Short abstract
The size and modular structure of versican and its gene suggest the existence of multiple splice variants. We have identified, cloned, and sequenced a previously unknown exon located within the noncoding gene sequence downstream of exon 8. This exon, which we have named exon 8β, specifies two stop-codons. mRNAs of the versican gene with exon 8β are predicted to be constitutively degraded by nonsense-mediated RNA decay. Here, we tested the hypothesis that these transcripts become expressed in a model of neuropathic pain.
Keywords: Versican, nerve injury, neuropathic pain, nonsense mediated RNA decay, eIF2α(PO4)2-, Isolectin B4
Introduction
The extracellular matrix molecule versican is a member of the lectican gene family; chondroitin sulfate carrying proteoglycans with an immunoglobulin-like hyaluronan-binding domain at their N-terminus and a C-type lectin-like binding motif at their C-terminus.1,2 Five splice variants of versican (V0, V1, V2, V3, and V4) have been identified so far and the existence of additional variants has been proposed.3 All five splice variants have identical N- and C-termini but vary with respect to the size of their protein backbone and the number of attached chondroitin sulfate chains.4–8
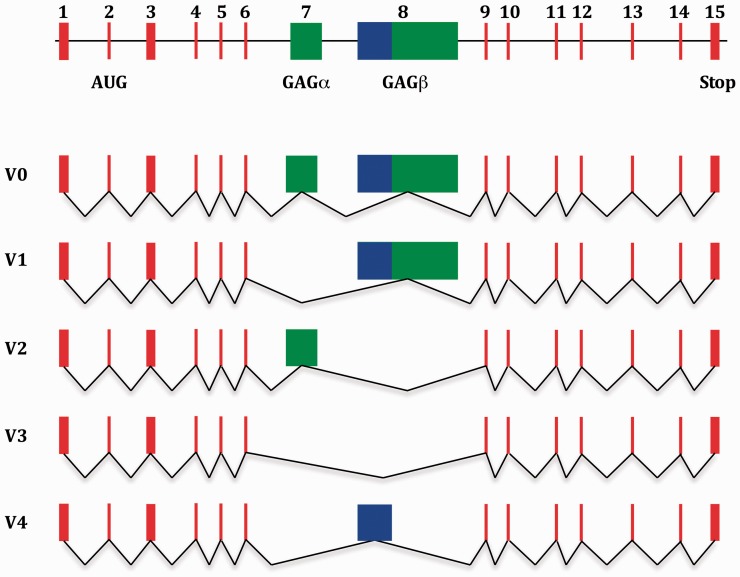
The versican gene (Vcan; Synonym: CSPG2), which is located on chromosome 2 in the rat genome,9 and on chromosome 5 in humans,10 consists of 15 exons that encompass almost 100 kilo bases (kb) of continuous DNA. V0, versican’s largest splice variant, is generated by the transcription and translation of its whole set of protein encoding exons. The other four variants are products of alternative splicing of exon 7 and (or within) exon 8, which encode versican’s glycosaminoglycan (GAG) attachment domains, GAG α and GAG β.8,11 Therefore, V0 carries both GAG attachment domains, GAG α and GAG β, V1 only GAG β, V2 only GAG α, and V3 no GAG domain, whereas V4 carries only a shortened GAG β-domain.2,8 (Figure 1).
Figure 1.
Schematic representation of the genomic structure of the versican gene, including exon structure of the five known splice variants. Its 15 exons are represented as colored rectangles. Exons 1 to 6 and 9 to 15 are represented by red rectangles. Exons 7 and 8 are represented by green rectangles. The part of exon 8 that is expressed in versican V4 is represented by a blue rectangle. The black line between the exons in the top illustration represents non-coding intervening sequences. The black lines that connect the exons in the representation of the different splice variants illustrate the splicing process through which the different versican variants are generated. Note that the start-codon for versican biosynthesis is located within exon 2. GAG: glycosaminoglycan.
We have previously demonstrated that the versican variants V0 to V3 are transcribed by sensory neurons, in dorsal root ganglia (DRG).12 We also demonstrated that the “enigmatic” IB4-reactivity of nonpeptidergic C-fiber nociceptors can be traced back to the covalent modification of Vcan V2 with α-D-galactopyranose residues.12,13 Here, we took a closer look at versican’s gene structure. Given the spatial dimensions of the Vcan gene and the modular structure of its corresponding gene products we hypothesized the existence of additional splice variants.
By searching expressed sequence tag (EST) databases for transcribed sequences that could be mapped to the genomic region of the human versican gene, we identified one clone (BI818462), which contains a cDNA sequence that could not be completely assigned to any of the known versican exons. Its nucleotide sequence is well conserved in the rat. Using oligonucleotide primers specific for the amplification of transcripts of the Vcan gene carrying this splice insertion we were able to clone 2 overlapping cDNA-fragments from RNA extracts of rat nervous tissue. The derived sequence of both clones provides evidence for the existence of a previously unknown exon. The new exon contains two stop codons. Spliced transcripts of the versican gene that include this exon should, therefore, be degraded by nonsense-mediated mRNA decay under normal circumstances.14 We explored the hypothesis that mRNAs containing this exon are being translated into a C-terminal truncated variant of versican under conditions of extreme cellular stress in the peripheral nervous system.
Material and methods
Animals
Experiments were performed on adult male Sprague Dawley rats (Crl:CD; 280–350 g, approximately 8–12 weeks old) because the pathophysiological changes associated with spared nerve injury demonstrate greater variability in female rats.15 Rats were obtained from Charles River Laboratories (Hollister, CA, USA) and housed in an AAALAC International accredited animal facility, the Laboratory Animal Resource Center of the University of California San Francisco, under a 12-hour light/dark cycle, with water and food available ad libitum. Animal care and use conformed to NIH guidelines. The University of California, San Francisco Institutional Animal Care and Use Committee approved all experimental protocols. Concerted effort was made to reduce the suffering and number of animals used.
Cloning of exon 8β
Total RNA from rat brain and spinal cord was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA) with the PureLink™ RNA mini kit (Life Technologies, Grand Island, NY, USA), according to the manufacturer’s instructions. The amount of RNA was quantified with a UV-spectrophotometer, and cDNA preparation carried out with 1 µg of total RNA/sample and the superscript III platinum one-step RT-PCR system (Life Technologies). The PCR primers (Invitrogen) used for the detection and amplification of the different versican fragments were, according to the National Center for Biotechnology Information database entry NM_001170558, as follows: Vcan_exon 4_for = 5′-GCG ACC AGC AGA TAC ACT CT-3′, Vcan_exon6_rev = 5′-ATC CGA CAG CCA GCC GTA AT-3′ [size of the amplification product = 435 bp, number of PCR cycles: 30]; Vcan_exon8_for= 5′-GGG TTG AGA CAA GTA TGG TAC CT-3′, Vcan_ exon8β_rev= 5′-GCA CTG ACG TTT CTT CTG CTA CA-3′ [size of the amplification product = 372 bp, number of PCR cycles: 40]; Vcan_exon8β_for = 5′-TGT AGC AGA AGA AAC GTC AGT GC-3′, and, Vcan_ exon11_rev = 5′-CAT GTA CGG CGA TGA GCA AAG TA-3′ [size of the amplification product = 360 bp, number of PCR cycles: 40].
