Abstract
Protein mutations can result in dysfunctional cell signaling pathways; therefore it is of significance to develop a robust platform for the detection of protein mutations. Here, we report that the channel of bacterial virus T7 DNA packaging motor is able to discriminate peptides containing a mixture of acidic (negatively charged) and basic (positively charged) amino acids. Peptides were differentiated based on their current signatures created by their unique charge compositions. In combination with protease digestion, peptides with the locational differences of single amino acid were also identified. The results suggest that the T7 motor channel has the potential for peptide differentiation, mutation verification, and analysis of protein sequence.
Keywords: Bacteriophage DNA packaging motor, T7 channel, Nanopore, Analysis of protein sequence, Detection of protein mutation, Biomotor
1. Introduction
The sequence and structure of proteins play critical roles in living systems. Sequence modification or mutation can lead to dysregulation of cell signaling, alternation of protein function and the development of diseases, including mental health problems, cancer, neurodegenerative, and autoimmune diseases [1–6]. Therefore, it is necessary to develop an easy and reliable method to study or detect protein mutations. Nanopore sensing, an emerging technology, provides an approach to detect or identify macromolecules at a single molecule level [7–10]. There are two major classes of nanopores, solid-state and biological nanopores. Compared with solid-state nanopores, biological nanopores are more homogenous and amendable owing to the intrinsic properties of proteins [11–15]. The principle of sensing using biological nanopores is that macromolecules pass through a channel inserted into an artificial membrane, resulting in the generation of electric signatures reflecting the shape, size and charge of analytes [8,11,12,16].
The feasibility of peptide and protein sensing using nanopore technology has been extensively investigated in many studies [17–28]. Solid-state nanopores, which are relatively easy to fabricate pores with large diameters, have been developed as a great platform for the study of protein detection [19,23,27,29], translocation dynamics of native proteins [25], protein-DNA interactions [29–31], as well as the structure calculations of proteins [20,32]. However, the pore size of almost all currently reported biological nanopores investigated for sensing purposes is smaller than 4 nm [11], making it difficult to investigate native protein structuring. Only several studies have been reported of the detection of small native protein, including Frac [17] and ClyA [18]. Due to smaller pore size of biological nanopores, biological nanopores are more viable candidates for the sensing of peptides and denatured proteins. For example, the translocation and differentiation of various peptides [22,33,34], detection of peptide oligomerization [24,35], discrimination of different types of amino acids [36,37], and others [38–40] have been previously investigated.
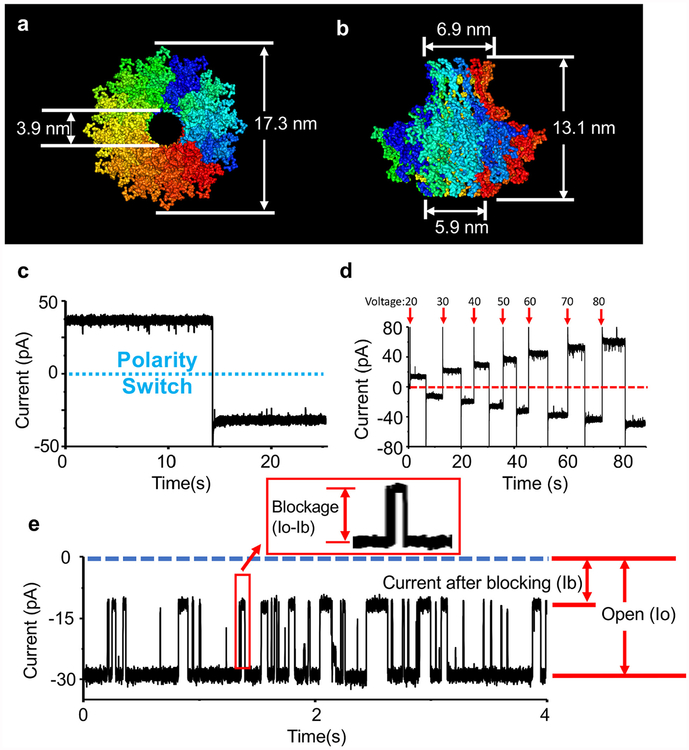
The DNA translocation channels from bacterial virus DNA packaging motors, such as phi29, SPP1, T3, T4, and T7, play an important role in the process of biogenesis or host infection [41–46]. These viral motor channels can be inserted into lipid membranes and utilized as a nanopore biosensor (Fig. 1) [47]. The application of viral motor channels now has been expanded to the sensing of RNA [48], chemicals [49], peptides [22,23] and proteins [50]. Recently, there was a great breakthrough for nanopore-based peptide sensing using T7 viral motor channel [36]. Poly-arginine peptides with different lengths were discriminated by the channel of bacterial virus T7 DNA packaging motor based on current blockage and dwell time. The T7 viral motor channel is assembled by multiple subunits (each subunit size: 59 K Da), and expected to have a similar pore size as phi29 channel. The narrow restriction is 3.9 nm based on the results of cryo-electron microscopy with a resolution of 12 Å(PDB:3J4A; Fig. 1a and b) [51,52]. In comparison with previously reported aerolysin nanopore, requiring a peptide concentration of several hundred or thousand nanomolar [37]; the T7 nanopore was able to distinguish peptides with different lengths at a few dozen nanomolar [36]. This allows for the detection of lower concentration peptides that may be seen in clinical applications and provide a more positive identification of peptide to electrical signal due to less blockage events. Herein, we applied T7 viral motor channel to distinguish the same-length peptides with the mixture of positively and negatively charged amino acids. In combination with a protease digestion assay, peptides with the locational difference of a single amino acid were able to be discriminated.
