U192. doi: 10.1038/nature11744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arioli T., Peng L., Betzner A.S., Burn J., Wittke W., Herth W., Camilleri C., Hofte H., Plazinski J., Birch R., et al. Molecular analysis of cellulose biosynthesis in Arabidopsis. Science. 1998;279:717–720. doi: 10.1126/science.279.5351.717. [DOI] [PubMed] [Google Scholar]
- 19.Cano-Delgado A., Penfield S., Smith C., Catley M., Bevan M. Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J. 2003;34:351–362. doi: 10.1046/j.1365-313x.2003.01729.x. [DOI] [PubMed] [Google Scholar]
- 20.Fagard M., Desnos T., Desprez T., Goubet F., Refregier G., Mouille G., McCann M., Rayon C., Vernhettes S., Höfte H. PROCUSTE1 Encodes a Cellulose Synthase Required for Normal Cell Elongation Specifically in Roots and Dark-Grown Hypocotyls of Arabidopsis. Plant Cell. 2000;12:2409–2423. doi: 10.1105/tpc.12.12.2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beeckman T., Przemeck G.K., Stamatiou G., Lau R., Terryn N., De Rycke R., Inze D., Berleth T. Genetic complexity of cellulose synthase a gene function in Arabidopsis embryogenesis. Plant Physiol. 2002;130:1883–1893. doi: 10.1104/pp.102.010603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Persson S., Paredez A., Carroll A., Palsdottir H., Doblin M., Poindexter P., Khitrov N., Auer M., Somerville C.R. Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2007;104:15566–15571. doi: 10.1073/pnas.0706592104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown D.M., Zeef L.A.H., Ellis J., Goodacre R., Turner S.R. Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell. 2005;17:2281–2295. doi: 10.1105/tpc.105.031542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Timmers J., Vernhettes S., Desprez T., Vincken J.P., Visser R.G.F., Trindade L.M. Interactions between membrane-bound cellulose synthases involved in the synthesis of the secondary cell wall. Febs Lett. 2009;583:978–982. doi: 10.1016/j.febslet.2009.02.035. [DOI] [PubMed] [Google Scholar]
- 25.Wang X., Cnops G., Vanderhaeghen R., De Block S., Van Montagu M., Van Lijsebettens M. AtCSLD3, a cellulose synthase-like gene important for root hair growth in arabidopsis. Plant Physiol. 2001;126:575–586. doi: 10.1104/pp.126.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hazen S.P., Scott-Craig J.S., Walton J.D. Cellulose synthase-like genes of rice. Plant Physiol. 2002;128:336–340. doi: 10.1104/pp.010875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burton R.A., Wilson S.M., Hrmova M., Harvey A.J., Shirley N.J., Stone B.A., Newbigin E.J., Bacic A., Fincher G.B. Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-beta-D-glucans. Science. 2006;311:1940–1942. doi: 10.1126/science.1122975. [DOI] [PubMed] [Google Scholar]
- 28.Liu X.L., Liu L.F., Niu Q.K., Xia C.A., Yang K.Z., Li R., Chen L.Q., Zhang X.Q., Zhou Y.H., Ye D. MALE GAMETOPHYTE DEFECTIVE 4 encodes a rhamnogalacturonan II xylosyltransferase and is important for growth of pollen tubes and roots in Arabidopsis. Plant J. 2011;65:647–660. doi: 10.1111/j.1365-313X.2010.04452.x. [DOI] [PubMed] [Google Scholar]
- 29.Saxena I.M., Lin F.C., Brown R.M. Cloning and sequencing of the cellulose synthase catalytic subunit gene of Acetobacter xylinum. Plant Mol. Biol. 1990;15:673–683. doi: 10.1007/bf00016118. [DOI] [PubMed] [Google Scholar]
