Abstract
The centrosome, composed of two centrioles surrounded by pericentriolar material, is the cell’s central microtubule-organizing center. Centrosome duplication is coupled with the cell cycle such that centrosomes duplicate once in S phase. Loss of such coupling produces supernumerary centrosomes, a condition called centrosome amplification (CA). CA promotes cell invasion and chromosome instability, two hallmarks of cancer. We examined the contribution of centriole overduplication to CA and the consequences for genomic stability in breast cancer cells. CEP135, a centriole assembly protein, is dysregulated in some breast cancers. We previously identified a short isoform of CEP135, CEP135mini, that represses centriole duplication. Here, we show that the relative level of full-length CEP135 (CEP135full) to CEP135mini (the CEP135full:mini ratio) is increased in breast cancer cell lines with high CA. Inducing expression of CEP135full in breast cancer cells increases the frequency of CA, multipolar spindles, anaphase-lagging chromosomes, and micronuclei. Conversely, inducing expression of CEP135mini reduces centrosome number. The differential expression of the CEP135 isoforms in vivo is generated by alternative polyadenylation. Directed genetic mutations near the CEP135mini alternative polyadenylation signal reduces the CEP135full:mini ratio and decreases CA. We conclude that dysregulation of CEP135 isoforms promotes centriole overduplication and contributes to chromosome segregation errors in breast cancer cells.
INTRODUCTION
Centrosomes are microtubule-organizing centers composed of a pair of centrioles surrounded by pericentriolar material (PCM; Brinkley, 1985). G1 phase cells have two centrioles that will each duplicate once during S phase (Vorobjev and Chentsov, 1982; Piel et al., 2000). This produces the two centrosomes that ensure bipolar spindle assembly and faithful chromosome segregation during mitosis (Nigg and Stearns, 2011; Firat-Karalar and Stearns, 2014). Controlled centriole duplication and cell division preserve the homeostasis of centrosome number (Tsou and Stearns, 2006a,b). Disruption of this homeostasis can result in too many centrosomes, a condition called centrosome amplification (CA; Nigg, 2002; Chan, 2011; Pihan, 2013). CA is defined as the presence of more than one or two centrosomes, depending upon the cell cycle state, and is detected in cells from low-grade premalignant lesions to advanced metastatic cancers including breast cancers (Lingle et al., 2002; Pihan et al., 2003; Martinho et al., 2009; Denu et al., 2016; Lopes et al., 2018; Marteil et al., 2018). CA contributes to chromosome instability through the formation of multipolar spindles. Centrosomes in multipolar spindles often cluster to form bipolar spindles capable of producing viable daughter cells (Quintyne et al., 2005; Ganem et al., 2009; Godinho et al., 2009). However, clustered multipolar spindles also produce merotelic microtubule–kinetochore attachments, giving rise to lagging chromosomes during anaphase that are subsequently packaged into micronuclei in the following interphase (Cimini, 2008; Ganem et al., 2009; Silkworth et al., 2009; Thompson et al., 2010; Thompson and Compton, 2011; Ly and Cleveland, 2017). This creates both aneuploidy and severe chromosome rearrangements through chromothripsis that occurs in micronuclei (Crasta et al., 2012; Zhang et al., 2015). By triggering chromosome instability, CA may initiate tumorigenesis (Levine et al., 2017). Furthermore, in a pathway independent of chromosome instability, CA increases the invasiveness of tumor cells, a hallmark of aggressive cancers (Sluder and Nordberg, 2004; Godinho et al., 2009, 2014; Arnandis et al., 2018).
CA can arise through defective cell division, centrosome fragmentation, and centriole overduplication (Salisbury et al., 2004; Duensing, 2005; Nigg, 2006; Duensing et al., 2007; Kleylein-Sohn et al., 2007; Brownlee and Rogers, 2013 ). While all of the above mechanisms can produce more cells in a cell population with CA, there is limited evidence as to which mechanisms occur in breast cancer cells (Salisbury et al., 2004; Duensing, 2005; Duensing et al., 2007; Nigg, 2006; Kleylein-Sohn et al., 2007; Brownlee and Rogers, 2013).
Centriole duplication initiates during late G1 phase when PLK4 and STIL concentrate at an asymmetric site on the mother centriole wall (Godinho et al., 2012; Firat-Karalar and Stearns, 2014; Ohta et al., 2014). Centriole assembly factors SAS-6, CEP135, and CPAP then self-assemble at this site during S phase into a structure known as the cartwheel, giving rise to the procentriole (Ohta et al., 2002; Carvalho-Santos et al., 2010; Lin et al., 2013; Firat-Karalar and Stearns, 2014; Hirono, 2014; Dahl et al., 2015). The number of procentrioles formed at the mother centriole determines the number of daughter centrioles that will form (Carvalho-Santos et al., 2010; Hirono, 2014). In normal cells, centriole assembly factors are tightly regulated such that the two existing centrioles give rise to a total of two daughter centrioles. Many breast cancer cells harbor excess centrioles, yet whether centriole assembly dysregulation is primarily responsible for amplified centrosomes and CA is not well understood. Importantly, the chromosomal locus of the centriole assembly factor CEP135 (4q12) is amplified and mutated in aggressive breast cancer patient samples, making CEP135 a candidate oncogene (Martinho et al., 2009; Yu et al., 2009; Johansson et al., 2011; Tuupanen et al., 2014). At least two isoforms of CEP135 are transcribed from this single locus. Full-length CEP135 (CEP135full) promotes the formation of daughter centrioles. Interestingly, a shorter CEP135 isoform, CEP135mini, represses centriole assembly, at least in part, by limiting centriolar localization of SAS-6 and CPAP (Dahl et al., 2015). The antagonistic functions of these two CEP135 isoforms suggest that their dysregulation may contribute to the increase in centriole numbers observed in breast cancer cells.
The CEP135 gene contains 26 exons in which all the exons comprise the CEP135full mRNA. In contrast, CEP135mini mRNA is composed of the first six exons and at least part of intron 6 where an in-frame stop codon terminates translation. This leads to a protein that is identical to the first 233 amino acids of CEP135full, but then harbors a unique 16 amino acid tail at its C-terminus (Dahl et al., 2015). We previously reported CEP135mini mRNA to be an alternative splice variant based on database reports (Dahl et al., 2015), yet the biogenesis of these isoforms has not yet been experimentally tested. Genome-wide gene expression studies and databases would normally distinguish between alternative splicing and alternative polyadenylation by identifying the location of transcription termination. However, due to the presence of genomically encoded poly(A)s at the 3′ end of intron 6, CEP135mini is an internal poly(A) priming candidate in these studies (Nam et al., 2002). Thus, it is unclear where the CEP135mini mRNA ends and whether the CEP135mini mRNA variant results from alternative splicing with intron 6 retention or alternative polyadenylation within intron 6.
We investigated CA in breast cancer cells and found that the occurrence of centriole overduplication events in which multiple daughter centrioles arise from a single mother centriole is detectable, but infrequent. Once a cell has too many centrioles, the amplified state is maintained by new centriole assembly from more than just two mother centrioles. We explored the level of CEP135 dysregulation in breast cancer cell lines and found that an abundance of the CEP135full isoform relative to the CEP135mini isoform may contribute to centriole overduplication and CA. Finally, the CEP135mini isoform is generated by alternative polyadenylation and is affected by nucleotide sequences within intron 6.
RESULTS
Increased centriole number, CA, and chromosome missegregation in aggressive breast cancer cells
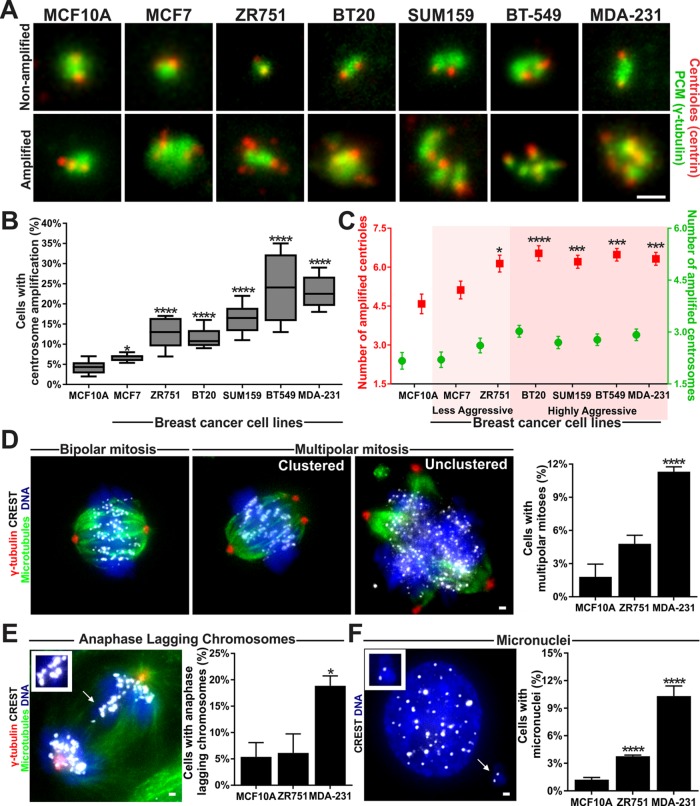
Breast cancers are classified into distinct subtypes based on their clinical prognosis (Dai et al., 2015). Luminal breast cancers typically have good prognoses and are less aggressive, whereas Her2+/basal breast cancers are highly aggressive with poor prognoses (Dai et al., 2015). We measured the frequency of cells with CA in various breast cancer cell lines. Cells with CA are conservatively defined as cells with numerically more than two centrosomes, regardless of their cell cycle stage (Figure 1A). By this conservative criterion, ∼5% of the cell population of normal-like, MCF10A breast cells have CA. Less aggressive breast cancer cell lines have 7–13% of cells with CA, whereas more aggressive breast cancer cell lines exhibit 12–24% of cells with CA (Figure 1B and Supplemental Figure S3A; Neve et al., 2006; Marteil et al., 2018). In addition to having a higher percentage of cells with CA, the cells with CA in more aggressive breast cancer types also have more centrioles and centrosomes per cell (Figure 1C and Supplemental Figure S1A). The excess number of centrioles in these cells with CA suggests that centriole duplication contributes to CA and that centrosome fragmentation is not a major mechanism by which centrosomes are amplified in these breast cancer cell lines. These results strengthen previous findings that the percentage of cells with CA in a cell population is greater in aggressive breast cancer cells (D’Assoro et al., 2002; Schneeweiss et al., 2003; Guo et al., 2007; Denu et al., 2016; Marteil et al., 2018).