Fig. 1.
Illustration and characterization of the channel of T7 bacterial virus DNA packaging motor. (a–b) Top (a) and side (b) view of T7 channel and its three dimensional size with 12 Å resolution (PDB:3J4A). (c) A single inserted T7 channel into a lipid bilayer membrane (Buffer: 0.15 M KCl, 5 mM HEPES, pH 7.4. Voltage: ± 50 mV). (d) Current trace of single T7 channel under different voltages (± 20 mV, ± 30 mV, ± 40 mV, ± 50 mV, ± 60 mV, ± 70 mV, ± 80 mV). The arrows (red color) indicate the applied potentials. (e) Illustration of the measurement of current blockage in this study. The blockage (%) is defined as (Io − Ib) ÷ Io ×100%. Ib represents the current after blocking with peptide translocation and Io represents the open channel current without analyte translocation. The recording time for the plotted data of blockage histogram and scatter plot in this study was 5 min.
2. Materials and methods
Materials.
All peptides in this study were synthesized and purified by Genescript. 1, 2-diphytanoyl-sn-glycero-3-phosphocholine (DPhPC) was obtained from Avanti Polar Lipids. Endoprotease Lys-C was purchased from New England Biolabs (NEB). All other chemicals were from Fisher Scientific, if not specified. A list of peptides below was used in this study (Table 1.):
R12: RRR RRR RRR RRR;
RD3: RRD RRR RRR RRR;
RE3: RRE RRR RRR RRR;
RMM: RRR RRR RKE RDR;
RK2: RKR RRR RRR RRR;
RK3: RRK RRR RRR RRR;
RK4: RRR KRR RRR RRR.
Table 1.
Peptides used in this study. Isoelectric point was calculated from the website (https://pepcalc.com/).
| Name | Sequence | Molecular weight (g/mol) | Isoelectric point (pI) | Net charge at pH 7 |
|---|---|---|---|---|
| R12 | RRRRRRRRRRRR | 1892.25 | 13.14 | 12 |
| RD3 | RRDRRRRRRRRR | 1851.15 | 12.75 | 10 |
| RE3 | RRERRRRRRRRR | 1865.18 | 12.75 | 10 |
| RMM | RRRRRRRKERDR | 1796.06 | 12.4 | 8 |
| RK2 | RKRRRRRRRRRR | 1864.23 | 13.1 | 12 |
| RK3 | RRKRRRRRRRRR | 1864.23 | 13.1 | 12 |
| RK4 | RRRKRRRRRRRR | 1864.23 | 13.1 | 12 |
Nanopore experiments.
The protein purification and liposome preparation of the channel of bacterial virus T7 DNA packaging motor were described previously (see Ref. [36]). A free-standing lipid bilayer was formed on the Teflon membrane, which separated the whole compartment to two sub-compartments, cis- and trans-chamber. A pair of Ag/AgCl were placed at the cis- and trans-chamber for the connection with patch clamp instruments. Both chambers were filled with conductance buffer (0.15 M KCl, 5 mM HEPES, pH 7.4). Bilayer Clamp Amplifier BC-535 (Warner Instruments) was connected to Axon Digi-Data 1440 A analog-digital converter (Molecular Devices). Data was recorded at bandwidth 1 KHz and sampling frequency 20 KHz by Clampex 10 (Molecular Devices). The Clampfit 10 (Molecular Devices), MOSAIC [53] and Origin 8 (Origin Lab Corporation) were used to read and analyze data. The blockage was normalized by the equation (Io − Ib) ÷ Io × 100% and Io represents the current without peptide translocation and Ib represents the current after blocking for peptide translocation. The blockage peak was fit by single or multiple Gaussian distributions. Cis-chamber buffer volume: 600 μl; trans-chamber buffer volume: 1800 μl.
Peptide digestion assay.
