Abstract
Background
Long non-coding RNAs (lncRNAs) participate in all cancer biology processes of cells. Although functions and associated mechanisms of lncRNAs have been proven in colorectal cancer (CRC), the roles of lncRNA X-inactive specific transcript (XIST) have not been clearly investigated in CRC.
Material/Methods
Expression of XIST was detected by quantitative real-time PCR (qRT-PCR) assay in CRC cell lines and 196 clinical samples. Correlations between XIST expression and CRC clinicopathological features were analyzed. Log-rank test and Kaplan-Meier test were performed to assess and compare the prognoses of patients with higher and lower expression of XIST. The multivariate Cox regression and univariate Cox regression were conducted to evaluate the risk factors for prognosis of CRC.
Results
lncRNA XIST was upregulated in CRC cells lines and tissues (p<0.05). Statistical analysis found high XIST expression was correlated with larger tumor size, N1, M1, and topography lymph node metastasis (TNM) III+IV stage of CRC. Moreover, higher expression of XIST could predict poor progression-free survival (PFS) and poor overall survival (OS) of CRC patients. The M1 stage and high expression of XIST were proven to be independent risk factors for poor prognosis (p<0.05).
Conclusions
XIST is upregulated in CRC and is significantly correlated with CRC clinical progression. lncRNA XIST overexpression predict poor PFS and poor OS for CRC patients. lncRNA XIST can be an independent risk factor for CRC prognosis, and could be a potential therapeutic target and prognostic biomarker for CRC patients.
MeSH Keywords: Colorectal Neoplasms; Prognosis; RNA, Long Noncoding
Background
Colorectal cancer (CRC) is the second most common cancer in females and the third in males, with an estimated 1.4 million new cases and about 693 900 deaths in 2012 [1]. In past decades, the mortality rate of CRC has been reduced by the development of new screening methods and treatment strategies. However, more than 10% of CRC patients are diagnosed at an advanced stage and about 30% of patients diagnosed at an early stage will develop metastatic disease [2]. Moreover, the 5-year survival rate differs greatly between CRC patients with early stage (90%) and advanced stage (12%). Presently, the AJCC TNM staging system is the most frequently used approach for the therapeutic selection and prognostic evaluation [3]. Nevertheless, the TNM staging system lacks sufficient sensitivity in predicting the recurrence rate of localized CRCs following radical surgical resection. Therefore, the underlying mechanisms of CRC must be further explored to find novel prognostic markers and therapeutic targets.
Long non-coding RNAs (lncRNAs), defined as non-protein-coding RNA molecules greater than 200 nt, are important members of the ncRNAs family. Next-generation sequencing has allowed detection of functional mutations of the non-coding genome. The pivotal role of lncRNAs in cell biology and disease progression is being revealed gradually [4–6]. lncRNAs drive many cancer phenotypes and provide signals for malignant transformation [7,8]. Recent evidence of the role and mechanisms of lncRNAs show these molecules as attractive targets for therapeutic intervention [4–8].
lncRNA X-inactive specific transcript (XIST) is considered to be the most important regulator (or gene) for X inactivation in mammals [9]. XIST plays critical roles in the processes of cancer cell proliferation, differentiation, and genomic maintenance [10]. Specifically, due to gene dysregulation caused by heterochromatin instability, XIST may function as an oncogenic molecule in cancer [11]. Moreover, expression of XIST has been proven to be associated with the progression of the cancers and was reported to be dysregulated in multiple non-sex-associated tumors [12–20]. Previous research has reported that XIST gene amplification could be detected in micro-satellite-unstable sporadic human CRC tissue in comparison with paired normal colorectal epithelium [21]. However, the detailed function of XIST in CRC has not been elucidated.
This study aimed to investigate the clinical significance of the lncRNA XIST in the pathogenesis of CRC. The expression of XIST was discovered and its functional role was analyzed. We also assessed the prognostic value of XIST in progression-free survival (PFS) and overall survival (OS) of CRC patients.
Material and Methods
Patients and samples
We initially included 220 patients, but 13 patients did not complete follow-up and 11 patients did not finish the imaging examinations. Therefore, we finally included 196 patients who were diagnosed as CRC and underwent radical surgery at Linyi People’s Hospital from June 2010 to March 2013. None of the enrolled patients had received radiochemical treatments prior to surgery. Cancer tissues and adjacent tissues were immediately isolated after surgery, frozen in liquid nitrogen, and stored at −80°C until use.