PCR products inserted into the pCR4™-TOPO vector (Invitrogen) were used to transform chemically competent Escherichia coli cells (TOP10; Invitrogen). Only clones that were verified by restriction analysis were selected for DNA sequencing (GenScript, New Jersey, NJ, USA).
Spared nerve injury
The spared nerve injury model, a well-established surgical model of neuropathic pain, was performed as previously described.16 Briefly, rats were anesthetized with 2.5% isoflurane (Henry Schein Animal Health, Dublin, OH, USA) in oxygen and s.c. injected with 2 mg/kg meloxicam (Metacam, Boehringer Ingelheim Vetmedica, St. Joseph, MO, USA). Rat corneas were protected with ophthalmic ointment (Artificial Tears, Rugby, Livonia, MI, USA) during the surgical procedure. The surgical site was infiltrated with 0.1 ml of 0.25% bupivacaine (Marcaine, Hospira, Lake Forest, IL, USA) immediately before incision. The skin on the lateral surface of the right thigh was incised and a dissection made through the biceps femoris muscle exposing the sciatic nerve and its three terminal branches. The tibial and common peroneal nerve branches were tightly ligated with 5–0 silk (Surgical Suture U.S.P., Henry Schein) and sectioned ∼1 mm distal to the ligation. A ∼2 mm segment of the distal nerve stump was then excised. Care was taken to avoid any contact, stretching, or damage of the intact sural nerve. Sham surgery consisted in the exposure of the sciatic nerve and its branches without any lesion. Muscle was sutured with 5–0 silk and skin closed with 5–0 nylon (Surgical Suture U.S.P., Henry Schein). Rats recovered from anesthesia in an individual cage with a heat source and were transferred to standard housing cages when fully awake. Once ambulatory, rats received a daily s.c. injection of meloxicam for the first two days after surgery. Rats were allowed to recover for one week before tissue harvesting.
Tissue harvesting and protein extraction
On post-surgery day 7, rats subjected to SNI or sham-surgery were euthanized by exsanguination while under isoflurane anesthesia. Ipsilateral L4 to L6 DRG were surgically removed, snap frozen on dry ice, and stored at −80°C until further processing. DRG were transferred into homogenization buffer [150 mM NaCl, 50 mM Tris-HCl pH 7.4, 2 mM EDTA, 2% sodium dodecylsulfate (SDS)] supplemented with a complete protease inhibitor cocktail (Roche Diagnostics Corp., Indianapolis, IN, USA) and manually homogenized with a plastic pestle. Proteins were solubilized by incubating the homogenate for 2 h at 25°C in an Eppendorf Thermomixer at 1400 r/min and then extracted by a 15 min centrifugation at 14,000 r/min, in an Eppendorf tabletop centrifuge. Protein concentration of the samples was determined using the micro bicinchoninic acid assay (BCA) protein assay kit (Pierce, Rockford, IL, USA) with bovine serum albumin (BSA) as a standard.
SDS-PAGE and Western blot analysis of eIF2α(PO4)2-immunoreactivity
40 µg of protein per sample was mixed with 4× sample buffer [62.5 mM Tris-HCl pH 6.8, 3% SDS, 10% glycerol, 0.025% bromphenol blue], denatured by shaking for 10 min at 500 r/min at 90°C in an Eppendorf Thermomixer and electrophoresed on a 4%–15% precast polyacrylamide gel (Biorad, Hercules, CA, USA) in 25 mM Tris buffer containing 192 mM glycine, and 0.1% SDS. Proteins were transferred onto a nitrocellulose (NC) membrane using the semidry method (transfer time 2 h at 1.5 mA*cm−2 with 47.9 mM Tris, 38.9 mM glycine, 0.038% SDS and 20% (v/v) methanol). The blotting membrane was saturated by shaking in Tris-buffered saline pH 7.4 containing 5% BSA and 0.1% Tween 20 (antibody dilution buffer) for 1 h at room temperature (RT). The blot was probed with a rabbit anti-eIF2α antibody (Cell signaling technology, catalog #9722S; 1:500 in antibody dilution buffer) at 4°C overnight, rinsed with TBST (three times at RT, 15 min each) and probed with a donkey anti-rabbit horseradish peroxidase conjugated antibody (GE healthcare, catalog#NA934V; 1:2500 in antibody dilution buffer) for 2 h at RT. The Western blot was rinsed with TBST (three times at RT, 15 min each) and the eIF2α immunoreactivity visualized with the enhanced chemiluminescence (ECL) detection kit (Pierce).
The Western blot was stripped by a 1 h incubation at RT with 2% SDS, 10 mM β-mercaptoethanol in 62.5 mM Tris-HCl pH 6.8, and washed extensively with TBST. The blotting membrane was saturated with TBST containing 5% BSA and probed with the rabbit anti-eIF2αPO42− antibody (Cell signaling technology, catalog #9722S; 1:500 in antibody dilution buffer) overnight at 4°C. The blot was rinsed with TBST (three times at RT, 15 min each), probed with a biotinylated anti-rabbit antibody (Jackson Immunoresearch, catalog# 111–065-003; 1:2500 in antibody dilution buffer) for 2 h at RT, rinsed with TBST (three times at RT, 15 min each), probed with streptavidin horseradish peroxidase (Sigma-Aldrich, catalog# S2438; 1:5000 in antibody dilution buffer) for 1 h at RT and rinsed with TBST (three times at RT, 15 min each). eIF2αPO42− immunoreactivity was visualized with the ECL detection kit (Pierce). Results were analyzed by computer-assisted densitometry and levels of eIF2αPO42− immunoreactivity were normalized with respect to the eIF2α immunoreactivity in each sample.