The digestion assay followed the protocol supplied with Lys-C. Several components were mixed on ice with a total volume 10 μl for digestion reaction in a PCR tube: peptide (1 μl 1 mg/ml), Lys-C enzyme (3 μl 100 ng/μl) and 6 μl dd H2O. Then the digestion reaction was incubated at 37 °C for 1 h and then it was heated at 95 °C for 10 min to inactivate enzyme using thermal cycler. Finally, the digestion reaction was cooled down to 4 °C before running nanopore experiments. The amount of added digested materials for nanopore experiments was 0.1 μl digestion reaction per 100 μl conductance buffer (0.15 M KCl, 5 mM HEPES, pH 7.4).
Lys-C control experiment.
The digestion assay followed the protocol supplied with Lys-C. Several components were mixed on ice with a total volume 10 μl for the reaction in a PCR tube: Lys-C enzyme (3 μl 100 ng/μl) and 7 μl ddH2O. Then the control reaction was incubated at 37 °C for 1 h and heated at 95 °C for 10 min to inactivate enzyme using thermal cycler. Finally, the control reaction was slowly cooled down to 4 °C. After single T7 channel was inserted into a lipid bilayer membrane, the control reaction with different volume was added and mixed in cis-chamber. The data was recorded under ± 50 mV. It was repeated for three times. Buffer: 0.15 M KCl, 5 mM HEPES, pH 7.4. Buffer volume in cis-chamber: 600 μl; Buffer volume in trans-chamber: 1800 μl.
Peptide translocation experiments.
Peptide was mixed with conductance buffer (0.15 M KCl, 5 mM HEPES, pH 7.4), or peptide was added and mixed after single T7 channel insertion. The electrical signals were recorded under −50 mV. All nanopore experiments for peptide translocation have been repeated for several times. The final concentration of tested peptides in this study was from 40 to 65 nM.
3. Results and discussion
3.1. The discrimination of peptides with the mixture of positively and negatively charged amino acids
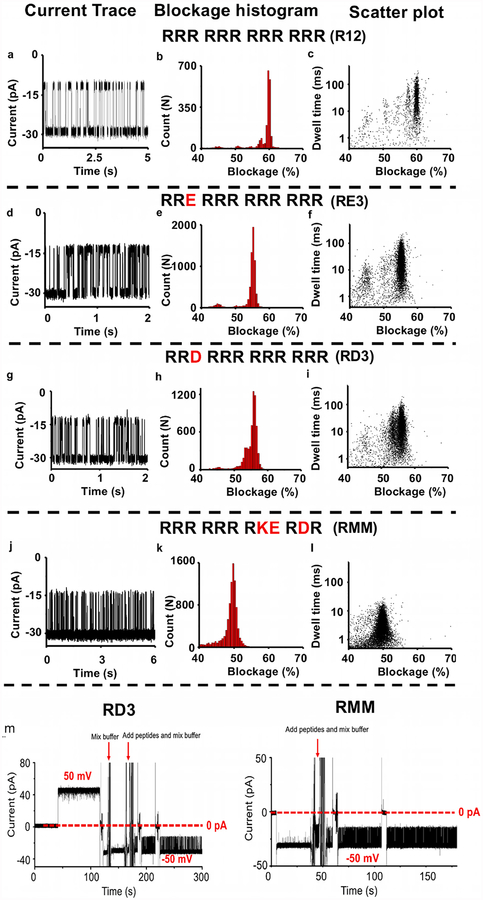
Our previous study shows that the channel of T7 DNA packaging motor can discriminate poly-arginine peptides with a single arginine difference based on different blockage peaks of each individual peptide under the buffer (0.15 M KCl, 5 mM HEPES, pH 7.4) [36]. Longer length peptides generate larger blockage peaks and longer dwell time; however, it was previously unknown whether T7 channel could discriminate peptides composed of a mixture of positively and negatively charged amino acids while retaining the same length. To answer this question, the translocation experiments for a variety of peptide mutations were tested using the T7 channel as a nanopore. In this study, we used the same buffer conditions as the previous study [36] containing 0.15 M KCl in which T7 channel was stable and quiescent under ± 50 mV over long periods of time and has relatively high insertion efficacy. Fig. 1 c shows an electrical current trace of a single T7 channel within the buffer containing 0.15 M KCl under ± 50 mV and Fig. 1 d shows the current trace for the T7 channel under different voltages. Varying peptides were mixed in conductance buffer or added after the insertion of single T7 channel. The method of measuring current blockage is presented in Fig. 1 e. In our study, the blockage (%) is defined as (Io ‒ Ib) ÷ Io × 100%. Ib represents the current after blocking during peptide translocation and Io represents the open channel current. The blockage peaks of R12 (RRR RRR RRR RRR; Fig. 2 a–c), RE3 (RRE RRR RRR RRR; Fig. 2 d–f), RD3 (RRD RRR RRR RRR; Fig. 2 g–i), and RMM (RRRRRRRKERDR; Fig. 2 j–l) were59.9% ± 0.4%, 55.2% ± 0.6%, 55.9% ± 0.8%, 49.7% ± 1.0%, respectively. The T7 channel itself did not cause these blockage peaks (Fig. 2 m). Fig. 2 m shows the full current traces of two examples, RD3 and RMM. After the insertion of single T7 channel under 50 mV, the voltage was switched to −50 mV. To avoid further insertions, cis-chamber buffer was mixed and disturbed. Then RD3 peptides were added and mixed in cis-chamber under the holding potential −50 mV; and a burst of signals representing RD3 were observed under −50 mV. The peptide RMM was added and mixed in cis-chamber under the holding potential −50 mV before the RMM signals were observed. These results show that a single mutation of either glutamic acid or aspartic acid within the arginine (RD3 and RE3) reduced the blockage peak from ~60% to ~55%. As more arginine amino acids within a peptide were changed to glutamic acid or aspartic acid (RMM), the blockage peaks were reduced further and further.