FIGURE 1:
Aggressive breast cancer cells exhibit increased CA and chromosome missegregation. (A) Nonamplified and amplified centrioles and centrosomes in breast cancer cell subtypes. Centrioles (centrin, red) and PCM (γ-tubulin, green) are labeled. (B) Percentage of the cell population with cells exhibiting CA. Interquartile range ± highest and lowest observations. (C) Mean number of centrioles (red) and centrosomes (green) in breast cancer cells that have amplified centrosomes. Cell lines are classified from less aggressive to highly aggressive. (D) Left panels, bipolar and multipolar mitoses in breast cancer cells. PCM (γ-tubulin, red), microtubules (α-tubulin, green), DNA (Hoechst 33342, blue) and kinetochores (α-CREST; grayscale) are labeled. Right panel, percentage of multipolar cells in the mitotic cell population. (E) Left panel, anaphase-lagging chromosomes in breast cancer cells. Cells are labeled as in D. Arrow and insets denote lagging chromosomes. Right panel, percentage of late anaphase cells with lagging chromosomes. (F) Left panel, micronuclei in breast cancer cells. Cells were stained for DNA (Hoechst 33342; blue) and kinetochores (CREST; white). Arrow and inset denote a micronucleus. Right panel, percentage of cells with micronuclei. Mean ± SEM. Statistical tests compare to MCF10A cells. Fisher’s exact test and Mann–Whitney U test. *, p < 0.05, ***, p < 0.005, and ****, p < 0.0005. Scale bars, 1 μm.
We next examined the functional consequences of CA on chromosome segregation in normal-like (MCF10A), less aggressive (ZR751), and highly aggressive (MDA-231) breast cancer cells, respectively. Aggressive breast cancer cells exhibit an increased percentage of cells with multipolar mitoses (Figure 1D and Supplemental Figure S1, B and C; Salisbury et al., 2004). Furthermore, CA was previously reported to promote the formation of anaphase-lagging chromosomes and micronuclei (Ganem et al., 2009; Crasta et al., 2012). Similarly, we observe more cells with anaphase-lagging chromosomes and micronuclei in cell populations that had greater CA (Figure 1, E and F). Thus, elevated centriole numbers are associated with CA and chromosome missegregation in breast cancer cells.
Centriole overduplication at amplified centrosomes
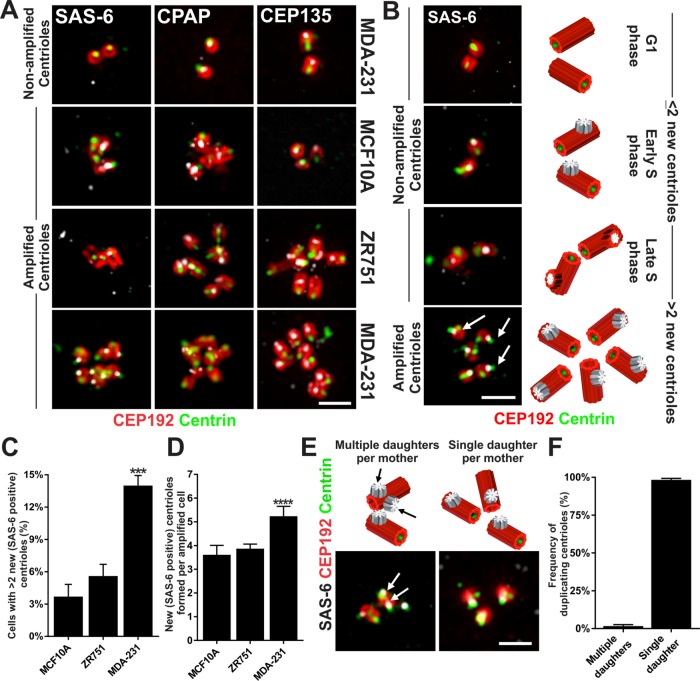
To investigate whether centriole overduplication contributes to the amplified centrioles in breast cancer cells, we examined the frequency of both complete and new centriole assembly in amplified centrosomes. Centriole proteins (Centrin, CEP135, CPAP, CEP192, CEP152, and CEP170) were visualized to determine whether the observed centrioles contain a full complement of representative centriole proteins. Most of the amplified centrioles are mature, as defined by the presence of CEP170, and have a complete complement of the above centriole proteins (Figure 2A and Supplemental Figure S2A). To quantify the frequency of centriole overduplication, we measured the percentage of cells with more than two centriole foci containing SAS-6. SAS-6 is present only in daughter centrioles until the cartwheel disassembles in late mitosis (Vorobjev and Chentsov, 1982; Strnad et al., 2007; Arquint and Nigg, 2014). Normally, G1 phase centrioles are devoid of SAS-6 foci, whereas S, G2, and early mitotic phase cells have two SAS-6 foci (Figure 2B). Compared to MCF10A cells, a larger proportion of MDA-231 cells have more than two new SAS-6–positive procentrioles (Figure 2C). Additionally, the number of newly formed procentrioles in MDA-231 cells with amplified centrosomes is greater than that for MCF10A and ZR751 cells (Figure 2D and Supplemental Figure S2B). Thus, amplified centrosomes in breast cancer cells contain more than two new centrioles.
FIGURE 2:
Centriole overduplication in breast cancer cells that have CA. (A) Nonamplified and amplified centrioles have a full complement of representative centriolar proteins. SAS-6 (left, grayscale), CPAP (middle, grayscale) and CEP135 (right, grayscale) are labeled relative to CEP192 (red) and centrin (green) and imaged using structured illumination microscopy (SIM). (B) New centriole assembly at nonamplified and amplified centrioles. Left panel, centrioles labeled for SAS-6 (grayscale), CEP192 (red), and centrin (green). Right panel, schematic of G1-, early and late S phase, and amplified centrioles. Arrows denote multiple new, SAS-6–positive procentrioles. (C) Percentage of the total cell population that have more than two new (SAS-6–positive) daughter centrioles. (D) Number of new centrioles (SAS-6 positive) assembled in cells that have amplified centrosomes. (E) Multiple and single daughter centrioles form from mother centrioles. Top panel, schematic of multiple and single daughter centrioles forming from mother centrioles. Bottom panel, representative images of centrioles labeled for SAS-6 (grayscale), CEP192 (red), and centrin (green) in S-phase MDA-231 cells that have overduplicated centrioles. Arrows denote multiple daughter centriole assembly events at a single mother centriole. (F) Relative frequency of duplication of multiple daughter centrioles compared with a single daughter centriole from a single mother centriole. (C, D) Statistical tests compare to MCF10A cells. Mean ± SEM. Fisher’s exact test and Mann–Whitney U test. ***, p < 0.005 and ****, p < 0.0005. Scale bars, 1 μm.
We next measured the frequency of a single mother centriole giving rise to more than one daughter centriole, indicative of a centriole overduplication event that would increase the number of centrioles in a cell. The number of SAS-6 foci at each mother centriole was quantified using structured illumination microscopy (SIM) imaging of MDA-231 cells that exhibit greater than two procentrioles (SAS-6 positive) per cell. Of the MDA-231 cells that have more than two SAS-6 foci, only 2% had multiple SAS-6 foci associated with a single mother centriole (Figure 2, E and F). Conversely, 98% of cells with more than two SAS-6 foci have only one daughter centriole per mother centriole. These data suggest that the formation of multiple daughter centrioles from a single mother centriole is detectable, but infrequent. Conversely, an existing centriole amplified state is maintained by single, new daughter centrioles assembling from each of the existing mother centrioles.
The CEP135full:mini ratio is elevated in centrosome-amplified breast cancer cells
It is not clear how centriole duplication is dysregulated to increase the frequency of cells with amplified centrosomes in the cell population. CEP135 is a centriole duplication factor whose isoforms, CEP135full and CEP135mini, perform opposing functions in controlling centriole assembly. CEP135full is important for cartwheel formation and promotes centriole assembly, while CEP135mini represses centriole assembly (Kleylein-Sohn et al., 2007; Dahl et al., 2015). Moreover, the chromosomal locus containing CEP135 (4q12) has an eightfold copy gain in aggressive breast cancer patient data sets, suggesting that expression of CEP135 isoforms is elevated (Finak et al., 2008; Martinho et al., 2009; Yu et al., 2009; Johansson et al., 2011; Glück et al., 2012; Tuupanen et al., 2014). CEP135full mRNA levels are also elevated in aggressive breast cancer cell lines (Supplemental Figure S3A; Neve et al., 2006; Barretina et al., 2012). This led us to ask how the two CEP135 mRNA isoforms with opposing functions are regulated in centrosome-amplified breast cancer cell lines.
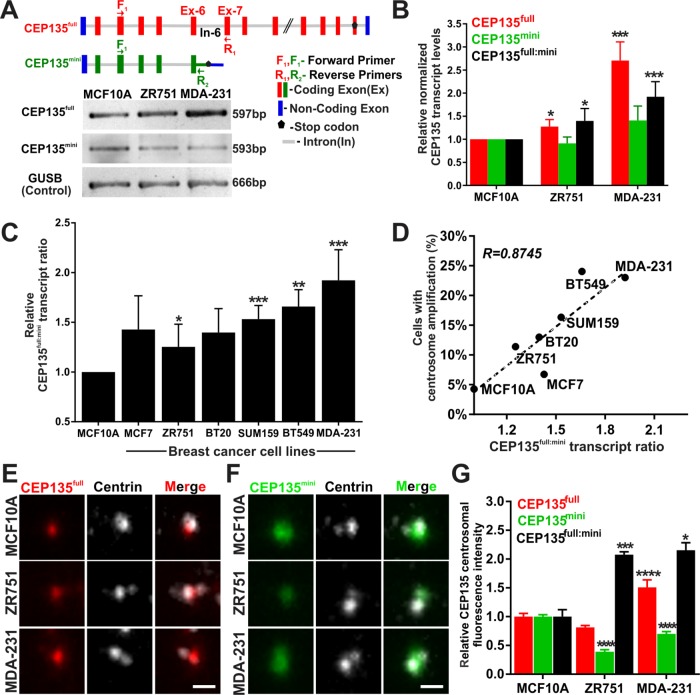
Transcription of CEP135full and CEP135mini mRNA initiates from the same promotor and both isoforms begin translation from the same start codon (genome.ucsc.edu and ensembl.org). The CEP135full mRNA contains 26 exons including two noncoding exons in its untranslated regions (UTRs). Our prior study that focused on the CEP135mini protein did not note these two noncoding exons (Dahl et al., 2015). In contrast to CEP135full mRNA, CEP135mini mRNA contains only the first six exons and at least part of intron 6 where its coding sequence terminates 48 nucleotides (16 amino acids) past the exon 6/intron 6 boundary. To evaluate the levels of these two CEP135 isoforms in breast cancer cells, we detected each isoform by reverse transcription PCR (RT-PCR) using primers directed to unique mRNA sequences (Figure 3A). CEP135full mRNA levels are elevated in MDA-231 cells relative to MCF10A cells. Although CEP135mini mRNA levels are also up-regulated, it is to a lesser degree than CEP135full mRNA levels (Figure 3, A and B). Given the opposing functions of the two isoforms, we hypothesize that the relative levels of these two isoforms are important for regulating centrosome number in breast cancer cells. To examine this, the relative mRNA levels of the CEP135full and CEP135mini isoforms (CEP135full:mini transcript ratio) were quantified in breast cancer cell lines (Figure S3, B and C). The CEP135full:mini ratio is greater in aggressive breast cancer cell lines (Figure 3C). Furthermore, the CEP135full:mini ratio correlates with the percentage of CA observed in these breast cancer cells (Pearson coefficient = 0.8745; Figure 3D).