The patients were followed up at an out-patient clinic, and progression of disease was assessed primarily by imaging examinations. Informed consent was obtained from all CRC patients. The protocol of this study was approved by the Ethics Committee of Linyi People’s Hospital, Linyi, China.
Cell lines and cell culture
A total of 6 CRC cell lines (LOVO, HT-29, HCT8, HCT116, SW480, and DLD1) and normal human colon epithelial cells (HCoEpics) were used to define the level of XIST in CRC. These cells were obtained from Shanghai Institute of Biological Sciences (Shanghai, China). The HT-29, LOVO, and DLD1 cells were cultured in RPMI-1640, and the HCT116, HCT8, and SW480 cells were cultured in DMEM, supplemented with 10% FBS (Gibco BRL. Co., Grand Island, NY) and 1% penicillin-streptomycin (Beyotime Biotech. Shanghai, China) in an atmosphere with 5% CO2 at 37°C.
Quantitative real-time PCR (qRT-PCR) assay
The RNAs were extracted from the CRC cancer cells or the cancer tissues using TRIzol reagents (Invitrogen, NY). Extracted total RNA (2 μg) was used as the template for complementary DNA (cDNA) reverse transcription using a PrimeScript RT Reagent Kit (Cat. No. RR037A, Takara Biotech, Dalian, China). According to the manufacturer’s instructions, qRT-PCR assay was conducted using SYBR Premix Ex Taq (Cat. No. DRR041A, Takara Biotech, Dalian, China) on an ABI 7500 system (Applied Biosystems, CA). Relative expression level of XIST was calculated as 2−ΔΔCT values, normalized to glyceraldehydes-3-phosphate dehydrogenase (GAPDH) gene. The sequences of the primers used in this study were: human XIST: 5′-GCATAACTCGGCTTAGGGCT-3′ (forward), anti-sense: 5′-TCCTCTGCCTGACCTGCTAT-3′(reverse); human GAPDH: sense: 5′-CTCTGC TCCTCCTGTTCGAC-3′ (forward), anti-sense: 5′-ACCAAATCCGTTGACTCCGA-3′ (reverse).
Statistical analysis
All statistical analyses were conducted with SPSS 17.0 software (SPSS, Inc., Chicago, IL). Data are represented as mean values ± standard deviations (mean ±SD). Statistical differences of qualitative data between groups were compared by χ2 test. The t test was performed to define the difference in quantitative data. Tukey’s post hoc test was used to validate the ANOVA for comparing measurement data between groups. Survival curves were calculated with Kaplan-Meier method, and differences between different groups were assessed with the log-rank test. The median of XIST expression across all samples was used as a cut-off (T/A values in this study) for the survival analysis. The cut-off values were determined by comparing XIST expression in CRC tissues with adjacent tissues. The negative cut-off value represented the lower expression of XIST compared to the cut-off value, and the positive cut-off value represented the higher expression of XIST. Univariate and multivariate Cox regression methods were used to evaluate the effects of variables on survival. p<0.05 indicates statistically significant differences.