Fig. 2.
Discrimination of peptides containing a mixture of positively and negatively charged amino acids. (a–l) The profiles of current trace, blockage histogram, scatter plot of R12 (RRR RRR RRR RRR; a-c), RE3 (RRE RRR RRR RRR; d-f), RD3 (RRD RRR RRR RRR; g-i) and RMM (RRR RRR RKE RDR; j-l). (m) Full current trace before and after the addition of RD3 (Left) and RMM (Right). The blockage histogram is fitted by single Gaussian distribution. Buffer: 0.15 M KCl, 5 mM HEPES, pH7.4. Voltage: −50 mV.
The physical/chemical properties of tested analytes can affect current blockages. When analytes with different size pass through the nanopore channel, they partially block the ionic flow through the nanopore, and result in different blocked currents. It is expected that the length of the charged peptides, sequence and structures of the peptides, which occupied a part of the narrowest portion of the channel, could affect the current signals. By analyzing the generated data, it was found that isoelectric point of analytes also played a role in current blockages. Each of the short peptides used in this study showed similar secondary structure, predicted by PEP-FOLD [54,55], while RE3 and RK3 (RK3: RRK RRR RRR RRR; Fig. 3c) showed significantly different current blockage events (55.2% ± 0.6% and 59.4% ± 0.5%, respectively). It seemed the difference in isoelectric points (pI) of RE3 and RK3 (12.75 and 13.1, respectively) was the main difference between the peptides as the molecular weights of RE3 and RK3 are very similar (1865 g/mol and 1864 g/mol, respectively). Additionally, R12 and RE3 displayed roughly a 5% difference of current blockage although these two peptides have molecular weights with 27 Da difference and pI with 0.39 difference. R12 and RK3 showed similar current blockages ~60%, although those two peptides show similar pIs (13.14 and 13.1) and 28 Da difference of molecular weight. These results indicate that peptide pI may play an important role in determining current blockages.
Fig. 3.
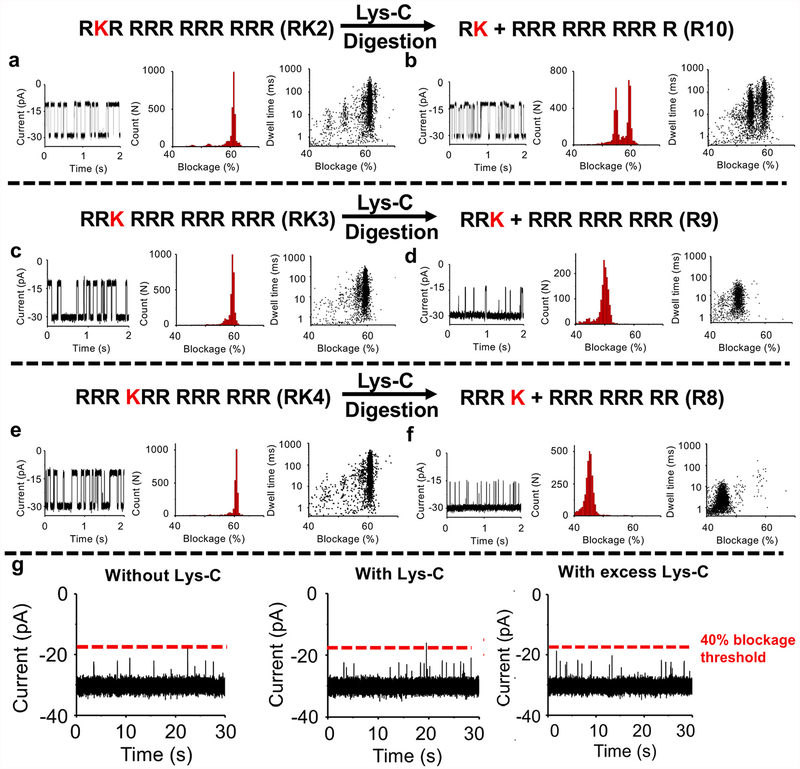
Identification and discrimination of three peptides (RK2 (RKR RRR RRR RRR), RK3 (RRK RRR RRR RRR) and RK4 (RRR KRR RRR RRR)) with a single amino acid mutation via protease Lys-C cleavage. A profile of ionic signatures (current trace, blockage histogram, scatter plot) of RK2 (a), RK3 (c) and RK4 (e) before Lys-C digestion were shown. A profile of ionic signatures (current trace, blockage histogram, scatter plot) of digested RK2 (b), RK3 (d) and RK4 (f) via Lys-C cleavage were shown. (g) Current traces of Lys-C control experiment. Left: Current trace before the addition of Lys-C control reaction. Middle: Sample current trace during 10 min post addition of the Lys-C control reaction (0.05 μl reaction per 100 μl conductance buffer). Right: Sample current trace during 19 min post addition of excess Lys-C control reaction (0.5 μl reaction per 100 μl conductance buffer). The line (red color) indicates the blocked current with 40% blockage. The amount of digested peptides in the chamber was 0.1 μl digestion reaction per 100 μl conductance buffer. The blockage histogram is fitted by single or two Gaussian distributions. Buffer:0.15 M KCl, 5 mM HEPES, pH 7.4. Voltage: −50 mV.