FIGURE 3:
CEP135 isoform transcript and protein levels are altered in breast cancer cells. (A) Top panel, CEP135full and CEP135mini genes. Red and green bars denote coding exons (Ex) of CEP135full and CEP135mini, respectively. Blue bars denote noncoding exons. Gray lines denote introns (In). F1 and R1 (red) and F1 and R2 (green) denote forward and reverse primers for CEP135full and CEP135mini, respectively. Black pentagons represent stop codons for translation termination. Bottom panels, CEP135full, CEP135mini, and control (GUSB) RT-PCR in breast cancer cells. (B) The relative CEP135full (red) and CEP135mini (green) transcript levels and the CEP135full:mini (black) ratio. (C) The CEP135full:mini transcript ratio in breast cancer cells. (D) Linear regression fit and Pearson coefficient (R) of the CEP135full:mini transcript ratio relative to the percentage of CA cells in the breast cancer cell populations. (E) CEP135full (red) and centrin (grayscale) at one centrosome of a G2 phase cell. (F) CEP135mini (green) and centrin (grayscale) at one centrosome of G2 phase cells. (G) Centrosomal CEP135full (red) and CEP135mini (green) protein fluorescence intensities per centrosome of a G2 phase cell and their ratios (black). (B, C, G) Statistical tests compared to MCF10A cells. Mean ± SEM. Student’s t test and Mann–Whitney U test. *, p < 0.05, **, p < 0.01, ***, p < 0.005, and ****, p < 0.0005. Scale bars, 1 μm.
To test whether the relative CEP135full and CEP135mini protein levels reflect the increased transcript ratios in breast cancer cells, CEP135full- and CEP135mini-specific antibodies were used to measure the fluorescence intensities of CEP135full and CEP135mini at centrosomes. Consistent with transcript levels, CEP135full protein levels at centrosomes are elevated in MDA-231 cells relative to MCF10A cells (Figure 3, E and G, and Supplemental Figure S3, D and F). CEP135mini protein levels at centrosomes are reduced in MDA-231 cells relative to MCF10A cells, despite a slight elevation in CEP135mini mRNA levels in MDA-231 cells (Figure 3, F and G, and Supplemental Figure S3, E and G). Surprisingly, the fluorescence intensity of both CEP135full and CEP135mini is lower in ZR751 cells relative to MCF10A cells (Figure 3, E–G and Supplemental Figure S3, D–G). However, consistent with the transcript ratios, the CEP135full:mini protein ratio is elevated in both ZR751 and MDA-231 cells compared with MCF10A cells (Figure 3G). These data indicate that the CEP135full:mini protein ratio is elevated at centrosomes in breast cancer cells.
Elevated CEP135full is sufficient to increase CA and chromosome missegregation
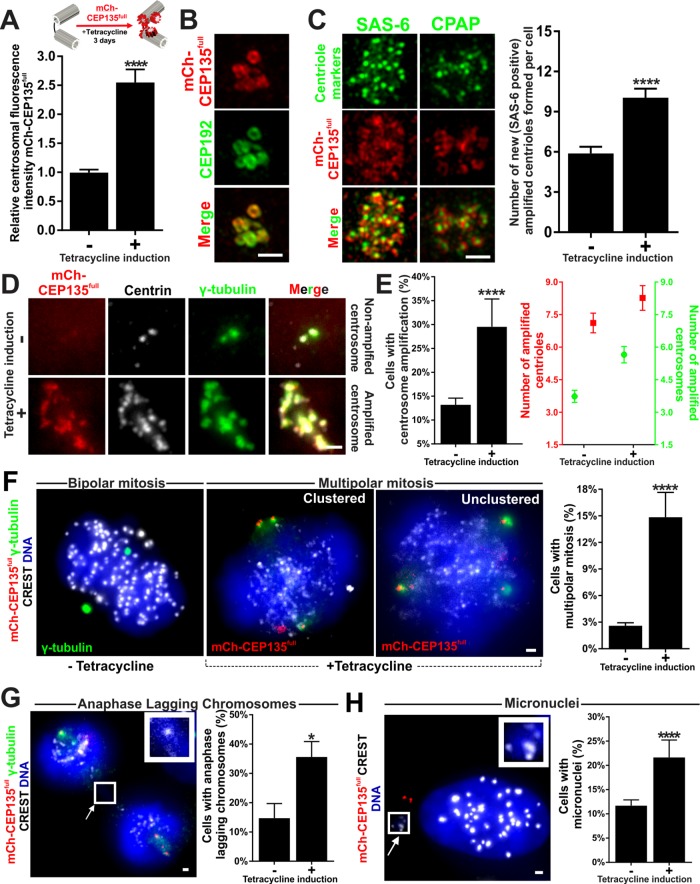
To examine the effects of elevated CEP135full expression in breast cancer cells, we engineered a stable tetracycline-inducible CEP135full MDA-231 cell line. Tetracycline treatment promotes exogenous expression of fluorescently labeled mCherry-CEP135full (hereafter mCh-CEP135full-Tet). We treated mCh-CEP135full-Tet MDA-231 cell lines with tetracycline for 3 d and quantified the number of centrosomes in these cells. The noninduced mCh-CEP135full-Tet cells had a reduced frequency of cells in the cell population with CA (14%) relative to wild-type MDA-231 cells (23%; Supplemental Figure S4A). We hypothesize that clonal selection of the mCh-CEP135full-Tet transfected MDA-231 cells selected for cells with reduced CA, thereby altering the homeostasis of CA. Tetracycline induces a 2.5-fold increase in mCh-CEP135full fluorescence intensity at centrosomes when compared with the noninduced cells (Figure 4A and Supplemental Figure S4B), and an approximately twofold increase in the number of cells with CA (Figure 4, D and E). The number of centrioles and centrosomes within the population of cells with amplified centrosomes was also greater in the induced mCh-CEP135full-Tet cell line (Figure 4E). This suggests that elevated mCh-CEP135full-Tet expression in breast cancer cells promotes new centrioles and centrosomes.
FIGURE 4:
Elevated CEP135full increases breast cancer cell CA and chromosome missegregation. (A) Top panel, schematic and timeline of exogenous mCh-CEP135full-Tet expression in MDA-231 cells. Bottom panel, relative centrosomal fluorescence intensity of mCh-CEP135full in noninduced and induced mCh-CEP135full-Tet MDA-231 cells. (B) SIM localization of mCh-CEP135full (red) and CEP192 (green) at centrioles in mCh-CEP135full-Tet cells. (C) Left panel, amplified centrioles in mCh-CEP135 full-Tet cells. mCh-CEP135full-Tet (red) cells were colocalized with SAS-6 (green) or CPAP (green). Right panel, mean number of new, daughter centrioles (SAS-6 positive) in noninduced and induced mCh-CEP135full-Tet CA MDA-231 cells. (D) Centrosomes in noninduced and induced mCh-CEP135 full-Tet cells. mCh-CEP135full-Tet (red) cells were stained for centrin (grayscale) and γ-tubulin (green). (E) Left panel, percentage of cells with CA in noninduced and induced mCh-CEP135full-Tet MDA-231 cells. Right panel, number of amplified centrioles (red) and centrosomes (green) in noninduced and tetracycline-induced mCh-CEP135full-Tet MDA-231 cells displaying CA. (F) Left panels, bipolar and multipolar mitoses in noninduced and tetracycline-induced mCh-CEP135full-Tet MDA-231 cells stained for PCM (γ-tubulin, green), DNA (Hoechst 33342, blue) and kinetochores (α-CREST, grayscale). Right panel, percentage of mitotic cells with more than two poles in noninduced and induced mCh-CEP135full-Tet cells. (G) Left panel, anaphase-lagging chromosomes in induced mCh-CEP135full-Tet (red) MDA-231 cells stained for DNA (Hoechst 33342, blue) and kinetochores (α-CREST, grayscale). Arrow and inset denote a lagging chromosome. Right panel, percentage of late anaphase cells with lagging chromosomes in noninduced and induced mCh-CEP135full-Tet MDA-231 late mitotic cells. (H) Left panel, micronuclei in induced mCh-CEP135full-Tet (red) MDA-231 cells stained for DNA (Hoechst 33342, blue) and kinetochores (α-CREST, grayscale). Arrow and inset denote a micronucleus. Right panel, percentage of interphase cells with micronuclei in noninduced and induced mCh-CEP135full-Tet MDA-231 cells. Mean ± SEM. Fisher’s exact test and Mann–Whitney U test. *, p < 0.05 and ****, p < 0.0005. Scale bars, 1 μm.