Results
lncRNA XIST expressions were enhanced in the CRC cells and tissues
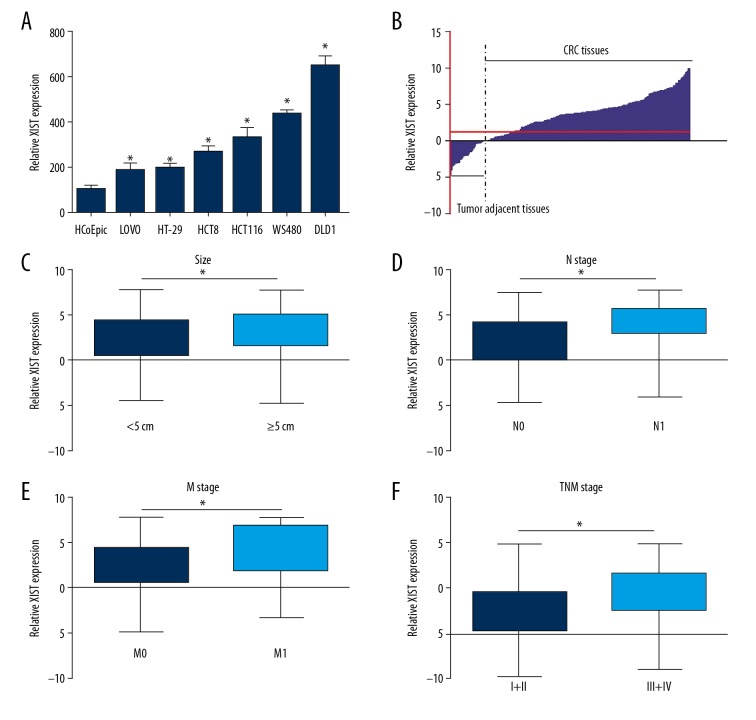
Expressions of lncRNA XIST were evaluated in the CRC cancer cells and cancer tissues to clarify the roles of XIST in the CRC. As shown in Figure 1A, XIST expression was notably higher in CRC cells than in normal human colon epithelial cells (HCoEpics). Expression of XIST was measured in 196 CRC tissues and paired adjacent tissues. The qRT-PCR results showed that XIST was significantly overexpressed in the CRC cancer tissues compared to the associated adjacent tissues (1.00±1.72 vs. 3.25±3.06, p<0.001) (Figure 1B). The upregulation of lncRNA XIST suggested that it may play an oncogenic role in CRC.
Figure 1.
lncRNA XIST was overexpressed in CRC and was associated with CRC clinical progression. XIST expression was evaluated using qRT-PCR assay in the CRC cells and the normal colon cells (A), CRC cancer tissues and the paired or associated tumor adjacent tissues (B), the CRC tumors with size more than or less than 5 cm (C), the CRC tumor samples with N0 state and N1 stage (D), CRC tumor samples with the M0 stage and M1 stage (E), and the CRC tumor samples with the TNM stage I +II and TNM stage III+IV (F). * p<0.05 was calculated with the t test.
Overexpression of lncRNA XIST was correlated with CRC progression
To further verify the role of XIST in CRC, the expression of XIST in patients with different clinicopathological features was detected by qRT-PCR assay. It was discovered that XIST displayed higher expression levels in tumor tissues with tumor size no less than 5 cm, N1 stage, M1 stage, and TNM stage III and IV compared with tumor size less than 5 cm (2.38±2.84 vs. 3.24±2.97, p<0.05), N0 stage (2.17±2.94 vs. 3.83±2.59, p<0.001), M0 stage (2.56±2.89 vs. 4.06±2.83, p<0.001), and TNM stage I and II (2.48±3.02 vs. 4.27±2.81, p<0.001), (Figure 1C–1F).
XIST promoted the clinical progression of CRC
We divided the CRC patients into a high XIST expression group (n=110) and a low XIST expression group (n=86) with the mean T/A value (3.26, determined by comparing XIST expression in CRC tissues with corresponding tumor adjacent tissues) serving as the cut-off score. The results also indicated that high expression of XIST was correlated with advanced N stage, M stage, and TNM stage and was associated with larger tumor sizes (Table 1, p<0.05). Together, these findings indicate XIST can promote the clinical progression of CRC.
Table 1.
Correlation between lncRNA XIST expression and clinicopathological characteristics of CRC patients.