Deglycosylation and Western blot analysis of Vcan immunoreactivity
1.5 mg of protein in homogenization buffer from DRG of SNI rats was transferred into 10 K cutoff centrifugal filter devices (Millipore, Burlington, MA, USA) and the buffer replaced through stepwise and repeated centrifugation at 10,000 r/min and the addition of bridging buffer [phosphate buffered saline pH 7.4 containing 1% Triton X-100 and 1% sodium deoxycholate and supplemented with 2x complete protease inhibitor cocktail (Roche)]. The proteins in bridging buffer were further replaced through stepwise and repeated centrifugation at 10000 r/min and the addition of deglycosylation buffer [0.1M NaxHxPO4-buffer pH 5.5 containing 0.05 M NaCl, 1% Triton X-100 and 1% Tween20 and supplemented with 2× complete protease inhibitor cocktail (Roche)]. The proteins in deglycosylation buffer were transferred into an Eppendorf tube, mixed with 50 U hyaluronidase (Sigma Aldrich, Saint Louis, MO, USA), and incubated for 2 h at 37°C in an Eppendorf Thermomixer. The proteins were transferred into a 10 k cutoff centrifugation filter device and the deglycosylation buffer with homogenization buffer exchanged as described earlier. The protein concentration of the sample was determined using the BCA protein assay kit (Pierce) with BSA as the standard. Sixty micrograms of protein per lane were electrophoresed onto a 4%–15% precast polyacrylamide gel and electro-blotted onto NC membrane afterwards. The membrane was stained with Ponceau S (0.1% Ponceau S in 5%(v/v) acetic acid) and, using a scalpel, the lanes were separated from each other by cutting the membrane into strips. The strips were transferred into a stripe box and blocked with blocking buffer for 1 h at RT. The strips were then probed with the different Vcan antibodies [mouse monoclonal 12C5 (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA); rabbit polyclonal anti-GAGα (Millipore, catalog#AB1032), rabbit polyclonal anti-GAGβ (Millipore, catalog#ABT1370), mouse monoclonal anti-Cterm. (Millipore, catalog#MABT161); all 1:500 in antibody dilution buffer] at 4°C overnight, rinsed with TBST (three times, 15 min each) and probed with the respective horseradish peroxidase conjugated secondary antibodies [anti-rabbit horseradish peroxidase (GE healthcare, catalog#NA934V), anti-mouse horseradish peroxidase (Pierce, catalog#1858413); 1:2500 in antibody dilution buffer] for 2 h at RT. The strips were rinsed in TBST (three times, 15 min each) and the Vcan immunoreactivities visualized with the ECL detection kit (Pierce).
Results
The EST, 603032745F1 is part of a V0/V1-based splice variant in rat nervous tissue
The human EST is part of a clone (GenBank entry: BI818462) that comprises 836 nucleotides. The nucleotide sequence of this clone can be matched with sequences of the 3′-end of exon 8, the unknown sequence insertion and nucleotide sequences of exons 9, 10, and 11. The sequence of this EST/clone suggests that additional splice variants of the versican gene exist.
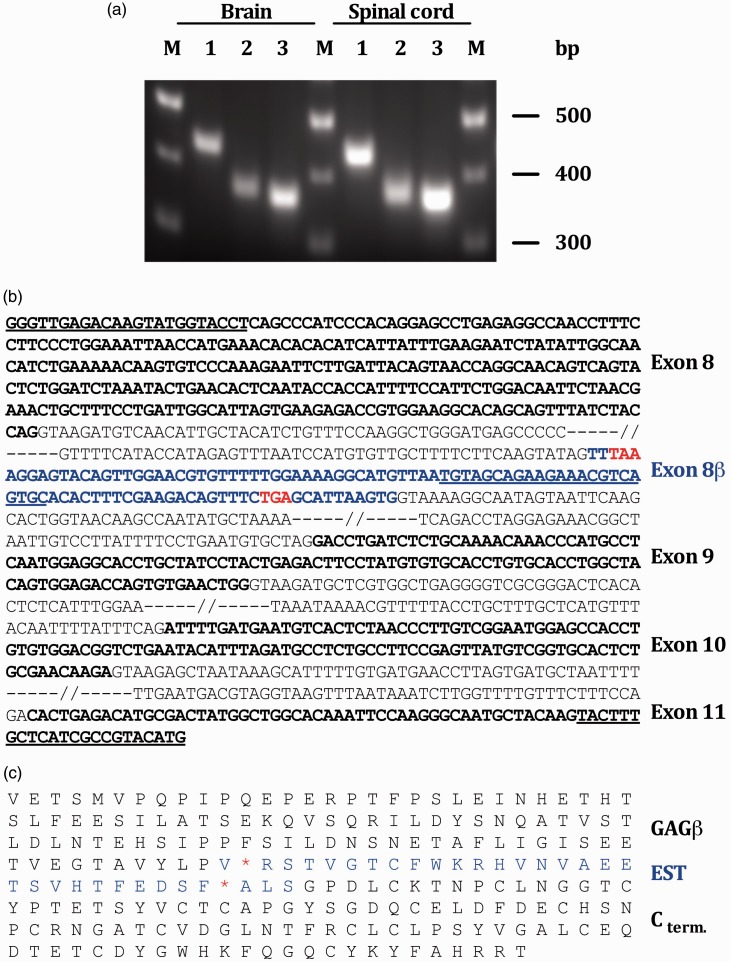
To determine whether this insertion represents an additional exon, several reverse transcriptase polymerase chain reactions (RT-PCRs) were performed. Using primers located within exon 8 and the unknown sequence insertion, we amplified a 372 bp cDNA-fragment of RNA extracts from rat brain and spinal cord (Figure 2(a)). The resulting PCR product from rat brain was inserted into the pCR4™-TOPO vector and amplified by propagation of E. coli cells into which the vector had been transformed. Several clones were checked by restriction analysis for the presence of the insert (data not shown) before one “positive” clone was selected for DNA sequencing.
Figure 2.
The EST 603032745F1/exon 8β is part of a V0- and/or V1-based mRNA. (a) 2 RT-PCRs were performed to define the spatial dimensions of exon 8β; a RT-PCR with primers located within exon 8 and exon 8β amplified a 372bp DNA-fragment (lane 2), and a RT-PCR with primers located within exon 8β and exon 11 amplified a 360bp DNA-fragment (lane 3). A control PCR with primers located within exons 4 and 6 [size = 435bp]—which should detect all currently known versican splice variants12—was performed to verify the quality of the RNA/cDNA preparation (lane 1). (b) Genomic context of exon 8β. Coding sequences (exons) are shown in bold letters. Non-coding, intervening sequences (introns) are shown in plain letters. Exon 8β is highlighted in blue letters. The primer sequences that were used for the amplification of cDNA-fragments including exon 8 are underlined; the top two primers were used to define the 5′-end of exon 8β, the bottom two primers were used to define its 3′-end. Of note, the two primers located within exon 8β are complementary to each other and of reverse orientation. (c) C-terminal amino acid sequence of the corresponding versican splice variant V5 (and a hypothetical variant V6). Two stop-codons (red asterisks) within the reading frame of exon 8β would terminate protein translation of the corresponding mRNA transcript downstream of exon 8, versicans GAGβ domain.
Another RT-PCR was performed to define the 3′ end of the putative new exon. Using primers located within the putative splice insertion and exon 11, we amplified a 360 bp cDNA-fragment from RNA of rat brain and spinal cord (Figure 2(a)). The resulting PCR product from rat brain was inserted into the pCR4™-TOPO vector and amplified by propagation in E. coli cells. Several clones were checked by restriction analysis, for the presence of the insert (data not shown), before one “positive” clone was selected for DNA sequencing.
From the combination of the two cDNA sequences, one can conclude that the splice insertion represents an additional exon within the Vcan gene. This exon—that we have named exon 8β, because it is located downstream of exon 8, which encodes versican’s GAG β domain—is made of 102 nucleotides or 33 codons (Figure 2(b)), of which 31 specify amino acids and 2 stop signals (Figure 2(c)).