To measure the formation of new centrioles in the induced mCh-CEP135full-Tet cell line, we visualized mCh-CEP135full, SAS-6, and CPAP. Surprisingly, mCh-CEP135full not only localizes to the centriole proximal end but also decorates the walls of some of the centrioles (Figure 4B and Supplemental Figure S4C). Induced mCh-CEP135full-Tet cells have an increased number of SAS-6–positive centrioles relative to the noninduced cells (Figure 4C). Furthermore, multiple SAS-6 foci per mother centriole were observed (Supplemental Figure S4D). However, it is not clear whether all SAS-6 foci represent daughter centrioles or whether overexpressed CEP135full can stabilize SAS-6 at mother centrioles. Regardless, these data suggest that elevated CEP135full levels in breast cancer cells are sufficient to increase centriole number and CA.
Because CEP135full expression is sufficient to increase the number of centrosomes in cells, we next asked whether CEP135full expression disrupts mitosis. Upon mCh-CEP135full-Tet induction, cells exhibit substantial cell death and an approximately fivefold increase in multipolar mitoses (Figure 4F). Accordingly, mCh-CEP135full-Tet–expressing cells have an increased incidence of anaphase-lagging chromosomes and micronuclei (Figure 4, G and H, and Supplemental Figure S4E). Thus, elevated CEP135full in breast cancer cells is sufficient to increase centriole overduplication, CA, and chromosome missegregation.
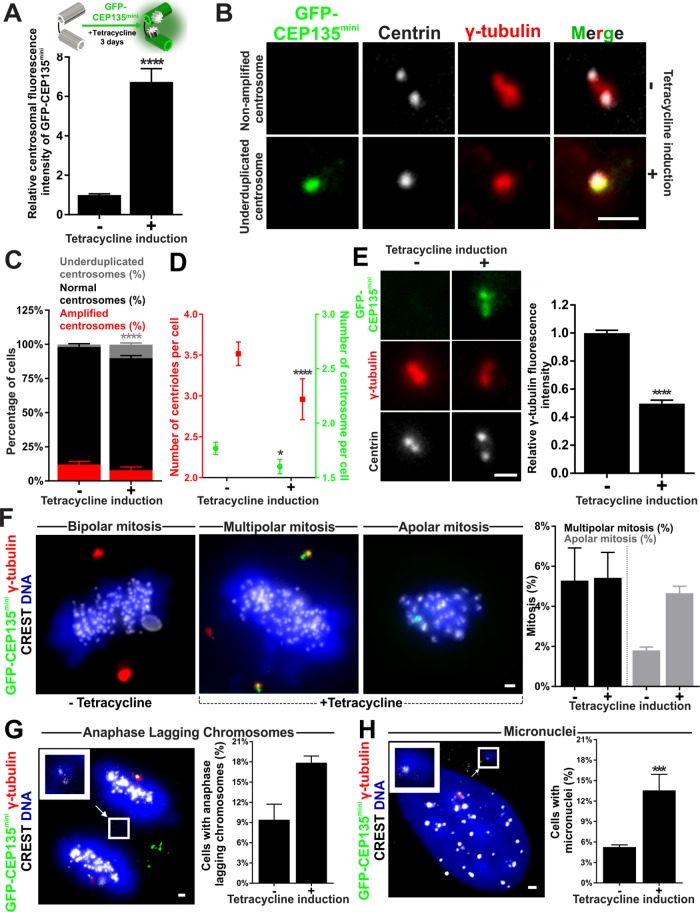
Elevated CEP135mini is sufficient to repress centrosome frequency
CEP135mini represses centriole assembly and limits the localization of centriolar proteins including SAS-6, CPAP, and the PCM protein, γ-tubulin (Dahl et al., 2015). Moreover, CEP135mini levels are regulated in a cell cycle–dependent manner such that levels are lowest during G1/S when centrioles duplicate. CEP135mini levels increase through the rest of the cell cycle, peaking at metaphase of mitosis. We suggest that this is to prevent promiscuous centriole duplication. Given that CEP135mini levels are reduced in breast cancer cells relative to CEP135full levels, we tested whether increasing CEP135mini could repress centriole overduplication. We expressed CEP135mini in MDA-231 cells using a stable tetracycline-inducible fluorescently labeled GFP-CEP135mini MDA-231 cell line (hereafter GFP-CEP135mini-Tet) and measured the frequency of cells with CA. Tetracycline induction for 3 d produced a 6.5-fold increase in centrosomal CEP135mini fluorescence intensity (Figure 5, A and B, and Supplemental Figure S5A). While CA and centrosome number are modestly decreased, GFP-CEP135mini-Tet expression resulted in an approximately threefold increase in the number of cells with underduplicated centrosomes and acentrosomal cells (Figure 5, C and D). Moreover, centrosomal γ-tubulin was reduced in GFP-CEP135mini-Tet–expressing cells (Figure 5E and Supplemental Figure S5B; Dahl et al., 2015). In summary, increased CEP135mini-Tet expression is sufficient to reduce centrosome number and γ-tubulin in breast cancer cells.
FIGURE 5:
Elevated CEP135mini is sufficient to decrease CA and alters chromosome segregation. (A) Top panel, schematic and timeline of tetracycline induction in GFP-CEP135mini-Tet MDA-231 cells. Bottom panel, relative centrosome fluorescence intensity of GFP-CEP135mini in noninduced and induced GFP-CEP135mini-Tet cells. (B) Nonamplified and underduplicated centrosomes in noninduced and induced GFP-CEP135mini-Tet cells. GFP-CEP135mini-Tet (green) cells stained for centrioles (centrin, grayscale) and PCM (γ tubulin, red). (C) Percentage of cells with underduplicated (gray), normal (black), and amplified (red) centrosomes in noninduced and induced GFP-CEP135mini-Tet cells. (D) Frequency of centrioles (red) and centrosomes (green) in the total population of noninduced and induced GFP-CEP135mini-Tet MDA-231 cells. (E) Left panel, nonamplified centrosomes in noninduced and induced GFP-CEP135mini-Tet cells. GFP-CEP135mini-Tet (green) cells stained for centrioles (centrin, grayscale) and PCM (γ tubulin, red). Right panel, relative fluorescence intensity of γ-tubulin in noninduced and tetracycline-induced GFP-CEP135mini-Tet cells. (F) Left panels, bipolar, multipolar, and apolar mitoses in noninduced and induced GFP-CEP135mini-Tet MDA-231 cells stained for PCM (γ-tubulin, red), DNA (Hoechst 33342, blue), and kinetochores (α-CREST, grayscale). Right panels, percentage of multipolar and apolar mitoses in noninduced and induced GFP-CEP135mini-Tet MDA-231 cells. (G) Left panel, anaphase-lagging chromosomes in induced GFP-CEP135mini-Tet (green) MDA-231 cells stained for PCM (γ-tubulin, red), DNA (Hoechst 33342, blue), and kinetochores (α-CREST, grayscale). Arrow and inset denote a lagging chromosome. Right panel, percentage of late anaphase cells with lagging chromosomes in noninduced and induced GFP-CEP135mini-Tet MDA-231 cells. (H) Left panel, micronuclei in induced GFP-CEP135mini-Tet MDA-231 cells stained for DNA (Hoechst 33342, blue) and kinetochores (α-CREST, grayscale). Arrow and inset denote a micronucleus. Right panel, percentage of interphase cells with micronuclei in noninduced and induced GFP-CEP135mini-Tet MDA-231 cells. Mean ± SEM. Fisher’s exact test and Mann–Whitney U test. *, p < 0.05, ***, p < 0.005, and ****, p < 0.0005. Scale bars, 1 μm.
We next examined the effect of CEP135mini overexpression on mitosis. Surprisingly, tetracycline-induced GFP-CEP135mini-Tet cells have the same frequency of multipolar divisions as noninduced cells (Figure 5F). However, more tetracycline-induced GFP-CEP135mini-Tet cells exhibit apolar mitotic divisions compared with noninduced controls, likely resulting from CEP135mini-induced centriole underduplication (Figure 5F and Supplemental Figure S5C). Elevated GFP-CEP135mini-Tet also increases the number of cells with anaphase-lagging chromosomes (Figure 5G). Consistent with the increased lagging chromosomes, more GFP-CEP135mini-Tet–expressing cells have micronuclei (Figure 5H). Thus, elevated CEP135mini expression in breast cancer cells reduces centrosome number but also disrupts normal mitotic chromosome segregation.
Mutations affecting CEP135mini alternative polyadenylation reduce the CEP135 full:mini ratio and CA
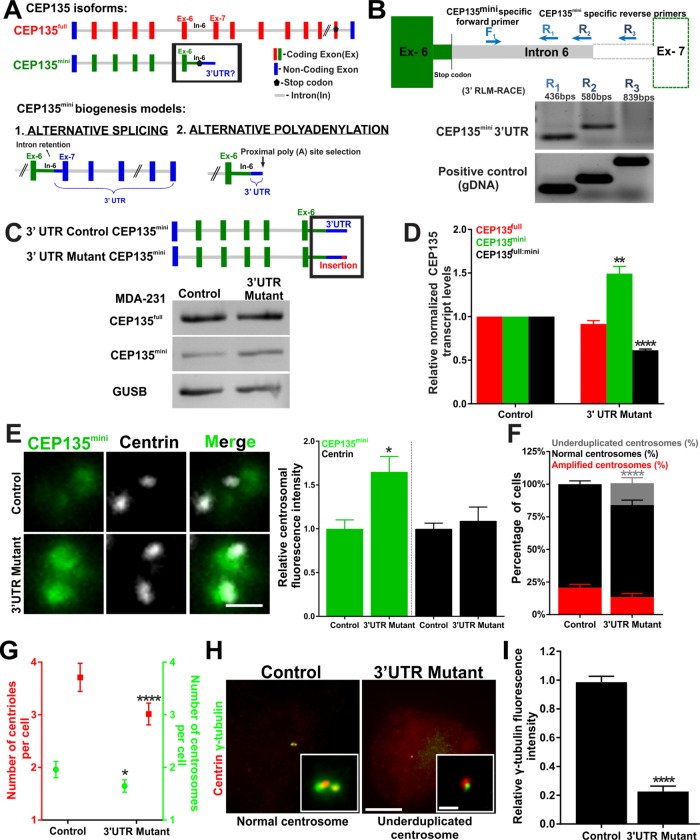
Dahl et al. (2015) showed that CEP135mini mRNA includes at least part of intron 6 where a translation termination codon is present (Figure 6A). On the basis of genome database annotations, we previously reported CEP135mini to be an alternative splice isoform (Dahl et al., 2015). However, the precise 3′ end of the CEP135mini mRNA was not investigated. Intron 6 of CEP135 contains a genomically encoded poly(A) tract and is an internal priming candidate in studies that utilize oligo d(T) to map mRNA 3′ ends. Database annotations do not accurately reflect the transcription termination for RNAs from such genes (Nam et al., 2002). Understanding the nature of the CEP135mini mRNA 3′ end could inform how the CEP135 isoforms are formed and regulated. Two plausible models for CEP135mini mRNA isoform generation are alternative splicing and alternative polyadenylation that both utilize overlapping machinery for RNA processing (Tian et al., 2007). Alternative splicing would promote CEP135mini formation through intron 6 retention. Alternatively, a proximal noncanonical poly(A) signal within intron 6 may be used for transcriptional termination (Figure 6A). To distinguish between these models, 3′ RNA-ligation–mediated RACE was performed on cytoplasmic RNA to enrich for mature messages. RT-PCR primers determined that CEP135mini’s mRNA terminates at a site between 580 and 839 nucleotides downstream from the stop codon (Figure 6B). Moreover, 3′READS+ data suggest there are three sites where poly(A) tracts are added in intron 6 (Hoque et al., 2013; Zheng et al., 2016). Among them, the first site is 802 base pairs downstream of the stop codon, which is consistent with our 3′ RACE data and suggests that CEP135mini’s 3′ UTR terminates 802 base pairs downstream from the stop codon in intron 6 (Supplemental Figure S6A). Two nonconsensus poly(A) signals near this site are AAUAUA and GAUAAA (Beaudoing, 2000). These results support the model that CEP135mini is an alternatively polyadenylated CEP135 isoform.