- 30.Saxena I.M., Brown R.M., Jr., Fevre M., Geremia R.A., Henrissat B. Multidomain architecture of beta-glycosyl transferases: implications for mechanism of action. J. Bacteriol. 1995;177:1419–1424. doi: 10.1128/jb.177.6.1419-1424.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong H.C., Fear A.L., Calhoon R.D., Eichinger G.H., Mayer R., Amikam D., Benziman M., Gelfand D.H., Meade J.H., Emerick A.W. Genetic organization of the cellulose synthase operon in Acetobacter xylinum. Proc. Natl. Acad. Sci. USA. 1990;87:8130–8134. doi: 10.1073/pnas.87.20.8130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pear J.R., Kawagoe Y., Schreckengost W.E., Delmer D.P., Stalker D.M. Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc. Natl. Acad. Sci. USA. 1996;93:12637–12642. doi: 10.1073/pnas.93.22.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Appenzeller L., Doblin M., Barreiro R., Wang H.Y., Niu X.M., Kollipara K., Carrigan L., Tomes D., Chapman M., Dhugga K.S. Cellulose synthesis in maize: isolation and expression analysis of the cellulose synthase (CesA) gene family. Cellulose. 2004;11:287–299. doi: 10.1023/B:CELL.0000046417.84715.27. [DOI] [Google Scholar]
- 34.Burton R.A., Shirley N.J., King B.J., Harvey A.J., Fincher G.B. The CesA gene family of barley. Quantitative analysis of transcripts reveals two groups of co-expressed genes. Plant Physiol. 2004;134:224–236. doi: 10.1104/pp.103.032904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joshi C.P., Bhandari S., Ranjan P., Kalluri U.C., Liang X., Fujino T., Samuga A. Genomics of cellulose biosynthesis in poplars. New Phytol. 2004;164:53–61. doi: 10.1111/j.1469-8137.2004.01155.x. [DOI] [PubMed] [Google Scholar]
- 36.Djerbi S., Lindskog M., Arvestad L., Sterky F., Teeri T.T. The genome sequence of black cottonwood (Populus trichocarpa) reveals 18 conserved cellulose synthase (CesA) genes. Planta. 2005;221:739–746. doi: 10.1007/s00425-005-1498-4. [DOI] [PubMed] [Google Scholar]
- 37.Nairn C.J., Haselkorn T. Three loblolly pine CesA genes expressed in developing xylem are orthologous to secondary cell wall CesA genes of angiosperms. New Phytol. 2005;166:907–915. doi: 10.1111/j.1469-8137.2005.01372.x. [DOI] [PubMed] [Google Scholar]
- 38.Wang L.Q., Guo K., Li Y., Tu Y.Y., Hu H.Z., Wang B.R., Cui X.C., Peng L.C. Expression profiling and integrative analysis of the CESA/CSL superfamily in rice. Bmc Plant Biol. 2010;10:282. doi: 10.1186/1471-2229-10-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sundari B.K.R., Dasgupta M.G. Isolation of developing secondary xylem specific cellulose synthase genes and their expression profiles during hormone signalling in Eucalyptus tereticornis. J. Genet. 2014;93:U403–U437. doi: 10.1007/s12041-014-0391-y. [DOI] [PubMed] [Google Scholar]
- 40.Guerriero G., Legay S., Hausman J.F. Alfalfa Cellulose Synthase Gene Expression under Abiotic Stress: A Hitchhiker’s Guide to RT-qPCR Normalization. PLoS ONE. 2014;9:e103808. doi: 10.1371/journal.pone.0103808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki S., Li L., Sun Y.H., Chiang V.L. The cellulose synthase gene superfamily and biochemical functions of xylem-specific cellulose synthase-like genes in Populus trichocarpa. Plant Physiol. 2006;142:1233–1245. doi: 10.1104/pp.106.086678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li M., Xiong G.Y., Li R., Cui J.J., Tang D., Zhang B.C., Pauly M., Cheng Z.K., Zhou Y.H. Rice cellulose synthase-like D4 is essential for normal cell-wall biosynthesis and plant growth. Plant J. 2009;60:1055–1069. doi: 10.1111/j.1365-313X.2009.04022.x. [DOI] [PubMed] [Google Scholar]