A C-terminal truncated versican variant can be detected in DRG extracts from spared nerve injury rats
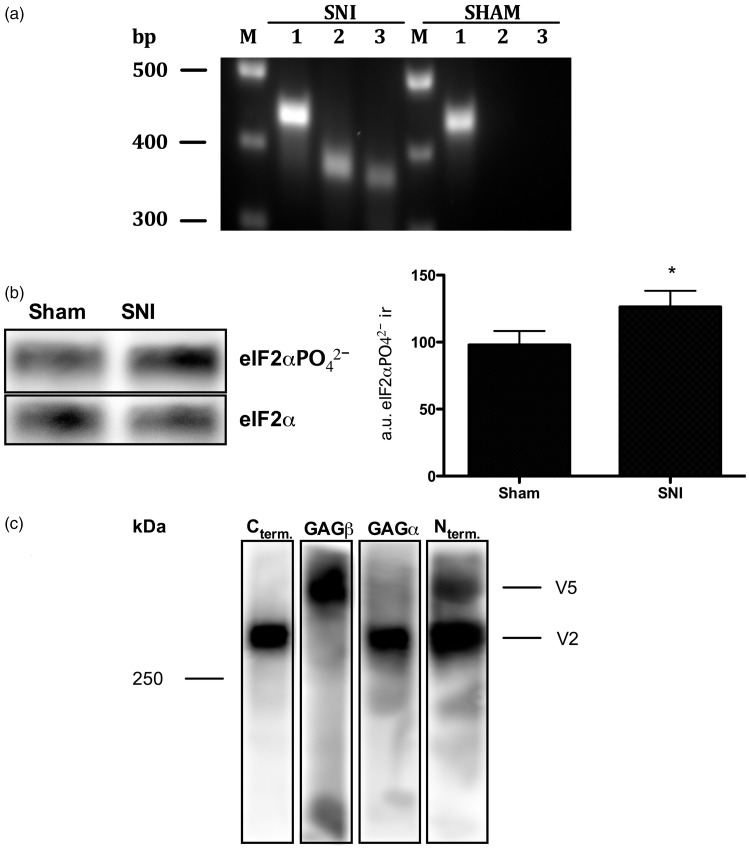
We have previously demonstrated that versican can bind glial-cell line derived neurotrophic factor (GDNF) and MCP-1 at sensory terminals,17,18 although the precise details of its molecular interaction with both molecules are not yet clear. GDNF and MCP-1 are of particular importance for sensory neurons in the event of nerve injury as GDNF promotes the survival of IB4-binding, nonpeptidergic C-fiber nociceptors,19–21 while MCP-1 attracts CCR2 expressing monocytes/macrophages to the site of injury.22,23 To analyze if the novel versican variant could also be transcribed in the peripheral nervous system, we performed a PCR on RNA extracts of DRG from rats subjected to the spared nerve injury (SNI) model for neuropathic pain.16 As shown in Figure 3(a), cDNA-fragments of the novel splice variant could be amplified within RNA extracts of SNI but not sham-operated rats.
Figure 3.
A novel Vcan variant is expressed in SNI rats. (a) A novel Vcan variant is transcribed in SNI but not sham control rats. PCRs on cDNA from rat DRG demonstrates that the new Vcan variant is also transcribed in spared nerve injury rats but that it is absent in sham-operated rats. 1: Vcan_exon4_for and Vcan_exon6_rev = 435bp; 2: Vcan_exon8_for and Vcan_exon8β_rev = 372bp; 3: Vcan_exon8β_for and Vcan_exon11_rev = 360bp. (b) ISR is activated in SNI rats. Analyzing the post-translational modification of eIF2α by Western blotting demonstrates a significant increase of eIF2αPO42− in DRG extracts from rats submitted to SNI compared to sham-surgery rats [eIF2αPO42− immunoractivity in SNI = 126 ± 12 arbitrary units; eIF2αPO42− immunoractivity in sham controls = 98 ± 10 arbitrary units; normalized to the reference protein eIF2α; P<0.05 (unpaired student’s t-test), a 28.4 ± 15.8% increase in the eIF2αPO42− immunoreactivity, N=9]. Note that the calculated molecular weight of eIF2α ± PO42− is 36 kDa (according to the UniProtKB database entry P68101). *P<0.05. (c) SNI rats express a C-terminally truncated Vcan variant. DRG extracts from SNI rats were analyzed by Western blotting using a set of four different antibodies, each of which is directed against one of versican’s four major structural and functional domains. Rat V2 is made of 1601 amino acids with a calculated molecular weight of ∼176 kDa.62 The discrepancy between its calculated and apparent molecular weight on the Western blot can be explained by its highly acidic character, which interferes with micelle formation during gel electrophoresis.63,64 The Vcan variant above V2 is composed of Vcan’s N-terminus and only the GAGβ domain. It’s domain composition and apparent molecular weight suggests that it is the C-terminal truncated Vcan variant predicted by the translation of V1-derived mRNAs carrying exon 8β.
mRNAs of the Vcan gene with exon 8β should be degraded under normal circumstances.14 However, axotomized neurons are experiencing extreme stress as they struggle for survival.24 Cells respond to stress with the integrated stress response (ISR), a genetic program that targets the production of proteins that protects the cell from harm.25 The ISR is often associated with the inhibition of nonsense-mediated RNA decay (NMD) given that the translation of a number of transcripts for stress-related proteins are controlled by NMD.26–28 Based on these findings, we analyzed if the novel splice variant could also become expressed in DRG from rats submitted to spared nerve injury.
First, we analyzed whether the phosphorylation of eIF2α, a molecular marker for the ISR, could be enhanced in SNI rats. Proteins extracted from DRG of SNI or sham-operated rats were analyzed by Western blotting. As shown in Figure 3(b), a comparison of the eIF2αPO42− immunoreactivity between SNI and sham-operated rats demonstrates a 28.4 ± 15.8% increase within protein extracts from SNI rats. We then analyzed whether the novel splice variant is expressed in SNI rats. Protein extracts from the DRG of SNI rats were deglycosylated, separated by gel electrophoresis, and analyzed by Western blotting, using a set of four different antibodies, each of which is specific to one of versican’s four major functional domains. As shown in Figure 3(c), two different versican variants could be detected. The lower one, with an apparent molecular weight of >250 kDa, is composed of versican’s N-terminus, its GAGα domain, and its C-terminus and most likely represents versican V2.12,13The second variant above V2 is comprised only of the N-terminus and the GAGβ domain and thus most likely represents a C-terminal truncated version of versican V1 (see Figures 1 and 4).
Figure 4.