FIGURE 6:
CEP135mini is an alternatively polyadenylated isoform, and mutations near the CEP135mini poly(A) signal reduce the CEP135full:mini ratio and centrosome number in breast cancer cells. (A) Top panel, schematic of CEP135full and CEP135mini genes. Red and green bars denote coding exons (Ex) of CEP135full and CEP135mini, respectively. Blue bars denote noncoding exons. Gray lines denote introns (In). Black pentagons denote translation stop codons. Bottom panel, alternate models for CEP135mini transcript biogenesis. (B) Top panel, schematic of the CEP135mini 3′UTR-specific forward primer (F1) and reverse primers (R1, R2, and R3) used to map the approximate end of the CEP135mini 3′UTR using 3′ RNA-ligated RACE. Bottom panel, RT-PCR and PCR. (C) Top panel, schematic of control and mutant 3′UTR CEP135mini transcripts. Red bar denotes the Cas9-mediated insertion site in the 3′UTR. Bottom panel, CEP135full, CEP135mini, and GUSB transcript levels in control and 3′UTR mutant MDA-231 cells. (D) CEP135full, CEP135mini, and CEP135full:mini transcript levels in control and 3′UTR mutant MDA-231 cells. (E) Left panels, centrosomal CEP135mini protein in control and 3′UTR mutant MDA-231 cells stained for CEP135mini (green) and centrin (grayscale). Right panel, quantification of CEP135mini and centrin fluorescence intensities in control and 3′UTR mutant MDA-231 cells. (F) Percentage of cells with underduplicated (gray), normal (black), and amplified (red) centrosomes in control and 3′UTR mutant MDA-231 cells. (G) Number of centrioles and centrosomes per cell in control and 3′UTR mutant MDA-231 cells. (H) Normal and underduplicated centrosomes in control and 3′UTR mutant MDA-231 cells stained for centrioles (centrin, red) and γ-tubulin (green). (I) Relative γ-tubulin fluorescence intensity in control and 3′UTR mutant MDA-231 cells. Mean ± SEM. Student’s t test, Fisher’s exact test, and Mann–Whitney U test. *, p < 0.05, **, p < 0.01, and ****, p < 0.0005. Scale bars, 1 μm.
Polyadenylation signal usage is often misregulated in cancers (DeRisi et al., 1996; Mayr and Bartel, 2009). Altered utilization of CEP135 poly(A) signals would affect CEP135full:mini transcript ratios in breast cancer cells. The usage of one poly(A) signal over another depends on both the relative strengths of the poly(A) signals and the juxtaposed sequences and transacting factors that bind to them (Moreira et al., 1995; Nunes et al., 2010; Di Giammartino et al., 2011). CEP135full utilizes a distal, consensus poly(A) signal for transcription termination, whereas CEP135mini utilizes a proximal nonconsensus poly(A) signal. We suggest that CEP135mini’s proximal nonconsensus poly(A) signal is weaker than CEP135full’s and is subject to regulation that controls the relative CEP135 isoform levels.
To test whether poly(A) signals regulate the CEP135full:mini ratio, we used CRISPR-Cas9 to attempt to insert an exogenous consensus poly(A) signal upstream of the endogenous CEP135mini nonconsensus poly(A) signals in MDA-231 cells (Supplemental Figure S6B; Levitt et al., 1989). However, cells with these mutations were not recoverable. When screening for clones with mutations in the 3′UTR of CEP135mini, we identified a mutant (CEP135mini-3′ UTR mutant) that exhibits a 1.5-fold increase in CEP135mini mRNA (Figure 6, C and D, and Supplemental Figure S6C). This produced a 40% decrease in CEP135full:mini ratio in MDA-231 cells and a corresponding increase in CEP135mini protein levels (Figure 6, D and E).
Consistent with the reduced CEP135full:mini ratio, the CEP135mini-3′UTR mutant cells exhibit a significant increase in the number of cells with underduplicated centrosomes and a modest decrease in the number of cells in the population with CA (Figure 6, F and G). Furthermore, the CEP135mini-3′UTR mutant cells exhibit a dramatic reduction in centrosomal γ-tubulin (Figure 6, H and I, and Supplemental Figure S6E). The CEP135mini-3′ UTR MDA-231 cells have an increased mitotic index and many of these cells have apolar mitoses (Supplemental Figure S6D). This is likely due to the significant increase in underduplicated centrosomes. Overall, these results support a model in which the nucleotide sequences adjacent to the alternative poly(A) site influence the CEP135full:mini ratio and regulate centrosome number and function.
DISCUSSION
Centriole overduplication provides a fundamental mechanism by which the frequency of amplified centrosomes increases in breast cancer cell lines. Centriole overduplication in breast cancer cells may be, in part, modulated by the regulated levels of two CEP135 isoforms: one that promotes centriole assembly and one that represses centriole assembly. These isoforms are generated by transcriptional termination either at the distal, canonical poly(A) signal or at a proximal noncanonical poly(A) signal. This suggests that precise control of transcription termination is required to prevent tumor-promoting events like centriole and centrosome overduplication.
Centriole overduplication is associated with CA in breast cancer
The level of CA in our studies of cultured breast cancer cells is stable through multiple passages of each cell line. This suggests that despite abnormalities in centrosome number, homeostatic mechanisms exist that maintain a specific level of amplified centrioles and centrosomes in a cell population. This likely reflects a balance between the initiation and propagation of centriole overduplication and the loss or death of cells with CA. Centriole rosette-like structures containing multiple daughter centrioles surrounding a single mother were reported in primary malignancies, suggesting a high frequency of multiple daughter centriole overduplication from a single mother centriole (Cosenza et al., 2017). We tested whether multiple daughter centrioles duplicate from a single mother centriole in breast cancer cells and observed a low frequency of these events. In the MDA-231 cell population that has more than two new centrioles, 2% of cells have multiple new centrioles from a single mother centriole. This represents a small fraction of the total population, which we estimated to be 0.3%, given that 13% of the MDA-231 cells are overproducing new (SAS-6–positive) centrioles. This low frequency corresponds with the stable levels of CA observed through multiple cell passages of breast cancer cell lines, as a high frequency of centriole overduplication at each cell division would produce increasing frequencies of cells with centriole overduplication as cell cultures are passaged. Furthermore, we did not observe rosette formations. Almost all new centrioles form in a once-and-only-once event where each existing centriole forms only one new centriole to maintain either normal or excess numbers of centrioles. We therefore suggest that most centriole overduplication events simply maintain existing CA in breast cancer cells (Figure 2, E and F). This indicates that breast cancer cells, despite having lost some aspects of centrosome number control, retain regulatory mechanisms that limit centriole duplication to a single daughter centriole for each mother centriole.
CEP135 isoform dysregulation promotes centriole overduplication in breast cancer
Consistent with CEP135full’s function as a positive regulator of centriole duplication, CEP135full levels are elevated in breast cancer cell lines with increased frequencies of cells with CA (Figure 3). Furthermore, expression of CEP135full is associated with new centriole formation and CA in breast cancer cells (Figure 4). One model for CEP135full-induced centriole formation is that increases in CEP135full act to stabilize one of its interacting partners, SAS-6, at newly formed centrioles, promoting both centriole assembly and nascent centriole stability (Figure 4; Matsuura et al., 2004; Jerka-Dziadosz et al., 2010; Lin et al., 2013). Moreover, CEP135full’s function is not limited to centriole formation. CEP135full has a microtubule-binding domain and affects microtubule stability and organization (Ohta et al., 2002; Carvalho-Santos et al., 2012; Lin et al., 2013; Kraatz et al., 2016). Consistent with this, overexpressed CEP135full decorates the centriole cylinder walls in addition to its conventional proximal-end localization (Figure 4B). Therefore, we hypothesize that increased CEP135full promotes ectopic procentriole formation by stabilizing additional SAS-6 foci.
In contrast to CEP135full, CEP135mini’s expression is not, or is minimally, up-regulated in breast cancer cells. Consistent with CEP135mini’s function as a negative regulator of centriole duplication, increased expression of CEP135mini in breast cancer cells produces a modest decrease in the number of cells with CA and a significant increase in centrosome underduplication (Figure 5). This suggests that increased CEP135mini levels in breast cancer cells repress centriole duplication leading to underduplicated centrosomes. It is interesting to consider how modulating CEP135mini could be used as a tool to reduce the number of cells with too many centrosomes in cancer. However, appropriate dosage routines of CEP135mini expression are required to limit the centrosome underduplication that we observed in these studies. Surprisingly, elevated CEP135mini levels increase anaphase-lagging chromosomes and micronuclei, two phenotypes commonly associated with cells that have amplified centrosomes (Figures 1 and 5). The severe loss of γ-tubulin in elevated CEP135mini cells disrupts centrosome microtubule nucleation (unpublished data) and may thereby disrupt mitotic progression. Indeed, maintaining the fine balance between the CEP135 isoforms is critical for the homeostasis of centrosome numbers and the proper segregation of chromosomes during mitosis.