- 43.Hepton A., Hodgson A.S., Bartholomew D.P., Paull R.E., Rohrbach K.G. The Pineapple: Botany, Production and Uses. CABI Publishing; Wallingford, UK: 2003. pp. 4895–4902. [Google Scholar]
- 44.Ming R., VanBuren R., Wai C.M., Tang H., Schatz M.C., Bowers J.E., Lyons E., Wang M.L., Chen J., Biggers E., et al. The pineapple genome and the evolution of CAM photosynthesis. Nat. Genet. 2015;47:1435–1442. doi: 10.1038/ng.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fang J.P., Miao C.Y., Chen R.K., Ming R. Genome-Wide Comparative Analysis of Microsatellites in Pineapple. Trop. Plant Biol. 2016;9:117–135. [Google Scholar]
- 46.Paull R., Chen N., Ming R., Wai C., Shirley N., Schwerdt J., Bulone V. Carbon Flux and Carbohydrate Gene Families in Pineapple. Trop. Plant Biol. 2016;9:200–213. [Google Scholar]
- 47.Wai C.M., Powell B., Ming R., Min X.J. Analysis of Alternative Splicing Landscape in Pineapple (Ananas comosus) Trop. Plant Biol. 2016;9:150–160. [Google Scholar]
- 48.Zhang X.D., Liang P.P., Ming R. Genome-Wide Identification and Characterization of Nucleotide-Binding Site (NBS) Resistance Genes in Pineapple. Trop. Plant Biol. 2016;9:187–199. [Google Scholar]
- 49.Zheng Y., Li T., Xu Z.N., Wai C.M., Chen K., Zhang X.T., Wang S.P., Ji B., Ming R., Sunkar R. Identification of microRNAs, phasiRNAs and Their Targets in Pineapple. Trop. Plant Biol. 2016;9:176–186. [Google Scholar]
- 50.Song S.W., Hao L.Y., Zhao P., Xu Y., Zhong N.Q., Zhang H.J., Liu N. Genome-wide Identification, Expression Profiling and Evolutionary Analysis of Auxin Response Factor Gene Family in Potato (Solanum tuberosum Group Phureja) Sci. Rep.-UK. 2019;9 doi: 10.1038/s41598-018-37923-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xing H.Y., Pudake R.N., Guo G.G., Xing G.F., Hu Z.R., Zhang Y.R., Sun Q.X., Ni Z.F. Genome-wide identification and expression profiling of auxin response factor (ARF) gene family in maize. Bmc Genomics. 2011;12:178. doi: 10.1186/1471-2164-12-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Han Y., Li X., Cheng L., Liu Y., Wang H., Ke D., Yuan H., Zhang L., Wang L. Genome-Wide Analysis of Soybean JmjC Domain-Containing Proteins Suggests Evolutionary Conservation Following Whole-Genome Duplication. Front. Plant Sci. 2016;7 doi: 10.3389/fpls.2016.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bailey T.L., Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 54.Azam S.M., Liu Y., Rahman Z.U., Ali H., Yan C., Wang L., Priyadarshani S.V.G.N., Hu B., Huang X., Xiong J. Identification, Characterization and Expression Profiles of Dof Transcription Factors in Pineapple (Ananas comosus L) Trop. Plant Biol. 2018;11:49–64. doi: 10.1007/s12042-018-9200-8. [DOI] [Google Scholar]
- 55.Farrokhi N., Burton R.A., Brownfield L., Hrmova M., Wilson S.M., Bacic A., Fincher G.B. Plant cell wall biosynthesis: genetic, biochemical and functional genomics approaches to the identification of key genes. Plant Biotechnol. J. 2006;4:145–167. doi: 10.1111/j.1467-7652.2005.00169.x. [DOI] [PubMed] [Google Scholar]
- 56.Burton R.A., Farrokhi N., Bacic A., Fincher G.B. Plant cell wall polysaccharide biosynthesis: real progress in the identification of participating genes. Planta. 2005;221:309–312. doi: 10.1007/s00425-005-1495-7. [DOI] [PubMed] [Google Scholar]