3.2. The discrimination of peptides with the locational difference of single amino acid
We investigated whether the channel of bacterial virus T7 DNA packaging motor can discriminate or identify peptides with the same amino acid, but at different locations within the peptide. Three peptides (RK2: RKR RRR RRR RRR; RK3: RRK RRR RRR RRR; RK4: RRR KRR RRR RRR) were tested for proof-of-concept study, and each individual peptide was driven through T7 channel by electric force (Fig. 3). Each peptide is of the same molecular weight, isoelectric point and net charge. It was found that T7 channel could not discriminate RK2 (Fig. 3a), RK3 (Fig. 3c) and RK4 (Fig. 3e) based on the similar blockage peaks of those three peptides, 61.2% ± 0.4%, 59.4% ± 0.5%, and 60.7% ± 0.4%, respectively. Thus, an alternative approach using protease digestion was taken for the discrimination of peptides with the locational difference of single amino acid detection.
Lys-C protease is an enzyme that specifically digests the carboxyl of lysine and utilized for peptide or protein digestion for the purpose of sequencing [56,57]. To discriminate RK2, RK3 and RK4, Lys-C digestion was performed for all three peptides and then the shorter, digested peptides were translocated through the T7 channel. Prior to the translocation experiments, individual peptides were incubated with Lys-C protease at 37 °C for 1 h and then the protease was inactive at 95 °C for 10 min and cooled down to 4 °C. As shown in Fig. 3, we observed differences in each of the newly-appeared blockage peaks of digested RK2 (Fig. 3b), digested RK3 (Fig. 3d) and digested RK4 (Fig. 3f):54.7% ± 0.5%, 50.1% ± 1.0%, and 45.3% ± 1.1%, respectively. The data for the digested peptides with different lengths are in agreement with the previous studies (see Ref. [36]). Peptides of less than eight amino acid could not be detected by T7 channel [36]. That being said, the signals of cleaved peptides of RK, RRK and RRRK could not be observed or detected. It was noted that there were two blockage peaks fitted by two Gaussian distributions for the digested RK2, 54.7% ± 0.5% and 59.5% ± 0.5%, indicating that RK2 was not fully digested. The 59.5% ± 0.5% blockage peak had the same profile of undigested RK2. Lys-C only as a control experiment was also tested and it was found that Lys-C protease did not produce any blockage signals than 40% (Fig. 3 g). The reaction containing Lys-C only was performed by using the same activation and inactivation procedure before nanopore experiments. Even when the total amount of Lys-C protease in the chamber was increased by five times (0.5 μl digestion reaction per 100 μl conductance buffer; Fig. 3 g, Right), it still did not affect the sensing results. Owing to high specificity of Lys-C protease, the lysine at different positions within peptides can be distinguished based on the changes of the blockage peaks before and after digestion assays.
Understanding the process of protein assembly into three dimensional structures is of high significance for a better understanding of protein function within cell signaling pathways. Point mutations of proteins at defined locations can result in failure of protein folding, assembly, and even functional activity. Proteins are generally composed of around twenty different amino acids; however, detection of individual amino acids is difficult due to different physical/chemical properties for each amino acid, including molecular weight, net charge, isoelectric point, solubility, hydrophilicity and hydrophobicity. Nanopore technology mainly relies on the translocation of analytes driven by electric force, and direct detection or sequencing of the proteins composed of twenty unique amino acids by nanopore platform remains highly challenging. Therefore, it is necessary to combine other technologies with the nanopore platform for peptide or protein sensing, such as host-guest interactions, chemical modification of proteins, enzymatic digestion assays, channel modifications. Further alternative approaches are required to be tested in the future.