Alternative polyadenylation in CEP135 isoform regulation
Two isoforms of CEP135 act antagonistically to control centriole duplication (Dahl et al., 2015). The CEP135 gene locus (chromosome locus 4q12) exhibits copy number gain in breast cancer patients predicting that both isoforms would be similarly up-regulated and would maintain the CEP135full:mini ratio (Yu et al., 2009). However, both CEP135 isoforms do not increase in the breast cancer cell lines examined. Inefficient use of the nonconsensus poly(A) signal is likely responsible for the low CEP135mini levels relative to CEP135full. However, a less efficient poly(A) signal on its own does not explain the increased ratio of CEP135full to CEP135mini in breast cancer cells. Additional levels of CEP135 isoform regulation must malfunction in breast cancer cells.
To study the direct impact of altered transcription termination, we engineered mutations near CEP135mini’s poly(A) signal to assess whether this would alter CEP135mini levels. Attempted insertions of a consensus poly(A) signal did not yield viable cells, suggesting that high levels of CEP135mini are lethal. This is consistent with our prior studies using transient expression of CEP135mini (Dahl et al., 2015). An MDA-231 cell line with inserted random DNA sequences near the poly(A) signal was isolated (Figure 6). The mutations in this cell line increase CEP135mini levels and reduce centrosome numbers. We hypothesize that the increased CEP135mini levels result from altered mRNA stability or changes to the strength or regulation of CEP135mini’s poly(A) signal. Either result is interesting and will be a target of future investigations to understand the mechanism by which the CEP135full:mini ratio is controlled. To our knowledge, this is the first demonstration that altered 3′-end formation in cancer cells regulates centrosome number. This is important because alternative splicing and polyadenylation are commonly dysregulated in cancer cells that exhibit CA (Mayr and Bartel, 2009; David and Manley, 2010).
In summary, the dysregulation of CEP135 isoforms in breast cancer cells contributes to the loss of centrosome number homeostasis and chromosome segregation errors. Differential levels of CEP135 isoforms are generated by the usage of alternative polyadenylation signals. These findings support the conclusion that alternative polyadenylation, which is commonly disrupted in cancers, regulates the assembly of macromolecular structures such as centrosomes.
MATERIALS AND METHODS
Cell culture
Breast cancer cell lines MCF10A, MCF7, ZR-75.1 (ZR751), BT-20, MDA-MB-231 (MDA-231), and SUM159PT were obtained from the University of Colorado Cancer Center Tissue Culture Core and BT-549 were obtained from the American Type Culture Collection. Mammalian tissue culture lines were all grown at 37°C with 5% CO2. MCF10A cells were received at passage 51 and were grown in DMEM/F12 (Invitrogen; #11330-032), 5% horse serum (Invitrogen; #16050-122), 20 ng ml−1 EGF (Invitrogen; #PHG0311), 0.5 mg ml−1 hydrocortisone (Sigma; #H-0888), 100 ng ml−1 cholera toxin (Sigma; #C-8052), 10 μg ml−1 insulin (Sigma; #I-1882), and 1% pen/strep (Invitrogen; #15070-063). MDA-MB-231 cells were received at passage 15, BT-20 were received at passage 11, and MCF-7 were received at passage 7. These lines as well as 293FT cells were grown in DMEM (Invitrogen; #11965-092), pen/strep (Invitrogen; #15070-063), and 10% fetal bovine serum (FBS; Gemini Biosciences). ZR-75.1 (ZR751) cells were received at passage 51, ZR-75.1 and BT-549 cells were grown in RPMI (Invitrogen; #11875-093), 10% FBS (Gemini Biosciences), and pen/strep (Invitrogen; #15070-063). SUM-159.PT (SUM159) were received at passage 10 and grown in Ham’s media (Invitrogen; #11765054), hydrocortisone (Sigma; #H-0888), pen/strep (Invitrogen; #15070-063), and 10% FBS (Gemini Biosciences). Cell lines were authenticated at the sources and tested negative for mycoplasma using the MycoAlert mycoplasma detection kit through the University of Colorado Cancer Center Tissue Culture Core. Cells were passaged and subcultured using trypsin (Invitrogen; #150901-046) when cultures reached 60–80% confluency.
Generation of mCherry-CEP135full-tet, GFP-CEP135mini-tet, and 3′UTR mutant cells
Lentivirus harboring tetracycline-inducible mCherry-CEP135full-Tet or GFP-CEP135mini-Tet was made by transfection of 293FT cells. 293FT cells were plated in 6-cm dishes and allowed to reach 50–70% confluency. Cells were then transfected with tetracycline-inducible mCherry-CEP135full-Tet or GFP-CEP135mini-Tet constructs, and second-generation lentivirus packaging plasmids (pMD2.G and psPAX2) using lipofectamine 2000 (Life Technologies; #11668019). 293FT media-containing virus was harvested and MDA-231 cells were infected for 24–48 h in the presence of 10 μg ml−1 (26.7 μM) polybrene. After a 24 h recovery, transduced cells were selected with puromycin at 2 μg ml−1 (4.24 μM) and were flow sorted to isolate and plate single cells into 96-well plates. mCh-CEP135full-Tet and GFP-CEP135mini-Tet cells were induced with tetracycline (Invitrogen; #550205) at 2.5 μg ml−1 (5.63 μM).
The 3′UTR of CEP135mini was edited using CRISPR/Cas9-mediated genome engineering in MDA-231 cells. The oligonucleotide with the consensus poly(A) signal donor sequences was introduced to edit the predicted poly(A) signals (Levitt et al., 1989). Cas9-EGFP containing the sgRNA was expressed from the pX458 construct. pX458 and the single-stranded consensus poly(A) signal donor sequences were cotransfected into MDA-231 cells. Cas9-GFP–positive cells were flow sorted into 96-well plates, and clones were screened using PCR with primers flanking the predicted poly(A) signals.
Transfections
MDA-231 cells at 50–80% confluence were transfected using lipofectamine 2000 (Invitrogen; #11668019). Plasmid DNA and plus reagent (Invitrogen; #11514015) were mixed at 1:1 and incubated for 5 min. This mixture was then combined with lipofectamine at a1:3 ratio. Complexes were diluted in Opti-MEM (Invitrogen; #31985062). After a 4-h incubation, the complexes were removed and the transfected cells were supplied with fresh media.
Immunofluorescence
Coverslips 12 mm in diameter were acid-washed and heated to 50°C in 100 mM HCl for 16 h. This was followed by washes with water, 50%, 70%, and 95% ethanol for 30 min each. Coverslips were coated with type 1 collagen (Sigma; #C9791), air-dried for 20 min in the laminar hood, and exposed to UV light for cross-linking of collagen for 20 min. Cells were cultured on collagen-coated coverslips to 55–70% confluence.
For centrosome immunofluorescence, cells were fixed with 100% methanol at −20°C for 8 min. Mitotic cells in Figure 1 were fixed with 4% paraformaldehyde (Electron Microscopy Sciences) for 4 min followed by 100% methanol at −20°C for 4 min. Mitotic cells in Figures 4 and 5 were fixed with 100% methanol at −20°C for 8 min to preserve the mCherry-CEP135full and GFP-CEP135mini fluorescence. Fixed cells were washed with phosphate-buffered saline (PBS)/Mg (1× PBS, 1 mM MgCl2), and then blocked with Knudsen buffer (1× PBS, 0.5% bovine serum albumin, 0.5% NP-40, 1 mM MgCl2, 1 mM NaN3) for 1 h. Cells were incubated overnight with primary antibodies diluted in Knudsen buffer at 4°C. Coverslips were washed with PBS three times at 5-min intervals. Secondary antibodies and Hoechst 33258 (10 μg ml−1; Sigma; #B2261) were diluted in Knudsen buffer and incubated for 1 h at room temperature. Coverslips were mounted using Citifluor (Ted Pella) and sealed with clear nail polish. Coverslips for SIM imaging were mounted using Prolong Gold (Life Technologies; #P10144) and sealed with clear nail polish.
Antibodies used for immunofluorescence are α-centrin (1:2000; 20H5; Abcam), α-γ-tubulin (1:1000; DQ-19; Sigma), α-CEP135full (1:5000; generous gift from T. K. Tang, Institute of Biomedical Sciences, Academia Sinica, Taipei, Taiwan), α-CEP135mini (1:2000; Dahl et al., 2015), α-SAS-6 (1:2000; Bethyl; A301-802A), α-α-tubulin (1:500; DM1A; Sigma), α-centromere derived from human calcinosis, Raynaud’s phenomenon, esophageal dysmotility, sclerodactyly, and telangiectasia (CREST) patient serum (1:2000; generous gift from J. DeLuca, Department of Biochemistry and Molecular Biology, Colorado State University, Fort Collins, CO), α-CPAP (1:350; Proteintech CENPJ 11517-1-AP), and α-CEP192 (1:2000; generous gift from A. Holland, Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine, Baltimore, MD). Alexa Fluor secondary antibodies were diluted to 1:1000 for all experiments (Molecular Probes).
Microscopy
Superresolution imaging in Figure 2 was acquired using Nikon SIM (N-SIM) with a Nikon Ti2 (Nikon Instruments; LU-N3-SIM) microscope equipped with a 100× SR Apo TIRF, NA 1.49 objective. Images were captured using a Hamamatsu ORCA-Flash 4.0 Digital CMOS camera (C13440).
The fluorescence imaging utilized for Figures 1, 4, 5, and 7 is identical to that described in Dahl et al. (2015). Briefly, images were acquired using a Nikon TiE (Nikon Instruments) inverted microscope stand equipped with a 100× PlanApo DIC, NA 1.4 objective. Images were captured using an Andor iXon EMCCD 888E camera or an Andor Xyla 4.2 CMOS camera (Andor Technologies). Images in Figure 3 were acquired using a Swept Field Confocal system (Prairie Technologies/Nikon Instruments) on a Nikon Ti inverted microscope stand equipped with a 100× Plan Apo λ, NA 1.45 objective. Images were captured with an Andor Clara CCD camera (Andor Technologies).
Nikon NIS Elements imaging software was used for image acquisition. Image acquisition times were constant within a given experiment and ranged from 50 to 400 ms, depending on the experiment. All images were acquired at ∼25°C. Images presented in most of the figures are maximum-intensity projections of the complete z-stacks. Exceptions include certain mitotic images that are constructed from selected z-planes to clearly distinguish kinetochores and lagging chromosomes.