| Parameters | No. | XIST | p Value | |
|---|---|---|---|---|
| Low (n=86) | High (n=110) | |||
| Sex | 0.944 | |||
| Male | 102 | 45 | 57 | |
| Female | 94 | 41 | 53 | |
| Age | 0.375 | |||
| <60 years | 86 | 39 | 47 | |
| ≥60 years | 110 | 47 | 73 | |
| Tumor location | 0.505 | |||
| Colon | 95 | 44 | 51 | |
| Rectum | 101 | 42 | 59 | |
| Differentiation grade | 0.601 | |||
| Well+moderate | 152 | 63 | 69 | |
| Poor | 44 | 23 | 21 | |
| Tumor size | 0.008 | |||
| <5 cm | 109 | 57 | 52 | |
| ≥5 cm | 87 | 29 | 58 | |
| T stage | 0.208 | |||
| T1+T2 | 104 | 50 | 54 | |
| T3+T4 | 92 | 36 | 56 | |
| N stage | 0.004 | |||
| N0 | 126 | 65 | 61 | |
| N1 | 70 | 21 | 49 | |
| M stage | 0.010 | |||
| M0 | 168 | 80 | 88 | |
| M1 | 28 | 6 | 22 | |
| TNM stage | 0.025 | |||
| I+II | 112 | 51 | 61 | |
| III+IV | 84 | 25 | 59 | |
The upregulated lncRNA XIST can predict poor prognosis of CRC
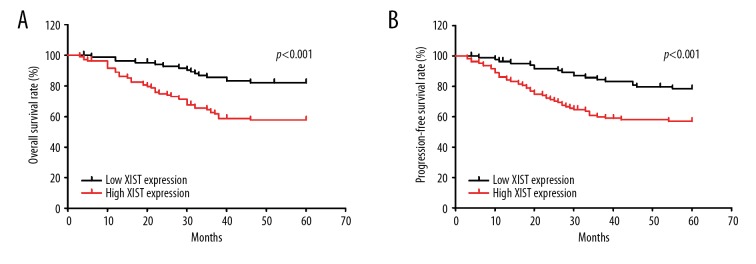
To clarify the prognostic and predictive values of lncRNA XIST in CRC, the log-rank test and the Kaplan-Meier test were performed to define the functions of lncRNA XIST in the prognosis of CRC. We found that high expression of XIST was correlated with poor PFS and poor OS in the CRC patients (Figure 2A, 2B). By using univariate analysis to evaluate the association of clinicopathological factors with survival, we found that N1 stage (HR=1.965, 95%CI=1.184–3.264, p=0.009), M1 stage (HR=5.694, 95%CI=3.226–10.048, p<0.001), TNM stage III and IV (HR=3.020, 95%CI=1.783–5.115, p<0.001), and high XIST expression (HR=1.284, 95%CI=1.143–1.442, p<0.001) were risk factors of poor OS (Table 2). These factors were also proven to be risk factors for poor PFS, N1 stage (HR=1.874, 95%CI=1.146–3.065, p=0.012), M1 stage (HR=5.164, 95%CI=2.952–9.036, p<0.001), TNM stage III and IV(HR=2.757, 95%CI=1.665–4.564, p<0.001), and high XIST expression (HR=1.242, 95%CI=1.115–1.384, p<0.001) (Table 2). Multivariate analysis of the above factors showed that M1 stage (HR=4.007, 95%CI=1.884–8.522, p<0.001) and high XIST expression (HR=1.197, 95%CI=1.064–1.364, p=0.003) were independent risk factors for poor OS (Table 2). The M1 stage (HR=3.725, 95%CI=1.768–7.845, p=0.001) and high XIST expression (HR=1.165, 95%CI=1.044–1.300, p=0.007) were proven to be the independent risk factors for poor PFS. In summary, the lncRNA XIST may be a potential prognostic biomarker for use in CRC patients (Table 2).
Figure 2.
High lncRNA XIST predicts poor prognosis of CRC patients. The overall survival rate (A) and progression-free survival rate (B) of patients with high XIST expression and low XIST expression. p<0.001 in A and B were analyzed by log-rank test.
Table 2.
Statistical analysis of risk factors for overall survival and progression-free survival of CRC patients.
| Parameters | Overall survival | Progression-free survival | ||||
|---|---|---|---|---|---|---|
| HR | 95%CI | p Value | HR | 95%CI | p Value | |
| Univariate analysis | ||||||
| Sex: Male vs. Female | 1.144 | 0.688–1.903 | 0.604 | 1.194 | 0.729–1.957 | 0.481 |
| Age: <60 years vs. ≥60 years | 0.990 | 0.594–1.650 | 0.970 | 0.947 | 0.576–1.555 | 0.947 |
| Tumor location: Colon vs. Rectum | 0.872 | 0.525–1.448 | 0.597 | 0.816 | 0.499–1.334 | 0.417 |
| Differentiation: Poor vs. well+moderate | 1.436 | 0.819–2.517 | 0.207 | 1.321 | 0.759–2.301 | 0.325 |
| Tumor size: ≥5 cm vs. 5 cm | 1.207 | 0.727–2.005 | 0.467 | 1.076 | 0.656–1.763 | 0.772 |
| T stage (T1+T2) vs. (T3+T4) | 0.847 | 0.508–1.412 | 0.524 | 0.862 | 0.526–1.413 | 0.556 |
| N stage: N1 vs. N0 | 1.965 | 1.184–3.264 | 0.009 | 1.874 | 1.146–3.065 | 0.012 |
| M stage: M1 vs. M0 | 5.694 | 3.226–10.048 | <0.001 | 5.164 | 2.952–9.036 | <0.001 |
| TNM stage: (III+IV) vs. (I+II) | 3.020 | 1.783–5.115 | <0.001 | 2.757 | 1.665–4.564 | <0.001 |
| XIST: High vs. low | 1.284 | 1.143–1.442 | <0.001 | 1.242 | 1.115–1.384 | <0.001 |
| Multivariate analysis | ||||||
| N stage: N1 vs. N0 | 1.261 | 0.539–2.952 | 0.593 | 1.216 | 0.521–2.836 | 0.651 |