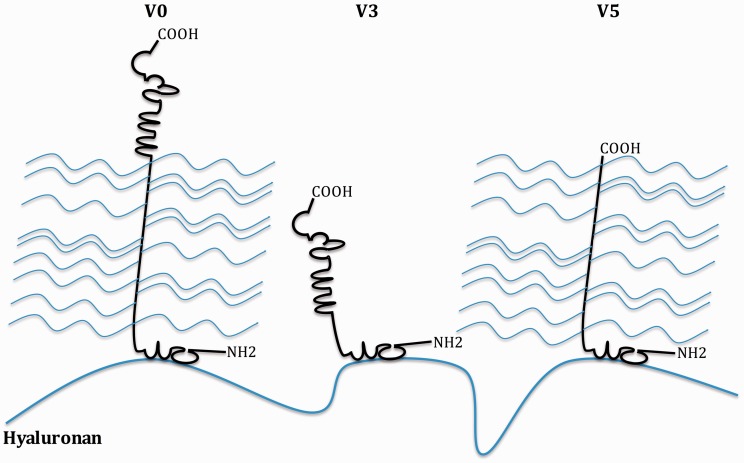
Schematic representation of the potential structure of the novel versican variant V5 and its structural differences from two of the “classic” splice variants, V0 and V3. Thick black lines represent the protein backbone. Thin blue lines represent the chondroitin sulfate side chains, which are attached to versican’s GAG domains, and the thick blue line represents hyaluronan. Versican V0 is comprised of three distinct structural domains, a hyaluronan-binding domain at its N-terminus, two GAG domains in the middle portion of the protein, and a selectin-like domain at its C-terminus. V3 lacks both GAG domains and therefore comprises only the N-terminal hyaluronan-binding domain and the C-terminal selectin-like domain. V5 lacks the C-terminal selectin-like domain and therefore would only be comprised of the N-terminal hyaluronan-binding domain and the GAGβ domain.
Discussion
We hypothesized that more than the five previously reported versican splice variants exist (see Figure 1). This hypothesis is based on the idea that the Vcan gene comprises 15 exons and that most of these exons encode self-contained structural and functional motifs that could—theoretically—be assembled to many more protein variants.1,11,29
Using the genomic sequence of human versican and the algorithm blastn at the National Library of Medicine Website we searched EST databases for cDNA sequences that are derived from the versican gene. We found a clone whose nucleotide sequence carries an uncharacterized splice insertion. Its nucleotide sequence could be matched with an intronic sequence downstream of exon 8 within the versican gene. The sequence of this splice insertion is well conserved in the rat.
Using RNA extracts from rat central nervous system (CNS) and PCR primers specific for the amplification of transcribed sequences of the versican gene that include this splice insertion, we generated two cDNA clones with overlapping oligonucleotide sequences. From the combination of these cDNA sequences, one can conclude that this splice insertion must represent an additional exon with 102 nucleotides (see Figure 2(b)). The exact position of this exon—for which we have chosen the name 8β because it is located downstream of exon 8, which encodes versican’s GAG β domain—within the linear sequence of the Vcan gene, according to the NIH database entry NC_005101, is 64107–64209.
Exon 8β is framed by classical splice sites: the AG di-ribonucleotide toward its 5′-end represents the splice acceptor site of the preceding intron and the GU di-ribonucleotide toward the 3′ end represents the splice donor site of the subsequent intron.30,31 (Figure 2(b)). Two additional consensus splice motifs required for the excision of the intronic sequence that lies between exons 8 and 8β, the branch site (consensus sequence YURAC, with Y= C or U and R= A or G; IUPAC nucleic acid notation) and the polypyrimidine tract (pyrimidine-rich stretch of 15–20 ribonucleotides),31,32 can be found 32 to 28 and 29 to 9 nucleotides upstream of exon 8β, respectively (Figure 2(b)).
Exon 8β specifies two stop-codons (Figure 2(b) and (c)). One could therefore predict that transcripts of the Vcan gene with exon 8β are subjected to NMD, an intrinsic quality control mechanism of eukaryotic cells that stimulates the degradation of spliced mRNAs with a premature termination codon (PTC).14 According to the exon-junction complex activation model, the most common model for NMD, does each additional stop-codon upstream of the regular termination codon in the open reading frame of a spliced mRNA, induce NMD if it lies more than 50–55 nucleotides in front of an exon–exon junction.33–35 Given that this so-called 50–55 nucleotide rule applies to transcripts of the Vcan gene with exon 8β we did not analyze whether the novel splice variant is expressed in the CNS. We did, however, analyze whether the novel versican variant could be transcribed in the peripheral nervous system of neuropathic rats. The idea for this experiment was based on the finding that traumatized sensory neurons undergo a phenotype switch; many of the molecules such as danger associated molecular pattern molecules, cytokines, growth factors, and stress hormones that are released by injured cells and tissue, including glial cells, leukocytes, and the hypothalamic-pituitary-adrenal gland axis, stimulate the transcription of genes that support the survival and recovery of the damaged neurons.21,36–39
Following the detection of mRNAs of the novel variant in the DRGs of SNI rats (Figure 3(a)), we analyzed whether the transcribed sequences are also being translated into protein. The idea for this experiment was based on the finding that (a) sensory neurons react to mechanical injury,40 inflammation,41 and toxic insults42 with the integrated stress response (ISR), a genetic program aiming to limit and reverse damage to cellular components and (b) that the ISR is often associated with the inhibition of NMD because some of the proteins manufactured by the cell in response to stress such as ATF-3 and ATF-4 are regulated by it.26–28 Our results demonstrate that the eIF2α-PO42− immunoreactivity, a molecular marker for the ISR, is increased in DRGs of SNI rats (Figure 3(b)). eIF2α, one of three subunits of eIF2, is a eukaryotic translation factor essential for the initiation of CAP-dependent protein synthesis.43 The phosphorylation of eIF2α by stress-related protein kinases is known to cause a drastic decrease in global protein translation and, at the same time, leading to the production of proteins that help the cell to limit and reverse any damage that is caused by potential stressors such as mechanical injury or reactive oxygen species.43–45
The detection of a C-terminal truncated Vcan variant in DRG of SNI rats (Figure 3(c)) suggests that NMD is inhibited in axotomized sensory neurons. It is likely that the inhibition of NMD in injured neurons is a consequence of ER stress and the activation of the “classical” PERK → eIF2α-PO42− → ATF4 → ATF3 signaling cascade,46–49 a hypothesis also supported by the fact that ATF3 and ATF4 are widely known to be up-regulated in injured sensory neurons.47,50,51
The translation of mRNAs of the versican gene that carry exon 8β could theoretically lead to the production of two different C-terminal truncated variants: a V0-derived variant that contains both GAG domains and a V1-derived variant that contains the GAGβ domain only (see Figures 1 and 4). While our Western blot results only provide evidence for the existence of the V1 based variant (Figure 3(c)), this doesn’t rule out the existence of the V0-based variant. Taking into account the nomenclature of the previously characterized splice variants,2,52 we propose to include the new versican variant as V5 into the literature.