Fluorescence intensity quantitation
Image analysis was performed using Python with the tiff file library for reading images and the NumPy library (https://pypi.org/project/tifffile/ and www.numpy.org/) for performing computations. A command-line Python script was written and utilized for this analysis. The code is available at http://thepearsonlab.com/image-analysis-routines.html. The script was given the path of the image as a command-line argument. The script first showed the image in a graphical user interface. For fluorescence intensity analysis at the centrosome, a 15-pixel square box was centered on the centrosome. The total intensity in this box was computed and divided by the area of the box. The background value of the image was computed by identifying four boxes outside the centrosome and dividing the total intensity in these boxes by the total area of these boxes. The final intensity at the centrosome was computed after subtraction of the background value.
Centriole and centrosome number counts
Cells were scored as amplified, nonamplified, and underduplicated based on centrin and γ-tubulin staining. Cells with greater than two γ-tubulin and four centrin foci were scored as amplified centrosomes. Nonamplified centrosomes have both one or two γ-tubulin and two or four centrin foci. Underduplicated centrosomes contain one centrin focus.
Cells were scored for duplication of multiple daughter centrioles versus a single daughter centriole based on SAS-6, centrin, and CEP192 staining. When a mother centriole (CEP192, centrin, and no SAS-6) had greater than one SAS-6 foci, it was scored as duplicating multiple daughter centrioles. When a mother centriole formed a single SAS-6 focus it was quantified as duplicating a single daughter centriole.
Chromosome missegregation counts
Mitotic cells that contain one, two, or greater than two poles labeled by γ-tubulin and microtubules (α-tubulin) were considered as mono-, bi-, and multipolar, respectively. Cells were considered to have lagging chromosomes when they had a kinetochore-positive chromosome in between the two completely separated anaphase chromosome masses.
Reverse-transcription PCR and quantitation
Cytoplasmic RNA from cancer cells at 55–70% confluence was harvested using the RNAeasy kit with DNase I treatment (Qiagen; #74104). Equal amounts of RNA were utilized for reverse-transcription and cDNA amplification using One-Step rtPCR (Invitrogen; #1257401). CEP135full transcripts were specifically detected using primers to exon 3 and exon 7 (CAAAATTATCTGCTGTGAAAGCTG and CCAAAGCAACTGACAGTCG). CEP135mini transcripts were specifically detected using the above primer that anneals in exon 3 and a primer specific to CEP135mini in intron 6 (ACCTATCTCAATCCCTACTATGCAA). The absolute CEP135full and CEP135mini transcript levels were normalized to GUSB (CATTCCTATGCCATCGTGTGG and GACACCGTGGAAATAGAAAGG).
3′ RNA-ligation–mediated RACE
Cytoplasmic RNA was harvested from cancer cells using the RNAeasy kit with DNase I treatment (Qiagen; #74104). Poly A+ RNA was enriched using poly(A) spin columns (NEB: #S1560S). 5′-Adenylation and phosphorylation of the adapter was performed using Mth RNA Ligase (NEB; #M2611A) and T4 Polynucleotide Kinase (NEB; #M0201S). Phosphorylated and adenylated adapter was ligated to poly A+ RNA using T4 RNA Ligase 2, truncated K227Q (NEB; #M0351S). cDNA was made using an adapter-specific reverse primer. To probe the 3′ end of CEP135mini, primers throughout the intron were utilized (Figure 6B).
Statistics and biological replicates
All center values represent means and error bars represent the standard error of the mean except for Figure 1B boxes, which represent the mean and interquartile range, while vertical lines indicate the highest and lowest observations. The number of biological replicates, number of cells utilized per replicate, and p values are described in Supplemental Table 1. Fisher’s exact test, Student’s two-tailed t test, and Mann–Whitney U test were used to assess statistical significance between means. Fisher’s test was utilized to examine the significance of contingency when data were classified into two or more categories. Student’s two-tailed unpaired t test was used to examine the significance between two normal distributions (equal variance assumed). Normality tests were performed on both the raw data and meta-data extracted from the replicates of raw data. A Shapiro–Wilk normality test and D’Agostino–Pearson omnibus normality test were utilized to examine the normality of data. A Shapiro–Wilk normality test was used when the number of samples was less than eight. When the number of samples was greater than eight, a D’Agostino–Pearson omnibus normality test was used. A Mann–Whitney U test was utilized to examine the significance on nonnormal distributions. Results were considered statistically significant with p values less than 0.05. p Values were denoted on figures according to the following values: *, p < 0.05; **, p < 0.01; ***, p < 0.005; and ****, p < 0.0005.
Supplementary Material
Acknowledgments
We thank Marisa Ruehle for discussions and comments on the manuscript and David Bentley, Tassa Saldi, and Domenico “Nick” Galati for helpful discussions. We thank Kristin Dahl and Heide Ford for constructs, Bharath Hariharan for assistance with image analysis, Aik Choon Tan for database expertise, Carol Sartorius for cell lines, and Jennifer DeLuca, Keith DeLuca, Andrew Holland, and Tang K. Tang for antibodies. This work was supported by the CU Tissue Culture Core and Flow Cytometry Core. C.G.P. is funded by the American Cancer Society (RSG-16-157-01-CCG), the National Institute of General Medical Sciences (GM099820), the Linda Crnic Institute, and the Boettcher Foundation.
Abbreviations used:
- CA
centrosome amplification
- nts
nucleotides
- PCM
pericentriolar material
- 3′ UTR
3′ untranslated region.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-10-0674) on February 27, 2019.
REFERENCES
- Arnandis T, Monteiro P, Adams SD, Bridgeman VL, Rajeeve V, Gadaleta E, Marzec J, Chelala C, Malanchi I, Cutillas PR, Godinho SA. (2018). Oxidative stress in cells with extra centrosomes drives non-cell-autonomous invasion. Dev Cell , 409–424.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arquint C, Nigg EA. (2014). STIL microcephaly mutations interfere with APC/C-mediated degradation and cause centriole amplification. Curr Biol , 351–360. [DOI] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV, Sonkin D, et al. (2012). The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature , 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoing E. (2000). Patterns of variant polyadenylation signal usage in human genes. Genome Res , 1001–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkley BR. (1985). Microtubule organizing centers. Annu Rev Cell Biol , 145–172. [DOI] [PubMed] [Google Scholar]
- Brownlee CW, Rogers GC. (2013). Show me your license, please: deregulation of centriole duplication mechanisms that promote amplification. Cell Mol Life Sci , 1021–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho-Santos Z, Machado P, Alvarez-Martins I, Gouveia SM, Jana SC, Duarte P, Amado T, Branco P, Freitas MC, Silva STN, et al. (2012). BLD10/CEP135 is a microtubule-associated protein that controls the formation of the flagellum central microtubule pair. Dev Cell , 412–424. [DOI] [PubMed] [Google Scholar]
- Carvalho-Santos Z, Machado P, Branco P, Tavares-Cadete F, Rodrigues-Martins A, Pereira-Leal JB, Bettencourt-Dias M. (2010). Stepwise evolution of the centriole-assembly pathway. J Cell Sci (Pt 9), 1414–1426. [DOI] [PubMed] [Google Scholar]
- Chan JY. (2011). A clinical overview of centrosome amplification in human cancers. Int J Biol Sci , 1122–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D. (2008). Merotelic kinetochore orientation, aneuploidy, and cancer. Biochim Biophys Acta Rev Cancer , 32–40. [DOI] [PubMed] [Google Scholar]
- Cosenza MR, Cazzola A, Rossberg A, Schieber NL, Konotop G, Bausch E, Slynko A, Holland-Letz T, Raab MS, Dubash T, et al. (2017). Asymmetric centriole numbers at spindle poles cause chromosome missegregation in cancer. Cell Rep , 1906–1920. [DOI] [PubMed] [Google Scholar]
- Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D. (2012). DNA breaks and chromosome pulverization from errors in mitosis. Nature , 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl KD, Sankaran DG, Bayless BA, Pinter ME, Galati DF, Heasley LR, Giddings TH, Pearson CG. (2015). A short CEP135 splice isoform controls centriole duplication. Curr Biol , 2591–2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Li T, Bai Z, Yang Y, Liu X, Zhan J, Shi B. (2015). Breast cancer intrinsic subtype classification, clinical use and future trends. Am J Cancer Res , 2929–2943. [PMC free article] [PubMed] [Google Scholar]
- D’Assoro AB, Barrett SL, Folk C, Negron VC, Boeneman K, Busby R, Whitehead C, Stivala F, Lingle WL, Salisbury JL. (2002). Amplified centrosomes in breast cancer: a potential indicator of tumor aggressiveness. Breast Cancer Res Treat , 25–34. [DOI] [PubMed] [Google Scholar]
- David CJ, Manley JL. (2010). Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev , 2343–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denu RA, Zasadil LM, Kanugh C, Laffin J, Weaver BA, Burkard ME. (2016). Centrosome amplification induces high grade features and is prognostic of worse outcomes in breast cancer. BMC Cancer , 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi J, Penland L, Brown PO, Bittner ML, Meltzer PS, Ray M, Chen Y, Su YA, Trent JM. (1996). Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet , 457–460. [DOI] [PubMed] [Google Scholar]
- Di Giammartino DC, Nishida K, Manley JL. (2011). Mechanisms and consequences of alternative polyadenylation. Mol Cell , 853–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duensing S. (2005). A tentative classification of centrosome abnormalities in cancer. Cell Biol Int , 352–359. [DOI] [PubMed] [Google Scholar]
- Duensing A, Liu Y, Perdreau SA, Kleylein-Sohn J, Nigg EA, Duensing S. (2007). Centriole overduplication through the concurrent formation of multiple daughter centrioles at single maternal templates. Oncogene , 6280–6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu A, et al. (2008). Stromal gene expression predicts clinical outcome in breast cancer. Nat Med , 518–527. [DOI] [PubMed] [Google Scholar]