4. Conclusions
The reengineered channel from bacterial virus T7 DNA bacteriophage motor was used for the discrimination of peptides composed of a mixture of acidic (negatively charged) and basic (positively charged) amino acids. Further, peptides with the locational difference of a single lysine amino acid can be detected and discriminated by measuring the changes of the current blockage peaks for each peptide before and after the addition of specific protease. These findings suggest that T7 channel can play an important role in the detection of protein mutation. Our results promote the insights into the studies of proteomics using nanopore technology.
Acknowledgements
We would like to thank Daniel W. Binzel and Dana Driver for their modifications of this manuscript. The research was supported by NIH grant R01 EB012135 and Oxford Nanopore Technologies Ltd.
Footnotes
Data availability statement
The data that supports the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of interest
P.G.’s Sylvan G. Frank Endowed Chair position in Pharmaceutics and Drug Delivery is funded by the CM Chen Foundation. P.G. is the consultant of Oxford Nanopore Technologies; the cofounder of Shenzhen P&Z Bio-medical Co. Ltd and its subsidiary US P&Z Biological Technology LLC, as well as cofounder of ExonanoRNA, LLC and its subsidiary Weina Biomedical LLC in Foshan.
References
- [1].McDaniell R, Warthen DM, Sanchez-Lara PA, Pai A, Krantz ID, Piccoli DA, et al. , NOTCH2 mutations cause Alagille syndrome, a heterogeneous disorder of the notch signaling pathway, Am. J. Hum. Genet 79 (1) (2006) 169–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Quist A, Doudevski I, Lin H, Azimova R, Ng D, Frangione B, et al. , Amyloid ion channels: a common structural link for protein-misfolding disease, Proc. Natl. Acad. Sci. U. S. A 102 (30) (2005) 10427–10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Katoh M, Katoh M, WNT signaling pathway and stem cell signaling network, Clin. Cancer Res 13 (14) (2007) 4042–4045. [DOI] [PubMed] [Google Scholar]
- [4].Logan CY, Nusse R, The Wnt signaling pathway in development and disease, Annu. Rev. Cell Dev. Biol 20 (2004) 781–810. [DOI] [PubMed] [Google Scholar]
- [5].Valastyan JS, Lindquist S, Mechanisms of protein-folding diseases at a glance, Dis Model Mech 7 (1) (2014) 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].le-Donne I, Aldini G, Carini M, Colombo R, Rossi R, Milzani A, Protein carbonylation, cellular dysfunction, and disease progression, J. Cell Mol. Med 10 (2) (2006) 389–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lee K, Park KB, Kim HJ, Yu JS, Chae H, Kim HM, et al. , Recent progress in solid-state nanopores, Adv. Mater 30 (42) (2018) 1704680. [DOI] [PubMed] [Google Scholar]
- [8].Haque F, Li J, Wu H-C, Liang X-J, Guo P, Solid-state and biological nanopore for real-time sensing of single chemical and sequencing of DNA, Nano Today 8 (2013) 56–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Deamer D, Akeson M, Branton D, Three decades of nanopore sequencing, Nat. Biotechnol 34 (5) (2016) 518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Dekker C, Solid-state nanopores, Nat. Nanotechnol 2 (2007) 209–215. [DOI] [PubMed] [Google Scholar]
- [11].Wang S, Zhao Z, Haque F, Guo P, Engineering of protein nanopores for sequencing, chemical or protein sensing and disease diagnosis, Curr. Opin. Biotechnol 51 (2018) 80–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Cao C, Long YT, Biological nanopores: confined spaces for electrochemical single-molecule analysis, Acc. Chem. Res 51 (2) (2018) 331–341. [DOI] [PubMed] [Google Scholar]
- [13].Venkatesan BM, Bashir R, Nanopore sensors for nucleic acid analysis, Nat. Nanotechnol 6 (2011) 615–624. [DOI] [PubMed] [Google Scholar]
- [14].Zhang Y, Chen Y, Fu Y, Ying C, Feng Y, Huang Q, et al. , Monitoring tetracycline through a solid-state nanopore sensor, Sci. Rep 6 (2016) 27959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chen K, Juhasz M, Gularek F, Weinhold E, Tian Y, Keyser UF, et al. , Ionic current-based mapping of short sequence motifs in single DNA molecules using solid-state nanopores, Nano Lett 17 (9) (2017) 5199–5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Liu L, Wu HC, DNA-based nanopore sensing, Angew Chem. Int. Ed. Engl 55 (49) (2016) 15216–15222. [DOI] [PubMed] [Google Scholar]