One would think that injured cells and tissues struggling for survival express primarily genes that are essential for survival and recovery. How does the new versican variant V5 fit into this category? By binding and activating MCP-1 versican’s GAG side chains could help to establish the chemotactic gradient that attracts CCR2 expressing leukocytes,18 which, in turn, will clear the injury site of dead cells and cellular debris.22,23 Versican’s GAG chains could also provide ample attachment sites for cell surface receptors such as CD44, β1-integrin, and TLR 2/6 and 4 on glial cells and leukocytes that help to restore tissue homeostasis after injury.52,53 The electron-rich GAG side chains are also known to: (a) protect cells against oxidative stress,54,55, a major contributor to neuropathic pain after nerve injury,56–60 and (b) bind GDNF,17 a survival factor for traumatized IB4(+)−, nonpeptidergic C-fiber nociceptors.19 And finally, versican could initiate neuronal regeneration by stimulating neurite outgrowth/axonal regeneration.61 Interestingly, this property seems to depend solely on versican’s GAG β domain and the interaction of its GAG chains with β1-integrins at the surface of regenerating neuronal processes.61
Taken together, we have confirmed the existence of a new exon within the genomic sequence of the versican gene and demonstrated that a novel versican splice variant, comprised only of the immunoglobulin-like hyaluronate binding N-terminus and a C-terminal GAG β domain, is expressed in DRG under neuropathic conditions. Our results suggest that the novel versican variant V5 is a potential survival factor for injured sensory neurons.
Author contribution
O Bogen designed experiments, analyzed data, and wrote the manuscript; O Bender and PA performed the experiments and analyzed the data, MK performed experiments, ST analyzed data, FH and JL supervised the study. All authors read and approved the manuscript.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was financially supported by the BMBF (Federal Ministry for Education and Research in Germany) and the NIH (National Institute of Health, AR075334).
References
- 1.Schwartz NB, Pirok EW, Mensch JR, Domowicz MS. Domain organization, genomic structure, evolution, and regulation of expression of the aggrecan gene family. Prog Nucleic Acid Res Mol Biol 1999; 62: 177–225. [DOI] [PubMed] [Google Scholar]
- 2.Wight TN. Versican: a versatile extracellular matrix proteoglycan in cell biology. Curr Opin Cell Biol 2002; 14: 617–623. [DOI] [PubMed] [Google Scholar]
- 3.Lemire JM, Braun KR, Maurel P, Kaplan ED, Schwartz SM, Wight TN. Versican/PG-M isoforms in vascular smooth muscle cells. Arterioscler Thromb Vasc Biol 1999; 19: 1630–1639. [DOI] [PubMed] [Google Scholar]
- 4.Zimmermann DR, Ruoslahti E. Multiple domains of the large fibroblast proteoglycan, versican. Embo J 1989; 8: 2975–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dours-Zimmermann MT, Zimmermann DR. A novel glycosaminoglycan attachment domain identified in two alternative splice variants of human versican. J Biol Chem 1994; 269: 32992–32998. [PubMed] [Google Scholar]
- 6.Zako M, Shinomura T, Ujita M, Ito K, Kimata K. Expression of PG-M(V3), an alternatively spliced form of PG-M without a chondroitin sulfate attachment in region in mouse and human tissues. J Biol Chem 1995; 270: 3914–3918. [DOI] [PubMed] [Google Scholar]
- 7.Ito K, Shinomura T, Zako M, Ujita M, Kimata K. Multiple forms of mouse PG-M, a large chondroitin sulfate proteoglycan generated by alternative splicing. J Biol Chem 1995; 270: 958–965. [DOI] [PubMed] [Google Scholar]
- 8.Kischel P, Waltregny D, Dumont B, Turtoi A, Greffe Y, Kirsch S, De Pauw E, Castronovo V. Versican overexpression in human breast cancer lesions: known and new isoforms for stromal tumor targeting. Int J Cancer 2010; 126: 640–650. [DOI] [PubMed] [Google Scholar]
- 9.Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, Scott G, Steffen D, Worley KC, Burch PE, Okwuonu G, Hines S, Lewis L, DeRamo C, Delgado O, Dugan-Rocha S, Miner G, Morgan M, Hawes A, Gill R, Celera Holt RA, Adams MD, Amanatides PG, Baden-Tillson H, Barnstead M, Chin S, Evans CA, Ferriera S, Fosler C, Glodek A, Gu Z, Jennings D, Kraft CL, Nguyen T, Pfannkoch CM, Sitter C, Sutton GG, Venter JC, Woodage T, Smith D, Lee HM, Gustafson E, Cahill P, Kana A, Doucette-Stamm L, Weinstock K, Fechtel K, Weiss RB, Dunn DM, Green ED, Blakesley RW, Bouffard GG, De Jong PJ, Osoegawa K, Zhu B, Marra M, Schein J, Bosdet I, Fjell C, Jones S, Krzywinski M, Mathewson C, Siddiqui A, Wye N, McPherson J, Zhao S, Fraser CM, Shetty J, Shatsman S, Geer K, Chen Y, Abramzon S, Nierman WC, Havlak PH, Chen R, Durbin KJ, Egan A, Ren Y, Song XZ, Li B, Liu Y, Qin X, Cawley S, Worley KC, Cooney AJ, D’Souza LM, Martin K, Wu JQ, Gonzalez-Garay ML, Jackson AR, Kalafus KJ, McLeod MP, Milosavljevic A, Virk D, Volkov A, Wheeler DA, Zhang Z, Bailey JA, Eichler EE, Tuzun E, Birney E, Mongin E, Ureta-Vidal A, Woodwark C, Zdobnov E, Bork P, Suyama M, Torrents D, Alexandersson M, Trask BJ, Young JM, Huang H, Wang H, Xing H, Daniels S, Gietzen D, Schmidt J, Stevens K, Vitt U, Wingrove J, Camara F, Albà M, Abril M, Guigo JF, Smit R, Dubchak A, Rubin I, Couronne EM, Poliakov O, Hübner A, Ganten N, Goesele D, Hummel C, Kreitler O, Lee T, Monti YA, Schulz J, Zimdahl H, Himmelbauer H, Lehrach H, Jacob H, Bromberg HJ, Gullings-Handley S, Jensen-Seaman J, Kwitek MI, Lazar AE, Pasko J, Tonellato D, Twigger PJ, Ponting S, Duarte CP, Rice JM, Goodstadt S, Beatson L, Emes SA, Winter RD, Webber EE, Brandt C, Nyakatura P, Adetobi G, Chiaromonte M, Elnitski F, Eswara L, Hardison P, Hou RC, Kolbe M, Makova D, Miller K, Nekrutenko W, Riemer A, Schwartz C, Taylor S, Yang J, Zhang S, Lindpaintner Y, Andrews K, Caccamo TD, Clamp M, Clarke M, Curwen L, Durbin V, Eyras R, Searle E, Cooper SM, Batzoglou GM, Brudno S, Sidow M, Stone A, Venter EA, Payseur JC, Bourque BA, López-Otín G, Puente C, Chakrabarti XS, Chatterji K, Dewey S, Pachter C, Bray L, Yap N, Caspi VB, Tesler A, Pevzner G, Haussler PA, Roskin D, Baertsch KM, Clawson R, Furey H, Hinrichs TS, Karolchik AS, Kent D, Rosenbloom WJ, Trumbower KR, Weirauch H, Cooper M, Stenson DN, Ma PD, Brent B, Arumugam M, Shteynberg M, Copley D, Taylor RR, Riethman MS, Mudunuri H, Peterson U, Guyer J, Felsenfeld M, Old A, Mockrin S, Collins S, Rat FG. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 2004; 428: 493–521. [DOI] [PubMed] [Google Scholar]