- Firat-Karalar EN, Stearns T. (2014). The centriole duplication cycle. Philos Trans R Soc Lond B Biol Sci , 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, Pellman D. (2009). A mechanism linking extra centrosomes to chromosomal instability. Nature , 278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glück S, Ross JS, Royce M, McKenna EF, Perou CM, Avisar E, Wu L. (2012). TP53 genomics predict higher clinical and pathologic tumor response in operable early-stage breast cancer treated with docetaxel-capecitabine ± trastuzumab. Breast Cancer Res Treat , 781–791. [DOI] [PubMed] [Google Scholar]
- Godinho S, Kwon M, Pellman D. (2009). Centrosomes and cancer: how cancer cells divide with too many centrosomes. Cancer Metastasis Rev , 85–98. [DOI] [PubMed] [Google Scholar]
- Godinho SA, Picone R, Burute M, Dagher R, Su Y, Leung CT, Polyak K, Brugge JS, Théry M, Pellman D, et al. (2012). Centriole maturation requires regulated Plk1 activity during two consecutive cell cycles. Nat Cell Biol , 855–865. [Google Scholar]
- Godinho SA, Picone R, Burute M, Dagher R, Su Y, Leung CT, Polyak K, Brugge JS, Théry M, Pellman D. (2014). Oncogene-like induction of cellular invasion from centrosome amplification. Nature , 167–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Gao M, Ma J, Xiao T, Zhao L, Gao Y, Pan Q. (2007). Analysis of the cellular centrosome in fine-needle aspirations of the breast. Breast Cancer Res , R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirono M. (2014). Cartwheel assembly. Philos Trans R Soc Lond B Biol Sci , 20130458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque M, Ji Z, Zheng D, Luo W, Li W, You B, Park JY, Yehia G, Tian B. (2013). Analysis of alternative cleavage and polyadenylation by 3′ region extraction and deep sequencing. Nat Methods , 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerka-Dziadosz M, Gogendeau D, Klotz C, Cohen J, Beisson J, Koll F. (2010). Basal body duplication in paramecium: the key role of Bld10 in assembly and stability of the Cartwheel. Cytoskeleton , 161–171. [DOI] [PubMed] [Google Scholar]
- Johansson I, Aaltonen KE, Ebbesson A, Grabau D, Wigerup C, Hedenfalk I, Rydén L. (2011). Increased gene copy number of KIT and VEGFR2 at 4q12 in primary breast cancer is related to an aggressive phenotype and impaired prognosis. Genes Chromosomes Cancer , 375–383. [DOI] [PubMed] [Google Scholar]
- Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA. (2007). Plk4-induced centriole biogenesis in human cells. Dev Cell , 190–202. [DOI] [PubMed] [Google Scholar]
- Kraatz S, Guichard P, Obbineni JM, Olieric N, Hatzopoulos GN, Hilbert M, Sen I, Missimer J, Gönczy P, Steinmetz MO. (2016). The human centriolar protein CEP135 contains a two-stranded coiled-coil domain critical for microtubule binding. Structure , 1358–1371. [DOI] [PubMed] [Google Scholar]
- Levine MS, Bakker B, Boeckx B, Moyett J, Lu J, Vitre B, Spierings DC, Lansdorp PM, Cleveland DW, Lambrechts D, et al. (2017). Centrosome amplification is sufficient to promote spontaneous tumorigenesis in mammals. Dev Cell , 313–322.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt N, Briggs D, Gil A, Proudfoot NJ. (1989). Definition of an efficient synthetic poly (A) site, Genes Dev , 1019–1025. [DOI] [PubMed] [Google Scholar]
- Lin Y-C, Chang C-W, Hsu W-B, Tang C-JC, Lin Y-N, Chou E-J, Wu C-T, Tang TK. (2013). Human microcephaly protein CEP135 binds to hSAS-6 and CPAP, and is required for centriole assembly. EMBO J , 1141–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingle WL, Barrett SL, Negron VC, D’Assoro AB, Boeneman K, Liu W, Whitehead CM, Reynolds C, Salisbury JL. (2002). Centrosome amplification drives chromosomal instability in breast tumor development. Proc Natl Acad Sci USA , 1978–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes CAM, Mesquita M, Cunha AI, Cardoso J, Carapeta S, Laranjeira C, Pinto AE, Pereira-Leal JB, Dias-Pereira A, Bettencourt-Dias M, Chaves P. (2018). Centrosome amplification arises before neoplasia and increases upon p53 loss in tumorigenesis. J Cell Biol , 2353–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly P, Cleveland DW. (2017). Rebuilding chromosomes after catastrophe: emerging mechanisms of chromothripsis. Trends Cell Biol , 917–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marteil G, Guerrero A, Vieira AF, De Almeida BP, Machado P, Mendonça S, Mesquita M, Villarreal B, Fonseca I, Francia ME, et al. (2018). Over-elongation of centrioles in cancer promotes centriole amplification and chromosome missegregation. Nat Commun , 1258.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinho O, Longatto-Filho A, Lambros MBK, Martins A, Pinheiro C, Silva A, Pardal F, Amorim J, MacKay A, Milanezi F, et al. (2009). Expression, mutation and copy number analysis of platelet-derived growth factor receptor A (PDGFRA) and its ligand PDGFA in gliomas. Br J Cancer , 973–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura K, Lefebvre PA, Kamiya R, Hirono M. (2004). Bld10p, a novel protein essential for basal body assembly in Chlamydomonas: localization to the cartwheel, the first ninefold symmetrical structure appearing during assembly. J Cell Biol , 663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. (2009). Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell , 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira A, Wollerton M, Monks J, Proudfoot NJ. (1995). Upstream sequence elements enhance poly(A) site efficiency of the C2 complement gene and are phylogenetically conserved. EMBO J , 3809–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam DK, Lee S, Zhou G, Cao X, Wang C, Clark T, Chen J, Rowley JD, Wang SM. (2002). Oligo(dT) primer generates a high frequency of truncated cDNAs through internal poly(A) priming during reverse transcription. Proc Natl Acad Sci USA , 6152–6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, et al. (2006). A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell , 515–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA. (2002). Centrosome aberrations: cause or consequence of cancer progression? Nat Rev Cancer , 815–825. [DOI] [PubMed] [Google Scholar]
- Nigg EA. (2006). Origins and consequences of centrosome aberrations in human cancers. Int J Cancer , 2717–2723. [DOI] [PubMed] [Google Scholar]
- Nigg EA, Stearns T. (2011). The centrosome cycle: centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol , 1154–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes NM, Li W, Tian B, Furger A. (2010). A functional human poly(A) site requires only a potent DSE and an A-rich upstream sequence. EMBO J , 1523–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Ashikawa T, Nozaki Y, Kozuka-Hata H, Goto H, Inagaki M, Oyama M, Kitagawa D. (2014). Direct interaction of Plk4 with STIL ensures formation of a single procentriole per parental centriole. Nat Commun , 5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T, Essner R, Ryu JH, Palazzo RE, Uetake Y, Kuriyama R. (2002). Characterization of Cep135, a novel coiled-coil centrosomal protein involved in microtubule organization in mammalian cells. J Cell Biol , 87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel M, Meyer P, Khodjakov A, Rieder CL, Bornens M. (2000). The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J Cell Biol , 317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pihan GA. (2013). Centrosome dysfunction contributes to chromosome instability, chromoanagenesis, and genome reprograming in cancer. Front Oncol , 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pihan GA, Wallace J, Zhou Y, Doxsey SJ. (2003). Centrosome abnormalities and chromosome instability occur together in pre-invasive carcinomas. Cancer Res , 1398–1404. [PubMed] [Google Scholar]
- Quintyne NJ, Reing JE, Hoffelder DR, Gollin SM, Saunders WS, Quintyne NJ, Reing JE, Hoffelder DR, Gollin SM, Saunders WS. (2005). Spindle multipolarity is prevented by centrosomal clustering. Science , 127–129. [DOI] [PubMed] [Google Scholar]
- Salisbury JL, D’Assoro AB, Lingle WL. (2004). Centrosome amplification and the origin of chromosomal instability in breast cancer. J Mammary Gland Biol Neoplasia , 275–283. [DOI] [PubMed] [Google Scholar]
- Schneeweiss A, Sinn H-P, Ehemann V, Khbeis T, Neben K, Krause U, Ho AD, Bastert G, Krämer A. (2003). Centrosomal aberrations in primary invasive breast cancer are associated with nodal status and hormone receptor expression. Int J Cancer , 346–352. [DOI] [PubMed] [Google Scholar]
- Silkworth WT, Nardi IK, Scholl LM, Cimini D. (2009). Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS One , e6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluder G, Nordberg JJ. (2004). The good, the bad and the ugly: the practical consequences of centrosome amplification. Curr Opin Cell Biol , 49–54. [DOI] [PubMed] [Google Scholar]
- Strnad P, Leidel S, Vinogradova T, Euteneuer U, Khodjakov A, Gönczy P. (2007). Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev Cell , 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Bakhoum SF, Compton DA. (2010). Mechanisms of chromosomal instability. Curr Biol , R285–R295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Compton DA. (2011). Chromosome missegregation in human cells arises through specific types of kinetochore-microtubule attachment errors. Proc Natl Acad Sci USA , 17974–17978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Pan Z, Ju YL. (2007). Widespread mRNA polyadenylation events in introns indicate dynamic interplay between polyadenylation and splicing. Genome Res , 156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou MFB, Stearns T. (2006a). Controlling centrosome number: licenses and blocks. Curr Opin Cell Biol , 74–78. [DOI] [PubMed] [Google Scholar]
- Tsou MFB, Stearns T. (2006b). Mechanism limiting centrosome duplication to once per cell cycle. Nature , 947–951. [DOI] [PubMed] [Google Scholar]
- Tuupanen S, Hänninen UA, Kondelin J, von Nandelstadh P, Cajuso T, Gylfe AE, Katainen R, Tanskanen T, Ristolainen H, Böhm J, et al. (2014). Identification of 33 candidate oncogenes by screening for base-specific mutations. Br J Cancer , 1657–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobjev IA, Chentsov YS. (1982). Centrioles in the cell cycle. I. Epithelial cells. J Cell Biol , 938–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Kanaan Y, Baed YK, Gabrielson E. (2009). Chromosomal changes in aggressive breast cancers with basal-like features. Cancer Genet Cytogenet , 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CZ, Spektor A, Cornils H, Francis JM, Jackson EK, Liu S, Meyerson M, Pellman D. (2015). Chromothripsis from DNA damage in micronuclei. Nature , 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Liu X, Tian B. (2016). 3′READS+, a sensitive and accurate method for 3′ end sequencing of polyadenylated RNA. RNA , 1631–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.