- [17].Huang G, Willems K, Soskine M, Wloka C, Maglia G, Electro-osmotic capture and ionic discrimination of peptide and protein biomarkers with FraC nanopores, Nat. Commun 8 (1) (2017) 935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Soskine M, Biesemans A, Moeyaert B, Cheley S, Bayley H, Maglia G, An engineered ClyA nanopore detects folded target proteins by selective external association and pore entry, Nano Lett 12 (9) (2012) 4895–4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Rosen Christian B., Single-molecule site-specific detection of protein phosphorylation with a nanopore, Nat. Biotechnol 32 (2014) 179–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Waduge P, Hu R, Bandarkar P, Yamazaki H, Cressiot B, Zhao Q, et al. , Nanopore-based measurements of protein size, fluctuations, and conformational changes, ACS Nano 11 (6) (2017) 5706–5716. [DOI] [PubMed] [Google Scholar]
- [21].Plesa C, Kowalczyk SW, Zinsmeester R, Grosberg AY, Rabin Y, Dekker C, Fast translocation of proteins through solid state nanopores, Nano Lett 13 (2) (2013) 658–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ji Z, Wang S, Zhao Z, Zhou Z, Haque F, Guo P, Fingerprinting of peptides with a large channel of bacteriophage Phi29 DNA packaging motor, Small 12 (33) (2016) 4572–4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhou Z, Ji Z, Wang S, Haque F, Guo P, Oriented single directional insertion of nanochannel of bacteriophage SPP1 DNA packaging motor into lipid bilayer via polar hydrophobicity, Biomaterials 105 (2016) 222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wang S, Zhou Z, Zhao Z, Zhang H, Haque F, Guo P, Channel of viral DNA packaging motor for real time kinetic analysis of peptide oxidation states, Biomaterials 126 (2017) 10–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yusko EC, Johnson JM, Majd S, Prangkio P, Rollings RC, Li J, et al. , Controlling protein translocation through nanopores with bio-inspired fluid walls, Nat. Nanotechnol 6 (4) (2011) 253–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Oukhaled A, Bacri L, Pastoriza-Gallego M, Betton JM, Pelta J, Sensing proteins through nanopores: fundamental to applications, ACS Chem. Biol 7 (12) (2012) 1935–1949. [DOI] [PubMed] [Google Scholar]
- [27].Larkin J, Henley RY, Muthukumar M, Rosenstein JK, Wanunu M, High-bandwidth protein analysis using solid-state nanopores, Biophys. J 106 (3) (2014) 696–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Fahie MA, Yang B, Mullis M, Holden MA, Chen M, Selective detection of protein homologues in serum using an OmpG nanopore, Anal. Chem 87 (21) (2015) 11143–11149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bell NA, Keyser UF, Digitally encoded DNA nanostructures for multiplexed, single-molecule protein sensing with nanopores, Nat. Nanotechnol 11 (7) (2016) 645–651. [DOI] [PubMed] [Google Scholar]
- [30].Hornblower B, Coombs A, Whitaker RD, Kolomeisky A, Picone SJ, Meller A, et al. , Single-molecule analysis of DNA-protein complexes using nanopores, Nat. Methods 4 (4) (2007) 315–317. [DOI] [PubMed] [Google Scholar]
- [31].Kowalczyk SW, Hall AR, Dekker C, Detection of local protein structures along DNA using solid-state nanopores, Nano Lett 10 (1) (2010) 324–328. [DOI] [PubMed] [Google Scholar]
- [32].Hu R, Rodrigues JV, Waduge P, Yamazaki H, Cressiot B, Chishti Y, et al. , Differential enzyme flexibility probed using solid-state nanopores, ACS Nano 12 (5) (2018) 4494–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Asandei A, Schiopu I, Iftemi S, Mereuta L, Luchian T, Investigation of Cu2+ binding to human and rat amyloid fragments Abeta (1–16) with a protein nanopore, Langmuir 29 (50) (2013) 15634–15642. [DOI] [PubMed] [Google Scholar]
- [34].Chavis AE, Brady KT, Hatmaker GA, Angevine CE, Kothalawala N, Dass A, et al. , Single molecule nanopore spectrometry for peptide detection, ACS Sens 2 (9) (2017) 1319–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sutherland TC, Long YT, Stefureac RI, Bediako-Amoa I, Kraatz HB, Lee JS, Structure of peptides investigated by nanopore analysis, Nano Lett 4 (7) (2004) 1273–1277. [Google Scholar]