- 10.Iozzo RV, Naso MF, Cannizzaro LA, Wasmuth JJ, McPherson JD. Mapping of the versican proteoglycan gene (CSPG2) to the long arm of human chromosome 5 (5q12-5q14). Genomics 1992; 14: 845–851. [DOI] [PubMed] [Google Scholar]
- 11.Shinomura T, Zako M, Ito K, Ujita M, Kimata K. The gene structure and organization of mouse PG-M, a large chondroitin sulfate proteoglycan. Genomic background for the generation of multiple PG-M transcripts. J Biol Chem 1995; 270: 10328–10333. [DOI] [PubMed] [Google Scholar]
- 12.Bogen O, Bender O, Löwe J, Blenau W, Thevis B, Schröder W, Margolis RU, Levine JD, Hucho F. Neuronally produced versican V2 renders C-fiber nociceptors IB4 -positive. J Neurochem 2015; 134: 147–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bogen O, Dreger M, Gillen C, Schröder W, Hucho F. Identification of versican as an isolectin B4-binding glycoprotein from mammalian spinal cord tissue. Febs J 2005; 272: 1090–1102. [DOI] [PubMed] [Google Scholar]
- 14.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem 2007; 76: 51–74. [DOI] [PubMed] [Google Scholar]
- 15.Pertin M, Gosselin RD, Decosterd I. The spared nerve injury model of neuropathic pain. Methods Mol Biol 2012; 851: 205–212. [DOI] [PubMed] [Google Scholar]
- 16.Decosterd I, Woolf CJ. Spared nerve injury: an animal model of persistent peripheral neuropathic pain. Pain 2000; 87: 149–158. [DOI] [PubMed] [Google Scholar]
- 17.Bogen O, Joseph EK, Chen X, Levine JD. GDNF hyperalgesia is mediated by PLCgamma, MAPK/ERK, PI3K, CDK5 and Src family kinase signaling and dependent on the IB4-binding protein versican. Eur J Neurosci 2008; 28: 12–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bogen O, Dina OA, Gear RW, Levine JD. Dependence of monocyte chemoattractant protein 1 induced hyperalgesia on the isolectin B4-binding protein versican. Neuroscience 2009; 159: 780–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zwick M, Davis BM, Woodbury CJ, Burkett JN, Koerber HR, Simpson JF, Albers KM. Glial cell line-derived neurotrophic factor is a survival factor for isolectin B4-positive, but not vanilloid receptor 1-positive, neurons in the mouse. J Neurosci 2002; 22: 4057–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munson JB, McMahon SB. Effects of GDNF on axotomized sensory and motor neurons in adult rats. Eur J Neurosci 1997; 9: 1126–1129. [DOI] [PubMed] [Google Scholar]
- 21.Matheson CR, Carnahan J, Urich JL, Bocangel D, Zhang TJ, Yan Q. Glial cell line-derived neurotrophic factor (GDNF) is a neurotrophic factor for sensory neurons: comparison with the effects of the neurotrophins. J Neurobiol 1997; 32: 22–32. [PubMed] [Google Scholar]
- 22.Gaudet AD, Popovich PG, Ramer MS. Wallerian degeneration: gaining perspective on inflammatory events after peripheral nerve injury. J Neuroinflammation 2011; 8: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Padi SS, Shi XQ, Zhao YQ, Ruff MR, Baichoo N, Pert CB, Zhang J. Attenuation of rodent neuropathic pain by an orally active peptide, RAP-103, which potently blocks CCR2- and CCR5-mediated monocyte chemotaxis and inflammation. Pain 2012; 153: 95–106. [DOI] [PubMed] [Google Scholar]
- 24.Hart AM, Terenghi G, Wiberg M. Neuronal death after peripheral nerve injury and experimental strategies for neuroprotection. Neurol Res 2008; 30: 999–1011. [DOI] [PubMed] [Google Scholar]
- 25.Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM. The integrated stress response. EMBO Rep 2016; 17: 1374–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet 2004; 36: 1073–1078. [DOI] [PubMed] [Google Scholar]
- 27.Gardner LB. Hypoxic inhibition of nonsense-mediated RNA decay regulates gene expression and the integrated stress response. Mol Cell Biol 2008; 28: 3729–3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karam R, Lou CH, Kroeger H, Huang L, Lin JH, Wilkinson MF. The unfolded protein response is shaped by the NMD pathway. EMBO Rep 2015; 16: 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naso MF, Zimmermann DR, Iozzo RV. Characterization of the complete genomic structure of the human versican gene and functional analysis of its promoter. J Biol Chem 1994; 269: 32999–33008. [PubMed] [Google Scholar]
- 30.Mount SM. Genomic sequence, splicing, and gene annotation. Am J Hum Genet 2000; 67: 788–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hastings ML, Krainer AR. Pre-mRNA splicing in the new millennium. Curr Opin Cell Biol 2001; 13: 302–309. [DOI] [PubMed] [Google Scholar]
- 32.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 2003; 72: 291–336. [DOI] [PubMed] [Google Scholar]
- 33.Hwang J, Kim YK. When a ribosome encounters a premature termination codon. BMB Rep 2013; 46: 9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lejeune F. Nonsense-mediated mRNA decay at the crossroads of many cellular pathways. BMB Rep 2017; 50: 175–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nickless A, Bailis JM, You Z. Control of gene expression through the nonsense-mediated RNA decay pathway. Cell Biosci 2017; 7: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lerch JK, Alexander JK, Madalena KM, Motti D, Quach T, Dhamija A, Zha A, Gensel JC, Webster Marketon J, Lemmon VP, Bixby JL, Popovich PG. Stress increases peripheral axon growth and regeneration through glucocorticoid receptor-dependent transcriptional programs. eNeuro 2017; 4: ENEURO.0246-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patodia S, Raivich G. Role of transcription factors in peripheral nerve regeneration. Front Mol Neurosci 2012; 5: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santoni G, Cardinali C, Morelli MB, Santoni M, Nabissi M, Amantini C. Danger- and pathogen-associated molecular patterns recognition by pattern-recognition receptors and ion channels of the transient receptor potential family triggers the inflammasome activation in immune cells and sensory neurons. J Neuroinflammation 