- 57.Dwivany F.M., Yulia D., Burton R.A., Shirley N.J., Wilson S.M., Fincher G.B., Bacic A., Newbigin E., Doblin M.S. The CELLULOSE-SYNTHASE LIKE C (CSLC) Family of Barley Includes Members that Are Integral Membrane Proteins Targeted to the Plasma Membrane. Mol. Plant. 2009;2:1025–1039. doi: 10.1093/mp/ssp064. [DOI] [PubMed] [Google Scholar]
- 58.Doblin M.S., Pettolino F.A., Wilson S.M., Campbell R., Burton R.A., Fincher G.B., Newbigin E., Bacic A. A barley cellulose synthase-like CSLH gene mediates (1,3;1,4)-beta-D-glucan synthesis in transgenic Arabidopsis. Proc. Natl. Acad. Sci. USA. 2009;106:5996–6001. doi: 10.1073/pnas.0902019106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bernal A.J., Yoo C.M., Mutwil M., Jensen J.K., Hou G., Blaukopf C., Sorensen I., Blancaflor E.B., Scheller H.V., Willats W.G. Functional analysis of the cellulose synthase-like genes CSLD1, CSLD2, and CSLD4 in tip-growing Arabidopsis cells. Plant Physiol. 2008;148:1238–1253. doi: 10.1104/pp.108.121939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nobles D.R., Romanovicz D.K., Brown R.M., Jr. Cellulose in cyanobacteria. Origin of vascular plant cellulose synthase? Plant Physiol. 2001;127:529–542. doi: 10.1104/pp.010557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Doblin M.S., Kurek I., Jacob-Wilk D., Delmer D.P. Cellulose biosynthesis in plants: from genes to rosettes. Plant Cell Physiol. 2002;43:1407–1420. doi: 10.1093/pcp/pcf164. [DOI] [PubMed] [Google Scholar]
- 62.Liepman A.H., Wilkerson C.G., Keegstra K. Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc. Natl. Acad. Sci. USA. 2005;102:2221–2226. doi: 10.1073/pnas.0409179102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dhugga K.S., Barreiro R., Whitten B., Stecca K., Hazebroek J., Randhawa G.S., Dolan M., Kinney A.J., Tomes D., Nichols S., et al. Guar seed beta-mannan synthase is a member of the cellulose synthase super gene family. Science. 2004;303:363–366. doi: 10.1126/science.1090908. [DOI] [PubMed] [Google Scholar]
- 64.Liepman A.H., Cavalier D.M. The CELLULOSE SYNTHASE-LIKE A and CELLULOSE SYNTHASE-LIKE C families: recent advances and future perspectives. Front. Plant Sci. 2012;3:109. doi: 10.3389/fpls.2012.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cocuron J.C., Lerouxel O., Drakakaki G., Alonso A.P., Liepman A.H., Keegstra K., Raikhel N., Wilkerson C.G. A gene from the cellulose synthase-like C family encodes a beta-1,4 glucan synthase. Proc. Natl. Acad. Sci. USA. 2007;104:8550–8555. doi: 10.1073/pnas.0703133104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Letunic I., Bork P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018;46:D493–D496. doi: 10.1093/nar/gkx922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu B., Jin J., Guo A.-Y., Zhang H., Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Y., Tang H., Debarry J.D., Tan X., Li J., Wang X., Lee T.H., Jin H., Marler B., Guo H., et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen P., Li Y., Zhao L., Hou Z., Yan M., Hu B., Liu Y., Azam S.M., Zhang Z., Rahman Z.U. Genome-Wide Identification and Expression Profiling of ATP-Binding Cassette (ABC) Transporter Gene Family in Pineapple (Ananas comosus(L.) Merr.) Reveal the Role ofAcABCG38 in Pollen Development. Front. Plant Sci. 2017;8:2150. doi: 10.3389/fpls.2017.02150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.