| M stage: M1 vs. M0 | 4.007 | 1.884–8.522 | <0.001 | 3.725 | 1.768–7.845 | 0.001 |
| TNM stage: (III+IV) vs. (I+II) | 1.146 | 0.405–3.244 | 0.798 | 1.159 | 0.417–3.219 | 0.778 |
| XIST: High vs. low | 1.197 | 1.064–1.346 | 0.003 | 1.165 | 1.044–1.300 | 0.007 |
Discussion
In recent years, the lncRNAs have attracted great attention due to their extensive involvement in cancer biology [7,22,23]. CRC-associated lncRNAs participate in modulation of the cancer-related bioactivities at both post-transcriptional levels and transcriptional levels [23]. Moreover, multiple signaling pathways have been discovered to mediate the function of lncRNAs in CRC. lncRNACCAT2 can trigger metastasis by regulating MYC-activated microRNAs (such as the miR-17-5p and the miR-20) by activating the Wnt signaling pathway [24]. Ellis et al. [25] reported that knockdown or downregulation for intronic-region in the lncRNACRNDE transcripts can affect insulin/IGF-associated signaling pathway gene expression. For evaluating the clinical significance of lncRNAs, numerous studies have examined the expression of lncRNAs in cancer tissues and its correlation with clinicopathological characteristics. For the primary roles and features of lncRNAs in cancers, a growing number of new lncRNAs are being discovered and characterized in recent years. However, to the best of our knowledge, no studies have investigated the significant effects of XIST in CRC until now.
The present study shows that lncRNA XIST was increased in the CRC cancer cells and cancer tissues. XIST overexpression was also discovered in CRC tissues with lager tumor size, N1 stage, M1 stage, and TNM stage III and IV. Furthermore, by analyzing the relationship between XIST expression and CRC clinicopathological features, patients in the high XIST expression group showed larger tumor size and more advanced N, M, and TNM stages. Further analysis with Kaplan-Meier analysis and log-rank test found that high XIST expression predicted poorer OS and shorter PFS. In addition, together with M1 stage, high XIST expression may be an independent risk factor of CRC poor prognosis. The above results suggest that lncRNA XIST might be a promising therapeutic target and a potential prognostic biomarker for use in CRC patients.
Recent studies have extended our understanding of the function of XIST in cancer metastasis, proliferation, apoptosis, and stem cell characteristics [12–20,26]. Rottenberg et al. [27] reported that high XIST expression is associated with cisplatin resistance in most tumors, and predicted shorter recurrence-free survival of HER2-negative stage III breast cancer patients treated with intensive platinum-based chemotherapy. Besides, high XIST and low 53BP1 expression predicted poor outcome after high-dose alkylating chemotherapy in patients with BRCA1-like breast cancer [28]. Moreover, the expression level of XIST demonstrated a significant association with the prognosis in patients with cervical squamous cell carcinoma [29], pancreatic cancer [19], NSCLC [18], nasopharyngeal carcinoma [16], and gastric cancer [13]. Mechanistically, XIST can influence cancer biology through various pathways. In glioma, lncRNA XIST knockdown or downregulation could inhibit the glioma angiogenesis and increase the permeability of the blood-tumor barrier by suppressing expression of FOXC1 and enhancing the levels of zonulaoccludens 2 (ZO-2) through upregulating the levels of miR-137 [30]. Previous studies [31–33] reported that the lncRNA can sponge miR. lncRNA XIST can also function as sponge of miR-449a, 133a [19], miR-34a-5p [16], miR-139-5p [15], and miR-133a [18], which could further modulate expression of the protein-coding genes. The mechanisms by which XIST exerts its functions in cancer remain obscure and deserve further exploration. The mechanisms by which XIST influences CRC clinical progression was not explored, and this needs to be verified in future studies.