2015; 12: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zigmond RE. gp130 cytokines are positive signals triggering changes in gene expression and axon outgrowth in peripheral neurons following injury. Front Mol Neurosci 2011; 4: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Melemedjian OK, Asiedu MN, Tillu DV, Sanoja R, Yan J, Lark A, Khoutorsky A, Johnson J, Peebles KA, Lepow T, Sonenberg N, Dussor G, Price TJ. Targeting adenosine monophosphate-activated protein kinase (AMPK) in preclinical models reveals a potential mechanism for the treatment of neuropathic pain. Mol Pain 2011; 7: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khoutorsky A, Sorge RE, Prager-Khoutorsky M, Pawlowski SA, Longo G, Jafarnejad SM, Tahmasebi S, Martin LJ, Pitcher MH, Gkogkas CG, Sharif-Naeini R, Ribeiro-da-Silva A, Bourque CW, Cervero F, Mogil JS, Sonenberg N. Sonenberg N. eIF2α phosphorylation controls thermal nociception. Proc Natl Acad Sci Usa USA 2016; 113: 11949–11954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Inceoglu B, Bettaieb A, Trindade da Silva CA, Lee KS, Haj FG, Hammock BD. Endoplasmic reticulum stress in the peripheral nervous system is a significant driver of neuropathic pain. Proc Natl Acad Sci Usa USA 2015; 112: 9082–9087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu B, Qian SB. Translational reprogramming in cellular stress response. Wires RNA 2014; 5: 301–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farley MM, Watkins TA. Intrinsic neuronal stress response pathways in injury and disease. Annu Rev Pathol Mech Dis 2018; 13: 93–116. [DOI] [PubMed] [Google Scholar]
- 45.Pavitt GD. Regulation of translation initiation factor eIF2B at the hub of the integrated stress response. Wires Rna 2018; 9: e1491. [DOI] [PubMed] [Google Scholar]
- 46.Li Z, Vuong JK, Zhang M, Stork C, Zheng S. Inhibition of nonsense-mediated RNA decay by ER stress. RNA 2017; 23: 378–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Larhammar M, Huntwork-Rodriguez S, Jiang Z, Solanoy H, Sengupta Ghosh A, Wang B, Kaminker JS, Huang K, Eastham-Anderson J, Siu M, Modrusan Z, Farley MM, Tessier-Lavigne M, Lewcock JW, Watkins TA. Dual leucine zipper kinase-dependent PERK activation contributes to neuronal degeneration following insult. Elife 2017; 6: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohtake Y, Matsuhisa K, Kaneko M, Kanemoto S, Asada R, Imaizumi K, Saito A. Axonal activation of the unfolded protein response promotes axonal regeneration following peripheral nerve injury. Neuroscience 2018; 375: 34–48. [DOI] [PubMed] [Google Scholar]
- 49.Goetz AE, Wilkinson M. Stress and the nonsense-mediated RNA decay pathway. Cell Mol Life Sci 2017; 74: 3509–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsujino H, Kondo E, Fukuoka T, Dai Y, Tokunaga A, Miki K, Yonenobu K, Ochi T, Noguchi K. Activating transcription factor 3 (ATF3) induction by axotomy in sensory and motoneurons: a novel neuronal marker of nerve injury. Mol Cell Neurosci 2000; 15: 170–182. [DOI] [PubMed] [Google Scholar]
- 51.Laedermann CJ, Pertin M, Suter MR, Decosterd I. Voltage-gated sodium channel expression in mouse DRG after SNI leads to re-evaluation of projections of injured fibers. Mol Pain 2014; 10: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu YJ, La Pierre DP, Wu J, Yee AJ, Yang BB. The interaction of versican with its binding partners. Cell Res 2005; 15: 483–494. [DOI] [PubMed] [Google Scholar]
- 53.Stephenson EL, Yong VW. Pro-inflammatory roles of chondroitin sulfate proteoglycans in disorders of the central nervous system. Matrix Biol 2018; 71–72: 432–442. [DOI] [PubMed] [Google Scholar]
- 54.Wu Y, Wu J, Lee DY, Yee A, Cao L, Zhang Y, Kiani C, Yang BB. Versican protects cells from oxidative stress-induced apoptosis. Matrix Biol 2005; 24: 3–13. [DOI] [PubMed] [Google Scholar]
- 55.Morawski M, Brückner MK, Riederer P, Brückner G, Arendt T. Perineuronal nets potentially protect against oxidative stress. Exp Neurol 2004; 188: 309–315. [DOI] [PubMed] [Google Scholar]
- 56.Wagner R, Heckman HM, Myers RR. Wallerian degeneration and hyperalgesia after peripheral nerve injury are glutathione-dependent. Pain 1998; 77: 173–179. [DOI] [PubMed] [Google Scholar]
- 57.Kim HK, Park SK, Zhou JL, Taglialatela G, Chung K, Coggeshall RE, Chung JM. Reactive oxygen species (ROS) play an important role in a rat model of neuropathic pain. Pain 2004; 111: 116–124. [DOI] [PubMed] [Google Scholar]
- 58.Naik AK, Tandan SK, Dudhgaonkar SP, Jadhav SH, Kataria M, Prakash VR, Kumar D. Role of oxidative stress in pathophysiology of peripheral neuropathy and modulation by N-acetyl-L-cysteine in rats. Eur J Pain 2006; 10: 573–579. [DOI] [PubMed] [Google Scholar]
- 59.Valek L, Kanngießer M, Häussler A, Agarwal N, Lillig CH, Tegeder I. Redoxins in peripheral neurons after sciatic nerve injury. Free Radic Biol Med 2015; 89: 581–592. [DOI] [PubMed] [Google Scholar]
- 60.Komirishetty P, Areti A, Gogoi R, Sistla R, Kumar A. Poly(ADP-ribose) polymerase inhibition reveals a potential mechanism to promote neuroprotection and treat neuropathic pain. Neural Regen Res 2016; 11: 1545–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu Y, Sheng W, Chen L, Dong H, Lee V, Lu F, Wong CS, Lu WY, Yang BB. Versican V1 isoform induces neuronal differentiation and promotes neurite outgrowth. MBoC 2004; 15: 2093–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bogen O. Identification of a versican V2-based splice variant as an IB4-binding marker at the surface of non-peptidergic, nociceptive C-fibers PhD Thesis, Department of Biology, Chemistry and Pharmacy, Free University of Berlin, Germany, 2006, pp. 52–54.
- 63.García-Ortega L, De los Ríos V, Martínez-Ruiz A, Oñaderra M, Lacadena J, Martínez del Pozo A, Gavilanes JG. Anomalous electrophoretic behavior of a very acidic protein: ribonuclease U2. Electrophoresis 2005; 26: 3407–3413. [DOI] [PubMed] [Google Scholar]
- 64.Graceffa P, Jancsó A, Mabuchi K. Modification of acidic residues normalizes sodium dodecyl sulfate-polyacrylamide gel electrophoresis of caldesmon and other proteins that migrate anomalously. Arch Biochem Biophys 1992; 297: 46–51. [DOI] [PubMed] [Google Scholar]