- [36].Ji Z, Kang X, Wang S, Guo P, Nano-channel of viral DNA packaging motor as single pore to differentiate peptides with single amino acid difference, Biomaterials 182 (2018) 227–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Piguet F, Ouldali H, Pastoriza-Gallego M, Manivet P, Pelta J, Oukhaled A, Identification of single amino acid differences in uniformly charged homopolymeric peptides with aerolysin nanopore, Nat. Commun 9 (1) (2018) 966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Rodriguez-Larrea D, Bayley H, Multistep protein unfolding during nanopore translocation, Nat. Nanotechnol 8 (4) (2013) 288–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Mereuta L, Roy M, Asandei A, Lee JK, Park Y, Andricioaei I, et al. , Slowing down single-molecule trafficking through a protein nanopore reveals intermediates for peptide translocation, Sci. Rep 4 (2014) 3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hoogerheide DP, Gurnev PA, Rostovtseva TK, Bezrukov SM, Real-time nanopore-based recognition of protein translocation success, Biophys. J 114 (4) (2018) 772–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hu B, Margolin W, Molineux IJ, Liu J, Structural remodeling of bacteriophage T4 and host membranes during infection initiation, Proc. Natl. Acad. Sci. Unit. States Am 112 (35) (2015) E4919–E4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kemp P, Garcia LR, Molineux IJ, Changes in bacteriophage T7 virion structure at the initiation of infection, Virology 340 (2005) 307–317. [DOI] [PubMed] [Google Scholar]
- [43].Plisson C, White HE, Auzat I, Zafarani A, Sao-Jose C, Lhuillier S, et al. , Structure of bacteriophage SPP1 tail reveals trigger for DNA ejection, EMBO J 26(15) (2007) 3720–3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Catalano CE, Cue D, Feiss M, Virus DNA packaging: the strategy used by phage lambda, Mol. Microbiol 16 (1995) 1075–1086. [DOI] [PubMed] [Google Scholar]
- [45].Lhuillier S, Gallopin M, Gilquin B, Brasiles S, Lancelot N, Letellier G, et al. , Structure of bacteriophage SPP1 head-to-tail connection reveals mechanism for viral DNA gating, Proc. Natl. Acad. Sci. U. S. A 106 (21) (2009) 8507–8512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Lowe J, Ellonen A, Allen MD, Atkinson C, Sherratt DJ, Grainge I, Molecular mechanism of sequence-directed DNA loading and translocation by FtsK, Mol. Cell 31 (4) (2008) 498–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wendell D, Jing P, Geng J, Subramaniam V, Lee TJ, Montemagno C, et al. , Translocation of double-stranded DNA through membrane-adapted phi29 motor protein nanopores, Nat. Nanotechnol 4 (2009) 765–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Geng J, Wang S, Fang H, Guo P, Channel size conversion of Phi29 DNA-packaging nanomotor for discrimination of single- and double-stranded nucleic acids, ACS Nano 7 (4) (2013) 3315–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Haque F, Lunn J, Fang H, Smithrud D, Guo P, Real-time sensing and discrimination of single chemicals using the channel of Phi29 DNA packaging nanomotor, ACS Nano 6 (2012) 3251–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Wang S, Haque F, Rychahou PG, Evers BM, Guo P, Engineered nanopore of Phi29 DNA-packaging motor for real-time detection of single colon cancer specific antibody in serum, ACS Nano 7 (2013) 9814–9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Agirrezabala X, Martin-Benito J, Valle M, Gonzalez JM, Valencia A,Valpuesta JM, et al. , Structure of the connector of bacteriophage T7 at 8A resolution: structural homologies of a basic component of a DNA translocating machinery, J. Mol. Biol 347 (5) (2005) 895–902. [DOI] [PubMed] [Google Scholar]
- [52].Cuervo A, Pulido-Cid M, Chagoyen M, Arranz R, Gonzalez-Garcia VA, Garcia-Doval C, et al. , Structural characterization of the bacteriophage T7 tail machinery, J. Biol. Chem 288 (36) (2013) 26290–26299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Forstater JH, Briggs K, Robertson JW, Ettedgui J, Marie-Rose O, Vaz C, et al. , MOSAIC: a modular single-molecule analysis interface for decoding multistate nanopore data, Anal. Chem 88 (23) (2016) 11900–11907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Maupetit J, Derreumaux P, Tuffery P, PEP-FOLD: an online resource for de novo peptide structure prediction, Nucleic Acids Res 37 (2) (2009) W498–W503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Shen Y, Maupetit J, Derreumaux P, Tuffery P, Improved PEP-FOLD approach for peptide and miniprotein structure prediction, J. Chem. Theory Comput 10 (10) (2014) 4745–4758. [DOI] [PubMed] [Google Scholar]
- [56].Raijmakers R, Neerincx P, Mohammed S, Heck AJ, Cleavage specificities of the brother and sister proteases Lys-C and Lys-N, Chem. Commun 46 (46) (2010) 8827–8829. [DOI] [PubMed] [Google Scholar]
- [57].Glatter T, Ludwig C, Ahrne E, Aebersold R, Heck AJ, Schmidt A, Large-scale quantitative assessment of different in-solution protein digestion protocols reveals superior cleavage efficiency of tandem Lys-C/trypsin proteolysis over trypsin digestion, J. Proteome Res 11 (11) (2012) 5145–5156. [DOI] [PubMed] [Google Scholar]