Conclusions
XIST is upregulated in CRC and is significantly correlated with CRC clinical progression. lncRNA XIST overexpression can predict poor PFS and poor OS for CRC patients. lncRNA XIST can also act as an independent risk factor for CRC prognosis in clinical practice. Thus, the lncRNA XIST might be considered as a promising therapeutic target and a potential prognostic biomarker for CRC.
Footnotes
Source of support: Departmental sources
Conflict of interest
None.
References
- 1.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Bockelman C, Engelmann BE, Kaprio T, et al. Risk of recurrence in patients with colon cancer stage II and III: A systematic review and meta-analysis of recent literature. Acta Oncol. 2015;54:5–16. doi: 10.3109/0284186X.2014.975839. [DOI] [PubMed] [Google Scholar]
- 3.Marisa L, de Reynies A, Duval A, et al. Gene expression classification of colon cancer into molecular subtypes: Characterization, validation, and prognostic value. PLoS Med. 2013;10:e1001453. doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang T, Wan CY, Mei XL, et al. Long non-coding RNA HULC promotes progression of bone neoplasms. Med Sci Monit. 2018;23:5754–60. doi: 10.12659/MSM.910220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maass PG, Luft FC, Bahring S. Long non-coding RNA in health and disease. J Mol Med. 2014;92:337–46. doi: 10.1007/s00109-014-1131-8. [DOI] [PubMed] [Google Scholar]
- 6.Liu LP, Gong YB. LncRNA-TCL6 promotes early abortion and inhibits placenta implantation via the EGFR pathway. Eur Rev Med Pharmacol Sci. 2018;22:7105–12. doi: 10.26355/eurrev_201811_16242. [DOI] [PubMed] [Google Scholar]
- 7.Schmitt AM, Chang HY. Long Noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–63. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartonicek N, Maag JL, Dinger ME. Long noncoding RNAs in cancer: Mechanisms of action and technological advancements. Mol Cancer. 2016;15:43. doi: 10.1186/s12943-016-0530-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown CJ, Ballabio A, Rupert JL, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 10.Agrelo R, Souabni A, Novatchkova M, et al. SATB1 defines the developmental context for gene silencing by XIST in lymphoma and embryonic cells. Dev Cell. 2009;16:507–16. doi: 10.1016/j.devcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weakley SM, Wang H, Yao Q, et al. Expression and function of a large non-coding RNA gene XIST in human cancer. World J Surg. 2011;35:1751–56. doi: 10.1007/s00268-010-0951-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang S, Chen B, Wang X, et al. Long non-coding RNA XIST regulates PTEN expression by sponging miR-181a and promotes hepatocellular carcinoma progression. BMC Cancer. 2017;17:248. doi: 10.1186/s12885-017-3216-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen DL, Ju HQ, Lu YX, et al. Long non-coding RNA XIST regulates gastric cancer progression by acting as a molecular sponge of miR-101 to modulate EZH2 expression. J Exp Clin Cancer Res. 2016;35:142. doi: 10.1186/s13046-016-0420-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang C, Wu K, Wang S, et al. Long non-coding RNA XIST promotes osteosarcoma progression by targeting YAP via miR-195-5p. J Cell Biochem. 2018;119:5646–56. doi: 10.1002/jcb.26743. [DOI] [PubMed] [Google Scholar]
- 15.Mo Y, Lu Y, Wang P, et al. Long non-coding RNA XIST promotes cell growth by regulating miR-139-5p/PDK1/AKT axis in hepatocellular carcinoma. Tumour Biol. 2017;39 doi: 10.1177/1010428317690999. 1010428317690999. [DOI] [PubMed] [Google Scholar]
- 16.Song P, Ye LF, Zhang C, et al. Long non-coding RNA XIST exerts oncogenic functions in human nasopharyngeal carcinoma by targeting miR-34a-5p. Gene. 2016;592:8–14. doi: 10.1016/j.gene.2016.07.055. [DOI] [PubMed] [Google Scholar]
- 17.Wang H, Shen Q, Zhang X, et al. The long non-coding RNA XIST controls non-small cell lung cancer proliferation and invasion by modulating miR-186-5p. Cell Physiol Biochem. 2017;41:2221–29. doi: 10.1159/000475637. [DOI] [PubMed] [Google Scholar]
- 18.Fang J, Sun CC, Gong C. Long noncoding RNA XIST acts as an oncogene in non-small cell lung cancer by epigenetically repressing KLF2 expression. Biochem Biophys Res Commun. 2016;478:811–17. doi: 10.1016/j.bbrc.2016.08.030. [DOI] [PubMed] [Google Scholar]
- 19.Wei W, Liu Y, Lu Y, et al. LncRNA XIST promotes pancreatic cancer proliferation through miR-133a/EGFR. J Cell Biochem. 2017;118:3349–58. doi: 10.1002/jcb.25988. [DOI] [PubMed] [Google Scholar]
- 20.Yao Y, Ma J, Xue Y, et al. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett. 2015;359:75–86. doi: 10.1016/j.canlet.2014.12.051. [DOI] [PubMed] [Google Scholar]
- 21.Lassmann S, Weis R, Makowiec F, et al. Array CGH identifies distinct DNA copy number profiles of oncogenes and tumor suppressor genes in chromosomal- and microsatellite-unstable sporadic colorectal carcinomas. J Mol Med. 2007;85:293–304. doi: 10.1007/s00109-006-0126-5. [DOI] [PubMed] [Google Scholar]
- 22.Sahu A, Singhal U, Chinnaiyan AM. Long noncoding RNAs in cancer: From function to translation. Trends Cancer. 2015;1:93–109. doi: 10.1016/j.trecan.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lisitsyn NA, Chernyi AA, Karpov VL, et al. A role of long noncoding RNAs in carcinogenesis. Mol Biol. 2015;49:500–7. doi: 10.7868/S0026898415040102. [DOI] [PubMed] [Google Scholar]
- 24.Ling H, Spizzo R, Atlasi Y, et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013;23:1446–61. doi: 10.1101/gr.152942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellis BC, Graham LD, Molloy PL. CRNDE, a long non-coding RNA responsive to insulin/IGF signaling, regulates genes involved in central metabolism. Biochim Biophys Acta. 2014;1843:372–86. doi: 10.1016/j.bbamcr.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 26.Wutz A. Gene silencing in X-chromosome inactivation: Advances in understanding facultative heterochromatin formation. Nat Rev Genet. 2011;12:542–53. doi: 10.1038/nrg3035. [DOI] [PubMed] [Google Scholar]
- 27.Rottenberg S, Vollebergh MA, de Hoon B, et al. Impact of intertumoral heterogeneity on predicting chemotherapy response of BRCA1-deficient mammary tumors. Cancer Res. 2012;72:2350–61. doi: 10.1158/0008-5472.CAN-11-4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schouten PC, Vollebergh MA, Opdam M, et al. High XIST and low 53BP1 expression predict poor outcome after high-dose alkylating chemotherapy in patients with a BRCA1-like breast cancer. Mol Cancer Ther. 2016;15:190–98. doi: 10.1158/1535-7163.MCT-15-0470. [DOI] [PubMed] [Google Scholar]
- 29.Kobayashi R, Miyagawa R, Yamashita H, et al. Increased expression of long non-coding RNA XIST predicts favorable prognosis of cervical squamous cell carcinoma subsequent to definitive chemoradiation therapy. Oncol Lett. 2016;12:3066–74. doi: 10.3892/ol.2016.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu H, Xue Y, Wang P, et al. Knockdown of long non-coding RNA XIST increases blood-tumor barrier permeability and inhibits glioma angiogenesis by targeting miR-137. Oncogenesis. 2017;6:e303. doi: 10.1038/oncsis.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang WC, Fu WM, Wang YB, et al. H19 activates Wnt signaling and promotes osteoblast differentiation by functioning as a competing endogenous RNA. Sci Rep. 2016;6:20121. doi: 10.1038/srep20121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao S, Zhao ZY, Wu R, et al. Prognostic value of long noncoding RNAs in gastric cancer: A meta-analysis. Onco Targets Ther. 2018;11:4877–91. doi: 10.2147/OTT.S169823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma L, Zhou Y, Luo X, et al. Long non-coding RNA XIST promotes cell growth and invasion through regulating miR-497/MACC1 axis in gastric cancer. Oncotarget. 2017;8:4125–35. doi: 10.18632/oncotarget.13670. [DOI] [PMC free article] [PubMed] [Google Scholar]